Abstract
This paper describes fabrication of a novel electrochemiluminescent (ECL) immunosensor array featuring capture-antibody-decorated single-wall carbon nanotube forests (SWCNT) residing in the bottoms of 10 µL wells with hydrophobic polymer walls. Silica nanoparticles containing [Ru(bpy)3]2+ and secondary antibodies (RuBPY-silica-Ab2) are employed in this system for highly sensitive two-analyte detection. Antibodies to PSA and IL-6 were attached to the same RuBPY-silica-Ab2 particle. The array was fabricated by forming the wells on a conductive pyrolytic graphite chip (1 × 1 in.) with a single connection to a potentiostat to achieve ECL. The sandwich immunoassay protocol employs antibodies attached to SWCNTs in the wells to capture analyte proteins. Then RuBPY-silica-Ab2 is added to bind to the captured proteins. ECL is initiated in the microwells by electrochemical oxidation of tripropyl amine (TprA), which catalytically reduces [Ru(bpy)3]2+ in the 100 nm particles, and is measured with a coupled charged device (CCD) camera. Separation of the analytical spots by the hydrophobic wall barriers enabled simultaneous immunoassays for two proteins in a single sample without cross-contamination. Detection limit (DL) for prostate specific antigen (PSA) was 1 pg mL−1 and for interleukin-6 (IL-6) was 0.25 pg mL−1 (IL-6) in serum. Array determinations of PSA and IL-6 in patient serum were well-correlated with single-protein ELISAs. These microwell SWCNT immunoarrays provide a simple, sensitive approach to detection of two or more proteins.
Keywords: Electrochemiluminescent immunosensor array, single-wall carbon nanotube forests, [Ru(bpy)3]2+-Doped Silica Nanoparticles, Cancer biomarkers
INTRODUCTION
The measurement of panels of biomarker proteins whose serum levels increase during the onset of cancer holds significant promise for early cancer detection and therapy monitoring.1–6 Proteins can be measured by methods including enzyme-linked immunosorbent assays (ELISA),7 radioimmunoassay,8 electrophoretic immunoassay9 and mass spectrometry-based proteomics.10,11 While reliable, these methods have limitations for clinical and point–of-care use, including cost, complex protocols, lengthy assay time, and/or difficulty in multiplexing. More recent approaches involve bead-based electrochemiluminescence (ECL), chemiluminescent, and fluorescence arrays,3,6 bio-barcode assays using oligonucleotide labels,5 and procedures utilizing surface plasmon resonance12 or electrochemistry.3,6 Typical detection limits (DL) for commercial bead-based assays are 1–10 pg mL−1 for various target proteins.6 Instruments and assay kits for these methods are relatively expensive and require trained operators.13 Alternatively, antibody microarrays for multiple proteins that employ high sensitivity multilabel formats have as yet unfulfilled promise for future automated point-of-care detection.3–6
Ultrasensitive protein immunoassays have been developed using antibody-coated magnetic beads to capture target proteins off-line. This strategy gave a DL of 14 fg mL−1 for serum prostate cancer biomarker prostate specific antigen (PSA)14 using fluorescent labels and optical fiber arrays,15 whereas a DL of 10 fg mL−116 was obtained using clustered magnetic particle labels in SPR for PSA in serum. Such sub-pg mL−1 DLs are required for some biomarker proteins,6 but have been elusive in multi-protein assays.
Electrochemical immunoarrays with nanostructured surfaces provide a relatively simple, inexpensive approach for detecting small panels of proteins.3,6 Such approaches usually require multielectrode microelectronic chips, as typified in papers by Wilson et al., who detected up to seven proteins at ng mL−1 levels using this approach.17,18 Similarly, Wong et al. described a 16-electrode chip19 to measure cancer biomarker protein interleukin (IL)-8 in saliva with a 7 pg mL−1 DL and IL-8 mRNA with DL of ~4 fM.20
We recently reported single-protein immunosensors featuring single-wall carbon nanotube (SWCNT) forests and gold nanoparticle (AuNP) electrodes combined with multilabel enzyme-antibody detection particles to achieve low- and sub-pg mL−1 DLs for PSA and interleukin-6 (IL-6).21–24 We used an array of four SWCNT electrodes with amperometric detection to simultaneously measure biomarker proteins PSA, prostate specific membrane antigen (PSMA), platelet factor-4 (PF-4) and IL-6 in cancer patient serum.25 Nanostructured electrode surfaces on these sensor platforms provide high functionalized surface areas for the attachment of large populations of capture antibodies.26 We recently interfaced 8-electrode nanostructured arrays with microfluidics and off-line protein capture using multiply labeled magnetic particles for multiplexed amperometric detection of PSA and IL-6 with sub-pg mL−1 DLs.27
