Abstract
Distinctive from that of the animal system, the basic plan of the plant body is the continuous formation of a structural unit, composed of a stem with a meristem at the top and lateral organs continuously forming at the meristem. Therefore, mechanisms controlling the formation, maintenance, and development of a meristem will be a key to understanding the body plan of higher plants. Genetic analyses of filamentous flower (fil) mutants have indicated that FIL is required for the maintenance and growth of inflorescence and floral meristems, and of floral organs of Arabidopsis thaliana. FIL encodes a protein carrying a zinc finger and a HMG box-like domain, which is known to work as a transcription regulator. As expected, the FIL protein was shown to have a nuclear location. In situ hybridization clearly demonstrated that FIL is expressed only at the abaxial side of primordia of leaves and floral organs. Transgenic plants, ectopically expressing FIL, formed filament-like leaves with randomly arranged cells at the leaf margin. Our results indicate that cells at the abaxial side of the lateral organs are responsible for the normal development of the organs as well as for maintaining the activity of meristems.
Keywords: Arabidopsis, FIL, zinc finger, HMG, nuclear protein, abaxial–adaxial development
Differing from the situation in animal systems, maintenance of the meristem activity throughout life is a key for the patterning of the plant body. The shoot apical meristem and the meristem of the main root are formed in embryonic development and kept active after germination. During vegetative growth, leaves are continuously formed in a strictly controlled fashion from the shoot apical meristem. After the plant shifts to reproductive growth, the meristem converts to an inflorescence meristem, which in turn forms floral meristems. The floral meristem generates floral organs at predetermined positions. Genetic and molecular studies are under way to help understand the genetic regulatory system supporting meristem activity: control of cell division, formation of lateral organ primordia, and maintenance of the meristem structure. Recent studies using Arabidopsis, snapdragon, and maize have started to unveil the genetic basis of these molecular mechanisms (Okada and Shimura 1994).
The leaf meristems are formed in a helical manner at the top of the inflorescence. When the leaf primordia begin to form, the petiole and flattened leaf blade are developed and the leaves show epinasty. Mature leaves show abaxial–adaxial polarity. The adaxial surface bears glossy, dark green epidermal cells, and produces many trichomes. On the contrary, the abaxial surface shows matte, gray–green epidermal cells and does not produce any trichomes. As the plant grows, the adaxial surface of the newly made leaves undergoes a gradual reduction in production of trichomes, and the abaxial surface starts to produce them. An Antirrhinum phantastica (phan) mutant produces filamentous leaves that are radially symmetrical with abaxial characters, and an Arabidopsis phabulosa-1d (phb-1d) dominant mutant shows adaxialized filamentous leaves. Analysis of these mutants suggests that the outgrowth of the leaf blade requires the juxtaposition of the abaxial–adaxial cell fate, and also suggests that the adaxial leaf environment is required for the development of the shoot apical meristem (Waites and Hudson 1995; MacConnell and Barton 1998).
The floral meristems are also formed in a helical manner at the top of the inflorescence. Determination of floral meristem identity and development of the floral meristem are known to be controlled by several genes. FILAMENTOUS FLOWER (FIL), LEAFY (LFY), APETALA1 (AP1), and CAULIFLOWER (CAL) genes are major players in this step (Bowman et al. 1993; Kempin et al. 1995; Weigel and Nilsson 1995; Sawa et al. 1999). The floral organ fate is determined by a combination of a set of ABC class genes (Weigel and Meyerowitz 1994).
By using a combined molecular and genetic approach to study the meristem activity that regulates the development of the plant body, we showed that the Arabidopsis FIL gene is responsible for normal floral development (Komaki et al. 1988; Okada and Shimura 1994; Sawa et al. 1999). Here we show the molecular nature of the FIL gene and its spatially controlled expression. We also report the phenotypes of the 35S::FIL plants. By determining the abaxial character, the results indicate that the FIL gene is responsible for the normal development of leaves and floral organs, and for the maintenance of meristem activity.
Results
Phenotypes of fil mutants
The fil mutant of Arabidopsis (former name, Fl54) shows a pleiotropic phenotype in the structure of the inflorescence with two different flower-related structures (Komaki et al. 1988; Okada and Shimura 1994; Sawa et al. 1999). The mutant generates clusters of filamentous structures (designated type B structures) and of flowers with floral organs of altered number and shape (type A flowers) (Fig. 1A,B,C,G,H). In view of their structural and developmental resemblance to peduncles, the filamentous structures are interpreted to be underdeveloped flowers that failed to form receptacles and floral organs. Therefore, it has been shown that FIL has a role in supporting the development of the floral meristem into a mature flower. FIL is also involved in the fate determination of the floral meristem, because a homeotic conversion from a flower to an inflorescence was observed when the fil mutation was combined with a homeotic mutation ap1 (Fig. 1E). In combination with a mutation of a floral meristem identity gene, LFY, the inflorescence formed heavily deformed structures such as filaments, bract-like structures, or pistil-like structures, but no flowers (Fig. 1F). Enhancement of the mutant phenotype detected in double mutants suggested that the FIL protein works coordinately with AP1 and LFY proteins, each of which is considered to function as a transcription factor. In addition to the fate determination and development of the floral meristem, FIL was also suggested to have a role in the formation of the inflorescence meristem.
Figure 1.
