Abstract
Background
Most bacteria can use various compounds as carbon sources. These carbon sources can be either co-metabolized or sequentially metabolized, where the latter phenomenon typically occurs as catabolite repression. From the practical application point of view of utilizing lignocellulose for the production of biofuels etc., it is strongly desirable to ferment all sugars obtained by hydrolysis from lignocellulosic materials, where simultaneous consumption of sugars would benefit the formation of bioproducts. However, most organisms consume glucose prior to consumption of other carbon sources, and exhibit diauxic growth. It has been shown by fermentation experiments that simultaneous consumption of sugars can be attained by ptsG, mgsA mutants etc., but its mechanism has not been well understood. It is strongly desirable to understand the mechanism of metabolic regulation for catabolite regulation to improve the performance of fermentation.
Results
In order to make clear the catabolic regulation mechanism, several continuous cultures were conducted at different dilution rates of 0.2, 0.4, 0.6 and 0.7 h-1 using wild type Escherichia coli. The result indicates that the transcript levels of global regulators such as crp, cra, mlc and rpoS decreased, while those of fadR, iclR, soxR/S increased as the dilution rate increased. These affected the metabolic pathway genes, which in turn affected fermentation result where the specific glucose uptake rate, the specific acetate formation rate, and the specific CO2 evolution rate (CER) were increased as the dilution rate was increased. This was confirmed by the 13C-flux analysis. In order to make clear the catabolite regulation, the effect of crp gene knockout (Δcrp) and crp enhancement (crp+) as well as mlc, mgsA, pgi and ptsG gene knockout on the metabolism was then investigated by the continuous culture at the dilution rate of 0.2 h-1 and by some batch cultures. In the case of Δcrp (and also Δmlc) mutant, TCA cycle and glyoxylate were repressed, which caused acetate accumulation. In the case of crp+ mutant, glycolysis, TCA cycle, and gluconeogenesis were activated, and simultaneous consumption of multiple carbon sources can be attained, but the glucose consumption rate became less due to repression of ptsG and ptsH by the activation of Mlc. Simultaneous consumption of multiple carbon sources could be attained by mgsA, pgi, and ptsG mutants due to increase in crp as well as cyaA, while glucose consumption rate became lower.
Conclusions
The transcriptional catabolite regulation mechanism was made clear for the wild type E. coli, and its crp, mlc, ptsG, pgi, and mgsA gene knockout mutants. The results indicate that catabolite repression can be relaxed and crp as well as cyaA can be increased by crp+, mgsA, pgi, and ptsG mutants, and thus simultaneous consumption of multiple carbon sources including glucose can be made, whereas the glucose uptake rate became lower as compared to wild type due to inactivation of ptsG in all the mutants considered.
Background
It is quite important to understand how the culture environment affects the cell metabolism. Among the culture environments, carbon source is by far important in practice. In particular, carbon catabolite repression has been paid recent attention [1], where most bacteria selectively consume substrates from a mixture of different carbon sources, and exhibit diauxic growth. Namely, most organisms consume glucose prior to consumption of other carbon sources. From the application point of view of utilizing lignocellulose for the production of biofuels etc., it is strongly desirable to ferment all sugars obtained by hydrolysis from lignocellulosic materials simultaneously [2]. In particular, simultaneous consumption of sugars would benefit the formation of bioproducts. Several attempts have, therefore, been made in the past, where ptsG was mutated for ethanol production [3] and for lactate production [4] from a mixture of glucose and xylose. Noting that the glucose consumption rate becomes low by a ptsG mutation and pts mutation [5], the gal regulon genes, which encode non-PTS transporter, were enhanced [6,7]. More recently, mgsA gene knockout which encodes the initial enzyme from DHAP to methylglyoxal pathway was considered for the simultaneous consumption of multiple sugars [8], but it is not explained on its mechanisms. Moreover, it has been shown that cAMP increases for pyk knockout mutant [9], but this may not be a significant contribution for the simultaneous consumption of a mixture of sugars, since the increase in cAMP is limited. Yet, another idea of co-fermentation strategy has been proposed, where this process uses two substrate-selective strains of E. coli, one of which is unable to consume glucose and the one which is unable to consume xylose for lactate production [10]. However, it may be difficult to analyze the mixed culture, since one cannot discriminate two strains, and one population may washout during continuous culture.
In the present study, we attempted to clarify the catabolic regulation mechanism of E. coli based on fermentation characteristics and selected gene transcript levels. In order to understand the catabolic regulation, the recognition and adjustment mechanisms must be understood in view of the relationships between global regulators and the metabolic pathway genes. In the catabolic regulation, cAMP-Crp complex plays an important role. The center for this regulatory network is the phosphoenolpyruvate (PEP): carbohydrate phosphotransferase systems (PTSs). These systems are involved in both transport and phosphorylation of carbohydrates for the regulation of the main metabolic pathways. The PTS in E. coli consists of two cytoplasmic proteins such as EI (enzyme I) and HPr (histidine-containing protein), as well as carbohydrate-specific EII (enzyme II) complexes. The unphosphorylated EIIAGlc inhibits the uptake of other non-PTS carbohydrates by a so-called inducer exclusion. Phosphorylated EIIAGlc activates the adenylate cyclase (Cya) and leads to an increase in the intracellular cAMP level, which then combines with Crp forming cAMP-Crp complex, and controls certain metabolic pathway genes [11-13].
In order to increase understanding of the catabolic regulation phenomenon, several continuous cultures were conducted for wild type E. coli at different dilution rates in the present study. Moreover, the effects of crp gene knockout and crp enhancement (crp+) on the metabolic regulation were also investigated. Since Crp activates ptsG, while Mlc represses it, we also investigated the effect of mlc gene knockout in addition to crp knockout and enhancement on the metabolism. In the present investigation, we also considered mgsA, pgi and ptsG mutants as well to clarify the mechanism of catabolite repression. Finally, the effects of those mutations on the simultaneous consumption of a mixture of carbon sources such as glucose and xylose were investigated.
