Abstract
Background
We previously created a serum-based algorithm that yielded excellent diagnostic accuracy in Alzheimer's disease. The current project was designed to refine that algorithm by reducing the number of serum proteins and by including clinical labs. The link between the biomarker risk score and neuropsychological performance was also examined.
Methods
Serum-protein multiplex biomarker data from 197 patients diagnosed with Alzheimer's disease and 203 cognitively normal controls from the Texas Alzheimer's Research Consortium were analyzed. The 30 markers identified as the most important from our initial analyses and clinical labs were utilized to create the algorithm.
Results
The 30-protein risk score yielded a sensitivity, specificity, and AUC of 0.88, 0.82, and 0.91, respectively. When combined with demographic data and clinical labs, the algorithm yielded a sensitivity, specificity, and AUC of 0.89, 0.85, and 0.94, respectively. In linear regression models, the biomarker risk score was most strongly related to neuropsychological tests of language and memory.
Conclusions
Our previously published diagnostic algorithm can be restricted to only 30 serum proteins and still retain excellent diagnostic accuracy. Additionally, the revised biomarker risk score is significantly related to neuropsychological test performance.
Key Words: Algorithm, blood-based; Alzheimer's disease; Diagnosis
Introduction
There are currently no rapid cost-effective means for providing routine screening of adults age 65 years and above for Alzheimer's disease (AD), which is the most common form of neurodegenerative dementia. Currently over 5.2 million Americans suffer from the disease, and this number is expected to reach 7.7 million in 2030. This represents more than a 50% increase from current prevalence rates [1] and means that, by mid-century, it is expected that one new case will develop every 33 s [1]. While advanced clinical, neuroimaging, and cerebrospinal fluid (CSF) analyses are quite accurate in detecting AD, they are cost prohibitive as large-scale screening measures. Additionally, these technologies and specialty services are not readily accessible to all (e.g. rural elders, ethnic minorities), which limits their usefulness as screeners for AD. A blood-based test, however, would provide a rapid cost-effective means of screening for AD at the population level, broadening access to care globally [2,3]. As part of a multi-stage process, such a blood test would provide an optimal initial screening tool that could be followed up by advanced clinical, neuroimaging, and/or CSF analyses [3] for screen-positive cases. Furthermore, an accurate and easily performed screen would create a cost-effective means of screening for therapeutic trials [4]. Historically, molecular biomarkers have focused on single molecules but as proteomic, genomic, and metabolomic technology improves, it has become increasingly feasible to develop classifiers based on many complex signatures of disease status.
While the search for blood-based biomarkers of AD has been largely unsuccessful for decades, recently there have been significant advancements. Ray et al. [5] analyzed a range of plasma-based proteins and developed an algorithm that accurately classified AD as well as predicted conversion from MCI to AD. More recently, Booij et al. [6] and Rye et al. [7] conducted a set of analyses utilizing gene expression arrays and found adequate (74%) to excellent (92%) overall diagnostic accuracy. Through analysis of the longitudinal cohort of the Texas Alzheimer's Research Consortium (TARC), we created a serum-based algorithm that yielded excellent diagnostic accuracy correctly classifying 95% of AD cases and controls [8]. The purpose of the current study was to refine and expand upon the validity of our algorithm. Specifically, we sought to: (1) determine if the addition of clinical labs (i.e. cholesterol, triglycerides, high-density lipoproteins, low-density lipoproteins, lipoprotein-associated phospholipase [Lp-PLA2], homocysteine, C-peptide) would increase the overall diagnostic accuracy of the algorithm, and (2) refine the algorithm to only the top 30 proteins and test the diagnostic accuracy of that briefer version. Many steps have been proposed for validation of AD biomarkers, one of which is to establish link between such putative biomarkers and cognitive functioning [9]; therefore, we also evaluated the link between the 30-protein version of the algorithm and neuropsychological test results.
Methods
Participants
Cross-sectional data from the 400 participants (197 AD cases, 203 controls) enrolled in the TARC utilized in our initial algorithm [8] were re-analyzed. The methodology of the TARC protocol has been described elsewhere [10,11]. Briefly, each participant undergoes an annual standardized assessment at one of the five participating sites that includes a medical evaluation, neuropsychological testing, an interview, and a blood draw. Diagnosis of AD is based on NINCDS-ADRDA criteria [12] and performed within normal limits on psychometric testing. Institutional Review Board approval is obtained at each site and written informed consent is obtained for all participants.
