Abstract
Antimicrobial peptides coupled to a ligand, receptor or antibody for a specific pathogenic bacteria could be used to develop narrow-spectrum pharmaceuticals with ‘targeted’ antimicrobial activity void of adverse reactions often associated with the use of broad-spectrum antibiotics. To assess the feasibility of this approach, in this study sheep myeloid antimicrobial peptide (SMAP) 28 was linked to affinity- and protein G-purified rabbit immunoglobulin G (IgG) antibodies specific to the outer surface of Porphyromonas gingivalis strain 381. The selective activity of the P. gingivalis IgG–SMAP28 conjugate was then assessed by adding it to an artificially generated microbial community containing P. gingivalis, Aggregatibacter actinomycetemcomitans and Peptostreptococcus micros. The specificity of the P. gingivalis IgG–SMAP28 conjugate in this mixed culture was concentration-dependent. The conjugate at 50 μg protein/mL lacked specificity and killed P. gingivalis, A. actinomycetemcomitans and P. micros. The conjugate at 20 μg protein/mL was more specific and killed P. gingivalis. This is an initial step to develop a selective antimicrobial agent that can eliminate a specific periodontal pathogen, such as P. gingivalis, from patients with periodontal disease without harming the normal commensal flora.
Keywords: Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, Peptostreptococcus micros, Cathelicidins, Targeted antimicrobial activity, SMAP28
1. Introduction
Antibiotics have the proven capacity to inhibit or kill microorganisms and thus are successfully used to treat many bacterial diseases in animals and humans [1,2]. Unfortunately, there are some problems. First, the widespread, indiscriminate or excessive use of many common antibiotics is thought to lead to the development of drug-resistant bacteria [2–4]. Second, antibiotic use in some individuals can result in a variety of ill effects [5]. Nausea, vomiting, abdominal discomfort, diarrhoea, allergic skin rashes and drug fever are induced by the penicillins, cephalosporins and other-related β-lactam antibiotics. Neurotoxicity, nephrotoxicity and vestibular toxicity are induced by the aminoglycosides. Infusion-site phlebitis is induced by the glycopeptides, and bone marrow toxicity is induced by chloramphenicol [5,6]. Finally, normal commensal flora are also susceptible to killing by antibiotics. Reduction in the types of bacteria and their concentrations leads to secondary Candida albicans infections on mucosal surfaces or to Clostridium difficile infections in the gastrointestinal tract [6].
Both local and systemic antibiotic therapy is used to treat periodontal disease [7,8]. Systemically administered antibiotics provide a clear clinical benefit; periodontal attachment levels are much better in patients post therapy compared with patients not receiving these agents [9,10]. Combined with non-surgical and surgical treatment, antibiotics also reduce the number of periodontal pathogens. However, many of the above described side effects still occur. Therefore, there is a need to develop a more ‘organism-specific’ targeted approach that kills specific periodontal pathogenic bacteria in complex polymicrobial environments, yet does not alter the composition or concentration of the normal commensal flora.
Antimicrobial peptides represent a new class of antibiotics that rapidly lyse or inhibit susceptible bacteria, are easy to synthesise in large quantities, work in synergy with other antimicrobial agents, and have very few side effects [11–13]. Because their antimicrobial activity is fast and their minimal inhibitory concentrations (MICs) are low for many susceptible bacteria, it is thought that antimicrobial peptides generally do not induce bacterial resistance [14]. However, some bacteria have proteolytic enzymes, positively charged moieties linked to teichoic acids, and positively charged moieties linked to lipopolysaccharides that allow them to resist the antimicrobial activity of these peptides [15–18]. Recently, Perron et al. [19] induced experimental resistance in Escherichia coli and Pseudomonas fluorescens to pexiganan, an analogue of magainin. Therefore, indiscriminate use of broad-spectrum antimicrobial peptides may not be prudent [19,20]. However, if antimicrobial peptides were coupled to a ligand or receptor for a specific pathogenic bacteria, they could be used at lower concentrations, would have narrow-spectrum or ‘targeted’ antimicrobial activity, and induce fewer side effects. Porphyromonas gingivalis, which is generally thought to be resistant to human β-defensins [21] and some antimicrobial peptides [22], may be a potential target.
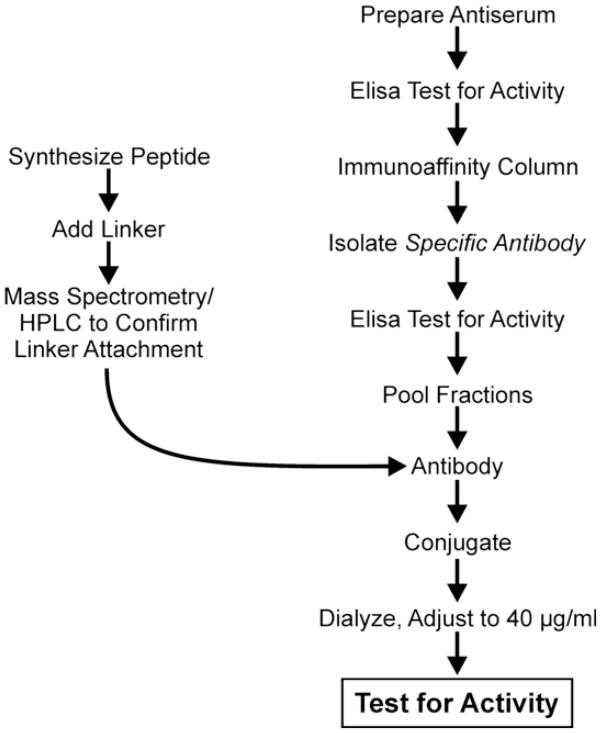
In this study, sheep myeloid antimicrobial peptide (SMAP) 28 was linked to affinity- and protein G-purified rabbit immunoglobulin G (IgG) antibodies specific to the outer surface of P. gingivalis strain 381 (Fig. 1) and it was demonstrated that it could selectively kill P. gingivalis in an artificially generated microbial community containing P. gingivalis, Aggregatibacter actinomycetemcomitans and Peptostreptococcus micros. This approach is an initial step for developing a selective antimicrobial agent capable of eliminating a specific periodontal pathogen, such as P. gingivalis, from patients with periodontal disease without harming the normal commensal flora.
