Abstract
Nuclear factor of activated T cells (NFATs) are crucial transcription factors that tightly control proinflammatory cytokine expression for adaptive immunity in T and B lymphocytes. However, little is known about the role of NFATs for innate immunity in macrophages. In this study, we report that NFAT is required for Toll-like receptor (TLR)-initiated innate immune responses in bone marrow-derived macrophages (BMMs). All TLR ligand stimulation including LPS, a TLR4 ligand, and Pam3CSK4, a TLR1/2 ligand, induced expression of TNF which was inhibited by VIVIT, an NFAT-specific inhibitor peptide. BMMs from NFATc4 knock-out mouse expressed less TNF than wild type. Despite apparent association between NFAT and TNF, LPS did not directly activate NFAT based on NFAT-luciferase reporter assay, whereas NF-κB was inducibly activated by LPS. Instead, macrophage exhibited constitutive NFAT activity which was not increased by LPS and was decreased by VIVIT. Immunocytochemical examination of NFATc1-4 of BMMs exhibited nuclear localization of NFATc3/c4 regardless of LPS stimulation. LPS stimulation did not cause nuclear translocation of NFATc1/c2. Treatment with VIVIT resulted in nuclear export of NFATc3/c4 and inhibited TLR-activated TNF expression, suggesting that nuclear residence of NFATc is required for TLR-related innate immune response. Chromatin immunoprecipitation (ChIP) assay using anti-RNA Polymerase II (PolII) antibody suggested that VIVIT decreased PolII binding to TNF gene locus, consistent with VIVIT inhibition of LPS-induced TNF mRNA expression. This study identifies a novel paradigm of innate immune regulation rendered by NFAT which is a well known family of adaptive immune regulatory proteins.
Keywords: Innate immunity, NFAT, Pathogen-associated molecular pattern, TLR, TNF, Signal transduction
1. Introduction
Innate immunity in macrophages is a powerful mechanism to recognize infection in a microorganism. During infection, macrophages secrete TNF and other pro-inflammatory cytokines. The fast acting and nonspecific nature of innate immunity is juxtaposed by the slow yet specific action of adaptive immunity. This non-specific detection is achieved by cellular membrane receptors on macrophages, for example, toll-like receptors (TLRs). TLRs detect common microorganism components, which are now collectively called pathogen-associated molecular patterns (PAMPs). According to current research at least 9 TLRs have been identified in humans and mice [1, 2] and each receptor is known to have specific target molecules. For example, TLR4 homodimer is the only receptor for E. coli-derived lipopolysaccharide (LPS) [3] and the TLR1/2 heterodimer detects triacylated lipoprotein, such as Pam3CSK4 [4-6]. Downstream events from TLRs have been well studied and they are known to share same pathway [1, 2]. In innate macrophage immunity, agonists bind to specific TLR resulting in activation of transcription factors, such as AP-1 and NF-kB, which in turn activate cytokine genes.
Nuclear factor of activated T cells (NFAT) is a transcription factor originally found in activated T lymphocytes [7]. NFAT function in T and B cells has been well studied in the context of the adaptive immune system [8-10]. The NFAT gene family consists of 5 members which include NFATc1, NFATc2, NFATc3, NFATc4, and NFAT5. NFATc1-c4 are under the control of calcium signaling [8-12], while NFAT5 is activated by osmotic stress [13]. In T or B cells, NFATc1-c4 are highly phosphorylated in the resting state i.e. inactivated. On the engagement of T or B cell receptor (TCR or BCR), the cytoplasmic calcium concentration is increased which causes calcineurin, a calcium-dependent phosphatase, to dephosphorylate cytoplasmic NFAT resulting in its activation. This activation mechanism is notable in that it is divergent to the common understanding of transcription factor activation. Dephosphorylated NFAT translocates to the nucleus and induces the transcription of target genes [8-10]. The activated NFAT is negatively regulated by re-phosphorylation activity of various nuclear NFAT kinases [14-20]. Re-phosphorylated NFAT is then quickly exported to the cytoplasm [10, 21]. Nuclear NFAT kinases not only complete a regulatory circuit but also serve to desensitize NFAT signaling to transient Ca2+ spikes such as those found during muscle contraction. As a result, NFAT activation requires prolonged calcium influx [22]. NFAT plays a central role in TNF expression resulting from B and T cell receptor engagement [22, 23]. Despite common expression in immune cells and being implicated in various signaling pathways and processes, there has been little to no reports on NFAT’s role in innate immunity [1, 2, 10].
Our study demonstrates that NFAT mediates TLR-initiated innate immune response by macrophages. We show that NFAT-specific inhibitor peptide, VIVIT, inhibited the expression of cytokines induced by all TLR ligands, including LPS and lipoprotein, suggesting that NFAT is commonly involved in the innate immune response through TLRs in macrophages. We also show that NFAT exhibits a basal activity independent of LPS stimulation in macrophage by subtype NFATc4 and perhaps by NFATc3 as well.
2. Materials and methods
2.1. Macrophage isolation from mouse bone marrow and culture
The TLR4−/− mouse strain, C57BL/10ScNJ, and the isogenic wild-type strain, C57BL/10ScSnJ [3], as well as the TLR2−/− strain B6.129-Tlr2tm1Kir/J and its corresponding isogenic wild-type strain, C57BL/6J [24], were purchased from The Jackson Laboratory. The NFATc4−/− strain was also purchased from The Jackson Laboratory [25]. This strain was maintained on a mixed C57BL/6 and 129S7 genetic background and we prepared F1 hybrid of C57BL/6J and 129X1/SvJ, both of which are also purchased from The Jackson Laboratory, as a control. Bone marrow-derived macrophages (BMMs) were prepared as described [26]. Briefly, the non-adherent fraction of mouse bone marrow cells was cultured in Minimum Essential Medium (MEM) α (Invitrogen) supplemented with 10 % fetal bovine serum (FBS; Gemini Bio), 1% antibiotic/antimycotic (Gemini Bio) and 10 ng/ml of recombinant mouse M-CSF (R&D). The resulting adherent cells were taken to represent purified BMMs. Purified primary BMMs were then cultured in serum-free MEMα supplemented with M-CSF, with or without 11R-VIVIT (EMD Biosciences), a cell-permeable peptide inhibitor of NFAT, for 1 hour [27, 28]. BMMs were then treated for 3 hours with 0.1 μg/ml each of ultrapure lipopolysaccharide (LPS; InvivoGen) or a synthetic bacterial lipoprotein, Pam3CSK4 (Pam; InvivoGen) [4]. Other TLR ligands are purchased as parts of Mouse TLR1-9 Agonist kit (InvivoGen). Animal experiments were approved by the Institutional Animal Care and Use Committee of Columbia University (Protocol No. AC-AAAA8363).