Biomarker Assays
Non-fasting blood samples were collected in serum-separating tubes during clinical evaluations, allowed to clot at room temperature for 30 min, centrifuged, aliquoted, and stored at −80°C in plastic vials. Batched specimens from either baseline or year-one follow-up exams were sent frozen to Rules Based Medicine (www.rulesbasedmedicine.com, Austin, Tex., USA) where they were thawed for assay without additional freeze-thaw cycles using their multiplexed immunoassay human Multi-Analyte Profile (humanMAP). Individual proteins were quantified with immunoassays on colored microspheres. Information regarding the least detectable dose, inter-run coefficient of variation, dynamic range, overall spiked standard recovery, and cross-reactivity with other humanMAP analytes were obtained from Rules Based Medicine. The humanMAP panel has evolved over time, therefore, the complete list of analytes can be found in appendix 1 of our prior publication that is located on the webpage of the TARC (http://www.txalzresearch.org).
Neuropsychological Testing
The TARC neuropsychology core battery consists of commonly utilized instruments in AD clinical/research settings and it overlaps largely with the NACC Uniform Dataset including digit span (WAIS-R, WAIS-III, WMS-R) [13], Trail-Making Test [14], WMS Logical Memory and Visual Reproduction (WMS-R and WMS-III) [13], Boston Naming Test (30- and 60-item versions) [15], verbal fluency (FAS) [15], Clock-Drawing Test [15], the American National Adult Reading Test (AMNART) [15], the Geriatric Depression Scale (GDS-30) [16], Mini-Mental State Examination (MMSE) [17], and ratings on the Clinical Dementia Rating scale (CDR) [18]. In order to equate scores and be consistent across tests, all raw scores (with the exception of the 4-point Clock-Drawing Test) were converted to scale scores based on previously published normative data [19,20,21]. For the Boston Naming Test, we recently published an independent study demonstrating the psychometric utility of an estimated 60-item BNT score that can be calculated from 30-item versions [22]; this estimated 60-item score was used for all 30-item administrations. Adjusted scale scores were utilized as dependent variables in analyses.
Statistical Analyses
Fisher's exact and Mann Whitney U tests were used to compare case versus controls for categorical variables (APOE ∊4 allele frequency, gender, race, or ethnicity) and continuous variables (age and education). In the first set of analyses, the original biomarker risk score [8] was applied to the test set along with demographic (age, gender, education) and clinical (total cholesterol, triglycerides, high-density lipoproteins, low-density lipoproteins, Lp-PLA2, homocysteine, C-peptide, APOE4 genotype) variables. Clinical variables were added to create a more robust diagnostic algorithm given the prior work documenting a link between such variables cognitive dysfunction and AD [23,24,25,26]. For the re-analysis of the biomarker risk score, we utilized only the 30 markers that were identified in our prior publication as the most important variables in the biomarker risk score (table 2). Random forest analyses were re-run on the training and test sets, as previously created. In the initial analyses, all analytes were log transformed then standardized, which was used in the current analyses. Analyses were performed using R (V 2.10) statistical software [27]. The random forest prediction model was performed using R package randomForest (V 4.5) [28], with all software default settings. The ROC curves were analyzed using R package. AUC was calculated using R package DiagnosisMed (V 0.2.2.2). Using the test set of (200 AD cases and controls), linear regression models were generated to examine the link between the biomarker risk score and neuropsychological scale scores; age, gender, and education were entered as covariates.
Table 2.
Biomarkers utilized in 30-protein version of risk score
| Thrombopoietin |
| Microphage inflammatory protein 1α |
| Eotaxin 3 |
| Tumor necrosis factor-α |
| Tenascin C |
| FAS |
| Fibrinogen |
| Interleukin-10 |
| Interleukin-7 |
| Cancer antigen 19.9 |
| Prostatic acid phosphatase |
| Apolipoprotein CIII |
| FAS ligand |
| C-reactive protein |
| Tissue inhibitor of metalloproteinases 1 |
| Angiopoietin 2 |
| Stem cell factor |
| Interleukin-5 |
| Lipoprotein(a) |
| α2-Macroglobulin |
| ACE CD143 angiotensin-converting enzyme |
| Monocyte chemotactic protein 1 |
| Ferritin |
| Pulmonary and activation-regulated chemmocine |
| Cancer antigen 125 |
| Von Willebrand factor |
| Carcinoembryonic antigen |
| Macrophage migration inhibitory factor |
| Creatine kinase MB |
| Pancreatic polypeptide |
Results
Demographic characteristics of the sample are provided in table 1. The cases were older (p < 0.001), achieved fewer years of education (p < 0.001), had lower MMSE scores (p < 0.001), higher CDR sum of boxes scores (p < 0.001), and were more likely to carry at least one copy of the APOE ∊4 allele (p < 0.001) than control participants. We have previously demonstrated that our biomarker risk score is significantly and independently associated with case status from demographic variables [8].