Results
Effect of dilution rate on the metabolic regulation in wild type E. coli
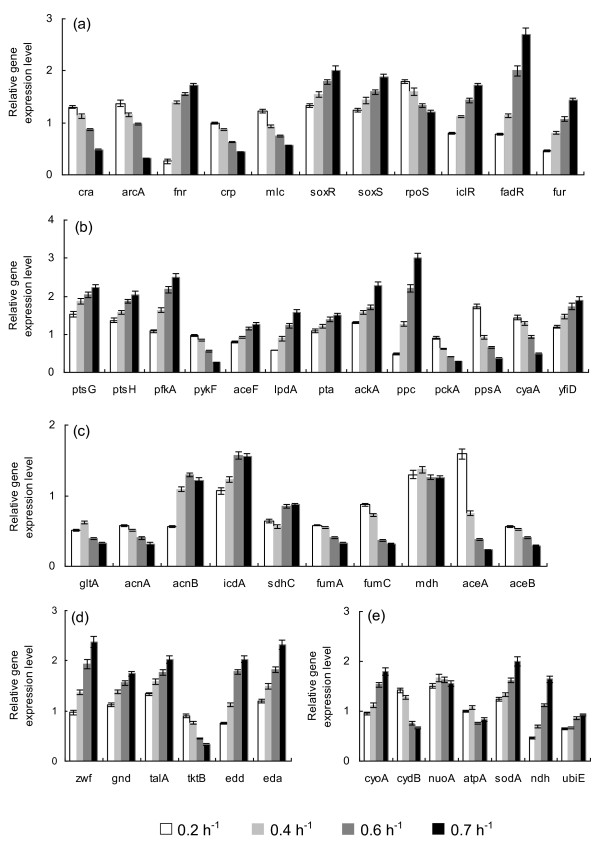
Table 1 shows the fermentation characteristics of the wild type E. coli for the continuous culture at different dilution rates, where it indicates that the specific glucose uptake rate, acetate production rate, and the specific CO2 evolution rate (CER) were increased as the dilution rate was increased. Figure 1 shows the effect of the dilution rate (the specific growth rate) on gene transcript levels, where it indicates that in accordance with the fermentation data of the increased specific glucose consumption rate, the transcript levels of ptsG, ptsH, and pfkA were increased as the dilution rate increased, where cra transcript level decreased and crp as well as mlc decreased accordingly (Additional file 1). The decrease in crp is also coincident with the decrease in cyaA which encodes Cya. The transcript levels of zwf, gnd, edd, and eda were increased as the dilution rate increased in accordance with the decrease in cra. The transcript level of ppc increased while pckA decreased as the dilution rate increased as evidenced by 13C-metabolic flux result (Additional file 2). Moreover, the transcript levels of fadR and iclR increased, and aceA and aceB decreased as the dilution rate increased. In accordance with the increase in the specific acetate production rate, the transcript levels of pta and ackA increased. TCA cycle genes such as gltA, acnA, fumA, C decreased, while acnB, icdA and lpdA increased (except the case of dilution rate at 0.7 h-1) as the dilution rate increased. In accordance with the increase in soxR/S transcript levels, zwf and sodA increased. In accordance with the decrease in the transcript level of arcA, the transcript level of cyoA increased, while cydB decreased as the dilution rate increased, where the latter phenomenon also coincided with the decrease in cra transcript level (Additional file 1). Further observation indicates that in accordance with the decrease in rpoS transcript level, tktB, acnA, fumC decreased, while fur increased as the dilution rate increased in accordance with the increase in soxR/S.
Table 1.
Effect of the dilution rate on fermentation characteristics of wild type E. coli
| Dilution rate (h-1) | Biomass conc. (g/L) | Glucose conc. (g/L) | Glucose uptake rates (mmol/g/h) | Acetate formation rates (mmol/g/h) | Biomass yield (g/g) | CER (mmol/g/h) | Carbon recovery |
|---|---|---|---|---|---|---|---|
| 0.2 | 1.45 ± 0.06 | ND* | 3.07 ± 0.13 | ND* | 0.37 ± 0.015 | 9.15 | 94% |
| 0.4 | 1.87 ± 0.09 | ND* | 4.75 ± 0.23 | 0.01 | 0.47 ± 0.023 | 11.61 | 98% |
| 0.6 | 2.0 ± 0.09 | ND* | 6.67 ± 0.3 | 0.88 ± 0.04 | 0.5 ± 0.023 | 13.17 | 98% |
| 0.7 | 1.93 ± 0.08 | ND* | 8.05 ± 0.34 | 1.33 ± 0.06 | 0.48 ± 0.02 | 15.83 | 97% |
ND*: not detectable, where glucose detectable limit was 0.038 g/l.
Figure 1.
The effect of dilution rate on the gene transcript levels of wild type E. coli. (a) Global regulator genes; (b) PTS, glycolysis, anaplerotic pathway, cyaA and yfiD genes; (c) TCA and glyoxylate pathway genes; (d) PP pathway genes; (e) Respiratory chain genes.
Additional file 2 shows the comparison of the metabolic flux distributions of E. coli at the dilution rates of 0.2 h-1, 0.4 h-1, 0.6 h-1 and 0.7 h-1, where it indicates that the metabolic pathway fluxes such as glycolysis, PP pathway, and anaplerotic pathway fluxes increased as the dilution rate increased, where those results are consistent with the gene transcript levels as stated above. Based on the flux result, the specific ATP production rate and the specific NADPH formation rate computed using Eqs. (1) and (2) as stated in Methods section are plotted with respect to the dilution rate in Additional file 3, which indicates that these are linearly correlated with the specific growth rate.
Effect of crp gene mutation on the metabolism
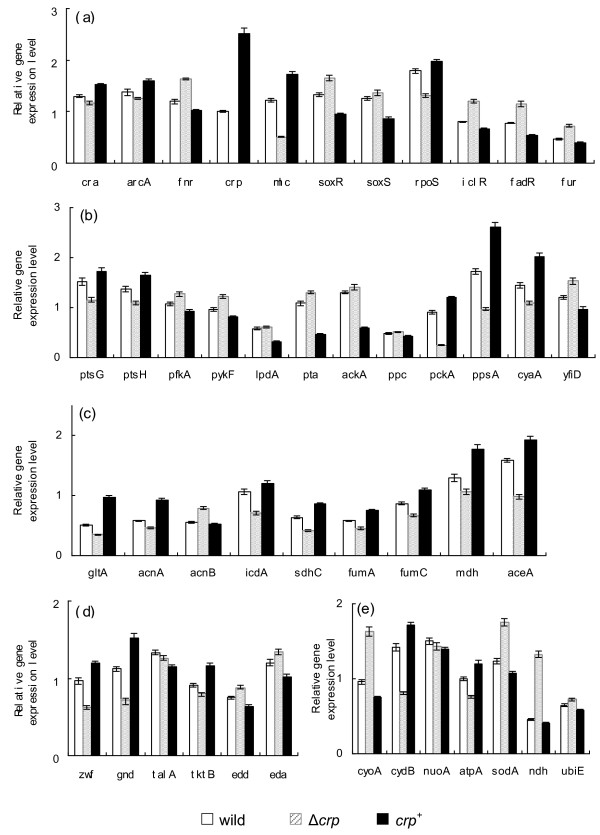
Table 2 shows the effect of crp gene knockout and crp enhancement (crp+) on the fermentation characteristics at the dilution rate of 0.2 h-1, where it indicates that the specific glucose uptake rate was lower and the cell concentration was higher as compared to the wild type, and the specific acetate production rate was higher for crp knockout mutant. In the case of crp+ mutant, the fermentation characteristics were similar to the wild type by considering the statistical significance. Figure 2a shows the transcript levels of crp knockout mutant and crp+ mutant as compared to wild type, where it indicates that mlc was down-regulated (P < 0.01) in crp knockout mutant, and up-regulated (P < 0.01) in crp+ mutant. Figure 2b indicates that ptsG and ptsH transcript levels changed in a similar fashion as mlc and crp, which corresponds to the lower glucose uptake rate for crp knockout mutant and similar to the wild type for crp+ mutant (Table 2). Figure 2d also indicates that acnA, gltA, fumA, mdh, pckA, sdhC all changed in a similar fashion as crp (Additional file 1). The cydB transcript level changed in a similar fashion as arcA, while cyoA gene changed in a reverse fashion (Additional file 1). The sodA and fur changed in a similar fashion as soxR/S. The tktB and fumC changed in a similar fashion as rpoS (Additional file 1). The changing patterns of aceA, icdA and pckA transcript levels were similar as cra, while reverse patterns may be seen for fadR and iclR.
Table 2.