2.2 Reverse Transcription (RT) real-time Polymerase Chain Reaction (PCR)
Total RNA was isolated from cells using an RNeasy Mini Kit (Qiagen). Single stranded cDNA was synthesized from total RNA with the SuperScript III system (Invitrogen). RT real-time PCR for each target was performed with LightCycler FastStart DNA MasterPLUS SYBR Green I (Roche) using the Realplex system (Eppendorf). Primer sets used are listed in “Supplemental Experimental Procedures”. The thermal cycling conditions consisted of preheating (10 min at 95°C) and 40 cycles of denaturation (15 sec at 95°C), annealing (15 sec at 60°C) and elongation (20 sec at 72°C). Gene expression levels were normalized against the GAPDH level.
2.3. Reporter Assay
Firefly luciferase reporter plasmids pNFAT-GL4 and pNFκB-GL4 were constructed by inserting a PCR-amplified promoter segment of pNFAT-TA-Luc or pNFκB-TA-Luc (Clontech) into pGL4.10 (Promega), respectively. VIVIT expression plasmid pGFP-VIVIT and control plasmid pEGFP-N1 were described previously [27]. Macrophage RAW264.7 cells (ATCC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% FBS and 1% antibiotic/antimycotic. Plasmids were introduced into RAW264.7 by FuGENE HD (Roche). Twenty four hours after transfection, cells were stimulated with LPS or calcium ionophore (“CaIO”) A23187 (EMD Biosciences) in serum-free media. After 6 hours of stimulation, luciferase activity was measured by the Dual-Glo Luciferase Assay System (Promega).
2.4. Calcium Imaging
BMMs were cultured in a 60 mm culture dish with serum-free, phenol red-free MEMα supplemented with 10 ng/ml M-CSF and 2 μg/ml fluo-4 AM (Invitrogen) for one hour. The cells were then washed and incubated for a further 30 minutes in fluo-4 free medium. Equal volume of media with 0.2 μg/ml LPS, 0.2μg/ml Pam or 10 μM A23187 was added before observing calcium-induced fluorescence with a Zeiss Axiovert 200 microscope (Carl Zeiss).
2.5. Immunocytochemistry
BMMs were cultured in 4-chamber slide glasses (BD) in serum-free MEMα and M-CSF for 1 hour, with or without 5 μM VIVIT. Cells were stimulated with 0.1 μg/ml LPS, 0.1 μg/ml Pam, or 5 μM CaIO for 30 minutes, fixed in phosphate-buffered 4 % paraformaldehyde, and then stained with one of the following four primary antibodies: mouse anti-NFATc1 (Abcam), mouse anti-NFATc2 (Abcam), rabbit anti-NFATc3 (Cell Signaling) or rabbit anti-NFATc4 (Santa Cruz). Either Alexa Fluor 488 rabbit anti-mouse IgG or Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) were used as secondary antibodies. Confocal fluorescent images were obtained with LSM510 upright confocal microscope (Carl Zeiss).
2.6. Chromatin Immunoprecipitation (ChIP) Assay
Primary BMMs were cultured in 15 cm dishes. Soluble chromatin was extracted following the manufacturer’s instructions (Upstate Biotechnologies) with modification in nuclear extraction. The cells were fixed with PBS with 1% formaldehyde, scraped and resuspended in buffer A (10 mM HEPES (pH 7.4), 10 mM KCl, 0.1 mM EDTA, 0.1 % Nonidet P-40 and Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific)) to burst cell membrane. The suspension was centrifuged for 10 min at 13,000 rpm at 4°C to spin down the nuclei. Isolated nuclei were resuspended with 1% SDS lysis buffer including protease inhibitor, and then sonicated 5 times each of 20 second at 100% duty cycle on a Bransson Sonifier 250 (Bransson). The samples were centrifuged for 10 min at 13,000 rpm at 4°C. 200μl of the sonicated cell supernatant was transferred to a new 2ml tube. The supernatants were precleared with salmon sperm DNA/Protein A agarose-50% slurry. The precleared 2mL of supernatant solution was incubated with RNA polymerase II antibody (Abcam) overnight at 4°C with constant agitation. Antibody-bound complexes were treated with salmon sperm DNA/Protein A agarose-50% slurry for 1 h at 4°C with rotation. The protein-DNA complex was eluted with buffer containing 1% SDS and 0.1M NaHCO3. Protein-DNA crosslinks were reversed and DNA was recovered by phenol/chloroform extraction, precipitated by ethanol, and resuspended with 30 μl Tris-EDTA (TE) buffer. 2 μl of recovered DNA was used as a template for PCR with TNF gene specific primers designed for the region (−224 to −30) of TNF promoter or coding and downstream regions. The primer sequences are described in “Supplemental Experimental Procedures”. The thermal cycling conditions consisted of preheating (5 min at 98°C) and 34 cycles of denaturation (20 sec at 98°C), annealing (20 sec at 60°C) and elongation (30 sec at 72°C). Input DNA refers to sonicated total DNA prior to immunoprecipitation with PolII antibodies.
2.7. Statistics
Each treatment group was done in triplicate per experiment and reported as mean ± standard deviation. Statistical analysis was performed by Student’s t-test to determine significance among treatment groups. Statistical significance was established at p ≥ 0.05.
