Abstract
We have established a one-hybrid screen that allows the in vivo localization of proteins at a functional Saccharomyces cerevisiae centromere. Applying this screen we have identified three proteins—Ctf19, Mcm21, and the product of an unspecified open reading frame that we named Okp1—as components of the budding yeast centromere. Ctf19, Mcm21, and Okp1 most likely form a protein complex that links CBF3, a protein complex directly associated with the CDE III element of the centromere DNA, with further components of the budding yeast centromere, Cbf1, Mif2, and Cse4. We demonstrate that the CDE III element is essential and sufficient to localize the established protein network to the centromere and propose that the interaction of the CDE II element with the CDE III localized protein complex facilitates a protein–DNA conformation that evokes the active centromere.
Keywords: Centromere, kinetochore, CBF3, Ctf19, Mcm21, one hybrid
The centromere and kinetochore are specialized chromosomal structures that fulfill several distinct functions to mediate high-fidelity chromosome segregation in mitosis and meiosis (Choo 1997a). Electron microscopy has revealed a differentiated structure of metaphase centromeres and kinetochores from higher eukaryotes (Choo 1997a). In contrast to higher eukaryotes, the centromere and kinetochore of Saccharomyces cerevisiae cannot be distinguished by electron microscopy. For this reason the terms kinetochore and centromere will be used synonymously for this organism.
Because the centromere of S. cerevisiae is less complex than the centromere of higher eukaryotes, it is ideal for molecular dissection and reconstitution. Most notably, the centromere DNA (CEN DNA), the chromosomal DNA that guides centromere formation, is only 125 bp in S. cerevisiae, whereas the CEN DNA of most other eukaryotes is very extensive (up to megabases) and highly repetitive (Choo 1997b). The S. cerevisiae CEN DNA consists of two partial palindromic consensus sequences, CDE I (8 bp) and CDE III (25 bp), which are separated by CDE II, a 78- to 86-bp-long stretch of A/T-rich DNA (Hegemann and Fleig 1993). CDE I is the binding site of a homodimer of Cbf1 (Hegemann and Fleig 1993), and CDE III binds CBF3, a protein complex, in vitro (Lechner and Carbon 1991). CDE I/Cbf1 are not essential for centromere function in vivo but increase chromosome segregation fidelity ∼10-fold (Hegemann and Fleig 1993). CDE III/CBF3 are indispensable for centromere function in vivo (Lechner and Ortiz 1996). Mutations in the central CCG triplet of CDE III inactivate the centromere in vivo and inhibit binding of CBF3 in vitro (Lechner and Ortiz 1996). Besides CDE III/CBF3, at least parts of CDE II are required to form a functional centromere in vivo (Schulman and Bloom 1993). The CBF3 complex originally was shown to contain the three components—Cbf3a, Cbf3b, and Cbf3c (Lechner and Carbon 1991)—that interact with the CEN DNA directly (Espelin et al. 1997). Corresponding genes identified later were named NDC10, CBF2, or CTF14 for Cbf3a (Doheny et al. 1993; Goh and Kilmartin 1993; Jang et al. 1993), CBF3B or CEP3 for Cbf3b (Lechner 1994; Strunnikov et al. 1995), and CTF13 for Cbf3c (Doheny et al. 1993). More recently a fourth component of CBF3, Cbf3d/Skp1, was identified (Connelly and Hieter 1996; Stemmann and Lechner 1996). Interaction of Cbf3d/Skp1 with Cbf3c is required to reconstitute the CBF3–CEN DNA complex from isolated CBF3 components in vitro and is part of the reconstituted CBF3–CEN DNA complex (Stemmann and Lechner 1996). Furthermore, Cbf3d/Skp1 was identified as a suppressor of a Cbf3c mutation (Connelly and Hieter 1996). Therefore, Cbf3d/Skp1 is essential for the formation of the CBF3–CEN DNA complex. According to Kaplan et al. (1997), the coexpression of Cbf3d/Skp1 with Cbf3c in insect cells promotes Cbf3c phosphorylation and results in an activated Cbf3c that can form CBF3–CEN DNA complexes in the absence of Cbf3d in vitro. In addition to its major role for centromere function in budding yeast, Cbf3d/Skp1 also functions as part of a protein ubiquitin–ligase, SCF (Skp1, a Cullin, and an F-box protein)(Bai et al. 1996; Skowyra et al. 1997) that promotes the degradation of cyclin-dependent kinase inhibitors or G1 cyclins (Krek 1998). SCF composition and function is conserved from yeast to higher eukaryotes (Krek 1998), as is reflected by the high conservation of Cbf3d/Skp1 among all eukaryotes examined (Connely and Hieter 1996). Currently, it is unclear whether the centromere function and the SCF function of Cbf3d/Skp1 overlap at any point.
The CBF3–CEN DNA complex most likely does not resemble an active kinetochore. In vitro this can be deduced from the observation that the CBF3–CEN complex is absolutely required but not sufficient to facilitate interaction between microtubules and CEN DNA (Sorger et al. 1994). Several proteins have been addressed as putative centromere proteins on the basis of genetic interactions with the CEN DNA or the CEN DNA-binding components Cbf1 and CBF3 (Hyman and Sorger 1995; Lechner and Ortiz 1996). Two of them, Mif2 and Cse4, recently have been localized to the centromere in vivo (Meluh and Koshland 1997; Meluh et al. 1998). Mif2, a protein that exhibits genetic interactions to CDE I, Cbf1, and Cbf3a (Meluh and Koshland 1995) exhibits homology with CENP-C, a protein of the mammalian kinetochore (Brown 1995). Cse4 exhibits genetic interaction with CDE II and shares considerable homologies with histone H3 and the mammalian centromere protein CENP-A (Stoler et al. 1995). Like CENP-A, Cse4 may therefore represent a specialized histone. Thus, the centromere of budding yeast and higher eukaryotes, although very different in size and complexity, may have common structural features that reflect their similar functional performance.
How do CEN–DNA-bound CBF3 and Cbf1 link up with Mif2 and Cse4 to assemble a centromere? To this date no physical interactions of Mif2 or Cse4 with CBF3 and Cbf1 have been revealed. Most likely unidentified centromere proteins provide the missing link. Here we describe such a missing link of the budding yeast centromere. Applying a one-hybrid approach, we have identified a protein complex consisting of Ctf19, Mcm21, and the product of the yeast ORF YGR179c, which clearly is a constituent of the S. cerevisiae centromere. This complex provides a physical link between the CBF3 complex, Mif2, Cse4, and Cbf1. We demonstrate further that the protein network established, with the exception of Cbf1, is localized to the CEN DNA exclusively via the CDE III element and provide evidence that the CDE II element is facilitating a distinct conformation of the centromere complex.
Results
Establishment of a one-hybrid system to identify kinetochore proteins
The one-hybrid system is a powerful tool to identify proteins that localize to specific DNA sequences in vivo (Li and Herskowitz 1993; Bourns et al. 1998). In general, the DNA sequence of interest is cloned upstream of a reporter gene and the resulting cassette is integrated into the yeast genome. The interaction of a protein fused to a transcriptional activation domain (AD) with the DNA sequence of interest will stimulate the expression of the reporter gene. Conversely, elevated expression of the reporter gene reveals the in vivo binding of an AD fusion protein to the DNA sequence of interest.
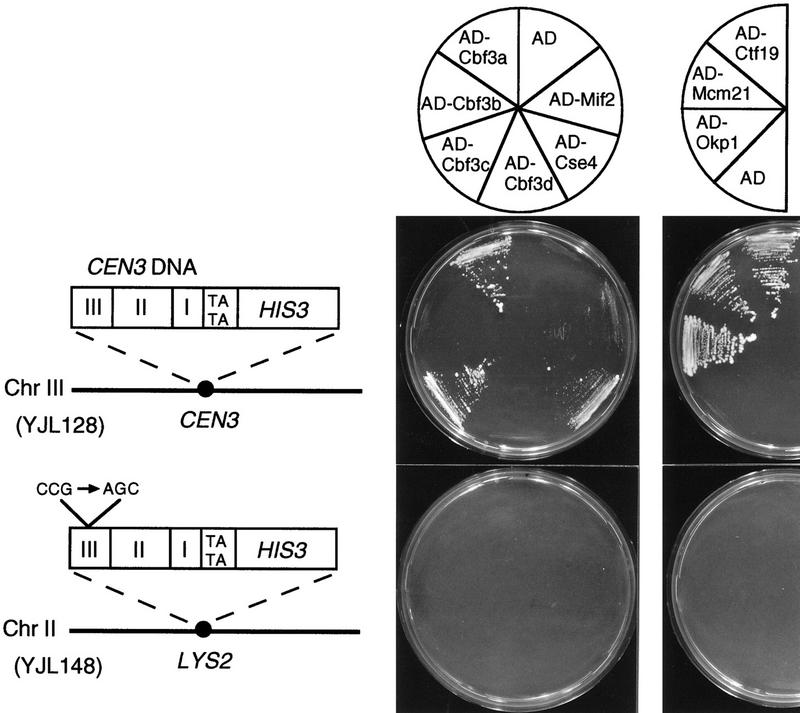
To assess whether a one-hybrid system could provide a method to identify undiscovered kinetochore proteins, we first investigated whether known kinetochore proteins like the CBF3 proteins, Mif2 or Cse4, could be localized at the CEN DNA by this method. A one-hybrid test strain was produced by replacing the endogenous CEN3 DNA of strain SFY526 with a DNA cassette that contained the CEN3 sequence upstream of a HIS3 reporter gene (Fig. 1). In the resulting strain, YJL128 (cen3::CEN3–HIS3), the reporter gene construct represents the only functional centromere on chromosome III. Thus, this approach avoids the formation of a dicentric chromosome. To address the specificity of a protein–CEN DNA interaction observed in YJL128 (cen3::CEN3–HIS3), we constructed a control strain, YJL148 (LYS2::cen3AGC–HIS3), as follows: CEN3 DNA, with the central CCG triplet of CDE III replaced by AGC, was cloned upstream of the HIS3 reporter gene. The CCG–AGC mutation in CDE III results in a nonfunctional CEN DNA (Hegemann and Fleig 1993). To maintain a functional copy of centromere DNA on the chromosome, the resulting reporter construct was integrated at a non-CEN locus (at the LYS2 locus) of SFY526. Mutations in the CCG triplet of CDE III strongly affect binding of the CBF3 complex in vitro (Ng and Carbon 1987; Lechner and Carbon 1991) and have been shown to interfere with the localization of Cbf3a and Mif2 to CEN DNA in vivo (Meluh and Koshland 1997). Therefore, if an AD fusion protein activated HIS3 expression in YJL128 (cen3::CEN3–HIS3) because of its specific interaction with the CDE III element of the reporter construct, the same fusion protein should not stimulate HIS3 expression in YJL148 (LYS2::cen3AGC–HIS3). Integration of the reporter constructs into the His− strain SFY526 led to His+ cells because of the basal HIS3 transcription from the HIS3 minimal promoter present in the reporter constructs. To identify events of activated HIS3 transcription, 3-aminotriazole (3-AT), an inhibitor of His3 activity, was added to agar plates in amounts that repressed growth due to basal HIS3 expression. These plates are referred to as 3-AT plates.
Figure 1.
Cbf3a, Cbf3c, Cse4, Ctf19, Mcm21, and Okp1 fused to the transcriptional activation domain of Gal4 (AD) stimulate the expression of HIS3 only when the reporter construct contains a functional CEN DNA. The reporter construct of strain YJL128 (cen3::CEN3–HIS3) containing wild-type CEN3 DNA and strain YJL148 (LYS2::cen3AGC–HIS3) containing a nonfunctional CEN3 DNA with a CCG to AGC mutation are presented. Growth of YJL128 (cen3::CEN3–HIS3) and YJL148 (LYS2::cen3AGC–HIS3) that express isolated AD (from pACT2) or the indicated AD fusion proteins (see Materials and Methods) on SD/His− medium containing 5 mm 3-AT is shown. AD–Ctf19, AD–Mcm21, and AD–Okp1 were identified by expressing AD fusion libraries in YJL128 (cen3::CEN3–HIS3) as described in the text.
