Abstract
The propagation of embryonic stem (ES) cells in an undifferentiated pluripotent state is dependent on leukemia inhibitory factor (LIF) or related cytokines. These factors act through receptor complexes containing the signal transducer gp130. The downstream mechanisms that lead to ES cell self-renewal have not been delineated, however. In this study, chimeric receptors were introduced into ES cells. Biochemical and functional studies of transfected cells demonstrated a requirement for engagement and activation of the latent trancription factor STAT3. Detailed mutational analyses unexpectedly revealed that the four STAT3 docking sites in gp130 are not functionally equivalent. The role of STAT3 was then investigated using the dominant interfering mutant, STAT3F. ES cells that expressed this molecule constitutively could not be isolated. An episomal supertransfection strategy was therefore used to enable the consequences of STAT3F expression to be examined. In addition, an inducible STAT3F transgene was generated. In both cases, expression of STAT3F in ES cells growing in the presence of LIF specifically abrogated self-renewal and promoted differentiation. These complementary approaches establish that STAT3 plays a central role in the maintenance of the pluripotential stem cell phenotype. This contrasts with the involvement of STAT3 in the induction of differentiation in somatic cell types. Cell type-specific interpretation of STAT3 activation thus appears to be pivotal to the diverse developmental effects of the LIF family of cytokines. Identification of STAT3 as a key transcriptional determinant of ES cell self-renewal represents a first step in the molecular characterization of pluripotency.
Keywords: Leukemia inhibitory factor (LIF), cytokine receptor, signaling, ES cells, tetracycline, episome
Embryonic stem (ES) cells are pluripotent cell lines derived by culture of preimplantation mouse embryos (Evans and Kaufman 1981; Martin 1981; Brook and Gardner 1997). At present, ES cells are the only nontransformed mammalian stem cells that can be continuously propagated in vitro. ES cell self-renewal is sustained by the cytokine leukemia inhibitory factor (LIF) (Smith and Hooper 1987; Smith et al. 1988; Williams et al. 1988). The effect of LIF is to inhibit differentiation and support proliferation of undifferentiated stem cells. However, the mechanisms underlying the maintenance of pluripotency during proliferative expansion remain elusive. We are attempting to define those signaling processes downstream of the LIF receptor complex that direct ES cell self-renewal. Elucidation of these principles will provide a molecular model for stem cell regulation in mammals. Insights provided by such a model should also be directly applicable to the extension of ES cell technology to nonmouse species.
The actions of LIF are mediated via heterodimerization of two members of the class I cytokine receptors, the low-affinity LIF receptor (LIF-R) and gp130 (Gearing et al. 1991; Gearing and Bruce 1992; Davis et al. 1993). The LIF-related cytokines, oncostatin M (OSM), cardiotrophin (CT-1), and ciliary neurotrophic factor (CNTF), act through the same receptor complex (in the case of CNTF, additionally including the CNTF-Rα subunit) and can similarly sustain ES cell self-renewal (Conover et al. 1993; Rose et al. 1994; Wolf et al. 1994; Yoshida et al. 1994; Pennica et al. 1995b). Furthermore, ES cells can also be derived and maintained using a combination of interleukin-6 and soluble interleukin-6 receptor (IL-6/sIL-6R) (Nichols et al. 1994; Yoshida et al. 1994). In this case, signaling is initiated via formation of gp130 homodimers without involvement of LIF-R (Murakami et al. 1993; Yoshida et al. 1994). Signals that emanate from gp130 are therefore sufficient for self-renewal.
gp130 mediates cellular responses to IL-6 and IL-11 in addition to the LIF-related cytokines (Kishimoto et al. 1994). All of these factors exert pleiotropic effects on diverse cell types in vitro and in vivo. In addition to ES cell self-renewal, stimulation of gp130 receptor complexes causes differentiation and growth inhibition in M1 myeloid leukemic cells (Tomida et al. 1984), induction of acute phase gene expression in hepatocytes (Baumann and Wong 1989), cholinergic differentiation of sympathetic neurons (Yamamori et al. 1989), survival of motor neurons (Li et al. 1995), proliferative and hypertrophic responses in cardiomyocytes (Hirota et al. 1995; Pennica et al. 1995a; Yoshida et al. 1996), and astrocyte differentiation of neuroepithelial progenitors (Bonni et al. 1997; Koblar et al. 1998).
Signaling processes downstream of gp130 are complex and are not yet fully characterized. Ligand-induced dimerization of the receptors (Davis et al. 1993; Murakami et al. 1993) leads to phosphorylation and activation of associated JAK tyrosine kinases (Narazaki et al. 1994; Stahl et al. 1994). The cytoplasmic domain of gp130 contains several tyrosine residues that are phosphorylated by the activated JAKs. These phosphotyrosine residues then interact with SH2 domain containing proteins that in turn themselves become targets for JAKs and possibly other nonreceptor tyrosine kinases. Consequences include activation of the Ras mitogen-activated protein (MAP) kinase (ERK) signaling cascade (Boulton et al. 1994; Yin and Yang 1994; Sheng et al. 1997) and of the STAT factors STAT1 and STAT3 (Lutticken et al. 1994; Stahl et al. 1995). STAT proteins are latent transcription factors that upon phosphorylation, dimerize and translocate to the nucleus where they activate target gene transcription (for review, see Ihle 1996). In myeloid leukemic M1 cells, activation of STAT3 appears to be the main effector of the differentiation response to IL-6 or LIF (Minami et al. 1996; Nakajima et al. 1996). STAT3 activation has also been adduced to mediate CNTF or LIF-induced differentiation of neuroepithelial precursors into astrocytes (Bonni et al. 1997).
In this study we have examined the receptor requirements for self-renewal signaling in ES cells and determined a critical contribution of STAT3 activation. In contrast to its role in somatic cells, activated STAT3 acts to suppress differentiation in ES cells.
Results
Granulocyte colony-stimulating factor receptor can signal ES cell self-renewal
Granulocyte colony-stimulating factor receptor (G-CSF-R) is a class I cytokine receptor that is evolutionarily related to gp130 and LIF-R (Gearing et al. 1991; Chambers et al. 1997). G-CSF-R is not present in ES cells. To begin delineating the signaling requirements for ES cell propagation, the capacity of these related receptors to sustain self-renewal was compared directly.