ECL is an electrode-driven luminescence process that presents a general sensing strategy not requiring a light source.28,29 In a typical scenario, Ru(bpy)32+ (RuBPY) labels produce ECL in a multi-step catalytic redox process featuring sacrificial reductant tripropylamine (TprA) to yield photoexcited [Ru(bpy)32+]* that emits light at 610 nm.29 An advantage of ECL for arrays is the lack of need for multi-electrode chips. Here we achieve ECL measurements on a simple carbon block electrode using RuBPY labels in an electrochemical cell, with a CCD camera to capture the light. We used a similar approach previously to measure relative reaction rates of metabolites with DNA in toxicity screening arrays. 30,31 Recently, we used such toxicity screening arrays with metabolic enzymes to validate genotoxicity screening by generating reactive metabolites of 11 different compounds.32
In a preliminary communication, we reported RuBPY-doped mesoporous silica nanoparticle labels with SWCNT forest sensors to develop single stand-alone immunosensors for accurate and sensitive detection of PSA in cancer patient serum.33 SWCNT forests feature ~30 nm terminally carboxylated nanotubes self-assembled in upright bundles on a thin Nafion-iron oxide layer.21,34,35 These upright, functionalized nanotubes provide a large conductive surface area for attachment of capture antibodies and an operating voltage window extending to the relatively high positive potentials needed to generate ECL from RuBPY. In the ECL immunosensor, PSA was captured from serum by antibodies on the sensor surface, then RuBPY-silica nanoparticles with surface secondary antibodies (Ab2) were added to bind to the PSA. ECL was measured by a photomultiplier tube at an applied potential of 0.95 V vs SCE in the presence of TPrA. The RuBPY-silica nanoparticles provide amplification via incorporation of thousands of RuBPY ions, and the sensor provided a DL of 40 pg mL−1 for PSA in serum.33
In the present work, we report fabrication of the first ECL array for multiple protein detection featuring microwells incorporating antibody-coated SWCNT forests surrounded by a hydrophobic polymer walls. The microwells localize up to 10 µL of sample solution over antibody-coated SWCNTs for protein capture. The array base is a simple 1 × 1 in. pyrolytic graphite block connected to a potentiostat, thus avoiding microelectronic fabrication. Light is measured with a coupled charged device (CCD) camera (Scheme 1). PSA and IL-6 were chosen as test analytes since they are both important biomarker proteins in serum for prostate cancer.6,14 The utility of this approach was demonstrated by detection of simultaneously in serum with sub-pg mL−1 DLs, and by accurate measurement of the two biomarkers in the serum of prostate cancer patients.
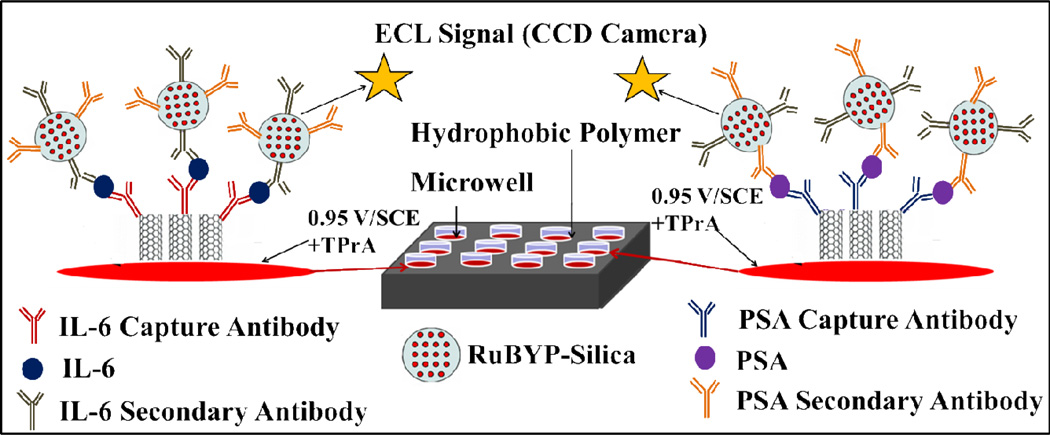
Scheme 1.
Representation of ECL immunoarray showing discrete wells in red on a 1 × 1 in. pyrolytic graphite chip (on left, black). SWCNT forests are surrounded by hydrophobic polymer (white) to make microwells on the chip. Wells are filled with sample solutions and SWCNT forests decorated with primary antibodies in the bottom of the well, capture the analyte proteins. RuBPY-silica nanoparticles with cognate secondary antibodies are added and bind to the captured protein analytes. Appropriate washing with blocking buffers minimizes non-specific binding. The chip is placed in an open top electrochemical cell, 0.95 V vs. SCE is applied, and ECL is detected by a CCD camera.
EXPERIMENTAL SECTION (full details in Supporting Information)
Chemicals and Materials
Lyophilized 99% bovine serum albumin (BSA), Tween-20, Tris(2,2'-bipyridyl)dichlororuthenium(II) hexahydrate, poly(diallydimethylammonium chloride) (PDDA) and poly acrylic acid (PAA) and tripropylamine (TPrA) were from Sigma/Aldrich. Single-walled carbon nanotubes (HiPco) were from Carbon Nanotechnologies, Inc. Monoclonal (mouse) primary anti-human prostate specific antigen (PSA) antibody (clone no. CHYH1), tracer secondary anti-PSA antibody (clone no. CHYH2), and PSA standards were from Anogen/Yes Biotech Lab, Ltd. DuoSet Human IL-6 containing monoclonal anti-human Interleukin-6 (IL-6) antibody, anti-human IL-6 antibody and human IL-6 standard were from R&D systems, Inc. Human serum and prostate serum samples from patients and controls were from Capital Biosciences. Immunoreagents were dissolved in pH 7.0 phosphate saline (PBS) buffer (0.01 M in phosphate, 0.14 M NaCl, 2.7 mM KCl) unless otherwise noted. 1-(3-(dimethylamino)-propyl)-3-ethylcarbodiimide hydrochloride (EDC) and N-hydroxysulfosuccinimide (NHSS) were dissolved in water immediately before use. PG block (1 × 1 in.) was obtained from Advanced Ceramics.
