Abstract
The c-Myb transcription factor is important for fetal hematopoiesis and has been proposed to mediate later stages of lymphocyte development. Using homozygous null c-Myb/Rag1 chimeric mice, we have determined that c-Myb plays an important role in the differentiation of macrophages and lymphocytes from precursor stem cells. We also determine that deletion of c-Myb leads to a complete block in early T cell development just before the oligopotent thymocyte matures into the definitive T cell precursor. These data indicate that c-Myb plays an important role at multiple stages of hematopoiesis and is required at an early stage of T cell development.
Keywords: c-Myb, T cell development, lineage commitment
The c-Myb transcription factor binds to cis-acting transcriptional control elements of genes critical for early hematopoiesis (Lipsick 1996; Ness 1996). Many of these genes, such as c-kit and c-myc (Cogswell et al. 1993; Ratajczak et al. 1998) are important for hematopoietic stem cell (HSC) proliferation and differentiation. These observations led to the hypothesis that c-Myb induces the expression of genes necessary for HSC lineage commitment. c-Myb may also have a critical role at later committed stages of hematopoietic cell development. Consensus c-Myb recognition sites have been identified in the promoters and enhancers of genes important in the regulation of late stages of lineage commitment (Lipsick 1996; Ness 1996). For example, c-Myb binding sites have been found in the transcriptional control elements of genes important in mediating T cell development and selection, indicating that c-Myb may be important in mediating these processes (Siu et al. 1992; Nakayama et al. 1993; Hernandez-Munain and Krangel 1994; Hsiang et al. 1995; M. Adlam, R.D. Allen, and G. Siu, in prep.).
T cells mature and acquire their antigenic specificity and self-major histocompatibility complex (MHC) restriction during a complex selection process in the thymus (Fowlkes and Pardoll 1989; Robey and Fowlkes 1994). The earliest committed T cell precursor that migrates to the thymus does not express the CD4 and CD8 accessory molecules and the T-cell antigen receptor (TCR), and is referred to as the double-negative (DN) thymocyte. DN thymocyte stages can be subfractionated into different developmental stages on the basis of their expression of other cell-surface markers (Godfrey and Zlotnik 1993; Godfrey et al. 1993). The most immature DN thymocyte is CD44loCD25−; although committed to the T cell lineage, these initial thymic immigrants are oligopotent and retain the ability to develop into T and B lymphocytes, NK cells, and dendritic cells (Guidos et al. 1989a,b; Wu et al. 1991; Shortman and Wu, 1996). The CD44loCD25− DN thymocyte subsequently commits to the T cell lineage and matures into the CD44+CD25− population and subsequently the CD44+CD25+ population, where it begins to rearrange its TCR β-chain genes (Godfrey et al. 1993). The rearrangement process continues while the thymocyte develops into the CD44−CD25+ cell; upon expressing a functional TCR β chain, the cell down-regulates CD25 and up-regulates expression of the CD4 and CD8 coreceptor molecules and the TCR. This latter stage, referred to as the double-positive (DP) stage, is where the thymocyte begins to undergo the repertoire selection process (Bevan et al. 1994; Nossal 1994; Fink and Bevan 1995). Surviving T cells will down-regulate either CD8, to become the mature CD4 single-positive (SP) T cell, or CD4, to become the mature CD8 SP T cell (Robey and Fowlkes 1994). c-Myb has been shown to induce the function of the promoter of the CD4 gene (Siu et al. 1992; Nakayama et al. 1993) and is one of three factors that induce the function of the stage-specific CD4 silencer (Kim and Siu 1998; H.K. Kim and G. Siu, in prep.; M. Adlam, R.D. Allen, and G. Siu, in prep.). In addition, c-Myb has been shown to induce the enhancers that mediate expression of the genes encoding the TCR γ and δ chains (Hernandez-Munain and Krangel 1994; Hsiang et al. 1995). All of these proteins are important at the later DP and SP stages of thymopoiesis.
Studying the role of c-Myb at later stages of lymphopoiesis using conventional targeted disruption techniques has been frustrated by the early lethality of homozygous null c-Myb mice, which die before definitive lymphopoiesis begins. Transgenic mice that overexpress a dominant-negative form of Myb in T cells have small thymuses with an expanded population of CD4−CD8+ thymocytes (Badiani et al. 1994) undergoing apoptosis (Taylor et al. 1996). To study directly the role of c-Myb in lymphocyte development, we generated and analyzed homozygous null c-Myb/Rag1 chimeric (Rag1−/− ↔ c-Myb−/−) mice. We determine that c-Myb is important for the proper development of lymphocytes and macrophages, and is required at a specific stage early in T cell development.
Results
Mature lymphocytes and macrophages do not develop from c-Myb−/− precursor cells
As discussed above, previous experiments have demonstrated that the homozygous disruption of the c-Myb gene results in death at days 13–15 of embryogenesis (Mucenski et al. 1991). Therefore, it was not possible to study the role of c-Myb in lymphopoiesis, as development of mature fetal lymphocytes occurs only later in embryogenesis. To avoid this problem, we utilized the Rag1−/− blastocyst chimera system (Chen et al. 1993). Because lymphocytes require functional antigen receptor gene rearrangements to develop, mice deficient in the recombination enzyme Rag1 cannot generate mature lymphocytes. In the Rag1−/− blastocyst chimera system, chimeric mice are generated by implanting c-Myb−/− ES cells into blastocysts generated from Rag1−/− mice. Resulting mice are chimeric in all tissues except for the lymphoid system, where lymphocytes that develop past the stage defined by antigen receptor gene rearrangement derive solely from the implanted ES cells. Eleven Rag1−/− ↔ c-Myb−/− mice were generated. We have used PCR analyses of genomic DNA as well as genomic blots to assay the chimerism in the brain, kidney, large intestine, liver, heart, tail, thymus, small intestine, lung, and skeletal muscle of these mice (data not shown). We can detect chimerism in each of these tissues in a subset of Rag1−/− ↔ c-Myb−/− mice, ranging from 2/11 for the brain to 7/11 for the liver and skeletal muscle; these data indicate that the c-Myb−/− ES cells are capable of contributing to the development of multiple somatic cell types.