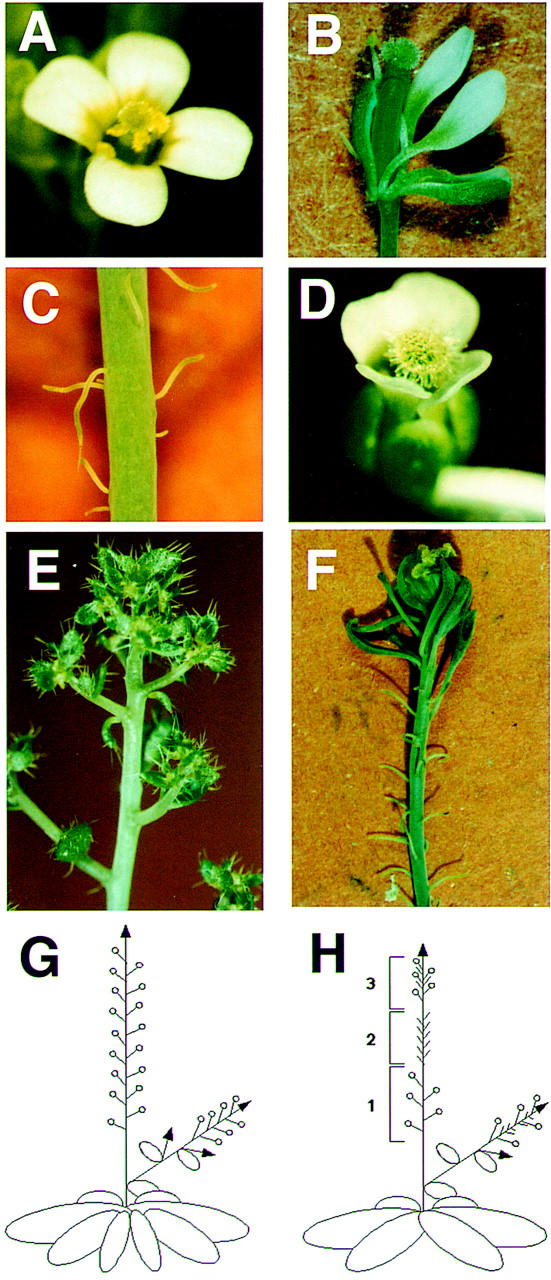
Inflorescence and flowers of Arabidopsis plants. (A) A wild-type flower; (B) the type-A flower has an aberrant number and arrangement of floral organs; (C) type-B structures are thin filaments interpreted as immature flowers that failed to develop floral organs; (D) a wild-type-looking flower of a transgenic plant carrying a TAC clone, TAC27M5; (E) a flower is converted homeotically to an inflorescence in the fil ap1 double mutant; (F) the fil lfy double mutant forms a cluster of filamentous structures and a cluster of filaments with a sepal-like organ but no type A flowers; (G) schematic drawing of inflorescence of the wild type; (H) schematic drawing of inflorescence of fil mutants. Regions 1–3 are clusters of type-A flowers, type-B structures, and a mixture of both, respectively.
Molecular isolation of the FIL gene
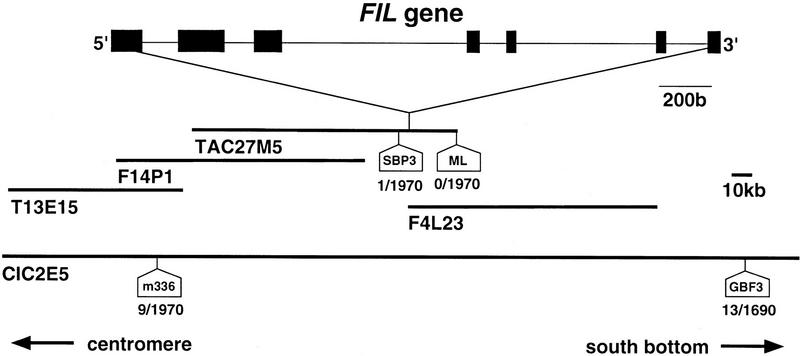
Cloning of the FIL locus was done by the map-based chromosome-walking procedure. The FIL locus was mapped to the lower part of chromosome 2 close to RFLP marker m336 (Fig. 2). A contiguous map covering a 400-kb region was made adjacent to the marker, and the FIL locus was pinpointed at the left end of a TAC clone, TAC27M5, between the two markers of SBP3 and ML. To confirm the presence of the FIL gene, we used the TAC clone to transform the fil-1 mutant and found wild-type flowers in three independent transgenic plants (Fig. 1D). Based on the nucleotide sequence of the clone, five ORFs were predicted in the 20-kb region between SBP3 and ML markers. By comparing the nucleotide sequences of the fil-1 mutant, fil-2 mutant, and wild type, we concluded that FIL is an ORF (TIGR accession no. F4L23.30) that encodes a protein of 229 amino acid residues in seven exons, because there was a single-base change in both mutants in one ORF but no sequence aberrations in the other ORFs.
Figure 2.
Genomic structure of the FIL region at the lower part of chromosome 2. FIL was isolated by map-based cloning and was mapped between markers SBP3 and ML on clone TAC27M5. Other BAC clones overlapping in the region are shown. Numbers below the molecular markers shown in white boxes indicates the recombination frequency between the marker and the FIL locus (no. of recombinant chromosomes/no. of chromosomes examined).
Characterization of fil alleles
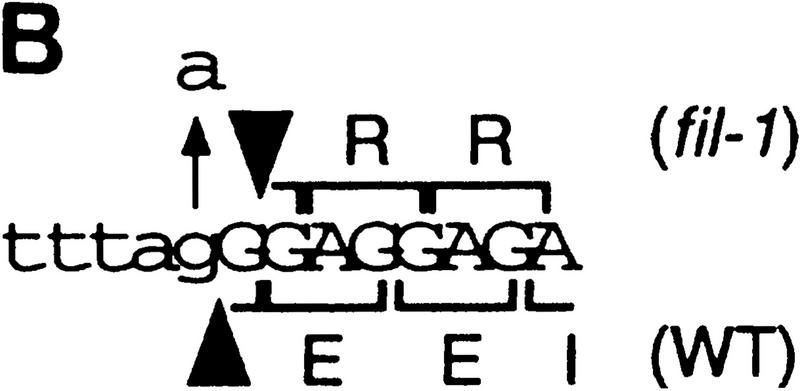
We isolated a cDNA clone corresponding to the ORF and confirmed the exon–intron organization of the gene predicted by computer annotation (Fig. 3A). In the fil-1 mutant, a nucleotide change from G to A was found at the splicing acceptor site of the fourth intron (Fig. 3A,B). The sequence of the fil-1 mRNA was confirmed by use of RT–PCR procedure, and it revealed that the fil-1 mutation formed a new splicing acceptor site at one base after the original site, resulting in the production of a putative short polypeptide of 165 amino acid residues (Fig. 3B). The change in the putative mutant protein (i.e., alteration of the 12-amino-acid sequence at the carboxy-terminal and the lack of 64 residues) may abolish the normal function of the FIL protein. On the other hand, the fil-2 mutant showed a base change from G to A at base 89, which caused an amino acid change from Cys to Tyr at residue 30 (Fig. 3 A).
