Abstract
The protozoan parasite Trypanosoma cruzi, which causes human Chagas' disease, exerts a variety of effects on host extracellular matrix (ECM) including proteolytic degradation of collagens and dampening of ECM gene expression. Exposure of primary human dermal fibroblasts to live infective T. cruzi trypomastigotes or their shed/secreted products results in a rapid down-regulation of the fibrogenic genes collagenIα1, fibronectin and connective tissue growth factor (CTGF/CCN2). Here we demonstrate the ability of a secreted/released T. cruzi factor to antagonize ctgf/ccn2 expression in dermal fibroblasts in response to TGF-ß, lysophosphatidic acid or serum, where agonist-induced phosphorylation of the mitogen-activated protein (MAP) kinases Erk1/2, p38 and JNK was also inhibited. Global analysis of gene expression in dermal fibroblasts identified a discrete subset of TGF-ß-inducible genes involved in cell proliferation, wound repair, and immune regulation that are inhibited by T. cruzi secreted/released factors, where the genes exhibiting the highest sensitivity to T. cruzi are known to be regulated by MAP kinase-activated transcription factors. Consistent with this observation, the Ets-family transcription factor binding site in the proximal promoter region of the ctgf/ccn2 gene (−91 bp to −84 bp) was shown to be required for T. cruzi-mediated down-regulation of ctgf/ccn2 reporter expression. The cumulative data suggest a model in which T. cruzi-derived molecules secreted/released early in the infective process dampen MAP kinase signaling and the activation of transcription factors that regulate expression of fibroblast genes involved in wound repair and tissue remodelling, including ctgf/ccn2. These findings have broader implications for local modulation of ECM synthesis/remodelling by T. cruzi during the early establishment of infection in the mammalian host and highlight the potential for pathogen-derived molecules to be exploited as tools to modulate the fibrogenic response.
Introduction
The kinetoplastid protozoan parasite Trypanosoma cruzi causes Chagas' disease in humans, a chronic and debilitating condition affecting several million individuals in Latin America. T. cruzi is transmitted by an insect vector which gains access to the host via breaches in the skin or through mucosal membranes, mainly conjunctival or gastric mucosa [1], [2]. As an obligate intracellular parasite that disseminates from initial infection sites to tissues such as heart and smooth muscle, T. cruzi undergoes multiple rounds of invasion, growth and egress from infected cells during the acute stage of infection. Very little is currently known regarding the early interactions between T. cruzi and its host that facilitate establishment of infection in vivo. Cellular models of T. cruzi infection have been very useful for defining the molecular and cellular events that regulate the early parasite-host cell interactions and host cell invasion. During its early interaction with mammalian host cells, trypomastigotes, the invasive forms of T. cruzi, trigger rapid changes in a number of cellular signaling pathways to facilitate the process of parasite entry into non-professional phagocytic cells (reviewed in [3], [4]). While these early signaling events have been relatively well-studied in the context of T. cruzi invasion, little is known regarding the impact of these parasite-induced signaling cascades downstream of the invasion process. Transcriptional profiling of T. cruzi-infected fibroblasts revealed that the earliest detectable changes triggered by infective T. cruzi trypomastigotes involve down-regulation of a small subset of genes including members of the CCN family (cyr61 and ctgf/ccn2) [5], which play important roles in angiogenesis and extracellular matrix (ECM) homeostasis [6]. T. cruzi-dependent dampening of ctgf/ccn2 expression occurs at both the mRNA and protein levels and is mediated by a secreted/released parasite factor that is capable of antagonizing TGF-ß-mediated induction of ctgf/ccn2 [7].
Connective tissue growth factor (CTGF/CCN2) is a 38 kDa secreted cysteine-rich heparin-binding glycoprotein [8] that promotes cell proliferation and co-operates with TGF-ß to promote myofibroblast differentiation and enhanced extracellular matrix (ECM) synthesis (reviewed in [9]). Dysregulation of CTGF/CCN2 expression leads to excessive scarring and fibrosis and this cytokine is over-expressed in a variety of tumors where CTGF/CCN2 levels correlate with disease progression [9]. As such there has been significant interest in CTGF/CCN2 as a therapeutic target for a number of disease states [10], [11], [12], [13]. Our finding that the human pathogen, Trypanosoma cruzi, releases a factor that inhibits TGF-ß-mediated expression of CTGF/CCN2 prompted further investigation into the mechanistic basis for this observation. CTGF/CCN2 expression is induced by diverse extracellular stimuli, including growth factors, cytokines and mechanical stress [9], [14], [15], [16], [17]. TGF-ß-stimulated expression of CTGF/CCN2 requires the activation of SMAD proteins and MAP kinases downstream of the TGF-ß receptor [9], [15]. Erk1/2 and p38 are generally associated with positive regulation of CTGF/CCN2 expression in different cell types [15], [18], [19], [20], whereas the role of JNK is more variable [15], [18], [20]. The expression of CTGF/CCN2 is also controlled via the activities of the ETS family of transcription factors. A functional Ets-binding site identified in the proximal ctgf/ccn2 promoter, spanning the region −91 to −84 bp upstream of the transcriptional start site, is bound by Ets-1 to promote TGF-ß-dependent induction of ctgf/ccn2 [21], while binding of the same site by Fli-1 negatively regulates ctgf/ccn2 expression in human fibroblasts [22]. The activity of many members of the ETS family is controlled through MAP kinase signaling, where the DNA-binding and trans-activation activities of ETS transcription factors are regulated by phosphorylation [23], [24].
In the present study, we demonstrate that T. cruzi-dependent abrogation of ctgf/ccn2 expression in human dermal fibroblasts is associated with inhibition of both basal and agonist-induced activation of MAP kinase signaling and requires the functional Ets-binding site in the proximal promoter of the ctgf/ccn2 gene. Expanding our analysis of the impact of T. cruzi released factors on TGF-ß-induced fibroblast gene expression we describe a discrete subset of agonist-inducible fibroblast genes that are sensitive to T. cruzi secreted/released factors. We report that the group of TGF-ß-inducible genes that exhibit the highest sensitivity to a T. cruzi secreted/released fraction are MAP kinase-regulated genes that function in wound repair, extracellular matrix remodelling and host response pathways. Collectively, these findings provide novel insights into early T. cruzi-host cell interactions. We demonstrate the ability of secreted/released parasite molecules to hinder agonist-induced host cell signaling pathways and gene expression. If similar events occur early in the T. cruzi infection process in vivo, it would be predicted that local inhibition of ECM synthesis would facilitate dissemination from initial sites of infection.
Results
A secreted/released trypanosome factor inhibits agonist-induced expression of CTGF/CCN2 in human dermal fibroblasts
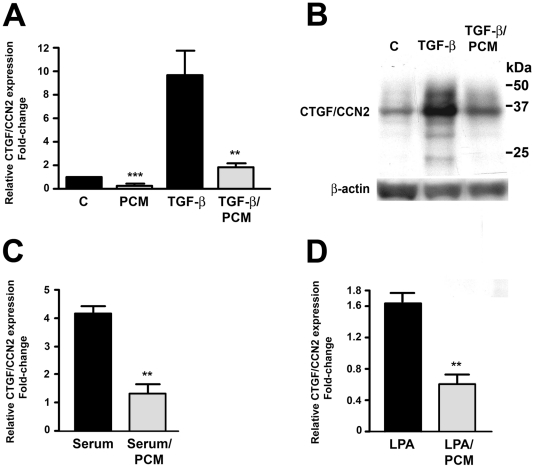
Connective tissue growth factor (CTGF/CCN2) expression is rapidly repressed in human dermal fibroblasts infected with the protozoan parasite, Trypanosoma cruzi, where the CTGF/CCN2-repressive activity was shown to be associated with a secreted/released trypanosome factor that is present in parasite-conditioned medium (PCM) [5] [7]. While the repressive factor released from live infective T. cruzi trypomastigotes into the medium (parasite-conditioned medium; PCM) [7] has not been identified, it is associated with a trypsin-sensitive, heat-labile, high molecular weight protein fraction of T. cruzi PCM enriched in GPI-anchored surface proteins (A. Mott and B. Burleigh, unpublished data). Here we show that treatment of human dermal fibroblasts (HFF) with T. cruzi PCM blocks induction of ctgf/ccn2 expression triggered by different agonists TGF-ß (Figure 1A, B), serum (Figure 1C) or lysophosphatidic acid (LPA) (Figure 1D). Thus, the ability of T. cruzi PCM to inhibit ctgf/ccn2 expression in response to distinct exogenous agonists suggests that signaling molecules primarily associated with TGF-ß receptor signaling such as the SMADs, are unlikely to be the downstream target of T. cruzi PCM leading to the repression of ctgf/ccn2.
