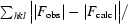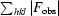The 1.6 Å resolution crystal structure of nitrilotriacetate monooxygenase component B (NTA-MoB) from M. thermoresistibile is presented, revealing a highly conserved C-terminal tail that may modulate the activity of NTA-MoB in mycobacteria.
Keywords: SSGCID, Mycobacterium thermoresistible, nitrilotriacetate monooxygenase component B, flavin reductases, oxidoreductases
Abstract
Mycobacterium tuberculosis belongs to a large family of soil bacteria which can degrade a remarkably broad range of organic compounds and utilize them as carbon, nitrogen and energy sources. It has been proposed that a variety of mycobacteria can subsist on alternative carbon sources during latency within an infected human host, with the help of enzymes such as nitrilotriacetate monooxygenase (NTA-Mo). NTA-Mo is a member of a class of enzymes which consist of two components: A and B. While component A has monooxygenase activity and is responsible for the oxidation of the substrate, component B consumes cofactor to generate reduced flavin mononucleotide, which is required for component A activity. NTA-MoB from M. thermoresistibile, a rare but infectious close relative of M. tuberculosis which can thrive at elevated temperatures, has been expressed, purified and crystallized. The 1.6 Å resolution crystal structure of component B of NTA-Mo presented here is one of the first crystal structures determined from the organism M. thermoresistibile. The NTA-MoB crystal structure reveals a homodimer with the characteristic split-barrel motif typical of flavin reductases. Surprisingly, NTA-MoB from M. thermoresistibile contains a C-terminal tail that is highly conserved among mycobacterial orthologs and resides in the active site of the other protomer. Based on the structure, the C-terminal tail may modulate NTA-MoB activity in mycobacteria by blocking the binding of flavins and NADH.
1. Introduction
Bacteria within the Mycobacterium genus include M. tuberculosis, the pathogen responsible for tuberculosis (TB), a disease which has infected millions worldwide (Anderton et al., 2006 ▶; Rylance et al., 2010 ▶). This highly contagious disease is responsible for three million deaths per year and highly regulated facilities are needed to study it owing to its ease of transmission. Over 120 species of Mycobacterium have been identified to date, many of which can cause disease, particularly in individuals with suppressed or compromised immunity (Neonakis et al., 2007 ▶). M. thermoresistibile is a non-tuberculous species of Mycobacterium which has had multiple pathology reports over the years, including a recent identification in an infected patient (Neonakis et al., 2009 ▶). As its name implies, M. thermoresistibile thrives at elevated temperatures and unlike most other mycobacteria it can survive in culture at 333 K for up to 4 h (Weitzman et al., 1981 ▶; Tsukamura, 1966 ▶). Although relatively rare, increasing evidence of its ability to infect humans and the fact that it shares many homologous genes with M. tuberculosis warrants further study of this pathogenic organism for possible therapeutic intervention (Weitzman et al., 1981 ▶; Kremer et al., 2002 ▶; Boloorsaz et al., 2006 ▶).
Mycobacteria are named for their ability to produce mycolic acid, and bacteria from this genus are capable of degrading a wide range of organic compounds (Savvi et al., 2008 ▶). Like other microorganisms, various mycobacteria can use small organic compounds such as nitrilotriacetate (NTA) as their sole source of nitrogen, carbon and energy, allowing rapid adaptation to varying conditions within the host (Uetz et al., 1992 ▶; Bally et al., 1994 ▶). Recent reports predicted that M. tuberculosis could subsist on alternative carbon sources during persistence within the human host, specifically during its macrophage infection period, where by nature it needs to endure glucose deficiency and an abundance of fatty acids. The complex repertoire of genes involved in lipid metabolism in Mycobacterium is thus a key factor in its strong pathogenicity (Van der Geize et al., 2007 ▶; Savvi et al., 2008 ▶). Nitrilotriacetate monooxygenase (NTA-Mo) is an oxidoreductase and a member of the family of two-component monooxygenases which initiates the oxidation of NTA under aerobic conditions (van Berkel et al., 2006 ▶). This enzyme is comprised of two parts: component A (NTA-MoA), which has monooxygenase activity and is responsible for the oxidative conversion of NTA to iminodiacetate (IDA) and glyoxylate, and component B (NTA-MoB), a flavin reductase which consumes NADH to reduce FMN to FMNH2, which is a required cofactor in the oxidization step. The combined NTA-Mo assembly as whole is categorized as a class C flavoprotein monooxygenase (van Berkel et al., 2006 ▶). The amino-acid alignment and the three-dimensional structure motif of NTA-MoB from M. thermoresistibile (MthNTA-MoB) associate it with a family of short-chain flavin reductases. This group of proteins exists in many eukaryotic and prokaryotic organisms, including all mycobacteria (Knobel et al., 1996 ▶). Here, we report the 1.6 Å resolution crystal structure of MthNTA-MoB, a homolog of Rv3567c from M. tuberculosis (Mtu) in a highly conserved family within Mycobacterium. At the time of writing, it is one of only five entries for M. thermoresistibile available in the Protein Data Bank (PDB), all of which have been solved by the Seattle Structural Genomics Center for Infectious Disease (SSGCID).