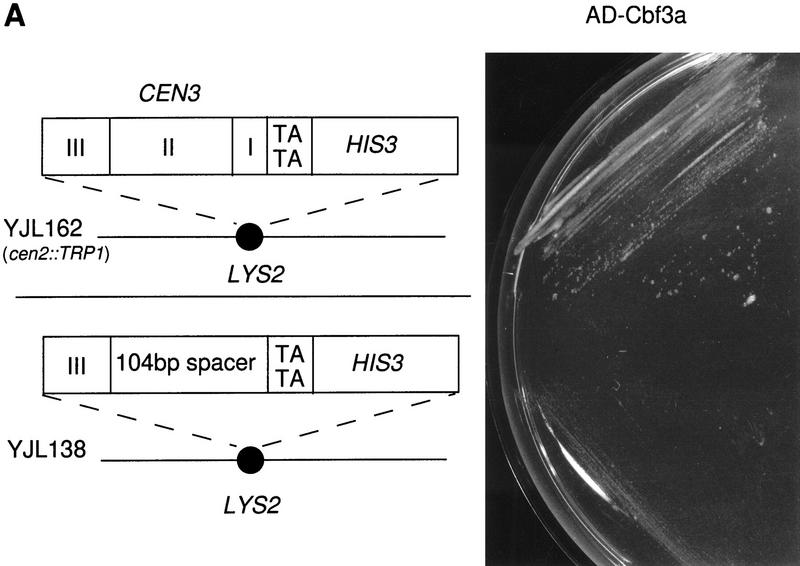
Fusion constructs consisted of the Gal4 transcriptional AD fused to the amino termini of the proteins tested. AD–Cbf3a, AD–Cbf3c, and AD–Cse4 supported strong growth of YJL128 (cen3::CEN3–HIS3) on 3-AT plates, whereas AD–Mif2 supported growth weakly (Fig. 1). The activation domain alone did not support growth on 3-AT plates, and none of the constructs supported growth of YJL148 (LYS2::cen3AGC–HIS3) cells on 3-AT plates. YJL128 (cen3::CEN3–HIS3) and YJL148 (LYS2::CEN3AGC–HIS3) contain the reporter construct in different chromosomal loci. To exclude that the observed results are due to a difference in the chromatin context at the two loci, we constructed a third strain, YJL162 (cen2::TRP1, LYS2::CEN3–HIS3), that harbors a CEN3–HIS3 construct at the LYS2 locus (on chromosome II) in addition to a CEN2 knockout. Similar to the results obtained with YJL128 (cen3::CEN3–HIS3), AD–Cbf3a and AD–Cbf3c support growth of YJL162 (cen2::TRP1, LYS2::CEN3–HIS3) on 3-AT plates (Fig. 7A, below; data not shown). Thus, the established one-hybrid system unambiguously detected the specific localization of known kinetochore proteins at a functional centromere in vivo. Furthermore, it revealed that Cse4 localizes to the centromere in a CDE III-dependent way. Therefore, these one-hybrid strains appear suitable for the identification of as yet unknown kinetochore proteins as well as for the confirmation of candidate kinetochore proteins.
Figure 7.
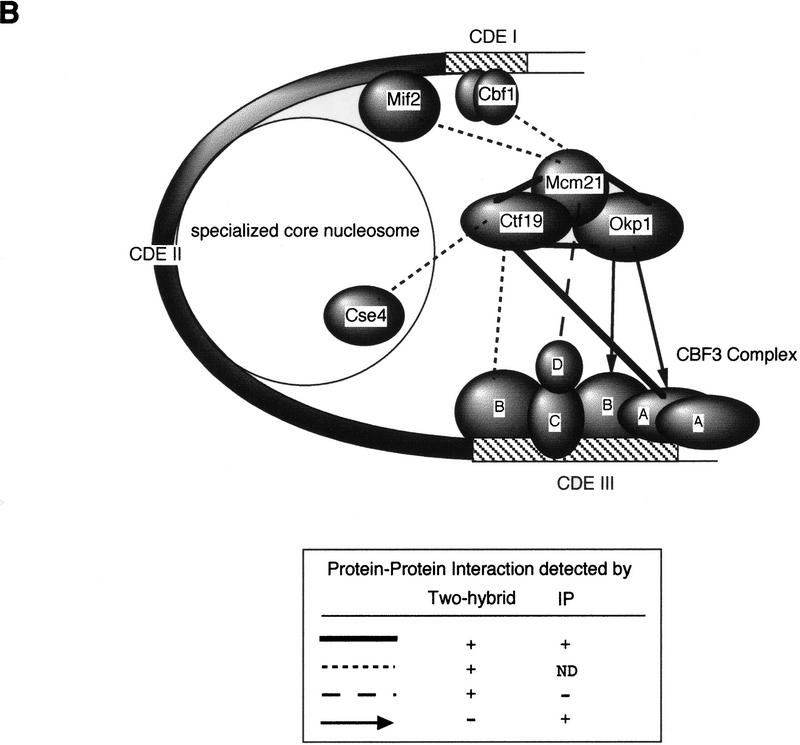
(A) Substitution of CDE I+II in a CEN3–HIS3 reporter construct interferes with AD–Cbf3a-promoted activation of HIS3 transcription. The reporter constructs of strain YJL162 (cen2::TRP1, LYS2::CEN3–HIS3) containing wild-type CEN3 DNA and strain YJL138 (LYS2::CDE III–HIS3) are presented. Growth of strains YJL162 (cen2::TRP1, LYS2::CEN3–HIS3) and YJL138 (LYS2::CDE III–HIS3) expressing AD–Cbf3a (pJO196) on SD/His− medium containing 5 mm 3-AT is shown. (B) S. cerevisiae kinetochore model. Solid lines, broken lines, and arrows represent protein–protein interactions revealed by experimental methods as indicated. For further explanation of the model, see text.
AD–Cbf3b and AD–Cbf3d did not support growth on 3-AT plates, although the transformants grew well on media that select for the plasmids. AD–Cbf3d may not be detected because it is not functional (as it cannot complement a cbf3d null mutation; data not shown). AD–Cbf3b, which is functional (it complements a cbf3b null mutation; data not shown) may not be detectable because the AD might be inaccessible.
Ctf19, Mcm21, and Okp1, the product of an uncharacterized ORF, are components of the budding yeast kinetochore
We applied the one-hybrid method to identify unknown proteins of the S. cerevisiae kinetochore. YJL128 (cen3::CEN3–HIS3) was transformed with two different S. cerevisiae plasmid libraries that overexpress AD fusion proteins. The first library contained genomic DNA, and the second, cDNA. Transformants were selected for growth on 3-AT plates, and plasmids of positive clones were sequenced. Several isolated plasmids contained the CBF3A gene. Besides Cbf3a, three proteins, Ctf19, Mcm21, and an unspecified ORF, YGR179c, named OKP1 for outer kinetochore protein, were identified. When expressed as AD fusions, these proteins supported growth of YJL128 (cen3::CEN3–HIS3) but not YJL148 (LYS2::CEN3AGC–HIS3) on 3-AT plates (Fig. 1). Ctf19, Mcm21, and Okp1 therefore localize to the CEN DNA in a CDE III-dependent way.
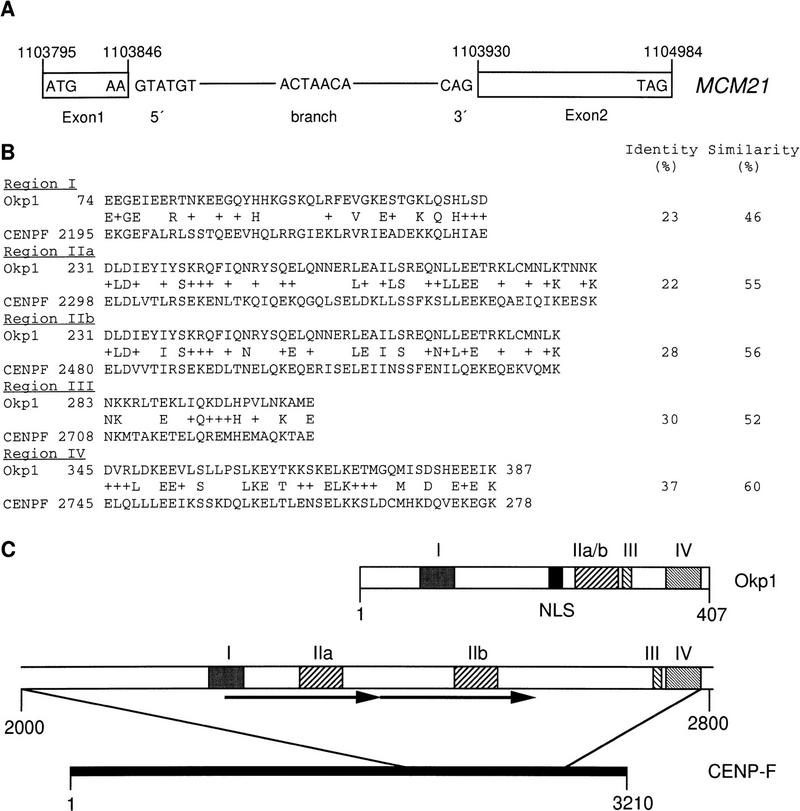
Ctf19 had been identified previously in a screen that detected compromised chromosome transmission fidelity (Ctf) (Spencer et al. 1990). A possible kinetochore function of Ctf19 had been proposed because of the high-dosage lethality of Cbf3c in a ctf19 mutant background (Kroll et al. 1996). Concurrently with our work, further genetic interactions between Ctf19 and the CBF3 components, as well as CEN chromatin immunoprecipitation (Chip) experiments, have established Ctf19 as a novel kinetochore protein (Hyland et al. 1999). Mcm21 had been identified in a mutant screen that detected defects in minichromosome maintenance (Mcm; Maine et al. 1984), but no connection to kinetochore functions had been established yet. Sequencing data of the plasmid that contained the MCM21 cDNA revealed that MCM21 translation starts at an ATG codon 434 bp upstream of the designated start ATG (SacchDB, Stanford, CA) and that MCM21 contains an 83-bp intron with S. cerevisiae characteristic 5′- and 3′-splice sites (Rymond and Rosbash 1992) that flank a putative branch sequence (Fig. 2A). Neither Ctf19 nor Mcm21 exhibits homologies to known proteins. However, Okp1 exhibits moderate homologies to CENP-F, a protein that localizes to the mammalian kinetochore in a cell cycle-dependent manner (Liao et al. 1995). Four regions of homology and a putative nuclear localization signal of Okp1 (amino acids 202–218) are presented in Figure 2, B and C.
Figure 2.
(A) Intron–exon structure of MCM21. 5′- and 3′-splice sites and a putative branch point are indicated. Arabic numerals refer to positions in Chromosome IV. (B) Regions of homology (I–IV) between Okp1 and CENP-F. (+) A conservative amino acid change. (C) Schematic representation of homology regions. Arrows indicate a direct repeat in CENP-F, and NLS denotes a putative nuclear localization signal. Roman numerals indicate regions of homology, as shown in B. Arabic numerals refer to amino acid positions.
ctf19, mcm21, and okp1 mutants exhibit a high rate of chromosome loss
We have analyzed ctf19, mcm21, and okp1 mutants with a colony-sectoring assay (Koshland and Hieter 1987) to support or establish the role of ctf19, mcm21, and okp1 in chromosome segregation. The mutant strains analyzed harbored an artificial chromosome carrying SUP11 that suppresses the ade2-101 mutation present in these cells. Loss of the artificial chromosome will result in cells that accumulate a red intermediate of the adenylate biosynthetic pathway. Consequently, frequent chromosome loss will lead to highly red-sectored colonies. Gene knockouts of CTF19, MCM21, and OKP1 were performed as described in Materials and Methods. As indicated elsewhere (SacchDB, Stanford), ctf19 null mutants are viable. Also, mcm21 null mutants are viable (see Materials and Methods). However, ctf19 and mcm21 null mutants exhibit a high sectoring phenotype (Fig. 3A). okp1 null mutants are not viable (see Materials and Methods) but okp1-5, a temperature-sensitive mutant generated as described in Materials and Methods, produced highly sectored colonies when grown at 34°C (Fig. 3A). Therefore, Ctf19, Mcm21, and Okp1 are involved in maintaining genetic stability, which is consistent with the kinetochore localization of these proteins. At 37°C okp1-5 arrests predominantly as large budded cells that contain one nucleus with replicated DNA (Table 1; Fig. 3B). This is indicative of an arrest at the G2/M transition of the cell cycle. Such an arrest could be mediated by a feedback control system that recognizes unsuccessful kinetochore spindle interaction (Gorbsky 1995), as has been proposed before for other kinetochore defects (Lechner and Ortiz 1996). However, this has to be determined.