Figure 1. T. cruzi PCM inhibits agonist-induced ctgf/ccn2 expression.
HFF were incubated with medium or T. cruzi PCM, alone or in combination with 5 ng/ml TGF-ß, 2 hours prior to mRNA harvest for quantitative real-time PCR analysis (A) or 24 hours prior to analysis of CTGF/CCN2 protein levels by western blot (B). ctgf/ccn2 mRNA levels were analyzed by quantitative real-time PCR following stimulation of HFF with serum (2% v/v) (C) or 10 µM LPA (D) for 2 hours in the presence or absence of T. cruzi PCM. Data is represented as the mean ± S.E. from 5 independent experiments carried out in duplicate. Statistical significance was assessed using the Student's t-test, (** p<0.01).
T. cruzi PCM inhibits agonist-induced MAP kinase activation
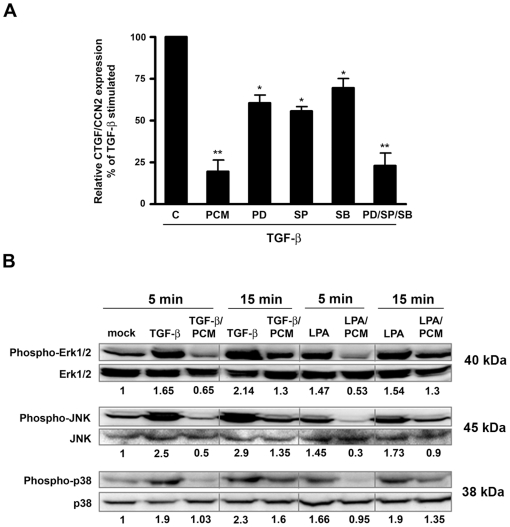
Given the well-established role of MAP kinase signaling in the regulation of ctgf/ccn2 expression in a variety of cell types [15], [18], [19], [20], [25], [26], Erk1/2, JNK and p38, were targeted in HFF with selective inhibitors to determine the impact on TGF-ß-inducible expression of ctgf/ccn2. Added individually, inhibitors of Erk1/2, JNK and p38 partially inhibited TGF-ß-dependent up-regulation of ctgf/ccn2 whereas in combination, the inhibitors were much more effective (Figure 2A) suggesting that different arms of the MAP kinase signaling pathway are involved in regulation of ctgf/ccn2 expression in HFF. As predicted from this result, stimulation of HFF with TGF-ß (5 ng/ml) or LPA (10 µM) promotes increased phosphorylation of Erk1/2, JNK and p38, which migrate on western blots at the expected molecular weights of 40 kDa, 45 kDa and 39 kDa respectively, where T. cruzi PCM abrogates both TGF-ß and LPA-induced phosphorylation of these kinases (Figure 2B). Combined, the data suggest that T. cruzi PCM exerts an inhibitory effect on ctgf/ccn2 expression by dampening MAP kinase signaling pathways.
Figure 2. T. cruzi PCM abrogates MAP kinase activation and decreased MAP kinase signaling results in inhibition of ctgf/ccn2 expression.
(A) HFF were stimulated with 5 ng/ml TGF-ß1 in the presence of medium, T. cruzi PCM or with 10 µM SP600125, 20 µM SB203580 or 50 µM PD98059 as indicated for 2 hours prior to mRNA harvest for quantitative real-time PCR analysis. MAP kinase inhibitor treatments were carried out following a 30-minute pre-incubation step. Data is represented as the mean ± s.e. from triplicate experiments (n = 4). Statistical significance was assessed using the Student's t-test (** p<0.01, * p<0.05). (B) Western blot of phospho-Erk, phospho-JNK and phospho-p38 normalized to total Erk, JNK and p38 respectively in lysates from HFF stimulated with 5 ng/ml TGF-ß1 or 10 µM LPA for 5 or 15 minutes in serum-free media or parasite-conditioned medium. Results for densitometric analysis are shown as numerical values below each panel and represented as phosphorylation relative to mock-treated controls (arbitrarily set to a value of 1.0).
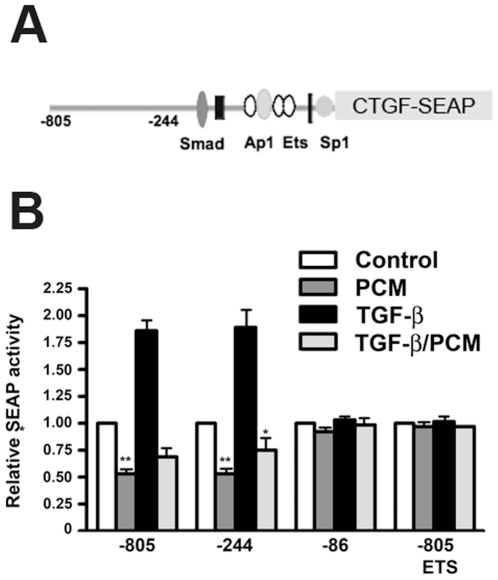
An Ets binding site in the proximal ctgf/ccn2 promoter is involved in T. cruzi-mediated repression of gene expression
The ctgf/ccn2 promoter/enhancer region contains several protein-binding sites that have been implicated in regulation of gene expression [21], [22], [27], [28], [29], [30]. Focusing on the proximal promoter region of ctgf/ccn2 (Figure 3A), we tested the impact of T. cruzi PCM treatment on basal and TGF-ß stimulated reporter activity using a series of ctgf/ccn2 promoter/SEAP reporter plasmids [21], [27] transfected into human dermal fibroblasts. As previously described, reporters containing functional Smad3 and Ets-binding sites: ie. spanning regions -805 to +17 bp (Figure 3B; −805) or −244 to +17 bp (Figure 3B ; −244) of the ctgf/ccn2 promoter were activated by TGF-ß whereas a severely truncated promoter/reporter spanning −86 to +17 bp of the ctgf/ccn2 promoter (Figure 3B; −86) failed to activate following TGF-ß stimulation. T. cruzi PCM treatment resulted in a significant decrease in basal activity of the −805 and −244 reporter constructs and blocked TGF-ß mediated activation of these reporters (Figure 3B). In contrast, basal expression of the reporter from the truncated construct (Figure 3B; −86) did not change in response to T. cruzi PCM. These data suggest that the Ets site present at −91 to −84 bp upstream of the transcriptional start site in the promoter is required to mediate the repressive effects of T. cruzi PCM on ctgf/ccn2 gene expression. In order to confirm this critical role for the Ets consensus site in PCM-mediated repression of ctgf/ccn2 expression, we examined the effect of T. cruzi PCM on the full-length reporter containing a mutated Ets site (Figure 3B; −805 ETS). As previously reported, a functional Ets binding site is required for TGF-ß-stimulated ctgf/ccn2 gene expression, and therefore this reporter is refractory to TGF-ß treatment (Figure 3B). Critically, a comparison between the response of the −805 and the mutant −805 ETS to PCM demonstrates the inability of PCM to repress activity in the absence of a functional Ets binding site (Figure 3B). We conclude from these experiments that a functional Ets binding site in the ctgf/ccn2 promoter is essential for TGF-ß dependent gene expression, and imparts sensitivity to T. cruzi-mediated inhibition of ctgf/ccn2 gene expression.
Figure 3. The Ets transcription factor binding site in the ctgf/ccn2 promoter is required for T. cruzi PCM-mediated repression of reporter expression.
SEAP reporter constructs driven by nucleotides −805 to +17 (−805), −244 to +17 (−244), −86 to +17 (−86) or −805 to +17 with a mutated ETS sequence (−805 ETS) representing the proximal ctgf/ccn2 promoter region (A) along with a ß-galactosidase containing control plasmid were transfected into HFF. (B) Serum-starved transfected cells were stimulated with medium alone or 5 ng/ml TGF-ß1 in the presence or absence of T. cruzi PCM for 6 hours. Relative reporter expression in treated cells relative to mock-treated controls is shown as the mean ± s.e. (n = 3). (*p<0.05, p<0.01).