Despite their ability to contribute to a wide variety of somatic tissues, c-Myb−/− ES cells were unable to repopulate most mature cells of the immune system. We cannot detect mature splenic B220+mIgM+ B cells in the Rag1−/− ↔ c-Myb−/− mice, nor can we detect mature splenic CD4 SP and CD8 SP T cells or CD4 SP, CD8 SP, and DP thymocytes (Fig. 1). Analysis of macrophages in Rag1−/− blastocyst chimeras is difficult as their development does not depend on functional Rag genes; thus, macrophages in the chimeric mice may originate from both the c-Myb−/− and the Rag1−/− lineages. To identify c-Myb−/− ES cell-derived macrophages, we took advantage of allelic differences in the Ly9 molecule between the c-Myb−/− cells and Rag1−/− cells. Hematopoietic cells from the Rag1−/− mice express the Ly9.2 allele, whereas cells originating from the c-Myb−/− cells express the Ly9.1 allele (Ledbetter and Herzenberg 1979). We can thus identify precursor cells originating from c-Myb−/− stem cells using an antibody that distinguishes between the two Ly9 isoforms. Splenocytes were harvested from Rag1−/− ↔ c-Myb−/−, Rag1−/− ↔ c-Myb+/−, and Rag1−/− mice as well as from +/+ 129 mice, which is the genetic background of the ES cell used to generate the c-Myb−/− cells (Mucenski et al. 1991). As expected, macrophages are Ly9.1+ in the 129 mice and Ly9.1− in the Rag1−/− mice (Fig. 2). The Mac-1+Gr-1− population in the Rag1−/− ↔ c-Myb+/− mice contain both Ly9.1+ and Ly9.1− cells, indicating that Rag1−/− ↔ c-Myb+/− ES cells are capable of repopulating the macrophage compartment. The macrophage population in the Rag1−/− ↔c-Myb−/− mice are Ly9.1−, indicating that the macrophages in these mice are derived solely from the Rag1−/− blastocyst cells, indicating that the deletion of the c-Myb gene leads to defective macrophage development. Thus, we conclude that c-Myb plays an important role in the development of both the lymphoid and macrophage lineages.
Figure 1.
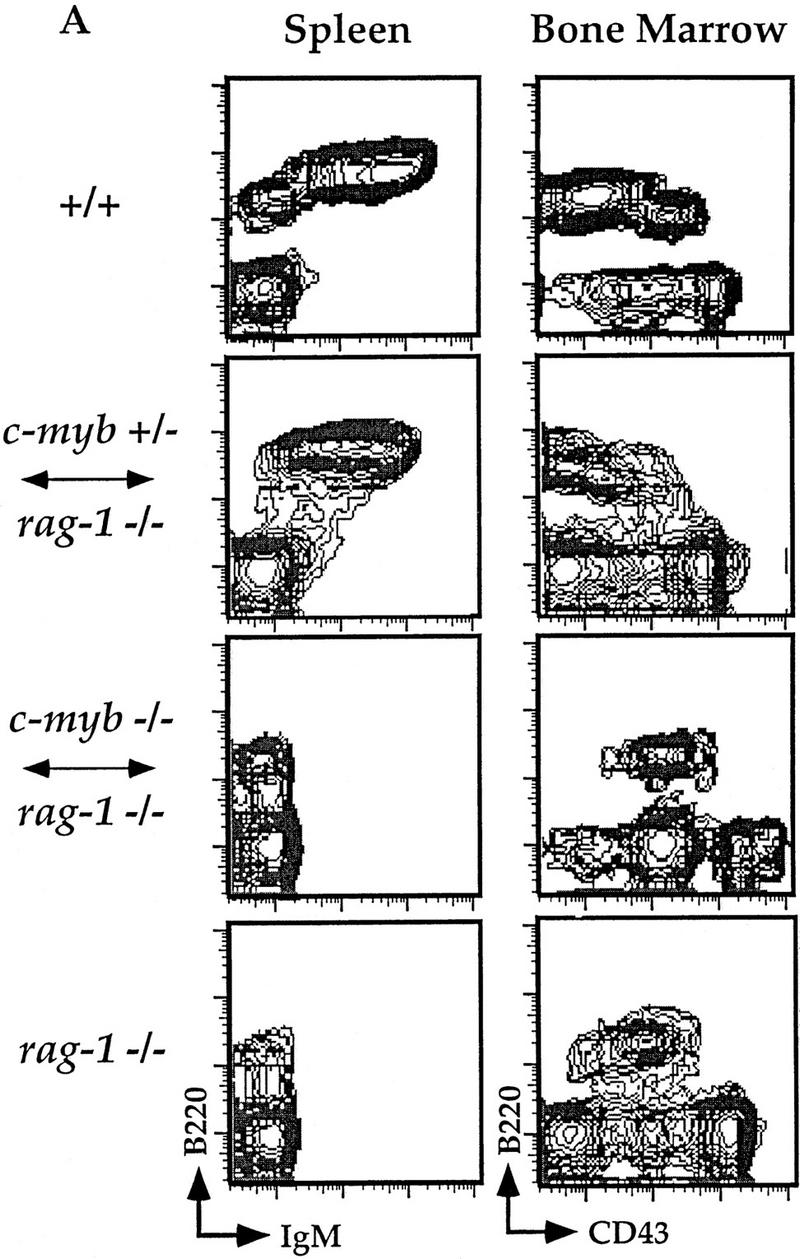
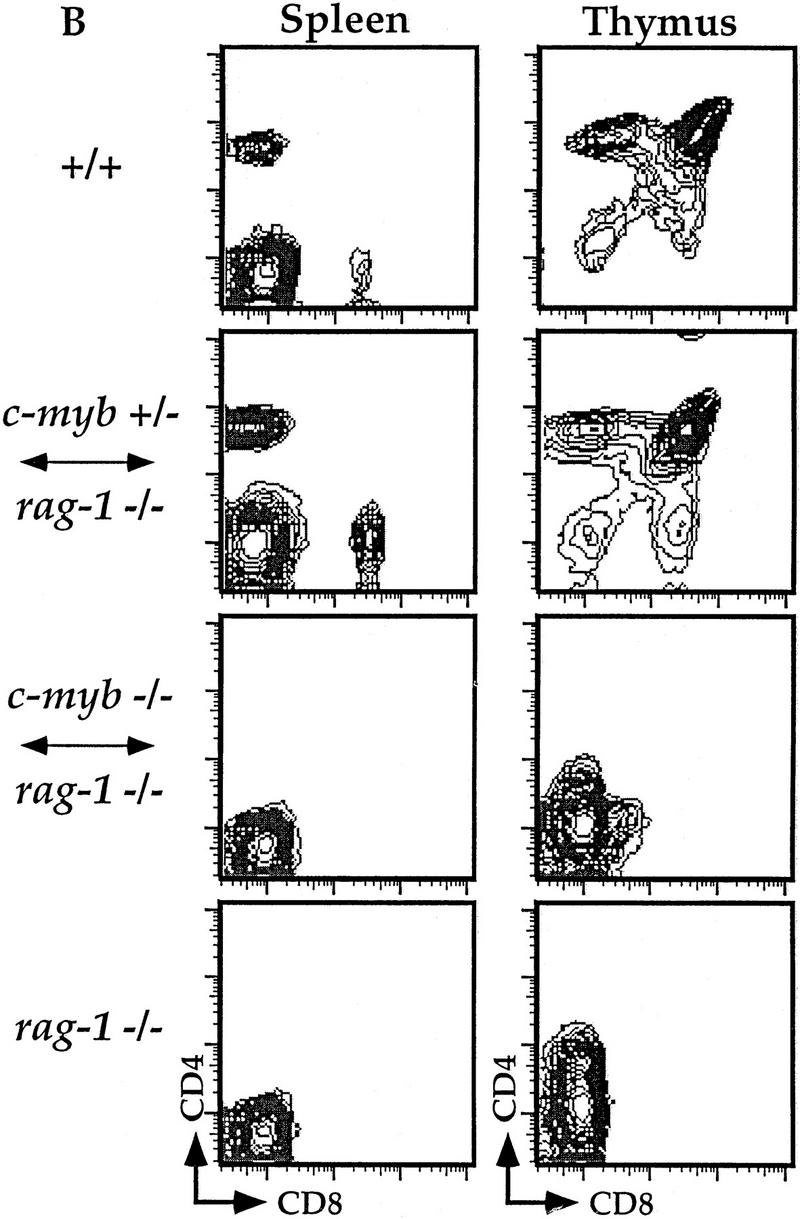
Flow cytometric analyses of B and T lymphocytes from Rag1−/− ↔ c-Myb−/− mice. Splenocytes and thymocytes were isolated from Rag1−/− ↔ c-Myb−/−, Rag1−/− ↔ c-Myb+/−, Rag1−/−, and 129 control mice and identified with combinations of B and T lineage-specific antibodies (see text for details). Chimera 2 was used as an example; similar results were obtained from all other chimeras (data not shown). (A) Splenic B cells and bone marrow B cell precursors; (B) splenic and thymic T cells.
