Abstract
Background
Despite the successful eradication of smallpox by the WHO-led vaccination programme, pox virus infections remain a considerable health threat. The possible use of smallpox as a bioterrorism agent as well as the continuous occurrence of zoonotic pox virus infections document the relevance to deepen the understanding for virus host interactions. Since the permissiveness of pox infections is independent of hosts surface receptors, but correlates with the ability of the virus to infiltrate the antiviral host response, it directly depends on the hosts proteome set. In this report the proteome of HEK293 cells infected with Vaccinia Virus strain IHD-W was analyzed by 2-dimensional gel electrophoresis and MALDI-PSD-TOF MS in a bottom-up approach.
Results
The cellular and viral proteomes of VACV IHD-W infected HEK293 cells, UV-inactivated VACV IHD-W-treated as well as non-infected cells were compared. Derivatization of peptides with 4-sulfophenyl isothiocyanate (SPITC) carried out on ZipTipμ-C18 columns enabled protein identification via the peptides' primary sequence, providing improved s/n ratios as well as signal intensities of the PSD spectra. The expression of more than 24 human proteins was modulated by the viral infection. Effects of UV-inactivated and infectious viruses on the hosts' proteome concerning energy metabolism and proteins associated with gene expression and protein-biosynthesis were quite similar. These effects might therefore be attributed to virus entry and virion proteins. However, the modulation of proteins involved in apoptosis was clearly correlated to infectious viruses.
Conclusions
The proteome analysis of infected cells provides insight into apoptosis modulation, regulation of cellular gene expression and the regulation of energy metabolism. The confidence of protein identifications was clearly improved by the peptides' derivatization with SPITC on a solid phase support. Some of the identified proteins have not been described in the context of poxvirus infections before and need to be further characterised to identify their meaning for apoptosis modulation and pathogenesis.
Keywords: viral Proteomics, Vaccinia Virus, MALDI-PSD-TOF MS, SPITC
Background
Despite remarkable progress in the control and treatment of infectious diseases, the problem of emerging and re-emerging pathogens is likely to be one of the main issues of medical science and public health in the twenty-first century [1]. In this respect viral diseases are of particular concern, because advances in the field of antiviral drugs have lagged behind those regarding bactericidal drugs and antibiotics. It was shown by the emergence of the severe acute respiratory syndrome (SARS) that new members of neglected virus families can cross into humans from unsuspected reservoirs, making rapid advances in our understanding of virus-host dynamics necessary [2]. In this regard poxviruses are of particular importance. Despite the success of the WHO-led smallpox eradication programme, other related human-pathogenic poxviruses remain a considerable threat. In particular, the 2003 outbreak of human monkeypox virus in the United States of America pointed out the imminence posed by zoonotic pox infections originating from animal-borne viruses [3]. Moreover, an increasing number of human cowpox virus infections, especially affecting younger people lacking smallpox vaccination, have been reported in Europe in recent years [4]. In addition to the threat of zoonoses, the relevance of a deeper understanding of the interactions of poxviruses with their host cells is pointed out by considering the classification of smallpox as a category A bioterrorism agent by the Centers for Disease Control and Prevention (CDC), especially since no acceptable treatment is available and the immunity in the population is declining [5].
In contrast to many other viruses, the ability of poxviruses to productively infect a given cell is not determined at the level of specific host receptors, but is regulated downstream of virus adsorption to the cell surface and virus entry by intracellular processes which for the most part are unexplored [6-10]. Since the permissiveness of orthopoxviruses is therefore dependent on the hosts proteome set, the identification of cellular proteins affected by the protein products of the so-called viral host-range genes with proteomic approaches will be a starting point for further functional protein analysis. The characterization of the Vaccinia Virus (VACV) virions' proteome has enlightened the protein equipment of the virus at the entry stage of infection [11]. Furthermore, virus-host dynamics have been analysed by yeast two-hybrid (Y2H) studies to identify interaction partners [12]. No closer look at the dynamic changes of the human proteome during a VACV infection has been taken yet on a global scale.
In this report an infection of human embryonic kidney (HEK) 293 cells with well-studied VACV was chosen as a model system. Uninfected HEK 293 cells serve as a control, while VACV-infected HEK 293 cells inactivated with UV enable the study of the influence of the virions' proteins on the cellular proteome. The investigation of infection effects on the expression of the cellular proteome was carried out in the late phase of virus replication. An offline-coupling of two-dimensional sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Matrix Assisted Laser Desorption/Ionisation Time of Flight Mass Spectrometry (MALDI-ToF MS) was used in a bottom-up approach. In-gel trypsinated proteins from relevant spots of the 2-D gels are unambiguously identified via Mascot database search of the peptide mass fingerprint (PMF) recorded in positive ion reflectron mode of the MALDI-ToF MS. Identification rate was improved by obtaining additional sequence data of selected peptides in post source decay (PSD) mode, whereas a considerably advanced data quality was achieved by N-terminally derivatising the peptides on a ZipTipμ-C18 solid phase support with 4-Sulfophenyl isothiocyanate (SPITC) prior to MS measurement.
Results
PSD spectra of SPITC derivatized peptides improve protein identification efficiency
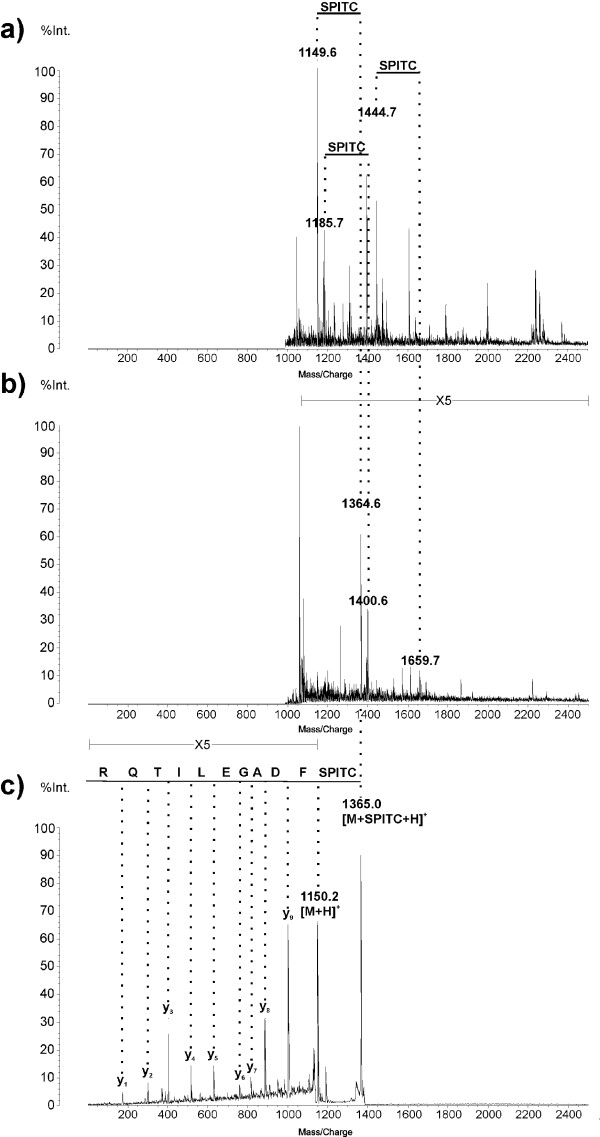
Improved protein identification by combined Mascot search of PMF and PSD spectra is shown exemplarily in Figure 1 for the spot C-5, human prohibitin (PHB). Database search of the underivatised PMF has not led to unambiguous protein identification. The best hits held protein scores between 55 and 57 at a threshold value for significance of 64. Among these, prohibitin is ranked third with a score of 56.
Figure 1.