G-CSF-R undergoes ligand-induced homodimerization to produce an active signaling complex. G-CSF responsiveness can therefore be conferred on cytoplasmic domains of heterologous receptors through construction of appropriate fusions. cDNAs encoding full-length G-CSF-R cDNA and fusions between the extracellular portion of G-CSF-R and the transmembrane and cytoplasmic region of gp130 or LIF-R were cloned into the expression vector pPCAGIZ. Plasmids were introduced into LIF-R-deficient ES cells to eliminate the contribution of autocrine LIF signaling (Rathjen et al. 1990) from subsequent analyses. In this and all other experiments, ES cells were grown without feeder layers (Smith 1991). Transfectants were selected and expanded in the presence of IL-6/sIL-6R, acting through endogenous gp130, to avoid any selective pressure for adaptation to the introduced receptor.
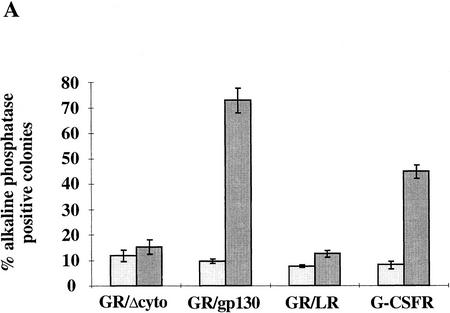
Stable transfectants were then plated at clonal density in the absence of cytokine or presence of IL-6/sIL-6R or G-CSF. The number of stem cell colonies generated was scored after 6 days. The data in Figure 1A show that the G-CSF-R/gp130 chimeric receptor sustained stem cell propagation in response to G-CSF. This result is anticipated from previous findings on the capacity of gp130 homodimers to signal self-renewal (Yoshida et al. 1994). The G-CSF-R/LIF-R chimera did not support formation of stem cell colonies despite higher levels of cell surface expression measured by radioligand binding (not shown). This is in line with previous reports that homodimerization of the LIF-R cytoplasmic domain results in quantitatively (Baumann et al. 1994a; Stahl et al. 1995) and qualitatively (Stahl et al. 1995) diminished activation of downstream pathways compared with LIF-R/gp130 heterodimerization or gp130 homodimerization. However, ES cells transfected with G-CSF-R did form stem cell colonies in response to G-CSF-R though with lower efficiency than cells expressing the G-CSF-R/gp130 chimera. This somewhat surprising finding corroborates similar data reported recently (Starr et al. 1997). Propagation of the G-CSF-R transfectants remained factor dependent, and the cells differentiated normally when deprived of cytokine.
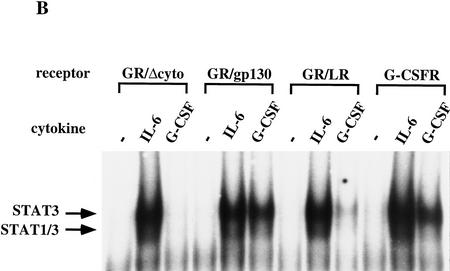
Figure 1.
ES cell self-renewal and induction of STAT DNA-binding activity mediated by G-CSF-R wild-type, truncated, and chimeric cytokine receptors. (A) Efficiency of clonal stem cell renewal in response to G-CSF measured by formation of alkaline phosphatase-positive colonies. (Light gray bars) −G-CSF; (dark gray bars) +G-CSF. Data are mean ± s.e.m. of triplicate determinations on single representative clones normalized to response to IL-6/sIL-6R. (B) Induction of STAT DNA binding by IL-6/sIL-6R and G-CSF determined by electophoretic mobility-shift assay. Cells were untreated or stimulated for 30 min with IL-6/sIL-6R or G-CSF (30 ng/ml). Nuclear extracts were prepared and assayed for SIE binding. Note the absence of detectable STAT1/STAT3 heterodimer complex on stimulation of full-length G-CSF-R.
The finding that G-CSF-R is competent to maintain the stem cell phenotype suggests that the signaling interactions essential for ES cell self-renewal are preserved between gp130 and G-CSF-R. Conserved features in the intracellular domains of these two receptors are not readily identifiable because of extensive sequence divergence. However, G-CSF-R contains a putative STAT binding site and is thought to signal primarily through activation of STAT3 (Shimozaki et al. 1997). Electrophoretic mobility-shift assays were performed to determine the induction of nuclear STAT DNA-binding activity by G-CSF in the various ES cell transfectants. Significant STAT3 activation was evident in ES cells transfected with expression vectors for the G-CSF-R/gp130 chimera or the full-length G-CSF-R. In contrast, ES cells expressing the G-CSF-R/LIF-R chimera showed only weak induction of STAT3 DNA-binding activity in response to G-CSF (Fig. 1B). Antibody supershift experiments (not shown) confirmed that the DNA-binding complex consisted predominantly of STAT3 homodimers with a minor component of STAT3/STAT1 heterodimer as described previously in ES cells and other systems (Hocke et al. 1995; Stahl et al. 1995; Starr et al. 1997). These observations pointed to a potentially critical role for STAT3 activation in mediation of the self-renewal signal.
STAT3 docking sites on gp130 are required to signal ES cell self-renewal
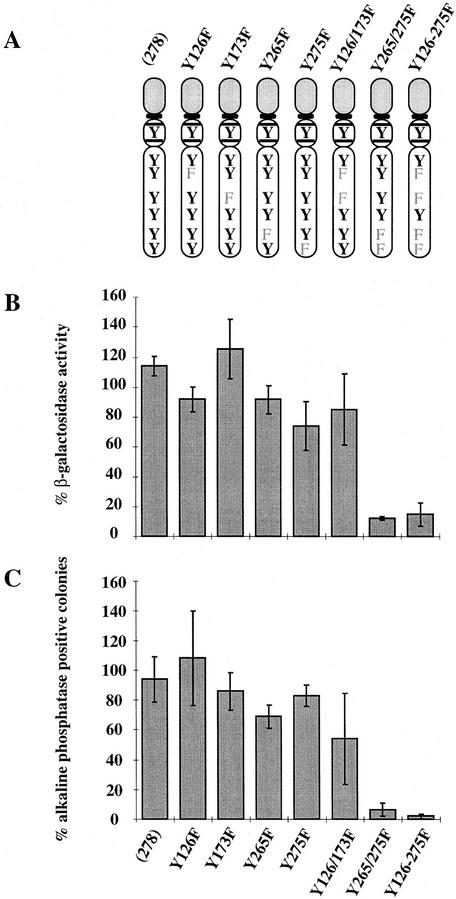
The cytoplasmic domain of mouse gp130 contains seven tyrosine residues. Four of these have been identified as phosphorylation-dependent sites of interaction with STAT3 (Stahl et al. 1995). Substitution of these tyrosine residues with phenylalanine in the context of the G-CSF-R/gp130 chimera was therefore used to determine their significance for self-renewal signaling. The modified chimeric receptor expression constructs were introduced into DO27 ES cells. These cells are LIF-deficient because of targeted deletion of both gene copies and, in addition, carry a β-galactosidase reporter integrated into one allele of the Oct-4 gene (C. Dani, I. Chambers, S. Johnstone, M. Robertson, B. Ebrahimi-Chahardahcherik, M. Saito, T. Taga, M. Li, T. Burdon, J. Nichols, and A.G. Smith, in prep.). This reporter is expressed only in undifferentiated ES cells (Mountford et al. 1994). Self-renewal was assayed both by measuring β-galactosidase activity in medium density cultures (Fig. 2B) and by scoring formation of alkaline phosphatase positive colonies at clonal density (Fig. 2C). Three independent transfectant clones were analyzed for each receptor. The data summarized in Figure 2 demonstrate that the presence of STAT3 docking sites is essential for stem cell propagation.
