Abstract
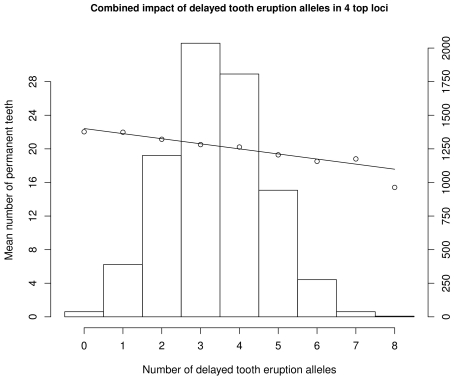
The sequence and timing of permanent tooth eruption is thought to be highly heritable and can have important implications for the risk of malocclusion, crowding, and periodontal disease. We conducted a genome-wide association study of number of permanent teeth erupted between age 6 and 14 years, analyzed as age-adjusted standard deviation score averaged over multiple time points, based on childhood records for 5,104 women from the Danish National Birth Cohort. Four loci showed association at P<5×10−8 and were replicated in four independent study groups from the United States and Denmark with a total of 3,762 individuals; all combined P-values were below 10−11. Two loci agreed with previous findings in primary tooth eruption and were also known to influence height and breast cancer, respectively. The two other loci pointed to genomic regions without any previous significant genome-wide association study results. The intronic SNP rs7924176 in ADK could be linked to gene expression in monocytes. The combined effect of the four genetic variants was most pronounced between age 10 and 12 years, where children with 6 to 8 delayed tooth eruption alleles had on average 3.5 (95% confidence interval: 2.9–4.1) fewer permanent teeth than children with 0 or 1 of these alleles.
Author Summary
While genome-wide association studies (GWAS) initially focused on the disease under investigation, additional findings in secondary traits have shown further benefits of having extensive phenotype data at hand. Using records from the nationwide dental registry for children and genotype data from two GWAS, we were able to identify four genomic loci associated with permanent tooth eruption in children. Two of the identified genomic regions had no previous GWAS findings, whereas two loci were reported in the context of primary dentition. A follow-up in an on-going GWAS showed that rs7924176 also plays a substantial role in primary dentition. During the age period of permanent tooth eruption many important developmental processes take place. Thus, we suggest following up the four reported SNPs in other growth-related traits to further elucidate the genetic background of maturation.
Introduction
Dental maturation is the process of exfoliation of primary teeth and eruption and calcification of permanent teeth that generally takes place between 6 and 13 years of age [1]. One commonly used measure of dental maturity is the number of permanent teeth erupted at a given age [2]. This is influenced by several factors, including gender [3]–[5], malnutrition [6], caries or trauma to the primary teeth [7], [8], ethnicity [9] and certain diseases [10]–[12]. In addition, many dental traits are known to be substantially influenced by genetic factors [13]–[15]. Although a recent study identified genetic variants for development of primary dentition in infancy [16], genetic factors influencing the eruption of permanent teeth have not been identified. To search for sequence variants associated with number of permanent teeth erupted, we carried out a GWAS in more than 5,100 women from the Danish National Birth Cohort (DNBC) [17], who had records in the nationwide dental registry for children (SCOR) and replicated the findings in more than 3,700 individuals from Denmark and the US (see Table S1 for description of study groups).
Results
We analyzed the association between the number of permanent teeth erupted and 521,741 SNPs in 5,104 women with dental records from their childhood, and identified four loci strictly fulfilling genome-wide significance (P<5×10−8, see Figures S1 and S2 for quantile-quantile and Manhattan plots). No other region showed a P-value<5×10−6. To confirm the observed associations we genotyped and tested the most significant SNP at each of the four loci in additional Danish samples, as well as tested for in silico replication in a US study of dental caries. In the 3,762 additional individuals all SNPs replicated with P<10−4 and the combined P-values were <10−11 (Table 1).
Table 1. Initial, replication, and combined results for number of permanent teeth erupted between age 6 and 14 years, analyzed as age-adjusted standard deviation scores averaged over multiple time points.
| SNP (effect/other allele) | rs12424086 C/T | rs4491709 T/C | rs2281845 T/C | rs7924176 G/A | ||||||||||||||||
| Study group | N | C freq. | Effect | SE | P-value | N | T freq. | Effect | SE | P-value | N | T freq. | Effect | SE | P-value | N | G freq. | Effect | SE | P-value |
| Initial stage | ||||||||||||||||||||
| DNBC I GWAS | 5099 | 0.177 | −0.118 | 0.020 | 1.11E-08 | 5088 | 0.707 | −0.109 | 0.017 | 6.51E-10 | 5097 | 0.381 | −0.105 | 0.016 | 1.31E-10 | 5100 | 0.426 | −0.094 | 0.016 | 6.58E-09 |
| Replication stage | ||||||||||||||||||||
| DNBC II | 2162 | 0.166 | −0.079 | 0.033 | 0.016 | 2171 | 0.717 | −0.087 | 0.026 | 9.70E-04 | 2172 | 0.382 | −0.099 | 0.025 | 5.43E-05 | 2172 | 0.420 | −0.119 | 0.024 | 5.18E-07 |
| DK Roskilde | 671 | 0.186 | −0.169 | 0.056 | 2.55E-03 | 690 | 0.698 | −0.105 | 0.045 | 0.019 | 691 | 0.400 | −0.165 | 0.042 | 1.09E-04 | 687 | 0.424 | −0.089 | 0.043 | 0.040 |
| USA | 668 | 0.202 | −0.130 | 0.067 | 0.051 | 669 | 0.732 | −0.106 | 0.060 | 0.078 | 669 | 0.392 | 0.065 | 0.056 | 0.242 | 668 | 0.449 | −0.160 | 0.051 | 1.80E-03 |
| DK Glostrup | 161 | 0.199 | −0.020 | 0.094 | 0.833 | 161 | 0.730 | −0.058 | 0.086 | 0.502 | 164 | 0.396 | −0.175 | 0.069 | 0.012 | 163 | 0.439 | −0.133 | 0.074 | 0.072 |
| Replication combined | 3662 | 0.177 | −0.100 | 0.025 | 6.29E-05 | 3691 | 0.715 | −0.091 | 0.021 | 9.09E-06 | 3696 | 0.388 | −0.099 | 0.019 | 2.06E-07 | 3690 | 0.426 | −0.120 | 0.019 | 1.25E-10 |
| All combined | 8761 | 0.177 | −0.111 | 0.016 | 2.30E-12 | 8779 | 0.711 | −0.102 | 0.013 | 2.16E-14 | 8793 | 0.384 | −0.103 | 0.012 | 8.03E-17 | 8790 | 0.426 | −0.105 | 0.012 | 5.64E-18 |
| Heterogeneity P-value | 0.528 | 0.947 | 0.015 | 0.678 | ||||||||||||||||
Effect is given as the change in mean standard deviation score, so all effect alleles result in lower numbers of permanent teeth erupted. Alleles refer to the forward strand.