Figure 3.
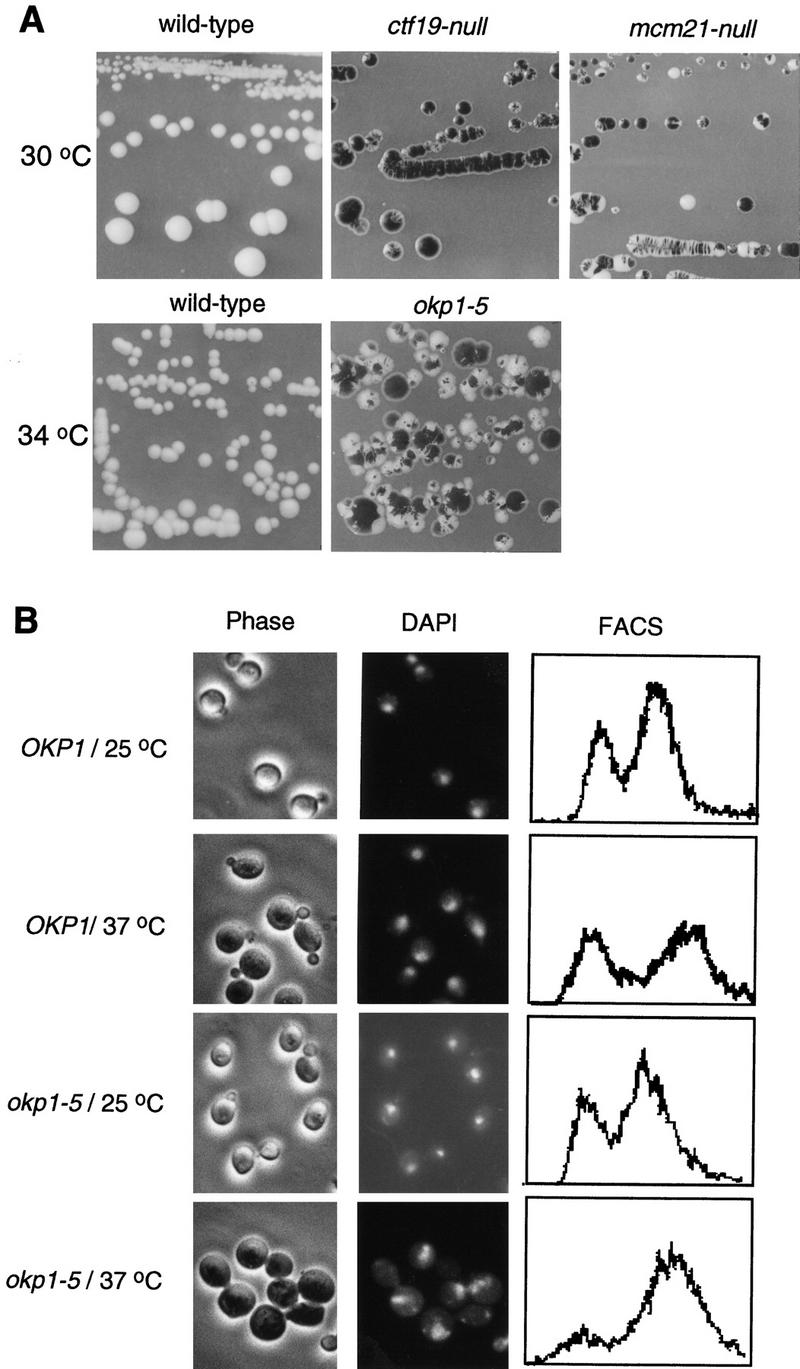
(A) ctf19, mcm21, and okp1 mutants exhibit elevated chromosome loss. Colony sectoring of wild-type or mutant strains harboring an artificial test chromosome on SD medium containing reduced amounts of adenine (Koshland and Hieter 1987) is shown. The temperature-sensitive okp1-5 mutant and a corresponding control strain were grown for 2 days at 34°C and then shifted to 28°C. (B) okp1-5 terminal phenotype. Strains as indicated were grown at 25°C to early logarithmic stage and than shifted to 37°C or kept at 25°C for 4 hr. Cellular morphology (phase contrast), nuclear staining (DAPI), and DNA content (FACS) are shown. okp1-5 arrests at the nonpermissive temperature predominantly as large budded cells with one nucleus and replicated DNA.
Table 1.
okp1-5 phenotype
|
Strain
|
Large budded cells (%)
|
Small/ nonbudded cells (%)
|
Total cells counted
|
|
|---|---|---|---|---|
|
1 nucleus
|
2 nuclei
|
|||
| OKP1/25°C | 12 | 11 | 77 | 341 |
| OKP1/37°C | 9 | 12 | 78 | 306 |
| okp1-5/25°C | 7 | 11 | 82 | 360 |
| okp1-5/37°C | 54 | 4 | 42 | 364 |
A putative protein complex consisting of Ctf19, Mcm21, and Okp1 interacts with the CBF3 complex as well as with Cse4, Mif2, and Cbf1
Potential protein–protein interactions between Ctf19, Mcm21, and Okp1 and with other components of the kinetochore were first investigated by the two-hybrid system (Fields and Song 1989). Two proteins, one as a fusion with the DNA-binding domain of Gal4 (BD), and the other as a fusion with the transcriptional AD of Gal4, were coexpressed in strain HF7c as indicated in Figure 4A. Interaction of the coexpressed fusion proteins results in the activation of HIS3 and lacZ transcription from reporter constructs present in HF7c. Consequently, a positive protein–protein interaction is revealed by the ability of the transformed strain to grow on 3-AT plates or by an increase in β-galactosidase activity. All possible combinations of Ctf19, Mcm21, and Okp1 with each other, with each of the CBF3 proteins, and with Cse4, Mif2, and Cbf1 were tested. All transformed strains grew well in media that selected for the plasmids. Protein combinations that resulted in strong growth on 3-AT plates when coexpressed in HF7c are presented in Figure 4A. As a negative control, positively identified fusion constructs were coexpressed with the appropriate two-hybrid vector that contained no insert. When β-galactosidase activity was assayed instead of HIS3 expression, identical results were obtained (data not shown). According to the two-hybrid results obtained, Ctf19, Mcm21, and Okp1 interact with each other. Furthermore, Ctf19 interacts with Cbf3a, Cbf3b, and Cse4; Mcm21 interacts with Cbf3d, Mif2, Cbf1, and itself.
Figure 4.
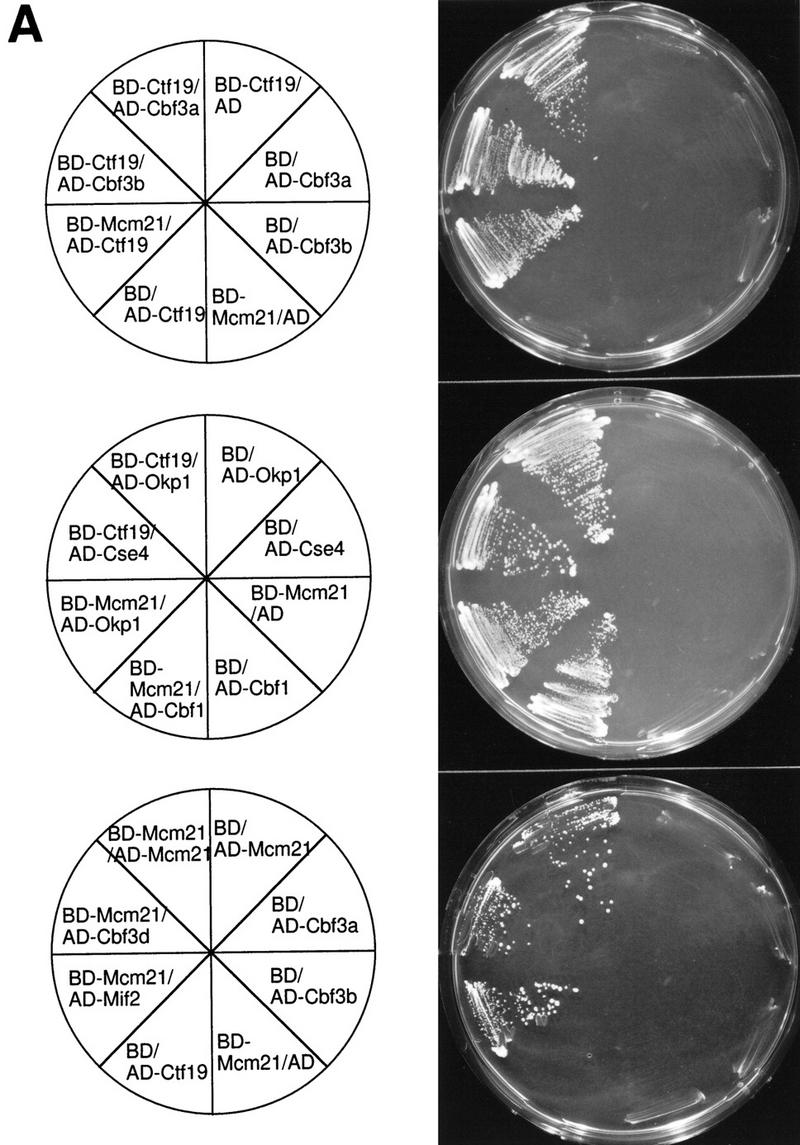
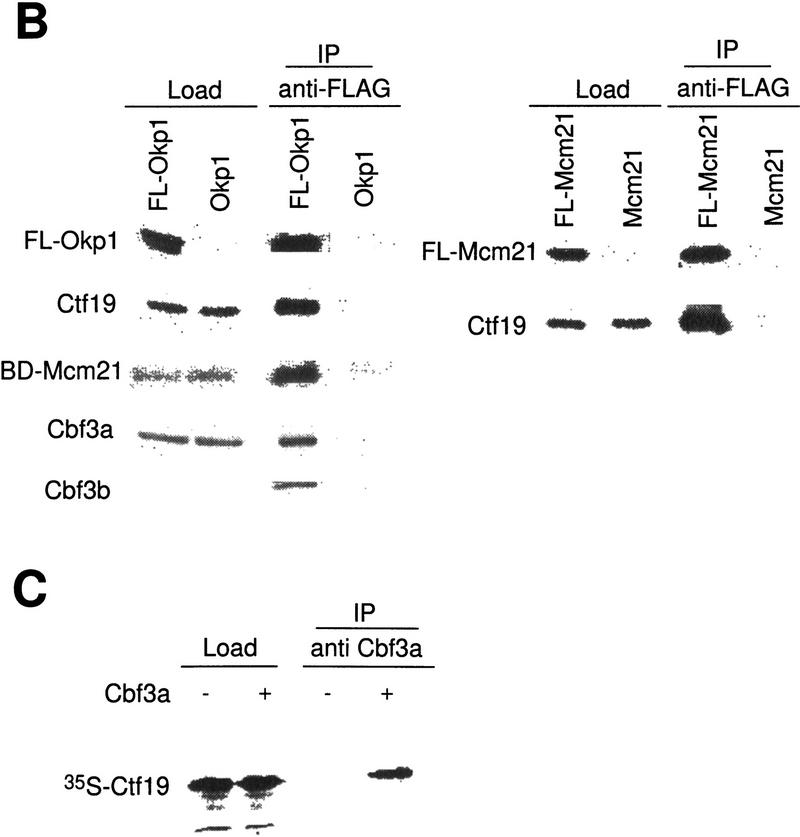
Protein–protein interactions involving Ctf19, Mcm21, Okp1, and other components of the S. cerevisiae centromere. (A) Two-hybrid analysis. The indicated fusion constructs containing the DNA-binding domain of GAL4 (BD) or the transcriptional activation domain of Gal4 (AD) were coexpressed in HF7c (for plasmid construction see Materials and Methods); growth on SD/His− medium containing 15 mm 3-AT is shown. As controls, coexpressions of corresponding fusion proteins with the appropriate two-hybrid vector containing no insert are shown. (B) For coimmunoprecipitation of Cbf3a, Cbf3b, and Ctf19 with Flag-tagged Okp1 (FL-Okp1), extracts from strain YJL132 (okp1::TRP1) harboring pJL471 (Flag–OKP1) were prepared. As a control, extracts from YJL19 were used that did not contain Flag-tagged Okp1. For coimmunoprecipitation of Mcm21 fused to the DNA-binding domain of Gal4 (BD-Mcm21) with Flag-tagged Okp1, extracts from YPH499 (Sikorski and Hieter 1992) harboring pJL471 (Flag–OKP1) and pJO508 (BD–MCM21) were prepared and as a control, extracts from YPH499 harboring only pJO508 (BD–MCM21) was used. For coimmunoprecipitation of Ctf19 with Flag-tagged Mcm21 (FL-Mcm21) extracts from YJL146 (mcm21::TRP1) harboring pJL505 (Flag–MCM21) were prepared and as a control, extracts from YJL19 were used. The extracts were immunoprecipitated with anti-Flag M2 agarose. Aliquots (0.5%–1.5% of the total load) of the extracts (Load) and aliquots (5% or 25% of the total load for the analysis of Flag-tagged proteins or coprecipitated proteins, respectively) of the immunoprecipitations (IPs) were subjected to Western analysis. Anti-Cbf3a, anti-Cbf3b, and anti-Ctf19 antibodies were used to detect corresponding proteins. Anti-HA antibody was used to detect BD–Mcm21, which contains an HA tag. The FL-Okp1 shown for Load and IP fractions corresponds to an experiment where the coprecipitation of Ctf19 and Cbf3a was revealed. For BD-Mcm21 or Cbf3b coimmunoprecipitation, the amount of FL-Okp1 detected in the Load and IP fractions was similar. Note that Cbf3b was undetectable in the loads. However, the same amount of total protein from the extract containing FL-Okp1 and of the control extract was subjected to immunoprecipitation. (C) 35S-Labeled Ctf19, synthesized in vitro, was incubated with or without 1 μg of purified Cbf3a–His6 expressed in Pichia pastoris (Stemmann and Lechner 1996). Subsequently, the samples were immunoprecipitated with anti Cbf3a antibody. Aliquots of the load (5% of the total) and IP fraction (30% of the total load) were subjected to SDS-PAGE and autoradiography.