Table 1.
Demographic characteristics of sample
| Total | AD (n = 197) | Control (n = 203) | p value | |
|---|---|---|---|---|
| Male, % | 33 | 34.5 | 32 | 0.67 |
| Age, years | 73.9 ± 9.3 | 77.4 ± 8.3 | 70.4 ± 8.9 | <0.0001 |
| Education, years | 14.8 ± 3.2 | 14.0 ± 3.5 | 15.5 ± 2.7 | <0.0001 |
| MMSE | 24.4 ± 6.8 | 19.2 ± 6.2 | 29.4 ± 0.9 | <0.001 |
| CDR sum of boxes score | 3.8 ± 5.0 | 7.8 ± 4.4 | 0.01 ± 0.05 | <0.001 |
| APOE4 carrier (yes/no) | 163/218 (19 unknown) | 110/71 (16 unknown) | 53/147 (3 unknown) | <0.0001 |
| Hispanic ethnicity, % | 9 | 3.6 | 5.4 | 0.47 |
| Race | ||||
| White | 377 | 187 | 190 | |
| Non-White | 23 | 10 | 13 | 0.67 |
Where indicated, data presented as means ± SD.
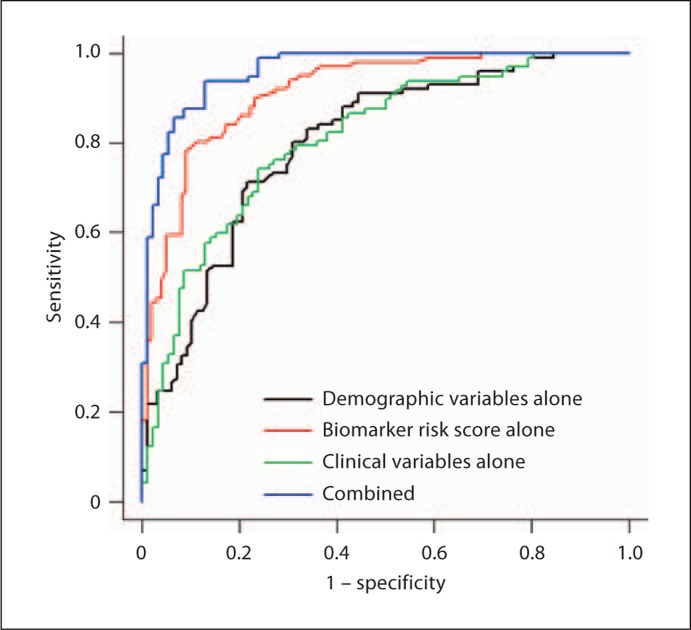
First, we added the clinical labs to the full diagnostic algorithm we previously generated. An optimal balance between sensitivity (SN) and specificity (SP) for the full algorithm utilizing the additional clinical data was found at a cut-score of 0.465. The overall diagnostic accuracy of the full algorithm with the additional clinical data improved somewhat with an observed AUC = 0.96, SN = 0.94, and SP = 0.87 (table 3; fig. 1).
Table 3.
Diagnostic accuracy of full algorithm plus new clinical variables
| AUC | Sensitivity | Specificity | |
|---|---|---|---|
| Demographic data only (cutoff 0.550) | 0.799 (0.741–0.858) | 0.713 (0.618–0.792) | 0.781 (0.692–0.854) |
| Clinical data only (cutoff 0.494) | 0.809 (0.747–0.870) | 0.742 (0.647–0.819) | 0.761 (0.664–0.836) |
| RBM data only (cutoff 0.476) | 0.914 (0.878–0.951) | 0.802 (0.714–0.868) | 0.889 (0.812–0.937) |
| Combined (cutoff 0.465) | 0.962 (0.937–0.986) | 0.938 (0.872–0.971) | 0.870 (0.786–0.924) |
Rules Based Medicine data only.
Fig. 1.
ROC for full algorithm.
