Abstract
The scs and scs′ elements were proposed to function as chromatin domain boundaries for the 87A7 heat shock locus in Drosophila melanogaster. Here we report the identification and characterization of SBP (scs binding protein), a component of the scs nucleoprotein complex. SBP binds specifically to a 24-bp region of scs in vitro and is associated with scs in vivo. Multiple copies of an oligonucleotide containing the SBP recognition sequence are capable of blocking enhancer–promoter interactions in transgene assays. Mutations in the oligonucleotide that disrupt SBP binding in vitro also eliminate enhancer-blocking activity in vivo. We show that SBP is encoded by the zeste-white 5 gene and that mutations in zeste-white 5 reduce the enhancer-blocking activity of the multimerized oligonucleotides.
Keywords: Chromatin domain boundary, heat shock locus, Drosophila, enhancer–promoter interaction
To fit within the small volume of the nucleus, the eukaryotic genome must be compacted into a complex nucleoprotein structure, chromatin. At the same time, specific regions of the genome must remain accessible to the large multiprotein complexes that are responsible for transcription, replication, recombination, and repair. One mechanism for satisfying these opposing requirements is the organization of the chromatin fiber into a series of discrete and topologically independent domains. The domain model was initially formulated based on cytological studies on specialized chromosomes like those found in Drosophila salivary glands (for review, see Grosveld and Schedl 1995). The salivary gland polytene chromosomes exhibit a highly reproducible banding pattern suggesting that the chromatin fiber is subdivided into a series of distinct domains. More recent support for the domain model has come from biochemical studies that have revealed marked differences in the properties of chromatin in adjacent regions of the chromosome. For example, the chromatin in the β-globin gene domain is highly sensitive to DNase I digestion and has a high level of histone acetylation, whereas the chromatin in the neighboring chromosomal DNA segments is resistant to DNase I digestion and is underacetylated (for review, see Felsenfeld et al. 1996). The domain model predicts the existence of boundary elements that define the endpoints of each domain. In addition to providing a mechanism for subdividing the chromatin fiber into discrete structural units, it is thought that the boundaries might have a role in protecting a gene within a given domain from the influences of regulatory elements in neighboring domains.
The idea that boundaries may have an insulating activity has been used to establish in vivo assays for these elements, and several putative boundaries from different organisms have been identified (for review, see Kellum and Elgin 1998; Udvardy 1999). These elements have a number of properties in common. They are able to prevent enhancers from activating a promoter when positioned between the enhancer and the promoter, but have no apparent effect on enhancer–promoter interactions when located upstream of the enhancer. This blocking activity is not limited to activation; boundary elements also shield a promoter from a silencer when positioned between the silencer and the promoter. In addition to blocking enhancers or silencers, boundary elements can insulate reporter genes from chromosomal position effects. Second, putative boundary elements have a distinct chromatin structure and contain one or more chromatin-specific nuclease hypersensitive sites. The nuclease hypersensitive sites are often detected tissue or cell type nonspecifically, suggesting that boundaries constitute a structural component of eukaryotic chromatin. Finally, there is evidence that boundaries delimit units of genetic activity. These genetic units can correspond to a single gene or can contain multiple genes that share common regulatory elements (e.g., β-globin locus) (Felsenfeld et al. 1996). In the Drosophila Bithorax Complex, boundaries are required to ensure the functional autonomy of the cis-regulatory domains that control the parasegment-specific expression of Abdominal-B (Mihaly et al. 1998). Even though many of these putative boundaries have similar characteristics, it remains to be determined whether there are quantitative and/or qualitative differences both in their mechanism of action and in their in vivo functions.
Although the number of insulating elements continues to grow, little is known about the proteins that mediate the insulating activity or how this activity might be related to the higher-order organization of the chromatin fiber. Thus far, only two proteins with insulator activity have been identified. The Su(Hw) protein is responsible for the enhancer-blocking activity of the gypsy retrotransposon in D. melanogaster (Geyer and Corces 1992). Although no genomic binding sites for Su(Hw) other than gypsy are known, the chromosomal distribution of Su(Hw) protein suggests that it is part of endogenous boundary elements (Spana et al. 1988). The second protein, BEAF (boundary element-associated factor), was originally identified as a protein component of the scs′ element (Zhao et al. 1995). Recently published observations indicate that BEAF is also part of a large number of additional elements that have insulating activity (Cuvier et al. 1998).
The scs and scs′ elements were proposed to function as boundaries of the 87A7 heat-shock domain in D. melanogaster. These elements, positioned several kb downstream from the 3′ end of the two divergently transcribed hsp70 genes, were originally identified as unusual chromatin structures: two prominent nuclease hypersensitive regions surrounding a nuclease resistant core (Fig. 1A) (Udvardy et al. 1985). In situ hybridization localized scs and scs′ to the proximal and distal edges of the 87A7 heat shock puff, respectively. Although distant from the hsp70 genes, the characteristic nuclease cleavage pattern of both scs and scs′ is altered on heat-shock induction. Accompanying the changes in their chromatin structure, topoisomerase II is recruited to scs and scs′, suggesting that these elements may have a role in facilitating the transcription-induced decondensation of the chromatin fiber at the heat-shock locus (Udvardy et al. 1985; Udvardy and Schedl 1993). Like other putative boundaries, scs is able to protect transgenes from position effect and to block enhancer–promoter interaction when placed between an enhancer and a promoter (Kellum and Schedl 1991, 1992). Deletion analysis showed that multiple, partially redundant sequence elements distributed in the nuclease hypersensitive regions of scs are responsible for its enhancer-blocking activity (Vazquez and Schedl 1994). Subfragments from scs containing only a subset of these elements are unable to block enhancer–promoter interactions when present in a single copy; however, blocking activity can be reconstituted with two copies of one of these subfragments or by combining two different subfragments (Vazquez and Schedl 1994).
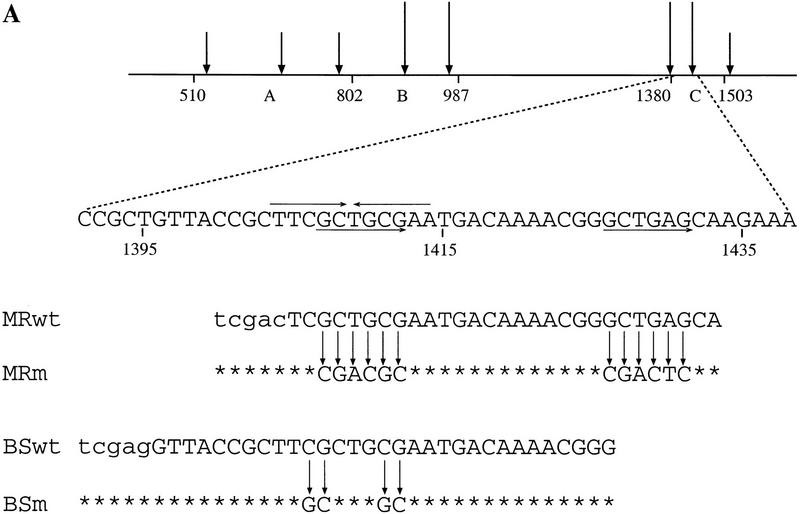
Figure 1.