Fig. 1.
An overview of the procedure used to prepare the Porphyromonas gingivalis IgG-SMAP28 conjugate. Antiserum to whole cells of P. gingivalis was prepared in rabbits. Specific cell surface antibodies were then isolated with an immunoaffinity column and specific IgG was isolated using a protein G column. SMAP28 was then synthesised and a maleimide linker was attached. SMAP28 with the maleimide linker was attached to the specific P. gingivalis IgG antibody and dialysed. The conjugate was then tested for specific antimicrobial activity. IgG, immunoglobulin G; SMAP28, sheep myeloid antimicrobial peptide 28.
2. Materials and methods
2.1. Synthesis of SMAP28
SMAP28 (RGLRRLGRKIAHGVKKYGPTVLRIIRIA-(NH2)) was synthesised by NeoMPS, Inc. (San Diego, CA) and suspended in 0.01 M sodium phosphate buffer (pH 7.2) with 145 mM NaCl (0.01 M phosphate-buffered saline (PBS) (pH 7.2)). Its purity was confirmed by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF-MS) (High Resolution Mass Spectrometry Facility, The University of Iowa, Iowa City, IA) and reversed-phase high-performance liquid chromatography (HPLC).
2.2. Bacterial species and growth conditions
Porphyromonas gingivalis strain 381 (obtained from Ann Progulske-Fox, Department of Oral Biology, University of Florida, Gainesville, FL), A. actinomycetemcomitans FDC-Y4 and P. micros were used. All three organisms were grown as previously described at 37 °C in an atmosphere containing 85% N2, 10% H2 and 5% CO2 [23]. Porphyromonas gingivalis strain 381 was grown in tryptic soy broth (Difco Laboratories, Detroit, MI) supplemented with 5 μg/mL hemin (Sigma, St Louis, MO) and vitamin K (Sigma). Aggregatibacter actinomycetemcomitans was grown in tryptic soy broth supplemented with 0.6% yeast extract (Difco Laboratories). Peptostreptococcus micros was grown in brain–heart infusion (Difco Laboratories) supplemented with 0.5% neopeptone and 5 μg/mL hemin.
2.3. Preparation of antiserum and isolation of P. gingivalis-specific IgG antibody
A water-in-oil emulsified, whole-cell bacterin was prepared. Each immunising 1 mL dose contained 1.0 × 108 colony-forming units (CFU) of P. gingivalis strain 381 and 25 μg of MDP in 140 mM NaCl with 0.3% formalin in a 50% oil emulsion of Freund’s incomplete adjuvant. Two rabbits were immunised seven times over a 13-week period (IMGENEX Corp., San Diego, CA). Pre-bleed serum samples were collected before immunisation and antiserum samples were collected at 9, 11 and 13 weeks.
Rabbit IgG antibody to P. gingivalis cell surface antigens was isolated by affinity chromatography using the Pierce AminoLink® Plus Immobilization Kit (Nos. 44894 and 20394; Pierce, Rockford, IL) to link formalin-fixed whole cells of P. gingivalis to the gel matrix. The coupling efficiency was estimated to be 33.9% for P. gingivalis whole cells. Porphyromonas gingivalis-specific IgG antibodies were isolated from the affinity column-eluted fractions concentrated using a HiTrap Protein G HP Cartridge (No. 17-0405-01; Pierce) and dialysed overnight against 0.01 M PBS (pH 7.2). Seven millilitres of 0.10 mg/mL (total protein 0.7 mg) of affinity-purified P. gingivalis IgG antibody was obtained. IgG isolated from pre-immune rabbit serum (IgGAb control) and IgG to P. gingivalis isolated from immune rabbit serum (PgAb control) were used as control solutions.
2.4. Whole cell enzyme-linked immunosorbent assay (ELISA) analysis
A whole-cell ELISA was performed at 26 °C as described by Zagursky et al. [24]. Briefly, a suspension of P. gingivalis in 140 mM NaCl with 0.3% formalin was adjusted to contain 1.0 × 108 CFU/mL in the spectrophotometer (0.108 optical density (O.D.) at 600 nm) and then 100 μL/well of this suspension was put into Immulon 1 microtitre wells and dried overnight. Non-specific antibody binding sites were blocked with 0.01 M Tris buffer containing 0.145 M NaCl, 1.0% fish gelatin and 0.05% blocking buffer and then two-fold dilutions of rabbit antiserum were added. After 60 min, all wells were washed twice with blocking buffer and bound rabbit IgG was detected with peroxidase-labelled goat anti-rabbit IgG antibody (1 μg/mL) (No. 074-1506; KPL, Inc., Gaithersburg, MD) using tetramethylbenzidine (TMB) peroxidase developing reagent (No. 50-85-05; KPL, Inc.) and TMB stop reagent (No. 50-85-05; KPL, Inc.). The O.D. values of the wells in the plate were determined at 450 nm (PowerWaveX; Bio-Tek Instruments, Inc., Winooski, VT) and values of the blank wells were subtracted from the test values. Linear regression of the log2 of dilution (x-axis) was plotted against the O.D. of the dilution (y-axis) to determine the slope of the line. Antibody titre was calculated as the log2 dilution value at the 0.1 O.D. intercept [25].