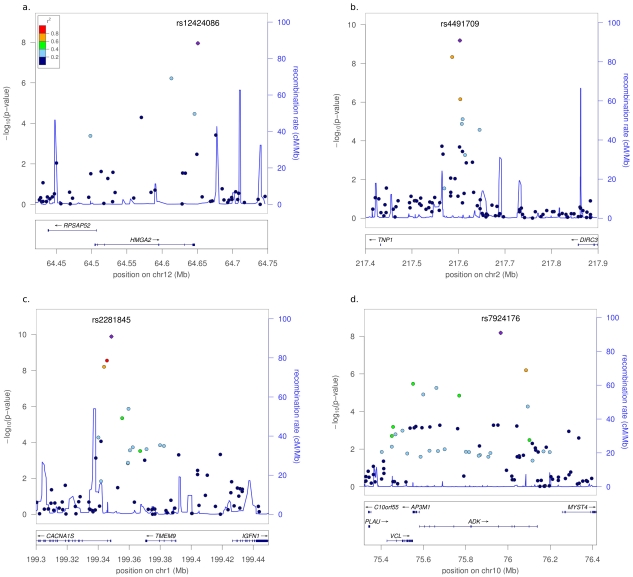
Interestingly, two of the variants have been previously reported to affect primary tooth development [16]. First, rs12424086 on chromosome 12q14.3 had a suggestive P-value (5×10−8<P<5×10−6) for number of primary teeth at age 1 (P = 3.64×10−6) [16]. This SNP is about 120 kb downstream of HMGA2 (see Figure 1 for genomic regions of the four associated SNPs) and is also in linkage disequilibrium (LD) with rs1042725 (r2 = 0.21 in HapMap Europeans, physical distance ∼6 kb), a SNP associated with adult and childhood height [18]. Second, rs4491709 on chromosome 2q35, is in LD with rs6435957 (r2 = 0.73 in the DNBC I study group, physical distance ∼17 kb), a SNP that showed suggestive evidence for an association with number of primary teeth at age 1 (P = 3.64×10−7) [16]. Furthermore, rs4491709 is in LD with rs13387042 (r2 = 0.34 in the DNBC I study group), which is associated with breast cancer [19]; the closest gene is TNP1 (160 kb telomeric). For the genomic regions around the two other SNPs there were no previous significant GWAS results. The third SNP, rs2281845, on chromosome 1q32.1 is just upstream of CACNA1S (voltage-dependent calcium channel, L type), a gene subject to mutation screens for malignant hyperthermia [20], [21] and periodic paralysis [22], [23]. The LD block containing rs2281845 extends to TMEM9 (transmembrane protein 9, physical distance 22 kb). The direction of the effect for rs2281845 in the US study group was - though not significant - opposite to the overall effect, which is most likely due to the relatively small sample size and greater variation in the phenotype (individual values are only based on one observation). The fourth SNP, rs7924176, on chromosome 10q22.2 is intronic in ADK (adenosine kinase), a gene that has been studied in the context of type 1 diabetes [24], and is located in a broader region showing linkage with Alzheimer's disease [25].
Figure 1. Plots of genetic regions associated with permanent tooth eruption.
The figure shows a) chromosome 12q14.3, b) chromosome 2q35, c) chromosome 1q32.1, and d) chromosome 10q22.2 (generated with LocusZoom [38]). Displayed are the recombination rates over the region and P-values for the analyzed SNPs, the colors indicate the r2 with the most significant SNP.
We also looked at all variants reportedly associated with tooth eruption in primary dentition (Table S2); among the 6 genome-wide significant SNPs, rs1956529 also showed a substantial effect on permanent tooth eruption, and three other loci had effects in the same direction with P-values between 0.01 and 0.17. The correlation between age at eruption of first tooth and permanent tooth eruption is evident (r = −0.408 in a subset of 1,442 children from the DNBC with questionnaire data on age at first tooth eruption and SCOR data on number of permanent teeth erupted; later ages at first tooth eruption going along with lower age-adjusted standard deviation scores (SDS) for number of permanent teeth erupted). We followed up all four SNPs associated with permanent tooth eruption in an on-going GWAS of primary dentition based on more than 6,000 individuals from the Avon Longitudinal Study of Parents and Children (ALSPAC) (Table S3 and Text S1). Rs7924176 also reached genome-wide significance in the analyses for number of primary teeth erupted at age 15 months and time to eruption of first tooth. The other three SNPs were at least nominally significant in the analysis of number of teeth erupted at age 15 months (P between 0.028 and 3.5×10−6), in all instances the same alleles associated with fewer teeth erupted in the respective dentition.