Effect of the specific gene mutation on the fermentation characteristics at the dilution rate of 0.2 h-1
| Strains | Biomass conc. (g/L) | Glucose conc. (g/L) | Glucose uptake rate (mmol/g/h) | Acetate formation rate (mmol/g/h) | Biomass yield (g/g) |
|---|---|---|---|---|---|
| BW25113 | 1.45 ± 0.06 | ND* | 3.07 ± 0.13 | ND* | 0.37 ± 0.015 |
| Δcrp | 1.57 ± 0.07 | ND* | 2.83 ± 0.13 | 0.35 ± 0.018 | 0.39 ± 0.018 |
| crp+ | 1.42 ± 0.07 | ND* | 3.12 ± 0.14 | ND* | 0.36 ± 0.018 |
| Δpgi | 1.6 ± 0.08 | ND* | 2.77 ± 0.14 | ND* | 0.4 ± 0.021 |
| Δmlc | 1.65 ± 0.08 | ND* | 2.69 ± 0.13 | ND* | 0.41 ± 0.019 |
| ΔmgsA | 1.55 ± 0.07 | ND* | 2.87 ± 0.14 | 0.1 ± 0.005 | 0.39 ± 0.018 |
ND*: not detectable, where glucose detectable limit was 0.038 g/l.
Figure 2.
Comparison of gene transcript levels of the wild type, crp knockout mutant, and crp+ mutant. (a) Global regulator genes; (b) PTS, glycolysis, anaplerotic pathway, cyaA and yfiD genes; (c) TCA and glyoxylate pathway genes; (d) PP pathway genes; (e) Respiratory chain genes. 1.
Effect of mlc gene knockout on the metabolism
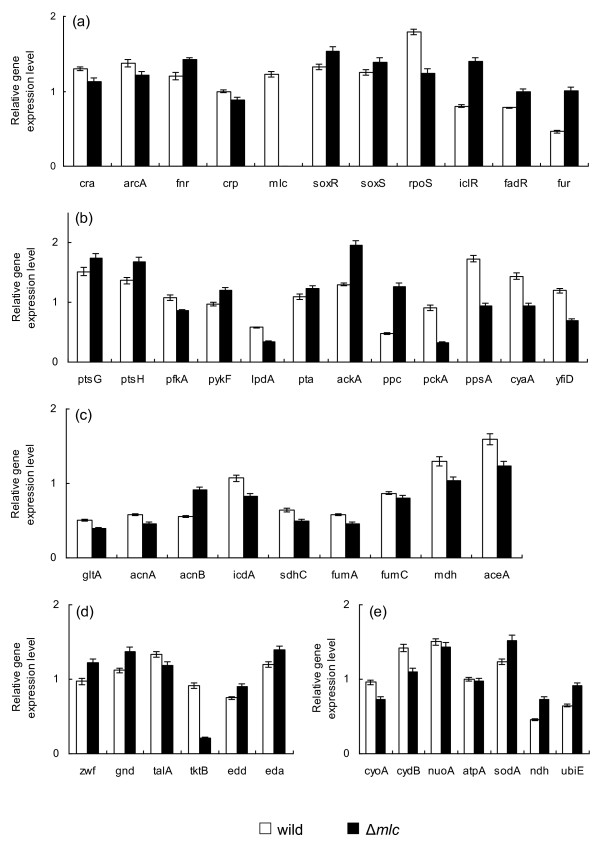
Table 2 also shows the fermentation characteristics of mlc mutant at the dilution rate of 0.2 h-1, where it indicates that the specific glucose uptake rate was lower than wild type. Figure 3 shows the transcript levels of mlc mutant as compared to those of the wild type, where the transcript level of crp decreased (P < 0.05) as compared to the wild type. Due to mlc gene knockout, the transcript levels of ptsG and ptsH increased (P < 0.05 and P < 0.01, respectively), which may be also coincided with the decrease in cra transcript level (P < 0.01). However, this does not cause the specific glucose consumption rate to increase, which may be partly due to decrease in pfkA gene transcript level (P < 0.01), while pykF increased (P < 0.01). The transcript level of rpoS decreased (P < 0.01) and tktB, fumC were down-regulated (P < 0.01 and P < 0.05, respectively) accordingly (Additional file 1). The transcript levels of soxR/S increased (P < 0.01 and P < 0.05, respectively), and sodA increased (P < 0.01) as compared to the wild type.
Figure 3.
Comparison of gene transcript levels of the wild type and mlc knockout mutant. (a) Global regulator genes; (b) PTS, glycolysis, anaplerotic pathway, cyaA and yfiD genes; (c) TCA and glyoxylate pathway genes; (d) PP pathway genes; (e) Respiratory chain genes.
Effect of mgsA and pgi gene knockout on the metabolism
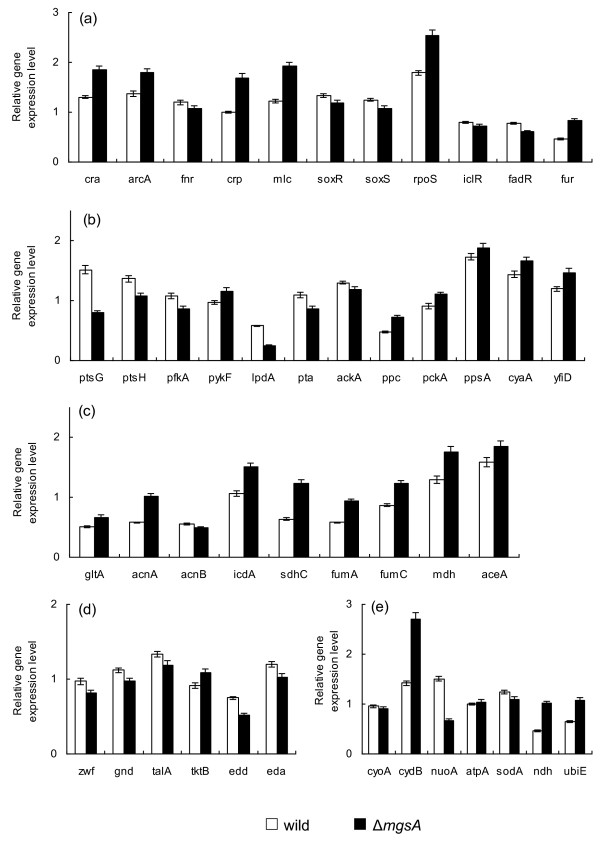
Table 2 also shows the effects of mgsA and pgi gene knockout on the fermentation characteristics, where it indicates that the glucose consumption rate decreased, and the cell yield increased. Figure 4 shows the transcript levels of mgsA mutant as compared to wild type, where it indicates that the transcript levels of cyaA and crp as well as mlc increased (P < 0.05, P < 0.01 and P < 0.01, respectively). The cra transcript level increased (P < 0.01) and thus the transcript levels of ptsG, ptsH and pfkA decreased (P < 0.01 for all genes), which is consistent with the decreased glucose consumption rate, while pckA, ppsA and aceA were all increased (P < 0.01, P < 0.05, P < 0.05, respectively). The increase in aceA transcript level (P < 0.05) coincided with the decrease in fadR and iclR transcript levels (P < 0.05 and P < 0.01, respectively).
Figure 4.
Comparison of gene transcript levels of the wild type and mgsA knockout mutant. (a) Global regulator genes; (b) PTS, glycolysis, anaplerotic pathway, cyaA and yfiD genes; (c) TCA and glyoxylate pathway genes; (d) PP pathway genes; (e) Respiratory chain genes.