Figure 3.

(A) Nucleotide sequence of the FIL cDNA clone and deduced amino acid sequence of the FIL protein (GenBank accession no. AF074948). Numbering was started from the putative translational initiation site. Positions separated by introns are shown by arrowheads. A zinc finger domain (amino acid residue 22–60) and a region homologous to the HMG-box domain (amino acid number 146–179) are underlined. (*) The position of the fil-1 mutation, which makes a shift of one base in the frame of exon 5. (↑ at nucleotide 89) The mutation in the fil-2 mutant causing a change in residue 30 from Cys to Tyr. (B) The frameshift caused by the fil-1 mutation. The G → A base substitution at the splicing acceptor site of the fourth intron makes a new splicing acceptor site 1 base downward. Novel splicing makes a short polypeptide (shown in A). (C) Amino acid sequence of the HMG domain. Partial sequence of the FIL protein is aligned to partial sequences of the rice homolog osFIL1 (GenBank accession no. AF098752), rice homolog osFIL2 (AF098753), human mitochondrial transcription factor 1 (mtTF1, S38413), and mouse testis-specific HMG (tsHMG, P40630). (*) The consensus sequence of the HMG-box domain (Parisi and Clayton 1991) (D) Amino acid sequence of the amino-terminal conserved region in the FIL protein and in two rice homologs and that of a putative zinc finger protein, LSD1 (GenBank accession no. ATU87833). (*) The position of the putative component of the zinc finger motif. In C and D, residues identical or similar to those of the FIL protein are boxed.
Sequence analysis of the FIL gene
The amino acid sequence deduced from the cDNA sequence showed a homology at the carboxy-terminal region to a part of a domain of the high mobility group (HMG) proteins. The HMG domain comprises ∼80 amino acid residues that form three α-helices (Reeves and Bustin 1996). Homology to the HMG domain was found in the FIL protein from amino acid 146 to 179 (underlined in Fig. 3A); this region corresponds to helix I and helix II. However, homology was not found in the region for helix III. A stretch of α-helices cannot form because there are four proline residues in the region (Fig. 3A).
To test whether similar FIL-like members are found in the Arabidopsis genome, we conducted genomic blot hybridization experiments using FIL cDNA as a probe and showed that there was more than one other gene with extensive sequential homology to FIL in the Arabidopsis genome (data not shown). Five genomic regions homologous to FIL were found in the database. In addition, two rice EST sequences were found in the database search. The sequences of these genes, designated osFIL1 and osFIL2, were highly conserved not only in the HMG domain but also in other regions (Fig. 3C,D). It is noteworthy that the amino acid substitution found in the fil-2 mutant is located in the conserved region at the amino terminus (Fig. 3D), suggesting that the cysteine at residue 30 plays an important role in the function of the FIL protein and its relatives.
Consistent with this assumption, the conserved region at the amino terminus showed a weak homology with the metal-binding region of the Cys2/Cys2-type zinc finger protein LSD1 (Fig. 3D; Dietrich et al. 1997). We propose that the four cysteines at residues 30, 33, 56, and 59 bind to a zinc atom and that the proline at residue 42 is located at the top of the finger structure in the FIL protein. We postulate that the replacement of the first cysteine with a tyrosine in the fil-2 mutation may cause a defect of the protein structure and, thus, loss of function.
Although the molecular mechanisms of the FIL protein are still unknown, these results indicate that FIL is a member of a new group in plants carrying a zinc finger and an HMG domain.
Content of zinc ion in the FIL protein
To determine whether an amino terminal region serves as a zinc finger domain or not, we used inductively coupled plasma (ICP) spectroscopy to detect the presence of zinc ions in the FIL protein. The sample of the protein from a bacterial preparation was purified by affinity chromatography. The zinc atoms, nonspecifically bound to the FIL protein, were removed during the purification by a column wash with 200 mm imidazole buffer, elution with 500 mm imidazole buffer, and dialysis with buffer containing 0.1 mm EDTA. The intensities of the ICP spectra were measured (Table 1). The results strongly suggest that the FIL protein contains one zinc ion and works as a zinc finger protein.
Table 1.
Zinc content in FIL protein analyzed with ICP spectroscopy
| Wavelength (nm)
|
213.856
|
202.548
|
206.200
|
|---|---|---|---|
| Zinc [sample/ background (ppm)] | 0.904/4 × 10−3 | 1.064/0 | 1.043/0 |
| Zinc (μm) | 13.85 | 16.28 | 15.96 |
| Protein (μm) | 16.9 | 16.9 | 16.9 |
| Zinc/protein | 0.82 | 0.96 | 0.94 |
Nuclear localization of the FIL protein
If the FIL protein works as a keeper of chromatin architecture or a member of a transcriptional complex, it would be expected to become localized in the nucleus. To examine the cellular location of the FIL protein, we transformed epidermal cells of an onion bulb with a chimeric gene made of the full-length FIL cDNA translationally fused to the glutathione S-transferase (GUS) coding region under the control of the 35S promoter of cauliflower mosaic virus (CaMV), as described previously (von Arnim and Deng 1994). As shown in Figure 4, the fused protein was found in the nucleus in all of the 48 samples tested, whereas GUS protein free from FIL protein did not show the nuclear enrichment of GUS staining in any of the 25 samples tested. This result suggests strongly that FIL protein is involved in the regulation of transcription of genes that control the formation and development of the meristem and floral organ primordia.
