Abstract
Telomerase is a ribonucleoprotein reverse transcriptase specialized for use of a sequence within its integral RNA component as the template for DNA synthesis. Telomerase adds telomeric simple sequence repeats to single-stranded primers in vitro or chromosome ends in vivo. We have investigated the sequences and structures of recombinant Tetrahymena thermophila telomerase RNA necessary for physical association and activity with the catalytic protein subunit expressed in rabbit reticulocyte lysate. In contrast with previous results using another reconstitution method, we find that phylogenetically conserved primary sequences and a phylogenetically nonconserved secondary structure are essential for telomerase RNA function. Telomerase RNA binding to the catalytic protein subunit requires sequences 5′ of the template and is highly sequence specific. Other telomerase RNA sequences are required for enzyme activity and proper template use but not for protein interaction affinity. In addition, we demonstrate that the production of active recombinant telomerase requires a factor in rabbit reticulocyte lysate that promotes ribonucleoprotein assembly. These studies demonstrate multiple functions for the telomerase RNA and indicate that recombinant telomerase activity requires more than the catalytic protein and RNA components of the enzyme that have been identified to date.
Keywords: telomerase, RNA, telomere, ribonucleoprotein, polymerase
Telomerase extends a substrate’s 3′ end by addition of the T+G strand of telomeric simple-sequence repeat DNA. The sequence specificity of DNA synthesis is directed by a template sequence within the intrinsic RNA component of the ribonucleoprotein (RNP) enzyme (Greider and Blackburn 1989; Yu et al. 1990). This makes telomerase a reverse transcriptase with an unprecedented mode of template recognition. Substrate recognition by telomerase is also atypical of other reverse transcriptases. Primers lacking any complementarity to the template can be elongated, albeit less efficiently than primers with a template-complementary 3′ end (for review, see Collins 1999). In vivo, telomeres are the preferred telomerase substrates although healing of broken chromosome ends without any telomeric repeat homology is also observed (for review, see Melek and Shippen 1996). In some cell types, telomerase activity at telomeres regulates cellular proliferative life span. Most cell cultures lacking telomerase activity experience a proliferation-dependent telomeric repeat attrition and ultimately stop dividing when telomeres become critically compromised (for review, see Harley and Villeponteau 1995). In fibroblasts, activation of telomerase is sufficient to confer indefinite proliferative capacity (Bodnar et al 1998). Thus, telomerase activity may function in vivo to govern a telomere-based mitotic clock.
The RNA subunit of telomerase has been identified in numerous organisms (for review, see Greider 1996). Large differences in length are observed among ciliate, yeast, and mammalian telomerase RNAs. Only a small percentage of the total telomerase RNA sequence serves as the template for telomeric repeat DNA synthesis. For the 148- to 209-nucleotide ciliate telomerase RNAs, conserved elements of primary sequence and secondary structure have been discerned by phylogenetic comparison (Romero and Blackburn 1991; Lingner et al. 1994; McCormick-Graham and Romero 1995, 1996). Direct assays of RNA accessibility to modification in the RNP (Zaug and Cech 1995) support the model for Tetrahymena thermophila telomerase RNA secondary structure shown in Figure 1. The function of the T. thermophila telomerase RNA has been investigated by RNP reconstitution either in vivo, following introduction of mutant telomerase RNA genes (Yu et al. 1990), or in vitro, following micrococcal nuclease (MNase) digestion of endogenous RNA and addition of an in vitro transcribed substitute (Autexier and Greider 1994). Activity assays with telomerase RNAs differing in their template sequences indicate that template-copying efficiency and fidelity depend on the template sequence in a complex manner (for review, see Collins 1999). These studies suggest that template function involves more than just base-pairing with substrate dNTPs.
Figure 1.
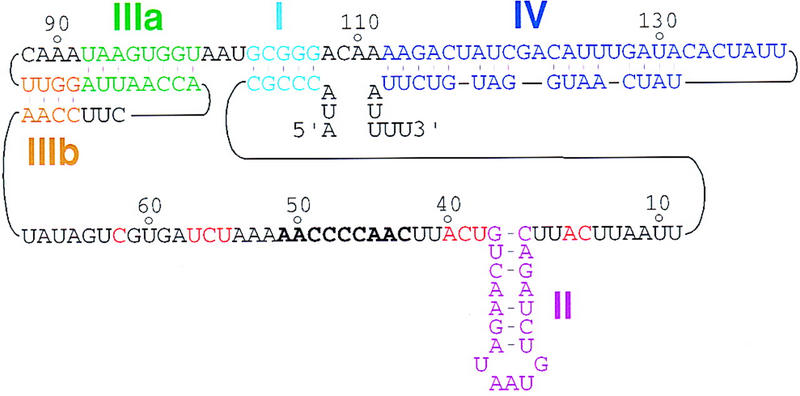
Telomerase RNA in T. thermophila. The sequence and structure of the T. thermophila telomerase RNA are shown as described previously (Greider and Blackburn 1989; ten Dam et al. 1991; Romero and Blackburn 1991; Zaug and Cech 1995). The representation here emphasizes the secondary structure elements and the primary sequences examined in this study. Stem I is in light blue, stem–loop II is in purple, the stem of stem–loop III as initially described is in green (IIIa), with pseudoknot formation producing a second stem in brown (IIIb), and stem-loop IV is in dark blue. The template region is in bold, and phylogenetically conserved sequences in the single-stranded, template-adjacent regions are in red. RNA 5′ and 3′ ends are indicated.
Protein components of telomerase RNPs have been discovered also (for review, see Nugent and Lundblad 1998). The telomerase reverse transcriptase or TERT subunit has been identified in species ranging from ciliates to mammals. The TERT protein is required for enzyme activity and contains the sequence motifs of a polymerase active site (Lingner et al. 1997; Nakamura and Cech 1998). Human or T. thermophila TERT expressed in rabbit reticulocyte lysate with the cognate telomerase RNA produces a telomerase activity that copies the correct six-nucleotide RNA template (Beattie et al. 1998; Collins and Gandhi 1998; Weinrich et al. 1997). Although recombinant T. thermophila TERT and telomerase RNA are sufficent to define the template sequence, not all substrates elongated by the endogenous enzyme are recognized by the recombinant telomerase RNA–TERT complex. Primer use by the recombinant enzyme is most consistent with an incomplete repertoire of substrate interaction sites (Collins and Gandhi 1998). Additional telomerase proteins have been characterized in T. thermophila and mammalian cells. The T. thermophila proteins p80 and p95 interact with telomerase RNA and TERT, and p95 binds single-stranded substrate DNAs (Collins et al. 1995; Collins and Gandhi 1998; Gandhi and Collins 1998). The mammalian TEP1 protein has sequence homology to p80 and also interacts with telomerase RNA and TERT (Harrington et al. 1997a,b; Nakayama et al. 1997). Surprisingly, none of the primary sequences of known telomerase proteins reveals any recognizable motif for interaction with nucleic acid. This may reflect novel types of interactions between telomerase proteins and telomerase RNA or substrate DNA.