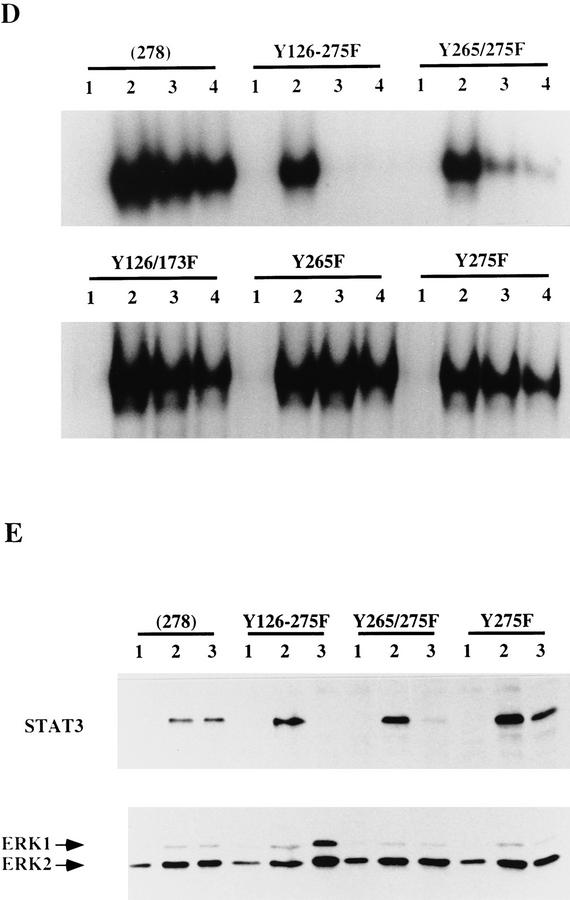
Figure 2.
Effect of mutating STAT3 interaction sites in gp130 on ES cell self-renewal and induction of STAT3 DNA-binding activity. (A) Schematic of the various chimeric receptors indicating the tyrosine–phenylalanine substitutions introduced into the wild-type (278) gp130 cytoplasmic domain. Numbering commences with the first residue of the 278-amino-acid intracellular domain of mouse gp130. The phenylalanine (F) for tyrosine (Y) substitutions in the four STAT3 docking sites are indicated. The additional three tyrosines do not interact with STAT3 (Stahl et al. 1995). (B) Stem cell renewal mediated by chimeric receptors in response to G-CSF measured by β-galactosidase expression from the Oct-4 locus. Data are mean ± s.e.m. for duplicate determinations on three independent clones normalized relative to response to IL-6/sIL-6R. (C) Efficiency of clonal stem cell renewal mediated by chimeric receptors in response to G-CSF measured by formation of alkaline phosphatase positive colonies. Data are mean ± s.e.m. for duplicate assays on three independent clones normalized relative to response to IL-6/sIL-6R. (D) Electrophoretic mobility-shift assay of induced STAT3 DNA binding. Transfected clones were left untreated (lane 1) or stimulated for 30 min with IL-6/sIL-6R (lane 2) or with G-CSF at 30 ng/ml (lane 3) or 3 ng/ml (lane 4). Nuclear extracts were assayed for SIE binding. (E) Immunoblot of STAT3 and ERK phosphorylation induced by G-CSF stimulation of chimeric receptors. Transfected clones were left untreated (lane 1) or were stimulated for 20 min with IL-6/sIL-6R (lane 2) or with G-CSF (lane 3). Immunoblots of cell lysates were probed sequentially with antibodies specific for the active phosphorylated forms of ERK and STAT3.
The intact gp130 cytoplasmic domain mediated a clear induction of SIE DNA-binding activity (Fig. 2D). Mutation of individual docking sites had no appreciable effect. However, mutation of all four sites eliminated both the self-renewal signal and the induction of STAT3 DNA-binding activity. Radioligand binding established that cell surface expression was not limiting for any of the receptors (not shown). To confirm that other signaling pathways are not impaired by mutation of the STAT3 docking sites, we examined activation of the ERK cascade. ERK activation requires receptor phosphorylation on tyrosine 118 by JAK kinases and recruitment of SHP2 (Stahl et al. 1995; Fukada et al. 1996). Figure 2E shows that the basal level of constitutive ERK activity was significantly enhanced by stimulation of chimeric receptors in all transfectants tested. In particular, the two receptors, Y265/275F and Y126-275F, which gave reduced activation of STAT3 and cannot signal self-renewal, mediated normal and heightened levels of ERK activation, respectively. Therefore, there is no general compromise in the signaling capacity of these molecules.
Interestingly, this data also indicates that the STAT3 sites in gp130 may not be equivalent in vivo. Specifically, mutation of the two adjacent carboxy-terminal STAT3 binding sites (Y265 and Y275) abolished selfrenewal signaling, whereas mutation of the two-membrane proximal sites had little effect. This difference correlated with the lower induction of STAT3 DNA-binding activity and the specific reduction in STAT3 phosphorylation relative to ERK phosphorylation (Figs. 2D,E) (see Discussion). Self-renewal thus appears to require an appreciable level of STAT3 activation.
Inhibition of STAT3 activation blocks self-renewal and promotes differentiation
The above findings indicated that STAT3 may play a key role in ES cell signaling. To assess directly the requirement for STAT3 activation in ES cell self-renewal, we exploited a dominant interfering mutant form of STAT3, STAT3F. In this mutant (Minami et al. 1996), the tyrosine residue at amino acid position 705 is mutated to phenylalanine. Phosphorylation of Tyr705 is required for dimerization and nuclear translocation. When expressed at high levels, STAT3F has been shown to block the activation of endogenous STAT3 in various cell types, possibly by titrating out receptor docking sites (Fukada et al. 1996; Minami et al. 1996; Nakajima et al. 1996; Bonni et al. 1997; Ihara et al. 1997).