Protein–protein interactions were analyzed further by coimmunoprecipitation (Fig. 4B,C). First, immunoprecipitates of Flag-tagged Okp1 and Mcm21 obtained from crude lysates were investigated (Fig. 4B). This revealed that Okp1, Ctf19, and Mcm21 are associated with each other. Quantitating the Western signals (data not shown) of load and precipitate fractions indicated that the degree of association is very high (∼30%–50%). We therefore speculate that Ctf19, Okp1, and Mcm21 form a protein complex. This complex interacts with CBF3 components: Okp1 is associated with Cbf3a and Cbf3b (Fig. 4B). These interactions were not detected by the two-hybrid system, presumably because the corresponding fusion constructs were not compatible with the assay. Conversely, the interactions between Ctf19 and Cbf3a or Cbf3b, as revealed by the two-hybrid assay, could not be detected unambiguously in crude lysates, probably because of the low amounts of Flag-tagged Ctf19 present in our extracts (data not shown). To examine Ctf19–Cbf3a interaction, we therefore synthesized Ctf19 in vitro. Immunoprecipitation with anti-Cbf3a antibodies revealed that Ctf19 coprecipitated with Cbf3a that had been added to the sample (Fig. 4C). Thus, the protein–protein interactions Okp1–Ctf19, Okp1–Mcm21, Ctf19–Mcm21, and Ctf19–Cbf3a that were revealed by the two-hybrid assay could be verified by coimmunoprecipitation (Fig. 7B, below). An interaction between Mcm21 and Cbf3d, as indicated by the two-hybrid assay, could not be verified unambiguously (data not shown). Other interactions revealed by the two-hybrid assay have yet to be analyzed by coimmunoprecipitation.
Neither technique allows a firm conclusion as to whether the observed interactions are direct. Nevertheless, the presented data clearly suggest that a putative complex consisting of Ctf19, Mcm21, and Okp1 links the CBF3 complex to Mif2, Cse4, and Cbf1, as summarized in Figure 7B.
An intact CDE III element is required and sufficient to localize Cse4, Ctf19, Mcm21, and Okp1 to the centromere in vivo
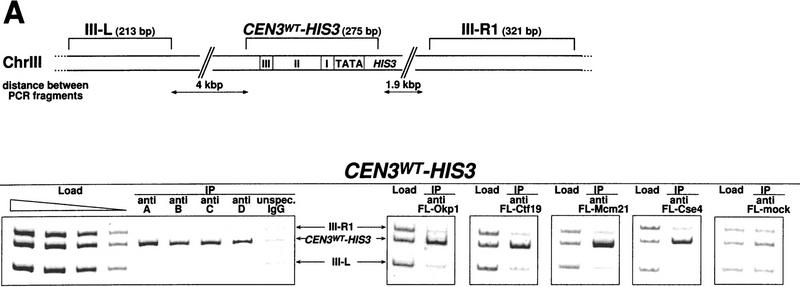
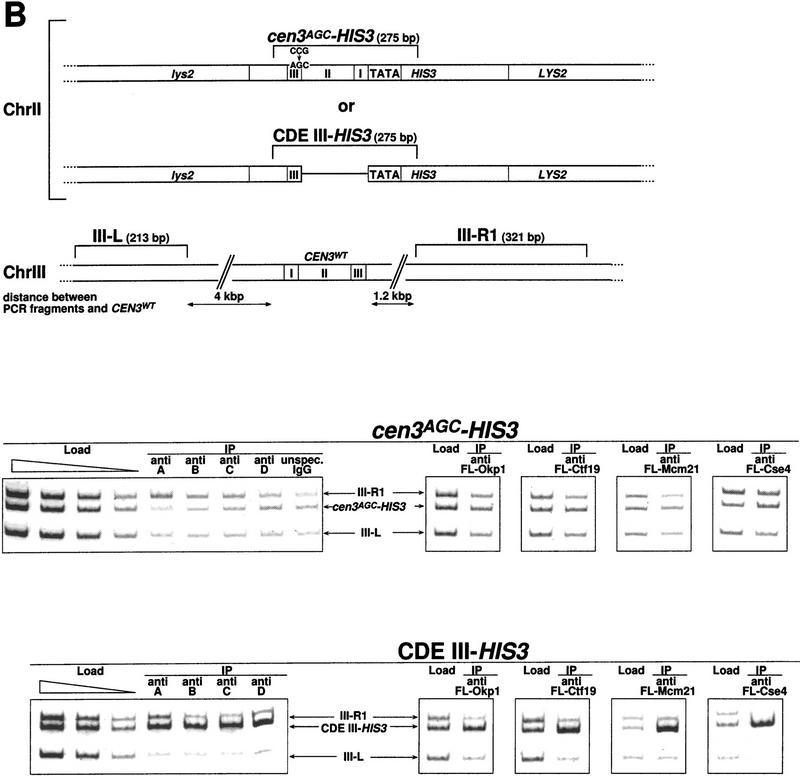
We used Chip to investigate the in vivo interaction of Cbf3b, Cbf3c, Cbf3d, Cse4, Ctf19, Mcm21, and Okp1 with CEN DNA in more detail. As a positive control, Cbf3a was assayed, as the in vivo localization of this protein to CEN DNA had been shown before (Meluh and Koshland 1997). Cells were treated with formaldehyde to cross-link cellular structures and then lysed and sonicated to shear the chromatin. Subsequently, the CBF3 proteins were immunoprecipitated with anti-CBF3 antibodies and protein A–Sepharose. Cse4, Ctf19, Mcm21, and Okp1 were expressed as Flag-tagged proteins and immunoprecipitated with anti-Flag agarose. After reversing the cross-links, coimmunoprecipitation of wild-type or mutant CEN3 DNA was probed by PCR.The analyzed wild-type CEN3 DNA included (1) CEN3–HIS3 at the CEN3 locus in strain YJL128 (cen3::CEN3–HIS3) (Fig. 5A); (2) CEN3 DNA of a wild-type unmarked CEN3 locus in the Cbf3a mutant strain ndc10-1 (Fig. 6); and (3) CEN16 in strain YPH499 (data not shown). Mutant CEN DNA consisted of the nonfunctional cen3AGC DNA integrated as part of the HIS3 reporter construct at the LYS2 locus in strain YJL148 (LYS2::CEN3AGC–HIS3) (Fig. 5B). In addition, a further construct was used in these experiments that contained CDE III plus pUC18 DNA replacing CDE II and CDE I upstream of HIS3 integrated at the LYS2 locus in strain YJL138 (LYS2::CDE III–HIS3) (Fig. 5B). To assess the specificity of the CEN DNA coprecipitation, the PCR mixture contained additional primers that resulted in the amplification of two noncentromeric control fragments. Furthermore, to exclude the possibility that CEN chromatin is preferentially precipitated with protein A–Sepharose or anti-Flag agarose, the following controls were performed: (1) Chip with protein A–Sepharose and extracts containing unspecific IgG (Fig. 5A,B, unspecific IgG) instead of CBF3 antibodies; and (2) Chip with anti-Flag agarose and extracts that contained no Flag-tagged proteins (Fig. 5A, anti FL-mock). To demonstrate that the set of primers resulted in equal amplification of the three template DNAs analyzed, preimmunoprecipitation samples (loads) were subjected to identical PCR analysis as the immunoprecipitates.
Figure 5.
CDE III is essential and sufficient to localize Cbf3a, Cbf3b, Cbf3c, Cbf3d, Okp1, Ctf19, Mcm21, and Cse4 to the CEN DNA in vivo as shown by Chip assays. (A) Association with wild-type CEN DNA, represented by CEN3WT–HIS3. Centromeric and noncentromeric loci analyzed are illustrated with corresponding sizes. Following in vivo cross-linking, extracts from strain YJL128 (cen3::CEN3–HIS3) without plasmids or YJL128 (cen3::CEN3–HIS3) harboring plasmids (see Materials and Methods) that expressed Flag-tagged versions of Okp1, Ctf19, Mcm21, and Cse4 were immunoprecipitated with anti CBF3 antibodies, unspecific IgG or anti Flag M2 agarose as indicated. (Anti Fl-mock) An extract from YJL128 (cen3::CEN3–HIS3) expressing no Flag-tagged proteins was immunoprecipitated with anti-Flag agarose. To test for specific copurification of CEN DNA, aliquots (∼0.016 μl of chromatin solution) of the extracts before immunoprecipitation (Load) and aliquots (∼5 μl of chromatin solution) of the immunoprecipitates (IPs) were analyzed by PCR. In the load dilution series, 0.1, 0.04, 0.016, and 0.0064 μl of chromatin solution were used. PCR mixtures were designed (see Table 2 for primers) to amplify the CEN DNA fragment in addition to two noncentromeric control fragments from chromosome III as indicated. (B) Association with nonfunctional CEN DNA, as represented by CEN3AGC–HIS3 and CDE III–HIS3. Centromeric and noncentromeric loci analyzed are illustrated with corresponding sizes. After in vivo cross-linking, extracts from strains YJL148 (LYS2::cen3AGC–HIS3), YJL138 (LYS2::CDE III–HIS3), or corresponding strains harboring plasmids, which express Flag-tagged versions of Okp1, Ctf19, Mcm21, and Cse4, were immunoprecipi tated as described in A. To test for specific copurification of cen3AGC DNA (inactive CDE III element) or the isolated CDE III DNA, aliquots (0.016 μl of chromatin solution) of the extracts before immunoprecipitation (Load) and aliquots (5 μl chromatin solution) of the immunoprecipitates were analyzed by PCR. In the load dilution series, 0.1, 0.04, 0.016, and 0.0064 μl of chromatin solution (for cen3AGC–HIS3) or 0.1, 0.03, and 0.008 μl of chromatin solution (for CDE III–HIS3) were used. PCR mixtures were designed (see Table 2 for primers) to amplify the cen3AGC or CDE III fragment in addition to two noncentromeric control fragments as indicated.