Figure 2.
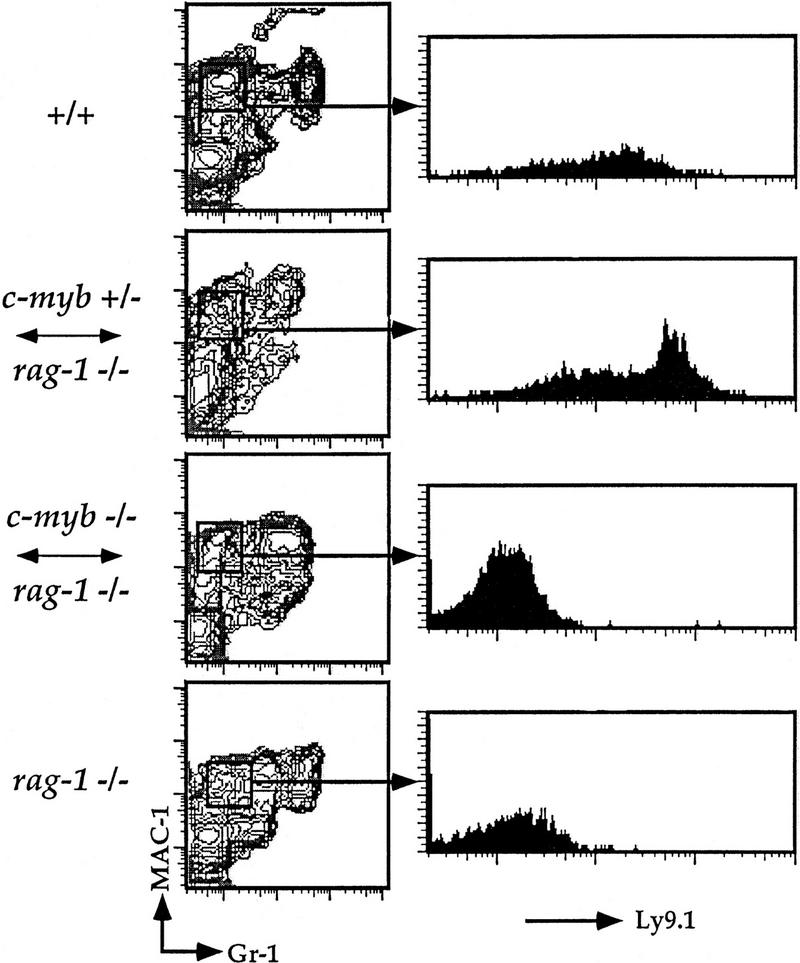
Flow cytometric analysis of splenic macrophages.Splenocytes were isolated from Rag1−/− ↔ c-Myb−/−, Rag1−/− ↔ c-Myb+/−, Rag1−/−, and 129 control mice and identified with antibodies against the macrophage lineage markers Mac-1 and Gr-1 and the c-Myb−/− cell-specific marker Ly9.1 (see text ). The mature macrophage population was gated on and analyzed for Ly9.1 expression as shown. Chimera 2 is shown; similar results were obtained from all other chimeras (data not shown).
Early thymocyte precursors develop from c-Myb−/− stem cells
Although the lack of detectable mature and immature lymphocytes in the Rag1−/− ↔ c-Myb−/− mice is consistent with an early pluripotent HSC defect, it is possible that the defect occurs after the stem cell commits to the lymphocyte lineage but before antigen receptor gene rearrangement. Because Rag1−/−-derived lymphocyte precursors can also develop to these stages, a block in development of the c-Myb−/− stem cells would be difficult to detect using flow cytometric analyses with the standard lineage markers. To address this issue, we determined the extent of Ly9 allelism in early B and T cell precursors isolated from the chimeric mice (Fig. 3; data not shown). We were unable to detect immature Ly9.1+B220+CD43+ B cells in the bone marrow in any of the Rag1−/− ↔ c-Myb−/− mice (data not shown), indicating that, as for macrophages, the c-Myb−/− ES hematopoietic stem cells cannot develop to the point of commitment to the B cell lineage. To analyze the early thymic precursor cells, we utilized antibodies to the CD44 and CD25 molecules. (Godfrey and Zlotnik 1993; Godfrey et al. 1993). Mice that have homozygous null mutations in their Rag genes have a developmental block at the CD44+/loCD25+ stages as the result of their inability to generate a successful rearrangement of their TCR genes[Godfrey et al. (1993); Fig. 3A]. Five of eleven Rag1−/−↔ c-Myc−/− mice contained cells derived from c-Myb−/− ES cells in the CD44loCD25− DNA population. As can be seen in Figure 3, we detect Ly9.1+CD44loCD25− DN thymocytes in the Rag1−/− ↔ c-Myb−/− chimeric mice, indicating this expanded population represents a stage that is blocked for further development (Fig. 3B). The presence of this early thymocyte originating from the c-Myb−/− ES cells and its failure to develop further indicate that c-Myb plays an essential role for the developmental progression of an early T cell precursor.