3. Results
3.1. Proinflammatory cytokine expression by BMMs in innate immune response requires NFAT activity
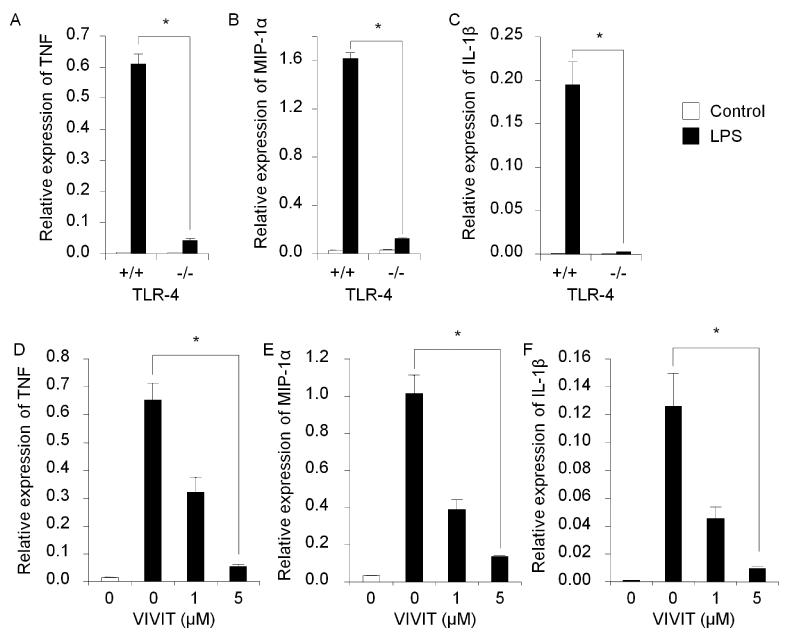
The importance of NFAT for cytokine expression has been reported in adaptive immunity by T and B lymphocytes [22, 23]. However, no similar function of NFAT has been reported in innate immunity by macrophages. To assess this, we interrupted the NFAT activity during the induction of innate immune response with E. coli LPS, which is a representative PAMP, i.e. a common microorganism component, and is widely used for model systems to study innate immunity. In this study we used specially prepared ultrapure E. coli LPS (see materials and methods) because we realized that generally available E. coli LPSs are contaminated with other bacterial components. In earlier tests we observed that some regular LPSs induced cytokine expression in TLR4−/− BMMs (data not shown) although TLR4 is only the receptor for E. coli LPS. We treated wild-type BMMs with ultrapure LPS and found expression of the proinflammatory cytokines TNF, MIP-1α and IL-1β, was induced as measured by RT real-time PCR (Fig. 1 A-C). This induction was almost completely abolished in TLR4−/− cells. These results indicate that cytokine production by BMMs is induced through TLR4 and that ultrapure E. coli LPS is a suitable inducer for innate immune response in our system.
Fig. 1.
LPS-induced cytokine expression in primary BMMs. RT real-time PCR showed relative mRNA expression levels for TNF (A), MIP-1α (B) and IL-1β (C). LPS induced cytokine expression in TLR4+/+ cells while demonstrating no cytokine induction in TLR4−/−. NFAT-specific inhibitor, VIVIT, inhibits gene expression of TNF (A), MIP-1α (B) and IL-1β (C) in BMMs stimulated with LPS. The relative mRNA expression levels were measured by RT real-time PCR with normalization to GAPDH levels. Symbol (*) denotes significant difference; p<0.05.
To interrupt the NFAT activity, we treated BMMs with VIVIT, which is an NFAT-specific peptide inhibitor and known to have excellent specificity [27-30]. We found that VIVIT blocked LPS-induced expression of TNF, MIP-1α and IL-1β in a dose dependent manner and near-complete inhibition was attained at a VIVIT concentration of 5 μM (Fig. 1 D-F). This suggests that NFAT is necessary for LPS-induced proinflammatory cytokine expression.
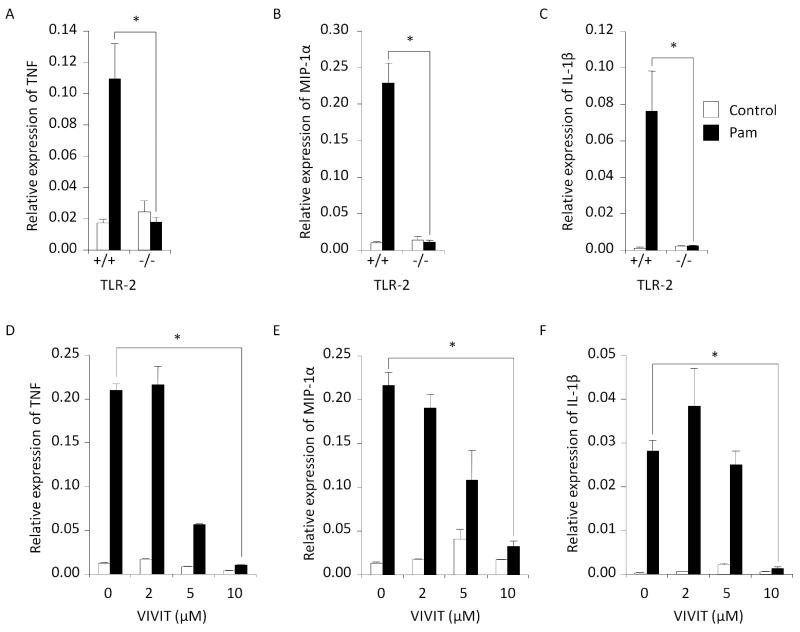
Macrophages express a wide variety of TLRs, each with their own ligands. However, many downstream mediators such as MyD88 are common to multiple TLR signaling pathways [1, 2]. We explored the possibility that NFAT is involved in proinflammatory cytokine expression through TLRs other than TLR4. BMMs were treated with a lipoprotein, Pam3CSK4 (Pam), which is a TLR1/2 heterodimer ligand [4]. We found that Pam treatment of wild-type BMMs induced expression of the pro-inflammatory cytokines TNF, MIP-1α and IL-1β, as measured by RT real-time PCR (Fig. 2 A-C); however, this induction was not observed in TLR2−/− BMMs. These data indicate Pam is functional. We then treated wild-type BMMs concurrently with Pam and VIVIT. It was found that VIVIT suppressed Pam-induced expression of TNF, MIP-1α and IL-1β with maximum blocking efficiency achieved at a VIVIT concentration of 10uM (Fig. 2 D-F). This result, which parallels our earlier result for LPS (Fig. 1 D-F), suggests that NFAT activity is necessary for Pam-induced pro-inflammatory cytokine expression through TLR2, just as well as LPS-induced expression through TLR4.
Fig. 2.
Cytokine gene expression by BMMs stimulated with synthesized lipoprotein, Pam3CSK4 (“Pam”). Pam induced TNF (A), MIP-1α (B) and IL-1β (C) only in TLR2-expressing cells. VIVIT inhibited expression of these cytokines (D-F). The relative mRNA expression levels were measured by RT real-time PCR with normalization to GAPDH levels. Symbol (*) denotes significant difference; p<0.05.