Figure 6.
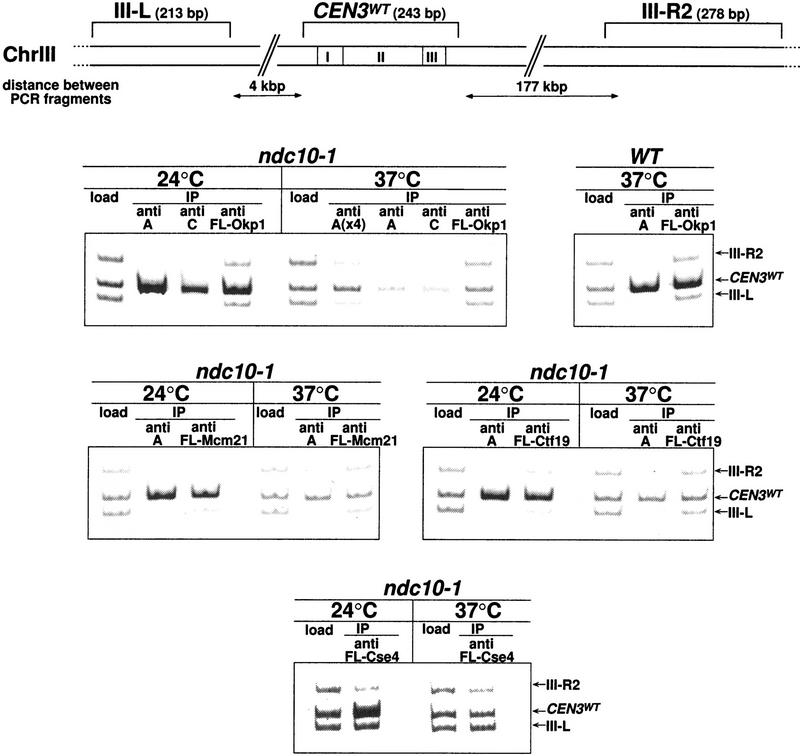
Localization of Cse4, Okp1, Ctf19 and Mcm21 to CEN DNA in vivo requires an intact CBF3 complex as shown by Chip. Centromeric and noncentromeric loci analyzed are illustrated with corresponding sizes. The ndc10-1 strain harboring a temperature-sensitive Cbf3a mutation, with or without plasmids that expressed Flag-tagged versions of Cse4, Okp1, Ctf19, and Mcm21, was grown at 24°C to early logarithmic stage before it was shifted to 37°C or kept at 24°C for 3 additional hr. A wild-type (WT) control strain (YPH499) was grown at 24°C and shifted to 37°C in the same way. After in vivo cross-linking, extracts of these strains were immunoprecipitated with anti-Cbf3a and anti-Cbf3c antibodies or with anti Flag M2 agarose as indicated. To test for the specific copurification of CEN DNA, aliquots of the extracts (Load; 0.016 μl of chromatin solution, with the exception of Fl-Cse4 load that contained 0.4 μl) and the immunoprecipitates [5 μl of chromatin solution; for A(x4), 20 μl] were analyzed by PCR. PCR mixtures were designed (see Table 2 for primers) to amplify the CEN DNA fragment in addition to two noncentromeric control fragments from chromosome III as indicated.
Wild-type CEN DNA (CEN3–HIS3, unmarked CEN3, and CEN16) specifically coimmunoprecipitated with all proteins tested regardless of whether asynchronous cultures (Figs. 5A and 6; data not shown) or cells treated with α factor (G1 arrest) or nocodazole (G2/M arrest; data not shown) were used. Okp1, Ctf19, and Mcm21 were therefore confirmed as kinetochore proteins by a second method independent of the one-hybrid approach. Furthermore, it was shown that like Cbf3a and Cbf3b, Cbf3c and Cbf3d also bind to the centromere in vivo, as would be expected from the in vitro data. Also, Cse4, as reported by Meluh et al. (1998), could be localized to the CEN DNA in vivo.
Interestingly, for all kinetochore proteins tested a wild-type CDE III element was sufficient for association with the CEN locus; CDE I and II were not required (Fig. 5B). cen3AGC DNA failed to coimmunoprecipitate with any of the kinetochore proteins tested (Fig. 5B). Thus, the CDE III element is not only sufficient but also essential for the kinetochore localization in all cases with the possible exception of Cbf1. It had been reported before (Meluh and Koshland 1997) that the kinetochore localization of Cbf3a and Mif2 requires CDE III exclusively, whereas Cbf1 can be only localized to a complete CEN DNA.
Okp1, Ctf19, Mcm21, and Cse4 require an intact CBF3–CEN DNA complex for centromere localization
Does the centromere localization of Ctf19, Mcm21, Okp1, and Cse4 depend on a CBF3–CEN complex? Flag-tagged versions of Okp1, Ctf19, Mcm21, and Cse4 were expressed in the background of ndc10-1, a temperature-sensitive Cbf3a mutation that disrupts the CBF3–CEN DNA complex formation in vitro (Sorger et al. 1995), and Chip assays with the anti-Flag antibody were performed. Wild-type CEN DNA and noncentromeric control DNA were analyzed by PCR as indicated (Fig. 6). After growing the ndc10-1 strain at the nonpermissive temperature, coimmunoprecipitation of CEN3 DNA with Cbf3a, Cbf3c, Okp1, Ctf19, Mcm21, and Cse4 was barely above the background. In contrast to this, coprecipitation of CEN3 DNA with theses proteins from ndc10-1 cells grown at the permissive temperature was very efficient (Fig. 6). Furthermore, coprecipitation of CEN3 DNA with Cbf3a and Okp1 was high in wild-type cells grown at 37°C. Therefore, as seen in vitro, the formation of the CBF3–CEN DNA complex is also compromised in vivo by the ndc10-1 mutation, and the CDE III localization of Okp1, Ctf19, Mcm21, and Cse4 depends on the existence of a CBF3–CEN DNA complex.
CDE I and CDE II may contribute to conformational changes necessary for a functional centromere/kinetochore
Given the data presented above and elsewhere (Meluh and Koshland 1997), none of the known kinetochore proteins (possibly with the exception of Cbf1) requires CDE II for centromere localization. So why is CDE II essential for centromere function? Because CDE II does not represent a consensus sequence, CDE II is believed to play a structural role (Cumberledge and Carbon 1987). To test this hypothesis, we took advantage of the fact that transcriptional activation depends on a particular DNA topology that positions transcription factors in the vicinity of the basal transcription apparatus (Grosschedl 1995). Interactions between nonadjacent DNA-bound proteins require a deformation of the DNA helix that is energetically unfavorable if the sites are >30 but <140 bp apart (Grosschedl 1995). For these proteins to interact, the deformation of the DNA helix has to be supported by an intrinsic DNA bend and/or chromatin factors that facilitate the helix deformation. The distance between the HIS3 promoter and CDE III in our one-hybrid constructs (see above) is only ∼100 bp. Nevertheless, proteins that localize to CEN3–HIS3 DNA, like AD–Cbf3a or AD–Cbf3c, were able to interact with the transcriptional machinery and activate HIS3 transcription in YJL128 (cen3::CEN3–HIS3; Fig. 1) and YJL162 (cen2::TRP1, LYS2::CEN3–HIS3; Fig. 7A; data not shown). In contrast to this, expression of AD–Cbf3a or AD–Cbf3c in YJL138 (LYS2::CDE III–HIS3), which harbors CEN3 DNA with the CDE II+I element replaced by spacer DNA of equal length, did not support growth on 3-AT plates (Fig. 7A; data not shown). This indicates that the transcriptional activation of HIS3 by these fusion proteins requires CDE I and CDE II to be the linker between the CDE III element and the HIS3 promoter. Therefore, CDE I and CDE II may facilitate a deformation of the DNA helix that is conducive with an interaction between AD–Cbf3a or AD–Cbf3c and the basal transcription complex. The results described above therefore support a kinetochore model with nonlinear CEN DNA as shown in Figure 7B. A multisubunit protein structure, including Cse4 and possibly other components of a putative core nucleosome, are anchored via the CBF3–CDE III complex. CDE II, but not control DNA, facilitates a DNA conformation that represents an active kinetochore, possibly in response to interactions with the CDE III localized protein complex.
YJL162 (cen2::TRP1, LYS2::CEN3–HIS3) exhibits a normal morphological phenotype, but the doubling time is extended by 30% in comparison to the corresponding wild-type strain. This could be due to a defect in the segregation of chromosome II, although it has not been verified. Nevertheless, the CEN3–HIS3 reporter construct provides a functional centromere, as the original centromere of chromosome II has been destroyed in this strain. YJL138 (LYS2::CDE III–HIS3) exhibits a wild-type morphology, reported likewise for a similar strain (Meluh and Koshland 1997). This indicates that the isolated CDE III bound protein complex does not elicit a spindle checkpoint signal that results in a preanaphase arrest.
Discussion
Identification of three new yeast kinetochore proteins: Ctf19, Mcm21, and Okp1
We have established a one-hybrid assay that allows the identification and in vivo localization of S. cerevisiae kinetochore proteins at active centromeres. Applying this assay we have identified three new kinetochore proteins, Ctf19, Mcm21, and Okp1. Subsequently we have confirmed the kinetochore localization and function of these proteins by three further lines of evidence. First, mutants of the three proteins exhibit severe chromosome loss; a temperature-sensitive mutant of Okp1, okp1-5, furthermore arrests at the G2/M transition of the cell cycle when shifted to the nonpermissive temperature, a phenotype that is consistent with a kinetochore defect. Second, several physical interactions between established kinetochore proteins and the newly identified proteins have been demonstrated. Third, in vivo cross-linking and subsequent chromatin immunoprecipitation revealed that Ctf19, Mcm21, and Okp1 localize to the CEN DNA in vivo. Moreover, a CDE III–CBF3 complex is required and sufficient for the centromere localization of the three new proteins.
Concurrently, Ctf19 has been independently identified as a kinetochore protein by Hyland et al. (1999). Complementary to our work, several genetic interactions between Ctf19 and components of CBF3 were revealed. Furthermore, these investigators describe that Ctf19 is important for kinetochore/microtubule interaction and that Ctf19 might have a second cellular role as a component of the spindle pole body.
Okp1 exhibits homology to CENP-F, a protein that associates with the mammalian centromere and might be involved in kinetochore maturation (Choo 1997c). The homology observed between Okp1 and CENP-F is not of low complexity (according to the BLASTP 2.0.6 program), and the extent of homology is similar to the one observed between Mif2 and CENP-C (Brown 1995). However, CENP-F localizes to the centromere only from prometaphase to early anaphase (Choo 1997c), whereas Okp1 (as well as Ctf19 and Mcm21) localize at the centromere during G1 and during G2/M. Furthermore, the region of CENP-F homologous to Okp1 lies within one of the two extensive coiled–coil domains predicted for CENP-F (Choo 1997c) and homologies between Okp1 and coiled–coil domains of other proteins (like myoglobin) exist (data not shown). Therefore one cannot exclude that the observed homologies could predominantly represent an inclination of Okp1 to form coiled–coil structures. However, a computer-based secondary structure analysis does not strongly predict an extensive coiled coil domains for Okp1. In conclusion, it is therefore unclear how comparable CENP-F and Okp1 are on a functional level.