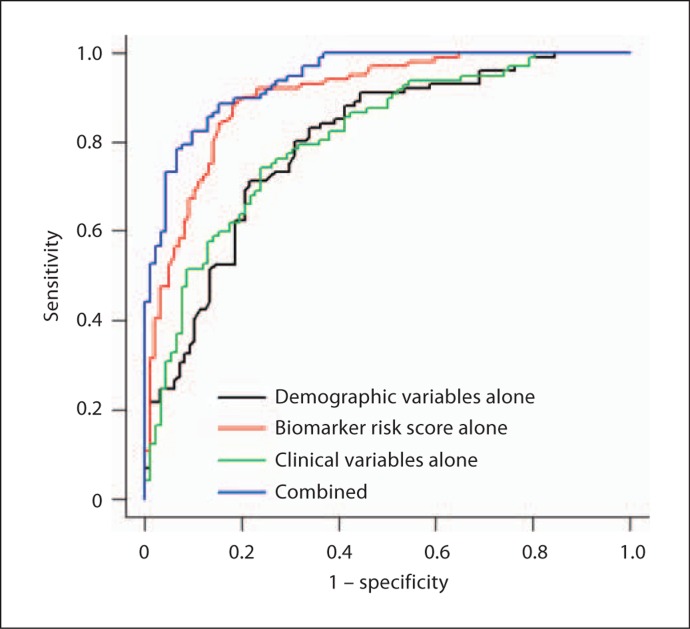
Next, we sought to determine if a 30-protein restricted biomarker-based algorithm would yield sufficient accuracy. The proteins included were selected from our original publication and represent the 30 markers that contributed most to the classification accuracy (table 2). The overall accuracy of the revised biomarker risk score (AUC = 0.91) was comparable to that found from the full protein risk score (tables 3 and 4). At the optimal cut-score of 0.426, there was an observed increase in SN with the 30-protein score (SN = 0.88) with an accompanying decrease in SP (0.82) (table 4; fig. 2). The overall accuracy of the revised algorithm that incorporated the 30-protein risk score, demographic characteristics, and the clinical data were excellent (AUC = 0.94, SN = 0.89, SP = 0.85).
Table 4.
Diagnostic accuracy of 30-protein-based algorithm
| AUC | Sensitivity | Specificity | |
|---|---|---|---|
| Demographic data only (cutoff 0.550) | 0.799 (0.741–0.858) | 0.713 (0.618–0.792) | 0.781 (0.692–0.854) |
| Clinical data only (cutoff 0.494) | 0.809 (0.747–0.870) | 0.742 (0.647–0.819) | 0.761 (0.664–0.836) |
| RBM data only (cutoff 0.426) | 0.905 (0.865–0.945) | 0.881 (0.804–0.931) | 0.818 (0.731–0.881) |
| Combined (cutoff 0.399) | 0.942 (0.913–0.972) | 0.887 (0.808–0.936) | 0.848 (0.761–0.907) |
Rules Based Medicine data only.
Fig. 2.
ROC for 30-protein-based algorithm.
Lastly, we sought to determine the link between the 30-protein risk score and cognitive performance. The 30-protein biomarker risk score was significantly related to global cognition (MMSE) as well as overall disease severity (CDR). It was also significantly related to the neuropsychological domains of executive functioning (Clock-, Trail-Making Test), language (Controlled Oral Word Association Test, Boston Naming Test), and memory (Wechsler Memory Scales story and figure, immediate and delayed recall) (table 5). Average scale scores for all neuropsychological tests are presented.
Table 5.
Results of regression analyses of the link between the 30-protein biomarker risk score and neuropsy-chological test results
| Total sample | AD cases | Controls | B (±SE) | P | |
|---|---|---|---|---|---|
| MMSE | 24.4 ± 6.8 | 19.2 ± 6.2 | 29.4 ± 0.9 | −16.64 ± 2.95 | <0.001 |
| CDRSB | 3.9 ± 5.0 | 7.9 ± 4.4 | 0.0 ± 0.1 | 15.00 ± 2.21 | <0.001 |
| CDR GS | 0.7 ± 0.8 | 1.3 ± 0.8 | 0.0 ± 0.0 | 2.50 ± 0.37 | <0.001 |
| CLOCK1 | 3.1 ± 1.2 | 2.4 ± 1.2 | 3.9 ± 0.4 | −2.94 ± 0.54 | <0.001 |
| COWAT | 9.6 ± 3.7 | 7.1 ± 3.1 | 11.6 ± 2.7 | −7.07 ± 1.76 | <0.001 |
| BNT | 9.3 ± 4.4 | 6.3 ± 3.8 | 11.9 ± 3.0 | −10.63 ± 2.05 | <0.001 |
| AMNART | 10.5 ± 3.9 | 8.8 ± 3.6 | 12.0 ± 3.5 | −4.44 ± 1.81 | 0.02 |
| Trails A | 8.4 ± 3.6 | 6.1 ± 3.1 | 10.3 ± 2.7 | −7.09 ± 1.78 | <0.001 |
| Trails B | 8.6 ± 4.1 | 4.8 ± 3.3 | 11.0 ± 2.5 | −7.85 ± 2.33 | 0.001 |
| Digit span | 10.0 ± 3.3 | 8.3 ± 3.0 | 11.7 ± 2.8 | −3.84 ± 1.56 | 0.01 |
| WMS LM I | 9.3 ± 5.4 | 4.0 ± 2.4 | 13.6 ± 2.8 | −16.46 ± 2.69 | <0.001 |
| WMS LM II | 9.3 ± 5.7 | 3.6 ± 1.8 | 14.0 ± 2.6 | −18.05 ± 2.78 | <0.001 |
| WMS VR I | 8.9 ± 4.9 | 4.6 ± 2.9 | 12.4 ± 3.2 | −14.04 ± 2.33 | <0.001 |
| WMS VR II | 9.6 ± 5.2 | 4.7 ± 2.3 | 13.6 ± 3.1 | −16.74 ± 2.39 | <0.001 |
Where indicated, data presented as means ± SD. B = Unstandardized beta; SB = sum of boxes score; GS = global score; COWAT = Controlled Oral Word Association Test scale score; BNT = Boston Naming Test scale score; AMNART = American North American Reading Test scale score; Trails A = Trail-Making Test part A scale score; Trails B = Trail-Making Test part B scale score; Digit Span scale score; WMS = Wechsler Memory Scale; LM = logical memory; VR = visual reproduction, all scale scores.
1 Clock-drawing test, raw scores (range of possible scores: 0–4).
Discussion