Fermentation characteristics of using multiple carbon sources
The modulation of catabolite regulation may be applied for the simultaneous consumption of multiple carbon sources originated from the hydrolysis of lignocellulose. Additional file 4a shows the batch cultivation result for wild type where the mixture of glucose and xylose was used as a carbon source, where it indicates that glucose was first consumed, while xylose was consumed after glucose was depleted due to catabolite repression in the wild type. Additional file 4b shows the comparison of batch cultures of wild type and crp+ mutant, where it indicates that although simultaneous consumption of both sugars could be attained, the substrate uptake rate became lower, which may be due to repression of pts by increased Mlc caused by the increased crp gene transcript level. Additional file 5 shows the comparison of wild type and its crp+ mutant for the continuous culture, which indicates that xylose tends to accumulate as the dilution rate increases for wild type (Additional file 5a) while both glucose and xylose concentrations are similar for crp+ mutant (Additional file 5b). The decreased glucose consumption rate for crp+ mutant as observed in the batch culture may have caused lower cell and acetate concentrations in the continuous culture (Additional file 5). Additional file 6 shows the continuous culture of ptsG mutant for the case of using a mixture of glucose and xylose. Although simultaneous consumption of both sugars can be attained at lower dilution, the glucose consumption rate became lower, and glucose concentration increased at higher dilution rate for ptsG mutant as illustrated by additional file 6. The similar phenomenon was observed for pgi mutant, where glucose consumption rate became lower (data not shown).
Discussion
The effect of dilution rate (the specific growth rate) on gene transcript levels (Figure 1) and fermentation characteristics (Table 1) may be explained as follows: As the dilution rate increased, the glucose concentration increased (though not detectable in Table 1), which caused cra transcript level to be decreased (Figure 1a). The crp transcript level also decreased as the dilution rate was increased, which may be due to decrease in phosphorylated EIIAGlc, caused by higher glucose concentration. This deactivated Cya (as implied by cyaA in Figure 1b), which in turn caused decrease in cAMP concentration, and thus cAMP-Crp or crp transcript level decreased (Figure 1a). Note that cAMP level starts to increase when glucose concentration becomes less than about 0.3 mM [14]. The increases in fadR and iclR (Figure 1a) also coincided with the increase in the glucose concentration as the dilution rate was increased. The decrease of rpoS (Figure 1a) may be also explained as less nutrient stress as the dilution rate was increased. The decrease of arcA transcript level and increases of soxR/S transcript levels may be due to higher dissolved oxygen concentration caused by lower cell concentration at higher dilution rate (Table 1).
Referring to Figure 5 and Additional file 1, some of the metabolic pathway transcript levels may be explained with respect to the change in global regulators. The decreases of aceA and aceB transcript levels coincided with the increase of those of iclR and fadR as the dilution rate was increased. This also corresponds to the flux result (Additional file 2). The decreases of the transcript levels of aceA, B, and pckA, ppsA, acnA (Figure 1 b, c) coincide with the decrease in the cra transcript level (Figure 1a), which also caused increase in eda, edd, pfkA, ptsH transcript levels. The increase of cyoA and decrease of cydB coincide with the decrease in arcA as the dilution rate was increased, where the decrease in cydB also coincides with the decrease in cra. Note that ppc transcript level increased as the dilution rate increased as confirmed by the flux result (Additional file 2), which may be due to increased F1,6BP. Although lpdA and aceF transcript levels increased as the dilution rate was increased, TCA cycle gene transcript levels such as gltA, acnA, fumA, fumC tended to decrease, while acnB and icdA (sdhC) transcript levels tended to increase. The increase in lpdA transcript level may be due to decrease in crp, where cAMP-Crp repressed such gene expression. The decreases of acnA and fumC transcript levels may be caused by the decrease in rpoS (Additional file 1). It has been shown that acnB is dominant for the exponential growth phase, while acnA is dominant at the stationary phase [15,16], which indicates that the continuous culture at higher dilution rate mimics the cell growth phase in the batch culture, while it mimics the stationary phase at low dilution rate. The increases in soxR/S caused zwf and sodA to be increased, whereas acnA and fumC decreased. The latter may be due to decrease in rpoS transcript level (Additional file 1). The increase in yfiD may be due to increase in fnr transcript level, though Fnr may be inactive form under aerobic condition. Upon complex formation of cAMP-Crp, it activates genes encoding the glucose PTS system, the TCA cycle and gluconeogenesis (Additional file 1). Although crp transcript level decreased as the dilution rate was increased (Figure 1a) as stated above, ptsG and ptsH transcript level increased (Figure 1b). This may be caused by the decrease in cra, where pfkA also increased.
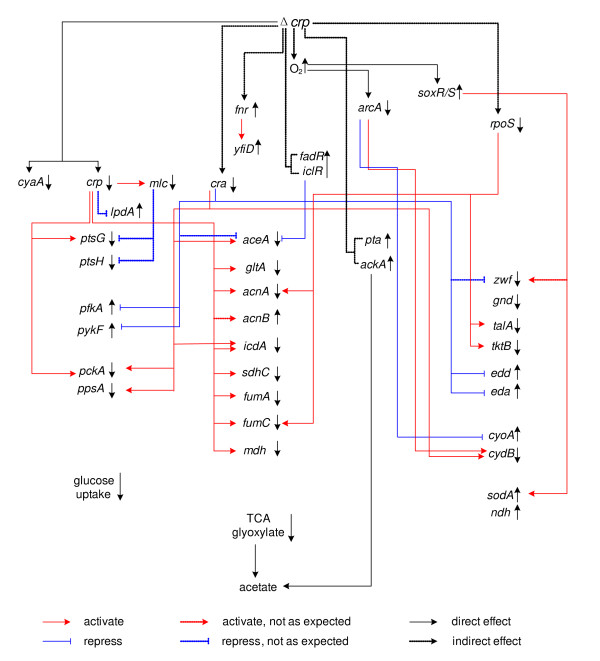
Figure 5.
The overall regulation of Δcrp mutant.
As stated above in the result section, the specific glucose consumption rate decreased in the crp knockout mutant, as compared to the wild type, being almost the same in crp+ mutant. The ptsG gene is activated by Crp, but also repressed by Mlc. Note that, the changing pattern of crp transcript level (Figure 2a) coincides with that of mlc (Figure 2a) as well as cyaA. It has been reported that there is a Crp binding region in the promoter region of mlc gene [17].
In the case of crp knockout mutant, the decrease in glucose consumption rate may be caused by down regulations of ptsG and ptsH (P < 0.01 and P < 0.01, respectively), whereas pfkA and pykF were up-regulated (P < 0.01 and P < 0.01, respectively). The former may be caused by crp knockout, while the latter may be due to down-regulation of cra (P < 0.01) (Figure 2a), where mlc might not be dominant. The crp knockout caused TCA cycle genes such as gltA, acnA, sdhC, fumA, and mdh to be down regulated (P < 0.01, P < 0.01, P < 0.01, P < 0.05 and P < 0.01, respectively). Figure 2a shows cra to be down-regulated (P < 0.01), which may be caused by higher glucose concentration, though it is less than detectable level. The down regulation of cra (P < 0.01) also caused icdA gene as well as gluconegogenic pathway genes pckA and ppsA to be down regulated (P < 0.01, P < 0.05 and P < 0.01, respectively) (Figure 2a, b, c). The decrease in glyoxylate pathway gene aceA (P < 0.01) (Figure 2c) may be caused by the increases of fadR and iclR (P < 0.01 and P < 0.01, respectively), which may be due to higher glucose concentration. The decreased activities of TCA cycle and glyoxylate pathway may have caused acetate to accumulate, where this is also reflected by the up-regulation of pta and ackA transcript levels (P < 0.01 and P < 0.05, respectively) (Figure 2b). The batch culture of Δcrp mutant indicates that the acetate formed during cell growth phase, could not be consumed during stationary phase (Additional file 7). The decreased transcript levels of acnA, fumC, and tktB (P < 0.01, P < 0.01 and P < 0.01, respectively) may also be due to down regulation of rpoS (P < 0.01), which may be due to less nutrient stress caused by the increase of glucose concentration. The sodA gene transcript level increased (P < 0.01) (Figure 2e), which is consistent with up-regulation of soxR/S (P < 0.01 and P < 0.05, respectively) (Figure 2a), which might be due to increase in oxygen concentration. The decrease of arcA (P < 0.05) (Figure 2a) is also reflected by this hypothesis, whereas its effect on TCA cycle genes may be minor and not dominant. The overall regulation mechanism is illustrated in Figure 5.