These findings imply that all the TLR receptor downstream may commonly require NFAT activity to lead pro-inflammatory cytokine expression. To examine that, we tested VIVIT effect in proinflammatory cytokine expression by BMMs under the treatment of ligands for all of the well established TLRs. As same as the cases of TLR4 and TLR1/2 ligands, all ligand effects were inhibited by VIVIT (Fig. 3). This strongly suggests that innate immune response by macrophage against all the TLR requires NFAT activity in common.
Fig. 3.
TNF gene expression in BMMs stimulated with various TLR agonists measured by RT real-time PCR. All agonists induced TNF gene expression and 5 μM VIVIT inhibited it in all cases. The relative mRNA expression levels were normalized to GAPDH levels. Symbol (*) denotes significant difference; p<0.05.
3.2. NFAT is constitutively active while NF-κB is inducibly activated by LPS stimulation
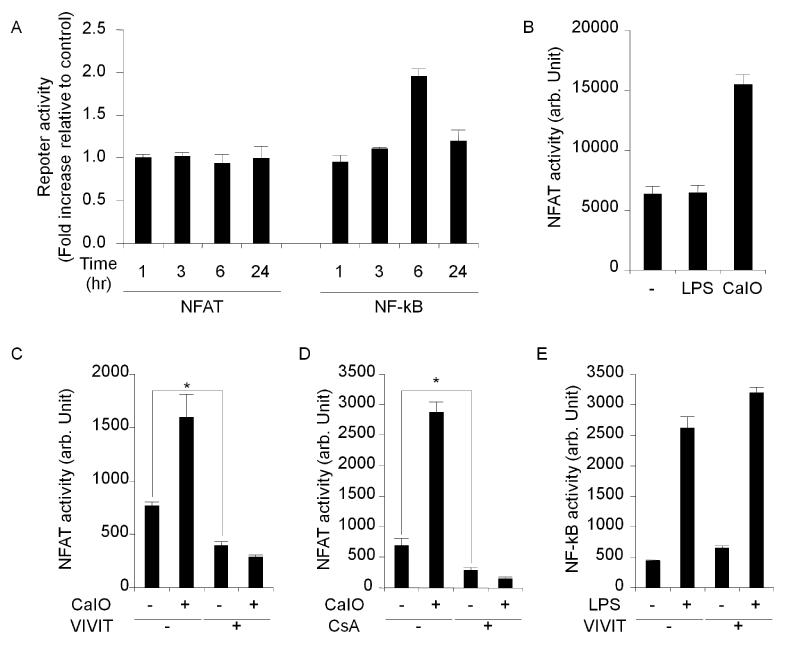
Next, we determined NFAT activity after stimulation by luciferase reporter assay. Due to technical difficulties with transfection, the luciferase assay of BMM cells could not be conducted. Instead, RAW264.7 cells, a mouse macrophage line, were used. We transfected RAW264.7 cells with firefly-luciferase reporter plasmids for either NFAT or NF-κB (pNFAT-GL4 and pNFκB-GL4 plasmids, respectively). NFAT and NF-κB activities were measured at various time points following LPS treatment (Fig. 4 A). As expected, a significant increase in NF-κB activity was observed 6 hours after LPS treatment. However, no changes in NFAT activity were observed. When calcium influx to cytoplasm was forcefully induced by a calcium ionophore A23187, NFAT activity was increased, thus indicating that the reporter system was functional (Fig. 4 B). Therefore, although NFAT is required to transduce the pro-inflammatory LPS signal, NFAT activity was not upregulated by LPS. Next, we investigated the effect of VIVIT on NFAT activity. We co-transfected RAW264.7 cells with both the reporter plasmid pNFAT-GL4 and either the VIVIT peptide expression plasmid pGFP-VIVIT or a vector control pEGFP-N1. We found that NFAT activity in cells co-transfected with pGFP-VIVIT was significantly lower than the control (Fig. 4 C). Thus, it revealed that RAW264.7 cells have a basal NFAT activity that is inhibited by VIVIT and it is not increased after LPS stimulation. VIVIT was found to strongly suppress NFAT activity, even in the presence of calcium ionophore (Fig. 4 C). In addition to VIVIT, which is a selective peptide inhibitor of NFAT, we also used a chemical calcineurin/NFAT-inhibitor, cyclosporine A [27]. We treated pNFAT-GL4 transfectants with calcium ionophore and cyclosporine A. As with VIVIT, cyclosporine A treatment strongly depressed NFAT activity, even in the presence of calcium ionophore (Fig. 4 D). Parallel control experiments were performed on pNFκB-GL4/pGFP-VIVIT and pNFκB-GL4/pEGFP-N1 co-transfectants. As predicted, NF-κB activity was increased by LPS treatment, and this activation was unaffected by NFAT-specific VIVIT (Fig. 4 E). This result suggests that the NFAT activity decrease was by a specific action of VIVIT, and not by general cellular toxicity. According to current research, LPS-induced cytokine expression in macrophages has been reported to be mediated by the transcription factor NF-κB as well as the ERK1/2 kinases [1, 2]. We independently confirmed that ERK1/2 inhibitor, PD98059, and the NF-κB inhibitor, SN50, inhibited LPS-induced cytokine expression in wild-type BMMs through RT real-time PCR (Fig. 5 A-F). These findings suggest that LPS-induced cytokines require not only inducible factors such as NF-κB but also non-inducible factors like NFAT. It is most probable that these factors act in concert to induce proinflammatory gene expression.
Fig. 4.
Luciferase reporter assays. Macrophage cell line RAW264.7 exhibits basal NFAT activity which is decreased by VIVIT. (A): Relative NFAT or NF-κB activity of LPS-stimulated cells normalized to non-stimulated cells. Unlike NF-κB, NFAT showed no changes in activity. (B): NFAT activity was upregulated by calcium ionophore but not LPS. (C): VIVIT peptide expressed through a co-transfected plasmid inhibited NFAT activation by calcium ionophore and further inhibited the basal activity of NFAT. (D): A chemical inhibitor of calcineurin, cyclosporine A, inhibited NFAT activity in a similar fashion to the VIVIT plasmid. (E): VIVIT did not inhibit NF-κB activity or affect the viability of the cells. Symbol (*) denotes significant difference; p<0.05.
Fig. 5.
Inhibition of cytokine gene expression in post-LPS stimulated primary BMMs by ERK1/2 and NF-κB inhibitors. ERK1/2 inhibitor: PD98059 (A-C) and NF-κB inhibitor: SN50 (D-F) were used. The relative mRNA expression levels were measured by RT real-time PCR with normalization to GAPDH levels. Symbol (*) denotes significant difference; p<0.05.