Figure 3.
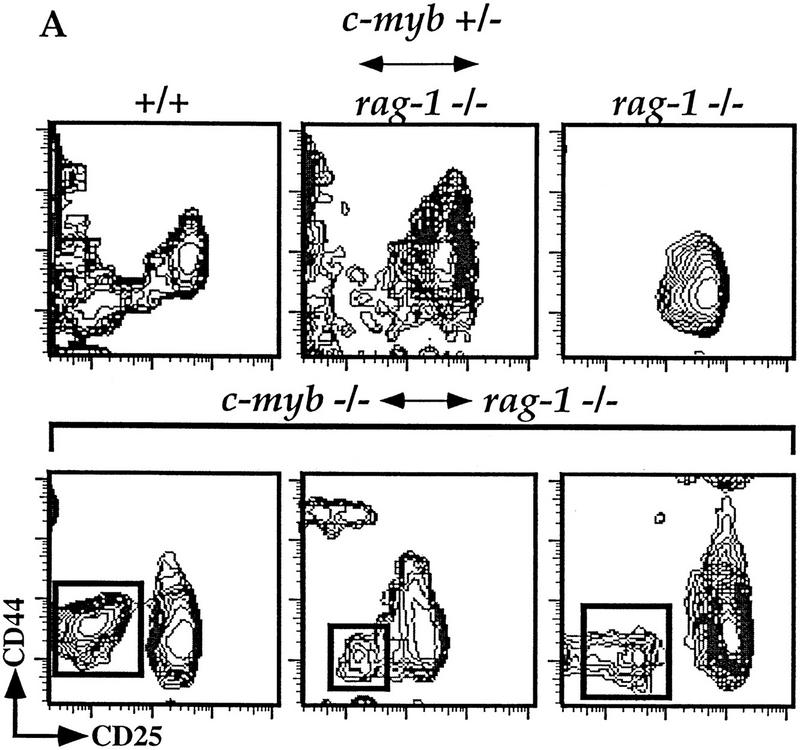
c-Myb−/− HSCs can generate early double-negative thymocytes. (A) Thymocytes were isolated from representative Rag1−/− ↔ c-Myb+/−, Rag1−/−, phenotype positive Rag1−/− ↔ c-Myb−/−, and 129 control mice and analyzed for surface expression of T-lineage-specific markers. Cells expressing any one of the CD8/CD3/B220/Mac-1/Gr-1/Ter-119 markers were excluded from the analysis and the remainder plotted based on their expression of CD25 (x-axis) and CD44 (y-axis). For the Rag1−/− ↔ c-Myb−/− mice, representative phenotype-positive data are shown. (B) Analysis gates drawn on the CD44loCD25− and CD44+/loCD25+ cells for analysis of the expression of the 129 lineage-specific surface marker Ly9.1 are shown.
The CD44loCD25− DN thymocytes in the Rag1−/− ↔ c-Myb−/− mice have germ-line TCR β-chain genes
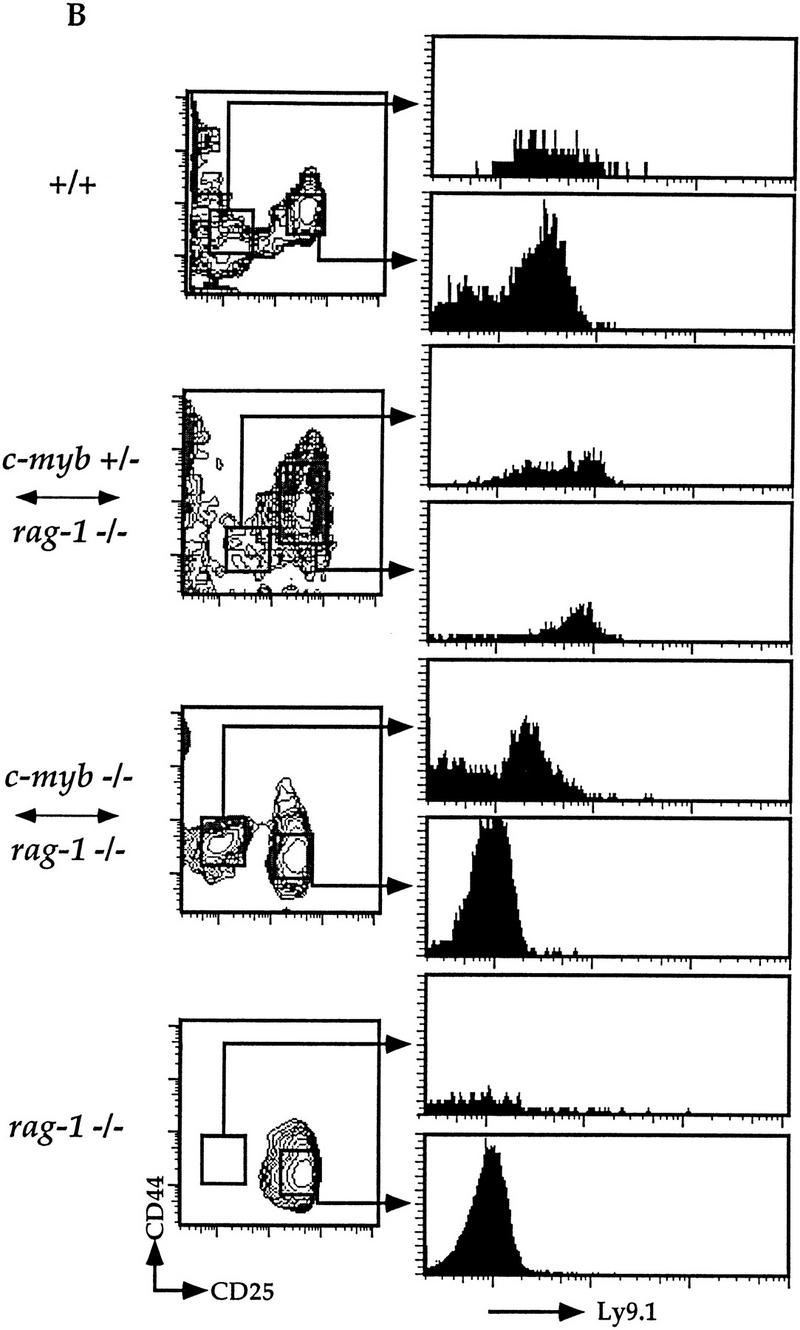
The expanded Ly9.1+CD44loCD25− thymocytes observed in the Rag1−/− ↔ c-Myb−/− mice may represent thymocyte precursors of two different developmental stages: the recent oligopotent immigrant from the bone marrow; or the late DN precursor to the DP thymocyte. Flow cytometry data using different early thymocyte markers indicate that the phenotype of this population is consistent with that of the early DN thymocyte (Fig. 3; data not shown); the lack of Ly9.1+CD44+CD25+ thymocytes in the Rag1−/− ↔ c-Myb−/− mice also supports the hypothesis that thymopoiesis is blocked previous to this step (Fig. 3B). However, it is still possible that this population represents the late DN thymocyte population that has modified marker expression due to the deletion of the c-Myb gene. To confirm that this population is the earliest thymic precursor, we analyzed thymocytes from the Rag1−/− ↔ c-Myb−/− mice for rearrangement of the TCR β-chain gene (Fig. 4). As discussed above, the earliest CD44lo/+CD25− DN thymocyte has not begun rearranging the TCR genes, whereas cells of all subsequent T cell developmental stages have at least partial Dβ–Jβ rearrangements. Thus, should the expanded CD44loCD25− DN population detected in the Rag1−/− ↔ c-Myb−/− mice represent the earliest DN precursor, we predict that we would only detect the germ-line configuration in the β-chain gene locus. Alternatively, should this population represent a late DN thymocyte ready to progress into the CD4−CD8loCD3− stage, we predict we would be able to detect Dβ–Jβ rearrangements.