2. Methods
2.1. Protein expression and purification
The gene for the full-length NTA-MoB protein (Target DB MythA.00250.a; GenBank accession No. HQ644138; NCBI YP_890259.1; A0R521 homolog) spanning residues 1–189 (‘ORF’) was amplified from M. thermoresistibile Tsukamura strain ATCC19527/NCTC10409 (genomic DNA and sequence information provided by Dr Christoph Grundner, Seattle Biomedical Research Institute) and cloned into a pAVA0421 vector encoding an N-terminal hexahistidine-affinity tag followed by the human rhinovirus 3C protease cleavage sequence (MAHHHHHHMGTLEAQTQGPGS-ORF; Alexandrov et al., 2004 ▶) by ligation-independent cloning (LIC; Aslanidis & de Jong, 1990 ▶). The plasmid construct for MthNTA-MoB (MythA.00250.a.A1) was transformed into Escherichia coli BL21 (DE3) Rosetta cells. An overnight culture was grown in LB broth at 310 K and was used to inoculate 2 l ZYP-5052 auto-induction medium, which was prepared as described by Studier (2005 ▶). MthNTA-MoB protein was expressed in a LEX bioreactor in the presence of antibiotics. After 24 h at 298 K, the temperature was reduced to 288 K for a further 60 h. The sample was centrifuged at 4000g for 20 min at 277 K and the cell paste was flash-frozen in liquid nitrogen and stored at 193 K.
For purification, the frozen cell pellet was thawed and completely resuspended in lysis buffer (20 mM HEPES pH 7.4, 300 mM NaCl, 5% glycerol, 30 mM imidazole, 0.5% CHAPS, 10 mM MgCl2, 3 mM β-mercaptoethanol, 1.3 mg ml−1 protease-inhibitor cocktail, 0.05 mg ml−1 lysozyme). The resuspended cell pellet was then disrupted on ice for 15 min with a Branson Digital 450D Sonifier (70% amplitude, with alternating cycles of 5 s pulse-on and 10 s pulse-off). The cell debris was incubated with 20 µl Benzonase nuclease at room temperature for 40 min. The lysate was clarified by centrifugation at 277 K with a Sorvall RC5 at 10 000 rev min−1 for 60 min. The clarified solution was filtered through a 0.45 µm syringe filter (Corning Life Sciences, Lowell, Massachusetts, USA). The lysate was purified by IMAC using a HisTrap FF 5 ml column (GE Biosciences, Piscataway, New Jersey, USA) equilibrated with binding buffer (25 mM HEPES pH 7.0, 300 mM NaCl, 5% glycerol, 30 mM imidazole, 1 mM TCEP) and eluted with 500 mM imidazole in the same buffer. MthNTA-MoB was concentrated without 3C protease cleavage of the hexahistidine tag. The concentrated pool was further resolved by size-exclusion chromatography (SEC) using a Superdex 75 26/60 column (GE Biosciences) equilibrated with SEC buffer (20 mM HEPES pH 7.0, 300 mM NaCl, 5% glycerol, 1 mM TCEP) attached to an ÄKTA FPLC system (GE Biosciences). Peak fractions were collected and pooled based on purity-profile assessment by SDS–PAGE. Concentrated pure protein in SEC buffer was flash-frozen in liquid nitrogen and stored at 193 K. The final concentration (68.9 mg ml−1) was determined by UV spectrophotometry at 280 nm and the final purity (>97%) was assayed by SDS–PAGE.
2.2. Crystallization
Crystallization trials were set up according to a rational crystallization approach (Newman et al., 2005 ▶) using the JCSG+ and PACT sparse-matrix screens from Emerald BioSystems and Molecular Dimensions, respectively. 0.4 µl protein solution (68.9 mg ml−1) was with mixed with an equal volume of precipitant and set up against 80 µl reservoir solution in sitting-drop vapor-diffusion format in 96-well Compact Jr plates from Emerald BioSystems at 289 K. Crystals grew in several conditions within 9 d, but the crystal used for data collection grew in the presence of 0.2 M magnesium chloride, 0.1 M MES pH 6.0 and 20% PEG 6000 (PACT condition B10).