Figure 4.

Subcellular localization of the FIL protein by biolistic bombardment of onion skin cells with GUS as a vital marker. (A,B) Transiently transformed with 35S promoter::GUS. (C,D) Transiently transformed with 35S promoter::FIL–GUS translational fusion construct. The same cells were stained with 2 mg/ml X-Gluc (A,C) or with 1 μg/ml DAPI (B,D). Scale bar, 100 μm.
Spatially controlled expression of the FIL gene
The temporal and spatial pattern of FIL expression was tested by in situ hybridization using FIL cDNA as a probe. The expression pattern was changed drastically according to the stage of the meristem. In the vegetative meristem, the expression of FIL was observed in the peripheral region expected to form a leaf primordium (Fig. 5A), and in a young leaf primordium. As the primordium developed, gene expression became restricted to two to four cell layers on the abaxial side of the leaf. In seedlings also, FIL was expressed on the abaxial side of cotyledons (data not shown). The FIL expression pattern in embryogenesis is not known. In the inflorescence meristem, FIL showed a similar expression pattern. The gene was expressed in a region in the peripheral zone expected to form a floral meristem (stage 0; Fig. 5B). In the floral meristem of stage 1, the expression was observed in the cell layers on the abaxial side (Fig. 5B). At stage 3, strong expression of FIL was observed in the sepal primordia but not in other regions (Fig. 5C). Similar to that in the leaf primordia, the expression of FIL in the sepal primordia was shifted to the abaxial side as they developed (Fig. 5D). The same spatial and temporal control of the expression pattern, expressing in the whole region of young primordia at the beginning but in cells on the abaxial side later, was observed in the other floral organs: petals, stamens, and carpels (Fig. 5D–F). Even in the stamen filaments, FIL was expressed on the abaxial side (Fig. 5F). The expression continued until the organs matured.
Figure 5.
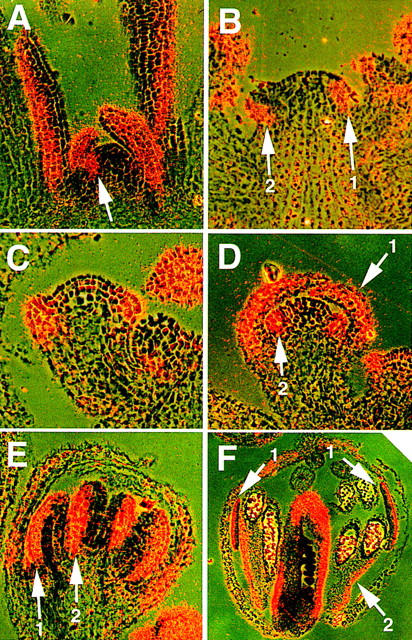
In situ localization of FIL mRNA in wild-type Arabidopsis plants. Photographs were taken with double exposures of the dark field with a red filter and the bright field without a color filter. (A) Vegetative meristem. FIL is expressed on the abaxial side of leaves, in young leaf primordia, and in a small region in the peripheral zone of the meristem under a young leaf primordium (arrow). (B) Inflorescence meristem with very young floral meristems. FIL is expressed in a region in the peripheral zone where the floral meristem will be formed (arrow 1) and in a floral meristem of stage 1 (arrow 2). (C) A stage-3 floral meristem. FIL is expressed in sepal primordia and in their base regions in the meristem. (D) A stage-5 floral bud. FIL is expressed on the abaxial side of sepals (arrow 1) and in stamen primordia (arrow 2). (E) A stage-7 floral bud. FIL is strongly expressed on the abaxial side of sepals, young stamens (arrow 1), and young carpels (arrow 2). (F) A stage-10 floral bud. FIL expression continues on the abaxial side of floral organs, including petals (arrows 1). Note that FIL is expressed on the abaxial side of the filaments of stamens (arrow 2).
Phenotypes of 35S::FIL plants
To examine FIL gene function in relation to spatially specific expression, we tested the phenotype of transgenic lines, overexpressing FIL under the control of the 35S CaMV promoter. All 20 independent plants showed aberrant structure in their leaves. Plants of heavy phenotype (15 plants) died after forming one to four rosette leaves that looked to be filamentous structures because they failed to develop lobes (Fig. 6C). Plants of mild phenotype (5 plants) formed 5–10 wrinkled leaves (Fig. 6F).
Figure 6.