[Ru(bpy)3]2+-doped silica nanoparticles were synthesized as described previously.33 Average particle size was 102±9 nm (Figure S1). The ECL detection particle was prepared by sequential layer-by-layer deposition of positively charged PDDA and negatively charged PAA onto RuBPY-silica particles. A 1:1 mixture of anti-IL6 secondary antibodies (IL6-Ab2) and anti-PSA secondary antibodies (PSA-Ab2) were then attached via EDC-NHSS amidization.33
Fabrication of ECL arrays
Prior to formation of SWCNT forests, the area around each analytical spot was inked by poly-butadiene PAP pens (5 mm tip width, Sigma Aldrich) on a 1 × 1 pyrolytic graphite (PG) chip to form a hydrophobic barrier capable of containing ~10 µL of solution. SWCNT forests were assembled in each of these microwells by first spotting thin layers of 10 µL of Nafion and 15 µL of Fe(OH)x (pH:1.7–1.8), followed by 5 µL of SWCNT solution (0.1 mg mL−1 in DMF).21,34,35 For AFM, SWCNT forest microwells were prepared on freshly-cleaved mica.
Array fabrication and measurements
The immunoassay was performed over each microwell on the PG chip. SWCNTs were incubated with 10 µL of 33 µg mL−1 PSA capture antibody (PSA-Ab1) and 100 µg mL−1 IL-6 capture antibody (IL-6-Ab1), which were activated by addition of 15 µL of freshly prepared 400 mM EDC and 100 mM NHSS in pure water. The array was then washed by shaking the ECL sensor on a platform shaker (New Brunswick Scientific) at 200 rpm once in 0.05% Tween-20/ PBS buffer (pH 7) and twice in PBS buffer (pH 7) for 3 minutes each. To minimize evaporation during incubations, the immunosensor area was covered by an inverted beaker that had been rinsed with water to increase humidity. The capture antibody /SWCNT sensors were then incubated sequentially with 10 µL of 2% BSA, 5 µL of antigen (PSA/IL-6) in undiluted calf serum and 5 µL of ECL bioconjugate. The bioconjugate features secondary antibodies for PSA and IL-6 attached to a RuBPY-silica nanoparticle. The PSA antibody will capture PSA antigens and the IL-6 antibody will capture Il-6 antigens.. Each addition mentioned above was followed by a washing step. For measurement of ECL, the array with captured analytes was placed in a 150-mL beaker filled to 60 mL with 100 mM TPrA, 0.05% tween 20 and 0.05% triton X-100 in pH 7.5 buffer in a dark box.30 The assembled array had a single connection to a potentiostat, with a cylindrical platinum mesh counter electrode placed directly above and around the perimeter of the array, and an Ag/AgCl reference electrode (Figure S3). A potential of 0.95 V versus Ag/AgCl was applied to the array electrode for 400 s using a CH Instruments model 1232 electrochemical analyzer. ECL light intensity was integrated by the CCD camera (Chem 1 Genius Bioimaging system). Data analysis and quantification was done using GeneSnap and GeneTools software provided by SynGene.
RESULTS AND DISCUSSION
Array fabrication and characterization
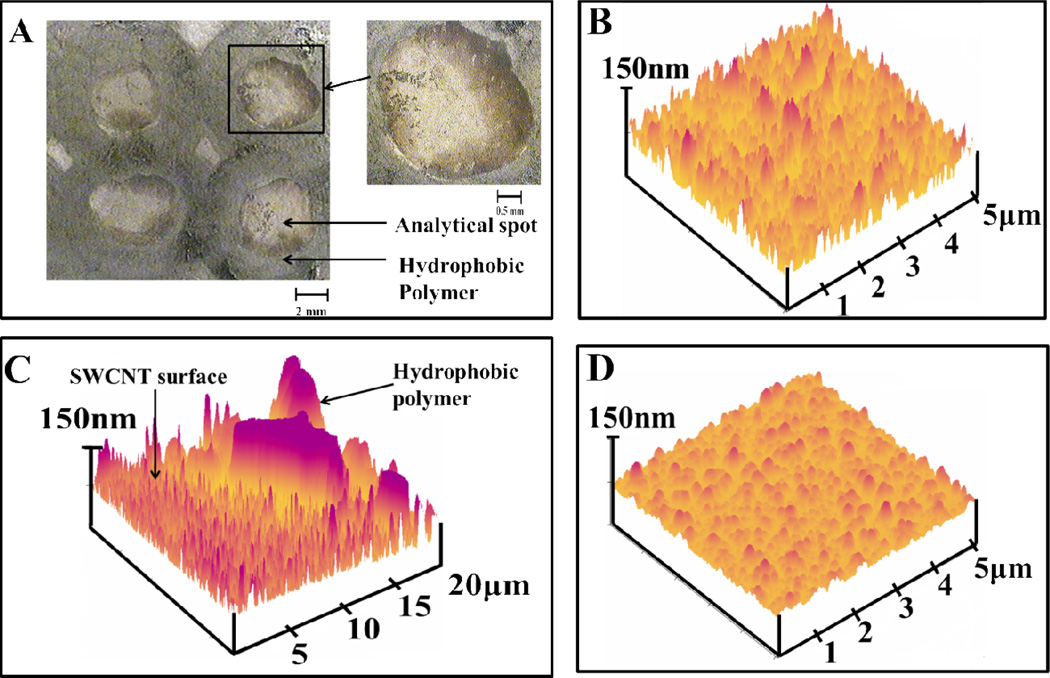
We prepared 12 to 16 evenly-spaced SWCNT forest spots on 1×1 in. pyrolytic graphite blocks. Each spot was surrounded with a hydrophobic barrier by “inking-on” poly(butadiene) using commercial PAP pens.36 These green-tinged polymer barriers create shallow microwells of ~2 mm diameter capable of holding up to 10 µL of sample (Scheme 1). Figure 1A shows an optical micrograph of 4 microwells with diameter ~2 mm on a PG block with a clear view of the light green hydrophobic polymer walls surrounding the SWCNT forests. The inset shows a larger view of a single microwell.