T. cruzi PCM inhibits the expression of a subset of MAP kinase-regulated fibroblast genes involved in wound repair
Results presented above demonstrate that the factor(s) that are secreted or shed by T. cruzi trypomastigotes antagonize MAP kinase signaling in HFF downstream of TGF-ß and LPA. To determine the global impact of T. cruzi PCM on both basal and TGF-ß stimulated gene expression in human dermal fibroblasts, transcriptomic analyses were conducted following exposure of HFF to TGF-ß (5 ng/ml) in the presence or absence of T. cruzi PCM for 3 hours. cDNA prepared from treated and control cells were hybridized to human Affymetrix arrays containing ∼47,000 probesets (HG_U_133, 2.0). Raw expression data from triplicate experiments was imported into, and processed using, Rosetta Resolver 7.0 as outlined in the Experimental Procedures. Genes with expression levels that differed by 2-fold or greater (p<0.01) between the different experimental groups were considered for further analysis.
Examining first the impact of the secreted/released T. cruzi fraction on fibroblast gene expression, we find 89 unique genes (in 121 probe-sets) consistently down-regulated in PCM-treated cells and 82 genes (in 107 probe-sets) up-regulated greater than 2-fold (Table S1). Many of the repressed genes are ‘immediate early’ genes that regulate transcription (egr-1, jun, myc, fos, KLF2, KLF6) and are involved in cell proliferation [31], [32], [33], [34], [35] and inflammation [36], [37], [38]. As predicted from our studies with ctgf/ccn2, T. cruzi PCM treatment also results in the down-regulation of genes that are involved in extracellular matrix synthesis and tissue remodeling including hyaluronan synthase 2 (HAS2), tenascin C (TNC) and the CCN family members ctgf/ccn2 and cyr61 [17].
T. cruzi PCM inhibited the expression of approximately 30% of the with TGF-ß-inducible genes in HFF (Table S2). The 59 genes appearing in Table 1 represent the annotated, non-redundant TGF-ß inducible genes for which expression was inhibited by 1.7-fold or greater by T. cruzi PCM. Among the most highly PCM-sensitive, TGF-ß inducible genes are those with known involvement in extracellular matrix remodeling, wound healing and angiogenesis including heparin-binding EGF-like growth factor (HBEGF), angiopoietin-like 4 (ANGPTL4), endothelin-1 (EDN1), tropomyosin 1 (TPM1), chondroitin 4-sulfotransferase (CHST11), early growth response 1 (egr-1), connective tissue growth factor (ctgf/ccn2) and cysteine-rich, angiogenic inducer, 61 (CYR61). In addition, interleukin 6 (IL-6), chemokine (C-C motif) ligand 2 (CCL2) and prostaglandin-endoperoxide synthase 2 (PTGS2), which have been grouped with PCM-sensitive genes involved in inflammation and immune regulation (Table 1), also play an important role in wound repair [39], [40], [41], [42]. In support of the proposed role for MAP kinase-inhibition in mediating the effects of T. cruzi PCM on fibroblast gene expression, the TGF-ß-inducible genes with the highest sensitivity to T. cruzi PCM-mediated repression in the microarray analysis (ie. those for which a 4-fold or greater inhibition of TGF-ß-mediated induction of target gene expression was observed: EGR1 (17-fold), HBEGF (13-fold), PTGS2 (8.2-fold), LIF (7-fold), EDN1 (5.7-fold), IL6 (5.5-fold), CYR61 (4-fold)) are known to be regulated by MAP kinase activated transcription factors such as ETS-family transcription factors, AP-1 and NF-κB in a variety of cell types [43], [44], [45], [46], [47], [48], [49]. Quantitative RT-PCR carried out to confirm the pattern of PCM sensitivity for several TGF-ß inducible genes (Figure 4) reveals consistency with the microarray data where T. cruzi PCM was shown to inhibit TGF-ß dependent induction of HBEGF, CTGF/CCN2, PTSG2 and Cyr61. In contrast, TGF-ß stimulated expression of SERPINE2, CDKN2B, SMAD7 and TSPAN2 were refractory to PCM treatment (Figure 4). These data highlight both the broader effect of T. cruzi-derived factors on fibroblast gene expression and reveal selectivity of the response as only a subset of TGF-ß-inducible genes were affected when cells were co-stimulated with T. cruzi PCM.
Table 1. T. cruzi PCM inhibits expression of a subset of TGFß-inducible genes.
| Accession | Gene Symbol | Gene Description | *TGF-ß | TGF-ß + |
| PCM | ||||
| ECM Remodeling/Angiogenesis | ||||
| M60278 | HBEGF | Heparin-binding EGF-like growth factor | 50.4 | 3.7 |
| AF169312 | ANGPTL4 | Angiopoietin-like 4 | 42.0 | 19.9 |
| J05008 | EDN1 | Endothelin-1 | 7.3 | 1.3 |
| BC020765 | SERPINE1 | PAI-1 | 6.7 | 3.9 |
| AI459194 | EGR1 | Early growth response 1 | 5.3 | 0.3 |
| M92934 | CTGF | Connective tissue growth factor | 4.2 | 2.4 |
| NM_007036 | ESM1 | Endothelial cell-specific molecule 1 | 3.4 | 0.9 |
| AI806905 | CHST11 | Chondroitin 4 sulfotransferase 11 | 2.9 | 1.7 |
| AI524125 | PCDH9 | Protocadherin 9 | 2.7 | 1.6 |
| BC036577 | DDAH1 | Dimethylarginine dimethylaminohydrolase 1 | 2.7 | 1.1 |
| AI694545 | PLXNA2 | Plexin A2 | 2.6 | 1.1 |
| AI860360 | TPM1 | Tropomyosin 1a | 2.5 | 1.3 |
| AI374739 | HAS2 | Hyaluronan synthase 2 | 2.3 | 1.2 |
| Y13786 | ADAM19 | ADAM metallopeptidase domain 19 | 2.2 | 1.3 |
| AB002373 | RUSC2 | RUN and SH3 domain containing 2 | 2.1 | 1.2 |
| AF003114 | CYR61 | Cysteine-rich, angiogenic inducer, 61 | 2.1 | 0.5 |
| Inflammation/Immunity | ||||
| NM_000600 | IL6 | IL-6 | 4.2 | 0.8 |
| NM_000963 | PTGS2 | Prostaglandin-endoperoxide synthase 2 | 3.1 | 0.4 |
| S69738 | CCL2 | Chemokine (C-C motif) ligand 2 | 2.6 | 0.9 |
| AI608902 | CD274 | CD274 antigen | 2.5 | 0.9 |
| U93091 | TLR4 | Toll-like receptor 4 | 2.1 | 0.9 |
| Cell growth/differentiation | ||||
| BC093735 | LIF | Leukemia inhibitory factor | 6.4 | 1.0 |
| AK000850 | NEDD9 | Crk-associated substrate related | 6.1 | 3.4 |
| NM_030751 | SNF1LK | SNF1-like kinase | 3.7 | 2.1 |
| H94882 | ANKRD15 | Ankryn repeat domain 15 | 3.7 | 1.8 |
| M13436 | INHBA | Inhibin, beta A | 3.3 | 1.0 |
| AK025317 | MAML2 | Mastermind-like 2 | 3.1 | 1.3 |
| AB011109.1 | NUAK1 | NUAK family, SNF1-like kinase | 3.0 | 1.4 |
| NM_005118 | TNFSF15 | Tumor necrosis factor superfamily 15 | 2.7 | 1.3 |
| AF329092 | DOC1 | Anaphase-promoting complex 1 | 2.4 | 0.8 |
| AL136919 | LRRC8C | Leucine rich repeat protein | 2.3 | 1.3 |
| Signaling | ||||
| AV686810 | RHOB | RhoB | 10.0 | 4.6 |
| AF087853 | GADD45B | Growth arrest and DNA-damage-inducible | 5.5 | 2.5 |
| BE302191 | STK38L | Serine/threonine kinase 38 like | 3.3 | 2.0 |
| AU144916 | IGFBP7 | Insulin-like growth factor binding protein 7 | 3.2 | 1.3 |
| NM_007368 | RASA3 | RAS p21 protein activator | 2.8 | 1.7 |
| NM_006823 | PKIA | Protein kinase cAMP-dependent, catalytic) inhbitor | 2.7 | 1.1 |
| AI139993 | SH3GLP3 | SH3-domain GRB2-like | 2.3 | 0.9 |
| AU157259 | SMURF2 | SMAD specific E3 ubiquitin protein ligase 2 | 2.2 | 1.1 |
| AB043703 | FZD8 | Frizzled homolog 8 | 2.2 | 0.8 |
| NM_004073 | PLK3 | Cytokine-inducible kinase | 2.2 | 0.8 |
| Transcriptional regulation | ||||
| NM_000399 | EGR2 | Early growth response 2 | 100 | 60 |
| BG326045 | BHLHB2 | Basic helix-loop-helix domain containing | 6.6 | 2.7 |
| M60721 | HLX1 | H2.0-like homeo box 1 | 4.0 | 2.1 |
| AL120562 | KLF7 | Kruppel-like factor 7 | 3.3 | 1.5 |
| BF436898 | ETV6 | Ets variant gene 6 (TEL oncogene) | 3.1 | 1.7 |
| NM_005251 | FOXC2 | MFH-1, mesenchyme forkhead 1 | 2.9 | 1.4 |
| NM_012099 | CD3EAP | CD3-epsilon-associated protein | 2.7 | 1.6 |
| AF247704 | NKX3-1 | NK3 transcription factor related | 2.5 | 1.3 |
| Other functions | ||||
| AW138350 | ICHTHYIN | Ichthyin | 33.1 | 17.8 |
| AW004016 | ST6GAL2 | Beta-galactosamide a-2,6-sialyltransferase 2 | 12.6 | 7.5 |
| AF153330 | SLC19A2 | Thiamine transporter | 5.6 | 2.6 |
| AK026466 | CYFIP2 | Cytoplasmic FMR1 interacting protein 2 | 5.0 | 2.5 |
| AK026905 | MICAL2 | Microtubule associated monoxygenase | 3.6 | 1.5 |
| NM_000427 | LOR | Loricrin | 3.2 | 1.3 |
| AF043472 | KCNS3 | Potassium voltage-gated channel | 2.7 | 1.4 |
| NM_020127 | TUFT1 | Tuftelin | 2.7 | 1.6 |
| AI628360 | NPR3 | Natriuretic peptide receptor C | 2.2 | 1.2 |
| AI091372 | AXUD1 | TGF-beta-induced apoptosis protein 3 | 2.2 | 1.3 |
Human foreskin fibroblasts were treated with medium or 5 ng/ml TGF- ß1 for 3 hours in the presence and absence of T. cruzi PCM.