In this report, we have used reconstitution of T. thermophila telomerase RNA and TERT in rabbit reticulocyte lysate to define residues of the telomerase RNA required for TERT binding and telomerase enzyme activity. Our reconstitution protocol differs from another in vitro telomerase reconstitution protocol, referred to in the following text as the MNase protocol, in several respects. In the MNase protocol, removal of telomerase RNA from partially purified endogenous enzyme by nuclease treatment leaves residual telomerase RNA fragments that would not be present in lysate reconstitution. Also, MNase protocol reconstitution could involve holoenzyme proteins in addition to the TERT subunit. In lysate but not by the MNase protocol, telomerase RNA reconstitution is highly sequence-specific. Similarly, in lysate but not by the MNase protocol, phylogenetically conserved and chemical modification-protected telomerase RNA sequences and structures are critical for telomerase activity. Thus, lysate reconstitution appears to provide a physiologically relevant indication of telomerase RNA function. We demonstrate that some residues of telomerase RNA are required for TERT binding, whereas other residues are not required for TERT binding but are required for enzyme activity. In addition, we show that a lysate factor is required to establish the stable association of telomerase RNA and TERT. This observation accounts for why MNase protocol reconstitution does not recreate a physiologically appropriate telomerase RNA–TERT interaction.
Results
Telomerase RNA reconstitution in lysate is sequence-specific
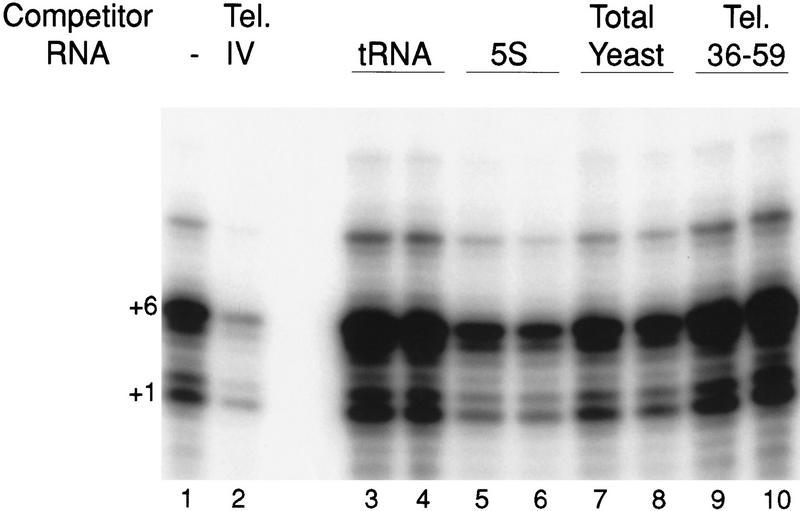
T. thermophila TERT can be coexpressed with telomerase RNA or combined with telomerase RNA after protein expression in lysate to produce an active recombinant enzyme (Collins and Gandhi 1998). Product accumulation is particularly strong using the primer (TG)8TTG, which aligns at the template 5′ end and copies one complete G3T2G repeat to produce products at primer +1 to +6. Longer product DNAs with a 6-nucleotide periodicity result from the limited, dGTP concentration-dependent repeat addition processivity characteristic of the recombinant enzyme. To address the specificity of recombinant telomerase RNA–TERT interaction, we expressed TERT in lysate and added various competitor RNAs before addition of the wild-type telomerase RNA. A similar study of telomerase RNA interaction specificity in MNase protocol reconstitution revealed previously that random RNAs added in only slight excess of the telomerase RNA can inhibit reconstitution of active telomerase strongly (Gandhi and Collins 1998). In contrast, telomerase activity assays of lysate-reconstituted telomerase reveal that the nonspecific competitor RNAs do not inhibit telomerase activity even when present at a 50-fold excess over telomerase RNA (Fig. 2). This is evident for competitor RNAs that do not inhibit reconstitution by the MNase protocol (tRNA, 5S RNA; lanes 3–6) and for competitor RNAs that do inhibit reconstitution by the MNase protocol (total yeast RNA; lanes 7,8). In fact, addition of most nonspecific competitor RNAs actually stimulates telomerase activity when low telomerase RNA concentrations are used (10 ng of telomerase RNA in Fig. 2). This derives most likely from binding of the competitors to lysate proteins that otherwise would sequester telomerase RNA from TERT.
Figure 2.
RNA sequence specificity of telomerase RNP reconstitution in lysate. Five microliters of lysate-expressed TERT was supplemented with 500 ng of the indicated competitor RNA and then with 10 ng of telomerase RNA. Samples were incubated at 30°C for 30 min then assayed for telomerase activity with the primer (TG)8TTG. (Lane 1) No competitor; (lane 2) a telomerase RNA variant missing stem–loop IV, which binds TERT but is less active than the wild-type RNA (see Results); (lanes 3,4) tRNA; (lanes 5,6) 5S RNA; (lanes 7,8) total yeast RNA; (lanes 9,10) a 24-nucleotide RNA oligonucleotide representing telomerase RNA residues 36–59 which have no activity (Collins and Gandhi 1998). In some experiments, total yeast RNA stimulated telomerase activity to the same level as tRNA (not shown). Products corresponding to primer +1 and +6 nucleotides are indicated.
We also investigated the competition of telomerase activity reconstitution by fragments of the telomerase RNA itself. We assayed two telomerase RNA variants that on their own have either reduced activity with wild-type TERT interaction affinity (a stem–loop IV deletion, see below) or no activity (a 24-nucleotide template-containing region spanning residues 36–59 not predicted to interact with TERT; Collins and Gandhi 1998 and see below). Although the stem–loop IV deletion telomerase RNA inhibits activity by competition with the wild-type RNA (Fig. 2, lane 2), the 24-nucleotide RNA containing the template and adjacent sequences does not (lanes 9,10). The shorter telomerase RNA variant, like the nonspecific RNA competitors, actually improves activity in assays with limiting full-length telomerase RNA. We conclude that reconstitution in lysate allows a greater specificity of telomerase RNA–TERT interaction than does reconstitution by the MNase protocol.