There is a significant need for a fast and cost-effective means of screening the rapidly growing elderly segment of the population. The ideal AD biomarker would come from blood [29], and we recently published a serum-based algorithm that yielded excellent diagnostic accuracy [8]. However, that algorithm utilized over 100 proteins in the original model. In order to become more cost efficient, such an algorithm would ideally require a more focused set of markers. We utilized variable importance estimates from Random Forest in our initial publication and utilized the 30 most important markers to create a refined algorithm. In the initial analyses, our protein biomarker risk score yielded an observed SN = 0.80, SP = 0.91, and AUC = 0.91 with the current 30-protein risk score being very comparable (SN = 0.88, SP = 0.82, AUC = 0.91). As can be seen from table 2, the 30 proteins in our biomarker portion of the algorithm cover a range of biological processes. It is our hypothesis that such a broad scope in the biomarker risk score will be necessary for the generalizability of the algorithm and approach to other populations. In fact, a lack of such breadth may be one reason for the failure of prior attempts to cross-validate.
There are several advantages to our approach. One of the recommended criteria proposed by the Consensus Report of the Working Group on Molecular and Biochemical Markers of Alzheimer's disease [30] was that biomarkers for AD have a SN and SP of >0.80. Even though the SP of our 30-protein risk score decreased to 0.82, the SN increased to 0.88, thereby providing a better balance across both estimates than the original biomarker risk score, which also meet the Consensus Working Group's criteria. The balance between SN and SP is another excellent feature of the current results as this also will provide a balance between positive and negative predictive power [31]. An additional advantage of our work is the direct comparison of the biomarker risk score to the diagnostic accuracy of common demographic and clinical data. Given the significant difference in age, gender, education, and APOE4 frequency between AD cases and controls in those at risk for this disease, one can classify a large number of individuals without the use of biomarker data (blood, imaging, CSF, genetic or otherwise). In fact, using only age, gender, and education, we find an AUC of 0.80. This may be somewhat inflated by the group differences in demographics in this study; however, the addition of demographic factors adds ecological validity to our methodology. In fact, Vemuri et al. [32] have shown that adding demographic factors to structural MRI diagnostics added to the overall accuracy of the models, even when cases and controls were matched by these variables. Others have also found that a multimodal approach to the search for biomarkers for AD is superior to any single method [33,34]; our method adds the modalities of demographics and clinical labs to the algorithm, which are more cost and time efficient than adding additional biomarker modalities. While it is not necessary that the biomarker surpass the accuracy provided by demographic and/or clinical labs, it is necessary that the biomarker add unique information to the overall accuracy thereby improving the utility of the approach. As such, presentation of the biomarker results in the absence of such comparisons should be considered inadequate. When examining our biomarker risk score, both of our serum-based protein risk scores (1) yield better overall diagnostic accuracy than demographic or clinical variables alone, (2) contributed significantly and independently to case status from demographic factors, and (3) the combination of all modalities yielded far superior results. The combined multimodal nature of our algorithm also increases the likelihood of utility across settings and populations, which we are currently testing.