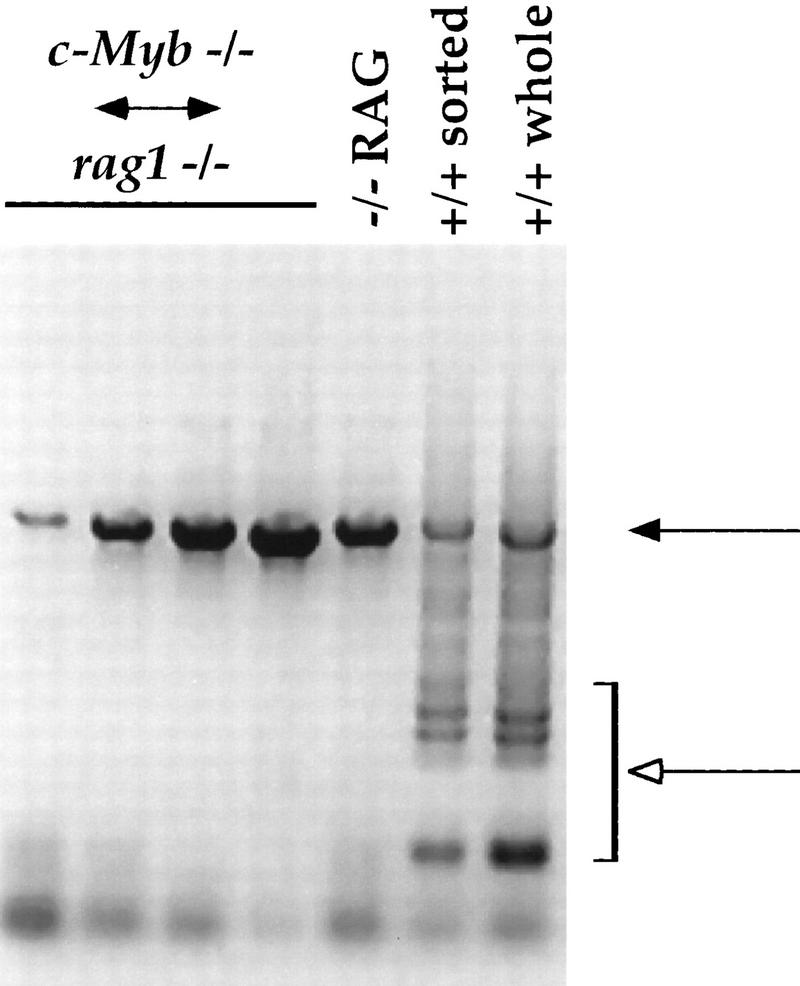
Figure 4.
TCRβ-chain gene rearrangement in thymocytes from the Rag1−/− ↔ c-Myb−/− mice. DNA was purified from thymuses from the Rag1−/− ↔ c-Myb−/− and Rag1−/− mice and from both whole thymus and sorted CD4−CD8−CD3− thymocytes from +/+ 129 mice and analyzed them for TCR β-chain gene rearrangement as described previously (Anderson et al., 1992). Results obtained from the Rag1−/− ↔ c-Myb−/− chimeras 1, 2, 9, and 11 (left to right) are shown and are representative of the other chimeras. (Solid arrow) Germ-line band; (open arrow) Dβ–Jβ-rearranged bands.
We purified DNA from the thymuses harvested from the Rag1−/− ↔ c-Myb−/− and the Rag1−/− mice, as well as from both whole thymus and sorted CD4−CD8−CD3− thymocytes from +/+ 129 mice and analyzed them for TCR β-chain gene rearrangement using a PCR-based method (Anderson et al. 1992). PCR products representing initial Dβ–Jβ rearrangements can be readily detected in either whole thymocytes or sorted CD4−CD8−CD3− cells from the +/+ 129 mouse (open arrow; Fig. 4). We cannot detect similar rearrangements in the Rag1−/− mouse, although the germ-line Dβ–Jβ can be readily detected (open and solid arrows, respectively; Fig. 4). These latter observations are consistent with the inability of these mice to rearrange their TCR and immunoglobulin genes due to the loss of the functional Rag1 gene. Interestingly, we also cannot detect Dβ–Jβ rearrangments in the thymuses of the Rag1−/− ↔ c-Myb−/− mice. However, in all chimeras we detect a PCR product that corresponds to the germ-line Dβ–Jβ band, demonstrating that the β-chain loci in the DN thymocytes in these mice are still in the germ-line configuration (solid arrow, Fig. 4). These data thus suggest that the TCR β-chain loci in the expanded population of CD44loCD25− DN thymocytes in the Rag1−/− ↔ c-Myb−/− mice are germ line, indicating further that this population represents the earliest DN thymocyte stage.