MALDI-ToF MS spectra of prohibitin. The peptide mass fingerprint of prohibitin is shown before (a) and after (b) solid phase supported SPITC derivatisation as well as the PSD spectrum of the m/z value 1364.6 along with the corresponding y-series representing the primary sequence (c).
After SPITC derivatization, the ten most intensive m/z values of the derivatised PMF were selected by an ion gate for PSD fragmentation. The amino acid sequence of selected peptides can be distinguished considering the series of y-fragments. For the peptide of m/z 1365.0 the sequence FDAGELITQR (Figure 1c), for the peptide of m/z 1401.1 the sequence DLQNVNITLR (data not shown) and for the peptide of m/z 1160.3 the sequence IFTSI[GE]DYDER (data not shown) was confirmed.
Sequencing enables the mapping of a peptide to a certain protein within the confidence interval of its m/z value and increases its Mascot score. If the sequenced peptides do not represent homologue sections in the human proteome even one peptide sequence can be significant. In case of prohibitin each of the three peptides alone was sufficient for unambiguous protein identification. The combined search of PMF and PSD spectra of SPITC-derivatised peptides identifies spot C-5 as being human PHB with a Mascot Ion Score of 237 at a threshold for significance of 34.
Enhanced sensitivity of SPITC derivatization on solid phase support
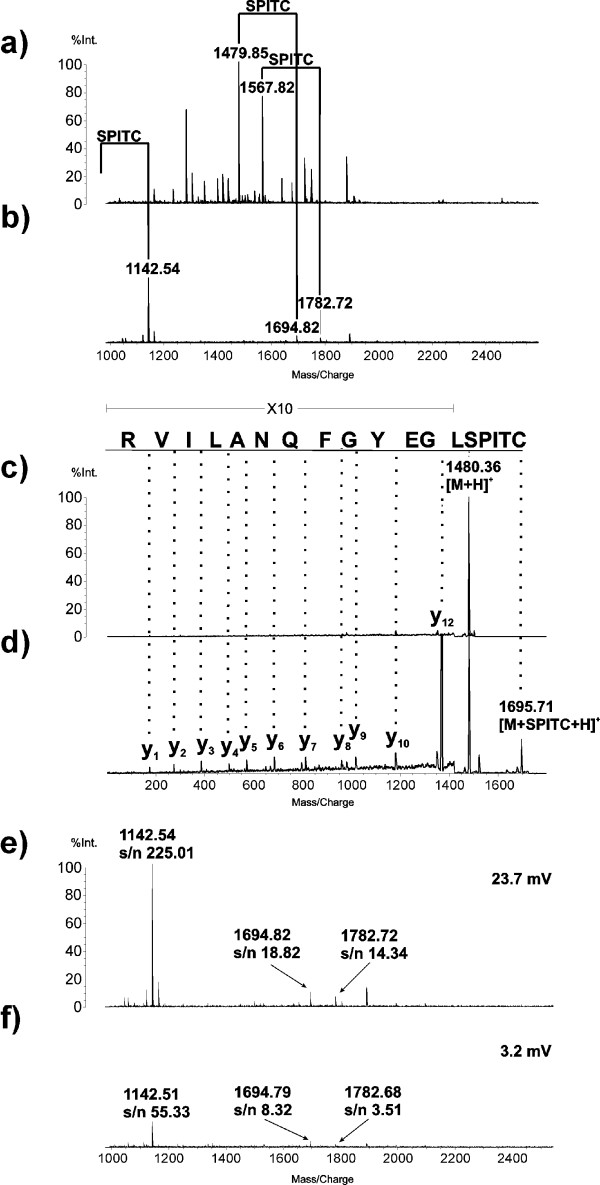
The peptide mass fingerprint of tryptic digested protein standard serum albumin (bovine) before and after SPITC derivatization is shown in Figure 2. The intensity is normalised on m/z value of 1479.9 of underivatised PMF spectra. The SPITC derivatization introduces a mass shift of 215 Da to the peptides and has strongly decreased their intensities. Many peptides could no longer be detected after derivatization, which negatively affects the sensitivity of protein identification. The underivatised protein standard was unambiguously identified with a MascotScore of 129 and a significant threshold of 61, but could no longer be identified from the PMF after SPITC derivatization.
Figure 2.
MALDI-ToF MS spectra of serum albumin (bos taurus). The pmf is faced before (a) and after (b) solid phase supported SPITC derivatization. The PSD spectra of m/z 1480,4 are shown before (c) and after (d) the derivatization along with the y-series. The pmfs of solid phase supported (e) and in solution (f) derivatization are opposed with the s/n ratios of identified peptides and the overall signal intensity charted.
The influence of the N-terminal sulfonation on the quality of PSD spectra is shown exemplarily in Figure 2c and 2d for the m/z value of 1480.4 of 10 pmol tryptic digested BSA. The intensity is normalised on the precursor ion of the underivatised PSD spectra. Despite the reduction of the precursor ions' intensity of 95%, the signal to noise (s/n) ratio of the peaks in the PSD spectra as well as the frequency of fragmentation were clearly increased by the N-terminal sulfonation. The improved coverage of the precursor ion y-series raised the Mascot Ion Score of the peptide with the mass of 1480.4 Da from 10 to 88.
The sulfonation of primary amines was done in aqueous solution at slightly alkaline pH value (pH = 8.6). Purification of peptides was carried out with ZipTipμ-C18 columns according to the principle of reversed phase chromatography. Generally, SPITC derivatization of peptides can be performed in solution prior to column binding as well as solid phase supported. Both sulfonation methods were modified compared to the literature with respect to signal intensity in order to enable protein identification with maximum sensitivity 15-16. Exemplarily, the resulting PMF spectra for in-solution trypsinated BSA has been compared with the solid phase supported approach concerning signal intensity, which is shown in Figure 2e/f. The signal to noise ratios (s/n) of m/z values used for identification were about 2 to 4 times higher for peptides derivatised on solid phase support than for the ones sulfonated in solution. The overall signal of PMF spectra in the mass range between 1000 and 2500 m/z is about 7 times larger for the standard derivatised on C18 columns. The improved s/n ratio and the increased signal intensity of the solid phase supported approach in PSD mode enlarges the coverage of the precursor ions' y-series and simplifies the automation of peak picking. The Mascot Score of the database search of ten accumulated PSD spectra was 176 for the solid phase supported approach and 139 for in solution sulfonation at a threshold value for significance of 33.
Comparative Proteome Analysis of VACV IHD-W infected HEK293 cells
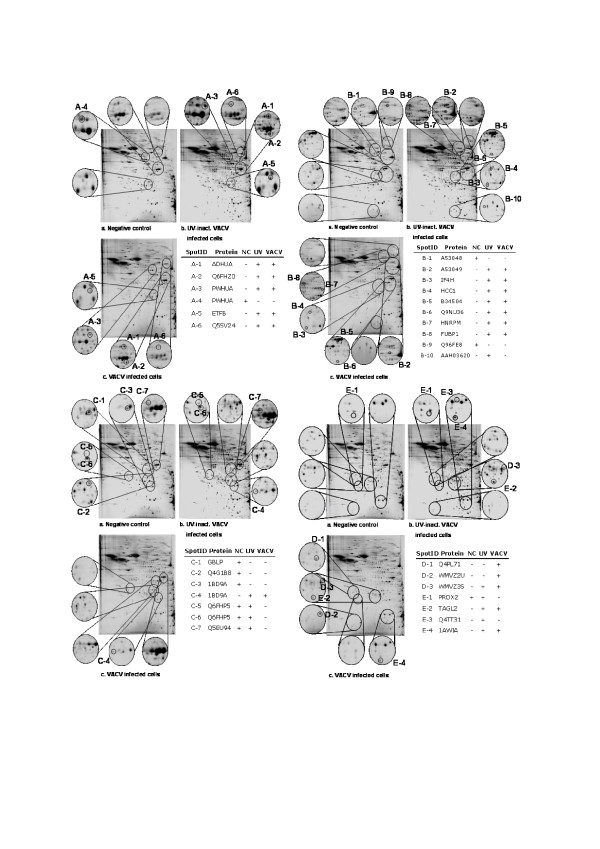
The comparative Proteome Analysis of infectious and UV-inactivated VACV IHD-W-infected HEK293 cells with non-infected control cells led to the identification of 24 modulated human proteins as well as 3 viral proteins. The proteins were identified via offline-coupling of 2D-SDS-PAGE (Figure 3) in the pI range of 4-9 and the mass range of 10-250 kDa with MALDI-PSD ToF MS and are classified according to their physiological role in the dynamic proteome. The five protein classes are proteins relevant for energy metabolism (A), proteins associated with gene expression and protein biosynthesis (B), proteins relevant for apoptosis (C), viral proteins (D) and other proteins (E). Proteins of the same class are numbered consecutively. The positions of the proteins identified in the 2DE gel are shown in different apertures in Figure 3. The molecular weight and pI values of these proteins are compared with the theoretical values in the SwissProt database (table 1). The entire protein denotations are listed in table 2.