TGF-ß1 treatment resulted in a ≥2-fold up-regulation of 298 unique genes (represented in 309 probe-sets) as compared to cells treated with medium. Genes highlighted above represent the 59 annotated, non-redundant TGF-ß-inducible genes for which expression is reduced by 1.7-fold or greater in the presence of T. cruzi PCM in 3 independent experiments. * Values represent the ratio of the normalized log2 intensities for treatment (TGF-ß or TGF-ß/PCM)/mock treated controls.
Figure 4. T. cruzi PCM inhibits the expression of a broader range of TGF-ß-inducible genes.
Quantitative real-time PCR analysis of the relative expression of representative PCM-sensitive and PCM-insensitive TGF-ß-inducible genes in HFF after 2 hours of treatment with 5 ng/ml TGF-ß in media or PCM. Genes analyzed: HBEGF, heparin-binding EGF-like growth factor; PTSG2, prostaglandin-endoperoxide synthase 2; CTGF, connective tissue growth factor; Cyr61, cysteine-rich angiogenic factor; SERPINE2, plasminogen activator inhibitor type 1, member 2; CDKN2B; cyclin-dependent kinase 4 inhibitor B; SMAD7, MAD, mothers against decapentaplegic homolog 7; TSPAN2, tetraspanin 2.
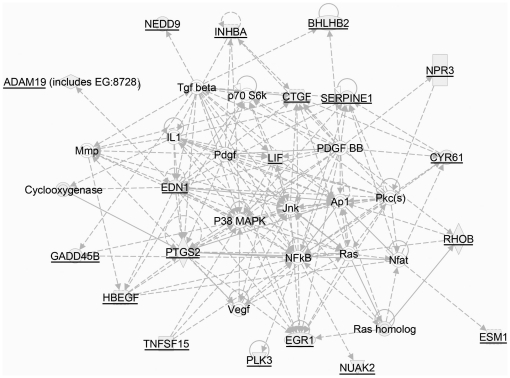
Biological network analysis conducted using the Ingenuity Pathway Analysis™ software program reveals a dense set of relationships between the subset of T. cruzi PCM sensitive TGF-ß-inducible genes and MAP kinases (Figure 5) where the top functions associated with this network include ‘cellular growth’, ‘cancer’ and ‘connective tissue development and function’. A comparative analysis of the top canonical pathways represented in the T. cruzi PCM-sensitive and PCM-insensitive TGF-ß inducible gene datasets are shown in Table S3. The top canonical pathways associated with the PCM-sensitive genes reveal 5 pathways with significant p-values (p<0.05) including ‘hepatic fibrosis/hepatic stellate cell activation’ (CTGF, FN1, TGFb2, EDN1, CCL2, IL-6) as the most significant pathway affected by the secreted/shed T. cruzi fraction. In contrast, when the PCM-insensitive group of TGF-ß-inducible genes was considered, the top functions involved ‘axonal guidance’ and ‘clathrin-mediated endocytosis signaling’ (Table S4) [43], [44], [45], [46], [47], [48], [49]. Collectively, the global expression data are supportive of a model in which infective T. cruzi trypomastigotes secrete or release factors that within minutes dampen MAP kinase signaling in human dermal fibroblasts. We propose that this dampening of host cell MAP kinase activity negatively impacts the activities of downstream transcription factors such as the ETS family of transcription factors known to be involved in the regulation of a number of genes involved in ECM synthesis and wound repair [50], [51] including ctgf/ccn2 gene expression [21], [22], [51].
Figure 5. Top biological network relevant to the subset of PCM-sensitive TGF-ß-inducible genes.
Biological network was generated through the use of Ingenuity Pathway Analysis software (Ingenuity® Systems, www.ingenuity.com) by uploading the group of TGF-ß-inducible genes that were highly sensitive to T. cruzi PCM. The analysis reports direct (solid lines) and indirect (dotted lines) relationships between genes as established in the literature. Thirteen (shaded symbols) of 15 genes entered into the analysis were included as focus genes in this network.
Discussion
This study demonstrates that mammalian-infective forms of Trypanosoma cruzi release a factor (or factors) that significantly impacts host cell signaling cascades, thereby altering the expression of a subset of genes involved in cell proliferation, wound repair and inflammation. Focusing on the pro-fibrogenic cytokine, CTGF/CCN2, a multifunctional secreted protein with a central role in wound repair and fibrosis [9], we demonstrate that a shed/secreted T. cruzi factor inhibits both basal and agonist-induced up-regulation of ctgf/ccn2 gene expression. In line with previous reports demonstrating a critical role for MAP kinases in regulating ctgf/ccn2 gene expression in response to pro-fibrogenic agonists [15], [17], [18], [19], [20], our data demonstrate a role for Erk1/2, JNK and p38 in TGF-ß-mediated ctgf/ccn2 expression in low passage primary HFF. Exposure of HFF to the shed/secreted T. cruzi fraction inhibits basal and agonist-dependent phosphorylation of Erk1/2, JNK and p38 and the induction of ctgf/ccn2 expression, a downstream target of MAP kinase activation [15]. Analysis of the global transcriptional response to T. cruzi PCM in HFF revealed significant repression of a subset of TGF-ß-inducible genes involved in cell proliferation, wound healing and inflammatory responses, where the genes most sensitive to T. cruzi-mediated inhibition (EGR1, HBEGF, PTGS2, LIF, EDN1, IL6, and CYR61) are known to be regulated by MAP kinase-activated transcription factors such as the ETS-family members and AP-1 [43], [44], [45], [46], [47], [48], [49]. In addition, genes encoding components of the AP-1 complex, c-jun and c-fos, as well as egr-1 were also rapidly down-regulated in T. cruzi PCM treated cells, suggesting the disruption of an important regulatory network centered around MAP kinase signaling, which is required for efficient expression of ctgf/ccn2 and a host of other TGF-ß-inducible genes. Consistent with the notion that T. cruzi might negatively regulate MAP kinase-activated transcription factors, we demonstrated that a functional Ets-binding site (spanning −91 and −84 bp) in the upstream ctgf/ccn2 promoter region, which is critical for TGF-ß stimulated expression of ctgf/ccn2 [15], [21], [22], is required for T. cruzi PCM repression of both basal and TGF-ß-stimulated reporter expression. Thus, we propose that the observed inhibition of agonist-induced phosphorylation of the MAP kinases, Erk, p38 and JNK, would impact the activation of MAP kinase-activated transcription factors such as Ets-1. Failure to assemble Ets transcription factor(s) at the Ets binding site, possibly in conjunction with other accessory proteins such as AP-1 [21], would prevent activation of the ctgf/ccn2 promoter in response to signals mediated by exogenous agonists such as TGF-ß [15]. Overall, our findings reinforce recent literature regarding the critical role for MAP kinase signaling in regulation of ctgf/ccn2 expression [9], [18], [20] as well as for the role of the Ets-binding site on the ctgf/ccn2 promoter [21], [22] in driving ctgf/ccn2 gene expression. Given that CTGF/CCN2 is a therapeutic target for the prevention and treatment of a number of disease states in which the expression of this critical cytokine is dysregulated [10], [11], [12], [13], our finding that the mammalian-infective forms of T. cruzi release a factor that interferes with agonist-dependent up-regulation of CTGF/CCN2 warrants further study.