2.3. Data collection and structure determination
A crystal was harvested, cryoprotected using precipitant solution supplemented with 20% glycerol and vitrified in liquid nitrogen. A 1.6 Å resolution data set was collected under a stream of liquid nitrogen on Advanced Light Source (ALS) beamline 5.0.2 as part of the ALS Collaborative Crystallography program (Table 1 ▶). The data were reduced with HKL-2000 (Otwinowski & Minor, 1997 ▶). The structure (Table 2 ▶) was solved by molecular replacement using Bacillus thermoglucosidasius A7 flavin reductase A2 protein-only dimer from molecules A and B of PDB entry 1rz0 (van den Heuvel et al., 2004 ▶) as a search model in Phaser (McCoy et al., 2007 ▶) from the CCP4 suite (Winn et al., 2011 ▶). The asymmetric unit was comprised of two independent dimers. The final model was obtained after numerous iterative rounds of refinement in REFMAC (Murshudov et al., 2011 ▶) and manual rebuilding in Coot (Emsley & Cowtan, 2004 ▶). The final model contained residues Ala3–Ala181 with no internal gaps for protomer A and a few additional protein residues for each of the other three protomers. In addition, the final model contained one glycerol molecule (bound to protomer D) and 548 water molecules. The structure was assessed and corrected for geometry and fitness using MolProbity (Chen et al., 2010 ▶). Data-collection results and structure-refinement statistics are listed in Tables 1 ▶ and 2 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest of 20 resolution shells.
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 48.5, b = 91.0, c = 165.6 |
| Wavelength (Å) | 1.000 |
| Resolution range (Å) | 50–1.6 (1.64–1.60) |
| No. of unique reflections | 97350 (7102) |
| Multiplicity | 8.2 (7.8) |
| Completeness (%) | 99.8 (99.9) |
| Rmerge† | 0.095 (0.428) |
| Mean I/σ(I) | 13.4 (5.4) |
R
merge = 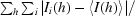
 .
.
Table 2. Refinement and model statistics.
Values in parentheses are for the highest of 20 resolution shells.
| Resolution range (Å) | 20–1.6 (1.64–1.60) |
| Rcryst† | 0.186 (0.180) |
| Rfree† | 0.217 (0.227) |
| R.m.s.d. bonds (Å) | 0.015 |
| R.m.s.d. angles (°) | 1.273 |
| Protein atoms | 6109 |
| Heteroatoms | 6 |
| Waters | 548 |
| Mean B factor (Å2) | 16.7 |
| Residues in favored region (%) | 97.6 |
| Residues in allowed region (%) | 100 |
| MolProbity‡ score [percentile] | 0.91 [100th] |
3. Results
3.1. Identification of MthNTA-MoB
Rv3567c (UniProt accession No. P96849) is a member of a large group of genes that are under the control of a ketosteroid regulon, kstR, which is a member of the tetracycline resistant-like family of transcriptional regulators (Kendall et al., 2007 ▶). Rv3567c is predicted to be involved in lipid catabolism; this gene has been shown to be inducible both in palmitic acid and when grown on cholesterol in Rhodococcus. sp strain RHA1, a soil bacterium related to M. tuberculosis (Van der Geize et al., 2007 ▶). Rv3567c clusters with about 28 genes specifically expressed in M. tuberculosis, among which Rv3569c (hsaD) has been shown to be essential for survival in primary murine macrophages by a transposon-site hybridization (TraSH) experiment in M. tuberculosis H37Rv. For these reasons, Rv3567c and related genes have been assigned to the cholesterol-degradation pathway (Van der Geize et al., 2007 ▶; Rengarajan et al., 2005 ▶). Although NTA-MoB tends to be highly conserved across Mycobacterium species, only one homolog was found in the UniProt database at the outset of this work: that from M. smegmatis (A0R521). A search for NTA-MoB homologs in M. tuberculosis in the TubercuList database (http://genolist.pasteur.fr/TubercuList/) resulted in a match to Rv3567c (Cole et al., 1998 ▶). The MthNTA-MoB protein consists of 189 amino-acid residues and is 82% identical (89% similar) to MtuNTA-MoB, which in turn matches 100% to Rv3567c in TubercuList. In addition, MthNTA-MoB is at least 80% identical to all other known Mycobacterium orthologs, including that from M. smegmatis (A0R521). During target selection, 21 additional paralogs of NTA-MoB were selected and have entered the SSGCID pipeline, including those from M. abscessus, M. avium, M. bovis, M. leprae, M. marinum, M. paratuberculosis, M. smegmatis and M. ulcerans. Among these 21 homologs, 12 were successfully cloned, expressed and purified and three formed crystals, one of which (MthNTA-MoB) diffracted to sufficiently high resolution for structure determination.