Discussion
c-Myb is important at multiple stages in hematopoiesis
Previous studies have demonstrated that c-Myb is necessary for the transition from fetal-type to adult-type erythropoeisis (Mucenski et al. 1991). In this study we present evidence that ES cells deficient for c-Myb cannot generate B cell or macrophage/monocyte precursors in the bone marrow. There are several possible explanations for these observations: (1) The c-Myb−/− HSC is present in normal numbers but is inhibited from further development into the B cell and macrophage/monocyte lineages but not the T cell lineage; and (2) it is possible that c-Myb−/− HSCs can develop but are at a selective disadvantage for developmental niches in the bone marrow. For example, should the c-Myb−/− HSC be unable to replicate efficiently, competition for the limited space in the bone marrow with Rag1−/− HSCs may inhibit the ability of the c-Myb−/− HSC to populate this compartment. Antisense experiments led to the proposal that c-Myb is important in the proliferation and differentiation of hematopoietic precursors (Clarke et al. 1988; Gewirtz and Calabretta 1988; McMahon et al. 1988; Todokoro et al. 1988; McClinton et al. 1990), which is consistent with the latter hypothesis but not the former. However, as we can detect further development of the c-Myb−/− HSCs into thymic stem cells in many of the Rag1−/− ↔ c-Myb−/− mice, it is clear that HSC development and maturation can occur to some extent in the absence of c-Myb. We thus believe that the absence of c-Myb leads at best to only a partial developmental block at early stages of HSC development. Thus, in contrast to the essential role that c-Myb has in early thymopoiesis and fetal erythropoiesis, c-Myb is not absolutely required for HSC maintenance and development.
Definitive roles for c-Myb in T cell development
Our data demonstrate that although Rag1−/− ↔ c-Myb−/− mice are capable of generating early hematopoietic precursors that seed the thymus, further development of these cells is blocked at the CD44loCD25− DN developmental stage, which is characterized by the beginning of definitive T cell differentiation. Thus, our data demonstrate an important role for c-Myb in the control of early T cell development. Although these cells eventually develop into T cells in vivo, cell transfer experiments have demonstrated that this population is capable of differentiating into T, B, NK, and dendritic cells, whereas subsequent DN thymocyte stages are completely committed to the T cell lineage (Guidos et al. 1989a,b; Wu et al. 1991; Shortman and Wu 1996). It is interesting to speculate that c-Myb plays a role in the developmental decision of an ES cell precursor to commit to the T cell lineage. In this model, the targeted disruption of c-Myb would prevent the development of the committed DN thymocyte, leading to the observed buildup of the early uncommitted precursor. Although several transcription factors have been demonstrated previously to be important in thymopoiesis, all are believed to be important either late in T cell development or in the multipotent ES cell and not at this stage of T cell commitment (Fig. 5). Using the Rag−/− blastocyst chimera approach, Ting et al. (1996) determined that Rag2−/− ↔ GATA-3−/− mice are deficient in T cell development, indicating that GATA-3 is important for the specification of the T cell lineage. However, using the same Ly9 allele marker system that we have utilized above, these workers could not detect Ly9.1+ DN thymocytes in the thymuses of these mice, indicating that the GATA-3−/− HSC could not seed the thymus at any level. These observations suggest that the block in the Rag2−/− ↔ GATA-3−/− mice is earlier than that seen in the Rag1−/− ↔ c-Myb−/− mice. However, because the expanded CD44loCD25− DN population in the Rag1−/− ↔ c-Myb−/− mice cannot mature further, it is difficult to define precisely their developmental potential; thus we cannot formally demonstrate that this population consists of the early uncommitted thymic ES cell. Nonetheless, this population phenotypically resembles this thymocyte precursor; thus, our data suggest that c-Myb may be playing a role in the committment of the ES cell to the T cell lineage.
Figure 5.
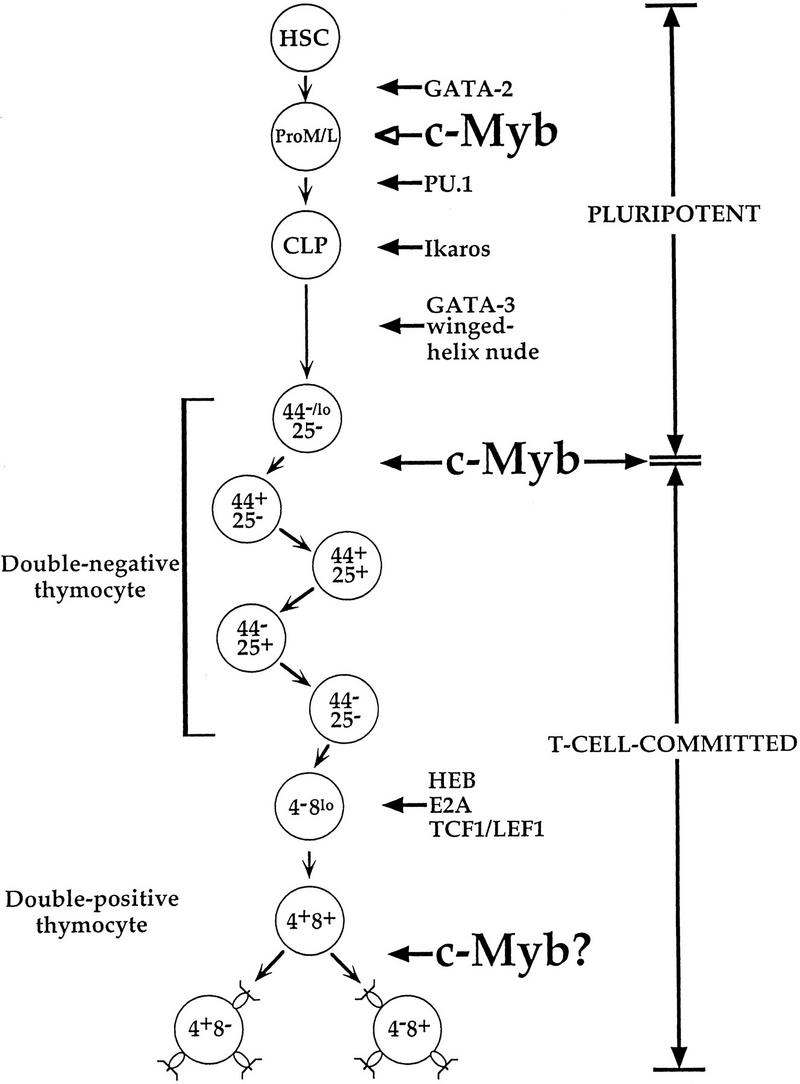
Transcription factors important in early and intermediate T cell development; data are from Glimcher and Singh (1999) and references therein.