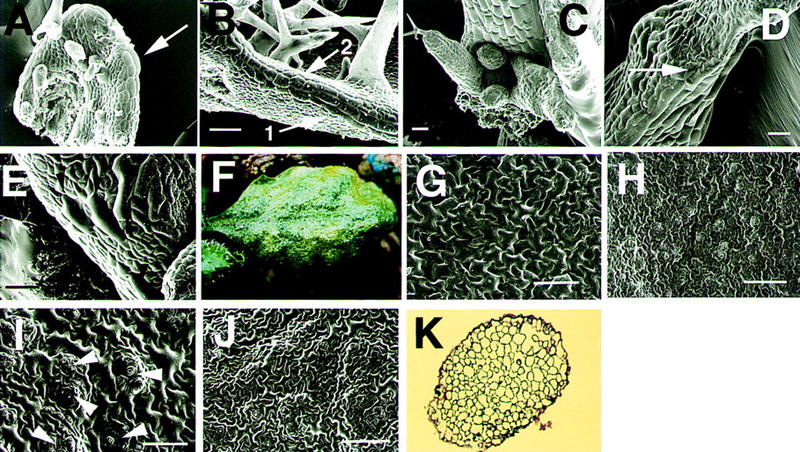
Morphology of the 35S::FIL transgenic plants. (A) A young wild-type leaf. Immature trichomes are formed on the adaxial surface. Thick and long cells (leaf margin cells) are observed at the margin (arrow). (B) A wild-type leaf. Trichomes are developing. Three files of leaf margin cells (arrow 1) are proliferating from the top to the bottom of the leaves. Arrow 2 indicates the proliferating front of the leaf margin cells. (C) A vegetative shoot apex of a transgenic plant forming filamentous leaves. A cotyledon at the bottom side was removed. (D) Enlarged picture of a filamentous leaf of the transgenic plant. No trichomes are observed. Formation of the leaf margin cells occurs only at the bottom of the leaf (arrow). (E) Enlarged picture of a young wrinkled leaf of a transgenic plant forming the leaf margin cells at random positions (arrowheads). (F) A developed wrinkled leaf of the transgenic plant. Epidermal cells are shown on the adaxial (G) and the abaxial (H) side of a wild-type leaf. (I) Epidermal cells on the adaxial side of the wrinkled leaf of a transgenic plant (F). Arrowheads indicate small patches of cells similar to the cells of the abaxial side. (J) Epidermal cells on the abaxial side of a wrinkled leaf (F). (K) A transverse section of a 35S::FIL filamentous leaf. Scale bars, 50 μm (A,B,D,E); 100 μm (C,G–J).
On the epidermis of wild-type leaves, thick and long cells are arranged longitudinally at the juxtaposition of abaxial and adaxial surfaces. The long cells (leaf margin cells) begin to develop from the top to the bottom of the margin before the trichomes mature (Fig. 6A). The leaf margin cells continue to proliferate to form three cell files (Fig. 6B). On leaves of the 35S::FIL plants, leaf margin cells did not develop normally. Even though the longitudinal length of the leaf was shorter than 300 μm, the leaf margin cells showed relatively normal development; although the leaves were narrow (Fig. 6C). When the leaves grew longer than 300 μm, they became wrinkled and filamentous (Fig. 6D). In those leaves, the leaf margin cells were not formed (Fig. 6D), or were arranged randomly (Fig. 6E). Vascular tissues were not differentiated in the filamentous leaves (Fig. 6K).
The first few rosette leaves of wild type possess trichomes on their adaxial surface but do not develop trichomes at the abaxial surface (Fig. 6A,B; Telfer et al. 1997). In the 35S::FIL plants, the filamentous leaves did not usually develop trichomes, suggesting that the epidermis of the filamentous leaves has a characteristic of the abaxial surface. Other features showing the difference of the abaxial and the adaxial surfaces, shape and color of epidermal cells, and the structure of vascular tissue were not clear in the filamentous leaves.
On the adaxial surface of wild-type leaves, large cells with the shape of jigsaw puzzle pieces are formed, but on the abaxial surface, small epidermal cells and a lot of stomata are produced (Fig. 6G,H). The adaxial surface of the wrinkled leaves of the 35S::FIL plants of mild phenotype showed several islands of small cells with many stomata, characteristic of the abaxial epidermal cells (Fig. 6I). The formation of the patches of abaxialized cells on the adaxial surface supports the model that the ec-topic expression of FIL promotes abaxialization. The epidermal cells on the abaxial surface (Fig. 6J) and internal tissues (data not shown) of the wrinkled leaves remained unaffected.
The abaxialization of plant tissues by the ectopic expression of the FIL gene indicates strongly that FIL is responsible for the determination of tissue identity of the abaxial side.
Discussion
We showed previously that the FIL gene regulates the flowering time and maintains and regulates the development of the inflorescence meristem, floral meristem, and floral organ primordia. In this work we have demonstrated that the FIL gene encodes a nuclear protein with zinc finger and HMG-related domains and that FIL expression occurs where the fil mutant has developmental defects, except for the inflorescence meristem. Unexpectedly, we found that FIL expression was restricted to the abaxial region.
Structure of the FIL protein
We revealed that the FIL gene encodes a protein including a zinc finger domain and a HMG-related domain. Compared with other HMG proteins in yeast, animals, and plants, the structure of the FIL protein is unique. In many cases of animal and plant HMG proteins, they have an acidic domain at the carboxyl terminus and often a basic domain at the amino terminus (Grasser 1995, 1998). The FIL protein has a long stretch of amino acid residues at the amino terminus of the HMG domain but lacks prominent basic or acidic domains, as well as helix III. The three-dimensional structure of the DNA-bound HMG domain of the human SRY protein shows that the three helices of the protein make a concave surface interacting with the minor groove of DNA bent 70–80°C (Werner et al. 1995). It would be interesting to examine whether the FIL protein can bind to double-stranded DNA or to a nucleosome and help to bend it.
FIL protein showed a weak homology to the zinc finger protein LSD1. LSD1 was classified as a member of the GATA1 family (Takatsuji 1998), whereas the FIL protein has longer spacing between the second and the third conserved cysteines compared with the GATA1 family, including the LSD1 protein; the FIL protein has only one finger. Although the sequence homology between the FIL protein and the Dof family is relatively low, Dof family members have spacing longer than that of the GATA1 family, and they have the ability to bind to DNA with only one finger (Takatsuji 1998). The zinc finger region of the FIL protein can thus be classified into the Dof family. The PBF gene of the Dof family in maize is known to show endosperm-specific expression, and PBF is known to bind to the prolamin box and to interact with Opaque2 of a bZIP-type transcription factor (Vicente-Carbajosa et al. 1997) . The interaction between the Dof family protein and b-ZIP-type transcription factors has also been found in the case of OBP1 of the Dof family protein and the OBF4, OBF5 proteins of b-ZIP-type transcription factors (Zhang et al. 1995). FIL protein may possibly function as a transcription factor with a b-ZIP protein in vivo.