Using conventional transfection approaches, we were unable to recover ES cell transfectants showing stable high-level expression of STAT3F. In parallel experiments, however, transfection of the LIF-independent embryonal carcinoma cell line P19 yielded multiple expressing clones. This suggested that blockade of STAT3 activation in ES cells specifically resulted in cell death, growth arrest, or differentiation. An alternative transfection and expression strategy was therefore adopted to enable characterization of the consequences of STAT3F expression. The approach, termed supertransfection, relies on expression of polyoma virus large T protein by the recipient ES cells and its interaction with a polyoma origin of replication present in the transfected DNA. This results in efficient episomal propagation of incoming plasmid (Gassmann et al. 1995). We have developed this system for efficent cDNA expression in ES cells (H. Niwa, I. Chambers, L. Forrester, M. Gassmann, and A.G. Smith, in prep.). The process yields at least 100-fold more stable transfectants than conventional transfection protocols. A second important advantage of episomal supertransfection is that the unpredictable effects of chromosomal integration are avoided, with the result that the level of expression is both stable and relatively uniform (H. Niwa, I. Chambers, L. Forrester, M. Gassmann, and A.G. Smith, in prep.).
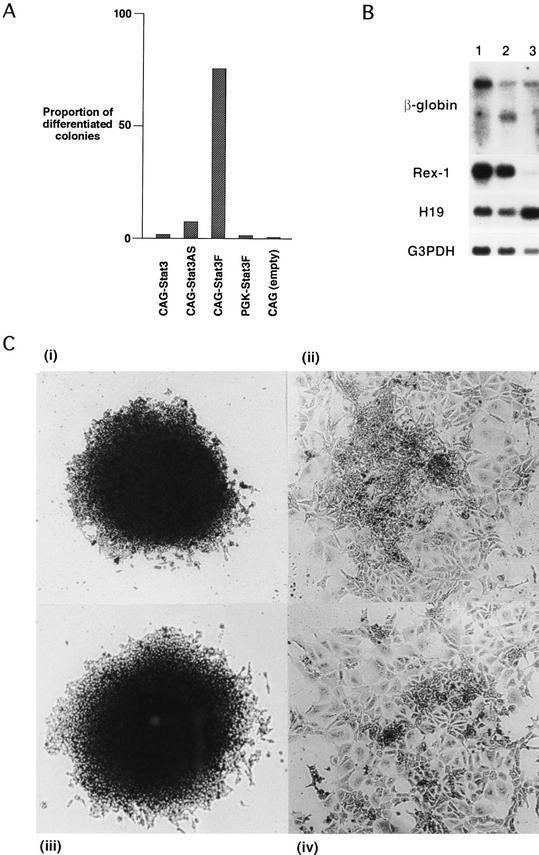
The STAT3F mutant cDNA was introduced into the supertransfection vector pHPCAG. The wild-type STAT3 coding sequence was also introduced, in both sense and antisense orientations. The three constructs were electroporated into MG1.19 cells that harbor a large T expression plasmid and can be supertransfected with constructs containing the polyoma origin (Gassmann et al. 1995). Supertransfectants were isolated by selection in hygromycin B for 8 days in the presence of LIF. Colonies were fixed, stained with Leishman’s reagent, counted, and scored for the presence of stem cells and differentiated cells. More than 95% of colonies obtained following supertransfection with control or wild-type STAT3 vector were stem cell colonies (Fig. 3A). A modest increase in the proportion of differentiated colonies was obtained with the antisense construct. The STAT3F vector, however, yielded predominantly differentiated colonies. A decrease in total number of colonies was also observed after supertransfection with STAT3F. This may reflect an early onset of differentiation that would produce very small clones that would not be scored. Alternatively, very high levels of STAT3F expression may also be toxic, though this has not been reported in other cell types. Morphologically, the differentiated STAT3F colonies closely resembled the differentiated colonies generated on culture of ES cells in the absence of LIF (Fig. 3C). Various other cDNAs have been expressed in ES cells using this system, with little or no effect on formation of stem cell colonies (data not shown). This suggested that the effect on differentiation was specifically attributable to expression of STAT3F.
Figure 3.
Induction of differentiation by expression of STAT3F in MG1.19 ES cells. (A) Proportion of differentiated colonies in LIF-supplemented medium resulting from supertransfection of STAT3, antisense STAT3, and STAT3F expression vectors. Colonies were fixed and stained with Leishman’s reagent after 8 days of selection, and the numbers of stem cell colonies and differentiated colonies were scored. (B) Marker gene expression in STAT3F supertransfectants. Expression of marker genes in pools of MG1.19 cells supertransfected with STAT3 (lane 1), STAT3 antisense (lane 2), and STAT3F (lane 3) expression vectors. Total RNA was prepared after 8 days of selection in LIF-supplemented medium, and 5-μg aliquots were analyzed by filter hybridization with β-globin, Rex-1, H19, and G3PDH probes. The β-globin probe detects all transgene mRNA species generated from pHPCAG, including an alternatively spliced product from the antisense contruct. (C) Photomicrographs of representative colonies 8 days after supertransfection with (i) STAT3, (ii) STAT3F, and (iii) empty expression vectors and selection in the presence of LIF, or (iv) induction of differentiation by culture in the absence of LIF for 8 days.
The differentiation induced by expression of STAT3F was examined further by expression analysis of the marker genes rex1 and H19. Rex-1 mRNA, which is specifically expressed in undifferentiated stem cells, was down-regulated in STAT3F supertransfectants. In contrast, H19 RNA, which is found at low levels in stem cells but is up-regulated during differentiation, was increased (Fig. 3B). A similar pattern of gene regulation is observed during differentiation of ES cells induced by withdrawal of LIF. These data confirm that the morphological differentiation triggered by STAT3F is accompanied by reprogramming of gene expression.
STAT3F was also expressed from the mouse phosphoglycerate kinase (pgk-1) promoter in the episomal vector pHPPGK. This vector gives at least 10-fold lower expression than pHPCAG (H. Niwa, I. Chambers, L. Forrester, M. Gassmann, and A.G. Smith, in prep.). In this case, there was no significant effect on either colony number or differentiation status of MG1.19 supertransfectants. A relatively high level of expression of the dominant interfering mutant therefore appears necessary to block self-renewal.