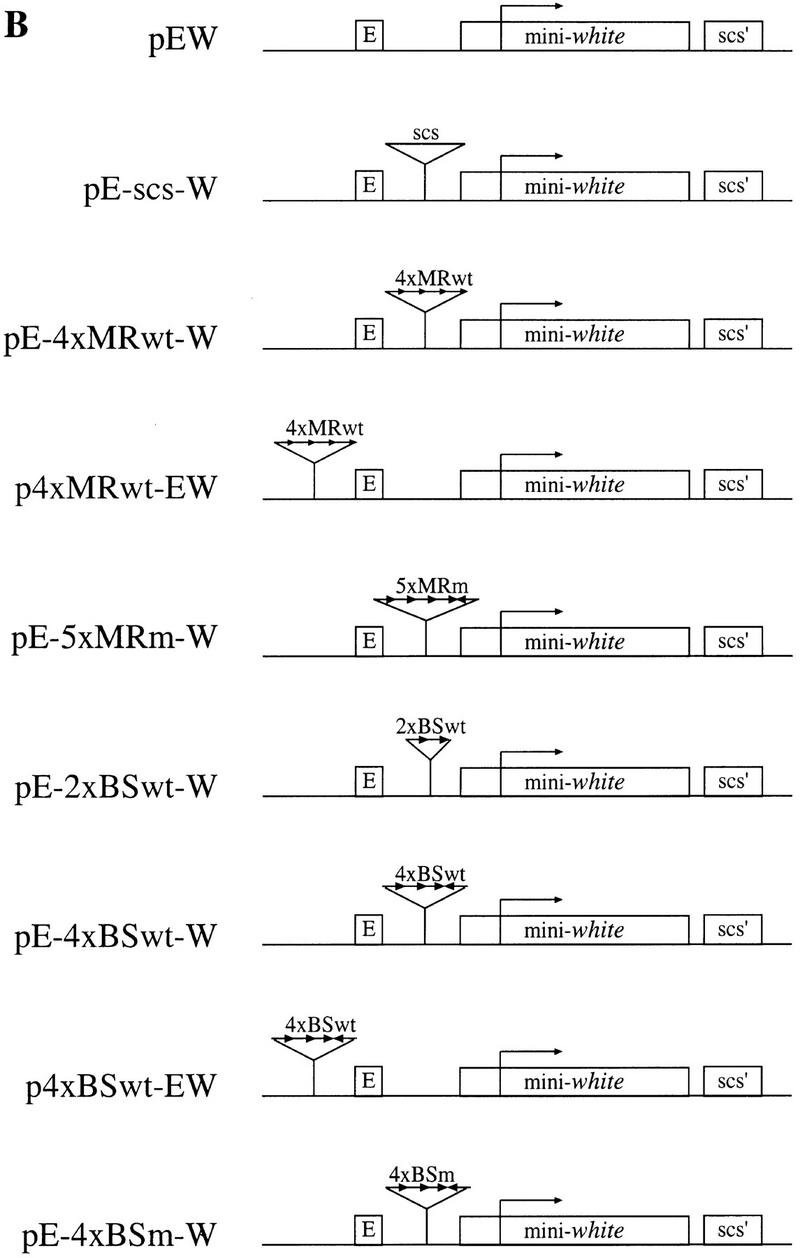
(A) Schematic representation of the scs element. Approximate positions of the major/minor nuclease hypersensitive sites within the scs element are marked by tall/short vertical arrows. Fragments A, B, and C contain sequence elements required for the enhancer-blocking activity of scs (Vazquez and Schedl 1994). Nucleotide positions are marked according to X63731 GenBank sequence file. The positions of the 5-bp inverted repeat and the 6-bp direct repeat are marked by horizontal arrows. The sequences of the MRwt, MRm, BSwt, and BSm oligonucleotides are aligned with their original positions in scs (the sequence of the restriction sites used for oligonucleotide multimerization is in lowercase type). (B) Structure of the white transgene constructs generated to test the in vivo enhancer-blocking activity of MRwt, MRm, BSwt, and BSm oligonucleotides. The basic pEW construct contains the white enhancer (E), mini-white gene, and scs′. scs′ was included to protect the 3′ end of the transgene from position effect. The relative orientation of the inserted oligonucleotides is indicated by the direction of the arrows.
With the goal of learning more about the mechanisms responsible for insulating activity, we have identified and characterized SBP (scs binding protein), a protein component of the scs nucleoprotein complex. SBP specifically binds in vitro to a 24-nucleotide sequence element that is positioned in one of the regions required for the enhancer-blocking activity of scs. Chromatin immunoprecipitations show that SBP is associated with the scs nucleoprotein complex in vivo. Our studies indicate that SBP contributes to the insulator function of scs. Thus, it is possible to partially recreate the enhancer-blocking activity of scs by multimerizing a short subfragment corresponding to the SBP-binding site. Consistent with the idea that SBP binding is necessary for the enhancer-blocking activity of the subfragment, a mutant version of the subfragment that is not recognized by SBP in vitro can not block enhancer–promoter interaction in vivo. We show that SBP is encoded by zeste-white 5 (zw5), an essential gene. To further support the idea that the SBP/Zw5 protein contributes to insulator function, we show that the enhancer-blocking activity of the multimerized subfragment is reduced by zw5 mutations.
Results
MRwt, a 35-bp fragment of scs is able to block enhancer–promoter interaction
In previous work we found that a single copy of scs subfragments (fragments A, B, and C in Fig. 1A) is unable to block enhancer–promoter interactions. However, we were able to reconstitute insulator activity by generating tandem repeats of these subfragments (Vazquez and Schedl 1994). These observations suggested that it might be possible to pinpoint some of the cis-acting elements that contribute to the insulator activity by multimerizing short oligonucleotide sequences from scs. As the starting point for our studies we focused on a 29-bp oligonucleotide derived from fragment C, MRwt (Fig. 1A). This sequence was selected because it is located within a major chromatin-specific nuclease hypersensitive region and because it contains two potentially interesting motifs, a 6-bp direct repeat (GCTGNG) and a 5-bp inverted repeat (TTCGC) (Fig. 1A).
We used the white enhancer-blocking assay to test the insulator activity of the MRwt oligonucleotide (Vazquez and Schedl 1994). In this assay, the test sequence is inserted either between the white enhancer and the mini-white reporter gene or upstream of the white enhancer (Fig. 1B). When positioned between the white enhancer and mini-white, an insulator interferes with enhancer–promoter interactions and reduces the expression of mini-white compared with that observed in transgenes lacking the insulator element. In the upstream position, an insulator has no effect on the communication between the white enhancer and the white promoter, and a high level of mini-white expression is observed. An insulator differs from a transcriptional repressor, which would be expected to reduce the expression of the mini-white reporter from both positions.
In the first experiment we generated a test construct with four tandem copies of the MRwt oligonucleotide inserted between the white enhancer and the mini-white reporter (pE-4×MRwt-W in Fig. 1B). Multiple independent transgenic lines were established with this test construct and their eye color was compared to lines carrying the control construct, pEW, which does not contain any insert (Fig. 2). As indicated in Table 1, we found that the pE-4×MRwt-W transgenic lines had a lighter eye color than the control pEW lines. In contrast, when the multimerized MRwt oligonucleotide was placed distal to the enhancer the eye color of the transgenic p4×MRwt-EW flies was the same or darker than that of the control pEW flies (see Fig. 2 and Table 1). These results indicate that the MRwt oligonucleotide has enhancer-blocking activity. It should be noted that the blocking activity of the multimerized MRwt oligonucleotide does not seem to be as strong as that of scs (Fig. 2, cf. pE-4×MRwt-W and pE-scs-W flies).
Figure 2.
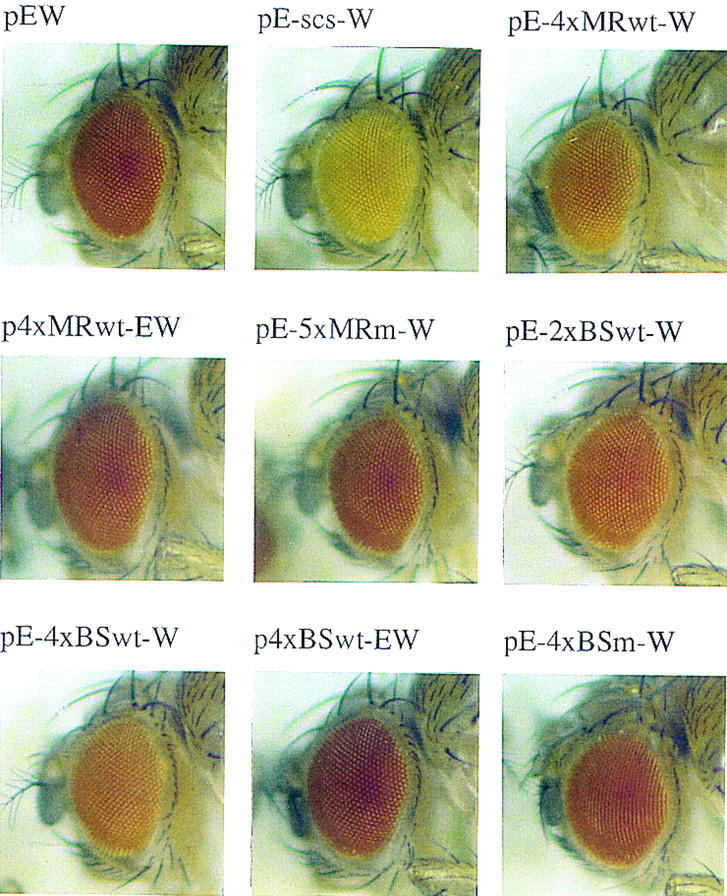
MRwt and BSwt oligonucleotides partially recreate the enhancer-blocking activity of scs. Eye-color phenotype of the various white transgene constructs. Heads of 1.5-day-old females from a representative line carrying a single copy of the indicated transgene are shown. (See Figure 1B for the structure of the transgenes.)