In the case of crp+ mutant, the ptsG and ptsH transcript levels were increased (P < 0.05 and P < 0.01, respectively), whereas pfkA and pykF were decreased (P < 0.01 and P < 0.01, respectively). It should be noted that crp gene enhancement caused increase in mlc gene (P < 0.01) (Figure 2a), and thus Mlc might have repressed ptsG and ptsH. This seems to be dependent on the culture condition. This might happen when glucose concentration is higher as will be shown for the batch culture. Different from the case of Δcrp mutant, TCA cycle genes and glyoxyalte pathway gene as well as gluconeogenic pathway genes were up-regulated. The activation of TCA cycle may have caused the decrease in cell yield.
In the case of mlc mutant (Figure 3), crp as well as cyaA transcript levels were also decreased (P < 0.05 and P < 0.01, respectively). The decreases of crp (P < 0.05) and cra (P < 0.01) both caused glucose uptake and glycolysis pathway genes (except pfkA) to be up-regulated, while gluconeogensis, TCA cycle and glyoxylate pathway genes were decreased. Although not reflected in the continuous culture (Table 2), the above phenomenon may cause more acetate formation as also implied by the up-regulations of pta and ackA genes (P < 0.05 and P < 0.01, respectively) (Figure 3b). In fact, this happened in the batch culture, and the acetate formed during the cell growth phase could not be consumed during the stationary phase (Additional file 8).
In the case of mgsA mutant (Figure 4), PTS may be repressed as shown by the down regulations of ptsG and ptsH as well as pfkA transcript level (P < 0.01 for all genes), which may be partly due to accumulation of G6P caused by mgsA gene knockout. A similar phenomenon may be seen also for pgi and pfk gene knockout (data not shown). The increase in G6P may cause mRNA of ptsG to be unstable [18], and the phosphorylation of glucose at EIICB by EIIAGlc-P might be disrupted, and this have caused crp (and therefore mlc) transcript level as well as cyaA and cra to be up-regulated (P < 0.01, (P < 0.01), P < 0.05 and P < 0.01, respectively), which in turn reduced the acetate formation as implied by the down-regulations of pta and ackA transcript levels (P < 0.01 and P < 0.05, respectively). The increases in cydB (P < 0.01) may be due to the increased transcript levels of cra and arcA (P < 0.01 and P < 0.01, respectively). The transcript level of rpoS is also increased (P < 0.01) for mgsA mutant, which might be due to stress imposed on catabolism.
The fermentation results of using a mixture of glucose and xylose indicate that simultaneous consumption of multiple carbon sources can be attatined by crp+, mgsA pgi (pfkA), and ptsG mutants. The mechanism for this phenomenon is due to increase in crp transcript level, which may be explained as the deficient ptsG expression for mgsA, pgi (pfkA) and ptsG mutants. This means that the glucose uptake rate may be decreased due to less activity of EIICBGlc encoded by ptsG, while Mlc represses the ptsG and ptsH for the case of crp+ mutant. In the case of deficient EIICBGlc, glucose transport may be made by GalP and MglBAC, and it is phosphorylated by ATP and converted to G6P, but this process may be less efficient than PTS. The increase in crp transcript level for those mutants may have caused TCA cycle as well as glyoxylate pathway to be activated, and thus less acetate may be formed.
In the case of pgi mutant, the glucose consumption rate became low, resulting in the significantly low cell growth rate [19,20]. The mgsA mutant may have caused G6P to be increased but its extent may be low as compared to pgi (pfkA) mutant, and thus the glucose uptake rate may retain higher but less than wild type. Although pyk mutant may cause G6P concentration to be increased [20,21], and thus cAMP tends to increase [9], this effect is minor and may not be able to attain simultaneous consumption of multiple sugars (data not shown).
Conclusion
The transcriptional catabolite regulation mechanism was made clear for wild type E. coli, and its crp, mlc, ptsG, pgi, and mgsA gene knockout mutants. The results indicate that catabolite repression can be relaxed, and crp as well as cyaA transcript levels can be increased by crp+, mgsA, pgi, and ptsG mutants, and thus simultaneous consumption of multiple carbon sources including glucose can be made, whereas the glucose uptake rate became lower as compared to wild type due to inactivation of ptsG in all the mutants considered.
Methods
Strains used and media composition
The strains used were E. coli BW25113 (F- λ- rph-1 ΔaraBADAH33 lacIq ΔlacZWJ16 rrnBT14 ΔrhaBADLD78 hsdR514) and its single gene knockout mutants lacking crp (JW5702) and mlc (JW1586), pgi (JW3985), mgsA (JW5129) and ptsG (JW1087), where those were obtained from Keio collection [22]. The crp enhanced (crp+) strain was constructed based on the method of Postai et al. [23] by introducing the mutation responsible for Crp phenotype which lies in the hinge region between the cAMP- and DNA-binding domains. The amino acid substitutions from Gly to Ser at 122 position altered the allosteric nature of Crp, which corresponds to crp*2 in Aiba et al. [24]. All the strains were first precultured in the Luria-Bertani medium. The second preculture and the main culture were carried out using M9 minimal medium containing 5 g/l of glucose for batch culture and 4 g/l for the continuous culture including 13C-labeling experiment together with the following components (per liter): 6.81 g Na2HPO4, 2.99 g KH2PO4, 0.58 g NaCl and 5.94 g (NH4)2SO4, unless otherwise stated for glucose concentration. The following components were filter sterilized and then added (per liter) with 1 ml of 1 M MgSO4 7H2O, 1 ml of 0.1 mM CaCl2 2H2O, 1 ml of 1 mg/L thiamine HCl and 10 ml of trace element solution containing (per liter): 0.55 g CaCl2 2H2O, 1.67 g FeCl3 6H2O, 0.1 g MnCl2 4H2O, 0.17 g ZnCl2 2H2O, 0.043 g CuCl2 2H2O, 0.06 g CoCl2 2H2O, and 0.06 g Na2MOO4 2H2O.
Culture condition
The first precultivation was performed by transferring 0.1 ml glycerol stock to a 10 ml L tube. After 8 h cultivation, the second precultivation was performed by transfering 1 ml of culture broth to a 100 ml T tube. The continuous culture was then conducted in a 2-l fermentor (Venus 25, Biott Co.) with a working volume of 1 l. The pH was controlled at 7.0 ± 0.1 using 2 N HCl or 2 N NaOH, and the temperature was set at 37°C. The air flow rate was 1 vvm (air volume/working volume/min), and the agitation speed was 400 rpm to maintain the dissolved oxygen concentration to be at 35%-40% (v/v) of air saturation [15]. The CO2 and O2 concentrations were monitored using an off-gas analyzer (BMJ-02 PI, ABLE Co., Japan). The dilution rates in the continuous culture were 0.2, 0.4, 0.6 and 0.7 h-1 for the wild type, and 0.2 h-1 for the mutants. The triplicate samples were collected at the steady state which was confirmed by the constant cell density and CO2 concentration in the off-gas. It generally took 3 residence times to achieve the steady state.
Analytical method
Bacterial growth was monitored by measuring the optical density of the culture broth at 600 nm (OD600) using a spectrophotometer (Ubet-30, Jasco, Tokyo, Japan). It was then converted to dry cell weight (DCW) based on the OD600-DCW relationship previously obtained [25]. Glucose, xylose and acetate concentrations in the medium were measured using commercially available kits (Wako Co., Osaka, Japan for glucose and xylose; Roche, Molecular Biochemical, Mannheim, Germany for acetate).