Effect of STAT3F on self-renewal is suppressed by coexpression of STAT3
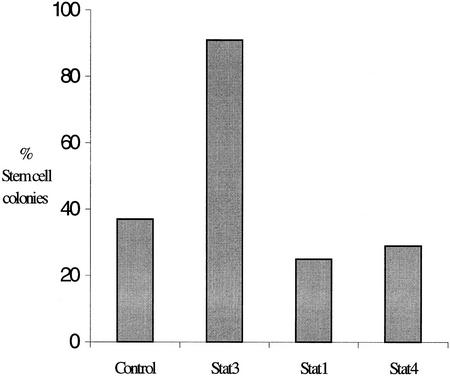
To test whether the induction of differentiation by expression of STAT3F was due to an inhibition of endogenous STAT3 activity, we attempted to rescue the stem cell phenotype by coexpression of wild-type STAT3 and also of STAT1 and STAT4. A STAT3F expression vector carrying a blasticidin resistance marker was cosupertransfected into MG1.19 cells with episomal constructs for expression of wild-type STATs and hygromycin resistance. Cosupertransfectants were isolated in medium containing both 20 μg/ml blasticidin S and 80 μg/ml of hygromycin B. The numbers of stem cell and differentiated colonies were scored after 8 days. As shown in Figure 4, only coexpression of wild-type STAT3 restored self-renewal in the presence of STAT3F. Transfection with STAT1 or STAT4 constructs alone had no effect on self-renewal in the absence of STAT3F (not shown) and did not alter differentiation induced by STAT3F. In the case of supertransfection with the CAG promoter STAT1 construct, the total number of colonies (stem plus differentiated) recovered was reduced, but the relative proportion of stem cell colonies versus differentiated cells was unaltered. This occurred in both the presence and absence of coexpression of STAT3F and suggests that high-level expression of STAT1 may be toxic to ES cells. By using the mouse PGK-1 promoter to drive lower levels of expression (H. Niwa, I. Chambers, L. Forrester, M. Gassmann, and A.G. Smith, in prep.), comparable numbers of colonies were recovered on transfection with the STAT1 as with the other constructs. In this case, again only the STAT3 construct showed any restoration of stem cell colonies, although to a lower degree than with the high-expression CAG vector (not shown). These data indicate that STAT3 has a specific function in ES cells that cannot be compensated by STAT1 or STAT4 (see Discussion).
Figure 4.
Cosupertransfection of STAT3F with wild-type STAT expression vectors. Proportions of undifferentiated stem cell colonies generated after cosupertransfection of MG1.19 ES cells with 10 μg of pBPCAGGS–STAT3F plus 10 μg of pHPCAG vector containing stuffer (control), STAT3, STAT1, or STAT4 inserts. After 8 days of selection with 80 μg/ml of hygromycin B plus 20 μg/ml of blasticidin S, colonies were fixed and stained with Leishman’s reagent.
Generation of an inducible STAT3F transgene integration in ES cells
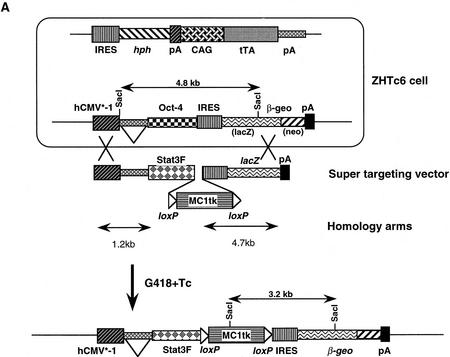
The effect of STAT3F expression on endogenous STAT3 activity could not be monitored directly in undifferentiated ES cells because ES cells expressing appreciable STAT3F constitutively could not be propagated. This required the generation of an inducible transgene. The tetracycline-regulatable system (tet-off) developed by Bujard and colleagues (Gossen and Bujard 1992) has been shown to confer inducibility on transgene expression in several cell types in culture and in the intact animal. However, it has proven problematic to establish this two-component system in ES cells. This is probably due to a combination of the relatively toxic effects of the tet repressor–VP16 fusion (tTA) and the tendency of ES cells to suppress expression of integrated transgenes (silencing). We have isolated previously an ES cell line, ZHTc6, that maintains stable production of effective but nontoxic levels of tTA from a gene trap integration (H. Niwa and A. Smith, in prep.). This cell line also contains a tetracycline-responsive hCMV*-1 transgene integrated at a favorable expression site. Expression of such transgenes is usually deregulated and/or mosaic in ES cells because of the sensitivity of the hCMV*-1 promoter to site of integration effects and silencing. However, transgene expression in line ZHTc6 is completely repressed in the presence of tetracycline but is activated in all cells on withdrawal of tetracycline as revealed by β-galactosidase reporter expression (H. Niwa and A. Smith, in prep.). Because of the low efficiency of establishing de novo transgene integrations with such favorable characteristics, we adopted a transgene substitution approach to generate an inducible STAT3F transgene.
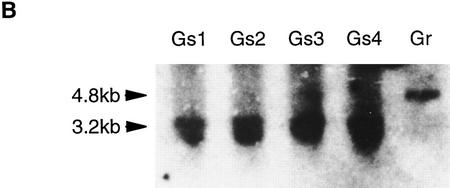
A targeting vector was designed for introduction of the STAT3F sequences into the hCMV*-1 locus by homologous recombination, using 5′ and 3′ sequences from the original transgenic construct as homology arms (Fig. 5A). In the presence of tetracycline, ZHTc6 cells are sensitive to G418 because the hCMV*-1 promoter is repressed. Advantage was taken of this by including a constitutive MC1 enhancer/promoter in the supertargeting vector to drive selectable marker expression. The absence of the neo sequence, however, requires that a legitimate recombination event with the resident transgene occur to confer G418 resistance. This powerful selection facilitated the isolation of targeted clones in which the STAT3F sequence was faithfully integrated 3′ to the hCMV*-1 promoter (Fig. 5B). In the continued presence of tetracycline, the targeted cells were maintained readily as undifferentiated stem cell colonies in the presence of LIF. Three clones, Gs1, Gs2, and Gs3, were then analyzed further.
Figure 5.
Generation of an inducible STAT3F transgene integration by supertargeting. (A) Schematic of supertargeting strategy for introduction of STAT3F into a tetracycline-regulatable expression site. ZHTc6 ES cells contain a tetracycline-regulated transgene comprising the hCMV*-1 promoter (Gossen and Bujard 1992), β-globin second intron, Oct-4 open reading frame (Okazawa et al. 1991), and IRESβgeopA selection marker (Mountford et al. 1994). Homologous recombination can be used to replace the Oct-4 sequence (supertargeting). Use of a truncated selection marker in the targeting vector facilitates the isolation of homologous recombinants. ZHTc6 cells were electroporated with the STAT3F–SuperKO vector and selected in G418 in the presence of tetracycline. G418-resistant clones were duplicated and screened for sensitivity against gancyclovir to enrich further for homologous recombinants. The option of excising the loxP-flanked MC1tk cassette by transient expression of Cre recombinase was not pursued. (B) Diagnosis of the supertargeting event in Gs ES cells. Gancyclovir-sensitive (Gs; lanes 1–4) and -resistant (Gr; lane 5) clones were analyzed by Southern hybridization. A 3.2-kb SacI fragment was detected with a probe from the 5′ end of lacZ in the Gs samples, indicative of the correct replacement of the Oct-4 cDNA sequence with STAT3F sequence. The Gr clone retained the 4.8-kb fragment diagnostic for the original Oct-4 transgene integration in ZHTc6 cells.