Table 1.
Eye color distribution of independently generated lines carrying the various white transgene constructs
|
|
|
Yellow
|
Orange
|
Dark orange
|
Red
|
Bright red
|
|---|---|---|---|---|---|---|
| pEW | 1 | 1 | ||||
| pE-scs-W | 2 | |||||
| pE-4×MRwt-W | (22) | 5 | 12 | 4 | 1 | |
| p4×MRwt-EW | (4) | 2 | 2 | |||
| pE-5×MRm-W | (11) | 1 | 2 | 5 | 3 | |
| pE-2×BSwt-W | (5) | 1 | 2 | 2 | ||
| pE-4×BSwt-W | (13) | 1 | 9 | 3 | ||
| p4×BSwt-EW | (15) | 1 | 3 | 9 | 2 | |
| pE-4×BSm-W | (11) | 1 | 1 | 4 | 4 | 1 |
Structure of the transgenes is shown in Fig. 1B. The number of independent transgenic lines generated for each construct is shown in parenthesis. Two representative lines of pEW and pE-scs-W were included as reference (Vazquez and Schedl 1994). The eye color phenotype was determined by visual examination of 1.5-day-old females with a single copy of the transgene.
The enhancer-blocking activity of the MRwt sequence could be due either to the binding of sequence-specific trans-acting factor(s) or to some unusual DNA structure generated by the tandem oligonucleotide array. To distinguish between these possibilities we tested a mutated version of the MRwt oligonucleotide in the white enhancer-blocking assay (MRm; Fig. 1A). The mutations introduced to the oligonucleotide disrupt both the direct and inverted repeat motifs (MRm in Fig. 1A) We inserted five copies of MRm between the white enhancer and mini-white and then isolated transgenic pE-5×MRm-W flies. The enhancer-blocking activity of the MRm oligonucleotide is diminished and the distribution of the eye-color phenotype of pE-5×MRm-W flies is similar to that of the control construct pEW (Fig. 2; Table 1).
SBP specifically binds MRwt in vitro
Because boundary activity in our mini-white assay system could be reconstituted by multimerizing the MRwt oligonucleotide but not the MRm oligonucleotide, we reasoned that the enhancer-blocking activity of the MRwt multimer is most likely due to the binding of a specific trans-acting factor(s). To identify this putative boundary protein(s), we screened an embryonic cDNA expression library with a radioactively labeled 4×MRwt multimer. The screen identified SBP, a protein that has eight, evenly spaced, C2H2-type zinc fingers in the carboxy-terminal half and an acidic amino-terminal half (Fig. 3A). The zinc finger domain is consistent with a sequence-specific DNA-binding protein, whereas the acidic amino-terminal domain could potentially have a role in protein–protein interactions. We failed to identify any protein in the database with significant similarity to SBP (residues corresponding to the zinc finger domains were excluded from the search).
Figure 3.
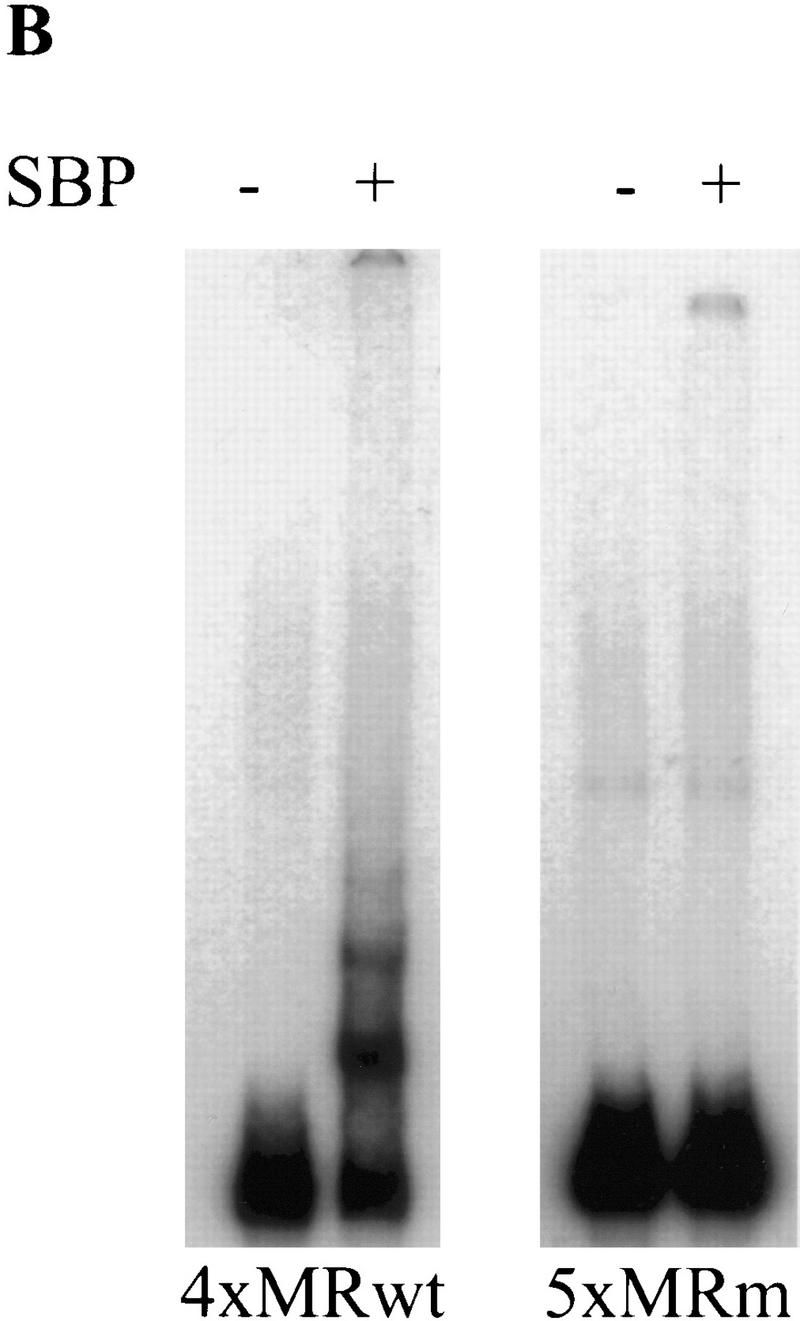
SBP specifically binds 4×MRwt in vitro. (A) Predicted amino acid sequence of the SBP protein. Residues corresponding to the eight evenly spaced C2H2-type zinc finger domains are underlined. Predicted amino acid substitutions found in the zw562jl and zw590 alleles are marked. (B) Recombinant SBP protein is able to bind 4×MRwt but not 5×MRm in vitro.
A prediction stemming from the results of the transgene experiments is that the insulator protein should recognize the MRwt but not the MRm oligonucleotide. To determine whether SBP conforms to this prediction, we used an in vitro mobility-shift assay to test the binding properties of the recombinant SBP protein. We found that SBP specifically binds 4×MRwt, but not 5×MRm, in the presence of nonspecific competitor (Fig. 3B). The formation of several different SBP:4×MRwt gel-shifted complexes suggests that multiple SBP molecules can bind simultaneously to a single 4×MRwt target. The disruption of SBP binding by the MRm point mutations argues that SBP recognizes a sequence element inside MRwt rather than a new sequence created by the multimerization of the oligonucleotides. The specific binding of SBP to MRwt is also seen in mobility-shift assays using a single-copy oligonucleotide as probe (data not shown). However, the SBP:1×MRwt complex is sensitive to disruption by nonspecific competitors. The weak binding of SBP to 1×MRwt is most likely explained by the subsequent finding that the SBP recognition sequence extends beyond the end of the MRwt oligonucleotide (see below).