2.5. SMAP28 and IgG conjugation
SMAP28 was conjugated to affinity-purified IgG antibody using the Controlled Protein–Protein Cross-Linking Kit (No. 23456; Pierce) as outlined in Fig. 1. This kit uses maleimide to link the amine groups of SMAP28 to the sulfhydryl groups of the IgG antibody. SMAP28 contains 11 free amine groups, of which 4 are available for linker attachment, 1 at each lysine or arginine residue.
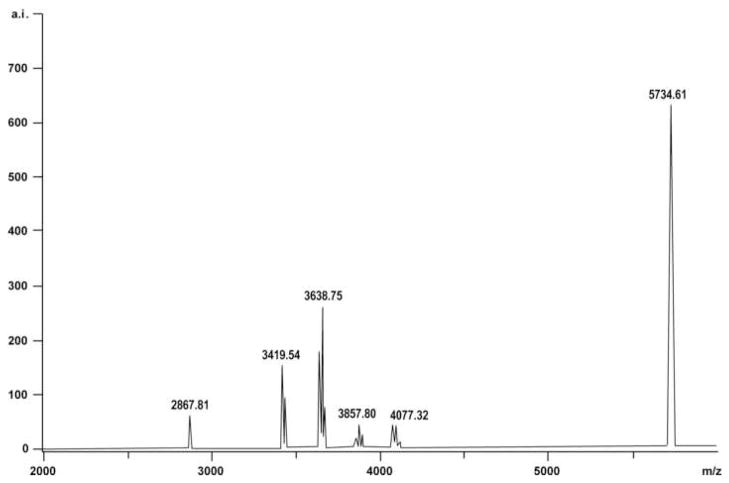
SMAP28 (1.0 mg/mL of 0.01 M PBS (pH 7.2)) was mixed with Sulfo-SMCC and incubated for 30 min at 26 °C. Unreacted maleimide reagents were removed using a Dextran Desalting Column. Peptide concentrations in eluted fractions were determined and those containing SMAP28 with maleimide linkers were pooled. MALDI-TOF-MS was used to confirm that maleimide linkers attached to SMAP28 (Fig. 2).
Fig. 2.
Matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry (MALDI-TOF-MS) showing that the maleimide linkers were successfully attached to sheep myeloid antimicrobial peptide (SMAP) 28. Up to four distinct peaks were seen. Each peak corresponded to the molecular mass of SMAP28 (i.e. 3198.95 Da) and the number of maleimide linkers attached (i.e. 219.24 Da). Predicted masses were 3418.19 Da (SMAP28 with one maleimide linker), 3637.43 Da (SMAP28 with two maleimide linkers), 3856.67 Da (SMAP28 with three maleimide linkers) and 4075.91 Da (SMAP28 with four maleimide linkers). Distinct peaks were seen at 3419.54, 3638.75, 3857.80 and 4077.32 m/z. Proteins with masses of 2867.81 Da and 5734.61 Da were used as internal calibration standards.
Two options (IIA and IIB) were used to prepare sulfhydryl-modified IgG antibody. In option IIA, the IgG antibody was suspended in 0.01 M PBS (pH 7.2), mixed with the kit conjugation/activation buffer and activated kit immobilised-reductant and incubated for 60 min at 37 °C. The mixture was centrifuged to pellet the kit immobilised-reductant. The supernatant contained the sulfhydryl-modified IgG antibody for the protein–protein cross-linking procedure with the SMAP28 with maleimide linkers.
In option IIB, sulfhydryl groups were artificially added by reaction with N-succinimidyl-S-acetylthioacetate (SATA). The IgG antibody was suspended in 0.01 M PBS (pH 7.2) (1.0 mL) and a 10 molar excess of SATA solution (SATA/dimethylformamide) was added and incubated for 20 min at 26 °C. Then, 100 μL of hydroxylamine HCl solution was added to the SATA-modified IgG antibody and the mixture was incubated for 120 min at 26 °C. Unbound reagents were removed by passing the de-protected sulfhydryl-IgG antibody solution over a desalting column. PBS–ethylene diamine tetra-acetic acid (EDTA) was used to elute sulfhydryl-IgG antibody and 1.0 mL fractions were collected. Protein concentrations were determined at A280 and fractions containing most of the sulfhydryl-IgG antibody were pooled. SMAP28 with maleimide linkers and sulfhydryl-IgG antibody were mixed in approximately equal molar amounts and incubated for 60 min at 26 °C.
Preliminary work showed that conjugation by option IIB was more effective and a P. gingivalis IgG–SMAP28 conjugate developed this way was more specific to P. gingivalis. Thereafter, the conjugate was prepared using option IIB and was called the P. gingivalis IgG–SMAP28 conjugate.