Given that both rs4491709 on chromosome 2q35 and rs1956529 on chromosome 14q24.1, which associated with tooth eruption in primary dentition [16], are close to breast cancer susceptibility loci, we checked all 19 SNPs associated with breast cancer in Caucasian study groups (P<5×10−8) [19], [26]–[32]. Only two of the other 18 loci were nominally significant (Table S4) without reaching a P<0.05/18. Thus, we could not find further evidence for a link to breast cancer.
Another trait of maturation, age at menarche, showed association with more than 30 SNPs in a recent GWA meta-analysis based on 87,802 women [33]. We checked the SNPs reported for age at menarche in our permanent tooth eruption GWAS (Table S5) and rs7821178 on chromosome 8q21.11 reached a P-value of 1×10−4, which is significant after adjustment for 42 SNPs tested. The allele increasing age at menarche also delayed tooth eruption, a pattern seen in all 6 SNPs with P<0.1 for permanent tooth eruption. This could be due to some genetic variants regulating maturation in a more general way, even though the correlation between age at menarche and permanent tooth eruption is modest (r = −0.095 in the combined DNBC I and DNBC II study groups, earlier age at menarche correlated with more permanent teeth erupted).
Results for the 180 SNPs recently reported to be associated with adult height [34] are given in Table S6. Apart from rs1351394, which is in LD with the identified SNP rs12424086 in the HMGA2 region, rs6473015 (P = 6.1×10−6) was also significant after adjusting for the number of SNPs tested for height. This SNP is on chromosome 8q21.11 within 90 kb of rs7821178 (r2 = 0.78), the age at menarche variant with low P-value for permanent tooth eruption. The correlation between permanent tooth eruption and adult height is modest (r = 0.074 in the combined DNBC I and DNBC II study groups, more permanent teeth erupted correlated with increased adult height) and there is no consistent direction among the height SNPs reaching nominal significance for permanent tooth eruption.
The correlation between age at menarche and adult height (r = 0.099, earlier age at menarche correlated with decreased adult height) is well known from the literature [35], but is opposite to what would be expected just based on the correlation results for permanent tooth eruption. Even though all correlations are modest, they underline that the interplay between these three growth and maturation traits is not straightforward.
To follow-up on the (potential) links to age at menarche, height and breast cancer, we contacted the latest published GWAS for these traits [26], [33], [34] and results for the reported four SNPs are displayed in Tables S7, S8, S9.
For age at menarche there is no indication that the four SNPs play a role (lowest P = 0.15) based on results from the meta-analysis with 87,802 women of European descent (Table S7).
For height, results based on 133,000 individuals showed the expected signal for rs12424086 in the HMGA2 region, and rs4491709 had a modest effect that reached nominal significance (P = 0.02, Table S8).
Currently, several GWAS on breast cancer have been conducted, with the latest having a combined 2,839 cases and 3,507 controls at the initial GWAS stage [26]. This group provided us with results for 1,693 cases vs. 5,588 controls (Table S9) and rs4491709 (which is in LD with the known breast cancer risk SNP rs6435957 (r2 = 0.73), reached nominal significance (P = 0.024), for the three other regions the lowest P was 0.096.
We investigated association with expression levels of nearby genes in an expression database for monocytes [36]. The four SNPs were not genotyped directly, but several SNPs in LD with rs2281845 showed significant association with expression of TMEM9, and the SNP with the strongest LD (rs6667912, r2 = 0.46 in HapMap Europeans) had a P-value of 6×10−29. Similarly, we observed association of rs7924176 with expression of ADK via rs1874152 (r2 = 0.71 in HapMap Europeans) at a P-value of 2×10−42.
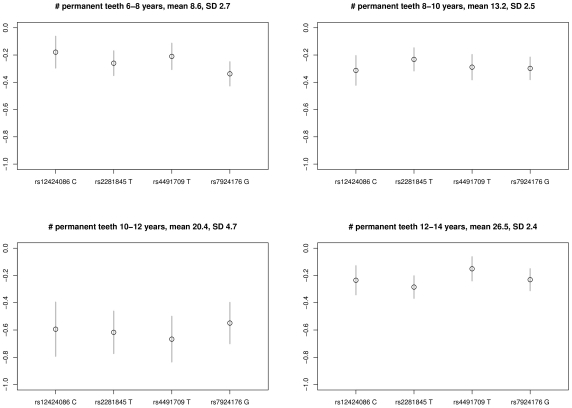
The women from the DNBC provided a relatively homogeneous group with similar mean number of individual observations. Therefore, we split the time period from age 6 to 14 years into four two-year periods to see the effect of the variants at different ages (Figure 2). All variants showed significant effects on the number of teeth erupted in all four age periods, including the age group 12 to 14 years in which the permanent dentition was nearly or fully erupted (mean number of erupted teeth is 26.5). The highest variation was observed in the age group 10 to 12 years (standard deviation (SD) 4.7 teeth), and went along with the strongest per allele effect estimates (−0.55 to −0.67 teeth). Considering the combined number of delayed tooth eruption alleles, the 4.8% of children with 6 to 8 delayed eruption alleles had on average 18.5 (SD 4.5) permanent teeth compared to 22.0 (SD 4.2) in the 6.3% of children with 0 or 1 risk allele (Figure 3). Effect estimates in the three other age categories ranged from −0.15 to −0.34 teeth per allele. The variance explained by the four variants combined ranged from 1.5% (age 12 to 14) to 3.0% (age 10 to 12).
Figure 2. Per allele effect of the four variants on number of permanent teeth erupted based on data for women from the DNBC I & II study groups at age 6–8 years (N = 5,865), 8–10 years (N = 6,548), 10–12 years (N = 6,919), and 12–14 years (N = 7,059).
Figure 3. Combined distribution of number of delayed permanent tooth eruption alleles and combined effects for all four identified SNPs at age 10–12 years (N = 6,919).