The in vitro formation of the CBF3–CEN DNA complex can be detected by electrophoretic mobility shift assays and DNA footprinting (Lechner and Carbon 1991). In contrast to this, no evidence for a CEN DNA-bound protein complex that contains Okp1, Ctf19, or Mcm21 could be detected by supershift experiments with extracts containing Flag-tagged versions of Ctf19, Mcm21, or Okp1 (J. Ortiz et al., unpubl.). The conditions applied to make the extracts might have been incompatible with maintaining the activity of some of the complex components. Alternatively, the complex might not be stable enough to be detected by the shift assay or might require a chromatin environment for stability.
A network of kinetochore proteins
Based on two-hybrid assays, protein–protein and protein–DNA immunoprecipitations, we describe a network of physical interactions that link the CBF3 complex to Cbf1, Mif2, and Cse4, via Okp1, Mcm21, and Ctf19 (Fig. 7B). It goes without saying that the presented network is in no way complete. First, we are not presuming that the described proteins represent the complete set of centromere proteins. Second, the network consist only of the minimal amount of interactions that could be detected. Protein–protein interactions identified by the two-hybrid assay were omitted when one of the fusion constructs in combination with the appropriate two-hybrid vector with no insert supported growth on 3-AT plates or β-galactosidase expression, even when the background detected was considerably below the actual signal. Moreover, some interactions might be missed by either of the methods applied. Third, some of the interactions presented may not be direct. None of our methods stringently excludes that the protein–protein interaction detected is mediated by a third protein.
The protein network is anchored to the CEN DNA via the CDE III-bound CBF3 complex. As a result of the extensive in vitro and in vivo evidence that CBF3 is an essential CDE III-bound kinetochore complex (Lechner and Ortiz 1996) and the in vivo localization of Cbf3a to CDE III (Meluh and Koshland 1997), it came as no surprise that Cbf3b and Cbf3c also can be localized to CDE III in vivo. However, as it was reported that a CBF3–CEN DNA complex can be formed in vitro in the absence of Cbf3d (Kaplan et al. 1997), the in vivo localization of Cbf3d was less predictable. Our data revealed that Cbf3d is part of the kinetochore in vivo. Whether Cbf3d has a transient role at the centromere has yet to be investigated.
A putative protein complex consisting of Ctf19, Mcm21, and Okp1 associates with CBF3 via Ctf19, Mcm21, and Okp1 and interacts with Cse4, Mif2, and Cbf1 via Ctf19 and Mcm21. Because Ctf19 and Mcm21 are not essential proteins, whereas Cse4 (Stoler et al. 1995) and Mif2 (Meeks-Wagner et al. 1986) are, Cse4 and Mif2 are predicted to be positioned in the kinetochore by additional interactions.
A kinetochore model
The original proposal that a specialized nucleosome is part of the S. cerevisiae kinetochore (Saunders et al. 1990) was supported further by the demonstration that the histone H3 homolog Cse4 is a component of the S. cerevisiae kinetochore (Stoler et al. 1995; Meluh et al. 1998). It is speculated that this nucleosome would have at least one histone H3 replaced by Cse4. In the most recent model (Meluh et al. 1998) the inner surface of the complete centromere DNA contacts the histone interface. This would leave the outer surface for the interaction with CBF3. Ctf19, Mcm21, and Okp1 have to assemble onto the centromere nucleosome via interactions with CBF3, Cbf1, Mif2, and Cse4. We have shown that Cse4 (and possibly a putative Cse4–histone complex) is localized to the CEN DNA via CDE III-bound CBF3, whereas CDE II and CDE I are not required. Furthermore removing 92 of the 131 amino acids that represent the unique amino terminus of Cse4 has no effect on the centromere localization of Cse4 (J. Ortiz et al., unpubl.). Thus, analogous to CENP-A (Sullivan et al. 1994), centromere localization of Cse4 via the CBF3–CDE III complexes may be dependent on Cse4 domains that are homologous to histone H3, such as the histone fold, and not on the unique amino terminus. Because these conserved domains confer most of the protein–DNA interactions observed in a nucleosome (Luger et al. 1997), anchoring Cse4 via these domains to the CDE III localized protein network might interfere with Cse4–DNA interactions. As a consequence, the specialized nucleosome may only wrap half the length of DNA a standard nucleosome does (Fig. 7B). Consequently, CDE III would not contact the histone interface and would be accessible for protein–DNA interaction on both sides of the helix, as observed in vitro (Espelin et al. 1997). Interactions between CBF3, Cbf1, Mif2, and Cse4 mediated by a putative protein complex containing Ctf19, Mcm21, and Okp1 would stabilize the structure and evoke a functional kinetochore.
Irrespective of the model, why do we need CDE II for an active centromere? CDE II is not required for the centromere localization of the known kinetochore proteins, with the possible exception of Cbf1. Instead, CDE II is thought to have a structural function, as it does not represent a consensus sequence but nevertheless is essential for centromere function (Cumberledge and Carbon 1987). As a consequence of this and the postulated centromere nucleosome, many kinetochore models presented to date include nonlinear CEN DNA. Nonetheless, the physical and genetic interactions observed between the kinetochore proteins may also be interpreted as a protein complex on a linear DNA. Furthermore, a standard nucleosome would not be dependent on CDE II. Our work provides in vivo evidence that CDE I+II allows a distinct CEN DNA conformation that control DNA cannot facilitate. As shown in the transcription activation assays, CDE III-localized AD–Cbf3a and AD–Cbf3c can only activate the downstream HIS3 reporter gene when the DNA that links CDE III and the reporter is CDE I+II; recent work showed that CDE II but not CDE I is sufficient (J. Ortiz et al., unpubl.). The effect of CDE II on transcriptional activation is not due to a specific helix phase established by the AT-rich DNA of CDE II. This was revealed by the observation that the transcriptional activation induced by AD–Cbf3a or AD–Cbf3c was not affected when an oligonucleotide consisting of 1.5 helix turns was integrated between the CEN DNA and the reporter gene (J. Ortiz et al., unpubl.). Assuming that AD–Cbf3a and AD–Cbf3c (both are functional fusion proteins, as they can complement null mutants; J. Ortiz et al., unpubl.) bind to isolated CDE III DNA as wild-type Cbf3a and Cbf3c do, we conclude that CDE II most likely facilitates DNA bending into a loop or a nucleosome-like structure that positions the basal transcription complex in the vicinity of CDE III-bound CBF3. Moreover, it was observed that transcription activation by CDE III-bound AD–Cbf3a and AD–Cbf3c, utilizing a CEN3–HIS3 reporter cassette [YJL128 (cen3::CEN3–HIS3)] was affected when the CDE III-localized protein complex was compromised (removal of Ctf19) (J. Ortiz et al., unpubl.). Therefore it is speculated that CDE II cannot form the postulated DNA conformation per se. Instead, the CEN DNA conformation is possibly the result of the interaction of CDE II with a protein interface that requires the CDE III-localized protein complex. This interface may be provided by Mif2, which contains a binding motif for AT-rich sequences (Brown et al. 1993), and by the putative Cse4–histone complex. As mentioned above, a standard nucleosome would not depend on CDE II, but a structure that assembles with half the length of DNA that a standard nucleosome contains (Fig. 7B) might only be stable in combination with AT-rich DNA.
CDE II is absolutely necessary for centromere function. This implies that the centromere conformation imposed by the presence of CDE II is essential. Possible explanations for the importance of the CDE II effect include the following. First, only CDE II might assemble a nucleosome-like structure at the S. cerevisiae kinetochore. This could make the kinetochore an integral part of higher order chromatin structures and provide stability. Second, CDE II-induced DNA conformation might allow protein–protein interactions that would not be possible otherwise. Third, a CDE II–protein interaction results in a conformational change and/or leads to the acquisition of further proteins.
As shown in the proposed model (Fig. 7B), there are 10 established kinetochore proteins to date. Furthermore, it is conceivable that the budding yeast kinetochore is constructed via a specialized centromere nucleosome and that the CDE II element may have a structural effect on chromatin conformation that evokes an active kinetochore. For these reasons, the S. cerevisiae kinetochore appears to be more complex than originally expected. On the other hand, we have more reasons to believe that the S. cerevisiae kinetochore may have a closer resemblance to the kinetochores of higher eukaryotes.
Materials and methods
Yeast strains with integrated reporter constructs
Strains with reporter gene constructs used for the one-hybrid screen and chromatin immunoprecipitation were produced as follows: YJL128 (cen3::CEN3–HIS3): A 1506-bp DrdI fragment of pRS413 (Sikorski and Hieter 1992) that contained the HIS3 ORF, including promoter and terminator sequences, was blunt-end cloned into SmaI of pUC18 resulting in pJL246. The BamHI–Mva1269I fragment of pJL246 was replaced by a PCR-generated fragment that contained the 5′ HIS3 ORF to the Mva1269I site plus 39 bp (minimal promoter) upstream of the start codon resulting in pJL400. The SmaI–XbaI fragment of pRN505 (Ng and Carbon 1987) containing CEN3 DNA was cloned into the HincII–XbaI sites of pJL400 resulting in pJL405. A PCR-generated DNA fragment that maps to the left of CEN3 on chromosome III (position 113261–113919) was blunt-end cloned into the SphI site of pJL405 resulting in pJL434 and a PCR-generated DNA fragment that maps to the right of CEN3 on chromosome III (position 114821–115487) was blunt-end cloned into the SacI site of pJL434 resulting in pJL437. The integration cassette was released from pJL437 by ClaI digestion and transformed into SFY526 (Harper et al. 1993) selecting for growth on histidine-free medium. Integration of the reporter construct at the CEN3 locus was verified by PCR.
YJL148 (LYS2::CEN3AGC–HIS3): The SacI–SphI fragment from pJL400 containing the HIS3 ORF plus minimal promoter and termination sequences was blunt-end cloned into the SmaI site of YDp-K (Berben et al. 1991) resulting in pJL469a. CEN3 DNA with the CCG triplet of the CDE III element replaced by AGC was produced by recombinant PCR (Innis et al. 1990) using pRN506 (Ng and Carbon 1987) as the template. The PCR product that included the polylinker sites of pRN506 was digested with XbaI and EcoRI and cloned into the XbaI–EcoRI sites of pJL469a resulting in pJL530. pJL530 was linearized within the LYS2 gene by StuI digestion and transformed into SFY526 selecting for growth on histidine- and lysine-free medium. Integration of the reporter construct was verified by PCR.
YJL138 (LYS2::CDE III–HIS3)
The SspI–BamHI fragment of pRN505 containing the CDE III element of CEN3 DNA was blunt-end cloned into the SmaI site of pUC18. In the resulting plasmid, pJL4, the remaining half of the SspI recognition sequence is adjacent to the BamHI site of the polylinker. Using pJL4 as a template, PCR was performed with the T7/T3 sequencing primer (AACAGCTATGACCATG) and the pUC18-2 primer (ATCAATCTAGAAGGCGATTAAGTTGGGTA) that introduces an XbaI site into the PCR product. After digestion with EcoRI and partial digestion with XbaI (there is an internal XbaI site in the PCR product) the DNA fragment containing CDE III plus pUC18 DNA was cloned into the EcoRI–XbaI sites of pJL469a. In the resulting plasmid, pJL476, the CDE III element is positioned at an identical distance from the HIS3 promoter as in the CEN3–HIS3 reporter construct (see above). pJL476 was linearized within the LYS2 gene by StuI digestion and transformed into SFY526 selecting for growth on histidine- and lysine-free medium. Integration into the LYS2 locus was verified by PCR.