3.3. LPS and Pam do not increase cytoplasmic Calcium ion level in macrophages
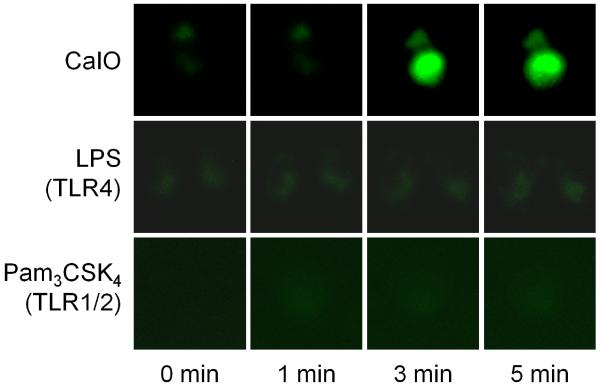
As LPS does not activate NFAT, the possibility emerges that LPS, and even other PAMPs, do not activate the entire NFAT pathway. NFAT activity is controlled by the cytoplasmic calcium ion concentration via the calcium-dependent phosphatase, calcineurin [11, 12]. For further confirmation that the basal NFAT activity is insensitive to TLR ligands, we stimulated BMMs with LPS or Pam and observed the time course of cytoplasmic calcium influx using a fluorescent calcium indicator, fluo-4. LPS or Pam had no noticeable effect on calcium-induced fluorescence, whereas the calcium ionophore caused a sharp increase (Fig. 6). This observation indicates that PAMP stimulation does not change the activity of the entire calcium/NFAT pathway.
Fig. 6.
Calcium imaging of BMMs. BMMs were pre-treated with fluo-4 AM to visualize cytoplasmic calcium. Calcium ionophore A23187 (CaIO, 5 μM) induced calcium ion increase in the cytoplasm whereas LPS (0.1 μg/ml) or Pam3CSK4 (0.1 μg/ml) did not.
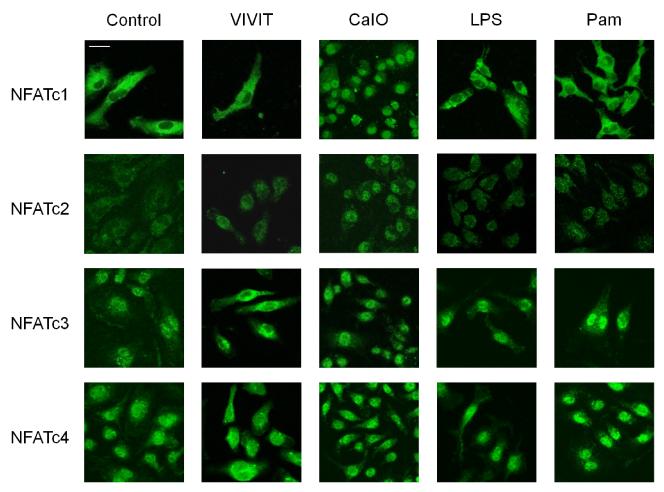
3.4. NFATc3 and c4 are localized to the nucleus under normal conditions
NFAT activation is triggered by calcineurin-catalyzed dephosphorylation leading to the unmasking of a nuclear localization signal (NLS) and subsequent nuclear translocation [21]. The study here showed that macrophages have constitutive NFAT activity in the absence of any stimulation. Therefore, we hypothesized that one or more NFAT family members may localize to the nucleus regardless of stimulation conditions. BMMs were immunostained with anti-NFAT antibodies after various treatments (Fig. 7). Among four of calcium controlled-NFAT members, NFATc3 and NFATc4 were confined to the nucleus without stimulation, while NFATc1 and NFATc2 were observed mainly in the cytoplasm. Consistent with prior observations, LPS or Pam treatment had no noticeable effect on NFAT localization, i.e., NFATc1 and NFATc2 stayed in cytoplasm and NFATc3 and NFATc4 stayed in nucleus. VIVIT expelled the nuclear NFATc3 and NFATc4 leaving no NFAT presence in the nucleus; thereby demonstrating the mechanism of VIVIT’s inhibitory action on NFAT. Moreover, only calcium ionophore treatment enabled all NFATs to localize in the nucleus. These results suggest that specific NFAT family members, namely c3 and c4, are responsible for basal NFAT activity in macrophages and are functional in proinflammatory cytokine expression, while NFATc1 and c2 family members are inactive.
Fig. 7.
Immunocytochemistry of BMMs using antibodies for NFATc1, c2, c3 and c4. In non-stimulated controls, NFATc1 and c2 were in the cytoplasm, while NFATc3 and c4 were in the nucleus. Calcium ionophore brought NFATc1 and c2 into nucleus. VIVIT treatment induced export of NFATc3 and c4 from the nucleus to the cytoplasm. LPS and Pam did not affect NFAT localization. Bar, 20 μm.
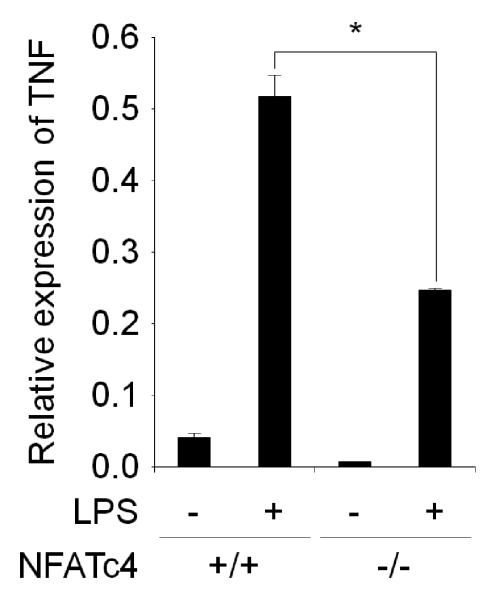
3.5. NFATc4 is responsible for TNF expression
As indicated in the immunostaining observation, NFATc3 and NFATc4 appear to be responsible for the NFAT activity that is required for the proinflammatory cytokine expression. To obtain genetic evidence about whether the NFATs are required for proinflammatory response, we prepared NFATc4−/− BMMs from a NFATc4 knockout mouse and measured the TNF level by RT real-time PCR. We decided to use NFATc4 knockout mice since they are reported to be viable and fertile [25] as well as being the only commercially available variety. Compared to control BMMs, NFATc4−/− BMMs showed significantly lower TNF expression (Fig. 8). These results corroborate that NFATc4’s responsibility in proinflammatory cytokine expression.