Permanent tooth eruption happens earlier in girls than in boys, so we checked for gender differences in the replication groups, but there was no indication of heterogeneous effects between sexes (results not shown). However, the low number of male individuals in the study means that we only have limited power to detect effect differences between sexes.
Discussion
We carried out the first GWAS for normal variation in the timing of permanent tooth eruption. Using longitudinal dental data between age 6 and 14 years, we identified four loci with robust associations. The main strengths of our study are: i) the comprehensive data set on dental exams during childhood, which allowed us to generate a mean SDS for the number of permanent teeth erupted, ii) the Danish replication groups with comprehensive phenotype data from the same database as the initial study, and iii) the substantial sample size of more than 3,700 individuals at the replication stage. The main limitations of the study are i) the lack of corresponding data on primary dentition, which is owed to the fact that there is usually no need to present children to the dentist in the time period where primary teeth erupt, ii) the lack of other growth and maturation traits, because such data are not collected in connection with visits to the dentist and are not readily available otherwise.
The connection of our study to primary tooth eruption is obvious with rs7924176 reaching genome-wide significance in the analysis of number of permanent teeth erupted at age 15 months in the ALSPAC data, and the three other loci being at least nominally significant. Looking at the 6 SNPs previously reported as genome-wide significant (P<5×10−8) for primary dentition, rs1956529 in the RAD51L1 region showed association with permanent dentition (p = 3.6×10−5) and three SNPs had P-values between 0.01 and 0.17, with the effect going in the same direction. Thus, maybe all these SNPs are relevant in both tooth eruption periods, just with substantial variation in the strength of association between periods. Children with data for both tooth eruption periods are necessary to see whether SNPs with strong effects on primary tooth eruption have an independent effect in permanent dentition. On the other hand, there was no association with permanent tooth eruption for the two correlated SNPs in the EDA region (both P>0.4 and effect in opposite direction), which means that there are also substantial differences between the genetic mechanisms driving primary and permanent tooth eruption.
The two identified SNPs previously reported in primary dentition are in LD with genome-wide significant SNPs in adult and childhood height (chromosome 12q14.3) and breast cancer (chromosome 2q35). Additional studies will be necessary to determine whether similar or different mechanisms explain the associations in these two regions.
We analyzed expression data for monocytes to investigate functional implications of the four SNPs. Rs2281845 and rs7924176 could be linked to expression of TMEM9 and ADK, respectively, with rs7924176 being intronic in ADK. However, the function of TMEM9 is unknown whereas ADK regulates the concentrations of extracellular adenosine and intracellular adenine nucleotides, providing no further insight into the potential role of these genes in tooth development.
Dental maturation was found to proceed rather independently of other forms of biological maturation in epidemiologic studies [1], [37] and our comprehensive comparisons with other GWAS findings for adult height and age at menarche showed only modest overlap between associated variants. Investigating permanent tooth eruption in 180 height loci and 42 age at menarche loci revealed three SNPs that were significant after adjustment for the number of variants tested for each trait, with the first signal being the height SNP in the previously discussed chromosome 12q14.3 region. The two other SNPs are correlated and located on chromosome 8q close to PXMP3 (peroxin 2), with GWAS findings for both age at menarche and adult height, suggesting that this region is of general importance in growth and maturation.
Age at menarche is currently the only pubertal trait with GWAS findings, leaving the genetic background of pubertal growth and maturation poorly understood. A focused investigation of the four SNPs in other puberty-related traits, e.g. skeletal maturation, Tanner score and growth spurt, could reveal whether these variants regulate maturation in a global way or are rather specific to dental maturation.
Methods
Study Groups
Basic characteristics of the study populations are provided as Table S1. For all study groups, all individuals with available information on permanent tooth eruption were analyzed, regardless of the phenotype they were recruited for.
DNBC I and II
The initial stage of the study (DNBC I, N = 5,104) was conducted in the context of the Danish National Birth Cohort, a project including mothers and their children from 101,042 pregnancies recruited in the years 1996–2002 [17]. Blood samples from mothers were obtained two times during pregnancy (around gestational weeks 8 and 24) and stored in a biobank at Statens Serum Institut as buffy coats. The genotype data were derived from two on-going GWAS of preterm birth [39] and obesity [40]. Additional 2,229 mothers (DNBC II) in the replication stage were recruited for studies of preeclampsia and psoriasis.
The study protocol was approved by the Danish Scientific Ethical Committee and the Danish Data Protection Agency for all subjects.
Denmark—Roskilde
The Danish study group from Roskilde (N = 695) included individuals drawn from the Danish Psychiatric Biobank, a national research platform that recruits individuals for gene-environment association studies [41]. DNA was obtained from whole blood. All participants have given written informed consent and the study protocol was approved by the Danish Scientific Ethical Committee and the Danish Data Protection Agency.
United States
The US replication sample (N = 669) included individuals recruited from the Center for Oral Health Research in Appalachia [42], [43] and the Iowa Fluoride Study [44]–[47], two initiatives designed to study oral health outcomes and included as part of the Gene Environment Association Studies consortium (GENEVA). DNA was obtained from blood, buccal swab, mouthwash, and saliva samples. All participants provided verbal assent with parental written consent, and all study protocols were approved by the pertinent Institutional Review Boards at the University of Pittsburgh, West Virginia University, and University of Iowa.
Denmark—Glostrup
The Danish study group from Glostrup (N = 169) included individuals recruited for a study of migraine and were selected from the Danish National Patient Register and from case files from neurological clinics [48]. All participants have given written informed consent and the study protocol was approved by the Danish Scientific Ethical Committee and the Danish Data Protection Agency.