Figure 3.
Identified protein spots in 2D gels. The spots of identified proteins are shown in the 2D gels of the negative control, HEK293 cells infected with VACV IHD-W and HEK293 incubated with UV inactivated VACV IHD-W concerning their classification. (+ identified, - not identified)
Table 1.
Molecular masses and pI values of identified proteins
| SpotID | Swiss Prot Accession # | Mascot Score |
# peptides (PSD) |
Sequence coverage [%] |
theor. MW [kDa] | MW [kDa] | Δ MW [kDa] | theor. pI | pI | ΔpI |
|---|---|---|---|---|---|---|---|---|---|---|
| A-1 | P04075 | 45(35) | 2 | 7 | 40 | 41 | -1 | 8.3 | 7.5 | 0.8 |
| A-2 | Q6FHZ0 | 179(36) | 2 | 8 | 36 | 34 | 2 | 8.9 | 7.6 | 1.3 |
| A-3 | P25705 | 132(35) | 3 | 6 | 60 | 50 | 10 | 9.2 | 6.3 | 2.9 |
| A-4 | P25705 | 94(35) | 3 | 5 | 60 | 49 | 11 | 9.2 | 5.8 | 3.4 |
| A-5 | P38117 | 86(36) | 2 | 9 | 28 | 26 | 2 | 8.3 | 6.5 | 1.8 |
| A-6 | Q5SV24 | 117(35) | 3 | 5 | 65 | 63 | 2 | 9.5 | 7.4 | 2.1 |
| B-1 | P41091 | 71(20) | 1 | 2 | 52 | 52 | 0 | 8.7 | 6.4 | 2.3 |
| B-2 | P41091 | 44(35) | 1 | 2 | 52 | 50 | 2 | 8.7 | 7.5 | 1.2 |
| B-3 | Q15056 | 95(39) | 1 | 5 | 27 | 26 | 1 | 6.9 | 6.0 | 0.9 |
| B-4 | HCC1 | 57(36) | 1 | 6 | 24 | 31 | -7 | 6.1 | 6.1 | 0.0 |
| B-5 | P22626 | 259(35) | 3 | 10 | 38 | 35 | 3 | 9.0 | 7.3 | 1.7 |
| B-6 | Q9NU36 | 48(33) | 1 | 4 | 29 | 27 | 2 | 9.1 | 7.2 | 1.9 |
| B-7 | HNRPM | 121(36) | 4 | 6 | 78 | 66 | 12 | 8.9 | 6.8 | 2.1 |
| B-8 | Q96AE4 | 140(36) | 3 | 5 | 68 | 73 | -5 | 7.2 | 5.9 | 1.3 |
| B-9 | Q96FE8 | 37(34) | 1 | 2 | 69 | 98 | -29 | 9.4 | 5.6 | 3.8 |
| B-10 | O60925 | 75(36) | 1 | 9 | 14 | 14 | 0 | 6.3 | 5.9 | 0.4 |
| C-1 | P63244 | 282(36) | 4 | 13 | 36 | 31 | 5 | 7.6 | 6.1 | 1.5 |
| C-2 | Q4G1B8 | 117(42) | 1 | 6 | 24 | 25 | -1 | 8.6 | 6.4 | 2.2 |
| C-3 | 1BD9A | 71(36) | 2 | 7 | 21 | 20 | 1 | 7.2 | 6.3 | 0.9 |
| C-4 | 1BD9A | 55(36) | 1 | 7 | 21 | 21 | 0 | 7.2 | 6.4 | 0.8 |
| C-5 | Q6FHP5 | 237(34) | 3 | 11 | 30 | 29 | 1 | 5.6 | 5.5 | 0.1 |
| C-6 | Q6FHP5 | 176(35) | 3 | 11 | 30 | 28 | 2 | 5.6 | 5.5 | 0.1 |
| C-7 | Q5EU94 | 95(41) | 1 | 4 | 33 | 37 | -4 | 4.7 | 6.9 | -2.2 |
| D-1 | Q4PL71 | 173(37) | 2 | 11 | 22 | 24 | -2 | 5.4 | 5.3 | 0.1 |
| D-2 | P20535 | 84(38) | 2 | 15 | 13 | 14 | -1 | 5.3 | 5.4 | -0.1 |
| D-3 | P20642 | 66(38) | 1 | 9 | 23 | 25 | -3 | 5.5 | 6.3 | -0.8 |
| E-1 | P32119 | 31(28) | 1 | 5 | 22 | 21 | 1 | 5.7 | 5.5 | 0.2 |
| E-2 | P37802 | 158(35) | 3 | 17 | 23 | 21 | 2 | 8.5 | 5.9 | 2.6 |
| E-3 | Q4TT31 | 37(35) | 1 | 6 | 19 | 17 | 2 | 8.7 | 6.8 | 1.9 |
| E-4 | 1AWIA | 92(36) | 1 | 10 | 15 | 14 | 1 | 8.5 | 6.7 | 1.8 |
Full names of identified proteins.