3.2. Comparison with other short-chain flavin reductases
A search of the Protein Data Bank (http://www.rcsb.org/pdb/) resulted in no proteins with a sequence similarity greater than 35% to MthNTA-MoB. The Pfam database (http://www.sanger.ac.uk/software/pfam) assigned MthNTA-MoB to the PF01613 family of proteins with the FMN-binding split-barrel motif at an E value of 8.7 × 10−40. This NADH:FMN oxidoreductase or flavin reductase family was first described in the early 1990s and exists in many organisms, primarily Gram-negative bacteria (Uetz et al., 1992 ▶; Blanc et al., 1995 ▶). These short-chain flavin reductases are involved in a variety of biological reactions and often act in concert with a flavin-dependent monooxygenase which oxidizes through the addition of molecular oxygen (Kirchner et al., 2003 ▶; Galán et al., 2000 ▶). However, they do not share sequence homology with flavin reductases found in E. coli or luminous bacteria or with the LuxG protein class found in lux operons (Nijvipakul et al., 2008 ▶).
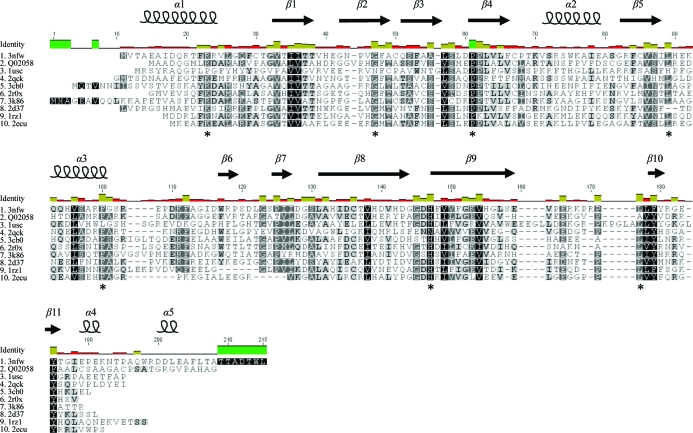
A sequence alignment of MthNTA-MoB with other members of the short-chain flavin reductase family is shown in Fig. 1 ▶. Sequences were selected either based on structural homology or owing to well reported functional similarity (Valton et al., 2008 ▶; Filisetti et al., 2003 ▶). MthNTA-MoB shares many conserved residues within this family of short-chain flavin reductases, including Arg12, Gly35, Pro48, Leu76, Phe88, His130 and Leu152 (Fig. 1 ▶). However, the overall sequence similarity is low, despite high E values in structural classification. Table 3 ▶ lists the amino-acid identities and structural similarities of MthNTA-MoB to a variety of other enzymes involved in the degradation of small organic compounds based on search results using DALI (Holm & Sander, 1993 ▶). In particular, the MthNTA-MoB structure bears marked homology to the phenol 2-hydroxylase component B of reduced flavin reductase from B. thermoglucosidasius (BtPheA2; PDB entry 1rz1; van den Heuvel et al., 2004 ▶).
Figure 1.
Protein sequence alignment of MthNTA-MoB (PDB entry 3nfw) with other short-chain flavin reductases. The secondary-structural elements of 3nfw are indicated and labelled above the aligned sequences. The similarity levels for each of the amino-acid positions are indicated using a grayscale, where darker shades indicate a higher degree of conservation. Strictly conserved residues are shown in black, while lighter grays and white indicate various degrees of low conservation and no conservation, respectively. Conserved residues shared with other short-chain flavin reductases are marked with an asterisk at the bottom of the column. Color legend for sequence identity: green, conserved; yellow, similar; red, unconserved. Details of the proteins aligned with 3nfw are shown in Table 3 ▶.
Table 3. Amino-acid identity and structural similarity across the top eight structural homologs of 3nfw available in the PDB.