2.6. HPLC
SMAP28 and the SMAP28 with maleimide linkers were separated on an Acclaim 300, C18, 3 μm analytical column (Dionex Corp., San Francisco, CA) using a Summit HPLC system (Dionex) with Chromeleon software and eluted with a gradient of acetonitrile (0–100%) in 0.1% trifluoroacetic acid. Fractions were collected in a FC 144 fraction collector, pooled with similar fractions of previous runs and dried overnight by rotary evaporation under vacuum at room temperature.
2.7. Spot blot
A dot-blot assay was used to confirm the presence both of the affinity-purified rabbit IgG antibody and of SMAP28 in the P. gingivalis IgG–SMAP28 conjugate. Briefly, 1.0 μL of the P. gingivalis IgG–SMAP28 conjugate and P. gingivalis IgG were placed on Immobilon-P transfer membranes, allowed to dry and soaked overnight at 26 °C in blocking buffer. Rabbit IgG antibody in the P. gingivalis IgG–SMAP28 conjugate was detected with peroxidase-labelled goat anti-rabbit antibody (1.0 μg/mL) (No. 074-1506; KPL, Inc.) using CN peroxidase developing reagent (No. 50-73-014; KPL, Inc.). SMAP28 in the P. gingivalis IgG–SMAP28 conjugate was detected first using goat anti-SMAP28 antibody (1:20 dilution; courtesy of Brian Tack) [25] and then peroxidase-labelled rabbit anti-goat antibody (1.0 μg/mL) (No. 14-13-06, KPL, Inc.) using 4CN peroxidase developing reagent (No. 50-73-01; KPL, Inc.).
2.8. Radial diffusion antimicrobial assay
The MICs of SMAP28 and SMAP28 with maleimide linkers were determined by the radial diffusion assay as previously described in our laboratory [23,26,27]. After incubation for 24–48 h, zones of inhibition were recorded with a Boley gauge as radial diffusion units (zone of inhibition–well diameter × 10). The x-intercept was obtained from the relationship between radial diffusion units versus log10 peptide concentration as determined after regression [23].
2.9. Porphyromonas gingivalis IgG–SMAP28 conjugate-specific killing in a mixed culture suspension
The specificity and activity of the P. gingivalis IgG–SMAP28 conjugate was determined in an artificially generated microbial community containing P. micros, A. actinomycetemcomitans and P. gingivalis. These organisms were grown for 48 h in their respective media at 37 °C under anaerobic conditions and diluted in reduced saline to a density containing ca. 1 × 108 CFU/mL (0.108 O.D. at 600 nm) (Spectronic 20D+; Thermo Fisher Scientific, Inc., Waltham, MA). Each culture was diluted 10-fold to contain 104 CFU/mL. Then, 2.0, 0.25 and 2.0 mL of P. gingivalis, A. actinomycetemcomitans and P. micros, respectively, was added to 5.75 mL of reduced anaerobic saline (e.g. constituting the 10−5 dilution of each) and diluted two-fold with the P. gingivalis IgG–SMAP28 conjugate and two control solutions, IgGAb control and PgAb control. At 0, 10, 20 and 30 min, 100 μL was removed and plated onto anaerobic blood agar (CDC formulation; Remel, Lenexa, KS) and incubated at 37 °C. After 9 days, bacterial colonies of differing morphologies, sizes and colour were counted by three different observers.
2.10. Statistical analyses
The experiment was divided into treatments at different time points: P. gingivalis IgG–SMAP28 conjugate, P. gingivalis antibody control (PgAb control) and pre-immune IgG antibody control (IgGAb control) at 0, 10, 20 and 30 min, defining 12 total treatments for the study. Each treatment had three plates and the experiments were performed on two different days using two different freshly prepared conjugates. Since day-to-day differences could affect the assays, each day was considered a block.
A multifactorial analysis of variance (ANOVA) model was used to analyse the effect of treatments, time and day on the number of CFU as well as to assess two- and three-way interactions. Logarithm transformations of the form log10 (CFU+1) were used as needed to conform to model assumptions such as normality and homoscedasticity. Adjustments for multiple comparisons utilised Tukey’s studentized range.
3. Results
3.1. Antimicrobial activity of SMAP28 and SMAP28 with maleimide linkers
MALDI-TOF-MS showed that up to four maleimide linkers were attached to SMAP28, likely at available lysine or arginine residues (Fig. 2). When separated by HPLC, the linker solution contained four broad peaks that eluted at 13.5 min (fraction 14), 14.3 min (fraction 15), 14.8 min (fraction 15) and 15.6 min (fraction 16).
Fraction 15 contained masses 3199.64, 3418.32 and 3637.90 m/z corresponding to SMAP28, SMAP28 with one maleimide linker, and SMAP28 with two maleimide linkers, respectively. Fraction 16 contained masses 3419.65, 3638.87 and 3860.11 m/z corresponding to SMAP28 with one maleimide linker, SMAP28 with two maleimide linkers, and SMAP28 with three maleimide linkers, respectively. In other runs, fraction 17 contained masses 3419.35, 3637.90 and 3860.03 m/z corresponding to SMAP28 with one maleimide linker, SMAP28 with two maleimide linkers, and SMAP28 with three maleimide linkers, respectively (not shown).
The antimicrobial activities of SMAP28 and SMAP28 with maleimide linkers were nearly identical, suggesting that the presence of the linkers did not affect antimicrobial activity. The MICs for native SMAP28 were 3.9, 5.9 and 5.0 μg/mL for E. coli ATCC 12795, Pseudomonas aeruginosa ATCC 47085 and Staphylococcus aureus ATCC 29213, respectively. The MICs for SMAP28 with maleimide linkers solution were 4.7, 4.7 and 4.4 μg/mL for E. coli, P. aeruginosa and S. aureus, respectively.