Full names of identified proteins
| SpotID | NC | UV | VACV | Accession # | Protein |
|---|---|---|---|---|---|
| A-1 | - | + | + | P04075 | Fructose-bisphosphate Aldolase A |
| A-2 | - | + | + | Q6FHZ0 | Malate dehydrogenase 2 |
| A-3 | - | + | + | P25705 | H+-transporting two-sector ATPase alpha chain precursor |
| A-4 | + | - | - | P25705 | H+-transporting two-sector ATPase alpha chain precursor |
| A-5 | - | + | + | P38117 | Electron transfer flavoprotein subunit beta (Beta-ETF) |
| A-6 | - | + | + | Q5SV24 | ATPase family, AAA domain containing 3A |
| B-1 | + | - | - | P41091 | Translation initiation factor eIF-2 gamma chain |
| B-2 | - | + | + | P41091 | Translation initiation factor eIF-2 gamma chain |
| B-3 | - | + | + | Q15056 | Eukaryotic translation initiation factor 4H (eIF-4H) |
| B-4 | - | + | + | HCC1 | Nuclear protein Hcc-1 (CIP29) |
| B-5 | - | + | + | P22626 | Heterogeneous nuclear ribonucleoprotein B1 |
| B-6 | - | + | + | Q9NU36 | Novel protein similar to small nuclear ribonucleoprotein polypeptide A |
| B-7 | - | + | + | HNRPM | Heterogeneous nuclear ribonucleoprotein M (hnRNP M) |
| B-8 | - | + | + | Q96AE4 | Far upstream element-binding protein 1 (FUSE-binding protein 1) |
| B-9 | + | - | - | Q96FE8 | Ewing sarcoma breakpoint region 1, isoform EWS (EWSR1 protein) |
| B-10 | - | + | - | O60925 | Prefoldin subunit 1 |
| C-1 | + | - | - | P63244 | Guanine nucleotide binding protein |
| C-2 | + | - | - | Q4G1B8 | PSMA8 protein |
| C-3 | + | - | - | 1BD9A | Phosphatidylethanolamine binding protein, chain A |
| C-4 | - | + | + | 1BD9A | Phosphatidylethanolamine binding protein, chain A |
| C-5 | + | + | - | Q6FHP5 | Prohibitin |
| C-6 | + | + | - | Q6FHP5 | Prohibitin |
| C-7 | + | + | - | Q5EU94 | Nucleophosmin |
| D-1 | - | - | + | Q4PL71 | Putative double-stranded RNA binding protein - vaccinia virus |
| D-2 | - | - | + | P20535 | 14K cell fusion protein - vaccinia virus |
| D-3 | - | + | + | P20642 | major core protein P4a - 23kDa - vaccinia virus |
| E-1 | - | + | + | P32119 | Peroxiredoxin-2 (Natural killer cell-enhancing factor B) |
| E-2 | + | + | - | P37802 | Transgelin-2 (SM22-alpha homologue) |
| E-3 | - | + | + | Q4TT31 | Sorting nexin 3 |
| E-4 | - | + | + | 1AWIA | Profilin, chain A |
NC = negative control,
UV = UV inactivated VACV IHD-W infected HEK293,
VACV = VACV IHD-W infected HEK293
+ identified, - not identified
Discussion
Protein identification via MALDI-PSD-ToF MS spectra of on solid phase support SPITC derivatized peptides
The recording of PSD spectra from m/z values of peptide mass fingerprints provides information about the peptides' primary structure and enables the unambiguous identification of proteins from few peptides with significant Mascot ion scores by an analysis of fragmentation patterns. The difficulties of protein identification with MALDI-PSD-ToF MS spectra are low s/n ratios of fragments, incomplete sequence coverage and complex fragmentation patterns as a cause of random charge stabilisation. The protonation of peptides during the MALDI process is mediated by carboxyl groups of the matrix and mainly stabilised at primary amine groups of the N-termini and the Lysine residues. The stabilised charge induces different fragmentations of the same ion. The derivatization of peptides with SPITC binds a negatively charged sulfonic acid to the peptides' N-terminus [13]. This negative charge neutralises the protonation of C-terminal fragments which are no longer detectable in mass spectrometry. Primarily, the PSD spectra of SPITC-derivatised peptides show fragments of the y-series, whose appearance among the N-terminal fragments is promoted by the large electron density around the peptide bond. The derivatization with SPITC enables the detection of complete y-series with good s/n ratios and offers the elucidation of the peptides' primary sequence. The unambiguous mapping of peptides with completely known primary sequences to proteins registered in databases is remarkably simplified and is expressed in increased Mascot scores [14]. However, the application of SPITC derivatization for protein identification from complex mixtures is limited by the reduction of the peptides' signal intensities following N-terminal sulfonation. This degradation of sensitivity could be decreased by an optimization of the reaction conditions. For increased sensitivity the N-terminal sulfonation of peptides bound on C18 solid phase support is preferable compared to an in solution approach since signal intensity is increased and quality of PSD spectra is raised.
Comparative Proteome Analysis of VACV IHD-W infected HEK293 cells
The comparison of the proteome analysis of VACV IHD-W-infected HEK293 cells with non-infected control cells 9 h post infection has enabled the identification of 24 human proteins, whose expression has been regulated by infection, as well as of 3 viral proteins. Since the genome of VACV IHD-W is not available, the identification of viral proteins occurred via sequence homologies of the VACV strains Western Reserve and Copenhagen. Afterwards peptide sequences were derived from spectra of 13 spots representing proteins regulated in their expression, whose database search gained no result. Single-base substitutions among different VACV strains, which led to amino acid replacement, might have resulted in different peptide mass fingerprints. PSD spectra are in correlation to the confidence interval of the m/z values of the precursor ion. Therefore, protein identification via Mascot Search is disabled by mass shifts of precursor ions resulting from an amino acid replacement. De novo sequencing of viral proteins could not be assured because of the polyproteonality of 2D-gel spots. The unambiguously identified differences in the expression profile of HEK 293 cells resulting from VACV IHD-W infection are discussed in the following sections.
Proteins relevant for energy metabolism
The infection of HEK293 cells with active and inactivated Vaccinia Virus IHD-W significantly affects the cells' energy metabolism. The expression of fructose-bisphosphate aldolase A (A-1), which acts as a central enzyme in the glycolysis by catalysing the retro-aldol cleavage of fructose-1,6-bisphosphate into dihydroxyacetone phosphate and glycerinaldehyde, is increased by both types of infection [15]. Also the enhanced expression of the enzyme malate dehydrogenase (A-2) is directly associated with a boost in the energy metabolism, which serves the viruses' need for energy for DNA replication. This enzyme is an integral part of the citric acid cycle by catalysing the oxidation of malate to fumarate [16]. The resulting reduction equivalent NADH/H+ transfers electrons to the electron transport chain which builds up a proton gradient at the inner mitochondrial membrane, resulting in ATP-synthesis catalysed by the H+-transporting two-sector ATPase (A-3, A-4) [17]. A modification of the α-chain precursor of the H+-transporting two-sector ATPase (A-3, A-4), resulting in spot migration, is triggered by both types of infection and has been detected as well as an overexpression of the mtDNA stabilising ATPase family AAA domain containing 3A (A-6) protein, whose existence has just been evidenced at transcript level [18]. Another energy metabolism-related protein whose expression is up-regulated by the VACV IHD-W infection is the βsubunit of the electron transfer flavoprotein (A-5) which acts as a specific electron acceptor for several dehydrogenases, e.g. malate dehydrogenase (A-2), and transfers them to the respiratory chain [19].
All changes detected in the human proteome profile caused by the infection which are related to energy metabolism indicate an enhancement of the metabolic rate of the glycolysis as well as of the oxidative phosphorylation in order to fulfil the viruses' energy need for replication.
Proteins associated with gene expression and protein biosynthesis
The expression of the eukaryotic translation initiation factor 4H (eIF-4H) (B-3) is up-regulated by infectious and inactivated VACV IHD-W. The protein eIF-4H stimulates protein biosynthesis, ATP hydrolysis and helicase activity of eIF-4A. Enzymatic investigations show that the affinity of eIF4A to RNA is increased two-fold and helicase activity four-fold by an interaction with eIF-4H [20]. The regulation of eIF-4H may contribute to an enhancement in protein biosynthesis which is required by the virus' need for protein expression.
Further on an increased expression of nuclear protein Hcc-1 (CIP29) (B-4) is denoted in both types of infected HEK293 cells. This protein possesses a binding domain for ss and dsDNA and is postulated to be part of the ribonuclein complex. Interactions with the RNA-helicases DDX39 and BAT 1 are verified which proves the influence of Hcc-1 on the transcription of DNA. The overexpression of Hcc-1 in HEK293 cells is known to decrease the cells' growth rate [21,22].
Several members of the spliceosome, heterogeneous nuclear ribonucleoprotein B1 (B-5), novel protein similar to small nuclear ribonucleo-protein polypeptide A (B-6), heterogeneous nuclear ribonucleoprotein M (B-7), are modulated in both types of VACV IHD-W-infected HEK2093 cells. This heterogeneous protein complex splices introns of the hnRNA. Since the poxvirus genome has no introns and splicing of mRNA is therefore obsolete, viral effectors may control the cells' protein biosynthesis by interfering with the cellular hnRNA processing [23,24].