Data based on the RCSB PDB (http://www.pdb.org/pdb/).
| PDB code | UniProt ID | Protein name | Organism | Amino-acid identity (%) | R.m.s.d. (Å) | Reference |
|---|---|---|---|---|---|---|
| 2rox | Q0I3S1 | Putative flavin reductase | Haemophilus somnus 129PT | 21.5 | 1.27 | — |
| 1rz1 | Q9LAG2 | Phenol 2-hydroxylase component B | Bacillus thermoglucosidasius A7 | 31 | 1.62 | van den Heuvel et al. (2004 ▶) |
| 2d37 | Q974C9 | Phenol 2-hydroxylase component B | Sulfolobus tokodaii strain 7 | 26 | 1.65 | Okai et al. (2006 ▶) |
| 2ecu | Q5SJP7 | Flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase polypeptide (L) | Thermus thermophilus HB8 | 27 | 1.73 | Kim et al. (2008 ▶) |
| 3cb0 | Q8YHT7 | 4-Hydroxyphenylacetate 3-monooxygenase | Brucella melitensis | 25.2 | 1.83 | Lawrence et al. (2008 ▶) |
| 1usc | P83818 | Styrene monooxygenase small component | Thermus thermophilus | 20 | 1.87 | — |
| 3k86 | 087008 | Chlorophenol-4-monooxygenase component 2 | Burkholderia cepacia | 26 | 2.05 | Webb et al. (2010 ▶) |
| 2qck | A0JVA7 | Flavin reductase domain protein | Arthrobacter sp. FB24 | 26 | 2.13 | — |
| n/a | Q02058 | Actinorhodin polyketide dimerase | Streptomyces coelicolor A3 | 32 | n/a | Filisetti et al. (2003 ▶), Valton et al. (2008 ▶) |
3.3. Three-dimensional structure of MthNTA-MoB
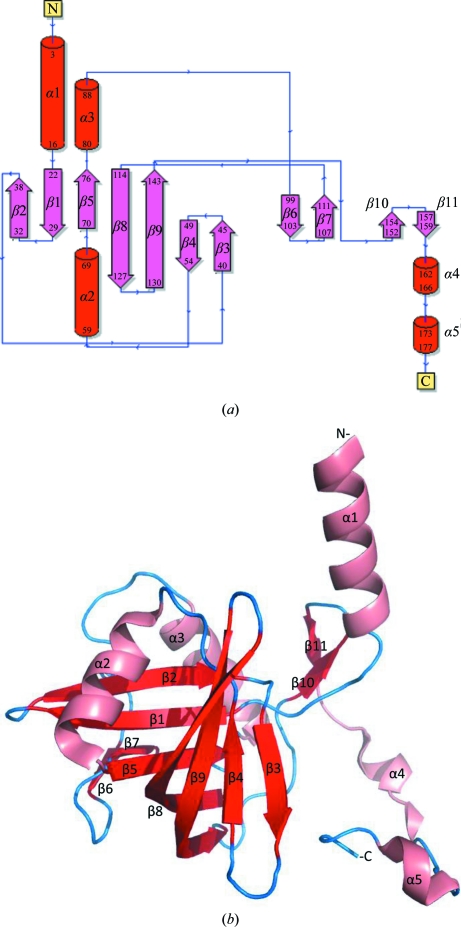
The native MthNTA-MoB protein crystallized in the orthorhombic space group P212121 and an apo structure was determined to a resolution limit of 1.6 Å (PDB entry 3nfw) with R cryst and R free factors of 0.186 and 0.217, respectively (Table 2 ▶). MthNTA-MoB consists of 11 β-strands and five α-helices, with seven antiparallel β-sheet core regions forming a split-barrel motif capped by an α-helix (Fig. 2 ▶). Consistent with observations from size-exclusion chromatography, the biological molecule observed within the crystal lattice is a dimer and the crystal structure contained two noncrystallographically related dimers. PISA (Krissinel & Henrick, 2007 ▶) calculated a total buried surface area of 8610 Å2 and a change in solvent free energy of −234 kJ mol−1 upon dimerization.
Figure 2.
Structural diagrams of 3nfw. (a) Secondary-structural topology diagram. Red cylinders indicate α-helices and pink arrows represent β-sheets. The beginning and ending amino-acid residue numbers are indicated inside each of the cylinders or arrows, together with the sequential numbers of each secondary-structural component. (b) Ribbon diagram of 3nfw. Pink spirals indicate α-helices and red arrows represent β-sheets. Secondary-structural elements and N- and C-terminal residues are indicated.