RNA secondary structure elements are differentially required for telomerase activity
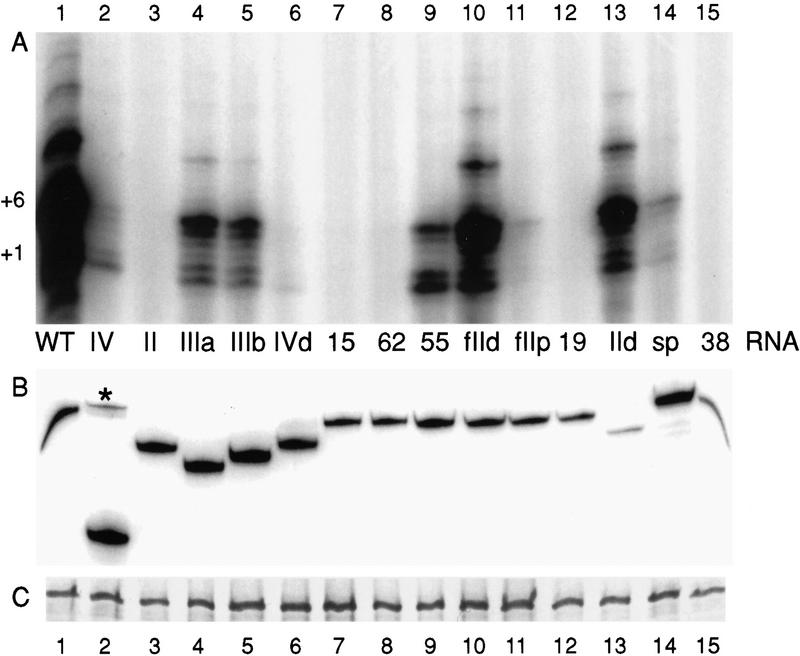
To investigate the telomerase RNA structural requirements for lysate reconstitution, we produced telomerase RNAs lacking individual elements of secondary structure based on the model shown in Figure 1. We created constructs expressing telomerase RNAs missing stem–loop II (purple), either stem–loop pairing of the pseudoknot element III (brown stem and intervening sequence or green stem and intervening sequence), stem–loop IV (dark blue), or only the distal end of stem–loop IV (Fig. 1; Table 1). These deletions remove 15 (II), 12 (IIIa), 17 (IIIb), 51 (IV), or 11 nucleotides (IVd) of the 159-nucleotide endogenous telomerase RNA (Table 1). Coexpression of TERT with telomerase RNA results in comparable expression levels of both protein and RNA components (∼1 ng/μl or 20 nm telomerase RNA and 2 ng/μl or 15 nm TERT; Collins and Gandhi 1998). Each telomerase RNA variant is expressed and accumulated, assayed by Northern blot hybridization with an oligonucleotide complementary to the template region (Fig. 3B). TERT also accumulates to a similar extent in each coexpression reaction (Fig. 3C).
Table 1.
Telomerase RNA variants
|
Telomerase RNA
|
Sequence alteration
|
Label
|
Activity coexpressa
|
Activity add-backa
|
Bindinga
|
|---|---|---|---|---|---|
| Wild type | none | WT | +++ | +++ | +++ |
| Delete stem–loop IV | truncate after 108 | IV | + | + | +++ |
| Delete stem–loop II | delete 21–35 | II | − | +b | − |
| Delete stem–loop IIIa | delete 77–98 | IIIa | ++ | ++ | +++ |
| Delete stem–loop IIIb | delete 70–86 | IIIb | ++ | ++ | +++ |
| Delete distal stem–loop IV | delete 130–140 | IVd | + | + | +++ |
| Alter conserved CA | CA15–16GU | 15 | − | +b | + |
| Alter conserved C | C62G | 62 | − | − | ++ |
| Alter conserved UCU | UCU55–57AGA | 55 | ++ | +++ | ++ |
| Flip distal stem–loop II | reverse 23–33 | fIId | +++ | +++ | +++ |
| Flip proximal stem–loop II | CAG19–21GUC | fIIp | + | ++b | ++ |
| CUG35–37GAC | |||||
| Unpair in proximal stem–loop II | C19A | 19 | − | +b | + |
| Tetraloop for distal stem–loop II | replace 22–34 with UUCG | IId | +++ | +++ | +++ |
| Insertion 5′ of stem–loop II | insert UU before C19 | sp | + | ++ | + |
| Alter conserved UCA | UCA38–40AGU | 38 | −/+ | + | ++ |
T. thermophila telomerase RNA variants used in this study. The sequence alteration and its abbreviated label are indicated. Telomerase activities obtained by coexpression of TERT and telomerase RNA (from Fig. 3) or by addition of excess telomerase RNA after protein synthesis (from Fig. 4) were quantitated by PhosphorImager analysis. Telomerase RNA binding to TERT was also quantitated by PhosphorImager analysis (from Fig. 5).
Data are represented as follows: (+++) 30 to >100% of wild type; (++) 10–29% of wild type; (+) <10% of wild type but detectable; (−) undetectable.
These values were reduced in other experiments.
Figure 3.
Reconstitution by coexpression of telomerase RNA and TERT. Coexpression reactions contained TERT and the indicated telomerase RNA variant. (A) An equal volume of each reaction was assayed for telomerase activity with the primer (TG)8TTG. Products corresponding to primer +1 and +6 nucleotides are indicated. (B) An equal volume of each reaction was analyzed for telomerase RNA by Northern blot hybridization. The asterisk indicates the ∼160-bp DNA fragment encoding the stem–loop IV deleted RNA, which does not encode a full-length telomerase RNA sequence (see Materials and Methods). (C) An equal volume of each lysate coexpression reaction was analyzed for TERT by SDS-PAGE and autoradiography.
Telomerase activity assays of coexpressed telomerase RNAs and TERT reveal that the wild-type telomerase RNA is most active (Fig. 3A, lane 1). Deletion of either alternate stem–loop pairing in the pseudoknot element III reduces activity (lanes 4,5), although neither stem was important for activity by MNase protocol reconstitution (Autexier and Greider 1998). The pattern of product DNA in lysate assays with these RNAs resembles the wild-type RNA pattern of product synthesis. In comparison, deletion of stem–loop IV both reduces total product synthesis and alters the pattern of product synthesis (lane 2). Notably, a smaller deletion of only the distal end of stem–loop IV results in greater loss of activity than does deletion of the entire stem–loop IV (lane 6). This may indicate that the stem IV sequences remaining in the distal deletion inhibit activity in a dominant fashion. For both the stem–loop IV deletion and stem–loop IV distal deletion RNAs, the product intensity corresponding to dissociation or pausing at the template 3′ end is greater than product intensity corresponding to dissociation or pausing at the template 5′ end (primer +1 > primer +6). In contrast, the wild-type RNA promotes more product accumulation derived from copying the complete template to its 5′ end (primer +6 > primer +1). By MNase protocol reconstitution, various truncations of stem–loop IV all reduced activity dramatically (Autexier and Greider 1998).
Our most unexpected finding was observed with the telomerase RNA lacking stem–loop II. Even with long exposures, little or no telomerase activity was detectable with this RNA (Fig. 2C, lane 3). In contrast, by MNase protocol reconstitution, a two-nucleotide larger deletion of residues 20–36 reduced telomerase activity only slightly (Autexier and Greider 1998). The sequence of stem–loop II is not conserved in evolution, and some ciliate species including Tetrahymena paravorax lack a stem–loop II element completely. Therefore, it was unclear why deletion of stem–loop II was more deleterious for activity than deletion of the other elements of secondary structure which are conserved evolutionarily. We assayed each of our telomerase RNA variants above by MNase protocol reconstitution with highly purified endogenous T. thermophila telomerase. Our MNase protocol reconstitution results parallel previous findings closely despite differences in the design of the deletion constructs (data not shown). We conclude that different regions of telomerase RNA are required for activity reconstituted by the two protocols.