It is unlikely that the sole role of c-Myb in T cell development is mediating early T cell progenitor development. As discussed above, c-Myb is an important transcription factor in the control of expression and function of a wide variety of different genes, including the CD4, ADA, TCRγ and δ chain, c-kit, and c-Myc genes (Siu et al. 1992; Cogswell et al. 1993; Hernandez-Munain and Krangel 1994; Hernandez-Munain et al. 1996; Ess et al. 1995; Hsiang et al. 1995; Ratajczak et al. 1998; M. Adlam, R.D. Allen, and G. Siu, in prep.); many of these genes are important at later stages of T cell development. For example, the CD4 and TCRγ- and δ-chain genes encode proteins whose expression are required for the T cell receptor-mediated selection process (Fowlkes and Pardoll 1989). In addition, overexpression of dominant–negative forms of Myb lead to perturbations in thymic development, implying a role for Myb proteins in late stages of thymopoiesis (Badiani et al. 1994; Taylor et al. 1996). Nonetheless, the extent of the role of c-Myb in thymic selection remains to be determined (Fig. 5). Use of conditionally targeted c-Myb mice will allow insight into the role of c-Myb as a potential mediator of proliferation or survival signals in the selecting and postselection thymocyte.
Materials and methods
RAG1−/− blastocyst complementation and implantation
Homozygous and heterozygous null c-Myb ES cells were kindly provided by M. Mucenski (Mucenski et al. 1991). Rag1−/− blastocysts were harvested from impregnated females at 3.5 dpc, microinjected with 15–18 c-Myb−/− or c-Myb+/− ES cells, and implanted into the uteri of pseudopregnant females as described previously (Chen et al. 1993). Chimerism in the pups was determined by coat color, and PCR and genomic blot analyses of tail DNA; a total of 11 chimeras were obtained. Chimerism in each tissue was determined by genomic PCR analyses, genomic blots, and flow cytometry. All mice were housed in the pathogen-free Cancer Center Animal Facility at Columbia University and sacrificed at 4–6 weeks of age for analysis.
Genomic DNA analyses
Genomic DNA was prepared from tissues and sorted cells by incubating overnight at 55°C in digestion buffer (50 mm Tris at pH 8.0, 100 mm NaCl, 100 mm EDTA, 1% SDS, 200 μg/μl proteinase K), extracting with phenol/chloroform/isoamyl alchohol, and resuspending the precipitated DNA in TE (10 mm Tris, 1 mm EDTA at pH 7.5). For PCR of the TCRβ-chain gene locus, primers 5′ of the Dβ2 region (Anderson et al. 1992) and 3′ of the Jβ2 cluster (Levin et al. 1993) were used to amplify DNA isolated from either whole or CD4−CD8−CD3− sorted thymocytes of chimeric and control mice. The amplification cycle (94°C for 60 sec, 62°C for 120 sec, 72°C for 120 sec) was repeated for 40 cycles, and products were size fractionated on a 1% agarose gel and visualized by ethidium bromide staining.
Flow cytometry
Cells were harvested and stored as described previously (Kim and Siu 1998). The following mAb reagents were obtained from PharMingen (San Diego, CA): α-CD8 (53-7.8), α-Ly9.1 (30C7), α-CD4 (GK1.5), α-CD25 (7D4), α-IgM (331); α-CD4 (GK1.5), α-CD3 (145-2C11), α-CD44 (IM7), α-CD43 (S7); α-Ly9.1 (30C7), α-CD25 (7D4), α-Gr-1 (RB6-8C5); α-Thy1.2 (53-2.1), α-B220 (RA3-6B2), α-Mac-1 (M1-70), α-CD4 (RM4-5), α-Ter-119, α-Gr-1 (RB6-8C5), α-CD8 (53-6.1), and α-CD3 (145-2C11). T cell populations in the thymus and spleen were identified with anti-CD3, anti-CD4, and anti-CD8. For DN T cells, thymocytes were stained with antibodies to CD8, CD3, CD44, CD25, B220, Mac-1, Gr-1, Ter-119, and Ly9.1. Cells expressing any one of the CD8/CD3/B220/Mac-1/Gr-1/Ter-119 markers were excluded and the remainder analyzed on the basis of their expression of CD25 and CD44. Mature B cells were identified in the spleen using anti-B220 and anti-IgM. B cell precursors were identified in bone marrow using anti-B220 and anti-CD43. Splenic macrophages and granulocytes were identified using Mac-1 and Gr-1 after excluding CD3-expressing cells from the analysis. Sorting for CD4−CD8−CD3− thymocytes was performed as described previously (Lam and Stall 1994).
Acknowledgments
We thank Monica Mendelson and Dan Ng for technical assistance, and Drs. Kathryn Calame, Chris Schindler, and Alexander Dent for critical review of the manuscript. This work was funded by grants from the National Institutes of Health (NIH) (AI34925) and the Irma T. Hirschl-Monique Weill Caulier Trust to G.S. and grants from the NIH (GM55985) and the Fogarty Foundation (TW02297) to T.P.B. R.D.A. was supported by NIH training grant T32AI07525.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL siu@cusiu3.cpmc.columbia.edu; FAX (212) 305-8013.