Figure 1.
Microscopy of microwells: A) Optical micrograph of 4 spots on a pyrolytic graphite array showing the light green hydrophobic polymer wall surrounding SWCNT forest spots. The inset shows a single SWCNT well surrounded by hydrophobic polymer. (B to D) are tapping mode atomic force microscope images of films on mica: (B) dense SWCNT forest in the bottom of an analytical well; (C) view showing the polymer wall and the adjacent SWCNT forest; D) SWCNT forest in the bottom of a microwell after covalent linkage of 2 nmol mL−1 anti-PSA antibody in pH 7.0 PBS buffer + 0.05% Tween-20 followed by washing with PBS buffer.
Tapping mode AFM images of 5 × 5 µm sections of microwells were acquired for arrays made on freshly cleaved flat mica (see Methods). Image of SWCNT forests in the well bottoms revealed a dense, spiky vertical assembly with surface roughness 22±1 nm and showed nearly full coverage of the underlying flat mica (Figure 1B). Images were similar to those reported previously for SWCNT forests on mica without the surrounding polymer walls, which had surface roughness 21±3 nm.26 AFM images taken near the interface of the SWCNT microwell bottom and the polymer wall showed ~200–300 nm differences in height (Figure 1C). After primary antibody (Ab1) was covalently linked onto the nanotube forests, the spiky SWCNT forest features disappear and a surface with decreased roughness of 12±1 nm was revealed (Figure 1D, Table 1). AFM images and roughness were similar to those of other antibody layers on SWCNT forests without the polymer wall. 21,26
Array reproducibility
Before proceeding to specific applications, the following criteria were pursued: (a) spot-to-spot reproducibility of ECL on an array within ±10%; (b) negligible cross-contamination during spotting and washing, and (c) minimal cross-reactivity of antibodies. In these studies, capture antibodies for PSA and IL-6 were attached onto the carboxylated ends of SWCNT forests in the bottoms of different wells by amidization21 (Scheme 1). This step was followed by incubation with 2% bovine serum albumin (BSA) to block nonspecific binding, and then incubation for 1 h with sample or standard containing analyte proteins in serum. Each solution of 10 µL or less was completely held in the wells until washing. Finally, a 5 µL dispersion of RuBPY-Silica - secondary antibody (Ab2) nanoparticles containing BSA and Tween-20/PBS buffer (pH 7) was incubated for 1 h with each spot. After washing with detergent and buffer, the array was placed into an open-top electrochemical cell, a potential of 0.95 V was applied, and ECL was collected with the CCD camera for 400 s, which gave an optimum difference between controls and analytes. The electrolyte solution contained TPrA, which is directly oxidized on the sensor surface facilitating a complex pathway involving [Ru(bpy)3]2+ that generates ECL.33