YJL162 (cen2::TRP1, LYS2::CEN3–HIS3)
The EcoRI–XbaI fragment of pJL530 was replaced by the EcoRI–XbaI fragment (CEN3WT) from pRN505 (Ng and Carbon 1987) resulting in pJL562. A DNA fragment of chromosome II (position 24244–26382) including CEN2 was amplified by PCR and blunt-end cloned into the EcoRI site of pBluescript SK− resulting in pJL569. The StuI–EcoRI fragment of pJL569 was replaced with the TRP1 gene resulting in pJL571. The integration cassette was released from pJL571 by StyI–SpeI digestion and cotransformed into SFY526, together with pJL562 that had been linearized by StuI digestion. Selecting for growth on lysine- and tryptophane-free medium resulted in strain YJL162. This strain has CEN2 replaced by TRP1 and harbors a CEN3–HIS3 reporter construct at the LYS2 locus, as was verified by PCR.
One-hybrid screen
YJL128 (cen3::CEN3–HIS3) was transformed with a S. cerevisiae genomic library in pGAD–C (James et al. 1996) or a S. cerevisiae cDNA library (gift from Steve Elledge, Department of Biochemistry, Baylor College of Medicine, Houston, Texas) in pACT2 (Li et al. 1994) selecting for growth on SD/His− medium (Guthrie and Fink 1991) plus 5 mm 3-AT. From 80 million transformants, spread out on 45 large (145/20-mm) plates, 120 clones were positively identified. Twenty-eight of these clones did not depend on the transformed plasmid for growth and were therefore discarded. Plasmids from 30 of the remaining clones were shuttled into Escherichia coli and the inserts were partly sequenced. Several clones derived from both libraries contained the OKP1 gene. The MCM21 gene was only obtained from the cDNA library, whereas CTF19 was only obtained from the genomic library. Furthermore, CBF3A and ATR1, which confers 3-AT resistance (Kanazawa et al. 1988), were only obtained from the genomic library. Southern analysis of plasmid inserts amplified by PCR showed that the remaining 62 clones harbored plasmids with one of the genes described above.
Gene disruptions
CTF19 disruption
A PCR product containing the CTF19 ORF with an NdeI site at the start codon and an XhoI site immediately after the stop codon was cloned into the EcoRV site of pBluescript II SK− resulting in pJL494. The EcoRV fragment of pJL494 was replaced by the TRP1 gene. After digestion with EcoRI and BglII the resulting plasmid, pJL507, was transformed into YJL19 [MATa ade2-101ochre trp1-Δ63 leu2-Δ1 ura3-52 his3-Δ200 lys2-801amber cyh2R (CF CEN6 URA3 SUP11 CYH2S)], which was derived by sporulation of YTF3, a gift from Johannes Hegemann, Institut für Mikrobiologie, Heinrich-Heine-Universität, Duesseldorf, Germany. Replacement of the wild-type CTF19 by the disruption cassette was verified by PCR. In the resulting strain, YJL142, the CTF19 ORF is disrupted after the first 282 bp.
MCM21 disruption
A PCR product containing the MCM21 ORF, including intron plus 380 bp upstream and 437 bp downstream sequences, was cloned into the EcoRV site of pBluescript II SK− resulting in pJL496. The NdeI fragment of pJL496 was replaced by the TRP1 gene, the resulting plasmid, pJL511, was digested with ClaI and EcoRI and transformed into YJL19 resulting in YJL146. As verified by PCR, gene replacement removed the complete MCM21 ORF.
OKP1 disruption
A PCR product containing the OKP1 ORF (YGR179c) plus 368 bp upstream and 452 bp downstream sequences was cloned into the SmaI site of pUC18 resulting in pJL465. Subsequently the SalI–SacI fragment containing the above PCR product was cloned into the SalI and SacI sites of pRS416 and pRS415 (Sikorski and Hieter 1992) resulting in pJL467 and pJL473, respectively. The SnaBI fragment of pJL465 was replaced by the TRP1 gene; the resulting plasmid, pJL466, was digested with HindII and SacI and transformed into the diploid strain YPH501 (Sikorski and Hieter 1992). As verified by PCR, gene replacement in the resulting strain YJL131 eliminated one complete copy of OKP1. To demonstrate that Okp1 is essential, YJL131 was transformed with pJL467 and subsequently sporulated. The resulting haploid strain, YJL132 (MATa OKP1::TRP1), contained pJL467, which provided a functional copy of OKP1. When streaked onto plates containing 5-fluoro-orotic acid (FOA), conditions that selected against the Ura+ phenotype established by pJL467, no growth could be detected. This demonstrates that OKP1 is essential because loss of pJL467 that contains the only functional copy of OKP1 cannot be tolerated.
Temperature-sensitive okp1-5 mutant
The complete OKP1 ORF was released from pJL473 (see above) by digestion with SnaBI. The remaining vector was mixed with a DNA fragment generated by error-prone PCR (Muhlrad et al. 1992) that contained the OKP1 ORF plus 368 bp upstream and 452 bp downstream sequences. DNA was denatured by incubation at 94°C for 4 min and reannealed by decreasing the temperature from 94°C to 45°C at a rate of 2°/min. After transformation into YJL132 harboring pJL467, single-stranded regions of mixed hybrids from vector DNA and PCR-generated DNA were healed in vivo. Transformants were regrown on SD medium containing FOA to select against pJL467. Subsequently, single colonies were streaked onto YPD plates (Guthrie and Fink 1991) and grown at 24°C or 37°C. Of 1050 clones that grew on FOA medium, 1 was temperature sensitive for growth at 37°C. The plasmid pJL499 harbored by this clone was isolated. For chromosomal integration, 376 bp of DNA starting 450 bp downstream of the OKP1 ORF with KpnI and EcoRI sites at the ends was generated by PCR. Utilizing the KpnI and EcoRI sites, this PCR product was cloned into the KpnI and EcoRI sites of pJL287, which contained the TRP1 gene as a DrdI fragment from pRS414 (Sikorski and Hieter 1992) blunt-end cloned into the SmaI site of pUC18. This resulted in pJL517. The KpnI–SalI fragment of pJL499 containing the OKP1 ORF plus flanking sequences was cloned into the BamHI–SalI site of pJL517 after filling in the BamHI and KpnI sites. The resulting plasmid pJL529 contains the OKP1 promoter and ORF with the temperature-sensitive mutation plus downstream sequences interrupted by the TRP1 gene. pJL529 was digested with SpeI and ClaI and transformed into YJL19 and transformants were tested for a temperature sensitive growth phenotype. Also the correct integration of the OKP1–TRP1 construct was verified by PCR. This resulted in the temperature-sensitive strain YJL158, which carries the okp1-5 mutation.
Plasmids that express fusion constructs
Plasmids that express proteins fused to the DNA-binding domain of Gal4 (BD) were constructed as follows: BD–Ctf19: A PCR product containing the CTF19 ORF with an NdeI site at the start codon and an XhoI site after the stop codon was cloned into the NdeI–SalI site of pAS1 (Harper et al. 1993) resulting in pJO501. BD–Mcm21: A PCR product containing the MCM21 ORF with an NdeI site at the start codon and an XhoI site after the stop codon was cloned into pAS1 resulting in pJO508.
Plasmids that expressed proteins fused to the Gal4 transcriptional activation domain (AD) were produced as follows: AD–Cbf3a: A DNA fragment that contained the complete CBF3A ORF plus downstream sequences derived from pWJ110-P (Jiang et al. 1993) by BamHI digestion and partial NdeI digestion was blunt-end cloned into the SmaI site of pACT2 (Li et al. 1994) resulting in pJO196. AD–Cbf3b: The NdeI–BamHI fragment containing the CBF3B ORF from pJL32 (Lechner 1994) was cloned into the SmaI site of pACT2 resulting in pJO197. AD–Cbf3c: The NdeI–BamHI fragment containing the CBF3C ORF from pJL36 (Stemmann and Lechner 1996) was cloned into the SmaI site of pACT2 resulting in pJO198. A PCR product containing the CBF3D ORF plus downstream sequences and a filled-in NdeI site at the start codon was blunt end cloned into the SmaI site of pACT2 resulting in pJO250. AD–Cse4: A PCR product that contained the CSE4 ORF as described by Stoler et al. (1995) with an NcoI site at the start codon and an XhoI site after the stop codon was cloned into the NcoI–SalI sites of pACT2 resulting in pJL484. AD-Mif2: A PCR product containing the MIF2 ORF with an NcoI site at the start codon and an XhoI site after the stop codon was cloned into the NcoI–SalI sites of pACT2 resulting in pJL486. AD–Cbf1: A PCR product containing the CBF1 ORF with an NdeI site at the start codon and an XhoI site after the stop codon was blunt-end cloned into the SmaI site of pACT2 resulting in pJO509a. AD–Ctf19, AD–Okp1, and AD–Mcm21 were isolated in the one-hybrid screen as the pGAD–C-based vectors pGAD–C/CTF19 and pGAD–C/OKP1, or the pACT2 based vector pACT2/MCM21.
Vectors that expressed Flag-tagged proteins were as follows: Flag–Ctf19: A PCR product that contained the ORF of Flag–Ctf19 (MDYKDDDDKH linked to the amino terminus of the CTF19 ORF) was blunt-end cloned into a NotI site of pNEV-N-Leu (Sauer and Stolz 1994). The resulting vector pJL497 contains an NcoI site at the start codon, an NdeI site at the first methionine codon of the CTF19 ORF, and an XhoI codon after the stop codon. For in vitro transcription, the NcoI–XhoI fragment of pJL497 was cloned into the NcoI–SalI sites of pSPUTK (Stratagene) resulting in pUW541. Flag–Mcm21: The NdeI–XhoI fragment from pJL497 was replaced by the NdeI–XhoI fragment from pJO508 resulting in pJL505. Flag–Okp1: A PCR product that contained an ORF coding for MDYKDDDDKRPQ fused to the amino terminus of Okp1 was blunt-end cloned into the NotI site of pNEV-N-Leu. The resulting vector pJL471 contains a NotI site 10 bp upstream of the first methionine codon of the OKP1 ORF and an XhoI site after the stop codon. Flag–Cse4: The NdeI–XhoI fragment from pJL497 was replaced by the NheI–XhoI fragment from pJL484 after filling in the NdeI and NheI sites resulting in pOS540.
To express His-tagged Ctf19 in E. coli the NdeI–XhoI fragment from pJL494 was cloned into the NdeI–XhoI sites of pET16b (Novagen), resulting in pJL515. To express His-tagged Cbf3d in E. coli, a PCR product containing the CBF3D ORF plus downstream sequences with an NdeI site at the start codon and an BamHI site after the downstream sequences was cloned into the NdeI–BamHI site of pET16b resulting in pOS233. Constructs that utilized PCR products were verified by sequencing.
Antibody preparation
For anti-Cbf3d and anti-Ctf19 antibody preparation, pOS233 or pJL515 was transformed into E. coli BL21(DE3). After induction with isopropylthiogalactoside, His10-tagged Cbf3d was purified under native conditions and His10-tagged Ctf19 was purified under denaturing conditions using NTA agarose (Qiagen) according to the manufacturer’s recommendations. Polyclonal rabbit anti sera production was performed by Dr. J. Pineda (Antikörper-Service, Berlin). Subsequently, antibodies were affinity purified as described (Lechner 1994).
Cytological procedures
The colony-sectoring assay was performed with strains YJL142 (MATa ctf19::TRP1 CF), YJL146 (MATa mcm21::TRP1 CF), and YJL158 (MATa okp1-5 CF) as described (Koshland and Hieter 1987).
For phase contrast, DAPI fluorescence microscopy, and FACS analysis, cells were grown to early logarithmic stage at room temperature and then shifted to 37°C for 4 hr. DAPI staining was performed as described (Sherman et al. 1986). For FACS analysis, 12 × 107 cells were washed in 1 ml of 0.2 m Tris (pH 7.5) and resuspended in 1 ml of 70% ethanol/0.2 m Tris (pH 7.5). After intensive vortexing, the samples were incubated at 4°C for 14 hr. Subsequently the cells were pelleted, washed, and resuspended in 1 ml of Tris (pH 7.5). Samples of 0.25 ml were incubated with RNase A at a final concentration of 4 mg/ml for 2 hr at 37°C. After addition of 5 ml of 0.2 m Tris (pH 7.5) the cells were collected by centrifugation, resuspended in 5 ml of a 5 μg/ml propidium iodide solution in 0.2 m Tris (pH 7.5), and subjected to FACS analysis.