3.2. Antimicrobial activity of the P. gingivalis IgG–SMAP28 conjugate
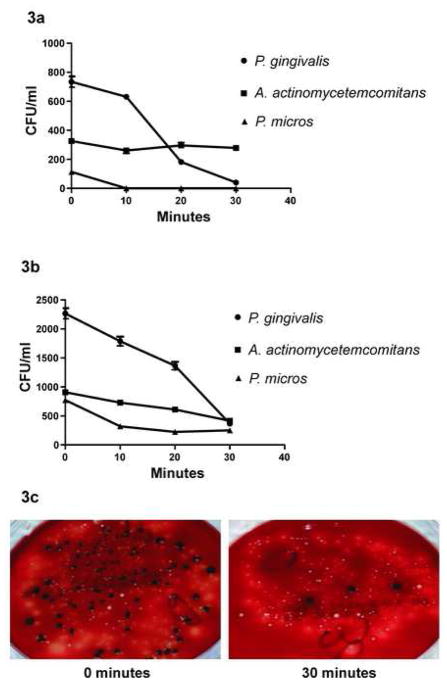
The effect of treatment with P. gingivalis IgG–SMAP28 conjugate, PgAb control or IgGAb control on the viability of P. gingivalis, A. actinomycetemcomitans and P. micros was assessed over four time periods. The P. gingivalis IgG–SMAP28 conjugate at 50 μg/mL final protein lacked specificity and killed P. gingivalis, A. actinomycetemcomitans and P. micros. The conjugate at 20 μg/mL final protein was more specific and the viability of P. gingivalis dropped at a faster rate than that of A. actinomycetemcomitans and P. micros (Table 1; Fig. 3).
Table 1.
Average rate of change in the number of colony-forming units over time and the Spearman rank correlation coefficient (ρ) to measure the strength and significance of the relationship. A simple linear regression model was used to determine these estimates by treatment and microorganism
| Microorganism | P. gingivalis IgG–SMAP28 conjugate | P. gingivalis IgG antibody control | IgG antibody isolated from pre-immune serum | |||
|---|---|---|---|---|---|---|
| ρ (P- value) | Average rate of change/10 min | ρ (P- value) | Average rate of change/10 min | ρ (P- value) | Average rate of change/10 min | |
| Porphyromonas gingivalis | −0.72 (0.0001) | 430 | −0.076 (0.81) | 15 | −0.42 (0.04) | 210 |
| Peptostreptococcus micros | −0.35 (0.095) | 102 | −0.089 (0.0001) | 83 | −0.42 (0.04) | 470 |
| Aggregatibacter actinomycetemcomitans | −0.33 (0.11) | 82 | −0.11 (0.73) | 10 | −0.13 (0.5) | 19 |
IgG, immunoglobulin G; SMAP28, sheep myeloid antimicrobial peptide 28.
Fig. 3.
Porphyromonas gingivalis IgG–SMAP28 conjugate-mediated killing of P. gingivalis in an artificially generated microbial community containing Aggregatibacter actinomycetemcomitans and Peptostreptococcus micros. (a,b) Two separate experiments show the decline of P. gingivalis after incubation with the conjugate. (c) At 30 min there was a clear reduction of small black P. gingivalis colonies. Colonies of A. actinomycetemcomitans and P. micros are still visible. The final concentration of conjugate protein was 20 μg/mL. IgG, immunoglobulin G; SMAP28, sheep myeloid antimicrobial peptide 28. CFU, colony-forming units.
For P. gingivalis, the P. gingivalis IgG–SMAP28 conjugate was active and there was a significant negative correlation with time in both experiments (P = 0.0001) (Table 1; Fig. 3). A significant negative correlation with time (P = 0.04) was also seen for P. gingivalis incubated in IgGAb control (Table 1). For P. gingivalis, there was a significant interaction between treatments and time (P < 0.0001). The treatment mean curves (Fig. 3) for the four time levels support this conclusion. The number of P. gingivalis CFU declined over time under all three treatments, but this relationship with time was significant under some treatments and not under others (Table 1).
Table 1 also lists the Spearman rank correlation coefficient (ρ) for this bacteria by treatment. Treatment with P. gingivalis IgG–SMAP28 conjugate showed a significant negative correlation with time (ρ = −0.72; P = 0.0001) and, similarly, the IgGAb control demonstrated a negative correlation with time (ρ = −0.42; P = 0.04). Using a simple linear regression line, the average rate of decline over time was estimated and is listed in Table 1. The average decline in the number of CFU for every 10 min for P. gingivalis was 430 (P. gingivalis IgG–SMAP28 conjugate), 15 (PgAb control) and 210 (IgGAb control).
For A. actinomycetemcomitans, the nature of time and treatment effects was not entirely consistent. The interaction between treatment and time was not significant (P = 0.61) in the first experimental replication but the interaction was significant (P = 0.0019) when run in the second experimental replication. The data for the first experimental replication did not show that time was a significant factor (P = 0.85) whilst treatment was significant (P = 0.02).
Tukey’s studentized range test demonstrated that treatment with the conjugate and the IgGAb control were significantly different. When the data were combined for the 2 days, the interaction between treatment and time became insignificant (P = 0.15). The Tukey’s studentized range test for the combined data indicated that all the pairwise comparisons between treatments were significant at the 5% level. The interaction between treatment, time and the day of the experiment for A. actinomycetemcomitans was not statistically significant (P = 0.09). Figures 3–5 shows the treatment mean curves for this bacterium.