Function of FIL in the floral meristem formation
The timing and region of FIL expression in the young floral meristem overlapped with that of LFY and AP1, which are known as floral meristem identity genes. LFY and AP1 were reported to begin their expression in early stages of floral meristems (Mandel et al. 1992; Weigel et al. 1992). The overlapping of gene expression supports our conclusion obtained from phenotype analysis of double mutants that the FIL protein may interact with LFY or AP1. Because LFY and AP1 proteins are considered to be transcription factors, it is likely that the FIL protein associates with LFY or AP1 to form a transcriptional complex. Localization of the FIL protein in the nucleus further supports this likelihood.
Because the structure of the inflorescence of the fil lfy double mutant was different from that of the fil ap1 double mutant (Fig. 1E,F), a transcriptional complex including FIL and LFY proteins and a complex including FIL and AP1 proteins may each control the expression of a different set of target genes.
There are other possibilities of how FIL interacts with LFY and AP1 in the determination of floral meristem identity. Distinctive phenotypes of fil, lfy, and ap1 mutants suggest that FIL may have a separate but partly overlapping function with LFY and AP1. It is also possible that the expression of FIL, LFY, and AP1 is mutually controlled. As we showed previously (Sawa et al. 1999), the expression of LFY could be promoted by FIL and the expression of AP1 could be induced by FIL in combination with LFY. FIL expression pattern in the lfy and ap1 mutant or lfy ap1 double mutant will reveal the functional relationship between FIL, LFY, and AP1 genes.
Function of FIL in floral organ formation
The expression of FIL was observed to continue from the very early to later stages of floral organogenesis, changing the region of gene expression according to the stage. This result, indicating that FIL is required in almost all stages of flower development, is consistent with the phenotype of the fil mutant, because we found structural and developmental abnormalities in all stages (Sawa et al. 1999). It is possible to speculate that FIL protein contributes to the accurate transcription of the genes whose expression is necessary in each stage of flower development by modifying the architecture of the upstream regulatory region of target genes.
Regulation of abaxial–adaxial polarity
The shift of expression of FIL to the abaxial side of young leaf and floral organs suggests strongly that FIL supports the growth and development of the abaxial side of the organs. The phenotypes of 35S::FIL plants support this idea.
The transgenic plants failed to form normal flat leaves with differentiated abaxial and adaxial surfaces. The aberrant leaf structure shows that the ectopic expression of FIL results in the appearance of the abaxial side characteristics with the reduction or loss of the adaxial side characteristics. The major phenotype of the transgenic plants was the formation of filamentous leaves with abaxial-looking tissue. The leaf margin cells, specifically those appearing at the juxtaposition of the abaxial and adaxial surfaces of wild-type leaves, were formed in a disturbed or abolished manner in young leaf primordia of the transgenic plants, suggesting that the juxtaposition of the abaxial and the adaxial sides induces the normally arranged development of the leaf margin cells. The aberrant distribution of the margin cells in the transgenic plants might reflect unstable formation of the abaxial–adaxial polarity. In addition, several patches of abaxialized epidermal cells were found on the adaxial surface of wrinkled leaves of the transgenic plants. Similar patchy formation of the abaxial epidermal cells on the adaxial surface of leaves was reported to occur in the phantastica (phan) mutant of snapdragon (Waites and Hudson, 1995). The PHAN gene was shown to be responsible for adaxialization of leaves.
The abaxialization observed in the 35S::FIL plants is a strong indication that FIL controls the identity of the abaxial side of the lateral organs. The phan mutant and the leafbladeless1 (lb1) mutant of maize are reported to form filamentous leaves with abaxialized cell organization (Waites and Hudson, 1995; Waites et al. 1998; Timmermans et al. 1998). Because the phan and the lb1 mutants are recessive, it is suggested that the genes are responsible for the determination of the adaxial side of the leaves. The PHAN gene was cloned and shown to encode a Myb protein (Waites et al. 1998). In addition, a dominant mutant of Arabidopsis, phb-1d, forms adaxialized filamentous leaves, indicating that PHB-1d is also responsible for the development of the adaxial side of leaves (MacConnell and Barton 1998). The molecular nature of PHB-1d is not known.
PHAN was shown to be expressed in the cells of lateral organ primordia but not restricted to the cells on the adaxial side as expected previously (Waites et al. 1998). To explain the role of PHAN in the development of adaxial character, it may be postulated that some other gene(s) expressed on the abaxial side repress PHAN function and support the development of the abaxial character on the abaxial side. Although the functional homolog of PHAN is not known in Arabidopsis, the expression pattern of the homolog could be the same as that of PHAN. If so, FIL may be the most probable candidate for the conceptual partner of the PHAN homolog that controls the development of the abaxial character, because the expression pattern of FIL is in line with this prediction. It is not clear, however, how the FIL protein would interfere with the function of the PHAN homolog. Recently the PINHEAD/TWILLE (PNH) gene of Arabidopsis was reported to encode a protein that has a homology with a mammalian translation factor, eIF2C, and with an Arabidopsis protein, ARGONAUTE 1 (AGO1), which is required for meristem development (Bohmert et al. 1998; Moussian et al. 1998; Lynn et al. 1999). PNH is speculated to be required for adaxial leaf development, because it was expressed in cells at the adaxial side of leaves and because the mutants homozygous for ago1 and heterozygous for pnh result in radialized rosette leaves and sepals (Lynn et al. 1999). The complementary expression patterns suggest that the expression of FIL and PNH is mutually repressed.
It was difficult to examine the effect of the ectopic expression of FIL in the development of inflorescence and floral organs, because most of the transgenic plants died after forming a few rosette leaves. Use of an inducible vector system will be required. An interesting question is whether the abaxial or the adaxial side is the default state of leaf tissues. Examination of the phb-1d mutant carrying 35S::FIL might hint at an answer.