Induced expression of STAT3F blocks ES cell self-renewal and causes differentiation
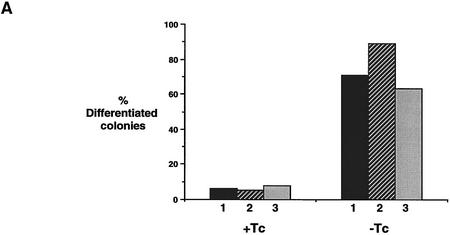
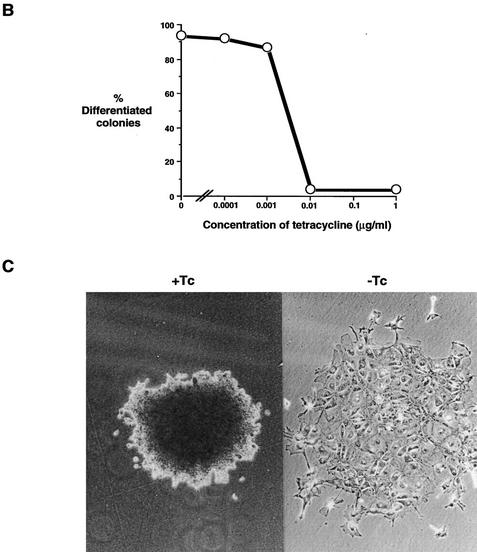
Withdrawal of tetracycline from Gs1, Gs2, or Gs3 cells resulted in the induction of differentiation in all three clones (Fig. 6A–C). Importantly, the efficiency of colony formation was not significantly different in the presence or absence of tetracycline, indicating that there is no toxic effect of STAT3F induction. The induced cultures differentiated over a 3- to 4-day time period, paralleling the behavior of parental ES cells on removal of LIF (Smith 1991). The differentiation response was confirmed by Northern hybridization analysis of Rex-1 and H19 transcripts (data not shown).
Figure 6.
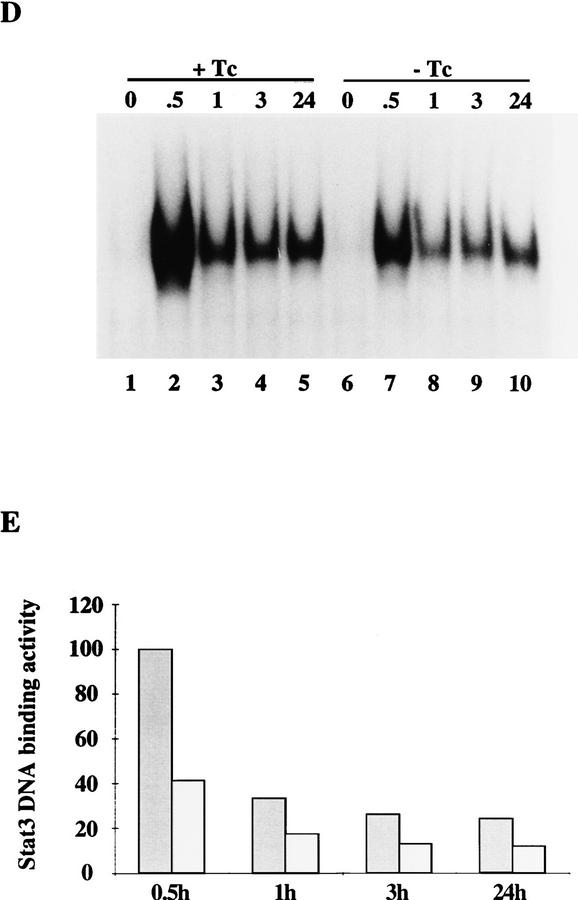
Induced expression of STAT3F causes ES cell differentiation and inhibits STAT3 activation. (A) Differentiation of Gs ES cells induced by withdrawal of tetracycline. Gs ES cells grown up in the presence of tetracycline were plated at clonal density (500 cells/60-mm dish) in LIF-supplemented medium in the presence or absence of tetracycline (1 μg/ml). After 6 days, colonies were fixed and stained with Leishman’s reagent. The histogram records the proportions of differentiated colonies for three independent clones, Gs1 (solid bars), Gs2 (hatched bars), and Gs3 (shaded bars). (B) Dose response curve of Gs2 cell differentiation. Gs2 ES cells were cultured as above in the presence of the indicated concentrations of tetracycline, then fixed, stained, and scored. (C) Photomicrographs of uninduced and induced Gs2 ES cells. Representative colonies of Gs2 cells cultured for 6 days in LIF-supplemented medium in the presence (+Tc) or absence (−Tc) of tetracycline (1 μg/ml) and then fixed and stained with Leishman’s reagent. (D) Mobility retardation assay of STAT3 DNA-binding activity in noninduced and induced Gs2 cells. Gs2 ES cells were cultured for 72 hr in the presence or absence of tetracycline. IL-6/sIL-6R was withdrawn for the final 24 hr, then restored for the indicated times. Nuclear extracts were prepared and assayed as described for SIE DNA-binding activity. (E) Quantitation of STAT3 SIE binding by PhosphorImager. (Shaded bars) +Tc; (open bars) −Tc.
Mobility retardation analysis was used to investigate directly STAT3 activation in STAT3F-expressing ES cells. The data in Figure 6D show that the level of STAT3 DNA-binding activity induced by gp130 stimulation was significantly lower in the presence of STAT3F. Quantitative PhosphorImager analysis confirmed a reduction of 50% or greater in the gel shift signal at all time points (Fig. 6E). The presence of residual STAT3 activity is consistent with the notion that a threshold level of active STAT3 is required to sustain self-renewal.
These findings confirm that expression of STAT3F in ES cells reduces gp130-mediated activation of STAT3, thereby blocking self-renewal and promoting differentiation.