References
- Anderson SJ, Abraham KM, Nakayama T, Singer A, Perlmutter RM. Inhibition of T cell receptor β-chain gene rearrangement by overexpression of the non-receptor protein tyrosine kinase p56lck. EMBO J. 1992;11:4877–4886. doi: 10.1002/j.1460-2075.1992.tb05594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes & Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- Bevan MJ, Hogquist KA, Jameson SC. Selecting the T cell receptor repertoire. Science. 1994;264:796–797. doi: 10.1126/science.8171333. [DOI] [PubMed] [Google Scholar]
- Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: An assay of gene function in lymphocyte development. Proc Natl Acad Sci. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Kukowska-Latallo JF, Westin E, Smith M, Prochownik EU. Constitutive expression of a c-Myb cDNA blocks Friend murine erythroleukemia cell differentiation. Mol Cell Biol. 1988;8:884–892. doi: 10.1128/mcb.8.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell JP, Cogswell PC, Kuehl WM, Cuddihy AM, Bender TP, Engelke U, Marcu KB, Ting JP. Mechanism of c-myc regulation by c-Myb in different cell lineages. Mol Cell Biol. 1993;13:2858–2869. doi: 10.1128/mcb.13.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ess KC, Whitaker TL, Cost GJ, Witte DP, Hutton JJ, Aronow BJ. A central role for a single c-Myb binding site in a thymic locus control region. Mol Cell Biol. 1995;5:5707–5715. doi: 10.1128/mcb.15.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink PJ, Bevan MJ. Positive selection of thymocytes. Adv Immunol. 1995;59:99–133. doi: 10.1016/s0065-2776(08)60630-6. [DOI] [PubMed] [Google Scholar]
- Fowlkes BJ, Pardoll D. Molecular and cellular events of T cell development. Adv Immunol. 1989;44:207–264. doi: 10.1016/s0065-2776(08)60643-4. [DOI] [PubMed] [Google Scholar]
- Gewirtz AM, Calabretta B. A c-Myb antisense oligonucleotide inhibits normal human hematopoiesis in vitro. Science. 1988;242:1303–1306. doi: 10.1126/science.2461588. [DOI] [PubMed] [Google Scholar]
- Glimcher LH, Singh H. Transcription factors in lymphocyte development—T and B cells get together. Cell. 1999;96:13–23. doi: 10.1016/s0092-8674(00)80955-1. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Zlotnik A. Control points in early T-cell development. Immunol Today. 1993;14:547–553. doi: 10.1016/0167-5699(93)90186-O. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8− triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- Guidos CJ, Weissman IL, Adkins B. Developmental potential of CD4−8− thymocytes. Peripheral progeny include mature CD4−8− T cells bearing α β T cell receptor. J Immunol. 1989a;142:3773–3780. [PubMed] [Google Scholar]
- ————— Intrathymic maturation of murine T lymphocytes from CD8+ precursors. Proc Natl Acad Sci. 1989b;86:7542–7546. doi: 10.1073/pnas.86.19.7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Munain C, Krangel MS. Regulation of the T-cell receptor δ enhancer by functional cooperation between c-Myb and core-binding factors. Mol Cell Biol. 1994;14:473–483. doi: 10.1128/mcb.14.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Munain C, Lauzurica P, Krangel MS. Regulation of T cell receptor δ gene rearrangement by c-Myb. J Exp Med. 1996;183:289–293. doi: 10.1084/jem.183.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang Y-H, Goldman JP, Raulet DH. The role of c-Myb or a related factor in regulating the T cell receptor γ gene enhancer. J Immunol. 1995;154:5195–5204. [PubMed] [Google Scholar]
- Kim HK, Siu G. The Notch pathway intermediate HES-1 silences CD4 gene expression. Mol Cell Biol. 1998;18:7166–7175. doi: 10.1128/mcb.18.12.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam KP, Stall AM. Major histocompatibility complex class II expression distinguishes two distinct B cell developmental pathways during ontogeny. J Exp Med. 1994;180:507–516. doi: 10.1084/jem.180.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter JA, Herzenberg LA. Xenogeneic monoclonal antibodies to mouse lymphoid differentiation antigens. Immunol Rev. 1979;47:63–90. doi: 10.1111/j.1600-065x.1979.tb00289.x. [DOI] [PubMed] [Google Scholar]
- Levin SD, Anderson SJ, Forbush KA, Perlmutter RM. A dominant-negative transgene defines a role for p56lck in thymopoiesis. EMBO J. 1993;12:1671–1680. doi: 10.1002/j.1460-2075.1993.tb05812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsick JS. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- McClinton D, Stafford J, Brents L, Bender TP, Kuehl WM. Differentiation of mouse erythroleukemia cells is blocked by late upregulation of a c-Myb transgene. Mol Cell Biol. 1990;10:705–710. doi: 10.1128/mcb.10.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon J, Howe KM, Watson RJ. The induction of Friend murine erythroleukemia differentiation is markedly affected by expression of a transfected c-Myb cDNA. Oncogene. 1988;3:717–720. [PubMed] [Google Scholar]
- Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott JWJ, Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Yamamoto R, Ishii S, Nakauchi H. Binding of c-Myb to the core sequence of the CD4 promoter. Int Immunol. 1993;5:817–824. doi: 10.1093/intimm/5.8.817. [DOI] [PubMed] [Google Scholar]
- Ness SA. The Myb oncoprotein: Regulating a regulator. Biochim Biophys Acta. 1996;1288:F123–F139. doi: 10.1016/s0304-419x(96)00027-3. [DOI] [PubMed] [Google Scholar]
- Nossal GJV. Negative selection of lymphocytes. Cell. 1994;76:229–239. doi: 10.1016/0092-8674(94)90331-x. [DOI] [PubMed] [Google Scholar]
- Ratajczak MZ, Perrotti D, Melotti P, Powzaniuk M, Calabretta B, Onodera K, Kregenow DA, Machalinski B, Gewirtz AM. Myb and ets proteins are candidate regulators of c-kit expression in human hematopoietic cells. Blood. 1998;91:1934–1946. [PubMed] [Google Scholar]
- Robey E, Fowlkes BJ. Selective events in T cell development. Annu Rev Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- Shortman K, Wu L. Early T lymphocyte progenitors. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- Siu G, Wurster AL, Lipsick JS, Hedrick SM. Expression of the CD4 gene requires a Myb transcription factor. Mol Cell Biol. 1992;12:1592–1604. doi: 10.1128/mcb.12.4.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D, Badiani P, Weston K. A dominant interfering Myb mutant causes apoptosis in T cells. Genes & Dev. 1996;10:2732–2744. doi: 10.1101/gad.10.21.2732. [DOI] [PubMed] [Google Scholar]
- Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- Todokoro K, Watson RJ, Higo H, Amanura H, Kuramochi S, Yanagisawa H, Ikawa Y. Downregulation of c-Myb gene expression is a prerequisite for erythropoietin-induced erythroid differentiation. Proc Natl Acad Sci. 1988;85:8900–8904. doi: 10.1073/pnas.85.23.8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Antica M, Johnson GR, Scollay R, Shortman K. Developmental potential of the earliest precursor cells from the adult mouse thymus. J Exp Med. 1991;174:1617–1627. doi: 10.1084/jem.174.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]