In the current analyses, the biomarker algorithm was also significantly related to neuropsychological status and disease severity. In fact, the biomarker risk score was most strongly associated with the cognitive domains of memory and language, which are among the first impacted by AD pathology. While this may be confounded by the fact that our cohort consisted of only AD cases and controls, such strong associations of the biomarker risk score with neuropsychological status and disease severity suggest utility of the algorithm to predict decline prospectively and we are working on those statistical models currently.
As can be seen from table 2, a large number of the proteins included in our biomarker algorithm are inflammatory in nature, which is consistent with our initial findings [8]. Therefore, it is possible that our biomarker algorithm is detecting a globally dysregulated inflammatory system (and other biological pathways) and future work will include non-AD disease groups (e.g. Parkinson's disease, Lewy body dementia, vascular dementia) for comparison purposes. There is a large body of literature documenting a significant link between inflammation and AD. In fact, in our prior work, we have proposed the existence of an inflammatory endophenotype of AD [8,35]. Such an endophenotype may explain inconsistent findings in the biomarker literature as well as the discrepancy between epidemiological studies demonstrating a protective effect of anti-inflammatory medications against AD development [36,37,38] and the failure of therapeutic trials using these compounds [39,40,41].
There are limitations to the current study. First, while the multiplex platform we utilized is superior to individual assay (e.g. ELISA) methodologies, we have not cross-validated the blood test on a separate platform. Additionally, we have not yet incorporated non-AD dementia cases into the analyses in an effort to determine the differential diagnostic utility of the algorithm. However, our neuropsychological findings that the biomarker risk score is most strongly related to the domains of language and memory may provide initial support for the notion of discriminative ability, though such analyses must be conducted. An additional limitation is the use of a clinic-based sample and our findings need to be tested in a population-based cohort. Lastly, our study is cross-sectional in nature and does not address the utility of the algorithm in predicting AD risk. Future work will include mild cognitive impairment cases as well as longitudinal data (controls, mild cognitive impairment, and AD) in order to determine the utility of the algorithm, or possibly the need for a separate algorithm, in predicting incident risk of AD.
Overall, the current results suggest that (1) the addition of standard clinical labs to the diagnostic algorithm yields increased overall accuracy, (2) the addition of clinical labs to the 30-protein algorithm (along with demographic data) results in excellent diagnostic accuracy, and finally (3) the biomarker risk score is a significantly associated with neuropsychological test scores, particularly memory and language function. While we must still apply our algorithm to an independent cohort, these analyses provide further support for our serum-based diagnostic algorithm for detecting AD.
Disclosure Statement
A patent has been filed in conjunction with Rules Based Medicine for the algorithm contained within this manuscript. The following authors are named on the patent: S.E.O.B., R.C.B., R.D.-A., G.X., P.M.A., J.S.R., R.S.D., and T.J.F.
Investigators from the Texas Alzheimer's Research Consortium: Baylor College of Medicine: Susan Rountree, Christie Ballantyne, Eveleen Darby, Aline Hittle, Aisha Khaleeg; Texas Tech University Health Science Center: Paula Grammas, Benjamin Williams, Andrew Dentino, Chuang Kuo Wu, Gregory Schrimsher, Parastoo Momeni, Larry Hill; University of North Texas Health Science Center: Janice Knebl, Lisa Alvarez, Douglas Mains; University of Texas Southwestern Medical Center: Roger Rosenberg, Ryan Huebinger, Janet Smith, Mechelle Murray, Tomequa Sears; University of Texas Health Sciences Center – San Antonio: Donald Royall, Raymond Palmer.