Although 35S::FIL transgenic plants showed a change in the organization of the abaxial–adaxial axis in their leaves, the leaves of the fil mutant exhibited normal differentiation. In addition, no recessive mutants in Arabidopsis have been reported to have defects in the abaxial character of the lateral organs. These results suggest the presence of a gene family sharing a function supporting the growth of the abaxial side of lateral organs. Southern blots and a database search revealed that there are at least five genes homologous to FIL on the Arabidopsis genome. A similar gene family also exists in the rice genome, because there are at least two FIL homologs of osFIL1 and osFIL2. It will be necessary to screen knockout plants carrying mutations in these genes and examine their phenotypes.
Regulation of the meristematic activity
The major phenotype of the fil mutant is abnormality in formation and development of meristems; however, FIL is not expressed in the central zone of vegetative or inflorescence meristems, but in the primordia of lateral organs formed in the peripheral zone of the meristems. In the case of the floral meristem, expression of FIL was observed at a very early stage but was limited in the primordia of floral organs (Fig. 5B,C). The results indicate that the formation and development of the ‘mother’ meristem can be controlled by primordia or meristem newly formed in the peripheral region. Namely, FIL can indirectly affect the growth of the mother meristem by controlling the growth of organ primordia, or the ‘child’ meristem. This idea is supported by recent reports on the Antirrhinum mutant phan (Waites et al. 1998) and the Arabidopsis mutant phb-1d (MacConnell and Barton 1998).
Regulation of the basic processes of plant development
The spatial and temporal expression pattern of FIL in leaf primordia is exactly the same in floral organ primordia. Considering the traditional concept that floral organs are modified leaves, the identical expression pattern in the lateral organs indicates that FIL may have a common function to support basic processes of organ development.
Formation and development of the meristem and lateral organ primordia are fundamental processes to maintain plant form. Our results may lead to further understanding of the molecular mechanism underlying these processes.
Materials and methods
Plant materials and growth condition
The fil-1 mutant (former name, FL54) was isolated from Arabidopsis thaliana Landsberg (La) erecta by ethylmethane sulfonate mutagenesis (Komaki et al. 1988; Okada and Shimura 1994; Sawa et al. 1999). Seeds were sown on the surface of vermiculite in small pots and incubated at 4°C for 3 days. Plants were grown in a laboratory room under continuous illumination of 50–100 μE/m2 per sec at 22°C.
Gene cloning
DNA markers used for chromosome walking were based on RFLP between Arabidopsis ecotypes Landsberg and Columbia (Col) . SBP3 was obtained from sequence data kindly released by Howard Goodman (Massachusetts General Hospital, Boston). The SBP3 marker is a single-base change of A (Col) to G (La) at 7562 bp from the stop codon of FIL. The m336 RFLP marker was given by Elliot Meyerowitz (California Institute of Technology, Pasadena). Other information on markers ML, GBF3, and nga168 were obtained from the Arabidopsis thaliana Genome Center. The genomic sequence of the FIL region can be found on the TIGR web site (http://www.tigr.org/tdb/at/atgenome/atgenome.html).
A genomic library of Arabidopsis (ecotype Landsberg) in λDASH vector (Stratagene) was screened with a probe of a PCR-amplified DNA fragment corresponding to the FIL gene sequence at 535–612 bp. The same probe was used to screen a cDNA library constructed from poly(A) RNA isolated from young flowers with λZAPII (Stratagene) vectors. Five clones were isolated from a library of ∼1.3 × 106 independent clones.
Sequencing
DNA fragments were subcloned into the pBluescript II plasmid vector (Stratagene), and sequenced by using ABI Prism Dye Terminator Cycle Sequencing Ready Reaction kits and an ABI Prism 310 Genetic analyzer from Perkin Elmer.
In situ hybridization
In situ hybridization test was performed as described (Drews et al. 1991). After hybridization, the sections were washed at room temperature for 40 min in 2× SSPE and 5 mm DTT, and additionally at 57°C for 20 min in 0.1× SSPE and 1 mm DTT. In the genomic Southern analysis, the FIL-related genes were not detected under the above washing condition (data not shown). Antisense and sense probes used for in situ hybridization were prepared by subcloning a cDNA fragment corresponding to the sequence at −138 to 661 bp into Bluescript SK( and KS( vectors (Stratagene). The obtained plasmids were named pFILA and pFILS, respectively. Labeled probes using [35S]UTP were prepared with T3 RNA polymerase using pFILA DNA linearized with XhoI or pFILS DNA linearized with XbaI as templates for antisense or sense probes, respectively.
Purification of the FIL protein
To express the FIL protein in bacteria, an expression plasmid was constructed by use of pET28a (Novagen, Inc.). The FIL gene was amplified by the PCR, and the obtained fragment was ligated into pET28a. The sequence of the FIL gene in the expression plasmid was confirmed by cycle sequencing. Expression of the FIL protein was induced in Escherichia coli BL21 (DE3) cells harboring the expression plasmid by the addition of IPTG. Cultivation of the E. coli transformants was carried out at 30°C. When the absorbance of the culture at 600 nm reached 0.8, 1 mm IPTG and 10 μm zinc acetate were added to the culture media, and the cultivation was then continued for an additional 5 hr. Purification procedure was carried out at 4°C. A 1-liter culture of E. coli cells were suspended in 50 ml of 10 mm Tris-HCl at pH 7.5, 0.1 mm EDTA, 5 mm β-mercaptoethanol, sonicated on ice for 2 min, and centrifuged at 15,000 rpm (27,000g) for 30 min. The supernatant was supplemented with 10 mm imidazole and then applied to a HiTrap chelating column with Ni2+ ions (Pharmacia Biotec). After the column had been washed with a solution containing 200 mm imidazole, 20 mm Na phosphate at pH 7.5, 0.5 m NaCl, and 5 mm β-mercaptoethanol, the FIL protein was eluted with a solution containing 500 mm imidazole, 0.5 m NaCl, 20 mm phosphate buffer at pH 7.5, and 5 mm β-mercaptoethanol. The elutant was dialyzed against a buffer containing 20 mm Tris-HCl (pH 9.0), 0.1 mm EDTA, 0.5 m NaCl, 1 mm DTT, and 20% glycerol. Prior to the dialysis, the buffer solution was passed through Chelex 100 resin (Bio-Rad) to remove the metal contaminants. The purity of the protein was analyzed by SDS-PAGE.