Genotyping and Quality Control
Genotyping for the DNBC I and the US group was performed with Illumina (Illumina, SanDiego, CA, USA) Human 660W-Quad (preterm birth) and 610-Quad (obesity and US) bead chips. Single SNP genotyping of rs2281845, rs4491709, rs7924176 and rs12424086 for the Danish replication samples was carried out at deCODE genetics using the Centaurus (Nanogen, Bothell, WA, USA) platform.
The initial GWAS was based on SNPs that passed quality control on both chips, SNPs were excluded based on a missing rate >2%, deviation from Hardy-Weinberg equilibrium (P<10−4) or minor allele frequency <0.5%, individuals with more than 5% missing genotypes were also excluded. The genotypes for the four selected SNPs in the replication stage did not show deviation from Hardy-Weinberg equilibrium or missing rates >2%.
Phenotype
Dental data for all Danish individuals were retrieved from the nationwide dental registry for children, SCOR, which was established in 1972 alongside the initiation of free municipal dental services to Danish children and adolescents from birth to the age of 18 years [49]. Data were reported annually for all children until January 1st 1993, from which date reporting was only mandatory for 5-, 7-, 12-, and 15-year-old children [50].
All participants for the US sample underwent intraoral examinations by trained dental clinicians to collect phenotype data including number of erupted permanent teeth.
Statistical Analysis
The study combined all observations between age 6 and 14 years (starting with the 6th and stopping with the 14th birthday), the time period when eruption of permanent dentition usually occurs. For each visit to the dentist the total number of permanent teeth (excluding third molars) was recorded, and regressed on age. The resulting residuals were then standardized, and for each individual the mean residual across all available records was used as phenotype. This approach was motivated by a recent study, which demonstrated that averaging over multiple records increases the power in quantitative trait analysis [51]. For the DNBC I study group, we limited analyses to SNPs passing quality control on both chips and analyzed the two subgroups (preterm birth study/obesity study) together. Initial GWAS analysis of mean standardized residuals was carried out by applying the Wald test under an additive genetic model (also for chromosome X since all analyzed individuals are female) as implemented in PLINK [52]. We selected only SNPs with P<5×10−8 for replication, because SNPs in other regions did not show P-values<5×10−6. With the initial GWAS reaching genome-wide significance, the replication was basically of technical nature, aiming to confirm the results for the four SNPs with a second genotyping method. The criterion for overall significance remained P<5×10−8, but the additional study groups should not show great heterogeneity in terms of effect estimates and therefore an improvement of the combined P-values was expected.
P-values from the studies based on chip-typed individuals were corrected by applying genomic control (estimated genomic inflation factors for the DNBC I and US study groups were 1.05 and 1.01, respectively). Combined effects and P-values were calculated using the inverse variance method as implemented in METAL [53]. Though the number of permanent teeth was not normally distributed and the distribution of mean standardized residuals remained somewhat skewed, parametric tests were nevertheless carried out to determine the effect of the variants. However, to protect against false positive findings due to the phenotype distribution, we additionally carried out a non-parametric analysis of our top SNPs from the four significant loci using Kruskal-Wallis tests in R and calculated combined P-values based on weighted Z-scores (Table S10). Despite the reduced power of the non-parametric analysis (which also does not account for the trend observed over the genotype groups), all SNPs reached genome-wide significance (P<5×10−8).
Population Structure and Relatedness
To control for possible population substructure, we performed multidimensional scaling analysis (as implemented in PLINK [52]) on the discovery data, using independent autosomal SNPs with missing call rates <1%, minor allele frequency >5%, and Hardy-Weinberg P-value>0.05. We utilized PLINK's LD pruning function to remove short and long-range LD. The resulting 23,111 SNPs were analyzed along with founder genotypes from 11 HapMap phase III reference populations. As expected most discovery samples clustered near the Caucasian populations from Utah and Tuscany, but 61 individuals fell more than 4 standard deviations away from the discovery set mean on one or more of the first five dimensions and were excluded from further analysis.
For all Danish replication sets, we obtained birthplace information from the Danish Civil Registry [54], and only included individuals who were born in Scandinavia and whose parents were not born outside of Europe. Data from the Danish Family Relations Database were used to exclude all individuals who were first or second degree relatives to an individual in the initial set or another subject already in the replication set.
The samples from the Denmark Roskilde and Glostrup groups were additionally genotyped on an ancestry informative microsatellite panel to confirm self-reported Danish European ethnicity, and individuals with <90% European ancestry were removed.
In the US study group we performed multidimensional scaling analysis to account for possible population substructure, resulting in the exclusion of 45 individuals. Recruitment was partly family-based (mainly sib ships), and we decided to keep one individual per pedigree based on the examination being closest to 10.0 years (the midpoint of the investigated age interval), leading to the exclusion of 132 individuals.
Imputation for the DNBC I Study Group
In order to allow for comparisons with previous GWAS on related phenotypes, we separately imputed genotypes for the two GWAS included in the DNBC I study group applying MACH [55]. The imputed genotypes were analyzed separately and meta-analyzed with METAL [53].
Web Resources
ALSPAC http://www.bristol.ac.uk/alspac/;
British 1958 Birth Cohort: http://www.b58cgene.sgul.ac.uk/;
DNBC: http://dnbc.dk/;
eQTL database: http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/;
GENEVA: http://www.genevastudy.org;
GIANT meta-analysis: http://www.broadinstitute.org/collaboration/giant/index.php/Main_Page; HapMap: http://hapmap.ncbi.nlm.nih.gov/;
Supporting Information
Quantile-quantile plot for GWAS of permanent tooth eruption between age 6 and 14 years (analyzed as age-adjusted standard deviation scores averaged over multiple time points) in 5,104 women from the DNBC I study group.
(TIF)
Manhattan plot for GWAS of permanent tooth eruption between age 6 and 14 years (analyzed as age-adjusted standard deviation scores averaged over multiple time points) in 5,104 women from the DNBC I study group.