Fig. 8.
TNF gene expression by BMMs from NFATc4 knockout mouse. TNF gene expression induced by LPS is significantly lower in NFATc4−/− BMMs than wild-type control. The relative mRNA expression levels were measured by RT real-time PCR with normalization to GAPDH levels. Symbol (*) denotes significant difference; p<0.05.
3.6. mRNA elongation on TNF gene is facilitated by NFAT
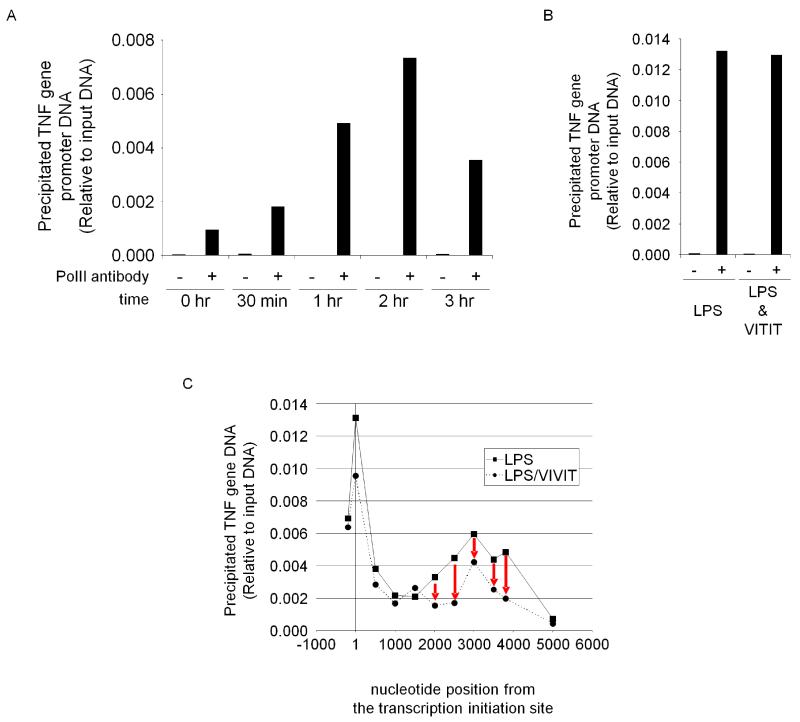
To obtain more insight about how NFAT functions, we performed chromatin immunoprecipitation (ChIP) for the mouse TNF promoter. The mouse promoter region was predicted by comparing human and mouse genome sequences (supplemental figure 1). We found mouse TNF promoter sequence is highly homologous to the human sequence [31]. LPS-treated primary BMMs were fixed with formaldehyde and nuclei were isolated. Following sonication, RNA polymerase II (PolII)-bound chromatin was immunoprecipitated using anti-PolII antibody. Precipitated DNA was detected by real-time PCR using primers for the mouse TNF promoter. PolII binding to the promoter showed LPS dependent increase in time course (Fig. 9 A). Unexpectedly, VIVIT did not hinder PolII binding to the promoter (Fig. 9 B), whereas VIVIT abolished mRNA synthesis effectively (Fig. 1 D-F). From this evidence we assumed that VIVIT stops mRNA synthesis in the middle of TNF gene. To find the position where the RNA synthesis pauses, we designed each set of PCR primers amplifying different part of TNF coding region and performed ChIP assay with the primer sets. LPS treatment showed PolII binding for the entire coding region (Fig. 9 C). However VIVIT-treated cells showed distinctively lower PolII binding from nt 2000 to the end of the sequence when compared to the control. This indicates that mRNA synthesis by PolII is stopped in the middle as a result of VIVIT treatment. Thus, we assumed that NFAT binds to the locus in the middle of the gene to help the elongation of the mRNA in the normal state due to the presence of the NFAT binding motif GGAAA around nucleotide position 2000. Current attempts at precipitating DNA fragments in the coding region with NFAT antibodies have been inconclusive, possibly due to antibody quality. Further bioinformatics analysis as well as optimized antibodies might be necessary to elucidate the function of NFAT in mRNA elongation.
Fig. 9.
Chromatin immunoprecipitation (ChIP) assay of TNF gene in BMMs with RNA polymerase II (PolII) antibody. (A): ChIP assay indicated that PolII binding to TNF gene promoter in BMMs is time-dependently increased upon LPS stimulation. (B): VIVIT did not inhibit PolII’s binding to TNF promoter. The result of 3 hr sample is shown. (C): ChIP “scanning” assay with primers targeting various region of TNF gene showed that PolII binding efficiency abruptly dropped with VIVIT treatment around nucleotide position 2000 which is in TNF coding region. Numbers on X-axis indicate nucleotide positions based on transcription initiation site of TNF gene. The leftmost plots are for the promoter region.
4. Discussion
In this study we identify a novel function of NFAT as a critical cellular signaling mediator in innate immunity, whereas NFAT function had only been discussed within the frame of adaptive immunity. This study demonstrates that NFAT activity is necessary for the induction of proinflammatory cytokine in BMMs during PAMP stimulation for any TLRs from TLR1 to TLR9. Furthermore, it was shown that macrophages have constitutive NFAT activity which does not change with PAMP stimulation. This is due to the fact that since NFATc3 and NFATc4 remain in the nucleus, they are able to achieve the constitutive NFAT activity regardless of PAMP stimulation. We confirmed the important role of NFATc4 in proinflammatory cytokine expression through macrophages from knockout mice. Based on our observations, we suggest following: NFATc3 and NFATc4 are constitutively active and localized within the nucleus, but can not singularly lead to the cytokine expression. NFAT requires activation of other independent factors, such as NF-κB and ERK1/2, via PAMP stimulation to lead to cytokine gene expression. Our study showed that NFAT is involved in the TNF transcription during innate immune response in macrophage cells. It is noteworthy that NFAT is constitutively active in macrophages which is different from T and B cells where NFAT is inducibly activated. The low constitutively active state of NFAT might be achieved by a basal level of cytoplasmic Ca2+ and concomitant calcineurin activity.