We had shown previously that infection with live T. cruzi trypomastigotes or parasite-conditioned medium (PCM) causes rapid dephosphorylation of Erk1/2 and sustained repression of extracellular matrix protein expression in dermal fibroblasts [7]. Focusing on the secreted/released T. cruzi fraction in this study, we demonstrate the broad impact of T. cruzi PCM on MAP kinase signaling where agonist-dependent activation of Erk1/2, p38 and JNK is thwarted, as is the expression of a subset of TGF-ß-dependent gene expression. Our findings are consistent with a recent study demonstrating that T. cruzi infection of primary cardiomyocytes renders infected cells refractory to TGF-ß-induced increases in fibronectin expression [52]. While these in vitro observations provide novel insights into the impact of infective T. cruzi trypomastigotes on host cell signaling and gene expression, the ultimate goal would be to measure these events and outcomes in the context of an in vivo infection. Given the critical role played by MAP kinases in cell proliferation, wound healing and host immunity [19], [53], [54], [55]. It is tempting to speculate that modulation of this signaling pathway early in the T. cruzi infection process could modulate the local host environment in favor of the parasite. If, as suggested by in vitro studies, ECM synthesis and host immune response pathways were to be repressed at local sites of T. cruzi infection, we would predict that these events would facilitate the early establishment of infection in the host. While this would be challenging to test in the context of ECM homeostasis, the concept of immune evasion early in the establishment of infection by T. cruzi trypomastigotes is not new [56]. There are several examples of soluble T. cruzi immunomodulatory molecules including the trypanosomal immunosuppressive factor (TIF) which represses T- and B-cell functions [57], [58], [59], [60] and a mucin-like protein expressed on the surface of epimastigotes and metacyclic trypomastigotes (AgC10) which inhibits LPS-mediated signaling and dendritic cell activation via MAP kinase [61]. A parasite glycoinositol-phospholipid [62] and another polypeptide antigen (Ag123) [63] have also been implicated in modulating the host immune response demonstrating that T. cruzi may have evolved several overlapping mechanisms for interfering with normal host immune function. The T. cruzi molecule(s) responsible for antagonizing TGF-ß signaling in dermal fibroblasts, that dampen fibrogenic and immune gene expression, have yet to be identified. Efforts focused on isolation of this heat-labile, trypsin-sensitive factor have revealed that it co-fractionates with the major parasite surface glycoproteins and elutes from an anion exchange column in a sharp peak with ∼200 mM NaCl (A. Mott and B. Burleigh, unpublished observations) and is likely a component of previously described small shed vesicles [64]. It will be of great interest to identify the molecular basis for the parasite-dependent repression to better assess its functional properties and to probe its potential as a novel antagonist of fibrogenic gene expression and potential immunosuppressive activity in vitro and in vivo.
Overall, this study highlights the ability of a trypomastigote-associated soluble factor to dampen basal and agonist-induced expression of a key regulator of ECM synthesis as well as genes involved in wound repair and host response to infection. As studies have clearly demonstrated, T. cruzi infection eventually elicits a robust inflammatory response in the host where elevated ECM synthesis and fibrosis are known to contribute to the pathophysiology of Chagas' disease (reviewed in [65]). The cellular responses to T. cruzi released/secreted factors studied here are immediate/early in nature and provide insights into potential processes that would be expected to occur at early time points of infection, where the parasite-mediated inhibition of host cell fibrogenic gene expression may serve a critical function in facilitating dissemination from the initial sites of infection to the circulation and peripheral organs during acute stages of infection.
Materials and Methods
Cell Culture and Parasite Maintenance
Human foreskin fibroblast (HFF) BJ cells were acquired from the American Tissue Culture Collection (Manassas, VA, USA) and maintained in DMEM +10% fetal bovine serum (FBS), 1% pen-strep, and 2 mM glutamine at 37°C in 5% CO2. Tissue culture-derived Trypanosoma cruzi trypomastigotes (Y strain) were generated by weekly passage in confluent monolayers of LLcMK2 cells in DMEM containing 2% FBS as described [5]. Trypomastigotes harvested from cell culture supernatants were washed 3 times in cold serum-free DMEM (SF-DMEM), resuspended in warm SF-DMEM at a concentration of 5×107 to 1×108 parasites per ml and incubated at 37°C in 5% CO2 for 14–18 hours. The resulting ‘parasite-conditioned medium’ (PCM) was clarified by centrifugation of parasites (1000xg) followed by passage through a 0.2 µm membrane. Prior to treatment of cells, PCM is adjusted to pH 7.4 by addition of a small volume of 1M Tris pH 8.0 and supplemented with heat-inactivated FBS (95°C, 15 minutes) to achieve a final concentration of 2%.
Quantitative Real-Time PCR
HFF were seeded in 6-well plates and grown to 80–90% confluence for 48 hours prior to treatment of cells as indicated. Following treatment, cells were washed with cold PBS and total cellular RNA was harvested using the RNeasy RNA isolation system (Qiagen, Valencia, CA, USA) according to manufacturer's instructions. 200 ng of DNaseI-treated RNA (Invitrogen, Carlsbad, CA, USA) was transcribed to cDNA using the iScript Reverse Transcription system (BioRad, Hercules, CA, USA). Multiplex quantitative real-time PCR was carried out using an ABI 7300 instrument. The amount of experimental cDNA was normalized to GAPDH levels present in the same sample. For quantification of CTGF/CCN2 mRNA each reaction included 20 nM of each primer (forward 5′-CTGCCCTCGCGGCTTA-3′ and reverse 5′GGACCAGGCAGTTGGC TCTA-3′) and 10 nM FAM-labeled CTGF/CCN2 probe (6FAM-ACACGTTTG GCCCAGACCCAACTATG-TAMRA). Primers and probes specific for human SMAD7, SERPINE-2, CDKN2B, Cyr61, HBEGF, PTSG2 and GAPDH gene assays were obtained from Applied Biosystems Inc. (Foster City, CA, USA).
Western blot Analysis
Relative expression of CTGF/CCN2 was determined in HFF that were serum-starved for 18 hours prior to treatment with 5 ng/ml TGF-ß for 24 hours in the presence or absence of PCM. Cells were rinsed in cold PBS, lysed directly in 2X Laemmli Buffer containing 5% ß-mercaptoethanol (BioRad, Hercules, CA, USA) and western blot analyses were performed. For MAP kinase activation, HFF were serum starved for 18 hours prior to treatment with 10 mM LPA or 5 ng/ml TGF-ß1 with or without PCM for 5 or 15 minutes. Anti-ß-actin antibodies were purchased from Sigma (St. Louis, MO, USA); anti-CTGF from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-p44/42, anti-phospho-p44/p42, anti-p38, anti-phospho-p38, anti-SAPK/JNK and anti-phospho-SAPK/JNK from Cell Signaling Technology (Danvers, MA, USA). The ECL Plus chemiluminescence kit (Amersham Pharmacia, Arlington Heights, IL, USA) was used for detection. Blots were scanned and band intensity was quantified using the software program UN-SCAN-IT gel (Silk Scientific Corporation, Orem, UT, USA).