SBP specifically binds to scs in vitro
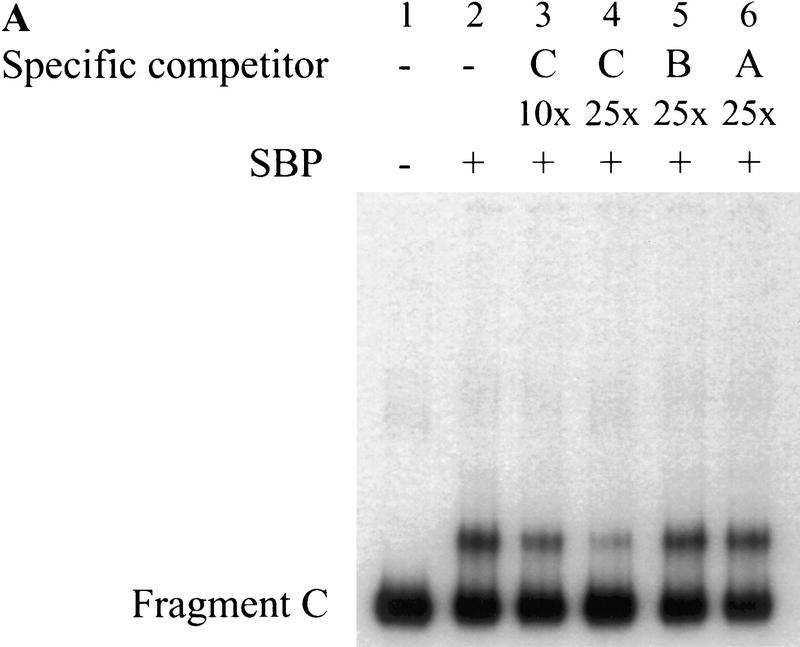
Although our experiments show that SBP binds to the MRwt oligonucleotide, it was important to establish that it also recognizes this sequence within scs. We found that recombinant SBP protein specifically binds fragment C, the scs fragment from which the MRwt oligonucleotide was derived (Fig. 4A, lane 2). This binding is specific because the formation of the SBP:fragment C complex is competed by increasing concentrations of cold fragment C, but not by fragments B or A (see Fig. 4A, lanes 3–6).
Figure 4.
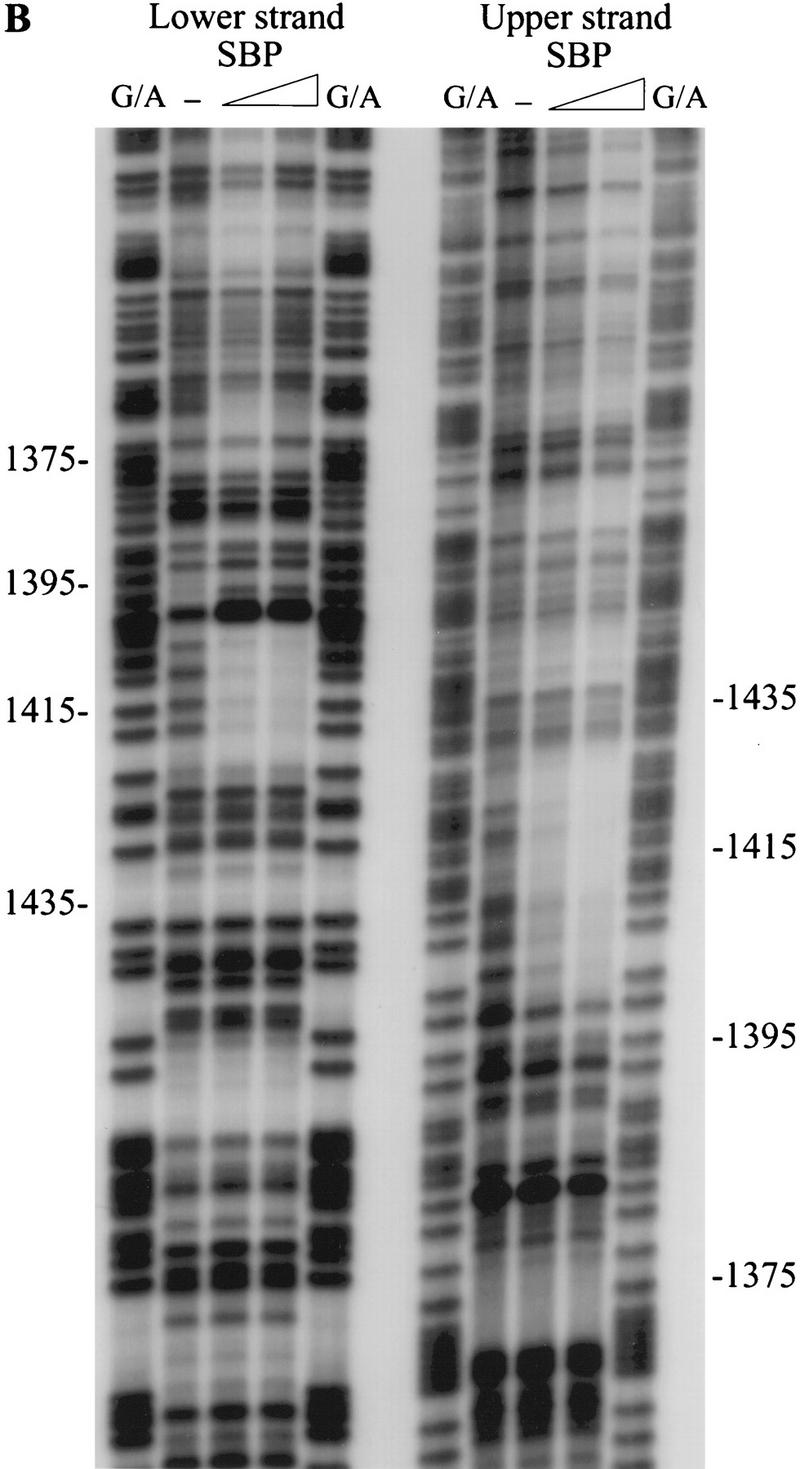
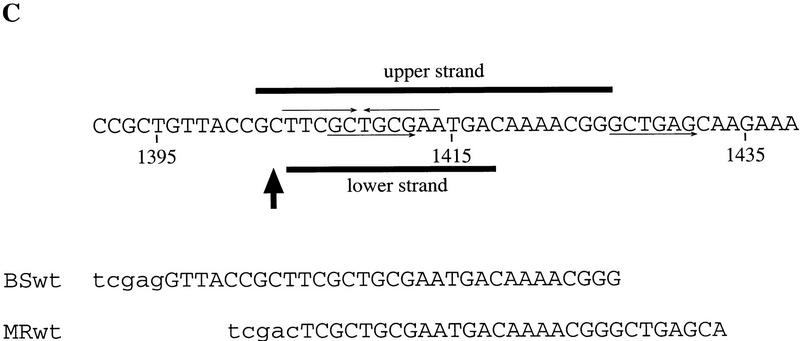
SBP specifically binds fragment C of scs in vitro. (A) Recombinant SBP protein was incubated with radioactively labeled fragment C in the presence of 50 μg/ml poly[d(I-C)] and the indicated amounts of specific cold competitors. DNA–protein complexes formed by SBP and fragment C were resolved by gel electrophoresis. (B) The binding site of SBP was determined by in vitro DNase I footprinting. (C) Position of the SBP binding site as determined by in vitro DNase I footprinting. The regions of the upper and lower strands protected from DNase I cleavage by SBP binding are marked by horizontal lines. The arrow corresponds to the position of a nuclease hypersensitive site induced by SBP binding.
DNase I footprinting was used to localize the SBP binding site in fragment C. As shown in Figure 4B, SBP binding induced several changes in the fragment C DNase I cleavage pattern. First, the upper strand was protected over a 24-bp sequence, whereas the lower strand was protected over a slightly shorter, 15-bp sequence (Fig. 4C). The 24-bp protected sequence in the upper strand overlaps, but is not precisely identical to, the MRwt oligonucleotide. At the 5′ end, the protected region extends 3 bp beyond the end of the MRwt sequence, whereas at the 3′ end it does not cover the sec-ond copy of the 6-bp repeat (Fig. 4C). Second, a set of nuclease hypersensitive sites was induced at the 5′ end of the major protected area on the lower strand. Third, at least one other minor protected site could be seen in a very AT-rich sequence on the lower strand (around position 1365). The significance of this weakly protected region is unclear, because corresponding change was not observed in the DNase I cleavage pattern on the upper strand.
SBP is able to block enhancer–promoter interaction
Although the results obtained with the MRwt and MRm oligonucleotides implicate SBP in enhancer-blocking activity, there are two caveats to this interpretation. First, the MRwt oligonucleotide does not correspond precisely to the SBP binding site. Second, MRm has mutations outside the SBP recognition sequence that could disrupt the binding of some unknown protein(s). To address these problems we designed and tested a new oligonucleotide, BSwt, that precisely corresponds to the SBP binding site (Fig. 1A).