(TIF)
Descriptive statistics of study groups. a) Basic description of study groups, and b) detailed distribution of number of exams by study group.
(DOC)
Results for GWAS of permanent tooth eruption between age 6 and 14 years in 5,104 women from the DNBC for all variants previously reported for primary dentition [16].
(DOC)
Primary tooth eruption analysis for the identified SNPs in the ALSPAC data. Results are presented for a) number of primary teeth erupted at age 15 months based on 6,609 individuals and b) time to eruption of first tooth based on 5,998 individuals.
(DOC)
Results from GWAS of permanent tooth eruption between age 6 and 14 years in 5,104 women from the DNBC for 19 variants previously reported with P<5×10−8 for breast cancer in Caucasians [19], [26]–[32].
(DOC)
Results for GWAS of permanent tooth eruption between age 6 and 14 years in 5,104 women from the DNBC for all 42 variants previously reported for age at menarche [33].
(DOC)
Results from GWAS of permanent tooth eruption between age 6 and 14 years in 5,104 women from the DNBC for 180 variants previously reported for adult height [34].
(DOC)
Age at menarche results for the four identified SNPs based on the GWAS meta-analysis with 87,802 women.
(DOC)
Height results for the four identified SNPs based on the GWAS meta-analysis with 183,727 individuals.
(DOC)
Breast cancer results for the four identified SNPs based on the recent British Breast Cancer GWAS.
(DOC)
ALSPAC GWAS for primary dentition.
(DOC)
Acknowledgments
We are extremely grateful to all individuals and families who took part in the study, as well as all people involved in recruiting and managing the study groups, especially the huge datasets available from the DNBC and the SCOR. We thank Hreinn Stefansson and Patrick Sulem for organizing and supervising the replication genotyping at deCODE genetics. ALSPAC especially thanks the midwives for their help in recruiting and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. We thank the Sample Logistics and Genotyping Facilities at the Wellcome Trust Sanger Institute and also 23andME for generating the ALSPAC GWA data. We are also thankful for the look-up of the four reported SNPs a) by the British Breast Cancer study (results provided by Nicholas Orr); b) the age at menarche meta-analysis within the ReproGen Consortium (results provided by John Perry); c) the height meta-analysis within the GIANT consortium (results provided by Timothy Frayling). This publication is the work of the authors and they will serve as guarantors for the contents of this paper.
Footnotes
TW declares having received consultancy and lecture fees from Lundbeck A/S.
This study was supported in part by grants from the Lundbeck Foundation (R19-A2059) and the Danish Medical Research Council (09-065592). Funding support for the Danish National Birth Cohort (DNBC) was provided by the Danish National Research Foundation, the Danish Pharmacists' Fund, the Egmont Foundation, the March of Dimes Birth Defects Foundation, the Augustinus Foundation, and the Health Fund of the Danish Health Insurance Societies. USA replication data were provided by the Center for Oral Health Research in Appalachia (a collaboration of the University of Pittsburgh and West Virginia University funded by NIDCR R01-DE 014899, PI MLM); the Iowa Fluoride Study and the Iowa Bone Development Study, funded by NIDCR R01-DE09551and R01-DE12101, respectively, PI SL). GWAS data for the DNBC obesity study (Genomics of Obesity in Young Adults) were funded by the Wellcome Trust (WT 084762MA). GWAS data for the preterm birth project DNBC samples and for the US dental caries project were generated within the GENEVA consortium with funding provided through the NIH Genes, Environment, and Health Initiative (GEI, preterm birth: U01HG004423; dental caries: U01DE018903). Assistance with genotype cleaning and general study coordination was provided by the GENEVA Coordinating Center (U01HG004446). Genotyping was performed at Johns Hopkins University Center for Inherited Disease Research, with support from the NIH GEI (U01HG004438) for the preterm birth project and from NIDCR for the dental caries project through the NIH contract to CIDR, contract number HHSN268200782096C. Collection of the phenotypic data for the dental caries study was funded by R01DE014899, R01DE009551, and R01DE012101. The DNBC preterm birth and US dental caries datasets are available from dbGaP. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental and Craniofacial Research, the National Human Genome Research Institute, or the National Institutes of Health. The Danish samples from Roskilde were sponsored by grants from the Lundbeck Foundation (R34-A3243), the Danish National Advanced Technology Foundation (001-2009-2), the Danish Council for Independent Research in Medical Sciences, and the Danish Psychiatric Research Foundation. The UK Medical Research Council, the Wellcome Trust, and the University of Bristol provide core support for ALSPAC. GF is funded by a Wellcome Trust four-year PhD studentship in molecular, genetic, and lifecourse epidemiology [WT083431MA]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Beunen GP, Rogol AD, Malina RM. Indicators of biological maturation and secular changes in biological maturation. Food Nutr Bull. 2006;27:S244–S256. doi: 10.1177/15648265060274S508. [DOI] [PubMed] [Google Scholar]
- 2.Hagg U, Taranger J. Dental development, dental age and tooth counts. Angle Orthod. 1985;55:93–107. doi: 10.1043/0003-3219(1985)055<0093:DDDAAT>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Helm S, Seidler B. Timing of permanent tooth emergence in Danish children. Community Dent Oral Epidemiol. 1974;2:122–129. doi: 10.1111/j.1600-0528.1974.tb01669.x. [DOI] [PubMed] [Google Scholar]