The function of NFAT in innate immunity has not received much attention thus far. It is probably due to the fact that macrophage NFAT activity is not increased in response to PAMPs, and that the inhibitors for calcineurin, e.g. cyclosporin A or FK506, have side effects that activate non-NFAT factors, such as MyD88, Trif, NF-κB and certain TLRs, which can increase cytokine expression independently of NFAT [32-36]. Calcineurin’s wide range of substrates makes it difficult to interpret its effects, especially when examining NFAT’s role in innate immunity whereas VIVIT’s specific NFAT inhibition first revealed sole NFAT function in innate immunity.
NFATc1-c4 were believed to have redundant functionality, however, it has recently been recognized that each member has a distinct role. Knockout mice of each NFAT family member exhibit differing phenotypic defects, possibly due to the different expression profiles of each NFAT among cell types [9, 10]. We showed that BMMs have nuclear NFATc3 and c4 while NFATc1 and c2 remain cytoplasmic and that only NFATc3 and NFATc4 are implicated in proinflammatory cytokine induction. Therefore the suggestion that at least two different types of regulation exist, one for NFATc1-c2 and one for NFATc3-c4, can be made. This is supported by the fact that both NFATc1-c2 and c3-c4 can be subdivided into subfamilies based on sequence homology (supplemental figure 2). Similar cases reported that among preexisting NFAT family members different NFATs are activated at different stages in T cell and skeletal muscle cell differentiation [37, 38]. This may be because each nuclear NFAT is selectively regulated by different kinases, although such possible specificity remains to be determined. Nuclear NFAT kinases re-phosphorylate NFAT to inactivate, leading to exportation. Certain studies have shown that specific kinases “prime” phosphorylation while other kinases can add additional phosphate groups on NFAT [10]. Dyrk1a has been suggested to be able to prime NFATc2 and c4 while CK1 is reported to prime NFATc2 and c3 [14, 15, 17, 18]. Furthermore, PKA has been supposed to prime NFATc1 [16]. Recently, mTOR and ERK5 have been suggested to function as priming kinases for NFATc4 [19] and GSK3 is known to add additional phosphate groups [20]. Thus, it is reasonable to make the assumption that a combination of priming NFAT kinases exist in macrophages that phosphorylate NFATc1 and NFATc2 while excluding NFATc3 and NFATc4. Differential nucleo-cytoplasmic exportation can be explained by differences in the nuclear export signal (NES) location of each NFAT protein as well. Exportin proteins bind to these NES to translocate proteins out of the nucleus. NFATc1 and c3 are known to have different NES locations. [39]. NFATc1 has it in the C-terminal domain, whereas NFATc3 does in the N-terminal domain where it overlaps with the calcineurin docking domain. Nuclear NFATc3 has been reported to bind to calcineurin and this prohibits the exportin protein Crm1 from binding and exportation of NFATc3 [40]. NFATc1 cannot participate in this competitive binding of calcineurin versus Crm1 and is easily exported from the nucleus. Other types of cells, such as macrophages, may have their own NES blocking factors. Interestingly, NFATc1 in skeletal muscle, but not NFATc3, shuttles freely between the cytoplasm and nucleus regardless of its phosphorylation state through an unknown mechanism [41]. Muscle cells may have a unique calcium-independent mechanism to regulate NFATc1 localization while avoiding the noise of short term calcium peaks generated by muscle contraction.
NFAT is also involved in non-TLR related stimuli as in the case of T and B cells [8-10]. In a previous study, we found that titanium nano-particle induced proinflammatory cytokine expression has NFAT dependence [26]. Although NFAT activity itself is not necessarily inducible during the activation process, its function is broadly required for proinflammatory cytokine expression.
This study demonstrates that NFAT is a constitutively active transcription factor which serves a critical role in the macrophage-mediated innate immune response. It is now being recognized that NFAT is expressed solely in vertebrates and has a central role in the development and control of an array of vertebrate-specific organs and systems [10]. Interestingly, while innate immunity has functioned in primitive organisms without NFAT [10], it has been increasingly portrayed as one of the key transcriptional cornerstones of vertebrate evolution. Constitutively active transcription factors generally have key roles in cellular function, for example well-characterized Pax6 in eye development [42, 43]. Thus, NFAT is a vital constitutively active transcription factor that can be utilized as a powerful regulator of innate immune activity.
Supplementary Material
Acknowledgment
We thank Dr. Anjana Rao and Dr. Fernando Macián for providing the pGFP-VIVIT plasmid and Dr. Robert Winchester, Dr. Sung-Wook Seo, Dr. Phillip Wong, Dr. Keiichi Inoue, Dr. Naoko Shimada and Mr. Kumar Nair for helpful discussion. This work was supported by National Institutes of Health grants R01EB006834 and R01AR056246 (to F.Y.L.).
Abbreviations
- NFAT
Nuclear factor of activated T cells
- NF-κB
nuclear factor of κ-light-chain-enhancer of activated B cells
- AP-1
activator protein 1
- ERK
extracellular signal-regulated kinase
- PolII
RNA polymerase II
- TNF
tumor necrosis factor
- MIP-1α
macrophage inflammatory protein 1α
- IL-1 β
interleukin 1β
- TLR
Toll-like receptor
- TCR
T cell receptor
- BCR
B cell receptor
- NLS
nuclear localization signal
- BMM
bone marrow-derived macrophage
- LPS
lipopolysaccharide
- PAMP
pathogen-associated molecular pattern
- CaIO
calcium ionophore
- CsA
cyclosporin A
- ChIP
chromatin immunoprecipitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
All authors have no conflicts of interest.