ICP spectroscopic analysis of the FIL protein
The content of zinc ion was analyzed by ICP spectroscopy. The concentration of the FIL protein was estimated from the UV absorption at 280 nm. The A280 value at 0.1% of 0.63 for FIL with a molecular weight of 25,779 was calculated by using 1576 m/cm for tyrosine (×7) and 5225 m/cm for tryptophan (×1) at 280 nm. ICP were measured and analyzed with an Optima 3000 (Perkin Elmer) spectrometer.
Construction of transgenes
The FIL cDNA fragment was ligated into a plasmid pTH2 (Chiu et al. 1996) downstream of the CaMV 35S promoter (Odell et al. 1985), and this plasmid was digested with EcoRI and BamHI to release the FIL cDNA connected to the CaMV 35S promoter. This fragment was ligated into a binary vector, pARK5mcs (a gift from Meiji-Seika, Kaisha, Ltd, Tokyo, Japan). We transformed Arabidopsis Columbia wild-type by the vacuum infiltration procedure with Agrobacterium strain C58. Transgenic plants were selected on the agar medium containing a 50 μg/ml kanamycin.
Scanning electron microscopy
For SEM, young leaves or vegetative shoot apexes of Columbia wild-type or 35S::FIL transgenic plants were fixed in carnoa liquid mixture (isoamyl acetate/ethanol = 1:3), and the samples were rinsed twice with ethanol. The samples were treated with 2% tannic acid in ethanol for 10 hr, rinsed twice with ethanol, and incubated in an ethanol/isoamyl acetate (=1:3) mixture for ∼15 min. In the last step, the samples were immersed in isoamyl acetate for 15 min and dried in liquid carbon dioxide. Individual flowers were removed from inflorescences and mounted on SEM stubs. The mounted specimens were coated with gold and observed with a scanning electron microscope (Nippon Denshi, type JSM-T20) at an accelerating voltage of 20 kV. The images were photographed on Fuji Neopan 120 film.
Acknowledgments
We thank Yoshiro Shimura for warm encouragement. We also thank Sumie Ishiguro, Toshiro Ito, Takuji Wada, Tokitaka Oyama, Miho Yamada, John Bowman, Kosuke Morikawa, and Takashi Araki for valuable discussions; Howard Goodman, Elliot Meyerowitz, Daisuke Shibata, Erick Richards, TIGR Arabidopsis thaliana database, Ohio State University Arabidopsis Biological Resource Center, and the University of Pennsylvania Arabidopsis thaliana Genome Center for providing us mapping data, TAC clones, sequence data or molecular markers for gene cloning. This work was supported by a grant (no. 08408031) from the program Grants-in-Aid for Scientific Research (A) and grants (06278103 and 10182101) from the program Grant-in-Aid for Scientific Research on Priority Areas of the Japanese Ministry of Education, Science, Culture and Sports, by funds from the Human Frontier Science Program, from Mitsubishi Foundation, and the Joint Studies Program for Advanced Studies from the Science and Technology Agency of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kiyo@ok-lab.bot.kyoto-u.ac.jp; FAX 81-75-753-4257.
References
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J, Alvarez J, Weigel D, Meyerowitz EM. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743. [Google Scholar]
- Chiu W-I, Niwa Y, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL. A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell. 1997;88:685–694. doi: 10.1016/s0092-8674(00)81911-x. [DOI] [PubMed] [Google Scholar]
- Drews GN, Bowman JL, Meyerowitz EM. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- Grasser KD. Plant chromosomal high mobility group (HMG) proteins. Plant J. 1995;7:185–192. doi: 10.1046/j.1365-313x.1995.7020185.x. [DOI] [PubMed] [Google Scholar]
- ————— HMG1 and HU proteins: Architectural elements in plant chromatin. Trends Plant Sci. 1998;3:260–265. [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF. Molecular basis of the cauliflower phenotype in Arabidopsis. Science. 1995;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- Komaki K, Okada K, Nishino E, Shimura Y. Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development. 1988;104:195–203. [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK. The PINHEAD/TWILLE gene acts pleiotropically in Arabidopsis development and has oberlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- MacConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jurgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate furing Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell JT, Nagy F, Chua N-H. Identification of DNA sequences required for the activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Genetic analyses of signalling in flower development using Arabidopsis. Plant Mol Biol. 1994;26:1357–1377. doi: 10.1007/BF00016480. [DOI] [PubMed] [Google Scholar]
- Parisi MA, Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- Reeves R, Bustin M. High-mobility-group chromosomal proteins: Architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- Sawa S, Ito T, Shimura Y, Okada K. FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. Plant Cell. 1999;11:69–86. doi: 10.1105/tpc.11.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuji H. Zinc-finger transcription factors in plants. Cell Mol Life Sci. 1998;54:582–596. doi: 10.1007/s000180050186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124:645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- Timmermans MCP, Schults NP, Jankovsky JP, Nelson T. Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development. 1998;125:2813–2823. doi: 10.1242/dev.125.15.2813. [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RT. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci. 1997;94:7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Waites R, Hudson A. phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development. 1995;121:2143–2154. [Google Scholar]
- Waites R, Selvadurai RN, Oliver IR, Hudson A. The PHANTASTICA gene encode a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell. 1998;93:779–789. doi: 10.1016/s0092-8674(00)81439-7. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X,Y sex reversal revealed from the three- dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- Zhang B, Chen W, Foley RC, Buttner M, Singh KB. Interaction between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell. 1995;7:2241–2252. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]