- 4.Leroy R, Bogaerts K, Lesaffre E, Declerck D. The emergence of permanent teeth in Flemish children. Community Dent Oral Epidemiol. 2003;31:30–39. doi: 10.1034/j.1600-0528.2003.00023.x. [DOI] [PubMed] [Google Scholar]
- 5.Eskeli R, Laine-Alava MT, Hausen H, Pahkala R. Standards for permanent tooth emergence in Finnish children. Angle Orthod. 1999;69:529–533. doi: 10.1043/0003-3219(1999)069<0529:SFPTEI>2.3.CO;2. [DOI] [PubMed] [Google Scholar]
- 6.Psoter W, Gebrian B, Prophete S, Reid B, Katz R. Effect of early childhood malnutrition on tooth eruption in Haitian adolescents. Community Dent Oral Epidemiol. 2008;36:179–189. doi: 10.1111/j.1600-0528.2007.00386.x. [DOI] [PubMed] [Google Scholar]
- 7.Leroy R, Bogaerts K, Lesaffre E, Declerck D. Impact of caries experience in the deciduous molars on the emergence of the successors. Eur J Oral Sci. 2003;111:106–110. doi: 10.1034/j.1600-0722.2003.00011.x. [DOI] [PubMed] [Google Scholar]
- 8.Korf SR. The eruption of permanent central incisors following premature loss of their antecedents. ASDC J Dent Child. 1965;32:39–44. [PubMed] [Google Scholar]
- 9.Chaillet N, Nystrom M, Demirjian A. Comparison of dental maturity in children of different ethnic origins: international maturity curves for clinicians. J Forensic Sci. 2005;50:1164–1174. [PubMed] [Google Scholar]
- 10.Lal S, Cheng B, Kaplan S, Softness B, Greenberg E, et al. Accelerated tooth eruption in children with diabetes mellitus. Pediatrics. 2008;121:e1139–e1143. doi: 10.1542/peds.2007-1486. [DOI] [PubMed] [Google Scholar]
- 11.Lehtinen A, Oksa T, Helenius H, Ronning O. Advanced dental maturity in children with juvenile rheumatoid arthritis. Eur J Oral Sci. 2000;108:184–188. doi: 10.1034/j.1600-0722.2000.108003184.x. [DOI] [PubMed] [Google Scholar]
- 12.Suri L, Gagari E, Vastardis H. Delayed tooth eruption: pathogenesis, diagnosis, and treatment. A literature review. Am J Orthod Dentofacial Orthop. 2004;126:432–445. doi: 10.1016/j.ajodo.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 13.Townsend G, Hughes T, Luciano M, Bockmann M, Brook A. Genetic and environmental influences on human dental variation: a critical evaluation of studies involving twins. Arch Oral Biol. 2009;54(Suppl 1):S45–S51. doi: 10.1016/j.archoralbio.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelsmaekers B, Loos R, Carels C, Derom C, Vlietinck R. The genetic contribution to dental maturation. J Dent Res. 1997;76:1337–1340. doi: 10.1177/00220345970760070201. [DOI] [PubMed] [Google Scholar]
- 15.Hughes TE, Bockmann MR, Seow K, Gotjamanos T, Gully N, et al. Strong genetic control of emergence of human primary incisors. J Dent Res. 2007;86:1160–1165. doi: 10.1177/154405910708601204. [DOI] [PubMed] [Google Scholar]
- 16.Pillas D, Hoggart CJ, Evans DM, O'Reilly PF, Sipila K, et al. Genome-wide association study reveals multiple loci associated with primary tooth development during infancy. PLoS Genet. 2010;6:e1000856. doi: 10.1371/journal.pgen.1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen J, Melbye M, Olsen SF, Sorensen TI, Aaby P, et al. The Danish National Birth Cohort–its background, structure and aim. Scand J Public Health. 2001;29:300–307. doi: 10.1177/14034948010290040201. [DOI] [PubMed] [Google Scholar]
- 18.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 20.Carpenter D, Ringrose C, Leo V, Morris A, Robinson RL, et al. The role of CACNA1S in predisposition to malignant hyperthermia. BMC Med Genet. 2009;10:104. doi: 10.1186/1471-2350-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levano S, Vukcevic M, Singer M, Matter A, Treves S, et al. Increasing the number of diagnostic mutations in malignant hyperthermia. Hum Mutat. 2009;30:590–598. doi: 10.1002/humu.20878. [DOI] [PubMed] [Google Scholar]
- 22.Lin SH, Hsu YD, Cheng NL, Kao MC. Skeletal muscle dihydropyridine-sensitive calcium channel (CACNA1S) gene mutations in chinese patients with hypokalemic periodic paralysis. Am J Med Sci. 2005;329:66–70. doi: 10.1097/00000441-200502000-00003. [DOI] [PubMed] [Google Scholar]
- 23.Ng WY, Lui KF, Thai AC, Cheah JS. Absence of ion channels CACN1AS and SCN4A mutations in thyrotoxic hypokalemic periodic paralysis. Thyroid. 2004;14:187–190. doi: 10.1089/105072504773297858. [DOI] [PubMed] [Google Scholar]
- 24.Lucarelli P, Scacchi R, Corbo RM, Palmarino R, Orsini M, et al. Genetic polymorphisms in juvenile-onset diabetes. Hum Hered. 1978;28:89–95. doi: 10.1159/000152948. [DOI] [PubMed] [Google Scholar]
- 25.Grupe A, Li Y, Rowland C, Nowotny P, Hinrichs AL, et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet. 2006;78:78–88. doi: 10.1086/498851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fletcher O, Johnson N, Orr N, Hosking FJ, Gibson LJ, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103:425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Humphreys K, Heikkinen T, Aittomaki K, Blomqvist C, et al. A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat. 2011;126:717–727. doi: 10.1007/s10549-010-1172-9. [DOI] [PubMed] [Google Scholar]
- 28.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elks CE, Perry JR, Sulem P, Chasman DI, Franceschini N, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onland-Moret NC, Peeters PH, van Gils CH, Clavel-Chapelon F, Key T, et al. Age at menarche in relation to adult height: the EPIC study. Am J Epidemiol. 2005;162:623–632. doi: 10.1093/aje/kwi260. [DOI] [PubMed] [Google Scholar]