Discussion
The primary cytoplasmic signal transduction event emanating from a ligand-activated LIF-R/gp130 complex in ES cells as in other cell types is considered to be transphosphorylation and activation of receptor-associated Janus kinases (JAKs) (Davis et al. 1993; Narazaki et al. 1994). The JAKs then phosphorylate tyrosine residues in the receptors, creating docking sites for SH2 domain-containing proteins, notably including the STAT factors STAT1 and STAT3 (Lutticken et al. 1994; Stahl et al. 1995). STAT proteins are themselves targets for phosphorylation by JAKs, which leads to their dimerization and translocation to the nucleus. Other signal transducing molecules can also be activated downstream of gp130, including insulin receptor substrate-1 (IRS-1), phosphoinositide-3 kinases (PI-3 kinase), nonreceptor tyrosine kinases such as Hck and Btk, the tyrosine phosphatase SHP2, and the mitogen-activated protein kinases ERK1 and ERK2 (Boulton et al. 1994; Ernst et al. 1994; Yin and Yang 1994; Argetsinger et al. 1995; Matsuda et al. 1995a,b). This modular signaling system has been assumed to underlie the diverse and pleiotropic effects of IL-6 and LIF-related cytokines in different cell types. A key issue therefore is to resolve the relative contribution of different signaling pathways in any given responsive cell type. A critical role has been ascribed to SHP2-mediated activation of the MAP kinase cascade in proliferation of BAF-BO3 cells (Fukada et al. 1996) and suppression of apoptosis in cardiomyocytes (Sheng et al. 1997). In contrast, the differentiation responses of myeloid M1 cells (Minami et al. 1996; Nakajima et al. 1996) and primary neural precursors (Bonni et al. 1997) are effected via activation of STAT3. Previous studies in ES cells have suggested that JAK–STAT signaling, ERK activation, and the nonreceptor tyrosine kinase Hck could all be involved in LIF signaling (Ernst et al. 1994, 1996; Narazaki et al. 1994; Hocke et al. 1995; Boeuf et al. 1997).
We initially investigated the ability of chimeric receptor constructs to signal ES cell self-renewal by isolation of stably expressing transfectants. The observation that G-CSF-R can support ES cell propagation drew attention to signaling features conserved between G-CSF-R and gp130, notably the induction of STAT3 DNA-binding activity. Combined substitutions of the tyrosine residues in the STAT3 binding sites of gp130 cytoplasmic domain were associated with different levels of STAT3 activation and indicated that a self-renewal signal is associated with a threshold of STAT3 activity. Moreover, the four STAT3 sites do not appear to act in either a redundant or simple cumulative manner. Both self-renewal signaling and induction of STAT3 DNA-binding activity were maintained on pairwise mutation of the two-membrane proximal STAT3 docking sites (Y126 and Y173) but not on mutation of the carboxy-terminal pair (Y265 and Y275) (see Fig. 2). This observation is somewhat unexpected as it has been shown previously that the isolated phosphopeptide sequences have equivalent STAT3 binding properties (Stahl et al. 1995) and that a truncated receptor with a single-membrane proximal STAT3 site (Y126) can efficiently induce STAT3-mediated differentiation of M1 cells (Yamanaka et al. 1996). It is important to note, however, that in the truncated receptor, sequences that mediate receptor internalization (Dittrich et al. 1996) have also been deleted with unpredictable consequences for signaling properties. Our findings indicate that in the normal context of the full-length receptor, the four STAT3 docking sites are not equivalent. The explanation for the reduced activity of the membrane proximal pair of sites is unclear though one possibility is that availability of Y126 may be influenced by interaction of SHP2 with Y118 (note enhanced ERK activation from Y126-275F chimera in Fig. 2E).
The finding that mutation of the STAT3 binding sites in the cytoplasmic domain of gp130 abolished the self-renewal signal prompted a direct investigation of the role of this transcription factor. New strategies were required to express the dominant interfering mutant STAT3F in ES cells. The methods we have deployed in this study enhance the experimental versatility and tractability of ES cells and establish new avenues for the characterization in vitro of gene functions involved in stem cell propagation, commitment, or differentiation. Because of the >100-fold increase in stable transfection efficiency and the relative homogeneity of expression (H. Niwa, I. Chambers, L. Forrester, M. Gassmann, and A.G. Smith, in prep.), episomal supertransfection provides a methodology for the screening and analysis of cDNAs whose expression is not compatible with ES cell self-renewal. The first demonstration of effective operation of the tetracycline regulation system in ES cells provides a complementary inducible expression approach. These two methods should find broad application in functional screening and in the genetic manipulation of lineage commitment and differentiation processes in ES cells.
Both constitutive expression of STAT3F following episomal supertransfection and induced expression from the regulatable chromosomal site inhibited self-renewal and resulted in differentiation. The episomal approach also allowed the specificity of the requirement for STAT3 to be established by coexpression of various STAT family members with STAT3F. The finding that STAT3 can restore self-renewal indicates that this factor serves a specific and nonredundant function in ES cell self-renewal in response to LIF. The evidence that STAT1 cannot compensate for STAT3 is noteworthy because STAT1 can be activated in response to LIF in ES cells, though to a much lesser extent than STAT3 (Starr et al. 1997). STAT1 may play little or no role in ES cell propagation. Induction of STAT1 DNA-binding activity was not evidently associated with self-renewal signaling from the various chimeric receptors used in this study (Figs. 1B and 2D). Furthermore, ES cells in which both alleles of the stat1 gene have been inactivated are phenotypically normal (Durbin et al. 1996).
A role for STAT3 in ES cell signaling has recently also been suggested by Boeuf et al. (1997) who reported the isolation of ES cell clones expressing STAT3F constitutively. These cells apparently showed an increased tendency to differentiate after 1 month or more in culture. The basis of this phenomenon is unclear because absence or blockade of LIF signaling results in complete differentiation within a few days (Smith et al. 1988; Williams et al. 1988; C. Dani, I. Chambers, S. Johnstone, M. Robertson, B. Ebrahimi-Chahardahcherik, M. Saito, T. Taga, M. Li, T. Burdon, J. Nichols, and A.G. Smith, in prep.). We were unable to establish conventional transfectants expressing significant levels of STAT3F. However, our data on both episomal and induced expression demonstrate that STAT3F rapidly and efficiently blocks ES cell self-renewal and triggers differentiation.
Our results establish that STAT3 activation is essential for LIF-R/gp130-mediated ES cell self-renewal. STAT3 activity is regulated by phosphorylation on both tyrosine and serine (Wen et al. 1995), and a constitutively active mutant has not been described. An isoform of STAT3, STAT3β, generated by alternative splicing, is reported to show sustained activation properties (Schaefer et al. 1995). ES cells supertransfected with a STAT3β vector remained LIF dependent (data not shown), however, indicating that this isoform does not substitute for activated STAT3 in ES cells. This may be because STAT3β appears to function by formation of heterodimers with c-Jun (Schaefer et al. 1995), and it is anticipated that the STAT3β/c-Jun complex regulates a distinct spectrum of target genes compared with the STAT3 homodimer. It is noteworthy, however, that expression of v-src in ES cells renders them LIF independent (Boulter et al. 1991). v-Src has been shown to associate with and cause constitutive activation of STAT3 (Cao et al. 1996).