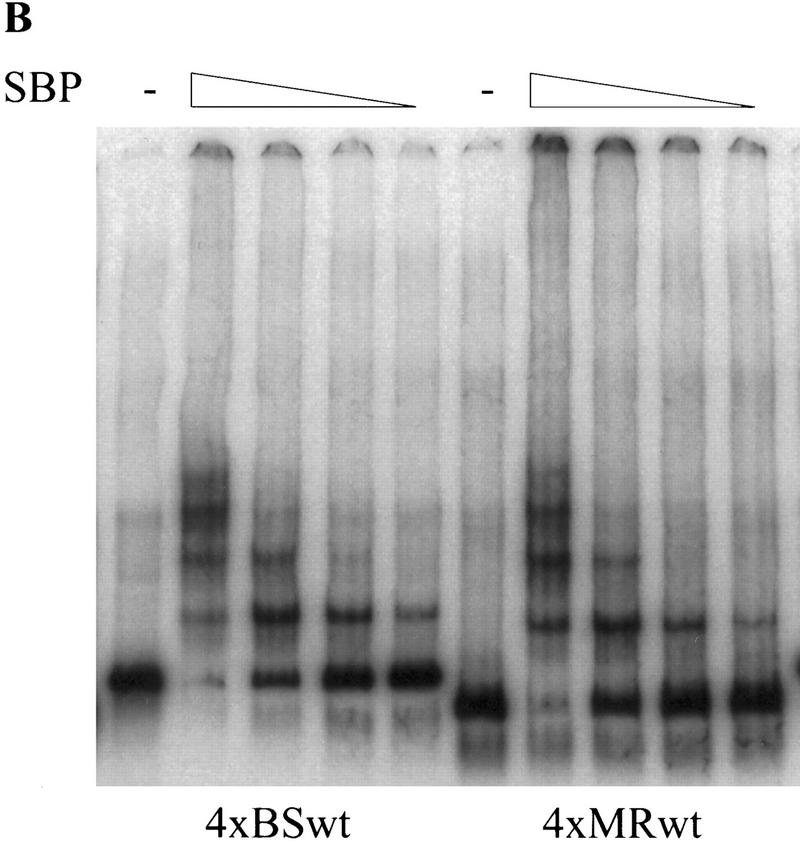
We found that SBP binds with high affinity to a single copy of the BSwt oligonucleotide in an in vitro mobility shift assay (Fig. 5A). The binding is eliminated by point mutations introduced into the BSwt oligonucleotide sequence (BSm in Fig. 1A; Fig. 5A). We were interested in comparing the relative binding affinity of SBP to multimerized versions of the BSwt and MRwt oligonucleotides. In the experiment shown in Figure 5B, a constant amount of either 4×MRwt or 4×BSwt probe was incubated with increasing amounts of SBP in the presence of nonspecific competitor. As was observed for the 4×MRwt probe (see above), SBP generates at least four distinct complexes with the 4×BSwt multimer. Although the overall pattern of the DNA–protein complexes is quite similar for the two probes, SBP appears to bind with slightly higher affinity to the 4×BSwt multimer. At each concentration of protein, the amount of the slowest migrating SBP:4×BSwt complex is discernibly greater than the corresponding SBP:4×MRwt complex.
Figure 5.
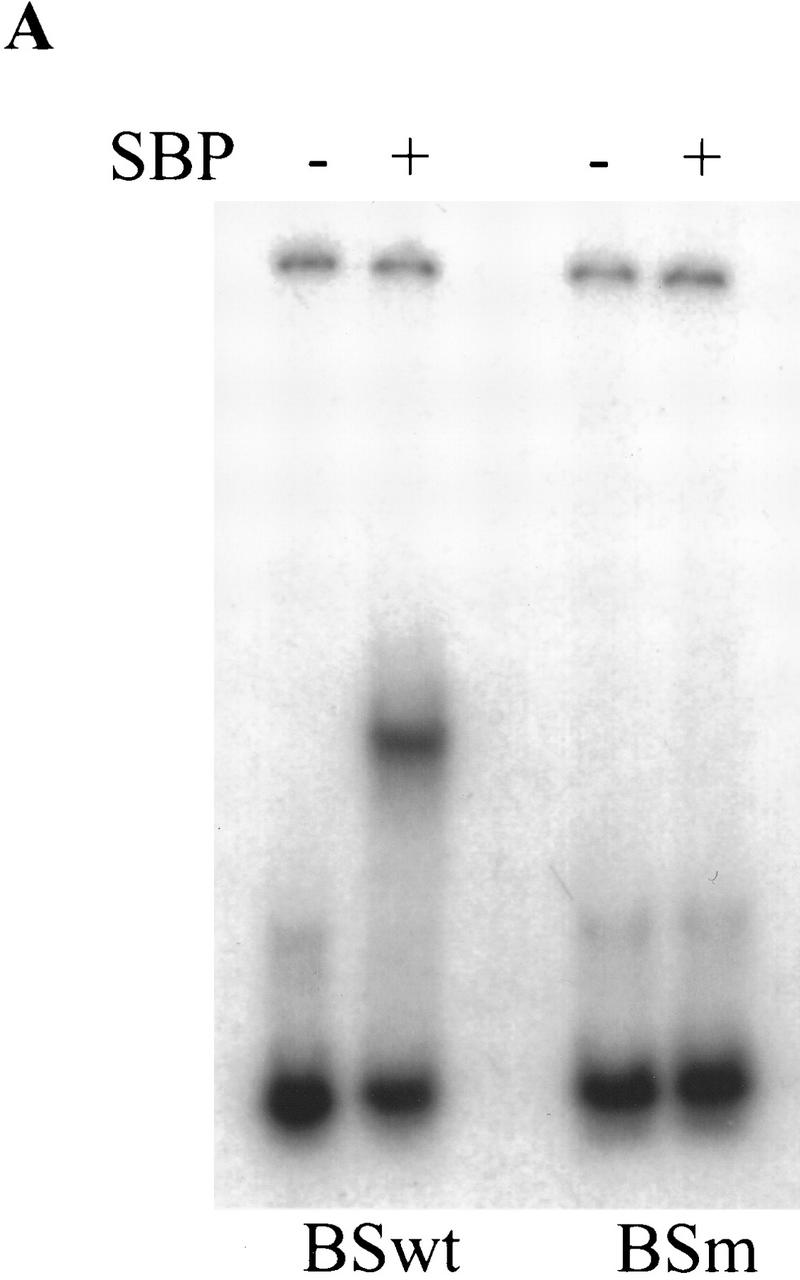
SBP binds 4×BSwt with slightly higher affinity than 4×MRwt. (A) SBP is able to form a stable complex in vitro with a single copy of BSwt, but not BSm. Recombinant SBP protein was incubated with radioactively labeled double-stranded BSwt or BSm oligonucleotides in the presence of 50 μg/ml poly[d(I-C)]. (B) SBP has a slightly higher affinity to 4×BSwt than 4×MRwt in vitro. Constant amounts of radioactively labeled 4×BSwt or 4×MRwt probes were incubated with increasing amounts of recombinant SBP protein in the presence of nonspecific competitor. The different DNA–protein complexes formed were resolved by gel electrophoresis.
To assess the enhancer-blocking activity of the SBP-binding sequence, we placed multiple copies of the BS oligonucleotide either between the enhancer and the promoter or upstream of the enhancer (Fig. 1B). Several independent transgenic lines were generated with each multimer construct and their eye-color phenotypes are summarized in Table 1 and Figure 2. We found that the 4×BSwt multimer interferes with enhancer–promoter interaction when placed between the enhancer and the promoter. This effect appears to be due to insulating activity because the multimerized BSwt oligonucleotide does not reduce mini-white expression when placed upstream of the enhancer (Fig. 2; Table 1). Consistent with the idea that SBP binding is required for the insulator function of the BSwt oligonucleotide, the mutant 4×BSm multimer has reduced enhancer-blocking activity (Fig. 2; Table 1). Finally, two copies of BSwt had little effect on white expression, suggesting that the insulating activity of SBP is dose dependent.
SBP is part of the scs nucleoprotein complex in vivo
Results described above showed that SBP specifically binds to a sequence element within scs in vitro. To determine whether SBP is also a component of the scs nucleoprotein complex in vivo, we used a chromatin immunoprecipitation assay developed by Orlando and Paro (1993). Affinity-purified polyclonal antibodies directed against the SBP protein (Fig. 6A) or control preimmune serum were used to immunoprecipitate formaldehyde cross-linked chromatin isolated from Drosophila S2 tissue culture cells. The DNA recovered from the control and anti-SBP immunoprecipitates was then probed with different fragments from the 87A7 heat shock locus.
Figure 6.