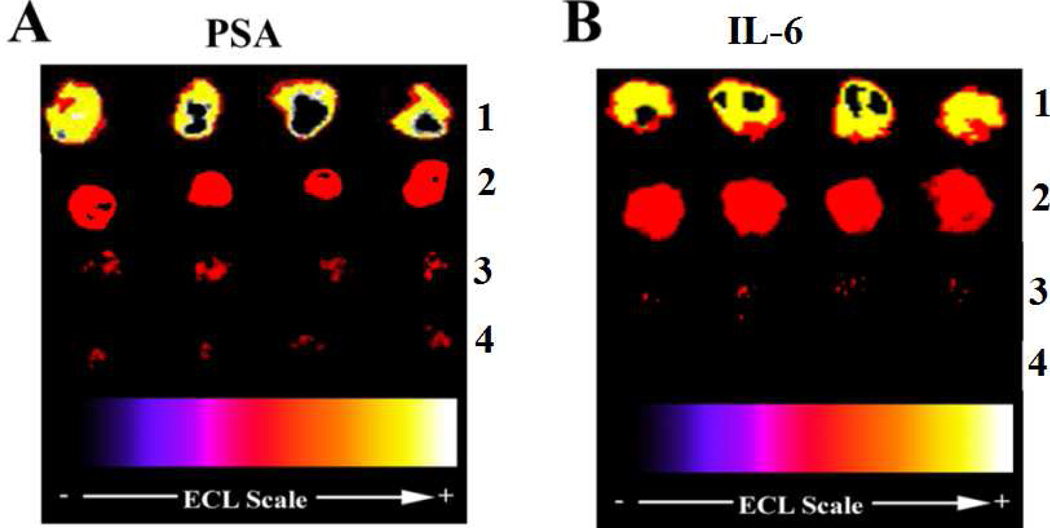
Reproducibility was assessed using assays to measure individual proteins in calf serum, which provides a good surrogate for human serum in immunoassays.21 Immunoassays on 16 spots for control with no PSA gave similar ECL responses (Supporting information, Figure S5) and measured relative ECL intensities were within ±5%. Immunoassays for PSA and IL-6 also had good reproducibility (Figure 2). ECL arrays reproducibly detected [PSA] (10 ng mL−1, 0.4 ng mL−1, 1 pg mL−1) and [IL-6] (2 ng mL−1, 0.2 ng mL−1, 0.1 pg mL−1) along with controls (0 PSA, 0 IL-6) (Figure 2). Relative ECL intensities for each antigen were reproducible within ±10% (Supporting information, Tables S3, S4, Figures S7, S9). Negligible cross contamination between spots occurs on the chip, and the antibodies used show no cross-reactivity with non-cognate proteins (Supporting information, Figures S10, S11).
Figure 2.
Examples of reproducible detection of PSA and IL-6 in calf serum using microwell arrays. ECL images obtained at 0.95 V vs Ag/AgCl in the presence of 0.05% Tween 20 + 0.05% Triton-X 100 + 100 mM TPrA in 0.2 M phosphate buffer (pH 7.5). CCD image of ECL array with 16 individual immunoassay spots for A) PSA at (1) 10 ng mL−1, (2) 0.4 ng mL−1, (3) 1 pg mL−1, 4) 0 pg mL−1 (control). B) IL-6 at (1) 2 ng mL−1, (2) 0.2 ng mL−1, (3) 0.1 pg mL−1, (4) 0 pg mL−1 (control).
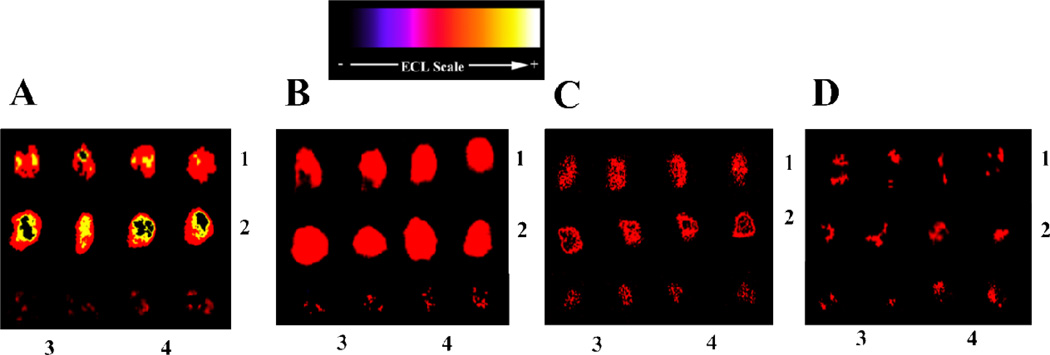
Next, we developed conditions to provide an acceptable dynamic range for PSA and IL-6 in mixtures. Emphasis was placed on developing assays that were sensitive over clinically relevant levels of PSA (~1 pg mL−1 to above 10 ng mL−1) and IL-6 (<1 pg mL−1 to >2 ng mL−1) in mixtures. The protocol utilized labels that featured antibodies to both PSA and IL-6 conjugated to the same RuBPY-silica nanoparticle. Analysis of the particles showed that ~26 secondary antibodies (PSA-Ab2 and IL-6-Ab2) are bound to a single nanoparticle, and each particle contains ~5.9×105 [[Ru(bpy)3]2+] molecules (Supporting information; Figures S2, S3). Thus, the RuBPY-silica particles are bound and can generate a high local concentration of electrochemiluminescent product in the shallow wells (~2 mm diam.). The Ab2/RuBPY-Silica particle ratio (26:1) drives the antibody-analyte binding to provide good capture efficiency. Digitally reconstructed, re-colored images for PSA and IL-6 mixtures on the same array are shown in Figure 3A–D. There was no significant difference between the ECL signals obtained using single or mixed antibodies attached to RuBPY-silica nanoparticles (Supporting information, Figure S12, Table S5).
Figure 3.