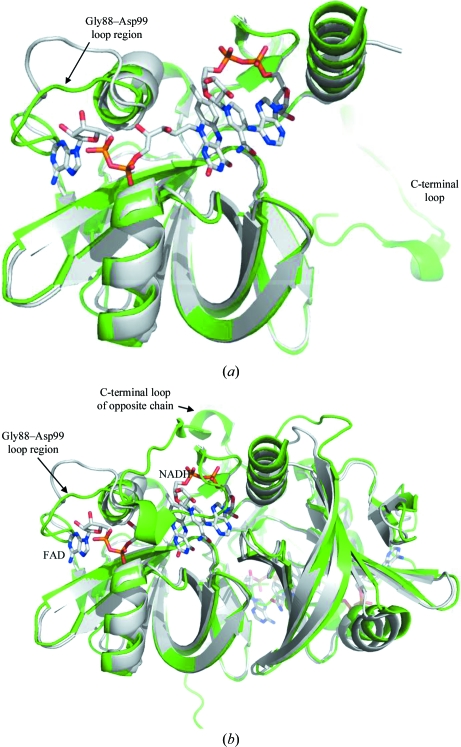
The spatial difference between Cα atoms of MthNTA-MoB and BtPheA2 is 0.98 Å, indicating a high degree of structural conservation for two proteins with only 32% sequence similarity (Fig. 3 ▶). Unlike the MthNTA-MoB structure, crystal data for BtPheA2 were acquired with the cofactor NADPH bound. According to structure-alignment results generated using the SSM (PDBe) server (http://www.ebi.ac.uk/msd-srv/ssm/cgi-bin/ssmserver), the loop region at amino-acid residues Gly88–Asp99 (between α-helix 3 and β-strand 6) has the lowest level of amino-acid residue alignment. This loop forms a critical part of the binding pocket recognized by FMN, with the analogous loop reported to be involved in binding either FAD or FMN in PheA2 and ferric reductase, respectively (van den Heuvel et al., 2004 ▶; Okai et al., 2006 ▶). This loop has been described as flexible in the absence of FMN but highly ordered in FMN-bound structures. In the MthNTA-MoB structure the loop region has the highest deviation from the structure of substrate-bound BtPheA2. While the average distance between superimposed pairs of residues ranges from 1.29 to 4.42 Å, with the greatest deviation at Ala96, the overall Cα r.m.s.d. is only 0.98 Å (Fig. 3 ▶ a).
Figure 3.
Cα trace of molecular superposition of 3nfw (green) and 1rz1 (gray). FAD and NADH molecules are shown bound to 1rz1. (a) Molecular overlay of the monomers, with the Gly88–Asp99 loop region and the C-terminal loop region indicated. (b) Molecular overlay of the dimers, with the Gly88–Asp99 loop region and the C-terminal loop region of the opposite chain and the FAD and NADH molecules bound to 1rz1 indicated.
The other region in which MthNTA-MoB deviates most from the BtPheA2 structure is in the C-terminal loop region from Lys165 to Thr182 (Fig. 3b). This region adopts the same conformation in all four protomers in the asymmetric unit and is flexible and adjacent to the NADH-binding cleft (Deng et al., 1999 ▶). It is expected to play an inhibitory role in substrate (flavin) and cofactor (NADH) binding (van den Heuvel et al., 2004 ▶). However, further structural data are necessary to confirm the interaction of the loop with cofactor bound to the same molecule or a neighboring molecule.
4. Discussion
The reaction mechanism of this class of enzymes has been a topic of discussion in recent years (Filisetti et al., 2003 ▶; van den Heuvel et al., 2004 ▶; Valton et al., 2008 ▶). One group proposed the ping-pong mechanism, while others described the reactions as following an ordered sequential mechanism. The flexible C-terminal region in the 3nfw structure could potentially play a role in inhibiting cofactor binding and/or release. However, without further evidence of a bound substrate or cofactor and a lack of functional analysis it is difficult to support either of the mechanisms one way or the nother.
Previous publications have suggested that lipid metabolism is fundamental to the unusual ability of M. tuberculosis to use fatty acids as a sole carbon source and survive in vivo (Van der Geize et al., 2007 ▶). Recent reports further predict that M. tuberculosis subsists on alternative carbon sources in the human host. Specifically, M. tuberculosis is able to persist in the glucose-deficient and fatty-acid-rich environment found in the macrophage stage of infection, thereby contributing to its unparalleled virulence (Savvi et al., 2008 ▶). The up-regulation of the Rv3567c group of enzymes during survival in macrophages indicated their involvement in cholesterol uptake, which links them to phagocytosis, a vital step in bacterial pathogenic metabolism (Van der Geize et al., 2007 ▶). An important role in cholesterol metabolism and cell survival could be the reason for the high conservation of these genes. Uncertainties remained in a previous discussion of the natural substrates of NTA-Mo (Uetz et al., 1992 ▶). It is possible that the structure of the synthetic NTA and its subsequent degradation products resemble some of the key metabolites in metabolism pathways such as the tricarboxylic acid (TCA) pathway, hence the analogous activity. Further experiments to compare the binding affinities of these compounds and their reaction mechanism may help to answer some of these questions.