Far upstream element-binding protein 1 (FUBP1) (B-8) stimulates the expression of transcription factor c-myc by binding onto far upstream element (FUSE) upstream of the c-myc promoter [25]. The protein is a known link between the apoptosis cascade and the c-myc oncogene, it further possesses helicase activity for dsDNA. Experiments with transfected cells show that a high FUBP1 expression increases the expression level of c-myc and in this way protects the cell from apoptosis, while caspase-mediated cleavage of FUBP1 induces apoptosis [26]. Both infectious and inactivated VACV IHD-W enhance the expression of FUBP1.
Ewing sarcoma breakpoint region 1 (B-9) is a multifunctional protein with essential functions in gene expression, signal transduction, and mRNA transport and processing. Mutations in the protein-expressing gene lead to the development of Ewing sarcomas and other tumours. The protein expression is suppressed after an infection with UV-inactivated as well as with active viruses [27].
Prefoldin subunit 1 (B-10) is part of a heterohexamer chaperon (prefoldin) which binds cytosolic chaperonin (c.CPN) and transports target proteins to the hexamer. It is also involved in the folding of nascent peptides. Gene deletions of prefoldin result in a dysfunction of the actin-tubulin cytoskeleton [28]. Prefoldin subunit 1 is exclusively identified in cells infected with UV-inactivated VACV.
The described alterations caused by the infection are results of the fact that the virus is taking control of the cells' gene expression as well as the protein biosynthesis and so aims for effective expression of viral proteins and the control of the cellular response.
Proteins relevant for apoptosis
Several viruses in general and poxviruses in particular are known to modulate the hosts' apoptosis pathways in order to avoid the antiviral defence mechanism at a cellular level [29]. This results in an altered expression profile of apoptosis-relevant genes caused by viral gene products. During VACV transcription double-stranded RNA (dsRNA) is synthesised, since the virus possesses overlapping genes on both strands of its DNA. The presence of dsRNA activates the dsRNA-dependent serine/threonine kinase (PKR) which phosphorylates the α-subunit of the translation initiation factor eIF-2 which is essential for protein biosynthesis. This phosphorylation inhibits the translation of mRNA and thereby induces apoptosis [30-33]. An altered modification on the γ-subunit of the translation initiation factor eIF-2 (B-1, B-2) caused by the infection was detected by a spot migration in the 2-DE gel the relevance of which has not yet been determined in the literature. The dsRNA-dependent PKR is usually inhibited by nucleophosmin 1 (C-7) which is ubiquitously expressed in human cells and translocates between nucleus and cytoplasm [34]. The downregulation of nucleophosmin 1 (C-7), which can not be detected in cells infected with active VACV IHD-W, suggests that it is a cellular response to the infection for the purpose of anti-viral defence. This immune response is undercut by the product of the VACV E3L gene, the putative double-stranded RNA binding protein (D-1) which binds dsRNA and inhibits apoptosis induction by preventing PKR from phosporylating the γ-subunit of the translation initiation factor eIF-2 [35,36]. The E3L gene product has been identified in HEK 293 cells infected with active VACV IHD-W, which is in correlation to proteome analysis of VACV virions that indicate that the putative double-stranded RNA binding protein (D-1) is not present in the virion but is expressed in the early replication phase.
The present proteome analysis has led to the identification of several further apoptosis-relevant proteins. The expression of two isoforms of prohibitin (C-5, C-6) is decreased after infection with active VACV IHD-W. Prohibitin regulates cell division, inhibits DNA synthesis and sensibilises cells for apoptosis by destabilising the mitochondrial membrane [37]. Beyond that, prohibitin enhances the transcription of p53 [38].
The infection of HEK293 cells with active and inactivated VACV IHD-W inhibits the expression of the protein Guanine nucleotide-binding protein subunit beta 2-like 1 (C-1) which is an acceptor for activated protein kinase C (PKC). PKC is then bound to the cytoskeleton and conveyed to its target proteins, the MARCKS proteins. The receptor is also engaged in the regulation of the SRC kinase which can inhibit apoptosis by a phosphorylation of caspase-8 [39,40].
The reduced expression of the proteasome subunit alpha type-7-like (C-2) can be seen as a further cellular defence strategy. The proteasome subunit alpha type-7-like (C-2) is part of the proteasome complex which cleaves peptides proteolytically at the amino acids Arg, Phe, Tyr, Leu and Glu [41]. The inhibition of the proteasome in tumour cell lines induces apoptosis.
The phosphatidylethanolamine-binding protein (C-3, C-4) is modified by an infection with active and inactivated VACV IHD-W, which results in an altered pI value. The protein binds to phosphatidylethanolamine which is presented at the surface of apoptotic cells, inhibits the Raf-kinase, prevents the activation of the MAP-Kinase-cascade and the antiapoptotic transcription factor NFκ-B [42,43]. It is to be postulated that the protein modification is associated with an altered physiological activity.
Viral proteins
In this comparative proteome analysis the 14 kDa cell fusion protein (D-2) is identified in HEK293 cells infected with active VACV IHD-W. Its presence is predicted according to Swissprot [44]. The variola homologue of the A27L gene is known to play an import role in virus penetration by fusing the outermost of the Golgi-derived membranes enveloping the virus with the cells' plasma membrane.
As expected, the major core protein P4a (D-3) of Vaccinia virions is further identified in both infected cell culture approaches [45].
Other proteins
Peroxiredoxin-2 (E-1) is an antioxidative enzyme which reduces hydrogen- and alkyl-peroxides and supports the antiviral activity of CD8(+) T-cells [46]. In contrast to the negative control, protein expression is enhanced by an infection with active and UV-inactivated viruses.
Transgelin-2 (SM22-alpha homologue) (E-2) has not been functionally characterised yet. The protein possesses an actin-binding domain and is overexpressed in cells infected with both infectious and inactivated VACV IHD-W. It seems as if human transgelin is packed into Vaccinia virions and transferred from cell to cell for an unknown reason [47]. It can be hypothesised that it may serve as an anchor for the virus to move along the cytoskeleton.
Sorting nexin 3 (E-3) has a binding domain (PX domain) for phospholipids and transports proteins within cells from organelles to membranes [48]. The protein expression is enhanced after infection of HEK293 cells with active and UV-inactivated VACV IHD-W.
Profilin (E-4) binds actin and participates in the build-up of the cytoskeleton. As a response of extracellular signals, high concentrations of profilin inhibit the Actin-polymerization which is enhanced by low profilin concentrations [49]. Both types of infection increase expression of human profilin, while VACV possesses a profilin homologue by itself.
Conclusions
Chemical assisted fragmentation (Caf) of peptides by N-terminally sulfonation with SPITC improves confidence in protein identification from PSD spectra. ZipTipμ-C18 columns solid phase-supported SPITC derivatization is preferable compared to an in-solution approach, since s/n ratios as well as signal intensities are increased, and so the sensitivity for recording high quality PSD spectra is enhanced. Proteome analysis of VACV IHD-W-infected HEK293 cells in the late replication phase shows the up-regulation of the energy metabolism and alteration of gene expression regulation. Further on the modulation of apoptosis which counteracts the cellular anti-viral response is demonstrated by the regulation of several proteins from different signalling pathways. Interestingly the effects of UV-inactivated viruses and infectious viruses on the hosts' proteome are quite similar. There is no difference between the identified alterations of the energy metabolism and just one differentially expressed protein associated with gene expression and protein biosynthesis (B-10). Therefore the up-regulation of the energy metabolism and alterations in the regulation of the cellular gene expression are results of virus entry and virions' proteins. However, the modulation of apoptosis is clearly affected by effects that correlate with infectious viruses. The absence of nucleophosmin in cells infected with infectious viruses, for example, may be the response on viral dsRNA resulting from transcription of viral DNA, as the competing apoptosis modulator putative double-stranded RNA binding protein (D-1) is not a part of the virion.