Microwell array images showing detection of PSA and IL-6 mixture in calf serum, obtained at 0.95 V vs Ag/AgCl using 0.05% Tween 20 + 0.05% Triton-X 100 + 100 mM TprA, pH 7.5. RuBPY-silica nanoparticles were used with antibodies attached for both proteins: (A) (1) 5 ng mL−1 PSA, (2) 1 ng mL−1 IL-6:, (B) (1) 0.4 ng mL−1 PSA, (2) 0.2 ng mL−1 IL-6, (C) 1) 40 pg mL−1 PSA, (2) 20 pg mL−1 IL-6, and D) (1) 1 pg mL−1 PSA, 2) 0.25 pg mL−1 IL-6. In all images, controls are indicated by duplicate spots (3) 0 pg mL−1 IL-6, and (4) 0 pg mL−1 PSA.
While semi-log calibration graphs were curved upward, they are quite usable for the determination of these proteins in the clinically relevant range in serum. The calibration curves vs. protein concentration are shown in Figure S13. For PSA, DL of 1 pg mL−1 was obtained as three times the standard deviation plus the ECL signal for the PSA-free control (Supporting information, Table S5). A slightly lower DL of 0.25 pg mL−1 was found for IL-6 and was only slightly above that for single biomarker detection (Supporting information, Table S5). The relative sensitivity for PSA (46.0 relative photon intensity-mL pg−1) and IL-6 (1954 relative photon intensity-mL pg−1) was obtained as slopes of the linear calibration graph in the lower range of antigen concentration (Figure S13, Table S5).
Analyses of patient serum
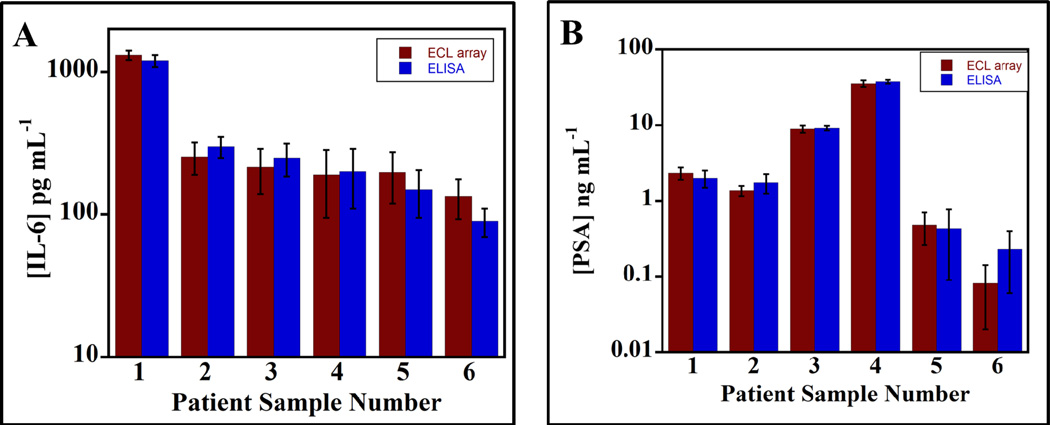
For validation of ECL arrays, PSA and IL-6 were determined in six human serum samples and compared with single-protein ELISAs (Figure 5). Samples were diluted 2-fold before assay, and if concentration was found to exceed the dynamic range of the calibration graphs, they were then diluted 15-fold. Calf serum was used as a diluent and all calibration standards were in calf serum. Samples 1 to 4 were from prostate cancer patients; samples 5 and 6 were from cancer-free patients. There was no significant difference in PSA and IL-6 values between the two methods as shown by t-tests at the 95% confidence level. ECL array results showed very good linear correlation plots with ELISA for all the serum samples with slopes close to 1.0, correlation coefficients of 0.999, and intercepts within experimental error of zero (Supporting Information, Figure S15 and Table S6).
Figure 5.
Comparison of simultaneous ECL array determinations of PSA and IL-6 in patient serum with individual ELISAs: (A) IL-6; (B) PSA. Error bars show standard deviations (n = 4). Samples 1 to 4 from prostate cancer patients; samples 5 and 6 were from cancer-free patients.
Results described above clearly show that arrays with SWCNT forests in microwells defined by polymer walls (Figure 1) coupled with ECL-producing nanoparticle labels (Scheme 1) are capable of simultaneous ultrasensitive detection of 2 proteins in serum. To help simplify protocols, the 100 nm RuBPY-silica detection particle had antibodies for both proteins attached. This approach provided detection of two cancer biomarker proteins in serum with high sensitivities and DLs of 1 pg mL−1 for PSA and 0.25 pg mL−1 for IL-6. These lower detection limits compared to our earlier ECL protein sensor were achieved by using a sensitive CCD camera and increased detection time to capture integrated light, i.e. 400 s as compared to 10 s used with a photomultiplier tube (PMT) in the single protein sensor.33 The dynamic range of the calibrations encompassed the clinically important levels of these proteins in normal and prostate cancer patient serum. Although calibrations were somewhat non-linear, they are quite adequate for simultaneous determinations of the two proteins. The ECL arrays were thus validated by analyzing serum samples from 4 prostate cancer patients and 2 cancer free patients, and results gave excellent correlations with standard single-protein ELISA analyses (Figures 5, S15 and Table S6).