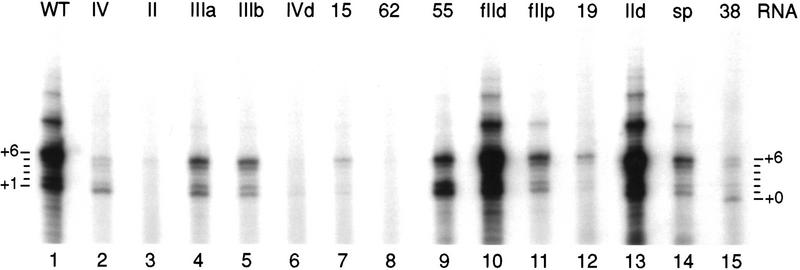
To address whether a higher concentration of telomerase RNA would rescue the deficiencies imposed by the losses of individual elements of secondary structure, we assayed lysate-expressed TERT with a large excess of purified telomerase RNA added after protein synthesis (Fig. 4). For the telomerase RNAs lacking individual elements of secondary structure, telomerase assembly by addition of excess purified RNA to lysate-expressed TERT yields the same relative activities and the same patterns of product DNA accumulation as assembly by RNA coexpression (cf. Figs. 3 and 4, lanes 1–6). For the wild-type telomerase RNA, maximal telomerase activity requires approximately stoichiometric telomerase RNA and TERT and is not stimulated or inhibited by titration of telomerase RNA concentration to a 200- to 1000-fold excess (Collins and Gandhi 1998; data not shown). Titration of the concentration of the secondary-structure deletion-variant telomerase RNAs added to lysate-expressed TERT, above approximately equimolar with TERT, also did not affect the level or the pattern of product DNA synthesis (data not shown).
Figure 4.
Reconstitution by addition of purified RNA to lysate-expressed TERT. All reactions contained the same TERT expression lysate and the indicated telomerase RNA variant. One hundred nanograms of purified RNA was added to 3 μl of TERT expression lysate, incubated for 30 min at 30°C, then assayed for telomerase activity with the primer (TG)8TTG. RNAs are in ∼40-fold molar excess of TERT. Products corresponding to primer +0, +1, and +6 nucleotides are indicated.
Phylogenetically conserved telomerase RNA sequences are important for activity
We next examined the requirement for phylogenetically conserved sequences of the telomerase RNA (red in Fig. 1; Table 1). T. thermophila telomerase RNA residues CA15–16, UCU55–57, and C62 are conserved evolutionarily among the 12 characterized Tetrahymena species telomerase RNAs but not among all ciliates (Romero and Blackburn 1991; Lingner et al. 1994; McCormickGraham and Romero 1995, 1996). Also, CA15–16 and C62 are accessible to chemical modification in the deproteinized telomerase RNA but not in the endogenous RNP(s), suggesting that they are protected by an RNP-specific RNA folding or RNA–protein interaction (Zaug and Cech 1995). In addition, T. thermophila telomerase RNA residues GUCA37–40 are conserved evolutionarily among all ciliate telomerase RNAs (Romero and Blackburn 1991; Lingner et al. 1994; McCormick-Graham and Romero 1995, 1996). These residues are protected partially from chemical modification specifically in the RNP(s) (Zaug and Cech 1995).
Telomerase RNA with the substitution CA15–16GU has wild-type RNA levels of activity when reconstituted by the MNase protocol (Autexier and Greider 1998) as does a telomerase RNA with substitution C62G (data not shown). In contrast, both substitutions almost completely abolish telomerase activity with lysate-expressed TERT. This result is evident for reconstitution by RNA coexpression (Fig. 3, lanes 7,8) or by addition of purified RNA (Fig. 4, lanes 7,8). A slight activity is observed only with high concentration of CA15–16GU telomerase RNA (cf. Figs. 3 and 4). Telomerase RNA with the substitution UCU55–57AGA has moderately reduced activity and an altered pattern of product DNA synthesis relative to the wild-type RNA, with more product accumulation from synthesis at the template 3′ end (primer +1 or +2) and less accumulation from the synthesis of multiple repeats (Figs. 3 and 4, lane 9).
Telomerase RNA with the substitution UCA38–40AGU is inhibited relative to the wild-type RNA in lysate reconstitution by RNA coexpression or by addition of purified RNA (Figs. 3 and 4, lane 15). Activity is increased at high RNA concentration (cf. Figs. 3 and 4). Telomerase RNA with the same substitution assayed by MNase protocol reconstitution resulted in about eightfold inhibition and more strikingly an extension of the 5′ template boundary (Autexier and Greider 1995). In assays of lysate-expressed TERT with UCA38–40AGU telomerase RNA and all four dNTPs, we find no alteration of the 5′ template boundary (M. Miller, J. Liu, K. Collins, unpubl.). However, this substitution does have the unique effect of stimulating product accumulation at primer size (primer +0 > primer +1 or +6). This is likely because of enhancement of the telomerase nucleolytic cleavage activity (Collins and Greider 1993). We conclude that some phylogenetically conserved residues of telomerase RNA are important for lysate reconstitution of telomerase activity, and that telomerase RNA residues protected from chemical modification in the endogenous RNP are most critical.
Specific telomerase RNA residues are required for TERT binding
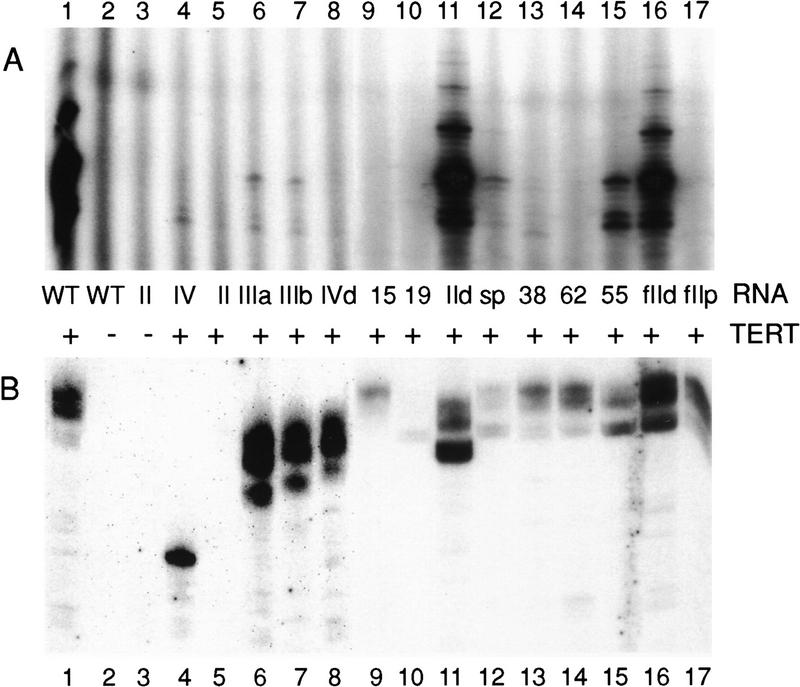
Previous work has not investigated which regions of the telomerase RNA are required directly for protein binding. To address this, we assayed the ability of the telomerase RNA variants to immunopurify with an amino-terminally hemagglutinin (HA)-tagged TERT. HA antibody was prebound to protein G–Sepharose, then the antibody resin was incubated with lysate-expressed TERT and purified telomerase RNA. In the assays shown in Figure 5, lysate-expressed, epitope-tagged TERT was incubated with an excess of each telomerase RNA variant then split to analyze telomerase activity and RNA–TERT interaction. The relative telomerase activities and product DNA patterns observed with the RNAs and epitope-tagged TERT (Fig. 5A) are indistinguishable from those observed with untagged protein (Fig. 4; data not shown). The antibody resin in the other half of each sample was washed extensively so that only telomerase RNA bound to the immobilized TERT was analyzed by Northern blot hybridization (Fig. 5B). Telomerase RNAs with a wild-type 3′ end transcribed from FokI-cut plasmid DNAs with purified T7 RNA polymerase appear as a sharp band and a slightly higher molecular mass RNA smear, likely derived from slippage of the polymerase on the homopolymeric sequence tract at the end of the template.