Transient Transfection and Reporter Assays
HFF were seeded in 6-well plates and grown to 50% confluence for 24 hours. Cells were transfected with 1 µg of experimental plasmid DNA and 0.05 µg of control plasmid by lipofection (FuGENE 6, Roche, Indianapolis, IN, USA) for 3–4 hours in DMEM +10% FBS. Cells were washed and incubated in DMEM +10% FBS for an additional 12–14 hours prior to incubation for 6 hours with DMEM-2, PCM prepared in DMEM-2, 5 ng/ml of TGF-ß1 in the presence and absence of PCM. Promoter/reporter constructs used contained ctgf/ccn2 promoter fragments spanning nucleotides −805 to +17 (−805), −244 to +17 (−244), −86 to +17 (−86) and a construct identical to 805 with a mutation in the Ets binding site (−805 ETS) corresponding to a GGAAT to TCCCG change in the consensus Ets binding motif located in the transcription enhancing factor (TEF) binding element located between −91 and −84. All constructs were kindly provided by Dr. A. Leask [21], [27]. Transfection efficiency was measured by co-transfection with pSV-ß-Galactosidase plasmid. Cells were harvested and SEAP and ß-galactosidase assays were performed using the Luminescent ß-galactosidase Reporter System or Great EscAPe SEAP Chemiluminescence Kit (Clontech, Mountain View, CA, USA) according to manufacturer's instructions.
DNA Microarray Hybridization and Analysis
HFF were mock-treated or stimulated with TGF-ß for 3 hours, in the presence or absence of T. cruzi PCM, rinsed 3 times with PBS. RNA was isolated with Trizol reagent (Invitrogen, Carlsbad, CA, USA) and concentrated using an RNeasy kit (Qiagen, Valencia, CA, USA). RNA integrity was evaluated employing an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), three independent samples for each treatment were processed and hybridized to individual HG_U133 plus 2.0 Affymetrix chips at the Harvard Medical School Biopolymers Facility. Arrays were scanned using a GeneChip® Scanner 3000 (Affymetrix, Santa Clara, CA, USA) and data files containing the unprocessed intensity values were imported into Rosetta Resolver Biosoftware 7.0. Data were pre-processed to reduce systematic errors (i.e. background subtraction and intra-array normalization). Through the application of the Affymetrix-specific error model within Rosetta Resolver [66] the data were transformed into profiles (i.e. scanned, imaged and normalized expression data from a microarray as described in http://www.rosettabio.com/tech/Data_processing_and_analysis_methods.pdf). Intensity ratios were calculated from the statistical combination of replicate profiles in order to increase the confidence in measurements and to obtain fold change values and associated p-values, using the control condition (cells treated with culture medium) as a baseline. Transcripts showing ≥2-fold change (p<0.01) with respect to matched controls were used to create gene lists and for analysis of biological networks using Ingenuity Pathway Analysis™ software (http://www.ingenuity.com). In accordance to the minimum information about microarray experiment (MIAME) guidelines, the complete raw and processed data files for each array are publicly available at the Gene Expression Omnibus (GEO) database repository (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE16416.
Supporting Information
Modulation of fibroblast gene expression by a secreted/released T. cruzi fraction. Human foreskin fibroblasts (HFF) were mock-treated or incubated with cell-free T. cruzi parasite-conditioned medium (PCM) for 3 hours and the isolated RNA was processed and used to hybridize HG_U133 plus 2.0 Affymetrix arrays. Ratios of the normalized log2 intensities for PCM vs medium were used to calculate the fold-change in HFF gene expression for each array and values from 3 biological replicates were averaged. HFF genes for which the transcript abundance changes ≥2-fold in either direction are included.
(XLSX)
Inhibition of TGF-ß-inducible gene expression by T. cruzi PCM. HFF were mock-treated or stimulated with 5 ng/ml TGF-ß1 for 3 hours in the presence or absence of T. cruzi PCM and cells were processed for microarray hybridization as described. Ratios of the normalized log2 intensities for TGF-ß vs medium and TGF-ß+PCM vs medium were used to calculate the fold-decrease in TGF-ß inducible gene expression that was observed in the presence of T. cruzi PCM. Only the non-redundant, annotated genes are shown with the exception of genes for which duplicates showed marked differences in the fold-change values.
(XLSX)
Canonical pathways enriched in the T. cruzi PCM-sensitive subset of TGF-ß inducible genes. TGF-ß-inducible genes sensitive to T. cruzi PCM (59 genes from Table 1) were analyzed using Ingenuity Pathway Analysis software. Canonical pathways were identified from the Ingenuity Pathways Analysis library of canonical pathways that were most significant to the data set and associated with a canonical pathway in Ingenuity's Knowledge Base. The significance of the association between the data set and the canonical pathway was measured in 2 ways: 1) A ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway is displayed. 2) Fisher's exact test was used to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone.
(XLS)
Canonical pathways enriched in the subset of TGF-ß inducible genes that is insensitive to T. cruzi PCM. TGF-ß-inducible genes that were insensitive to T. cruzi PCM (192 genes; Table S2) were analyzed using Ingenuity Pathway Analysis software and canonical pathways were identified from the Ingenuity Pathways Analysis library as described for Table S3.
(XLS)
Acknowledgments
We are grateful to Colby Wells for maintenance of T. cruzi cultures.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by National Institutes of Health R01 HL073227 (http://www.nhlbi.nih.gov/) and Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases (BAB) (http://www.bwfund.org/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Giddings OK, Eickhoff CS, Smith TJ, Bryant LA, Hoft DF. Anatomical route of invasion and protective mucosal immunity in Trypanosoma cruzi conjunctival infection. Infect Immun. 2006;74:5549–5560. doi: 10.1128/IAI.00319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshida N. Trypanosoma cruzi infection by oral route: how the interplay between parasite and host components modulates infectivity. Parasitol Int. 2008;57:105–109. doi: 10.1016/j.parint.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 3.Epting CL, Coates BM, Engman DM. Molecular mechanisms of host cell invasion by Trypanosoma cruzi. Exp Parasitol. 2010;126:283–291. doi: 10.1016/j.exppara.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mott GA, Burleigh BA. The role of host cell lysosomes in Trypanosoma cruzi invasion. Subcell Biochem. 2008;47:165–173. doi: 10.1007/978-0-387-78267-6_13. [DOI] [PubMed] [Google Scholar]
- 5.de Avalos SV, Blader IJ, Fisher M, Boothroyd JC, Burleigh BA. Immediate/Early Response to Trypanosoma cruzi Infection Involves Minimal Modulation of Host Cell Transcription. Journal of Biological Chemistry. 2002;277:639–644. doi: 10.1074/jbc.M109037200. [DOI] [PubMed] [Google Scholar]
- 6.Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol. 2009;41:771–783. doi: 10.1016/j.biocel.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Unnikrishnan M, Burleigh BA. Inhibition of host connective tissue growth factor expression: a novel Trypanosoma cruzi-mediated response. FASEB Journal. 2004;18:1625–1635. doi: 10.1096/fj.04-1554com. [DOI] [PubMed] [Google Scholar]
- 8.Grotendorst GR. Connective Tissue Growth Factor: A Mediator of TGF-beta Action on Fibroblasts. Cytokine & Growth Factor Reviews. 1997;8:171–179. doi: 10.1016/s1359-6101(97)00010-5. [DOI] [PubMed] [Google Scholar]