RNA preparation
Total RNA was isolated from E. coli cells by Qiagen RNeasy Mini Kit (QIAGEN K.K., Japan) according to the manufacture's recommendation. The quantity and purity of RNA were determined by the optical density measurements at 260 and 280 nm and by 1% formaldehyde agarose gel electrophoresis. The primer sequence for respective genes used in this study were reported elsewhere [19]. Criteria for the design of the gene-specific primer pairs were followed according to [26]. The primers used in this study were synthesized at Hokkaido System Science Co. (Sapporo, Hokkaido, Japan). In all cases, the primer-supplied company confirmed the purity and absolute specificity of the primers.
cDNA synthesis and PCR amplification
RT-PCR reactions were carried out in a Takara PCR Thermal Cycler (Takara TP240, Japan) using Qiagen One-step RT-PCR Kit (QIAGEN K.K., Japan). The reaction mixture was incubated for 30 min at 50°C for reverse transcription (cDNA synthesis) followed by 15 min incubation at 95°C for initial PCR activation. Then, the process was subjected to 30 cycles of amplification which consisted of a denaturing step (94°C for 1 min), an annealing step (approximately 5°C below melting temperature of primers for 1 min) and an extension step (72°C for 1 min), and finally the reaction mixture was incubated for 10 min at 72°C for final extension. To check for nucleic acid contamination, one negative control was run in every round of RT-PCR. This control lacks the template RNA in order to detect possible contamination of the reaction components. 5 ml of amplified products were run on a 1% agarose gel. Gels were stained with 1 μg/ml of ethidium bromide, photographed using a Digital Image Stocker (DS-30, FAS III, Toyobo, Osaka, Japan) under UV light and analyzed using Gel-Pro Analyzer 3.1(Toyobo, Osaka, Japan) software. Although the PCR products obtained for all the genes showed the predicted sizes on agarose gel, the identity of amplified fragments of some genes was demonstrated by DNA sequencing. In order to determine the optimal amount of input RNA, the two-fold diluted template RNA was amplified in RT-PCR assays under identical reaction conditions to construct a standard curve for each gene product. When the optimal amount of input RNA was determined for each gene product, RT-PCR was carried out under identical reaction conditions to detect differential transcript levels of genes. The gene dnaA, which encodes for DnaA, a replication initiation factor in E. coli and is not subjected to variable expression, i.e. abundant expression at relatively constant rate in most cells, was used as an internal control for the RT-PCR determinations. To calculate the standard deviation, RT-PCR was independently performed three times for each gene under identical reaction condition. To ensure that the observed changes were statistically significant, the measurements were made in triplicate, and the Student's t-test was applied.
13C-labeling experiments
The labeling experiments were started after the steady state was confirmed. The unlabeled (naturally labeled) feeding medium was replaced by an identical medium containing 3.2 g/l of unlabeled glucose, 0.4 g/l [U-13C] glucose, and 0.4 g/l [1-13C] glucose. Biomass samples were taken after another two residence times, where 200 mL of culture broth was taken for the measurement of biomass and extracellular metabolite concentrations, and the remaining 800 mL of the culture broth was processed for gas chromatography-mass spectrometry (GC-MS) analysis. The biomass sample was kept on ice for 2-3 min, and the sample was centrifuged at 6,000 rpm at 2°C for 15 min [27]. The cell pellets were washed three times with 20 mM Tris-HCl at pH 7.6 and suspended in 10 mL of 6 M HCl. The mixture was then hydrolyzed at 105°C for 15 h in a sealed glass tube. During acid hydrolysis, tryptophan and cysteine were oxidized, and glutamine and asparagine were deaminated. The hydrolysate was evaporated to dryness. The dried material was dissolved in Milli-Q water and filtered through a 0.22-μm pore-size filter and evaporated to dryness. About 1.5 ml acetonitrile was added in the dried hydrolyte and incubated at room temperature overnight. After the color of liquid was turned to slight yellow, it was filtrated through 0.22-μm pore-size filter. The filtrate was then derivatized by N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide (Aldrich, USA) at 110°C for 30 min and was transferred to GC-MS sample tube for analysis [27].
Metabolic flux analysis
Preparation of biomass hydrolysates and recording of the GC-MS spectra (PerkinElmer, Germany) were made as described previously [27,28]. The program Turbomass Gold (PerkinElmer, Germany) was used for peak assignment and MS data processing. The main idea is to perform isotopomer balance on carbon atoms in order to track the fate of the labeled carbon atoms from the substrate [29]. Isotopomer balance enables us to determine the isotopomer distributions of the intracellular metabolites in the central metabolic network. Since the isotopomer distributions of amino acids can be inferred from the isotopomer distributions of their corresponding precursors, the GC-MS signals for the amino acids can then be simulated. A set of flux distributions is then determined by minimizing the differences between the experimental and simulated data. For the estimation of GC-MS signals using isotopomer balance, three types of corrections were made to take into account the effects of natural abundance, non-steady state condition, and skewing effect. First, the isotopomer distributions of the input substrate were corrected for natural abundance in the unlabeled glucose and impurity in the labeled glucose. Second, the isotopic steady-state condition is normally not attained at the time of harvesting biomass. Thus, the simulated data have to be corrected based on the actual harvesting time by assuming first-order washout dynamics. Finally, the simulated GC-MS data were corrected for the natural isotope abundance of O, N, H, Si, S, and C atoms in the derivatizing agent using the correction matrices.
The detailed flux calculation is given previously [27]. Briefly, the best-fit intracellular fluxes were estimated by minimization of the deviation between experimental GC-MS data and the estimated values, using the iterative scheme in the minimization procedure. A set of intracellular fluxes (including net fluxes and exchange fluxes) that gives the minimum deviation could be taken as the best fit. The stage optimization was made by both local search and with global search such as Genetic algorithm [27].
Estimation of the specific ATP production rate and specific NAPH production rate
The ATP production is made either by substrate level phosphorylation and oxidative phosphorylation, where the reducing power of NADH and FADH2 can contribute in generating ATP via oxidative phosphorylation. The pathways involved in electron transport and oxidative phosphorylation have variable stoichiometry due to the use of different dehydrogenases and cytochromes. Then the specific ATP production rate may be estimated by the following equation:
| (1) |
where OPNADH and OPFADH2 is the specific ATP production rates via oxidative phosphorylation, and those may be estimated by
| (1a) |
and
| (1b) |
where (P/O) and (P/O)are the P/O ratios for NADH and FADH2, respectively.
Here, we set (P/O) to be 2 and (P/O)' to be 1.
The specific NADPH production rate vNADH may also be estimated by:
| (2) |
Abbreviations
Metabolites
3PG: 3-Phosphoglyceric acid; 6PG: 6-Phosphogluconolactone; AKG: 2-Keto-D-gluconate; AcCoA: Acetyl-CoA; AcP: Acetylphosphate; ASP: Aspartate; ADP: Adenosine diphosphate; ATP: Adenosine-5'-triphosphate; AMP: Adenosine monophosphate; cAMP: Cyclic AMP; CIT: Citrate; DHAP: Dihydroxyacetone phosphate; E4P: Erythrose 4-phosphate; F1,6-BP: Fructose 1,6-bisphosphate; F6P: Fructose 6-phosphate; FUM: Fumarate; G6P: Glucose-6-phosphate; GAP: Glyceraldehyde 3-phosphate; GLX: Glyoxylate; ICIT: Isocitrate; NAD/NADH: Nicotinamide adenine dinucleotide; NADP/NADPH: Nicotinamide adenine dinucleotide phosphate; MAL: Malate; OAA: Oxaloacetate; P: Phosphate; PEP: Phosphoenolpyruvate; PYR: Pyruvate; R5P: Ribulose 5-phosphate; Ru5P: Ribose 5-phosphate; S7P: Sedoheptulose 7-phosphate; SUCCoA: Succinyl-CoA; SUC: Succinate; X5P: Xylulose 5-phosphate.