Figure 5.
Telomerase RNA binding to TERT. Lysate-expressed, epitope-tagged TERT [(+) TERT lanes] or an equal volume of mock lysate synthesis reaction lacking the plasmid encoding TERT [(−) TERT lanes] was mixed with 100 ng of the indicated telomerase RNA variant. Samples were incubated at 30°C for 30 min after which a fraction of the sample was removed for a telomerase activity assay with the primer (TG)8TTG (A). The remainder of the sample was mixed with HA antibody–protein G–Sepharose. Bound material was washed repeatedly and analyzed by Northern blot hybridization for telomerase RNA (B) and by SDS-PAGE and autoradiography to verify equivalent protein recovery (not shown). Telomerase RNAs with a wild-type 3′ end transcribed from FokI-cut plasmid DNAs with purified T7 RNA polymerase appear as a sharp band and a slightly higher molecular mass RNA smear, likely derived from slippage of the polymerase on the homopolymeric sequence tract at the end of the template. Note that the background of RNA binding to antibody resin in the absence of TERT is minimal (lanes 2,3). Parallel recovery of RNAs was verified by addition of an internal control RNA to samples before RNA extraction (not shown). Immunopurification supernatants were also analyzed by Northern blot hybridization to verify RNA stability during the antibody incubation (not shown).
In the presence of TERT, wild-type telomerase RNA is recovered with the immunopurified material (Fig. 5, lane 1). In contrast, in the presence of a mock lysate synthesis reaction not containing TERT, the antibody resin does not bind significant levels of wild-type or variant telomerase RNAs (lanes 2–3; data not shown). Of the telomerase RNA variants lacking individual elements of secondary structure, both of the pseudoknot region and both of the stem–loop IV region deletion RNAs are not significantly inhibited in binding to TERT (Fig. 5, lanes 4, 6–8; additional data not shown). In contrast, no TERT binding above background is evident for the stem–loop II deletion telomerase RNA (Fig. 5, lane 5). Thus, a dramatic decrease in TERT binding is likely to be responsible for the activity deficit of this telomerase RNA variant.
We similarly assayed TERT binding by the other telomerase RNA variants (Fig. 5). The substitution CA15–16GU inhibits both activity and TERT binding dramatically (lane 9). The substitution UCA38–40AGU inhibits telomerase activity strongly but inhibits TERT binding only moderately (lane 13). The substitution C62G inhibits activity completely but inhibits TERT binding only moderately (lane 14). The substitution UCU55–57AGA inhibits activity moderately and inhibits TERT binding also moderately (lane 15). Thus, telomerase RNAs altered in phylogenetically conserved sequences demonstrate a range of deficiencies: moderate activity inhibition with moderately reduced TERT binding (UCU55–57), strong activity inhibition with moderately reduced TERT binding (C62 and UCA38–40), and strong activity inhibition with strongly reduced TERT binding (CA15–16). These results suggest that CA15–16 is critical for TERT binding. Also, although UCA38–40AGU, UCU55–57AGA, and C62G RNAs demonstrate roughly comparable TERT binding, their activity deficits are not comparable even at the same RNA concentration used to measure binding (activity of UCU55–57AGA RNA > UCA38–40AGU, C62G RNAs). Therefore, the major impact of C62G and UCA38–40AGU substitutions on telomerase activity is not attributable to a decrease in TERT binding affinity.
Stem–loop II has sequence, structure, and positioning specificity
From the results described above, telomerase RNA binding to TERT was inhibited most severely by deletion of stem–loop II. To determine why this region of telomerase RNA appeared to be the most significant determinant of TERT binding, we assayed additional telomerase RNA variants with finer alterations of the stem–loop II sequence and structure. To investigate the relative significance of primary sequence and secondary structure, we flipped the template-proximal 3-bp stem sequences or the template-distal 3-bp stem and loop sequence (see Fig. 1; Table 1). The template-distal alteration has no discernable influence on activity or TERT binding (Figs. 3 and 4, lanes 10; Fig. 5, lane 16). In contrast, the template-proximal stem switch decreases activity and TERT binding (Figs. 3 and 4, lanes 11; Fig. 5, lane 17). Likely because of the TERT-binding inhibition, as observed for CA15–16GU and UCA38–40AGU RNAs, activity is increased at high RNA concentration (cf. Figs. 3 and 4). We also deleted the template-distal region of stem–loop II entirely, replacing it with a heterologous tetraloop. This substitution removed the mismatched A residues that were unchanged from wild-type in both previous flip RNAs (see Fig. 1; Table 1). The tetraloop substitution, like the distal stem–loop II flip, has activity and TERT binding indistinguishable from the wild-type RNA (Figs. 3 and 4, lanes 13; Fig. 5, lane 11). These results indicate that neither the sequence nor the structure of the template-distal stem–loop II region is important, whereas the sequence of the template-proximal stem is important for TERT binding and telomerase activity.
The most template-proximal base pair of stem–loop II includes the phylogenetically conserved residue G37. Although this guanosine is base-paired to close stem II in 11 of the 12 characterized Tetrahymena species telomerase RNAs and in all telomerase RNAs characterized from Glaucoma and Paramecium species, it is conserved in primary sequence but not base-paired in ciliate telomerase RNAs that lack stem–loop II. To determine whether base-pairing of G37 is required for activity with the T. thermophila RNA, we assayed the substitution C19A (see Fig. 1; Table 1). This RNA retains the conserved G37 but can not engage it in a base-pairing interaction. We find that the C19A substitution inhibits telomerase activity and TERT binding almost completely (Figs. 3 and 4, lanes 12; Fig. 5, lane 10). Weak activity is observed only at high RNA concentration (cf. Figs. 3 and 4).