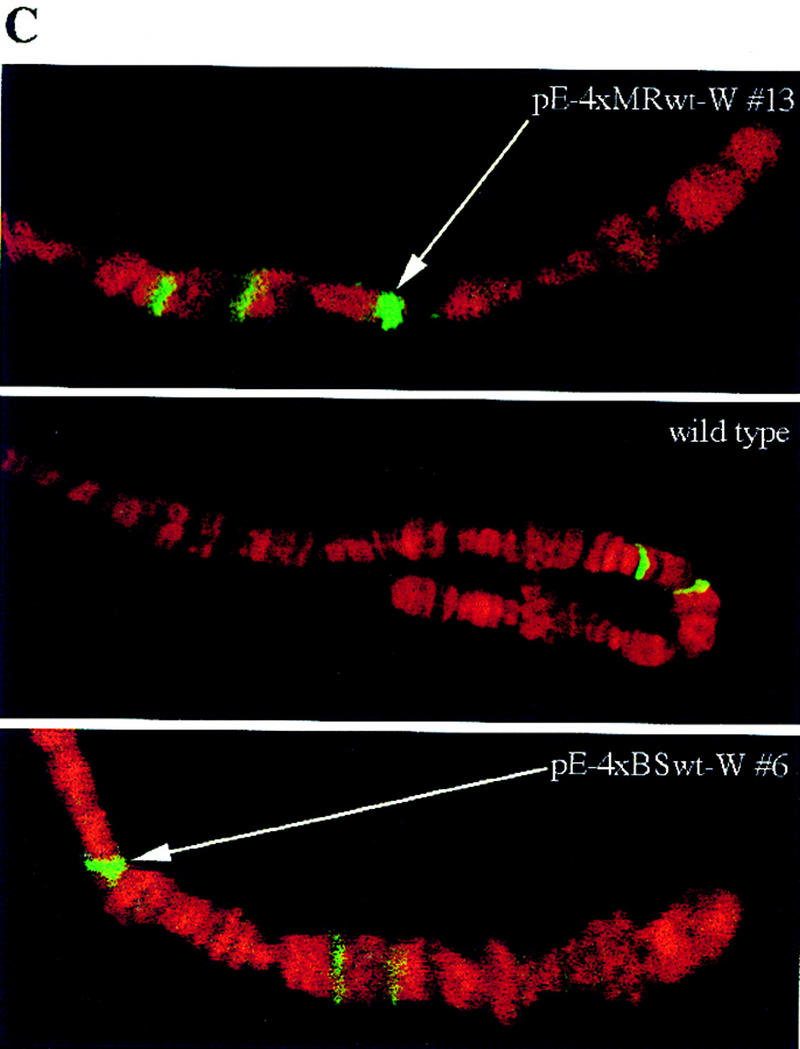
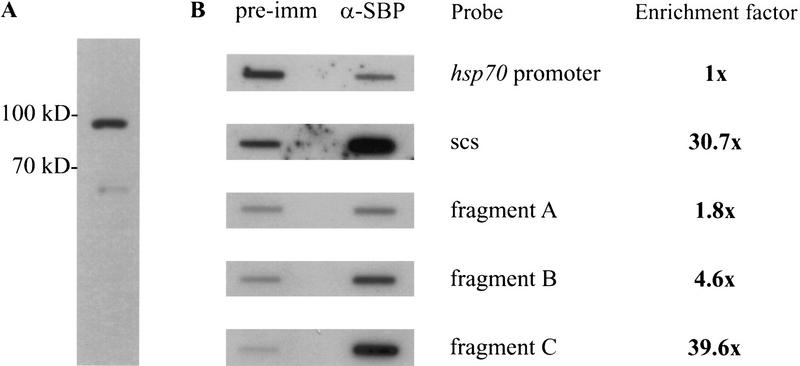
SBP binds to fragment C of scs in vivo. (A) Affinity-purified polyclonal anti-SBP antibody recognizes a protein with 90-kD apparent molecular weight on a Western blot. The faint lower band may represent a breakdown product or a weakly cross-reacting protein. The Western blot analysis was performed on total embryo extract. (B) Fragment C is specifically enriched in the anti-SBP chromatin IP. In vivo cross-linked chromatin was immunoprecipitated with affinity-purified anti-SBP antibody. The DNA recovered from the pellet was transferred to a nylon filter. The relative abundance of various parts of the 87A7 locus in the chromatin-immune pellet was determined by hybridizing the filter with the appropriate radioactive probes. Signal intensity was quantified on the PhosphorImager (Molecular Dynamics, Inc.). Enrichment is defined as the ratio of the SBP-specific signal to the preimmune serum signal. The data was normalized to the ratio of signals detected for the hsp70 promoter region. (C) SBP binds to the multimerized MRwt and BSwt oligonucleotides in vivo. Polytene chromosomes of transgenic and wild-type larvae immunostained with affinity-purified anti-SBP antibody are shown. The green and red colors mark the distribution of SBP and DNA, respectively. The transgenic lines are indicated in the photograph in each case. The new, transgene-specific SBP-binding sites are marked by arrows.
In the first experiment we used probes for scs and the hsp70 promoter region. After normalizing the signals for the hsp70 promoter region, we found that the scs region is 30-fold enriched in the SBP-specific immune pellet (Fig. 6B). To further pinpoint the SBP binding site in vivo, we used three probes from the scs region, fragments A, B, and C (see Fig. 1A). As can be seen in Figure 6B, fragment C, which contains the in vitro SBP-binding site, is enriched nearly 40-fold in the SBP-specific pellet. Although fragments A and B are also enriched in the SBP immunoprecipitates, the enrichment factor is inversely correlated with the distance of the two fragments from fragment C. Presumably, the modest enrichment of these two adjacent fragments reflects the size distribution of the DNA in the sonicated, formaldehyde cross-linked chromatin. These findings indicate that SBP is part of the scs nucleoprotein complex in vivo and argue that SBP is likely to bind to the same site in fragment C in vivo as it does in vitro.
If the enhancer-blocking activity of the multimerized MRwt and BSwt oligonucleotides depends on SBP binding, then SBP protein should be localized to the pE-4×MRwt-W and pE-4×BSwt-W transgenes in vivo. To test this prediction we immunostained salivary gland polytene chromosomes prepared from transgenic and wild-type larvae with affinity-purified anti-SBP antibody. If SBP binds to the oligonucleotide array, there should be one new anti-SBP antibody-stained site in chromosomes prepared from transgenic animals. We examined four transgenic lines inserted on the X chromosome, two pE-4×MRwt-W lines and two pE-4×BSwt-W lines. In each case, we found SBP protein at a single new site that is not present in X chromosomes prepared from nontransgenic larvae. This is illustrated for two of the lines in Figure 6C. These results are consistent with the idea that SBP is responsible for the enhancer-blocking activity of MRwt and BSwt.
The zw5 gene encodes the SBP protein
It is important to identify the gene encoding SBP to understand the in vivo function of the protein. In situ hybridization of the SBP cDNA to salivary gland polytene chromosomes mapped the SBP gene to the 3B2-4 region of the X chromosome. The cytological localization has been independently confirmed by sequence data from the European Drosophila Genome Project. We could place the SBP transcription unit between fs(1)Yb and cramped by analyzing the available genomic sequences (FlyBase 1998). There are two known lethal complementation groups in this interval, zw5 and zw11. To determine if either of them encodes SBP, we attempted to rescue the lethality of these mutations by constitutively expressing the full-length SBP cDNA under the control of the hsp83 promoter. Only mutations in the zw5 gene were rescued by the hsp83:SBP cDNA transgene.
Using information generated by the European Drosophila Genome Project we PCR amplified genomic fragments corresponding to the two available zw5 alleles, zw562jl (Judd et al. 1972) and zw590 (this laboratory; see Materials and methods), from hemizygous mutant males rescued by the hsp83:SBP cDNA transgene. The molecular nature of the alleles was determined by sequencing the genomic fragments. zw562jl contains a missense mutation (R14G) in the amino-terminal region of the protein that could potentially disrupt protein–protein interactions by changing the overall charge of this region of the protein. zw590 carries two missense mutations in the coding region (S100P and H436Y). One of these mutations (H436Y) disrupts the fifth zinc finger domain by replacing a histidine with a tryptophan (Fig. 3A).
The enhancer-blocking activity of the MRwt oligonucleotide is reduced in zw5 mutant background
If the Zw5 (SBP) protein is responsible for the enhancer-blocking activity of the multimerized 4×BSwt and 4×MRwt oligonucleotides, then this blocking activity should be eliminated by loss-of-function mutations in the zw5 gene. However, zw5 is an essential gene and it is not possible to test blocking activity in the complete absence of the Zw5 protein. Instead we asked whether insulator activity is sensitive to a reduction in zw5 gene dose. Because Zw5 binds with slightly lower affinity to the multimerized MRwt oligonucleotide than to the multimerized BSwt oligonucleotide, we reasoned that the blocking activity of the MRwt oligonucleotide might be somewhat more sensitive to a reduction in the dose of the zw5 gene than the BSwt oligonucleotide. Therefore we decided to examine not only the pE-4×BSwt-W transgenes but also the original pE-4×MRwt-W.