- 9.Shi-Wen X, Leask A, Abraham D. Regulation and function of connective tissue growth factor/CCN2 in tissue repair, scarring and fibrosis. Cytokine & Growth Factor Reviews. 2008;19:133–144. doi: 10.1016/j.cytogfr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Denton CP, Abraham DJ. Transforming growth factor-beta and connective tissue growth factor: key cytokines in scleroderma pathogenesis. Curr Opin Rheumatol. 2001;13:505–511. doi: 10.1097/00002281-200111000-00010. [DOI] [PubMed] [Google Scholar]
- 11.Hofmeister V, Schrama D, Becker JC. Anti-cancer therapies targeting the tumor stroma. Cancer Immunol Immunother. 2008;57:1–17. doi: 10.1007/s00262-007-0365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alapati D, Rong M, Chen S, Hehre D, Rodriguez MM, et al. CTGF Antibody Therapy Attenuates Hyperoxia-Induced Lung Injury in Neonatal Rats. Am J Respir Cell Mol Biol. 2011. [DOI] [PubMed]
- 13.Adler SG, Schwartz S, Williams ME, Arauz-Pacheco C, Bolton WK, et al. Phase 1 study of anti-CTGF monoclonal antibody in patients with diabetes and microalbuminuria. Clin J Am Soc Nephrol. 2010;5:1420–1428. doi: 10.2215/CJN.09321209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaqour B, Goppelt-Struebe M. Mechanical regulation of the Cyr61/CCN1 and CTGF/CCN2 proteins. Febs J. 2006;273:3639–3649. doi: 10.1111/j.1742-4658.2006.05360.x. [DOI] [PubMed] [Google Scholar]
- 15.Leask A, Holmes A, Black CM, Abraham DJ. Connective Tissue Growth Factor Gene Regulation. Journal of Biological Chemistry. 2003;278:13008–13015. doi: 10.1074/jbc.M210366200. [DOI] [PubMed] [Google Scholar]
- 16.Muehlich S, Cicha I, Garlichs CD, Krueger B, Posern G, et al. Actin-dependent regulation of connective tissue growth factor (CTGF). Am J Physiol Cell Physiol. 2007. 00552.02006. [DOI] [PubMed]
- 17.Muehlich S, Schneider N, Hinkmann F, Garlichs CD, Goppelt-Struebe M. Induction of connective tissue growth factor (CTGF) in human endothelial cells by lysophosphatidic acid, sphingosine-1-phosphate, and platelets. Atherosclerosis. 2004;175:261–268. doi: 10.1016/j.atherosclerosis.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Black SA, Palamakumbura AH, Stan M, Trackman PC. Tissue-specific Mechanisms for CCN2/CTGF Persistence in Fibrotic Gingiva. J Biol Chem. 2007;282:15416–15429. doi: 10.1074/jbc.M610432200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Blom IE, Sa S, Goldschmeding R, Abraham DJ, et al. CTGF expression in mesangial cells: Involvement of SMADs, MAP kinase, and PKC. 2002;62:1149–1159. doi: 10.1111/j.1523-1755.2002.kid567.x. [DOI] [PubMed] [Google Scholar]
- 20.Utsugi M, Dobashi K, Ishizuka T, Masubuchi K, Shimizu Y, et al. C-Jun-NH2-Terminal Kinase Mediates Expression of Connective Tissue Growth Factor Induced by Transforming Growth Factor-{beta}1 in Human Lung Fibroblasts. Am J Respir Cell Mol Biol. 2003;28:754–761. doi: 10.1165/rcmb.4892. [DOI] [PubMed] [Google Scholar]
- 21.Van Beek J, Kennedy L, Rockel J, Bernier S, Leask A. The induction of CCN2 by TGFbeta1 involves Ets-1. Arthritis Research & Therapy. 2006;8:R36. doi: 10.1186/ar1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakerakanti SS, Kapanadze B, Yamasaki M, Markiewicz M, Trojanowska M. Fli1 and Ets1 Have Distinct Roles in Connective Tissue Growth Factor/CCN2 Gene Regulation and Induction of the Profibrotic Gene Program. J Biol Chem. 2006;281:25259–25269. doi: 10.1074/jbc.M600466200. [DOI] [PubMed] [Google Scholar]
- 23.Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 24.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 25.Liu B, Yu J, Taylor L, Zhou X, Polgar P. Microarray and phosphokinase screenings leading to studies on ERK and JNK regulation of connective tissue growth factor expression by angiotensin II 1a and bradykinin B2 receptors in Rat1 fibroblasts. Journal of Cellular Biochemistry. 2006;97:1104–1120. doi: 10.1002/jcb.20709. [DOI] [PubMed] [Google Scholar]
- 26.Shi-wen X, Howat SL, Renzoni EA, Holmes A, Pearson JD, et al. Endothelin-1 Induces Expression of Matrix-associated Genes in Lung Fibroblasts through MEK/ERK. J Biol Chem. 2004;279:23098–23103. doi: 10.1074/jbc.M311430200. [DOI] [PubMed] [Google Scholar]
- 27.Holmes A, Abraham DJ, Chen Y, Denton C, Shi-wen X, et al. Constitutive Connective Tissue Growth Factor Expression in Scleroderma Fibroblasts Is Dependent on Sp1. J Biol Chem. 2003;278:41728–41733. doi: 10.1074/jbc.M305019200. [DOI] [PubMed] [Google Scholar]
- 28.Engel ME, McDonnell MA, Law BK, Moses HL. Interdependent SMAD and JNK Signaling in Transforming Growth Factor-beta -mediated Transcription. J Biol Chem. 1999;274:37413–37420. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- 29.Fu M, Zhang J, Zhu X, Myles DE, Willson TM, et al. Peroxisome Proliferator-activated Receptor gamma Inhibits Transforming Growth Factor beta -induced Connective Tissue Growth Factor Expression in Human Aortic Smooth Muscle Cells by Interfering with Smad3. J Biol Chem. 2001;276:45888–45894. doi: 10.1074/jbc.M105490200. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Feng X-H, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-[beta]-induced transcription. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 31.Heasley LE, Han S-Y. JNK Regulation of Oncogenesis. Molecules and Cells. 2006;21:167–173. [PubMed] [Google Scholar]
- 32.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and krüppel-like factor family of transcription factors in cell growth regulation and cancer. Journal of Cellular Physiology. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 33.Wagner EF. Functions of AP1 (Fos/Jun) in bone development. Annals of the Rheumatic Diseases. 2002;61:40ii–42. doi: 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: Sibling Rivals for Control of Cancer Cell Metabolism and Proliferation. Cancer Cell. 2007;12:108–113. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fahmy RG, Dass CR, Sun L-Q, Chesterman CN, Khachigian LM. Transcription factor Egr-1 supports FGF-dependent angiogenesis during neovascularization and tumor growth. Nature Medicine. 2003;9:1026–1032. doi: 10.1038/nm905. [DOI] [PubMed] [Google Scholar]
- 36.Johnson GL, Nakamura K. The c-jun kinase/stress-activated pathway: Regulation, function and role in human disease. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2007;1773:1341–1348. doi: 10.1016/j.bbamcr.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi J-H, Park S-W, Kapadia R, Vemuganti R. Role of transcription factors in mediating post-ischemic cerebral inflammation and brain damage. Neurochemistry International. 2007;50:1014–1027. doi: 10.1016/j.neuint.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, et al. KLF2 Is a Novel Transcriptional Regulator of Endothelial Proinflammatory Activation. J Exp Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore BB, Kolodsick JE, Thannickal VJ, Cooke K, Moore TA, et al. CCR2-Mediated Recruitment of Fibrocytes to the Alveolar Space after Fibrotic Injury. Am J Pathol. 2005;166:675–684. doi: 10.1016/S0002-9440(10)62289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vuolteenaho K, Moilanen T, Moilanen E. Non-Steroidal Anti-Inflammatory Drugs, Cyclooxygenase-2 and the Bone Healing Process. Basic & Clinical Pharmacology & Toxicology. 2008;102:10–14. doi: 10.1111/j.1742-7843.2007.00149.x. [DOI] [PubMed] [Google Scholar]
- 41.Stenson WF. Cyclooxygenase 2 and wound healing in the stomach. Gastroenterology. 1997;112:645–648. doi: 10.1053/gast.1997.v112.agast970645. [DOI] [PubMed] [Google Scholar]
- 42.Nian M, Lee P, Khaper N, Liu P. Inflammatory Cytokines and Postmyocardial Infarction Remodeling. Circulation Research. 2004;94:1543–1553. doi: 10.1161/01.RES.0000130526.20854.fa. [DOI] [PubMed] [Google Scholar]
- 43.Marshall AK, Barrett OP, Cullingford TE, Shanmugasundram A, Sugden PH, et al. ERK1/2 signaling dominates over RhoA signaling in regulating early changes in RNA expression induced by endothelin-1 in neonatal rat cardiomyocytes. PLoS One. 2010;5:e10027. doi: 10.1371/journal.pone.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarthy SA, Chen D, Yang BS, Garcia Ramirez JJ, Cherwinski H, et al. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol Cell Biol. 1997;17:2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JM, Adam RM, Peters CA, Guthrie PD, Sun Z, et al. AP-1 mediates stretch-induced expression of HB-EGF in bladder smooth muscle cells. Am J Physiol. 1999;277:C294–301. doi: 10.1152/ajpcell.1999.277.2.C294. [DOI] [PubMed] [Google Scholar]