The stem–loop II deletion and C19A telomerase RNAs are both strongly inhibited in TERT binding. Two features are shared by these RNAs which otherwise have nonoverlapping sequence alterations: (1) loss of C19–G37 base pairing and (2) the potential for altered positioning of sequences 5′ and 3′ of stem II. The template-proximal stem II flip RNA has a G19–C37 base pair instead of a C19–G37 base pair yet retains more ability to interact with TERT. Thus, it seemed possible that a stem II-dependent positioning of adjacent regions, maintained in the template-proximal stem II flip RNA, was important independent of the requirement for a C19–G37 base pair. To test this hypothesis, two uridines were inserted immediately 5′ of C19 to create the potential for altered positioning without disruption of the C19–G37 base pair (see Fig. 1; Table 1). This spacing-altered RNA is inhibited substantially in both telomerase activity and TERT binding (Figs. 3 and 4, lanes 14; Fig. 5, lane 12). However, activity with this RNA is greater than with the stem–loop II deletion or C19A RNAs, supporting the conclusion that the C19–G37 base pair contributes to telomerase RNA function directly in addition to maintaining a proper positioning of flanking sequences.
Lysate stimulates TERT–RNA interaction
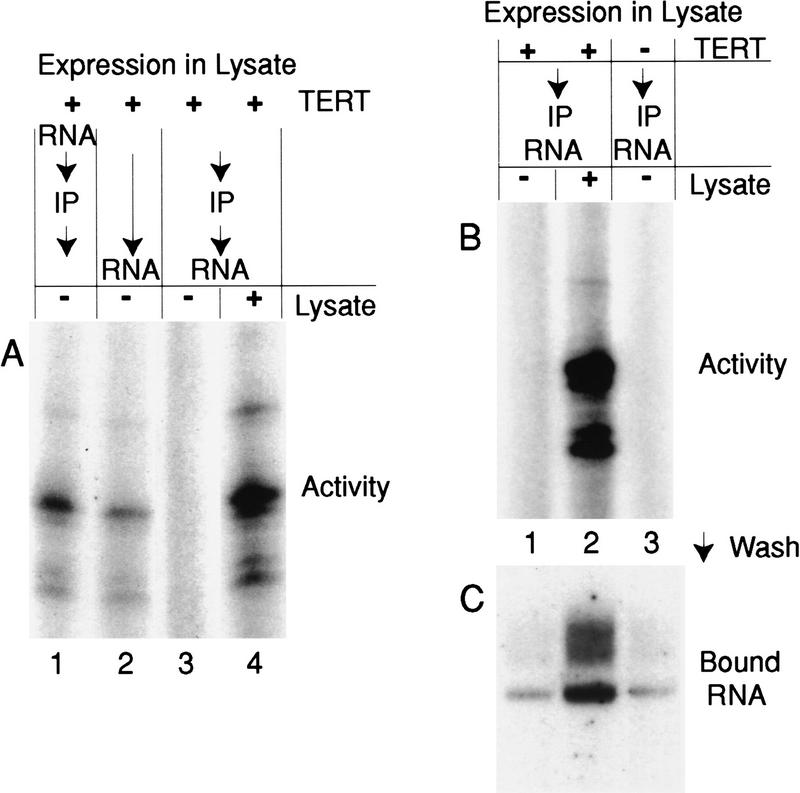
Because TERT expressed in lysate demonstrates a more specific and, based on the requirement for phylogenetically conserved sequences, an apparently more physiologically relevant association with telomerase RNA than can be reconstituted from purified endogenous telomerase proteins by the MNase protocol, we investigated the role of the lysate in our reconstitution assay. If TERT is incubated with telomerase RNA in lysate then immunopurified on HA antibody–protein G–Sepharose, the telomerase activity recovered by purification (Fig. 6A, lane 1) is close to the amount obtained from addition of telomerase RNA to the same volume of original expression lysate (lane 2). Immunopurification typically recovers 20%–25% of TERT (data not shown). The increase in specific activity with purification likely derives from a loss of misfolded or aggregated TERT unable to bind HA antibody. If TERT is immunopurified from its expression lysate before RNA addition, much less or no activity is observed, even when purified TERT is normalized to the level present in the original expression lysate (Fig. 6A, lane 3). However, readdition of fresh lysate to purified TERT and RNA (Fig. 6A, lane 4) but not addition of boiled lysate (data not shown) restores telomerase activity to near maximal specific activity. We conclude that although lysate is not required continuously for telomerase activity, it is required for at least one process other than protein synthesis.
Figure 6.
A role for lysate in recombinant telomerase RNP assembly. (A) Lysate-expressed, epitope-tagged TERT was left alone or immunopurified on HA antibody–protein G–Sepharose either after (lane 1) or before (lanes 3,4) addition of 5 μg total yeast RNA and 100 ng telomerase RNA and incubation at 30°C for 30 min. Telomerase RNA was also added to the original TERT-expression lysate and assembled at the same time as the purified TERT samples (lane 2). Two microliters of fresh lysate was added to the sample in lane 4 before incubation. Samples were assayed for telomerase activity with the primer (TG)8TTG. (B,C) Lysate-expressed, epitope-tagged TERT (lanes 1,2) and a mock synthesis reaction lacking the TERT expression plasmid (lane 3) were immunopurified on HA antibody–protein G–Sepharose. Samples were supplemented with 5 μg of total yeast RNA, 100 ng of telomerase RNA, and where indicated with 2 μl of fresh lysate before assembly at 30°C for 30 min. Half of each sample was analyzed for telomerase activity with the primer (TG)8TTG (B) and half was washed thoroughly and analyzed by Northern blot hybridization for telomerase RNA that remained associated with antibody-bound TERT (C).
To additionally investigate the role of lysate, we addressed whether lysate is required to assemble a telomerase RNA–TERT RNP. We immunopurified lysate-expressed TERT or a mock lysate synthesis reaction lacking TERT using HA antibody–protein G–Sepharose. Purified samples corresponding to equivalent lysate reaction volumes were supplemented with excess telomerase RNA and either fresh lysate or no lysate, then assayed for telomerase activity and TERT–RNA association. As described above, purified TERT and purified RNA with lysate added back can catalyze strong telomerase activity (Fig. 6B, lane 2) whereas purified TERT and purified RNA without lysate cannot (lane 1). When TERT–RNA association is assayed with the same samples, telomerase RNA can stably bind to purified TERT with lysate added back (Fig. 6C, lane 2). The amount of telomerase RNA bound to purified TERT in the absence of lysate (Fig. 6C, lane 1) is similar to the background level of RNA bound nonspecifically to antibody resin in the absence of TERT (lane 3). The presence of lysate actually reduces background binding of telomerase RNA to antibody resin, further demonstrating that the RNA associated with TERT in the presence of lysate is specifically bound (data not shown). We conclude that lysate is required for the assembly of a stable, recombinant telomerase RNP.
Discussion
Telomerase RNA structure and function
Telomerase is unique as a reverse transcriptase in its use of a region of the enzyme RNA component as the source of template information. Because of this specialization, the telomerase RNA must contain at least two functionally important motifs: a binding site or sites for telomerase proteins and a template sequence to direct dNTP polymerization. The template itself is an unlikely candidate for a high-affinity protein binding site because it is accessible to chemical modification in the RNP (Zaug and Cech 1995) and can be altered without preventing RNP assembly in vitro or in vivo (for review, see Collins 1999). Here, we have shown that telomerase RNA binding to TERT requires a region of the RNA 5′ of the template, spanning from CA15–16 to the C19–G37 base pair. Within this small region, both RNA sequence and sequence positioning are important. Although some sequence alterations outside this region compromise telomerase RNA–TERT interaction, none inhibit interaction to the same extent as the alterations 5′ of the template (summarized in Table 1). Large regions of telomerase RNA including stem–loop IV, either stem–loop pairing of the pseudoknot element III, and the distal end of stem–loop II can be deleted without significant reduction in RNA–protein interaction.