Acknowledgements
This study was made possible by the TARC funded by the state of Texas through the Texas Council on Alzheimer's Disease and Related Disorders. Investigators at the University of Texas Southwestern Medical Center at Dallas also acknowledge support from the UTSW Alzheimer's Disease Center NIH, NIA grant P30AG12300. We would like to thank Dr. Christie Ballantyne and his lab at Baylor College of Medicine for measuring Lp-PLA2 and homocysteine. The investigations at Baylor's Alzheimer's Disease and Memory Disorders Center were supported by the Cynthia and George Mitchell Foundation.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alzheimer's Association 2008 Alzheimer's disease facts and figures. Alzheimers Dement. 2008;4:110–133. doi: 10.1016/j.jalz.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 2.O'Bryant S, Xiao G, Barber R, Reisch J, Doody R, Fairchild T, Adams P, Waring S, Diaz-Arrastia R. Texas Alzheimer's Research Consortium: A serum protein-based algorithm for the detection of Alzheimer's disease. Arch Neurol. 2010;67:1077–1081. doi: 10.1001/archneurol.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider P, Hampel H, Buerger K. Biological marker candidates of Alzheimer's disease in blood, plasma, and serum. CNS Neurosci Ther. 2009;15:358–374. doi: 10.1111/j.1755-5949.2009.00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thal LJ, et al. The role of biomarkers in clinical trials for Alzheimer disease. Alzheimer Dis Assoc Disord. 2006;20:6–15. doi: 10.1097/01.wad.0000191420.61260.a8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ray S, et al. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 6.Booij BB, et al. A gene expression pattern in blood for the early detection of Alzheimer's disease. J Alzheimers Dis. 2011;23:109–119. doi: 10.3233/JAD-2010-101518. [DOI] [PubMed] [Google Scholar]
- 7.Rye PD, et al. A novel blood test for the early detection of Alzheimer's disease. J Alzheimers Dis. 2011;23:121–129. doi: 10.3233/JAD-2010-101521. [DOI] [PubMed] [Google Scholar]
- 8.O'Bryant SE, et al. A serum protein-based algorithm for the detection of Alzheimer disease. Arch Neurol. 2010;67:1077–1081. doi: 10.1001/archneurol.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mueller SG, et al. Ways toward an early diagnosis in Alzheimer's disease: The Alzheimer's Disease Neuroimaging Initiative (ADNI) Alzheimers Dement. 2005;1:55–66. doi: 10.1016/j.jalz.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waring S, O'Bryant SE, Reisch JS, Diaz-Arrastia R, Knebl J, Doody R, Texas Alzheimer's Research Consortium The Texas Alzheimer's Research Consortium longitudinal research cohort: Study design and baseline characteristics. Texas Pub Health J. 2008;60:9–13. [Google Scholar]
- 11.O'Bryant SE, et al. Brain-derived neurotrophic factor levels in Alzheimer's disease. J Alzheimers Dis. 2009;17:337–341. doi: 10.3233/JAD-2009-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKhann D, Drockman D, Folstein M, et al. Clinical diagnosis of Alzheimer's disease: Report of the NINCDS-ADRDA Work Group. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 13.Wechsler D. Wechsler Memory Scale. ed 3. San Antonio: The Psychological Corporation; 1997. [Google Scholar]
- 14.Lezak MD, Howieson DB, Loring DW. Neuropsychological Assessment. ed 4. Oxford: Oxford University Press; 2004. [Google Scholar]
- 15.Strauss E, Sherman EMS, Spreen O. A Compendium of Neuropsychological Tests: Administration, Norms, and Commentary. ed 3. Oxford: Oxford University Press; 2006. [Google Scholar]
- 16.Yesavage JA, Brink TL, Rose TL, et al. Development and validation of a geriatric depression screening scale: a preliminary report. J Psychiatr Res. 1983;17:37–49. doi: 10.1016/0022-3956(82)90033-4. [DOI] [PubMed] [Google Scholar]
- 17.Folstein MF, Folstein SE, McHugh PR. ‘Mini-Mental State’: A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 18.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules (see comment) Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 19.Ivnik R, Malec JF, Smith GE, Tangalos EG, Petersen RC, Kokmen E, Kurkland LT. Mayo's Older Americans Normative Studies: WAIS-R norms for age 56 to 97. Clin Neuropsychol. 1992;6:1–30. [Google Scholar]
- 20.Ivnik RJ, Malec JF, Smith GE, Tangalos EG, Petersen RC. Neuropsychological tests' norms above age 55 COWAT, BNT, MAE Token, WRAT-R Reading, AMNART, STROOP, TMT and JLO. Clin Neuropsychol. 1996;10:262–278. [Google Scholar]
- 21.Tulsky D, Zhu J, Ledbetter M. WAIS-III – WMS-III Technical Manual. San Antonio: The Psychological Corporation; 1997. [Google Scholar]