- 46.Zhang X, Zhang J, Yang X, Han X. Several transcription factors regulate COX-2 gene expression in pancreatic beta-cells. Mol Biol Rep. 2007;34:199–206. doi: 10.1007/s11033-007-9085-3. [DOI] [PubMed] [Google Scholar]
- 47.Yeo SJ, Yoon JG, Yi AK. Myeloid differentiation factor 88-dependent post-transcriptional regulation of cyclooxygenase-2 expression by CpG DNA: tumor necrosis factor-alpha receptor-associated factor 6, a diverging point in the Toll-like receptor 9-signaling. J Biol Chem. 2003;278:40590–40600. doi: 10.1074/jbc.M306280200. [DOI] [PubMed] [Google Scholar]
- 48.Pendurthi UR, Ngyuen M, Andrade-Gordon P, Petersen LC, Rao LV. Plasmin induces Cyr61 gene expression in fibroblasts via protease-activated receptor-1 and p44/42 mitogen-activated protein kinase-dependent signaling pathway. Arterioscler Thromb Vasc Biol. 2002;22:1421–1426. doi: 10.1161/01.atv.0000030200.59331.3f. [DOI] [PubMed] [Google Scholar]
- 49.Han JS, Macarak E, Rosenbloom J, Chung KC, Chaqour B. Regulation of Cyr61/CCN1 gene expression through RhoA GTPase and p38MAPK signaling pathways. Eur J Biochem. 2003;270:3408–3421. doi: 10.1046/j.1432-1033.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- 50.Tien Hsu MT, Watson DennisK. Ets proteins in biological control and cancer. Journal of Cellular Biochemistry. 2004;91:896–903. doi: 10.1002/jcb.20012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trojanowska M. Ets factors and regulation of the extracellular matrix. Oncogene. 2000;19:6464–6471. doi: 10.1038/sj.onc.1204043. [DOI] [PubMed] [Google Scholar]
- 52.Calvet CM, Oliveira FO, Araujo-Jorge TC, Pereira MC. Regulation of extracellular matrix expression and distribution in Trypanosoma cruzi-infected cardiomyocytes. Int J Med Microbiol. 2009;299:301–312. doi: 10.1016/j.ijmm.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Alcorn JF, Guala AS, van der Velden J, McElhinney B, Irvin CG, et al. Jun N-terminal kinase 1 regulates epithelial-to-mesenchymal transition induced by TGF-beta1. J Cell Sci. 2008;121:1036–1045. doi: 10.1242/jcs.019455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saika S, Okada Y, Miyamoto T, Yamanaka O, Ohnishi Y, et al. Role of p38 MAP kinase in regulation of cell migration and proliferation in healing corneal epithelium. Invest Ophthalmol Vis Sci. 2004;45:100–109. doi: 10.1167/iovs.03-0700. [DOI] [PubMed] [Google Scholar]
- 55.Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 56.Padilla AM, Simpson LJ, Tarleton RL. Insufficient TLR activation contributes to the slow development of CD8+ T cell responses in Trypanosoma cruzi infection. J Immunol. 2009;183:1245–1252. doi: 10.4049/jimmunol.0901178. [DOI] [PubMed] [Google Scholar]
- 57.Van Overtvelt L, Vanderheyde N, Verhasselt V, Ismaili J, De Vos L, et al. Trypanosoma cruzi infects human dendritic cells and prevents their maturation: inhibition of cytokines, HLA-DR, and costimulatory molecules. Infect Immun. 1999;67:4033–4040. doi: 10.1128/iai.67.8.4033-4040.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Majumder S, Kierszenbaum F. Trypanosoma cruzi immunosuppressive factor decreases the interleukin-2 mRNA level in cultured normal activated human lymphocytes. Infect Immun. 1995;63:4546–4549. doi: 10.1128/iai.63.11.4546-4549.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kierszenbaum F, Sztein MB, Beltz LA. Decreased human IL-2 receptor expression due to a protozoan pathogen. Immunol Today. 1989;10:129–131. doi: 10.1016/0167-5699(89)90246-6. [DOI] [PubMed] [Google Scholar]
- 60.Kierszenbaum F, Sztein MB. Mechanisms underlying immunosuppression induced by Trypanosoma cruzi. Parasitol Today. 1990;6:261–264. doi: 10.1016/0169-4758(90)90187-9. [DOI] [PubMed] [Google Scholar]
- 61.Van Overtvelt L, Andrieu M, Verhasselt V, Connan F, Choppin J, et al. Trypanosoma cruzi down-regulates lipopolysaccharide-induced MHC class I on human dendritic cells and impairs antigen presentation to specific CD8(+) T lymphocytes. Int Immunol. 2002;14:1135–1144. doi: 10.1093/intimm/dxf077. [DOI] [PubMed] [Google Scholar]
- 62.Gomes NA, Previato JO, Zingales B, Mendonca-Previato L, DosReis GA. Down-regulation of T lymphocyte activation in vitro and in vivo induced by glycoinositolphospholipids from Trypanosoma cruzi. Assignment of the T cell-suppressive determinant to the ceramide domain. J Immunol. 1996;156:628–635. [PubMed] [Google Scholar]
- 63.Hansen DS, Villacres-Eriksson M, Akerblom L, Hellman U, Segura E, et al. An immunoaffinity-purified Trypanosoma cruzi antigen suppresses cellular proliferation through a TGF-beta-mediated mechanism. Scand J Immunol. 1998;47:509–516. doi: 10.1046/j.1365-3083.1998.00340.x. [DOI] [PubMed] [Google Scholar]
- 64.Goncalves MF, Umezawa ES, Katzin AM, de Souza W, Alves MJM, et al. Trypanosoma cruzi: Shedding of surface antigens as membrane vesicles. Experimental Parasitology. 1991;72:43–53. doi: 10.1016/0014-4894(91)90119-h. [DOI] [PubMed] [Google Scholar]
- 65.Marin-Neto JA, Cunha-Neto E, Maciel BC, Simoes MV. Pathogenesis of Chronic Chagas Heart Disease. Circulation. 2007;115:1109–1123. doi: 10.1161/CIRCULATIONAHA.106.624296. [DOI] [PubMed] [Google Scholar]
- 66.Weng L, Dai H, Zhan Y, He Y, Stepaniants SB, et al. Rosetta error model for gene expression analysis. Bioinformatics. 2006;22:1111–1121. doi: 10.1093/bioinformatics/btl045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Modulation of fibroblast gene expression by a secreted/released T. cruzi fraction. Human foreskin fibroblasts (HFF) were mock-treated or incubated with cell-free T. cruzi parasite-conditioned medium (PCM) for 3 hours and the isolated RNA was processed and used to hybridize HG_U133 plus 2.0 Affymetrix arrays. Ratios of the normalized log2 intensities for PCM vs medium were used to calculate the fold-change in HFF gene expression for each array and values from 3 biological replicates were averaged. HFF genes for which the transcript abundance changes ≥2-fold in either direction are included.
(XLSX)
Inhibition of TGF-ß-inducible gene expression by T. cruzi PCM. HFF were mock-treated or stimulated with 5 ng/ml TGF-ß1 for 3 hours in the presence or absence of T. cruzi PCM and cells were processed for microarray hybridization as described. Ratios of the normalized log2 intensities for TGF-ß vs medium and TGF-ß+PCM vs medium were used to calculate the fold-decrease in TGF-ß inducible gene expression that was observed in the presence of T. cruzi PCM. Only the non-redundant, annotated genes are shown with the exception of genes for which duplicates showed marked differences in the fold-change values.
(XLSX)
Canonical pathways enriched in the T. cruzi PCM-sensitive subset of TGF-ß inducible genes. TGF-ß-inducible genes sensitive to T. cruzi PCM (59 genes from Table 1) were analyzed using Ingenuity Pathway Analysis software. Canonical pathways were identified from the Ingenuity Pathways Analysis library of canonical pathways that were most significant to the data set and associated with a canonical pathway in Ingenuity's Knowledge Base. The significance of the association between the data set and the canonical pathway was measured in 2 ways: 1) A ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway is displayed. 2) Fisher's exact test was used to calculate a p-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone.
(XLS)
Canonical pathways enriched in the subset of TGF-ß inducible genes that is insensitive to T. cruzi PCM. TGF-ß-inducible genes that were insensitive to T. cruzi PCM (192 genes; Table S2) were analyzed using Ingenuity Pathway Analysis software and canonical pathways were identified from the Ingenuity Pathways Analysis library as described for Table S3.
(XLS)