We found that the eye color of the pE-4×BSwt-W transgenic flies did not change discernibly in a zw5 heterozygous mutant background. However, as shown in Figure 7, the eye color of the pE-4×MRwt-W transgenic flies becomes darker when crossed into zw5 heterozygous mutant background. This effect is not allele specific, because it is observed with two independently isolated zw5 alleles. In addition, it does not depend on the insertion site of the transgene as a darkening of the eye color was observed in multiple pE-4×MRwt-W lines. These findings indicate that a twofold reduction in the SBP protein level is sufficient to compromise the enhancer-blocking activity of 4×MRwt and provide strong evidence that zw5 is necessary for the insulator activity of this oligonucleotide. (Note: Because scs has functionally redundant elements in fragments A and B, even the complete elimination of the Zw5 protein should have no effect on the blocking activity of the intact scs element.)
Figure 7.
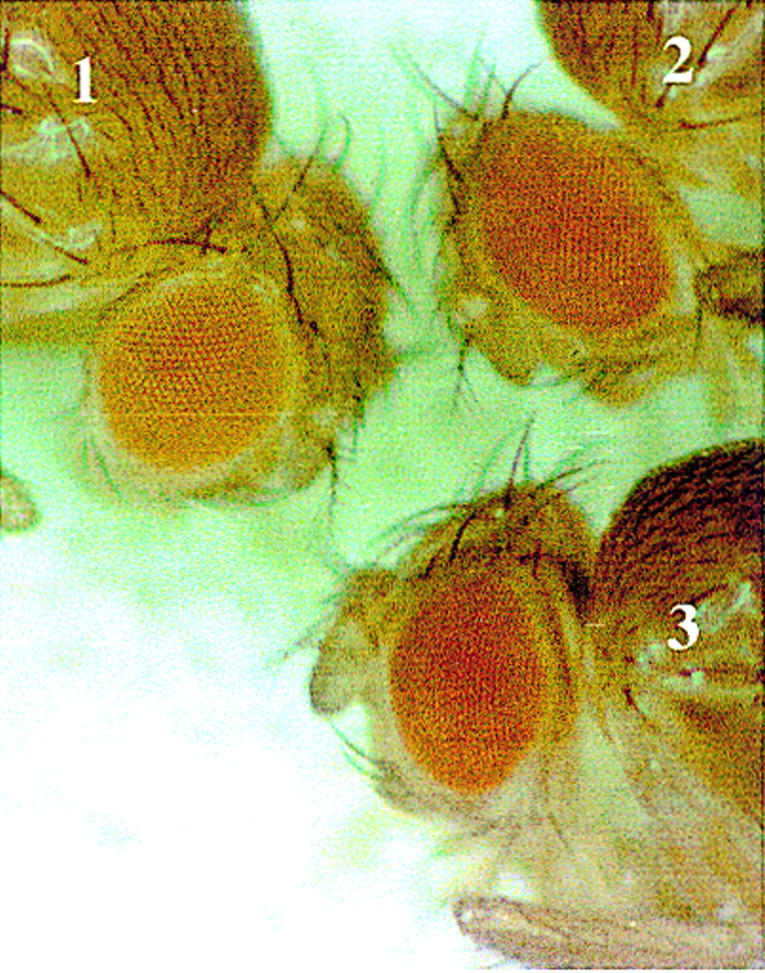
The enhancer-blocking activity of the MRwt oligonucleotide is reduced in zw5 heterozygous mutant background. Eye-color phenotype of the pE-4×MRwt-W transgenic construct in zw5 heterozygous mutant background. The genotypes of the flies are as follows: (1) w1/w1; P[pE-4×MRwt-W]/+; (2) zw562jl w1/w1; P[pE-4×MRwt-W]/+; (3) zw590 w1/w1; P[pE-4×MRwt-W]/+.
Discussion
Zw5 is able to block enhancer–promoter interaction
Two lines of evidence argue that the Zw5 protein is responsible for the enhancer-blocking activity of the 4×BSwt and 4×MRwt multimers. First, mutations in the BSwt and MRwt oligonucleotides that eliminate Zw5 protein binding in vitro compromise their enhancer-blocking activity in vivo. Although it remains possible that these mutations interfere with the binding of some as-yet-unidentified protein(s), the simplest interpretation of the result is that the loss of enhancer-blocking activity is attributable to the disruption in Zw5 protein binding. The second line of evidence comes from the effect of zw5 mutations on the enhancer-blocking activity of the multimerized MRwt oligonucleotide. Because zw5 mutations are cell autonomous lethal, we were limited to testing whether a twofold reduction in Zw5 protein levels would reduce the insulating activity of BSwt and MRwt. The zw5 gene is not haploinsufficient, and we observed no effects on the blocking activity of the 4×BSwt multimer. However, the blocking activity of the 4×MRwt multimer is compromised in zw5 heterozygous mutant animals. This effect is not allele specific nor is it dependent on the transgene’s site of insertion. It seems likely that the blocking activity of the MRwt oligonucleotide is reduced in the zw5 heterozygous mutant background because it does not contain a complete Zw5 binding site. Although the missing nucleotides have only a marginal effect on Zw5 binding to the 4×MRwt multimer in vitro, this reduction in affinity would appear to be sufficient to make the blocking activity of the MRwt multimer sensitive to the level of Zw5 protein.
Although the 4×BSwt and 4×MRwt multimers can block enhancer–promoter interactions, the insulator activity of these reiterated Zw5 binding sites is not equivalent to that of the intact scs element. One explanation for this discrepancy is that the dose of the Zw5 protein is not sufficient to give strong insulation. In the case of the gypsy insulator, for example, the strength of the blocking activity has been shown to depend on the number of reiterated Su(Hw) binding sites. The correlation between the number of Su(Hw) binding sites in the element and the mutagenic effects of the gypsy retrotransposon was first noted by Peifer and Bender (l988) in their studies on revertants of gypsy-induced mutations in the Ultrabithorax locus of the Bithorax Complex. Subsequent experiments have shown that gypsy insulators consisting of five Su(Hw) binding sites have only minimal blocking activity (Hagstrom et al. 1996). Hence, it is possible that four tandem copies of the Zw5 binding site are not sufficient to generate a level of blocking activity equivalent to that of the intact gypsy or scs elements. Consistent with dose dependence, the two Zw5 binding sites in the 2×BSwt multimer have little or no blocking activity. An alternate possibility is that the blocking activity of the Zw5 protein is inherently limited. In this view, it would be the combination of Zw5 with other as-yet-unidentified scs binding proteins that gives the strong insulating activity of the intact scs element. In this case, arrays consisting of more than four copies of the Zw5 binding site would not be expected to have a great deal more insulating activity than the 4×BSwt multimer.
The molecular nature of the zw5 alleles indicates that blocking activity of the Zw5 protein requires not only the zinc finger domains, but also the amino-terminal domain. We presume that the carboxy-terminal zinc finger domain is required for binding to the Zw5 recognition sequence. The amino-terminal portion of the Zw5 protein presumably functions in some key protein–protein interactions. Either it could directly prevent promoter activation by contacting proteins associated with the enhancer (or promoter) or it could recruit additional factors that actually have the insulating activity. Besides Zw5 there are two other known chromatin proteins, BEAF and Su(Hw), that are able to block enhancer–promoter interactions. Our failure to find regions of homology shared by Zw5, Su(Hw), and BEAF outside the zinc finger domain raises the possibility that they achieve enhancer-blocking through somewhat different molecular mechanisms. This idea is further supported by the lack of genetic interaction between zw5 and mod(mdg4), a gene implicated in mediating the activity of Su(Hw) (M. Gaszner and P. Schedl, unpubl.).
zw5 is required for cell proliferation and differentiation
Although our results argue that the Zw5 protein is required for the enhancer-blocking activity of the multimerized BSwt and MRwt oligonucleotides, the Zw5 protein may have a range of activities in the fly. Previous genetic studies indicate that zw5 is required for an essential cellular function (Zimm 1992). Homozygous mutant clones of strong zw5 alleles cannot be recovered, whereas clones generated with hypomorphic zw5 alleles have both cell-autonomous and cell-nonautonomous phenotypes (Shannon et al. 1972). As would be expected from the clonal analysis, strong loss of function mutations in zw5 are recessive lethal. Homozygous mutant animals arrest development at the first-instar larval stage and die after a few days. Given the results of the clonal analysis, it seems unlikely that the Zw5 protein is dispensable during embryogenesis. Instead, a more likely explanation for the postembryonic lethal phase of zw5 mutants is that there is a substantial maternal contribution of the zw5 gene product. Consistent with this idea, a similar late lethal phase has been observed for animals homozygous mutant in the Trithorax-like (Trl) gene, which encodes an essential gene product, the GAGA factor (Farkas et al. 1994). In this case, when the maternal contribution of the Trl gene is eliminated, the mutant animals die during early embryogenesis. Weak zw5 alleles have been reported to give rise to hemizygous mutant adults. The surviving males are sterile and display a variety of eye, bristle, and wing phenotypes indicating that the zw5 mutation has a pleiotropic effect on development. Because mutations in genes encoding generic chromosomal proteins, like the GAGA factor, exhibit a similar range of phenotypic effects, the spectrum of phenotypes observed in zw5 mutants would be consistent with the idea that the gene has a role in chromatin architecture. However, additional functions in gene regulation or replication cannot be excluded.