5. Conclusion
MthNTA-MoB is 82% identical (89% similar) to MtuNTA-MoB, which matches 100% to Rv3567c in TubercuList. Rv3567c and related cluster of genes, some of which are essential, have been assigned to the cholesterol-degradation pathway in M. tuberculosis. We have reported the 1.6 Å resolution structure of MthNTA-MoB, one of the first protein structures to be reported for this organism. Structure comparisons characterize it as a member of the short-chain flavin reductase family with a high confidence level. The overall structure and conserved amino-acid residues align well with those of other proteins, despite low sequence homology. Two of the loop regions showed deviations in a structural overlay with other members of the family, including those in which the cofactor is bound. The density was sufficient to allow modelling of the long loop at the C-terminus, a region that is often absent in structures obtained using crystallographic data owing to a high degree of flexibility, and may provide additional insight into its role in modulating activity via access for cofactor binding.
Supplementary Material
PDB reference: NTA-MoB, 3nfw
Acknowledgments
The authors thank the whole SSGCID team. This research was funded under Federal Contract No. HHSN272200700057C from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services. The Berkeley Centre for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences and the US Department of Energy under Contract No. DE-AC02-05CH11231. We thank Dr Christoph Grundner for providing genomic information on M. thermoresistibile Tsukamura strain, for helpful discussions and valuable insights into the TubercuList database.
References
- Alexandrov, A., Vignali, M., LaCount, D. J., Quartley, E., de Vries, C., De Rosa, D., Babulski, J., Mitchell, S. F., Schoenfeld, L. W., Fields, S., Hol, W. G., Dumont, M. E., Phizicky, E. M. & Grayhack, E. J. (2004). Mol. Cell. Proteomics, 3, 934–938. [DOI] [PubMed]
- Anderton, M. C., Bhakta, S., Besra, G. S., Jeavons, P., Eltis, L. D. & Sim, E. (2006). Mol. Microbiol. 59, 181–192. [DOI] [PubMed]
- Aslanidis, C. & de Jong, P. J. (1990). Nucleic Acids Res. 18, 6069–6074. [DOI] [PMC free article] [PubMed]
- Bally, M., Wilberg, E., Kühni, M. & Egli, T. (1994). Microbiology, 140, 1927–1936. [DOI] [PubMed]
- Berkel, W. J. van, Kamerbeek, N. M. & Fraaije, M. W. (2006). J. Biotechnol. 124, 670–689. [DOI] [PubMed]
- Blanc, V., Lagneaux, D., Didier, P., Gil, P., Lacroix, P. & Crouzet, J. (1995). J. Bacteriol. 177, 5206–5214. [DOI] [PMC free article] [PubMed]
- Boloorsaz, M. R., Khalilzadeh, S., Farnia, P., Hakimi, S. S. & Velayati, A. A. (2006). Tanaffos, 5, 61–63.
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cole, S. T. et al. (1998). Nature (London), 393, 537–544.
- Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B. III, Snoeyink, J., Richardson, J. S. & Richardson, D. C. (2007). Nucleic Acids Res. 35, W375–W383. [DOI] [PMC free article] [PubMed]
- Deng, Z., Aliverti, A., Zanetti, G., Arakaki, A. K., Ottado, J., Orellano, E. G., Calcaterra, N. B., Ceccarelli, E. A., Carrillo, N. & Karplus, P. A. (1999). Nature Struct. Biol. 6, 847–853. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Filisetti, L., Fontecave, M. & Nivière, V. (2003). J. Biol. Chem. 278, 296–303. [DOI] [PubMed]
- Galán, B., Díaz, E., Prieto, M. A. & García, J. L. (2000). J. Bacteriol. 182, 627–636. [DOI] [PMC free article] [PubMed]
- Heuvel, R. H. van den, Westphal, A. H., Heck, A. J., Walsh, M. A., Rovida, S., van Berkel, W. J. & Mattevi, A. (2004). J. Biol. Chem. 279, 12860–12867. [DOI] [PubMed]
- Holm, L. & Sander, C. (1993). J. Mol. Biol. 233, 123–138. [DOI] [PubMed]
- Kendall, S. L., Withers, M., Soffair, C. N., Moreland, N. J., Gurcha, S., Sidders, B., Frita, R., Ten Bokum, A., Besra, G. S., Lott, J. S. & Stoker, N. G. (2007). Mol. Microbiol. 65, 684–699. [DOI] [PMC free article] [PubMed]
- Kim, S.-H., Hisano, T., Iwasaki, W., Ebihara, A. & Miki, K. (2008). Proteins, 70, 718–730. [DOI] [PubMed]
- Kirchner, U., Westphal, A. H., Müller, R. & van Berkel, W. J. (2003). J. Biol. Chem. 278, 47545–47553. [DOI] [PubMed]
- Knobel, H. R., Egli, T. & van der Meer, J. R. (1996). J. Bacteriol. 178, 6123–6132. [DOI] [PMC free article] [PubMed]
- Kremer, L., Guérardel, Y., Gurcha, S. S., Locht, C. & Besra, G. S. (2002). Microbiology, 148, 3145–3154. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Lawrence, A. D., Deery, E., McLean, K. J., Munro, A. W., Pickersgill, R. W., Rigby, S. E. & Warren, M. J. (2008). J. Biol. Chem. 283, 10813–10821. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Neonakis, I. K., Gitti, Z., Kontos, F., Baritaki, S., Petinaki, E., Baritaki, M., Zerva, L. & Spandidos, D. A. (2009). Indian J. Med. Microbiol. 27, 264–267. [DOI] [PubMed]
- Neonakis, I. K., Gitti, Z., Kourbeti, I. S., Michelaki, H., Baritaki, M., Alevraki, G., Papadomanolaki, E., Tsafaraki, E., Tsouri, A., Baritaki, S., Krambovitis, E. & Spandidos, D. A. (2007). Scand. J. Infect. Dis. 39, 875–879. [DOI] [PubMed]
- Newman, J., Egan, D., Walter, T. S., Meged, R., Berry, I., Ben Jelloul, M., Sussman, J. L., Stuart, D. I. & Perrakis, A. (2005). Acta Cryst. D61, 1426–1431. [DOI] [PubMed]
- Nijvipakul, S., Wongratana, J., Suadee, C., Entsch, B., Ballou, D. P. & Chaiyen, P. (2008). J. Bacteriol. 190, 1531–1538. [DOI] [PMC free article] [PubMed]
- Okai, M., Kudo, N., Lee, W. C., Kamo, M., Nagata, K. & Tanokura, M. (2006). Biochemistry, 45, 5103–5110. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Rengarajan, J., Bloom, B. R. & Rubin, E. J. (2005). Proc. Natl Acad. Sci. USA, 102, 8327–8332. [DOI] [PMC free article] [PubMed]
- Rylance, J., Pai, M., Lienhardt, C. & Garner, P. (2010). Lancet Infect. Dis. 10, 886–892. [DOI] [PMC free article] [PubMed]
- Savvi, S., Warner, D. F., Kana, B. D., McKinney, J. D., Mizrahi, V. & Dawes, S. S. (2008). J. Bacteriol. 190, 3886–3895. [DOI] [PMC free article] [PubMed]
- Studier, F. W. (2005). Protein Expr. Purif. 41, 207–234. [DOI] [PubMed]
- Tsukamura, M. (1966). J. Gen. Microbiol. 45, 253–273. [DOI] [PubMed]
- Uetz, T., Schneider, R., Snozzi, M. & Egli, T. (1992). J. Bacteriol. 174, 1179–1188. [DOI] [PMC free article] [PubMed]
- Valton, J., Mathevon, C., Fontecave, M., Nivière, V. & Ballou, D. P. (2008). J. Biol. Chem. 283, 10287–10296. [DOI] [PMC free article] [PubMed]
- Van der Geize, R., Yam, K., Heuser, T., Wilbrink, M. H., Hara, H., Anderton, M. C., Sim, E., Dijkhuizen, L., Davies, J. E., Mohn, W. W. & Eltis, L. D. (2007). Proc. Natl Acad. Sci. USA, 104, 1947–1952. [DOI] [PMC free article] [PubMed]
- Webb, B. N., Ballinger, J. W., Kim, E., Belchik, S. M., Lam, K.-S., Youn, B., Nissen, M. S., Xun, L. & Kang, C. (2010). J. Biol. Chem. 285, 2014–2027. [DOI] [PMC free article] [PubMed]
- Weitzman, I., Osadczyi, D., Corrado, M. L. & Karp, D. (1981). J. Clin. Microbiol. 14, 593–595. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: NTA-MoB, 3nfw