In part, the identified proteins have not yet been described in the context of poxvirus infections and need to be further characterised using virological methods to identify their meaning for apoptosis modulation and pathogenesis.
Methods
Chemicals and equipment
All of the electrophoresis apparatus (IPGphor II, Multiphor II chamber Electrophorese, EPS 3500 power supply) were purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). IPG DryStrips and Pharmalyte pH 3-10 were obtained from GE Healthcare (Chalfont St Giles, Great Britain). Acrylamide, bis-acrylamide, iodoacetamide (IAA), thiourea, dithiothreitol (DTT), 3-[(3-Cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS), a-cyano-4-hydroxycinnamic acid (CHCA), acetonitrile (ACN), trifluoroacetic acid (TFA), 4-sulfophenyl isothiocyanate (SPITC), biammonium citrate, bovine serum albumin (BSA), sodium dodecyl sulfate (SDS), Trypsin Profile IDG Kit and the ProteoSilver Plus Silver Stain Kit were bought from Sigma-Aldrich (Taufkirchen, Germany). Tris (hydroxymethyl) aminomethane (TRIS) and solvents used (dichloromethane, ethanol, isopropanol) were supplied in analytical form by Merck (Darmstadt, Germany). Dulbecco's Modified Eagle Medium (DMEM) as well as foetal bovine serum (FBS) were purchased from Gibco (Eggenstein, Germany). TRIzol® Reagent was purchased from Invitrogen (Carlsbad, USA). Ultrapure water was generated by a water purification system and used throughout for standard and sample preparation.
Cell Culture and Proteome Extraction
HEK 293 cells (ATCC-CRL-1573™) were cultured in three 150 cm2 cell culture flasks with 25 ml culture media (DMEM containing 5% FBS [v/v] and 4 mM L-glutamine) up to a density of 2.3 × 107 cells/flask at 37°C and 5% CO2 in an incubator. Cells in one flask were infected with VACV IHD-W (ATCC-VR-1441™) and in the other one with UV-inactivated VACV IHD-W with a multiplicity of infection (MOI) of 5. Cells in the third flask were cultured as a negative control. 1 h post infection the infection media (11.5 × 107 plaque forming units [PFU] in 10 ml culture media/flask) was exchanged for fresh 25 ml culture media. Proteomes of all three cell culture approaches were purified 9 h post infection by use of TRIzol® Reagent (Invitrogen) according to the manufacturer's instructions. Dried proteome pellets were stored at -20°C until further use.
UV inactivation of VACV IHD-W
Virus stock solution containing 11.5 × 107 PFU was centrifuged for 1 h at 21,000 × g and 4°C. The virus pellet was resuspended in 1 ml phosphate buffered saline (PBS), transferred into a 6-well plate and inactivated by UV irradiation of 0.05 W/m2 in a StrataLinker 2400. Success of inactivation was checked by immunofluorescence staining of infected cells.
Two-Dimensional Polyacrylamide Gel Electrophoresis: 2D-SDS-PAGE
The two-dimensional polyacrylamide gel electrophoresis with immobilised pH gradient was performed according to the procedure by Görg et al [50]. Initially, 100 μg dried protein was solubilised in 400 μl rehydration solution (8 M urea, 2 M thiourea, 16 mM Chaps, 20 mM DDT, 0.5 vol.-% Pharmalyte 3-10, 10 μg bromphenol blue) and loaded onto isoelectric focusing strips with a linear gradient (pH 3-10, L = 18 cm) for 12 h. The rehydration occurred up to a thickness of 0.5 mm for 12 h passively and for 2 h actively, before the isoelectric focusing was done with the following parameters in 8 steps: S1: 100 V 14 h, S2: 100 V to 1800 V 30 min, S3: 1800 V 1 h, S4: 1800 V to 6000 V 40 min, S5: 6000 V 2 h, S6: 6000 V to 8000 V 15 min, S7: 8000 V 3 h, S8: 8000 V 5 h. For equilibration, the proteins were reduced for 15 min at 60°C with solution A (10 vol.-% 0.5 M Tris-HCl, pH 6.8, 6 M urea, 30 vol.-% glycerol, 140 mM SDS, 20 mM DDT) and then alkylated with solution B (10 vol.-% 0.5 M Tris-HCl, pH 6.8, 6 M urea, 30 vol.-% glycerol, 140 mM SDS, 240 mM iodoacetamid, bromphenol blue) for 15 min at 25°C in the dark. In the second dimension the proteins were separated in a 12-%-polyacrylamide gel (250 × 200 × 1 mm3) at 4 W, 600 V and 30 mA for 14 h at 12°C in a horizontal system. The size marker was RotiMark® 10-150 kDa (Roth, Karlsruhe, Germany) and a Tris-glycine buffered system according to Laemmli (1970) was used [51]. After electrophoresis, the proteins were silver stained with the ProteoSilver Plus Silver Stain Kit (Sigma-Aldrich) according to the manufacturer's instructions.
Gel image analysis
Differences in the protein expression profile of the 2-D gels were identified by use of the image analysis software Melanie 7.0 (Swiss Institute for Bioinformatics). A quantitative analysis was disclaimed since silver staining is not an end point method. A match set was created from scanned gel images in order to compare protein spots across gels. The gel containing the best resolution and most spots was chosen as the master gel. Landmark spots were defined and manual editing of the matching procedure was performed to improve the automated matching results. Finally, the mismatched spots representing differences in protein expression were manually checked and selected for Mass Spectrometric analysis.
In-gel trypsination
Differences in the protein expression profile of the 2-D gels were identified by use of the image analysis software Melanie 7.0 (Swiss Institute for Bioinformatics, Lausanne, Switzerland, http://www.expasy.org/melanie). A quantitative analysis was disclaimed since silver staining is not an end-point method. Spots representing differences in protein expression were excised as dice with 1 mm edge length each, which were destained with the ProteoSilver™ Plus Silver Stain Kit (Sigma-Aldrich) according to the manufacturer's instructions. Gel dice were then dried in a SpeedVac after dehydration in 100 μl ACN. The dried dice were swollen in 10 μl reaction buffer (25 mM ammonium bicarbonate [NH4HCO3] pH 8.0) containing 500 ng trypsin (Trypsin Profile IGD Kit, Sigma-Aldrich) for 15 min at 4°C, before another 40 μl reaction buffer was added. The tryptic digest was incubated overnight at 37°C. The proteolytic peptide containing supernatant was transferred into a new reaction tube. Peptides remaining in the gel dice were extracted by adding 20 μl 1:1 ACN/0.1%TFA for 30 min. Pooled peptides were evaporated to dryness in a SpeedVac for further analysis.
In-solution trypsination of serum albumin (bos taurus)
Reduction of disulfide bonds was done by addition of 2 μl 2 M dithiotreitol (DTT) to 200 μl 0.1 μg/μl BSA in 25 mM NH4HCO3 pH 8.0 at 60°C for 30 min. Subsequently free thiol groups were carboxyamidomethylated by adding 6 μl 1 M IAA at 25°C for 30 min in the dark. Excessive iodoacetamide (IAA) was converted with 1 μl 2 M DTT at 60°C for 15 min before the proteolytic digest was induced by addition of 1 μg trypsin (1:20 w/w) and incubated for 16 h at 37°C. For SPITC derivatization the desired amount of protein was transferred to a microcentrifuge tube and evaporated to dryness in a SpeedVac.