Materials and methods
Plasmids, transgenic lines, and mutant stocks
The pEW construct was generated and described as pRW by Vazquez and Schedl (1994). Unique XhoI and XbaI restriction sites were used to insert test fragments upstream of the enhancer or between the enhancer and the promoter, respectively. The pEW and pE-scs-W transgenic lines were generated and described by Vazquez and Schedl (1994). P-element-mediated transformation was performed according to standard protocols (Pirrotta 1988). Fly stocks were maintained at room temperature. The eye-color phenotype was determined by visual examination of 1.5-day-old females with a single copy of a transgene. The zw562jl allele was obtained from the Bloomington Drosophila Stock Center, and the zw590 allele was generated by EMS mutagenesis in our laboratory.
cDNA expression library screen
D. melanogaster, embryo 5′-STRECH cDNA Library (Clontech Laboratories, Inc.) was screened with a radioactively labeled DNA fragment containing four copies of the MRwt oligonucleotide following the protocol of C. Parker (Prakash et al. 1992). The screen identified a single clone with a 2-kb cDNA insert that contains the entire coding region for SBP.
In vitro binding assays
The 6×His-tagged full-length SBP protein was expressed in Escherichia coli using the QIAexpress (Qiagen, Inc.) system. Inclusion bodies containing the recombinant SBP protein were washed with 5 m urea, solubilized in 5 m urea/100 mm DTT, and refolded according to Jaenicke and Rudolph (1991). After the refolded protein was concentrated on a MonoQ column (Pharmacia), it was dialyzed into buffer S (80 mm KCl, 50 mm Tris-HCl at pH 8.0, 10 μm ZnCl2, 0.5 mm EDTA, 1 mm DTT, 1% Triton X-100, 10% glycerol) and stored at −70°C in small aliquots. Fragments A (415–781), B (696–1020), and C (1273–1577) were generated by PCR amplification followed by agarose-gel purification. Numbers in parenthesis show the position of each fragment according to X63731 GenBank sequence file. Gel mobility-shift assays were performed according to Zhao et al. (1995). Briefly, recombinant SBP protein, end-labeled probe (20,000 cpm), and poly[d(I-C)] (50 μg/ml final concentration) nonspecific competitor were incubated for 25 min at room temperature in 20 μl of 1× binding buffer (100 mm KCl, 10 mm HEPES–KOH at pH 7.6, 5 mm MgCl2, 0.1 mm EDTA, 0.1 mm DTT, 5% glycerol). DNA–protein complexes were subsequently resolved by gel electrophoresis on a 5% acrylamide/0.5× TBE/2.5% glycerol gel at 4°C. DNase I footprinting on fragment C was done as described by Hart et al. (1997).
Chromatin immunoprecipitation
Anti-SBP rabbit polyclonal antibody was generated and affinity purified according to standard protocols (Harlow 1988). 6×HIS-tagged full-length recombinant SBP was used for immunization and affinity purification. Chromatin immunoprecipitation was done as described by Orlando and Paro (1993). Slot-blot hybridization signals were determined using a PhosphorImager (Molecular Dynamics, Inc.).
Immunostaining of polytene chromosomes
Polytene chromosome were fixed and prepared for immunostaining according to Andrew and Scott (1994) with a small modification. Third-instar larva salivary glands were dissected in 1× PBS/1% Tween 20 and fixed for 2 min in 3.7% formaldehyde/50% acetic acid/1× PBS/1% Tween 20. Control experiments showed that the omission of the 3.7% formaldehyde fixation step did not change the staining pattern. Distribution of Zw5 was visualized using Alexa-488 (Molecular Probes, Inc.) conjugated goat anti-rabbit secondary antibody. DNA was stained with propidium iodide.
Acknowledgments
We thank past and present members of the laboratory, especially G. Deshpande, for stimulating discussions. We acknowledge G. Calhoun for her excellent technical assistance. This work was supported by a National Institutes of Health grant to P.S. M.G. was supported by a Charlotte Elizabeth Procter Fellowship. J.V. received support from the Swiss Science Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pschedl@molbio.princeton.edu; FAX (609) 258-1028
References
- Andrew DJ, Scott MP. Immunological methods for mapping protein distributions on polytene chromosomes. Methods Cell Biol. 1994;44:353–370. doi: 10.1016/s0091-679x(08)60923-1. [DOI] [PubMed] [Google Scholar]
- Cuvier O, Hart CM, Laemmli UK. Identification of a class of chromatin boundary elements. Mol Cell Biol. 1998;18:7478–7486. doi: 10.1128/mcb.18.12.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Chromatin structure and gene expression. Proc Natl Acad Sci. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase. FlyBase: A Drosophila database. Nucleic Acids Res. 1998;26:85–88. doi: 10.1093/nar/26.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes & Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- Grosveld F, Schedl P. Boundaries and domains. In: Elgin SCR, editor. Chromatin structure and gene expression. New York, NY: IRL Press; 1995. pp. 172–196. [Google Scholar]
- Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes & Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hart CM, Zhao K, Laemmli UK. The scs′ boundary element: Characterization of boundary element-associated factors. Mol Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenicke R, Rudolph R. Folding proteins. In: Creighton TE, editor. Protein structure—a practical approach. New York, NY: IRL Press; 1991. pp. 192–223. [Google Scholar]
- Judd BH, Shen MW, Kaufman TC. The anatomy and function of a segment of the X chromosome of Drosophila melanogaster. Genetics. 1972;71:139–156. doi: 10.1093/genetics/71.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R, Elgin SC. Chromatin boundaries: Punctuating the genome. Curr Biol. 1998;8:R521–R524. doi: 10.1016/s0960-9822(07)00337-5. [DOI] [PubMed] [Google Scholar]
- Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- ————— A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D, Zimm G. The genome of Drosophila melanogaster. San Diego, CA: Academic Press, Inc.; 1992. [Google Scholar]
- Mihaly J, Hogga I, Barges S, Galloni M, Mishra RK, Hagstrom K, Muller M, Schedl P, Sipos L, Gausz J, Gyurkovics H, Karch F. Chromatin domain boundaries in the Bithorax complex. Cell Mol Life Sci. 1998;54:60–70. doi: 10.1007/s000180050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Peifer M, Bender W. Sequences of the gypsy transposon of Drosophila necessary for its effects on adjacent genes. Proc Natl Acad Sci. 1988;85:9650–9654. doi: 10.1073/pnas.85.24.9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. In: Rodriguez RL, Denhardt DT, editors. Vectors: A survey of molecular cloning vectors and their uses. Boston, MA: Butterworth; 1988. pp. 437–456. [Google Scholar]
- Prakash K, Fang XD, Engelberg D, Behal A, Parker CS. dOct2, a Drosophila Oct transcription factor that functions in yeast. Proc Natl Acad Sci. 1992;89:7080–7084. doi: 10.1073/pnas.89.15.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon MP, Kaufman TC, Shen MW, Judd BH. Lethality patterns and morphology of selected lethal and semi-lethal mutations in the zeste-white region of Drosophila melanogaster. Genetics. 1972;72:615–638. doi: 10.1093/genetics/72.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana C, Harrison DA, Corces VG. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes & Dev. 1988;2:1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- Udvardy A. Dividing the empire: Boundary chromatin elements delimit the territory of enhancers. EMBO J. 1999;18:1–8. doi: 10.1093/emboj/18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- Udvardy A, Schedl P. The dynamics of chromatin condensation: Redistribution of topoisomerase II in the 87A7 heat shock locus during induction and recovery. Mol Cell Biol. 1993;13:7522–7530. doi: 10.1128/mcb.13.12.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J, Schedl P. Sequences required for enhancer-blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 1994;13:5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]