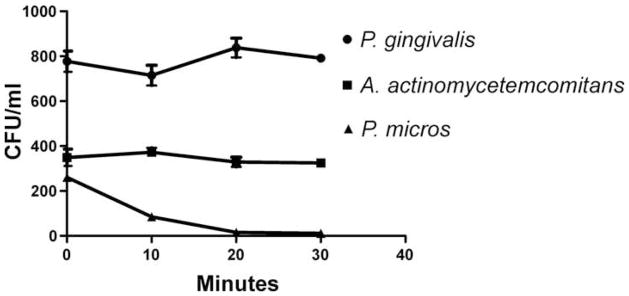
Fig. 5.

Viability of Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans and Peptostreptococcus micros incubated in the immunoglobulin G (IgG) antibody isolated from pre-immune serum. (a) In one experiment, the viability P. gingivalis and P. micros declined over the 30-min period, whereas the viability of A. actinomycetemcomitans did not. (b) In another experiment the bacterial concentrations of all three were increased. The viability of P. micros declined over the 30-min period, whereas the viability of P. gingivalis and A. actinomycetemcomitans did not. The final concentration of protein was 20 μg/mL. CFU, colony-forming units.
For P. micros, the interaction between time and treatment was not statistically significant (P = 0.22) in the first experimental replication but it was significant (P = 0.003) in the second experimental replication. The treatment mean curves for P. micros are shown in Figs 3–5. When the data for the 2 days were combined and analysed, the interaction between time and treatment became non-significant (P = 0.07), whilst both time and treatment were significant factors in the CFU. The three-way interaction between time, treatment and the day experiments were performed was not significant (P = 0.51).
Tukey’s studentized range test shows a significant difference (at the 5% level) between treatment with IgGAb control, P. gingivalis IgG–SMAP28 conjugate and PgAb control. Treatments with P. gingivalis IgG–SMAP28 conjugate and PgAb control were not significantly different at the 5% level. Peptostreptococcus micros showed a decline in bacterial counts over time for all four treatments (Figs 3–5). The Spearman correlation coefficient ρ (for P. micros) is listed in Table 1. The P. micros count in the IgGAb control (ρ = −0.42; P = 0.04) and PgAb control (ρ = −0.089; P = 0.0001) showed a significant negative correlation with time but this negative correlation was not significant for P. micros in the P. gingivalis IgG–SMAP28 conjugate (ρ = −0.35; P = 0.095). Using a simple linear regression line, the estimate of average rate of decline in CFU over time was evaluated and is listed in Table 1. This rate of decline for every 10 min is 102 (P. gingivalis IgG–SMAP28 conjugate), 83 (PgAb control) and 470 (IgGAb control).
4. Discussion
SMAP28 was conjugated to an IgG antibody specific to the outer surface of P. gingivalis via a maleimide linker. When added to an artificially generated microbial community containing P. gingivalis, A. actinomycetemcomitans and P. micros, the antimicrobial activity of the P. gingivalis IgG–SMAP28 conjugate for P. gingivalis was dependent upon the conjugation procedure and the concentration of the conjugate. At 50 μg/mL final protein, the conjugate lacked specificity and rapidly killed all three bacteria. At 20 μg/mL final protein, the conjugate was more specific and killed P. gingivalis. The two-factor ANOVA showed a strong interaction between treatment, time and experiment for all three bacteria.
Antimicrobial peptides can be directed to the surface of select microorganisms. This delivery system works well and has been used by other investigators. Peschen et al. [28] used a Fusarium spp.-specific antibody linked to antifungal peptides to protect plants against a Fusarium oxysporum f.sp. matthiolae infection. Qiu et al. [29] used a staphylococcal AgrD1 pheromone linked with the channel-forming domain of colicin Ia to kill meticillin-sensitive and -resistant S. aureus, but not Staphylococcus epidermidis or Streptococcus pneumoniae. Similarly, Qiu et al. [30] used an enterococcal cCF109 pheromone fused with the channel-forming domain of colicin Ia to kill vancomycin-resistant Enterococcus faecalis. Vancomycin-resistant E. faecalis-infected mice survived when treated with this peptide, but untreated control mice did not survive [30].
Although the concept in this study is feasible, additional work is needed (a) to reduce the variability in the antimicrobial activity of the conjugate, (b) to identify other carriers and (c) to define the mechanism of antimicrobial activity. For example, we observed variability in attaching both the maleimide linker and P. gingivalis IgG antibody to SMAP28 that likely induced some experiment-to-experiment variability in the antimicrobial activity of the conjugate seen in Figs 3–5. Furthermore, we could not determine the number of SMAP28 peptides bound to the P. gingivalis-specific antibodies nor their binding locations. It is likely that they did not bind to the Fab binding sites.
Other carriers may be more reliable. Future work will focus on identifying other carrier proteins as well as refining the specificity of the carrier peptide–SMAP28 conjugate for P. gingivalis. Besides antibodies, ligands could include fragments of hemin [31], peptides of proline-rich protein 1 [32–34], portions of Streptococcus gordonii SspB [35], fragments of glycoproteins [36] or fragment of lactoferrin [37].