N-terminal peptide derivatization with 4-sulfophenyl isothiocyanate on ZipTipμ-C18 columns
Dried peptides were resolved in 5 μl 0.1% TFA, 0.5 μl of which were mixed with 0.5 μl matrix (10 mg α-Cyano-4-hydroxycinnamicacid [CHCA] in 1 ml ACN/0.1% TFA [1:1 v/v]) on a stainless steel carrier and were air-dried. Prior to the recording of peptide mass fingerprints, the spots were washed with 1.5 μl 10 mM biammonium citrate three times. The remaining peptide solution was bound to a ZipTipμ-C18 (Millipore) pipette tip which was wetted with 10 μl ACN/0.1% TFA (1:1 v/v) and washed twice with 10 μl 0.1% TFA. Subsequently the ZipTipμ-C18 was loaded with 10 μl 5 mg/ml 4-Sulfophenyl isothiocyanate (SPITC) in 20 mM NaHCO3 pH 8.6 and incubated at 55°C for 1 h in a circulating air oven. Derivatised peptides were washed five times with 0.1% TFA and three times with 10 mM biammonium citrate prior to elution with 1 μl ACN/0.1% TFA (1:1 v/v) directly onto the MALDI target. 0.5 μl matrix (10 mg CHCA in 1 ml ACN/0.1% TFA [1:1 v/v]) was added and the spot was air-dried.
N-terminal peptide derivatization with 4-sulfophenyl isothiocyanate in aqueous solution
Dried peptides were resolved in 10 μl derivatization solution (10 mg/ml 4-Sulfophenyl isothiocyanate [SPITC] in 20 mM NaHCO3 pH 8.6) and incubated at 60°C for 30 min. Purification, elution and co-crystallization of derivatised peptides was done by use of ZipTipμ-C18 (Millipore) pipette tips as described above.
MALDI-ToF curved field reflectron (CFR)
The mass spectrometric investigations have been carried out using an AXIMA-CFRPLUS (Kratos Analytical, Manchester, UK). Peptide mass fingerprints were recorded in positive ion reflectron mode with a setting of the laser intensity of 2.2 μJ and a maximum laser-repetition rate of 5.0 Hz at a pulse extraction of 1500 m/z (Delay-time). Each recorded spectrum was the sum range of 200 profiles, accumulated from five laser shots. Ten m/z values greater than 1200 were automatically selected with descending intensity by the Launchpad Software (Kratos Analytical) for post source decay (PSD) measurement. 100 profiles accumulated from ten laser shots were accumulated for one PSD spectrum which was recorded in positive ion reflectron mode with a laser intensity of 4.0 μJ and a maximal laser-repetition rate of 5.0 Hz at a pulse extraction of 1500 m/z (delay-time). An external 5-points calibration was performed by using appropriate peptide standard (calibration Mix [Proteomix] 500-3500 Da [LaserBio Labs]) prior to each measurement. A combined data file of m/z values of the PMF and PSD spectra was sent to the ExPAsy Server (Swiss Institute for Bioinformatics) and synchronised with the SwissProt database for protein identification. Peaks from the raw data were picked using MascotDistiller (Matrix Science, Boston, USA) with a minimal signal/noise of 3 and a peak width of 0.3 m/z values were annotated with a mass tolerance of +/-0.3 Da in MS1 and +/-0.7 Da in PSD mode. Carbamidomethylated cystein residues and SPITC derivatised N-termini were set as fixed modifications, oxidised methionine residues as a variable modification.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JD carried out the two-dimensional gel electrophoresis, image analysis as well as the mass spectrometric measurements. SB established the method for fragmentation of SPITC derivatized peptides and executed the database searches for protein identification. The manuscript was drafted by SB and JD in equal parts. The cell culture part of this study was performed by DB, whereas KD supported the image analysis and database searches. RB financed the analytical part of the study. HL coordinated the study and gave relevant contributions to the technical background. AN conceived the study, and participated in its design. All authors read and approved the final manuscript.
Contributor Information
Sebastian Bartel, Email: SebastianBartel@gmx.de.
Joerg Doellinger, Email: DoellingerJ@rki.de.
Kai Darsow, Email: Kai.Darsow@bvt.cbi.uni-erlangen.de.
Daniel Bourquain, Email: BourquainD@rki.de.
Rainer Buchholz, Email: Rainer.Buchholz@bvt.cbi.uni-erlangen.de.
Andreas Nitsche, Email: NitscheA@rki.de.
Harald A Lange, Email: Harald.Lange@ttnc.de.
References
- Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB, See RH, Brunham RC. Rapid response research to emerging infectious diseases: lessons from SARS. Nature. 2004;2:602–607. doi: 10.1038/nrmicro930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giulio DB, Eckburg PB. Human monkeypox: an emerging zoonosis. Lancet Infect Dis. 2004;4:15–25. doi: 10.1016/S1473-3099(03)00856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorou RM, Papavassiliou VG, Pierroutsakos IN. Cowpox virus infection: an emerging health threat. Curr Opin Infect Dis. 2008;21:153–156. doi: 10.1097/QCO.0b013e3282f44c74. [DOI] [PubMed] [Google Scholar]
- Kortepeter MG, Parker GW. Potential biological weapons threats. Emerg Infect Dis. 1999;5:523–527. doi: 10.3201/eid0504.990411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco R, Sisler JR, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J Virol. 1993;67:3319–3325. doi: 10.1128/jvi.67.6.3319-3325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CS, Hsiao JC, Chang YS, Chang W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J Virol. 1998;72:1577–1585. doi: 10.1128/jvi.72.2.1577-1585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao JC, Chung CS, Chang W. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J Virol. 1998;72:8374–8379. doi: 10.1128/jvi.72.10.8374-8379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao JC, Chung CS, Chang W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J Virol. 1999;73:8750–8761. doi: 10.1128/jvi.73.10.8750-8761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CL, Chung CS, Heine HG, Chang W. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J Virol. 2000;74:3353–3365. doi: 10.1128/JVI.74.7.3353-3365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JD, Chen TS, Gagnier CR, Vemulapalli S, Maier CS, Hruby DE. Pox proteomics: mass spectrometry analysis and identification of Vaccinia virion proteins. Virology J. 2006;3:10. doi: 10.1186/1743-422X-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Villa NY, Rahman MM, Smallwood S, Shattuck D, Neff C, Dufford M, Lanchbury JS, Labaer J, McFadden G. Analysis of vaccinia virus-host protein-protein interactions: validations of yeast two-hybrid screenings. J Proteome Res. 2009;8(9):4311–4318. doi: 10.1021/pr900491n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Nie S, Mi W, Wang XC, Liang SP. De novo sequencing of tryptic peptides sulfonated by 4-sulfophenyl isothiocyanate for unambiguous protein identification using post-source decay matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:191–198. doi: 10.1002/rcm.1280. [DOI] [PubMed] [Google Scholar]
- León IR, Neves- Ferreira AG, Valente RH, Mota EM, Lenzi HL, Perales J. Improved protein identification efficiency by mass spectrometry using N-terminal chemical derivatization of peptides from Angiostrongylus costaricensis, a nematode with unknown genome. J Mass Spectrom. 2007;42:1363–1374. doi: 10.1002/jms.1324. [DOI] [PubMed] [Google Scholar]
- Sibley JA, Lehninger AL. Determination of aldolase in animal tissues. J Biol Chem. 1949;177(2):859–872. [PubMed] [Google Scholar]