Protien (enzyme)
Ack: Acetate kinase; AKGDH: 2-Keto-D-gluconate dehydrogenase; 6PGDH: 6-Phosphogluconate dehydrogenase; EI: Eyme I; EIIA: Ezyme IIA; EIICB: Ezyme IIB; EI-P: Posphorylated state of EI; EIIA-P: Posphorylated state of EIIA; EIICB-P: Posphorylated state of EIICB; G6PDH: Glucose-6-phosphate dehydrogenase; GAPDH: Glyceraldehyde 3-phosphate dehydrogenase; ICDH: Isocitrate dehydrogenase; HPr: Histidine-containing protein; HPr-P: Phorylated state of HPr; Mdh: Malate dehydrogenase; Mez: Malic enzyme; Pck: Phosphoenolpyruvate carboxykinase; PDH: Pyruvate dehydrogenase; Pfk: Phosphofructokinase; Pyk: Pyruvate kinase; SDH: Succinate dehydrogenase; UQ: Ubiquinone;
Gene
aceA: Isocitrate lyase gene; aceB: Malate synthase A gene; aceE, F: Pyruvate dehydrogenase gene; ackA: Acetate kinase gene; acnA/B: Aconitase gene; acs: Acetyl-coenzyme A synthetase; arcA/B: Anoxic redox control protein; atpA: ATP synthase; cra: Catabolite repressor/activator; crp: Cyclic AMP receptor; crr: Catabolite repressor; cyaA: adenylate cyclase gene; cyoABCDE: Cytochrome o oxidase gene; cydAB: Cytochrome d oxidase gene; edd: 6-Phosphogluconate dehydrogenase gene; eda: 2-Keto-3-deoxy-6-phosphogluconate aldolase gene; fadR: Fatty acid metabolism regulator; fnr: Fumarate/nitrate reduction transcriptional regulator; fumA, B, C: Fumarase gene; fur: Fe-responsive gene regulator; gltA: Citrate synthase gene; gnd: 6-phosphogluconate dehydrogenase gene; icdA: Isocitrate dehydrogenase A gene; iclR: Isocitrate lyase regulator; lpdA: Dihydrolipoamide dehydrogenase gene; mdh: Malate dehydrogenase gene; mgsA: Methylglyoxal synthase gene; mlc: DNA-binding transcriptional repressor; ndh: NADH dehydrogenase gene; nuoABCEFGHIJKLMN: NADH:ubiquinone oxidoreductase; pckA: Phosphoenolpyruvate carboxykinase gene; pfkA: 6-Phosphofructokinase I gene;
pgi: Glucose-6-phosphate isomerase gene; pgk: Phosphoglycerate kinase gene; ppc: Phosphoenolpyruvate carboxylase gene; ppsA: Phosphoenolpyruvate synthase gene; pta: Phosphate acetyltransferase gene; ptsG, ptsHI: Pts gene; pykA, F: Pyruvate kinase gene; rpoS: RNA polymerase sigma factor; sdhC: Succinate dehydrogenase; sodA: Speroxide dismutase; soxR/S: Superoxide stress regulon; sdhABCD: Succinate dehydrogenase gene; sucA: 2-Oxoglutarate decarboxylase; sucB: Dihydrolipoamide succinyltransferase; sucC: Succinyl-CoA ligase gene; sucD: Succinyl-CoA synthetase gene; talAB: Transaldolase gene; tktAB: Transketolase gene; tpiA: Triose phosphate isometase; ubiE: Uuinone/menaquinone biosynthesis methyltransferase gene; yfid: Autonomous glycyl radical cofactor; zwf: Glucose 6-phosphate dehydrogenase gene.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RY carried out fermentation, GC-MS and RT-PCR experiments, assayed, made statistical analysis, YH carried out fermentation experiments, DS carried out fermentation and GC-MS experiments, KN constructed crp+ mutant and prepared other single gene knockout mutants. YQ discussed on the result obtained, and participated in preparing the manuscript, KS considered experimental design, analyzed the result, and prepared manuscript. All authors read and approved the final manuscript.
Supplementary Material
Global regulators and their regulated genes.
The effect of dilution rate on the metabolic flux distribution. All fluxes are given as absolute values (mmol/g/h) and as values relative to the specific glucose consumption rate (in parentheses), values in < > indicate exchange coefficients.
The effect of dilution rate (specific growth rate) on the specific ATP production rate (3a) and specific NADPH production rate (3b). Filled square symbols are from the present study, open triangle symbols are from [30], while open circle symbols from [31].
Batch fermentation result of using a mixture of glucose and xylose as a carbon source. (a) wild type, (b) crp+ mutant.
Continuous fermentation result of using a mixture of glucose and xylose as a carbon source. (a) wild type, (b) crp+ mutant.
Continuous fermentation result of using a mixture of glucose and xylose as a carbon source for ptsG mutant.
Batch fermentation result of using glucose as a carbon source for Δcrp.
Batch fermentation result of using glucose as a carbon source for mlc mutant.
Contributor Information
Ruilian Yao, Email: yaoruilian@126.com.
Yuki Hirose, Email: yuki8621@hotmail.co.jp.
Dayanidhi Sarkar, Email: sarkardayanidhi5@gmail.com.
Kenji Nakahigashi, Email: knakahig@sfc.keio.ac.jp.
Qin Ye, Email: qye@ecust.edu.cn.
Kazuyuki Shimizu, Email: shimi@bio.kyutech.ac.jp.
Acknowledgements
This research was supported by Strategic International Cooperative Program, Japan Science and Technology Agency (JST).