References
- [1].Akira S, Uematsu S, Takeuchi O. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- [2].Akira S. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(4):143–156. doi: 10.2183/pjab.85.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Science. 1998;282(5396):2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- [4].Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. Proc Natl Acad Sci U S A. 2000;97(25):13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alexopoulou L, Thomas V, Schnare M, Lobet Y, Anguita J, Schoen RT, Medzhitov R, Fikrig E, Flavell RA. Nat Med. 2002;8(8):878–884. doi: 10.1038/nm732. [DOI] [PubMed] [Google Scholar]
- [6].Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. J Immunol. 2002;169(1):10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- [7].Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Science. 1988;241(4862):202–205. doi: 10.1126/science.3260404. [DOI] [PubMed] [Google Scholar]
- [8].Hogan PG, Chen L, Nardone J, Rao A. Genes Dev. 2003;17(18):2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- [9].Horsley V, Pavlath GK. J Cell Biol. 2002;156(5):771–774. doi: 10.1083/jcb.200111073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu H, Peisley A, Graef IA, Crabtree GR. Trends Cell Biol. 2007;17(6):251–260. doi: 10.1016/j.tcb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- [11].Flanagan WM, Corthesy B, Bram RJ, Crabtree GR. Nature. 1991;352(6338):803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- [12].Clipstone NA, Crabtree GR. Nature. 1992;357(6380):695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- [13].Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Proc Natl Acad Sci U S A. 1999;96(5):2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Arron JR, Winslow MM, Polleri A, Chang CP, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, Yamasaki N, Miyakawa T, Francke U, Graef IA, Crabtree GR. Nature. 2006;441(7093):595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- [15].Gwack Y, Sharma S, Nardone J, Tanasa B, Iuga A, Srikanth S, Okamura H, Bolton D, Feske S, Hogan PG, Rao A. Nature. 2006;441(7093):646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- [16].Sheridan CM, Heist EK, Beals CR, Crabtree GR, Gardner P. J Biol Chem. 2002;277(50):48664–48676. doi: 10.1074/jbc.M207029200. [DOI] [PubMed] [Google Scholar]
- [17].Zhu J, Shibasaki F, Price R, Guillemot JC, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Cell. 1998;93(5):851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]
- [18].Okamura H, Garcia-Rodriguez C, Martinson H, Qin J, Virshup DM, Rao A. Mol Cell Biol. 2004;24(10):4184–4195. doi: 10.1128/MCB.24.10.4184-4195.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yang TT, Yu RY, Agadir A, Gao GJ, Campos-Gonzalez R, Tournier C, Chow CW. Mol Cell Biol. 2008;28(10):3489–3501. doi: 10.1128/MCB.01847-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Beals CR, Sheridan CM, Turck CW, Gardner P, Crabtree GR. Science. 1997;275(5308):1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- [21].Okamura H, Aramburu J, Garcia-Rodriguez C, Viola JP, Raghavan A, Tahiliani M, Zhang X, Qin J, Hogan PG, Rao A. Mol Cell. 2000;6(3):539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- [22].Scharenberg AM, Humphries LA, Rawlings DJ. Nat Rev Immunol. 2007;7(10):778–789. doi: 10.1038/nri2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Oh-hora M, Rao A. Curr Opin Immunol. 2008;20(3):250–258. doi: 10.1016/j.coi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wooten RM, Ma Y, Yoder RA, Brown JP, Weis JH, Zachary JF, Kirschning CJ, Weis JJ. J Immunol. 2002;168(1):348–355. doi: 10.4049/jimmunol.168.1.348. [DOI] [PubMed] [Google Scholar]
- [25].Wilkins BJ, De Windt LJ, Bueno OF, Braz JC, Glascock BJ, Kimball TF, Molkentin JD. Mol Cell Biol. 2002;22(21):7603–7613. doi: 10.1128/MCB.22.21.7603-7613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Minematsu H, Shin MJ, Celil Aydemir AB, Seo SW, Kim DW, Blaine TA, Macian F, Yang J, Young-In Lee F. Ann N Y Acad Sci. 2007;1117:143–150. doi: 10.1196/annals.1402.026. [DOI] [PubMed] [Google Scholar]
- [27].Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Science. 1999;285(5436):2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- [28].Noguchi H, Matsushita M, Okitsu T, Moriwaki A, Tomizawa K, Kang S, Li ST, Kobayashi N, Matsumoto S, Tanaka K, Tanaka N, Matsui H. Nat Med. 2004;10(3):305–309. doi: 10.1038/nm994. [DOI] [PubMed] [Google Scholar]
- [29].Yu H, Sliedregt-Bol K, Overkleeft H, van der Marel GA, van Berkel TJ, Biessen EA. Arterioscler Thromb Vasc Biol. 2006;26(7):1531–1537. doi: 10.1161/01.ATV.0000225286.30710.af. [DOI] [PubMed] [Google Scholar]
- [30].Yu H, van Berkel TJ, Biessen EA. Cardiovasc Drug Rev. 2007;25(2):175–187. doi: 10.1111/j.1527-3466.2007.00011.x. [DOI] [PubMed] [Google Scholar]
- [31].Tsytsykova AV, Goldfeld AE. Mol Cell Biol. 2002;22(8):2620–2631. doi: 10.1128/MCB.22.8.2620-2631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Conboy IM, Manoli D, Mhaiskar V, Jones PP. Proc Natl Acad Sci U S A. 1999;96(11):6324–6329. doi: 10.1073/pnas.96.11.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tsai EY, Falvo JV, Tsytsykova AV, Barczak AK, Reimold AM, Glimcher LH, Fenton MJ, Gordon DC, Dunn IF, Goldfeld AE. Mol Cell Biol. 2000;20(16):6084–6094. doi: 10.1128/mcb.20.16.6084-6094.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kang YJ, Kusler B, Otsuka M, Hughes M, Suzuki N, Suzuki S, Yeh WC, Akira S, Han J, Jones PP. J Immunol. 2007;179(7):4598–4607. doi: 10.4049/jimmunol.179.7.4598. [DOI] [PubMed] [Google Scholar]
- [35].Jennings C, Kusler B, Jones PP. Innate Immun. 2009;15(2):109–120. doi: 10.1177/1753425908100928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Biswas G, Anandatheerthavarada HK, Zaidi M, Avadhani NG. J Cell Biol. 2003;161(3):507–519. doi: 10.1083/jcb.200211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Adachi S, Amasaki Y, Miyatake S, Arai N, Iwata M. J Biol Chem. 2000;275(19):14708–14716. doi: 10.1074/jbc.275.19.14708. [DOI] [PubMed] [Google Scholar]
- [38].Abbott KL, Friday BB, Thaloor D, Murphy TJ, Pavlath GK. Mol Biol Cell. 1998;9(10):2905–2916. doi: 10.1091/mbc.9.10.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kiani A, Rao A, Aramburu J. Immunity. 2000;12(4):359–372. doi: 10.1016/s1074-7613(00)80188-0. [DOI] [PubMed] [Google Scholar]
- [40].Zhu J, McKeon F. Nature. 1999;398(6724):256–260. doi: 10.1038/18473. [DOI] [PubMed] [Google Scholar]
- [41].Shen T, Liu Y, Cseresnyes Z, Hawkins A, Randall WR, Schneider MF. Mol Biol Cell. 2006;17(4):1570–1582. doi: 10.1091/mbc.E05-08-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Quiring R, Walldorf U, Kloter U, Gehring WJ. Science. 1994;265(5173):785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- [43].Halder G, Callaerts P, Gehring WJ. Science. 1995;267(5205):1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.