The SWCNT microwell arrays achieved as good or better DLs than bead-based ECL systems and other commercial bead based assays that typically have DLs in the range 1–10 pg.6 A major advantage of the microwell ECL array is that no multi-electrode chip is needed, and the SWCNT wells are easily fabricated using wet chemistry with no specialized technologies. Hydrophobic polymer walls around the analytical wells, drawn with a PAP pen, effectively sequester up to 10 µL of sample or washing solutions directly above the Ab1-SWCNT that capture proteins in the well bottom, eliminating cross-contamination and improving reproducibility. It is likely that these microwells might be possible to fabricate more reproducibly and at higher density, by inexpensive ink-jet materials printing.37 A contrasting microbead-based immunoassay platform featuring microwells at the end of optical fiber bundles that can detect three proteins simultaneously by individually imaging fluorescently encoded microbeads by ECL was recently described, but is as yet in development stages and does not currently approach DLs reported here.38
In summary, ECL arrays with SWCNT forest microwells can achieve highly sensitive and accurate detection of two cancer biomarker proteins in patient serum. Novel features of the assay system include hydrophobic polymer surrounding antibody-SWCNT forest bioconjugates that capture protein analytes, and attachment of secondary antibodies for both proteins to the same RuBPY-silica detection particle. This system requires only simple wet chemistry for fabrication, and an inexpensive potentiostat and CCD camera for measurements. Detection limits for IL-6 and PSA on the ECL arrays were equivalent to or better than commercial bead-based protein measurement systems. These arrays should be readily adaptable to interfacing with microfluidics for simultaneous detection of up to 10 proteins. These goals are currently being pursued in our laboratory.
Supplementary Material
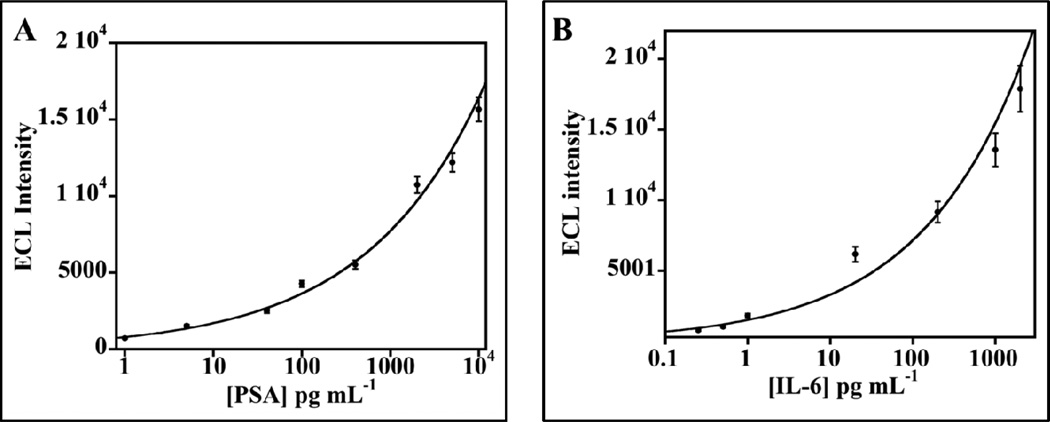
Figure 4.
Calibration of array responses accumulated over 400 s against clinically relevant concentration of PSA ([PSA]) and IL-6 ([IL-6]) in calf serum: A) Influence of PSA concentration on ECL signal. Control is without PSA. B) Influence of IL-6 concentration on ECL signal. ECL intensity for each antigen was plotted after subtracting ECL for protein-free controls. Error bars show standard deviations (n =4).
Acknowledgements
This work was supported by PHS grant ES013557 and in part by PHS grant ES03154 from NIEHS/NIH. The authors thank Drs. Vyomesh Patel and Silvio Gutkind for helpful suggestions concerning patient samples.