Several variant telomerase RNAs demonstrate a reduced or altered telomerase activity without corresponding inhibition of TERT binding affinity. The RNA sequence alterations most deleterious for activity independent of an effect on TERT binding are C62G, deletion of stem–loop IV, and UCA38–40AGU (summarized in Table 1). The UCA38–40AGU telomerase RNA has somewhat reduced TERT binding ability but a much greater deficit in telomerase activity. Previous work expressing mutant telomerase RNA genes in vivo has also suggested that nontemplate regions of telomerase RNA are important for activity. The Glaucoma chattoni telomerase RNA expressed in T. thermophila assembles into a telomerase RNP with apparently low and aberrant activity despite an identical template region (Bhattacharyya and Blackburn 1997). Also, a stem–loop region of the Kluyveromyces lactis telomerase RNA distant from the template is required for catalytic activity but not normal RNP mobility on a nondenaturing gel (Roy et al. 1998) and the template-distal 3′ end of the human telomerase RNA includes sequences important for activity but not RNP stability (Mitchell et al. 1999). We conclude that the telomerase RNA must have a minimum of three functional motifs: the template, the TERT binding site, and additional sequences required to form a catalytically competent polymerase active site. The RNA sequences required for activity independent of templating and TERT binding could serve a variety of roles, including involvement in substrate binding, active site conformation, or catalysis. It will be interesting to distinguish between these possibilities in future studies.
The requirement for lysate
The reconstitution of telomerase RNA and TERT in lysate has a high RNA sequence specificity not observed for telomerase RNA reconstitution with endogenous telomerase proteins by the MNase protocol. This specificity difference and the lysate reconstitution requirement for phylogenetically conserved and chemical modification-protected residues of the telomerase RNA suggest that the interaction of telomerase RNA and TERT occurs in vivo as in lysate. The fact that a lysate component is required to establish the stable interaction of telomerase RNA and TERT may explain why our results were not discovered in earlier studies. In the MNase protocol, telomerase RNA is added back to partially purified, nuclease-treated, endogenous telomerase proteins. If a physiologically appropriate interaction of telomerase RNA and TERT does not occur spontaneously under standard in vitro conditions, MNase protocol reconstitution would not create a physiologically appropriate telomerase RNP. In lysate reconstitution, the lysate facilitates a physiological telomerase RNA–TERT interaction.
In our studies, we found that purified TERT combined with purified telomerase RNA does not form a stable telomerase RNP unless lysate is added back. This differs from MNase protocol reconstitution, in which telomerase RNA added back to partially purified endogenous telomerase proteins does form an RNP that is functional in vitro. It may be that telomerase RNA is recruited to an MNased p80/p95/TERT complex by interaction with p80/p95, which are absent in the lysate system. Indeed, the RNA sequence specificity of telomerase activity reconstitution by the MNase protocol resembles the RNA-binding specificity of the T. thermophila p80/p95 complex (Gandhi and Collins 1998). Thus, these proteins rather than TERT may be responsible for the RNA sequence specificity of MNase protocol reconstitution. This hypothesis would explain why telomerase RNA sequence substitutions that inhibit TERT binding and telomerase activity dramatically by lysate reconstitution have little or no effect on telomerase activity by MNase protocol reconstitution (for example, the CA15–16GU substitution). It would also explain how the UCA38–40AGU telomerase RNA could alter template boundaries differently in telomerase RNPs created by the two reconstitution protocols, because the reconstituted template would have different interactions with TERT. Because neither p80 nor p95 is required for telomerase RNP assembly or telomerase activity in vivo (M. Miller and K. Collins, unpubl.), a p80/p95-directed telomerase RNP assembly by MNase protocol reconstitution in vitro is unlikely to reflect the endogenous requirements for telomerase RNA function. Also, because p80 and p95 do not interact directly with TERT in lysate (data not shown), we are unable to test the influence of these proteins on telomerase RNA–TERT interaction.
In yeast and mammals, the biogenesis of telomerase RNPs occurs as a multistep assembly process with concerted RNA processing (Chapon et al. 1997; Mitchell et al. 1999). In contrast, there is no evidence suggesting the presence of a specific telomerase RNP assembly pathway in ciliates, which can have telomerase RNPs of relatively smaller size (Collins et al. 1995; Greene and Shippen 1998; Lingner and Cech 1996). It will be interesting to investigate whether the lysate component that is required for telomerase RNA–TERT assembly in vitro, or other factors performing similar biochemical roles, are required for ciliate telomerase RNP assembly in vivo. The lysate can be fractionated to substantially enrich for a catalytic telomerase RNP assembly activity (C. Lai and K. Collins, unpubl.), suggesting that direct identification of the lysate component will be possible.
Materials and methods
Telomerase RNAs and proteins
Constructs for expression of telomerase RNA derivatives were created from pT7159, in which the wild-type RNA is fused to a 5′ leader of three extra guanosines to enhance transcription efficiency (Autexier and Greider 1994). Mutagenesis for all constructs except the stem–loop IV deletion was done by standard oligonucleotide-directed protocols, using single-stranded or double-stranded DNA templates (Ausubel et al. 1996; Gandhi and Collins 1998). The construct encoding the stem–loop IV deletion RNA is a cloned PCR product in which the reverse primer included an EcoRI site overlapping endogenous RNA positions 107–108. No telomerase RNA sequence 3′ of position 108 is present in the plasmid. All plasmids were sequenced after mutagenesis. The sequence changes for all constructs are listed in Table 1.
For RNA expression, plasmids for all constructs except the stem–loop IV deletion were digested with the multicutter FokI to generate a run-off transcript with an RNA 3′ end identical to the endogenous T. thermophila telomerase RNA. The stem–loop IV deletion construct plasmid was digested with two single-cutters EcoRI and PstI, and the ∼160-bp fragment containing the coding information was gel-purified prior to transcription. T7 transcription of this plasmid produces a telomerase RNA truncated after position 108 with an additional 3′ AUU. Restriction enzyme-digested telomerase RNA plasmids were purified by extraction and precipitation. For production of purified telomerase RNA added back to lysate, RNAs were transcribed with T7 RNA polymerase and purified by DNase digestion, extraction, and precipitation as described previously (Autexier and Greider 1994). RNAs were examined by gel electrophoresis to verify their length and purity. Competitor RNAs were purchased and prepared, or in the case of the 24-nucleotide telomerase RNA chemically synthesized, as described previously (Collins and Gandhi 1998).