SMAP29 has a disordered structure in aqueous buffers but has significant helicity in membrane-like environments, including sodium dodecyl sulphate (SDS) micelles, lipopolysaccharide dispersions and trifluoroethanol buffer systems [38]. A structure determined by nuclear magnetic resonance (NMR) indicated that residues 8–17 are helical, residues 18–19 form a hinge and residues 20–28 form an ordered hydrophobic segment. We do not know whether attachment of linkers or proteins to SMAP28 would alter its helical structure or impair its ability to insert into microbial membranes. It is likely the maleimide linkers are attached to lysine or arginine residues on the hydrophilic face of the SMAP28 α helix. IgG, linked to the hydrophilic face via maleimide, would likely allow the peptide to still form an α helix and insert into the bacterial membrane. Characterising the peptide antimicrobial activity with and without the linked P. gingivalis-specific IgG antibody will give insight on how to reduce the broad-spectrum antimicrobial activity of this conjugate at higher concentrations and increase the specific targeted pathogen antimicrobial activity in a mixed microbial culture.
In summary, SMAP28 linked to affinity- and protein G-purified rabbit IgG antibodies could selectively kill P. gingivalis in an artificially generated microbial community. This approach is an initial step for developing a selective antimicrobial reagent that could be used to eliminate specific periodontal pathogens, such as P. gingivalis, from patients with periodontal disease without harming their normal commensal population.
Fig. 4.
Viability of Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans and Peptostreptococcus micros incubated in the P. gingivalis immunoglobulin G (IgG) antibody control solution. The final concentration of protein was 20 μg/mL. CFU, colony-forming units.
Acknowledgments
The authors are grateful to James D. Herd for preparation of the figures and to Elizabeth A. Schmitt (Cairn Communications, Mahtomedi, MN) for critically reading the manuscript. We are grateful to Abbey R. Ackermann and Deborah E. Dietrich for their technical help and to Brian Tack for the goat anti-SMAP28 antibody.
Funding: This work was supported by funds from grant number R01 DE014390 from the National Institute of Dental and Craniofacial Research, National Institutes of Health (NIH) and subject to the NIH Public Access Policy.
Footnotes
Competing interests: None declared.
Ethical approval: Not required.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bud R. Antibiotics: the epitome of a wonder drug. BMJ. 2007;334(Suppl 1):s6. doi: 10.1136/bmj.39021.640255.94. [DOI] [PubMed] [Google Scholar]
- 2.Khardori N. Antibiotics—past, present, and future. Med Clin North Am. 2006;90:1049–76. doi: 10.1016/j.mcna.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Rice LB, Sahm D, Bonomo RA. Mechanisms of resistance to antibacterial agents. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH, editors. Manual of clinical microbiology. 8. Washington, DC: ASM Press; 2003. pp. 1074–101. [Google Scholar]
- 4.Smith DL, Harris AD, Johnson JA, Silbergeld EK, Morris JG., Jr Animal antibiotic use has an early but important impact on the emergence of antibiotic resistance in human commensal bacteria. Proc Natl Acad Sci USA. 2002;99:6434–9. doi: 10.1073/pnas.082188899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gleckman RA, Borrego F. Adverse reactions to antibiotics. Clues for recognizing, understanding, and avoiding them. Postgrad Med. 1997;101:97–8. 101–4, 107–8. doi: 10.3810/pgm.1997.04.198. [DOI] [PubMed] [Google Scholar]
- 6.Yao JDC, Moellering RC. Antibacterial agents. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH, editors. Manual of clinical microbiology. 8. Washington, DC: ASM Press; 2003. pp. 1039–73. [Google Scholar]
- 7.Slots J, Ting M. Systemic antibiotics in the treatment of periodontal disease. Periodontol 2000. 2002;28:106–76. doi: 10.1034/j.1600-0757.2002.280106.x. [DOI] [PubMed] [Google Scholar]
- 8.Preshaw PM. Systemic antibiotics in the management of chronic periodontitis. Eur J Prosthodont Restor Dent. 2004;12:63–9. [PubMed] [Google Scholar]
- 9.Haffajee AD, Socransky SS, Gunsolley JC. Systemic anti-infective periodontal therapy. A systematic review. Ann Periodontol. 2003;8:115–81. doi: 10.1902/annals.2003.8.1.115. [DOI] [PubMed] [Google Scholar]
- 10.Herrera D, Sanz M, Jepsen S, Needleman I, Roldan S. A systematic review on the effect of systemic antimicrobials as an adjunct to scaling and root planing in periodontitis patients. J Clin Periodontol. 2002;29(Suppl 3):136–59. doi: 10.1034/j.1600-051x.29.s3.8.x. [DOI] [PubMed] [Google Scholar]
- 11.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–50. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 12.Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–7. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 13.Marr AK, Gooderham WJ, Hancock RE. Antibacterial peptides for therapeutic use: obstacles and realistic outlook. Curr Opin Pharmacol. 2006;6:468–72. doi: 10.1016/j.coph.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 14.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–95. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 15.Groisman EA. How bacteria resist killing by host-defense peptides. Trends Microbiol. 1994;2:444–8. doi: 10.1016/0966-842x(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 16.Gunn JS. Bacterial modification of LPS and resistance to antimicrobial peptides. J Endotoxin Res. 2001;7:57–62. [PubMed] [Google Scholar]