References
- 1.Wulfkuhle JD, Liotta LA, Petricoin EF. Nat. Rev. Cancer. 2003;3:267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 2.Kulasingam V, Diamandis EP. Nat. Clin. Prac. Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 3.Wang J. Biosens. Bioelectron. 2006;21:1887–1892. doi: 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 4.Kingsmore SF. Nat. Rev. Drug Discov. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giljohann DA, Mirkin CA. Nature. 2009;462:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rusling JF, Kumar CV, Patel V, Gutkind JS. Analyst. 2010;135:2496–2511. doi: 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voller A, Bartlett A, Bidwell DE. J. Clin. Pathol. 1978;31:507–520. doi: 10.1136/jcp.31.6.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldsmith SJ. Semin. Nucl. Med. 1975;5:125–152. doi: 10.1016/s0001-2998(75)80028-6. [DOI] [PubMed] [Google Scholar]
- 9.Schmalzing D, Nashabeh W. Electrophoresis. 1997;18:2184–2193. doi: 10.1002/elps.1150181209. [DOI] [PubMed] [Google Scholar]
- 10.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 11.Hawkridge AM, Muddiman DC. Ann. Rev. Anal. Chem. 2009;2:265–277. doi: 10.1146/annurev.anchem.1.031207.112942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HJ, Wark AW, Corn RM. Analyst. 2008;133:975–983. doi: 10.1039/b717527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Healy DA, Hayes CJ, Leonard P, McKenna L, O’Kennedy R. Trends Biotechnol. 2007;25:125–131. doi: 10.1016/j.tibtech.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Lilja H, Ulmert D, Vickers AJ. Nature Rev. Cancer. 2008;8:268–278. doi: 10.1038/nrc2351. [DOI] [PubMed] [Google Scholar]
- 15.Ghindilis AL, Atanasov P, Wilkins M, Wilkins E. Biosens. Bioelectron. 1998;13:113–131. doi: 10.1016/s0956-5663(97)00031-6. [DOI] [PubMed] [Google Scholar]
- 16.Krishnan S, Mani V, Kumar CV, Rusling JF. Angew. Chem. Int. Ed. 2011;50:1175–1178. doi: 10.1002/anie.201005607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat. Biotech. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 18.Wilson MS, Nie WY. Anal. Chem. 2006;78:2507–2513. doi: 10.1021/ac0518452. [DOI] [PubMed] [Google Scholar]
- 19.Wei F, Liao W, Xu Z, Yang Y, Wong DT, Ho CM. Small. 2009;5:1784–1790. doi: 10.1002/smll.200900369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei F, Patel P, Liao W, Chaudhry K, Zhang L, Arellano-Garcia M, Hu S, Elashoff D, Zhou H, Shukla S, Shah F, Ho CM, Wong DT. Clin Cancer Res. 2009;15:4446–4452. doi: 10.1158/1078-0432.CCR-09-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X, Munge B, Patel V, Jensen G, Bhirde A, Gong JD, Kim SN, Gillespie J, Gutkind JS, Papadimitrakopoulos F, Rusling JF. J. Am. Chem. Soc. 2006;128:11199–11205. doi: 10.1021/ja062117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mani V, Chikkaveeraiah BV, Patel V, Gutkind JS, Rusling JF. ACS Nano. 2009;3:585–594. doi: 10.1021/nn800863w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malhotra R, Patel V, Vaque JP, Gutkind JS, Rusling JF. Anal. Chem. 2010;82:3118–3123. doi: 10.1021/ac902802b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munge BS, Krause CE, Malhotra R, Patel V, Gutkind JS, Rusling JF. Electrochem. Comm. 2009;11:1009–1012. doi: 10.1016/j.elecom.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chikkaveeraiah BV, Bhirde A, Malhotra R, Patel V, Gutkind JS, Rusling JF. Anal. Chem. 2009;81:9129–9134. doi: 10.1021/ac9018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malhotra R, Papadimitrakopoulos F, Rusling JF. Langmuir. 2010;26:15050–15056. doi: 10.1021/la102306z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chikkaveeraiah BV, Mani V, Patel V, Gutkind JS, Rusling JF. Biosens. & Bioelectron. 2011;26:4477–4483. doi: 10.1016/j.bios.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debad JB, Glezer EN, Leland JK, Sigal GB, Wholstadter J. In: Electrogenerated Chemiluminescence. Bard AJ, editor. NY: Marcel Dekker; 2004. pp. 359–396. [Google Scholar]
- 29.a) Forster RJ, Bertoncello P, Keyes TE. Annu. Rev. Anal. Chem. 2009;2:359–385. doi: 10.1146/annurev-anchem-060908-155305. [DOI] [PubMed] [Google Scholar]; b) Miao WJ. Chem. Rev. 2008;108:2506–2553. doi: 10.1021/cr068083a. [DOI] [PubMed] [Google Scholar]; c) Richter MM. Chem. Rev. 2004;104:3003–3036. doi: 10.1021/cr020373d. [DOI] [PubMed] [Google Scholar]
- 30.Hvastkovs EG, So M, Krishnan S, Bajrami B, Tarun M, Jansson I, Schenkman JB, Rusling JF. Anal. Chem. 2007;79:1897–1906. doi: 10.1021/ac061975q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnan S, Bajrami B, Hvastkovs EG, Choudhary D, Schenkman JB, Rusling JF. Anal Chem. 2008;80:5279–5285. doi: 10.1021/ac800763r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan S, Zhao L, Schenkman JB, Rusling JF. Anal. Chem. 2011;83:2754–2760. doi: 10.1021/ac200050n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sardesai NP, Pan S, Rusling JF. Chem. Commun. 2009:4968–4970. doi: 10.1039/b909220j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SN, Rusling JF, Papadimitrakopolous F. Adv. Mater. 2007;19:3214–3228. doi: 10.1002/adma.200700665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rusling JF, Yu X, Munge BS, Kim SN, Papadimitrakopoulos F. Single-Wall Carbon Nanotube Forests in Biosensors. In: Davis J, editor. Engineering the Bioelectronic Interface. UK: Royal Soc. Chem.; 2009. pp. 94–118. [Google Scholar]
- 36.Wu H, Huo Q, Varnum S, Wang J, Liu G, Nie Z, Liu J, Lin Y. Analyst. 2008;133:1550–1555. doi: 10.1039/b719810h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen GC, Krause CE, Sotzing GA, Rusling JF. Phys. Chem. Chem. Phys. 2011;13:4888–4894. doi: 10.1039/c0cp01755h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deiss F, LaFratta CN, Symer M, Blicharz TM, Sojic N, Walt DR. J. Am. Chem. Soc. 2009;131:6088–6089. doi: 10.1021/ja901876z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.