An epitope-tagged TERT expression construct was created from the TERT-coding region reconstructed as a synthetic gene and expressed in pCITE4a (Collins and Gandhi 1998). Linkers encoding HA and 6-histidine (HIS) tags were inserted at the start codon of p133CITE to create pHH133CITE, in which the HA tag follows the HIS tag. CITE plasmids were included in coupled transcription/translation reactions at a final concentration of ∼20 ng/μl, with or without ∼15 ng/μl telomerase RNA plasmid. Expression in lysate followed the manufacturer’s protocol (Promega TNT) with [35S]methionine used to radiolabel the synthesized proteins.
Proteins were analyzed by SDS–PAGE after addition of sample buffer and heat denaturation. RNAs were analyzed after extraction and precipitation, using Northern blot hybridization with a 5′ end-labeled oligonucleotide complementary to the telomerase RNA template (oligo 3; Greider and Blackburn 1989). Any blot including UCU55–57AGA RNA was hybridized at relaxed stringency.
Recombinant telomerase activity
Activity assays for recombinant TERT were modified from conditions used for the endogenous enzyme because of different primer and nucleotide requirements (Collins and Gandhi 1998). In the assays here, all samples were brought to 20 μl final assay volume with a maximum of 5 μl of the coupled transcription/translation reaction or fresh lysate. In assays with competitor RNAs, competitors were added prior to addition of telomerase RNA. Assay samples were adjusted with 10× assay buffer to final concentrations of 50 mm Tris acetate, 10 mm spermidine, 5 mm β-mercaptoethanol, and 2 mm MgCl2 (pH 8.0). Radiolabeled [α-32P]dGTP was supplemented with unlabeled dGTP to a final concentration of 3 μm to promote repeat addition processivity. Unlabeled TTP was present at 500 μm and primer (TG)8TTG was present at 1.0 μm. Reactions were allowed to proceed at 30°C for 1 hr, at which time product DNA was extracted, precipitated, and analyzed by denaturing acrylamide gel electrophoresis in 0.6× TBE.
Immunopurification
Protein G–Sepharose (Pharmacia) was preblocked and prebound to HA antibody in binding buffer (20 mm Tris HCl at pH 8.0, 1 mm MgCl2, 10% glycerol, 0.1 m NaCl) with 10 μg/ml BSA, 20 μg/ml tRNA, and 160 ng antibody per microliter of packed resin. For experiments in Figure 6, total yeast RNA was also included in the blocking solution. Antibody-bound protein G–Sepharose was washed with binding buffer after blocking. RNA-containing samples for immunopurification were diluted to 100 μl with binding buffer made to 10 μg/ml BSA and 20 μg/ml tRNA then mixed at 4°C overnight. Bound material was washed thoroughly in binding buffer and resuspended to a 50% slurry. For immunopurification of TERT alone, expression lysate was diluted to 500 μl in binding buffer with blocking agents, bound overnight to antibody resin, washed, and resuspended to the volume of slurry equal to the initial volume of lysate before purification.
Acknowledgments
We thank James Mitchell, Leena Gandhi, and Jesse Liu for telomerase RNA expression constructs; James Mitchell and Leena Gandhi for MNase protocol reconstitution data; and Carla Schultz for the epitope-tagged TERT expression construct. We also thank Donald Rio and members of the Collins laboratory for helpful comments on the manuscript. This work was funded by grants from the National Institutes of Health (GM54198) and the Burroughs Wellcome Fund (New Investigator Award in the Pharmacological Sciences).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kcollins@socrates.berkeley.edu; FAX (510) 642-6062.
References
- Ausubel F, Brent R, Kingston R, Moore D, Seidman F, Smith J, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley; 1996. [Google Scholar]
- Autexier C, Greider CW. Functional reconstitution of wild-type and mutant Tetrahymena telomerase. Genes & Dev. 1994;8:563–575. doi: 10.1101/gad.8.5.563. [DOI] [PubMed] [Google Scholar]
- ————— Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes & Dev. 1995;9:2227–2239. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- ————— Mutational analysis of the Tetrahymena telomerase RNA: Identification of residues affecting telomerase activity in vitro. Nucleic Acids Res. 1998;26:787–795. doi: 10.1093/nar/26.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie TL, Zhou W, Robinson MO, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya A, Blackburn EH. A functional telomerase RNA swap in vivo reveals the importance of nontemplate RNA domains. Proc Natl Acad Sci. 1997;94:2823–2827. doi: 10.1073/pnas.94.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C-P, Morin G, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Chapon C, Cech T, Zaug A. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997;3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- Collins K. Ciliate telomerase biochemistry. Annu Rev Biochem. 1999;68:187–218. doi: 10.1146/annurev.biochem.68.1.187. [DOI] [PubMed] [Google Scholar]
- Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K, Greider CW. Nucleolytic cleavage and non-processive elongation catalyzed by Tetrahymena telomerase. Genes & Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- Collins K, Kobayashi R, Greider CW. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- Gandhi L, Collins K. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes & Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene EC, Shippen DE. Developmentally programmed assembly of higher order telomerase complexes with distinct biochemical and structural properties. Genes & Dev. 1998;12:2921–2931. doi: 10.1101/gad.12.18.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;66:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Harley CB, Villeponteau B. Telomeres and telomerase in aging and cancer. Curr Opin Genet Dev. 1995;5:249–255. doi: 10.1016/0959-437x(95)80016-6. [DOI] [PubMed] [Google Scholar]
- Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Program E, Bass MB, Arruda I, Robinson MO. A mammalian telomerase-associated protein. Science. 1997a;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- Harrington L, Zhou W, McPhail T, Oulton R, Yeung DSK, Mar V, Bass MB, Robinson MO. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes & Dev. 1997b;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc Natl Acad Sci. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes & Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A single telomerase RNA is sufficient for the synthesis of variable telomeric DNA repeats in ciliates of the genus Paramecium. Mol Cell Biol. 1996;16:1871–1879. doi: 10.1128/mcb.16.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melek M, Shippen DE. Chromosome healing: Spontaneous and programmed de novo telomere formation by telomerase. BioEssays. 1996;18:301–308. doi: 10.1002/bies.950180408. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Cech TR. Reversing time: Origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: A gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Lundblad V. The telomerase reverse transcriptase: Components and regulation. Genes & Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- Roy J, Fulton TB, Blackburn EH. Specific telomerase RNA residues distant from the template are essential for telomerase RNA function. Genes & Dev. 1998;12:3286–3300. doi: 10.1101/gad.12.20.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam E, van Belkum A, Pleij K. A conserved psuedoknot in telomerase RNA. Nucleic Acids Res. 1991;19:6951. doi: 10.1093/nar/19.24.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrich SL, Pruzan R, Ma L, Ouellette M, Tesmer VM, Holt SE, Bodnar AG, Lichsteiner S, Kim NW, Trager JB, Taylor RD, Carlos R, Andrews WH, Wright WE, Shay JW, Harley CB, Morin GB. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- Yu G, Bradley JD, Attardi LD, Blackburn EH. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Cech TR. Analysis of the structure of Tetrahymena nuclear RNAs in vivo: Telomerase RNA, the self-splicing rRNA intron, and U2 snRNA. RNA. 1995;1:363–374. [PMC free article] [PubMed] [Google Scholar]