- Witt I, Kronau R, Holzer H. The Isoenzymes of malate dehydrogenase and their regulation in Saccharomyces cerevisiae. Biochim Biophys Acta. 1966;128(1):63–73. [PubMed] [Google Scholar]
- Champagne E, Martinez LO, Collet X, Barbaras R. Ecto-F1Fo ATP synthase/F1 ATPase: metabolic and immunological functions. Curr Opin Lipido. 2006;17:279–284. doi: 10.1097/01.mol.0000226120.27931.76. [DOI] [PubMed] [Google Scholar]
- Martin S. Nucleotide Sequence. Submitted (JAN-2009) to the EMBL/GenBank/DDBJ databases 2009, Cited for: NUCLEOTIDE SEQUENCE
- Colombo I, Finocchiaro G, Garavaglia B, Garbuglio N, Yamaguchi S, Frerman F, Berra B, Didonato S. Mutations and polymorphisms of the gene encoding the beta-subunit of the electron transfer flavoprotein in three patients with glutaric acidemia type II. Hum Mol Genet. 1994;3:429–435. doi: 10.1093/hmg/3.3.429. [DOI] [PubMed] [Google Scholar]
- Richter NJ, Rogers GW, Hensold JO, Merrick WC. Further. Biochemical and Kinetic Characterization of Human Eukaryotic Initiation Factor 4H. J Biol Chem. 1999;274:35415–35424. doi: 10.1074/jbc.274.50.35415. [DOI] [PubMed] [Google Scholar]
- Leaw CL, Ren EC, Choong ML. Hcc-1 is a novel component of the nuclear matrix with growth inhibitory function. Cell Mol Life Sci. 2004;61(17):2264–2273. doi: 10.1007/s00018-004-4205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Pelus LM. Growth inhibitory effect of Hcc-1/CIP29 is associated with induction of apoptosis, not just with G2/M arrest. Cell Mol Life Sci. 2005;62(13):1526–1527. doi: 10.1007/s00018-005-5093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozu T, Heinrich B, Schaefer KP. Structure and expression of the gene (HNRPA2B1) encoding the human hnRNP protein A2/B1. Genomics. 1995;25:365–371. doi: 10.1016/0888-7543(95)80035-K. [DOI] [PubMed] [Google Scholar]
- Jurica MS, Licklider LJ, Gygi SP, Grigorieff N, Moore MJ. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/S1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, Levens D. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- Jang M, Park BC, Kang S, Chi SW, Cho S, Chung SJ, Lee SC, Bae KH, Park SG. Far upstream element-binding protein-1, a novel caspase substrate, acts as a cross-talker between apoptosis and the c-myc oncogene. Oncogene. 2009;28(12):1529–1536. doi: 10.1038/onc.2009.11. [DOI] [PubMed] [Google Scholar]
- Embree LJ, Azuma M, Hickstein DD. Ewing Sarcoma Fusion Protein EWSR1/FLI1 Interacts with EWSR1 Leading to Mitotic Defects in Zebrafish Embryos and Human Cell Lines. Cancer Res. 2009;69(10):4363–4371. doi: 10.1158/0008-5472.CAN-08-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/S0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- O'Brien V. Viruses and apoptosis. J Gen Virol. 1998;79:1833–1845. doi: 10.1099/0022-1317-79-8-1833. [DOI] [PubMed] [Google Scholar]
- Rice AP. Interferon mediated, double-stranded RNA-dependent proteine kinase is inhibited in extracts from vaccinia virus-infected cells. J Virol. 1984;50:229–236. doi: 10.1128/jvi.50.1.229-236.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García MA, Meurs EF, Esteban M. The dsRNA proteine kinase PKR: virus and cell control. Biochimie. 2007;89(6-7):799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Gil J, Esteban M. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis. 2000;5(2):107–114. doi: 10.1023/A:1009664109241. [DOI] [PubMed] [Google Scholar]
- Whitaker-Dowling P, Youngner JS. Characterization of a specific kinase inhibitory factor produced by vaccinia virus which inhibits the interferon induced proteine kinase. Virology. 1984;137:171–181. doi: 10.1016/0042-6822(84)90020-5. [DOI] [PubMed] [Google Scholar]
- Pang Q, Christianson TA, Koretsky T, Carlson H, David L, Keeble W, Faulkner GR, Speckhart A, Bagby GC. Nucleophosmin Interacts with and Inhibits the Catalytic Function of Eukaryotic Initiation Factor 2 Kinase PKR. J Biol Chem. 2003;278:41709–41717. doi: 10.1074/jbc.M301392200. [DOI] [PubMed] [Google Scholar]
- Chang HW, Watson JC, Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis SA, Damaso CRA, Moussatche N. Vaccinia virus (strain IOC) putative double-stranded RNA binding protein. Submitted (MAY-2005) to the EMBL/GenBank/DDBJ databases. 2005. Cited for: NUCLEOTIDE SEQUENCE.
- Merkwirth C, Dargazanli S, Tatsuta T, Geimer S, Löwer B, Wunderlich FT, von Kleist-Retzow J-C, Waisman A, Westermann B, Langer T. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008;22(4):476–488. doi: 10.1101/gad.460708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro G, Dasgupta P, Rastogi S, Joshi B, Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278(48):47853–47861. doi: 10.1074/jbc.M305171200. [DOI] [PubMed] [Google Scholar]
- Lutz-Nicoladoni C, Letschka T, Leitges M, Villunger A, Baier G. Essential role of PKC[delta] in apoptosis induction of mouse thymocytes. Am J Immunol. 2005;1(1):14–20. doi: 10.3844/ajisp.2005.14.20. [DOI] [Google Scholar]
- Curs S, Rufini A, Stagni V, Condò I, Matafora V, Bachi A, Bonifazi AP, Coppola L, Superti-Furga G, Testi R, Barilà D. Src kinase phosphorylates Caspase-8 on Tyr380: a novel mechanism of apoptosis suppression. EMBO J. 2006;25:1895–1905. doi: 10.1038/sj.emboj.7601085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The MGC Project Team. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emoto K, Toyama-Sorimachi N, Karasuyama H, Inoue K, Umeda M. Exposure of Phosphatidylethanolamine on the Surface of Apoptotic Cells. Exp Cell Res. 1997;232(2):430–434. doi: 10.1006/excr.1997.3521. [DOI] [PubMed] [Google Scholar]
- Odabaei G, Chatterjee D, Jazirehi AR, Goodglick L, Yeung K, Bonavida B. Raf-1 kinase inhibitor protein: structure, function, regulation of cell signaling, and pivotal role in apoptosis. Adv Cancer Res. 2004;91:169–200. doi: 10.1016/S0065-230X(04)91005-6. [DOI] [PubMed] [Google Scholar]
- UniProtKB/Swiss-Prot P20535 (VFUS_VACCC) Integrated into UniProtKB/Swiss-Prot. 1991.
- Goebel SJ, Johnson GP, Perkus ME, Davis SW, Winslow JP, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- Golaz O, Hughes GJ, Frutiger S, Paquet N, Bairoch A, Pasquali C, Sanchez J-C, Tissot J-D, Appel RD, Walzer C, Balant L, Hochstrasser DF. Plasma and red blood cell protein maps: update 1993. Electrophoresis. 1993;14:1223–1231. doi: 10.1002/elps.11501401183. [DOI] [PubMed] [Google Scholar]
- Manes NP, Estep RD, Mottaz HM, Moore RJ, Clauss TR, Monroe ME, Du X, Adkins JN, Wong SW, Smith RD. Comparative proteomics of human monkeypox and vaccinia intracellular mature and extracellular enveloped virions. J Proteome Res. 2008;7(3):960–968. doi: 10.1021/pr070432+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Hortsman H, Seet L, Wong SH, Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- Gieselmann R, Kwiatkowski DJ, Janmey PA, Witke W. Distinct biochemical characteristics of the two human profilin isoforms. Eur J Biochem. 1995;229(3):621–628. doi: 10.1111/j.1432-1033.1995.tb20506.x. [DOI] [PubMed] [Google Scholar]
- Görg A, Obermaier C, Boguth G, Harder A, Scheibe B, Wildgruber R, Weiss W. The current state of two-dimensional electrophoresis with immobilized pH gradients. Electrophoresis. 2000;21:1037–1051. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1037::AID-ELPS1037>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]