- 17.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Gotz F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–10. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 18.Roland KL, Esther CR, Spitznagel JK. Isolation and characterization of a gene, pmrD, from Salmonella typhimurium that confers resistance to polymyxin when expressed in multiple copies. J Bacteriol. 1994;176:3589–97. doi: 10.1128/jb.176.12.3589-3597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perron GG, Zasloff M, Bell G. Experimental evolution of resistance to an antimicrobial peptide. Proc Biol Sci. 2006;273:251–6. doi: 10.1098/rspb.2005.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubert C. Nature News. Nature Publishing Group; 2005. Microbes overcome natural antibiotic. Might we create superbugs that resist our bodies’ ancient defences? [Google Scholar]
- 21.Shelburne CE, Coulter WA, Olguin D, Lantz MS, Lopatin DE. Induction of β-defensin resistance in the oral anaerobe Porphyromonas gingivalis. Antimicrob Agents Chemother. 2005;49:183–7. doi: 10.1128/AAC.49.1.183-187.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guthmiller JM, Vargas KG, Srikantha R, et al. Susceptibilities of oral bacteria and yeast to mammalian cathelicidins. Antimicrob Agents Chemother. 2001;45:3216–9. doi: 10.1128/AAC.45.11.3216-3219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joly S, Maze C, McCray PB, Jr, Guthmiller JM. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J Clin Microbiol. 2004;42:1024–9. doi: 10.1128/JCM.42.3.1024-1029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zagursky RJ, Ooi P, Jones KF, Fiske MJ, Smith RP, Green BA. Identification of a Haemophilus influenzae 5′-nucleotidase protein: cloning of the nucA gene and immunogenicity and characterization of the NucA protein. Infect Immun. 2000;68:2525–34. doi: 10.1128/iai.68.5.2525-2534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalfa VC, Jia HP, Kunkle RA, McCray PB, Jr, Tack BF, Brogden KA. Congeners of SMAP29 kill ovine pathogens and induce ultrastructural damage in bacterial cells. Antimicrob Agents Chemother. 2001;45:3256–61. doi: 10.1128/AAC.45.11.3256-3261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen CJ, Burnell KK, Brogden KA. Antimicrobial activity of Substance P and Neuropeptide Y against laboratory strains of bacteria and oral microorganisms. J Neuroimmunol. 2006;177:215–8. doi: 10.1016/j.jneuroim.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Steinberg DA, Lehrer RI. Designer assays for antimicrobial peptides. Disputing the ‘one-size-fits-all’ theory. Methods Mol Biol. 1997;78:169–86. doi: 10.1385/0-89603-408-9:169. [DOI] [PubMed] [Google Scholar]
- 28.Peschen D, Li HP, Fischer R, Kreuzaler F, Liao YC. Fusion proteins comprising a Fusarium-specific antibody linked to antifungal peptides protect plants against a fungal pathogen. Nat Biotechnol. 2004;22:732–8. doi: 10.1038/nbt970. [DOI] [PubMed] [Google Scholar]
- 29.Qiu XQ, Wang H, Lu XF, et al. An engineered multidomain bactericidal peptide as a model for targeted antibiotics against specific bacteria. Nat Biotechnol. 2003;21:1480–5. doi: 10.1038/nbt913. [DOI] [PubMed] [Google Scholar]
- 30.Qiu XQ, Zhang J, Wang H, Wu GY. A novel engineered peptide, a narrow-spectrum antibiotic, is effective against vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 2005;49:1184–9. doi: 10.1128/AAC.49.3.1184-1189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson W, Olczak T, Genco CA. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J Bacteriol. 2000;182:5737–48. doi: 10.1128/jb.182.20.5737-5748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kataoka K, Amano A, Kuboniwa M, Horie H, Nagata H, Shizukuishi S. Active sites of salivary proline-rich protein for binding to Porphyromonas gingivalis fimbriae. Infect Immun. 1997;65:3159–64. doi: 10.1128/iai.65.8.3159-3164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amano A, Shizukuishi S, Horie H, Kimura S, Morisaki I, Hamada S. Binding of Porphyromonas gingivalis fimbriae to proline-rich glycoproteins in parotid saliva via a domain shared by major salivary components. Infect Immun. 1998;66:2072–7. doi: 10.1128/iai.66.5.2072-2077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amano A, Nakamura T, Kimura S, et al. Molecular interactions of Porphyromonas gingivalis fimbriae with host proteins: kinetic analyses based on surface plasmon resonance. Infect Immun. 1999;67:2399–405. doi: 10.1128/iai.67.5.2399-2405.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung WO, Demuth DR, Lamont RJ. Identification of a Porphyromonas gingivalis receptor for the Streptococcus gordonii SspB protein. Infect Immun. 2000;68:6758–62. doi: 10.1128/iai.68.12.6758-6762.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hellstrom U, Hallberg EC, Sandros J, Rydberg L, Backer AE. Carbohydrates act as receptors for the periodontitis-associated bacterium Porphyromonas gingivalis: a study of bacterial binding to glycolipids. Glycobiology. 2004;14:511–9. doi: 10.1093/glycob/cwh073. [DOI] [PubMed] [Google Scholar]
- 37.Kalfas S, Andersson M, Edwardsson S, Forsgren A, Naidu AS. Human lactoferrin binding to Porphyromonas gingivalis, Prevotella intermedia and Prevotella melaninogenica. Oral Microbiol Immunol. 1991;6:350–5. doi: 10.1111/j.1399-302x.1991.tb00506.x. [DOI] [PubMed] [Google Scholar]
- 38.Tack BF, Sawai MV, Kearney WR, et al. SMAP-29 has two LPS-binding sites and a central hinge. Eur J Biochem. 2002;269:1181–9. doi: 10.1046/j.0014-2956.2002.02751.x. [DOI] [PubMed] [Google Scholar]