Two-hybrid analysis
Plasmids that expressed AD and BD fusion proteins, as described above and indicated in Figure 4A were cotransformed into HF7c (Feilotter et al. 1994) selecting for growth on SD/Leu−/Trp− medium. Transformants were streaked onto SD/His− plates containing 15 mm 3-AT (Fields and Song 1989) and grown for 3 days at 30°C.
Immunoprecipitation
Extracts from YJL131 harboring pJL471 grown in YPD medium were used to analyze coimmunoprecipitation of Cbf3a, Cbf3b, and Ctf19 with Flag-tagged Okp1. Extracts from YPH499 harboring pJL471 and pJO508 grown in SD/Leu−/Trp− were used to analyze coimmunoprecipitation of BD–Mcm21 with Flag-tagged Okp1. Extracts from YJL146 harboring pJL505 grown on SD/Leu− were used to analyze coimmunoprecipitation of Ctf19 with Flag-tagged Mcm21. Extracts were prepared from cells grown to late logarithmic phase as described (Schultz et al. 1997). Triton X-100 was added to 1%, and 250 μl of extracts with a protein concentration of ∼30 mg/ml was incubated with 20 μl of anti-Flag Sepharose (M2, Eastman-Kodak) for 3–14 hr at 4°C. Subsequently, the beads were washed four times in lysis buffer (Schultz et al. 1997) and proteins were eluted by boiling in 40 μl of SDS sample buffer. Western analysis was performed using anti-Flag M2 antibody (Eastman-Kodak), anti-HA antibody (BabCo), anti-Cbf3a antibody (Stemmann and Lechner 1996), anti-Cbf3b antibody (Lechner 1994), and anti-Ctf19 antibody (see above).
To analyze Ctf19 coimmunoprecipitation with Cbf3a, 35S-labeled Ctf19 was synthesized by in vitro transcription of pUW541 with SP6–RNA polymerase followed by translation in reticulocyte lysate (Promega) according to the manufacturer’s recommendation. Subsequently 1 μg of purified Cbf3a–His6 (Stemmann and Lechner 1996) was added to 50 μl of the translation mix. After incubation for 30 min at room temperature, 200 μl of lysis buffer (Schultz et al. 1997) plus 1% Triton X-100, 5 μg of purified anti-Cbf3a antibody, and 10 μl of protein A–Sepharose was added. The samples were incubated, washed, and eluted as above and subjected to SDS-PAGE following autoradiography.
Chromatin immunoprecipitation
In vivo cross-linking and chromatin immunoprecipitation was performed with 109 cells as described (Hecht et al. 1998). To assay the in vivo interaction of Cbf3a, Cbf3b, Cbf3c, and Cbf3d in YJL128 (cen3::CEN3–HIS3), YJL138 (LYS2::CDE III–HIS3), YJL148 (LYS2::cen3AGC–HIS3), YPH499 (Sikorski and Hieter 1992) or ndc10-1 (Goh and Kilmartin 1993), and Flag-tagged Okp1 in YJL132 (okp1::TRP1) harboring pJL471, cells were grown to 2 × 107 cells/ml in YPD medium (Figs. 5 and 6). To assay Flag-tagged Ctf19, -Mcm21, -Okp1, and -Cse4 in YJL128 (cen3::CEN3–HIS3), YJL138 (LYS2::CDE III–HIS3), YJL148 (LYS2::cen3AGC–HIS3), YJO157, and ndc10-1 harboring corresponding plasmids (pJL497, pJL505, pJL471, pOS540), cells were grown to 1 × 107 cells/ml under selective conditions in SD medium. For arrest at the nonpermissive temperature, ndc10-1 was grown at 25°C to 0.5 × 107 cells/ml and then shifted to 37°C for 3 hr. Immunoprecipitations were performed with polyclonal rabbit antibodies anti-Cbf3a, anti-Cbf3b, anti-Cbf3c (Stemmann and Lechner 1996) and anti-Cbf3d or with anti-Flag M2 agarose (Eastman-Kodak). Oligonucleotides used to amplify CEN DNA fragments and control fragments are summarized in Table 2.
Table 2.
PCR primers used in the Chip assay
|
Fragment amplifieda
|
Fragment size (bp)
|
Primers used
|
Primer origin
|
|---|---|---|---|
| CEN3WT–HIS3 | 275 | AACTTCCACCAGTAAACGTTTC | flanking CEN3 |
| CTTCGTTTATCTTGCCTGCTC | HIS3 | ||
| CEN3AGC–HIS3 | 275 | as above | |
| CDEIII–HIS3 | 275 | as above | |
| CEN3WT | 243 | AACTTCCACCAGTAAACGTTTC | flanking CEN3 |
| GATCAGCGCCAAACAATATGG | flanking CEN3 | ||
| III–L | 213 | ACTTTGGCTTTCCGCTCGTG | Chromosome III |
| GAAAGTCTTCTAGAGTTACAGG | Chromosome III | ||
| III–R1 | 321 | GACCAGCATGTAGGAAGGTG | Chromosome III |
| ACATTGATAAATTGCTCTCACCA | Chromosome III | ||
| III–R2 | 278 | ATCGCATTTCTTTTCGTCCACA | Chromosome III |
| TTGGACGGGGATCGTTCGTA | Chromosome III |
Acknowledgments
We thank P. James for the S. cerevisiae genomic library, S. Elledge for the S. cerevisiae cDNA library, S. Klein and J. Hegemann for performing the FACS analysis, K. Hyland and P. Hieter for communicating results before publication, S. Strahl-Bolsinger for technical advice concerning the Chip assay, and U. Weihe for performing some of the Chip assays and immunoprecipitations. This work was supported by a grant from the Deutsche Forschungsgemeinschaft.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Johannes.Lechner@vkl.uni-regensburg.de; FAX 0941-943-2855.
References
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:265–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. The YDp plasmids: A uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- Bourns BD, Alexander MK, Smith AM, Zakian VA. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol Cell Biol. 1998;18:5600–5608. doi: 10.1128/mcb.18.9.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MT. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 1995;160:111–116. doi: 10.1016/0378-1119(95)00163-z. [DOI] [PubMed] [Google Scholar]
- Brown MT, Goetsch L, Hartwell LH. Mif2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KHA. Structural organization and general properties of the higher eukaryotic centromere. In: Choo KHA, editor. The centromere. Oxford, UK: Oxford University Press; 1997a. pp. 77–96. [Google Scholar]
- Choo KHA. Centromere DNA of higher eukaryotes. In: Choo KHA, editor. The centromere. Oxford, UK: Oxford University Press; 1997b. pp. 97–142. [Google Scholar]
- Choo KHA. Centromere DNA of higher eukaryotes. In: Choo KHA, editor. The centromere. Oxford, UK: Oxford University Press; 1997c. pp. 143–253. [Google Scholar]
- Connelly C, Hieter P. Budding Yeast SKP1 encodes an evolutionary conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumberledge S, Carbon J. Mutational analysis of meiotic and mitotic centromere function in Saccharomyces cerevisiae. Genetics. 1987;117:203–212. doi: 10.1093/genetics/117.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feilotter HE, Hannon GJ, Ruddel CJ, Beach D. Construction of an improved host strain for two-hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–247. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Goh P, Kilmartin JV. NDC10: A gene involved in chromosome segregation in S. cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky GJ. Kinetochores, microtubules and the metaphase checkpoint. Trends Cell Biol . 1995;5:143–148. doi: 10.1016/s0962-8924(00)88968-0. [DOI] [PubMed] [Google Scholar]
- Grosschedl R. Higher-order nucleoprotein complexes in transcription: analogies with site-specific recombination. Curr Opin Cell Biol. 1995;7:362–370. doi: 10.1016/0955-0674(95)80091-3. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR, editors. Guide to yeast genetics and molecular biology. New York, NY: Academic Press; 1991. [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hecht, A., S. Strahl-Bolsinger, and M. Grunstein. 1999. Mapping chromosomal interaction sites of non-histone proteins: Crosslinking studies in yeast. Methods (in press). [DOI] [PubMed]
- Hegemann JH, Fleig UN. The centromere of budding yeast. BioEssays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- Hyland, K.M., J. Kingsbury, D. Koshland, and P. Hieter. 1999. Ctf19: A novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and the mitotic spindle. J. Cell Biol. (in press). [DOI] [PMC free article] [PubMed]
- Hyman AA, Sorger PK. Structure and function of kinetochores in budding yeast. Annu Rev Cell Dev Biol. 1995;11:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols. A guide to methods and applications. San Diego, CA: Academic; 1990. [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Lechner J, Carbon J. Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J Cell Biol. 1993;121:513–519. doi: 10.1083/jcb.121.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa S, Driscoll M, Struhl K. ATR1, a Saccharomyces cerevisiae gene encoding a transmembrane protein required for aminotriazole resistance. Mol Cell Biol. 1988;8:664–673. doi: 10.1128/mcb.8.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan KB, Hyman AA, Sorger P. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- Koshland D, Hieter P. Visual assay for chromosome ploidy. Methods Enzymol. 1987;155:351–372. doi: 10.1016/0076-6879(87)55024-8. [DOI] [PubMed] [Google Scholar]
- Krek W. Proteolysis and the G1-S transition: The SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the S. cerevisiae kinetochore complex, CBF3. EMBO J. 1994;13:5203–5211. doi: 10.1002/j.1460-2075.1994.tb06851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Carbon J. A 240 kdal multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Lechner J, Ortiz J. The S. cerevisiae kinetochore. FEBS Lett. 1996;389:70–74. doi: 10.1016/0014-5793(96)00563-7. [DOI] [PubMed] [Google Scholar]
- Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Li L, Elledge SJ, Peterson CA, Bales ES, Legerski RJ. Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maine GT, Sinha P, Tye B-K. Mutants of S. cerevisiae defective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D, Wood JS, Garvik B, Hartwell LH. Isolation of two genes that affect mitotic chromosome transmission in S. cerevisiae. Cell. 1986;44:53–63. doi: 10.1016/0092-8674(86)90484-8. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes & Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Ng R, Carbon J. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4522–4534. doi: 10.1128/mcb.7.12.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymond BC, Rosbash M. Yeast pre-mRNA splicing. In: Jones EW, Pringle JR, Broach JR, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 143–192. [Google Scholar]
- Sauer N, Stolz J. SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker’s yeast and identification of the histidine-tagged protein. Plant J. 1994;6:67–77. doi: 10.1046/j.1365-313x.1994.6010067.x. [DOI] [PubMed] [Google Scholar]
- Saunders MJ, Yeh E, Grunstein M, Bloom K. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol Cell Biol. 1990;10:5721–5727. doi: 10.1128/mcb.10.11.5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman IG, Bloom K. Genetic dissection of centromere function. Mol Cell Biol. 1993;13:3156–3166. doi: 10.1128/mcb.13.6.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MC, Hockman DJ, Harkness TAA, Garinther WI, Altheim BA. Chromatin assembly in a yeast whole-cell extract. Proc Natl Acad Sci. 1997;94:9034–9039. doi: 10.1073/pnas.94.17.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. DAPI staining protocol. In: Sherman F, Fink GR, Hicks JB, editors. A laboratory course manual for methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. p. 153. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1992;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge J, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Sorger PK, Severin FF, Hyman AA. Factors required for the binding of reassembled yeast kinetochores in microtubules in vitro. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Doheny KF, Hieter P, Kopski KM, Huffaker TC, Hyman AA. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc Natl Acad Sci. 1995;92:12026–12030. doi: 10.1073/pnas.92.26.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O, Lechner J. The Saccharomyces cerevisiae kinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO J. 1996;15:3611–3620. [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes & Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Kingsbury J, Koshland D. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J Cell Biol. 1995;128:749–760. doi: 10.1083/jcb.128.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]