References
- Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- Zaldivar J, Nielsen J, Olsson J. Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl Microbiol Biotechnol. 2001;56:17–34. doi: 10.1007/s002530100624. [DOI] [PubMed] [Google Scholar]
- Nichols NN, Dien BS, Bothast RJ. Use of catabolite repression mutants for fermentation of sugar mixtures to ethanol. Apple Microbiol Biotechnol. 2001;56:120–125. doi: 10.1007/s002530100628. [DOI] [PubMed] [Google Scholar]
- Dien BS, Nichols NN, Bothast RJ. Fermentation of sugar mixtures using Escherichia coli catabolite repression mutants engineered for production of L-lactic acid. J Ind Microbiol Biotechnol. 2002;29:221–227. doi: 10.1038/sj.jim.7000299. [DOI] [PubMed] [Google Scholar]
- Hernández-Montalvo V, Valle F, Bolivar F, Gosset G. Characterization of sugar mixtures utilization by an Escherichia coli mutant devoid of the phosphotransferase system. Appl Microbiol Biotechnol. 2001;57:186–191. doi: 10.1007/s002530100752. [DOI] [PubMed] [Google Scholar]
- Flores N, Flores S, Escalante A, Anda Rd, Leal L, Malpica R, Georgellis D, Gosset G, Bolívar F. Adaptation for fast growth on glucose by differential expression of central carbon metabolism and gal regulon genes in an Escherichia coli strain lacking the phosphoenolpyruvate:carbohydrate phosphotransferase system. Metab Eng. 2005;7:70–87. doi: 10.1016/j.ymben.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Anda DE, Lara AR, Hernández V, Hernández-Montalvoa V, Gosset G, Bolívar F, Ramírez OT. Replacement of the glucose phosphotransferase transport system by galactose permease reduces acetate accumulation and improves process performance of Escherichia coli for recombinant protein production without impairment of growth rate. Metab Eng. 2006;8:281–290. doi: 10.1016/j.ymben.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Yamano LP, York SW, Shanmugam KT, Ingram LO. Deletion of methylglyoxal synthase gene (mgsA) increased sugar co-metabolism in ethanol-producing Escherichia coli. Biotechnol Lett. 2009;31:1389–1398. doi: 10.1007/s10529-009-0011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham DS, Liu Z, Domagalski N, Koepsel RR, Ataai MM, Domach MM. Pyruvate kinase-deficient Escherichia coli exhibits increased plasmid copy number and cyclic AMP levels. J Bacteriol. 2009;191:3041–3049. doi: 10.1128/JB.01422-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiteman MA, Lee SA, Altman R, Altman E. A substrate-selective co-fermentation strategy with Escherichia coli produces lactate by simultaneously consuming xylose and glucose. Biotechnol Bioeng. 2009;102:822–827. doi: 10.1002/bit.22103. [DOI] [PubMed] [Google Scholar]
- Bettenbrock K, Fischer S, Kremling A, Jahreis K, Sauter T. A quantitative approach to catabolite repression in Escherichia coli. J Biol Chem. 2006;281:2578–2584. doi: 10.1074/jbc.M508090200. [DOI] [PubMed] [Google Scholar]
- Kremling A, Gilles ED. The organization of metabolic reaction networks: II. signal processing in hierarchical structured functional units. Metab Eng. 2001;3:138–150. doi: 10.1006/mben.2000.0175. [DOI] [PubMed] [Google Scholar]
- Kremling A, Fischer S, Sauter T, Bettenbrock K, Gilles ED. Time hierarchies in the Escherichia coli carbohydrate uptake and metabolism. Biosystems. 2004;73(1):57–71. doi: 10.1016/j.biosystems.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Ferenci T. Hungry bacteria - definition and properties of a nutritional state. Environ Microbiol. 2001;3:605–611. doi: 10.1046/j.1462-2920.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- Rahman M, Hassan MR, Shimizu K. Growth phase-dependent changes in the expression of global regulatory genes and associated metabolic pathways in Escherichia coli. Biotechnol Lett. 2008;30:853–860. doi: 10.1007/s10529-007-9621-1. [DOI] [PubMed] [Google Scholar]
- Rahman M, Shimizu K. Altered acetate metabolism and biomass production in several Escherichia coli mutants lacking rpoS-dependent metabolic pathway genes. Mol Biosyst. 2008;4(2):160–169. doi: 10.1039/b712023k. [DOI] [PubMed] [Google Scholar]
- Shin W, Lim S, Seok YJ, Ryu S. Heat shock RNA polymerase (Eσ32) is involved in the transcription of mlc and crucial for induction of the Mlc regulon by glucose in Escherichia coli. J Biol Chem. 2001;276:25871–25875. doi: 10.1074/jbc.M101757200. [DOI] [PubMed] [Google Scholar]
- Morita T, El-Kazzaz W, Tanaka Y, Inada T, Aiba H. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J Biol Chem. 2003;278:15608–15614. doi: 10.1074/jbc.M300177200. [DOI] [PubMed] [Google Scholar]
- Kabir MD, Shimizu K. Gene expression patterns for metabolic pathway in pgi knockout Escherichia coli with and without phb genes based on RT-PCR. J Biotechnol. 2003;105:11–31. doi: 10.1016/S0168-1656(03)00170-6. [DOI] [PubMed] [Google Scholar]
- Toya Y, Ishii N, Nakahigashi K, Hirasawa T, Soga T, Tomita M, Shimizu K. 13C-metabolic flux analysis for batch culture of Escherichia coli and its pyk and pgi gene knockout mutants based on mass isotopomer distribution of intracellular metabolites. Biotechnol Prog. 2010;26(4):975–992. doi: 10.1002/btpr.420. [DOI] [PubMed] [Google Scholar]
- Siddiquee KAZ, Arauzo-Bravo MJ, Shimizu K. Effect of a pyruvate kinase (pykF-gene) knockout mutation on the control of gene expression and metabolic fiuxes in Escherichia coli. FEMS Microbiol Lett. 2004;235:25–33. doi: 10.1111/j.1574-6968.2004.tb09563.x. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 inframe, single gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G, Kolisnychenko V, Bereczki Z, Blattner F. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acid Res. 1999;27:4409–4415. doi: 10.1093/nar/27.22.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H, Nakamura T, Mitani H, Mori H. Mutations that alter the allosteric nature of cAMP receptor protein of Escherichia coli. EMBO J. 1985;12:3329–3332. doi: 10.1002/j.1460-2075.1985.tb04084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Shimizu K. Effect of fadR gene knockout on the metabolism of Escherichia coli based on analyses of protein expressions, enzyme activities and intracellular metabolite concentrations. Enzyme Microbiol Technol. 2006;38:512–520. doi: 10.1016/j.enzmictec.2005.07.019. [DOI] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Vol. 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, NY; 2001. [Google Scholar]
- Zhao J, Shimizu K. Metabolic flux analysis of Escherichia coli K12 grown on 13C-labeled acetate and glucose using GC-MS and powerful flux calculation method. J Biotechnol. 2003;101:101–117. doi: 10.1016/S0168-1656(02)00316-4. [DOI] [PubMed] [Google Scholar]
- Zhao J, Baba T, Mori H, Shimizu K. Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metab Eng. 2004;6(2):164–174. doi: 10.1016/j.ymben.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Szyperski T. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids-an efficient analytical tool to investigate intermediary metabolism. Eur J Biochem. 1995;232:433–448. doi: 10.1111/j.1432-1033.1995.tb20829.x. [DOI] [PubMed] [Google Scholar]
- Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, Hirasawa T, Naba M, Hirai K, Hoque A, Ho PY, Kakazu Y, Sugawara K, Igarashi S, Harada S, Masuda T, Sugiyama N, Togashi T, Hasegawa M, Takai Y, Yugi K, Arakawa K, Iwata N, Toya Y, Nakayama Y, Nishioka T, Shimizu K, Mori H, Tomita M. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- Yang C, Hua Q, Baba T, Mori H, Shimizu K. Analysis of Escherichia coli anaplerotic metabolism and its regulation mechanisms from the metabolic responses to altered dilution rates and phosphoenolpyruvate carboxykinase knockout. Biotechnol Bioeng. 2003;84:129–144. doi: 10.1002/bit.10692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Global regulators and their regulated genes.
The effect of dilution rate on the metabolic flux distribution. All fluxes are given as absolute values (mmol/g/h) and as values relative to the specific glucose consumption rate (in parentheses), values in < > indicate exchange coefficients.
The effect of dilution rate (specific growth rate) on the specific ATP production rate (3a) and specific NADPH production rate (3b). Filled square symbols are from the present study, open triangle symbols are from [30], while open circle symbols from [31].
Batch fermentation result of using a mixture of glucose and xylose as a carbon source. (a) wild type, (b) crp+ mutant.
Continuous fermentation result of using a mixture of glucose and xylose as a carbon source. (a) wild type, (b) crp+ mutant.
Continuous fermentation result of using a mixture of glucose and xylose as a carbon source for ptsG mutant.
Batch fermentation result of using glucose as a carbon source for Δcrp.
Batch fermentation result of using glucose as a carbon source for mlc mutant.