- 22.Hobson V, Hall JR, Harvey M, Cullum CM, Lacritz L, Massman PJ, Waring SC, O'Bryant SE. An examination of the Boston Naming Test: calculation of an ‘estimated’ 60-item score from 30- and 15-item scores in a cognitively impaired population. Int J Geriatr Psychiatry. 2011;26:351–355. doi: 10.1002/gps.2533. [DOI] [PubMed] [Google Scholar]
- 23.Dickstein DL, et al. Role of vascular risk factors and vascular dysfunction in Alzheimer's disease. Mt Sinai J Med. 2010;77:82–102. doi: 10.1002/msj.20155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piazza F, et al. Increased tissue factor pathway inhibitor and homocysteine in Alzheimer's disease. Neurobiol Aging. 2010 doi: 10.1016/j.neurobiolaging.2010.02.016. E-pub ahead of print. [DOI] [PubMed] [Google Scholar]
- 25.Okereke OI, et al. A profile of impaired insulin degradation in relation to late-life cognitive decline: a preliminary investigation. Int J Geriatr Psychiatry. 2009;24:177–182. doi: 10.1002/gps.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Oijen M, et al. Plasma Abeta(1–40) and Abeta(1–42) and the risk of dementia: a prospective case-cohort study. Lancet Neurol. 2006;5:655–660. doi: 10.1016/S1474-4422(06)70501-4. [DOI] [PubMed] [Google Scholar]
- 27.R Development Core Team: R: A language and environment for statistical computing. 2009. www.R-project.org
- 28.Breiman L. Random forests. Machine Learn. 2001;45:5–32. [Google Scholar]
- 29.Graff-Radford NR, et al. Association of low plasma Abeta42/Abeta40 ratios with increased imminent risk for mild cognitive impairment and Alzheimer disease (erratum appears in Arch Neurol 2007;64:1246) Arch Neurol. 2007;64:354–362. doi: 10.1001/archneur.64.3.354. [DOI] [PubMed] [Google Scholar]
- 30.Anonymous, Consensus report of the Working Group on ‘Molecular and Biochemical Markers of Alzheimer's Disease’. The Ronald and Nancy Reagan Research Institute of the Alzheimer's Association and the National Institute on Aging Working Group (see comment) (erratum appears in Neurobiol Aging 1998;19:285) Neurobiol Aging. 1998;19:109–116. [PubMed] [Google Scholar]
- 31.O'Bryant SE, Lucas JA. Estimating the predictive value of the Test of Memory Malingering: an illustrative example for clinicians. Clin Neuropsychol. 2006;20:533–540. doi: 10.1080/13854040590967568. [DOI] [PubMed] [Google Scholar]
- 32.Vemuri P, et al. Alzheimer's disease diagnosis in individual subjects using structural MR images: validation studies. NeuroImage. 2008;39:1186–1197. doi: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang D, et al. Multimodal classification of Alzheimer's disease and mild cognitive impairment. NeuroImage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brys M, et al. Magnetic resonance imaging improves cerebrospinal fluid biomarkers in the early detection of Alzheimer's disease. J Alzheimers Dis. 2009;16:351–362. doi: 10.3233/JAD-2009-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Bryant SE, et al. Decreased C-reactive protein levels in Alzheimer disease. J Geriatr Psychiatry Neurol. 2010;23:49–53. doi: 10.1177/0891988709351832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.In't Veld BA, Ruitenberg A, Hofman A, Launer LJ, van Duijn CM, Stijnen T, et al. Nonsteroidal antiinflammatory drugs and the risk of Alzheimer's disease. New Engl J Med. 2001;345:1515–1521. doi: 10.1056/NEJMoa010178. [DOI] [PubMed] [Google Scholar]
- 37.Anthony JC, et al. Reduced prevalence of AD in users of NSAIDs and H2 receptor antagonists: the Cache County study. Neurology. 2000;54:2066–2071. doi: 10.1212/wnl.54.11.2066. [DOI] [PubMed] [Google Scholar]
- 38.Etminan M, Gill S, Samii A. Effect of non-steroidal anti-inflammatory drugs on risk of Alzheimer's disease: systematic review and meta-analysis of observational studies. BMJ. 2003;327:128. doi: 10.1136/bmj.327.7407.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aisen PS, et al. Neither rofecoxib nor naproxen slows cognitive decline in people with mild-to-moderate Alzheimer's disease. Evidence-Based Healthcare. 2003;7:200–201. [Google Scholar]
- 40.Thal LJ, et al. A randomized, double-blind, study of rofecoxib in patients with mild cognitive impairment. Neuropsychopharmacology. 2005;30:1204–1215. doi: 10.1038/sj.npp.1300690. [DOI] [PubMed] [Google Scholar]
- 41.Adapt Research Group et al. Naproxen and celecoxib do not prevent AD in early results from a randomized controlled trial. Neurology. 2007;68:1800–1808. doi: 10.1212/01.wnl.0000260269.93245.d2. [DOI] [PubMed] [Google Scholar]