- 36.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, et al. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demirjian A, Buschang PH, Tanguay R, Patterson DK. Interrelationships among measures of somatic, skeletal, dental, and sexual maturity. Am J Orthod. 1985;88:433–438. doi: 10.1016/0002-9416(85)90070-3. [DOI] [PubMed] [Google Scholar]
- 38.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornelis MC, Agrawal A, Cole JW, Hansel NN, Barnes KC, et al. The Gene, Environment Association Studies consortium (GENEVA): maximizing the knowledge obtained from GWAS by collaboration across studies of multiple conditions. Genet Epidemiol. 2010;34:364–372. doi: 10.1002/gepi.20492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nohr EA, Timpson NJ, Andersen CS, Davey SG, Olsen J, et al. Severe obesity in young women and reproductive health: the Danish National Birth Cohort. PLoS One. 2009;4:e8444. doi: 10.1371/journal.pone.0008444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen T, Olsen L, Lindow M, Jakobsen KD, Ullum H, et al. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS One. 2007;2:e873. doi: 10.1371/journal.pone.0000873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X, Shaffer JR, Weyant RJ, Cuenco KT, DeSensi RS, et al. Genes and their effects on dental caries may differ between primary and permanent dentitions. Caries Res. 2010;44:277–284. doi: 10.1159/000314676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polk DE, Weyant RJ, Crout RJ, McNeil DW, Tarter RE, et al. Study protocol of the Center for Oral Health Research in Appalachia (COHRA) etiology study. BMC Oral Health. 2008;8:18. doi: 10.1186/1472-6831-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franzman MR, Levy SM, Warren JJ, Broffitt B. Tooth-brushing and dentifrice use among children ages 6 to 60 months. Pediatr Dent. 2004;26:87–92. [PubMed] [Google Scholar]
- 45.Marshall TA, Levy SM, Broffitt B, Warren JJ, Eichenberger-Gilmore JM, et al. Dental caries and beverage consumption in young children. Pediatrics. 2003;112:e184–e191. doi: 10.1542/peds.112.3.e184. [DOI] [PubMed] [Google Scholar]
- 46.Levy SM, Warren JJ, Broffitt B. Patterns of fluoride intake from 36 to 72 months of age. J Public Health Dent. 2003;63:211–220. doi: 10.1111/j.1752-7325.2003.tb03502.x. [DOI] [PubMed] [Google Scholar]
- 47.Warren JJ, Levy SM, Kanellis MJ. Dental caries in the primary dentition: assessing prevalence of cavitated and noncavitated lesions. J Public Health Dent. 2002;62:109–114. doi: 10.1111/j.1752-7325.2002.tb03430.x. [DOI] [PubMed] [Google Scholar]
- 48.Anttila V, Stefansson H, Kallela M, Todt U, Terwindt GM, et al. Genome-wide association study of migraine implicates a common susceptibility variant on 8q22.1. Nat Genet. 2010;42:869–873. doi: 10.1038/ng.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helm S. Recording system for the Danish Child Dental Health Services. Community Dent Oral Epidemiol. 1973;1:3–8. doi: 10.1111/j.1600-0528.1973.tb01055.x. [DOI] [PubMed] [Google Scholar]
- 50.Hansen I, Foldspang A, Poulsen S. Use of a national database for strategic management of municipal oral health services for Danish children and adolescents. Community Dent Oral Epidemiol. 2001;29:92–98. doi: 10.1034/j.1600-0528.2001.290203.x. [DOI] [PubMed] [Google Scholar]
- 51.Rasmussen-Torvik LJ, Alonso A, Li M, Kao W, Kottgen A, et al. Impact of repeated measures and sample selection on genome-wide association studies of fasting glucose. Genet Epidemiol. 2010;34:665–673. doi: 10.1002/gepi.20525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedersen CB, Gotzsche H, Moller JO, Mortensen PB. The Danish Civil Registration System. A cohort of eight million persons. Dan Med Bull. 2006;53:441–449. [PubMed] [Google Scholar]
- 55.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2006;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantile-quantile plot for GWAS of permanent tooth eruption between age 6 and 14 years (analyzed as age-adjusted standard deviation scores averaged over multiple time points) in 5,104 women from the DNBC I study group.
(TIF)
Manhattan plot for GWAS of permanent tooth eruption between age 6 and 14 years (analyzed as age-adjusted standard deviation scores averaged over multiple time points) in 5,104 women from the DNBC I study group.
(TIF)
Descriptive statistics of study groups. a) Basic description of study groups, and b) detailed distribution of number of exams by study group.
(DOC)
Results for GWAS of permanent tooth eruption between age 6 and 14 years in 5,104 women from the DNBC for all variants previously reported for primary dentition [16].
(DOC)
Primary tooth eruption analysis for the identified SNPs in the ALSPAC data. Results are presented for a) number of primary teeth erupted at age 15 months based on 6,609 individuals and b) time to eruption of first tooth based on 5,998 individuals.
(DOC)
Results from GWAS of permanent tooth eruption between age 6 and 14 years in 5,104 women from the DNBC for 19 variants previously reported with P<5×10−8 for breast cancer in Caucasians [19], [26]–[32].
(DOC)
Results for GWAS of permanent tooth eruption between age 6 and 14 years in 5,104 women from the DNBC for all 42 variants previously reported for age at menarche [33].
(DOC)
Results from GWAS of permanent tooth eruption between age 6 and 14 years in 5,104 women from the DNBC for 180 variants previously reported for adult height [34].
(DOC)
Age at menarche results for the four identified SNPs based on the GWAS meta-analysis with 87,802 women.
(DOC)
Height results for the four identified SNPs based on the GWAS meta-analysis with 183,727 individuals.
(DOC)
Breast cancer results for the four identified SNPs based on the recent British Breast Cancer GWAS.
(DOC)
ALSPAC GWAS for primary dentition.
(DOC)