The p42/p44 MAP kinase pathway (ERK1 and ERK2) has been reported to be activated by LIF in ES cells as in other cell types (Ernst et al. 1996; Boeuf et al. 1997). The Ras–ERK cascade is coupled to gp130 via the adaptor molecule SHP2 (Fukada et al. 1996; Yamanaka et al. 1996). SHP2 interacts with activated gp130 at phosphorylated tyrosine residue 118 (Stahl et al. 1995). Significantly, mutation of this residue does not inhibit self-renewal signaling in ES cells (T. Burdon, I. Chambers, C. Stracey, J. Nichols, and A.G. Smith, in prep.). Furthermore, the MEK inhibitor PD098059 (Dudley et al. 1995) that specifically blocks activation of the ERK kinases does not inhibit stem cell colony formation in response to LIF (T. Burdon, I. Chambers, C. Stracey, J. Nichols, and A.G. Smith, in prep.). Thus, although contributions of other pathways are not precluded, STAT3 appears to play a central role in ES cell self-renewal. The underlying importance of STAT3 is further attested to by the finding that homozygous disruption of the Stat3 gene in mice is associated with early embryonic lethality (Takeda et al. 1997).
It is striking that the role of STAT3 in propagation of the undifferentiated pluripotential phenotype of ES cells contrasts with previously characterized functions as an effector of somatic cell differentiation. Dominant interfering mutants of STAT3 have been shown to block macrophage differentiation of myeloid M1 cells induced by IL-6 or LIF (Minami et al. 1996; Nakajima et al. 1996) or by GCSF (Shimozaki et al. 1997). STAT3 activation has similarly been shown to mediate IL-6- or LIF-induced astrocytic differentiation of primary cortical neuroepithelial cells (Bonni et al. 1997). Recently it has also been shown that STAT3 is activated by hepatocyte growth factor and mediates epithelial tubulogenesis (Boccaccio et al. 1998). STAT3 thus has distinct effects in different cell types. A common theme, however, may be the regulation of genes that determine cell identity. The diverse effects of the LIF/IL-6 family of cytokines on cellular differentiation and gene expression appear to reflect cell-type specific effects of active STAT3. In the context of stem cell propagation, the key issue now is to identify transcriptional targets of STAT3 in ES cells and to illuminate the relationship between STAT3 and the essential ES cell-specific transcription factor Oct-4.
Materials and methods
Cell culture and transfection
ES cells were maintained in the absence of feeder cells in Glasgow modification of Eagle medium (GMEM) supplemented with fetal calf serum, 2-mercaptoethanol, and LIF (Smith 1991). CGR8 (Mountford et al. 1994) and MG1.19 (Gassmann et al. 1995) ES cells have been described elsewhere. DO27 ES cells have had both copies of the lif gene inactivated by homologous recombination and the IRESβgeo selection marker/reporter inserted into the oct4 gene as described (C. Dani, I. Chambers, S. Johnstone, M. Robertson, B. Ebrahimi-Chahardahcherik, M. Saito, T. Taga, M. Li, T. Burdon, J. Nichols, and A.G. Smith, in prep.). LRKOh34 ES cells have targeted disruptions in both copies of the lifr gene (M. Li, I. Chambers, J. Nichols, and A.G. Smith, in prep.) and are maintained in medium in which LIF is substituted with IL-6 (50 ng/ml) and soluble IL-6 receptor (5% CHO-5E7 conditioned medium; Yasukawa et al. 1990). For conventional transfection with pPCAGIZ vectors, 1 × 107 cells were electroporated with 100 μg of linearized plasmid DNA at 800 V and 3 μF in a 0.4-cm cuvette using a Bio-Rad gene pulser and then selected in the presence of zeocin (Invitrogen). For transfection of episomal vectors (supertransfection), 5 × 106 MG1.19 cells were electroporated with 20 μg of supercoiled plasmid DNA at 200 V and 960 μF and then cultured in the presence of either 80 μg/ml hygromycin B (Boehringer Mannheim) or 4–20 μg/ml blasticidin S (Waken Seiyaku), or both hygromycin plus blasticidin for cosupertransfection.
Generation of tetracycline regulatable transgenes in ES cells
ZHTc6 ES cells were derived from CGR8 ES cells (Mountford et al. 1994) and will be described in detail elsewhere (H. Niwa and A.G. Smith, in prep.). They carry a targeted integration of IRESzeo in one Oct3/4 allele. They also carry a gene trap integration of an IREShph:CAGtTA construct that confers stable expression of the tetracycline-responsive tTA transactivator and a randomly integrated hCMV*-1–Oct4–IRESβgeopA transgene. These cells were routinely maintained in the presence of 10 μg/ml zeocin and 1 μg/ml tetracycline-HCl (Sigma).
The hCMV*-1–Oct–4-IRESβgeopA transgene is comprised of the tetracycline-inducible promoter hCMV*-1 derived from pUHD10-3 (Gossen and Bujard 1992), rabbit β-globin second intron, full-length Oct-4 cDNA, and IRESβgeopA unit (Mountford et al. 1994). pSuperKO (see Fig. 5A) contains the hCMV*-1 and rabbit globin sequences as the 5′ homology arm and the IRESlacZ cassette as 3′ arm. Intervening are a stuffer sequence with XhoI and SfiI cloning sites and a loxP-flanked MC1tk cassette (Mansour et al. 1988). The STAT3F cDNA was introduced as a SalI fragment between the XhoI sites. For gene targeting, 2 × 107 cells were electroporated with 100 μg linearized SuperKO–STAT3F plasmid DNA at 800 V and 3 μF and then selected in the presence of 200 μg/ml G418 (GIBCO BRL) and 1 μg/ml tetracycline-HCl. Targeted clones were maintained in the continuous presence of tetracycline-HCl.
Plasmid construction
DNA manipulations were performed by standard procedures (Sambrook et al. 1989). Full details of plasmid constructions are available on request. The full-length mouse G-CSF-R cDNA (pJ17) was provided by Shigekazu Nagata (Fukunaga et al. 1990), and the G-CSF-R/LIF-R chimeric receptor construct (Baumann et al. 1994b) was provided by Steve Ziegler. G-CSF-R/gp130 chimeric receptor constructs were generated by fusing the coding sequence for the extracellular domain of human G-CSF-R (Baumann et al. 1994b) to an EcoRI fragment encoding the transmembrane domain and the entire cytoplasmic region of mouse gp130 cDNA (Hibi et al. 1990). Phenylalanine substitutions were introduced into the intracellular domain of gp130 by PCR overlap mutagenesis (Higuchi et al. 1988). PCR products were substituted into the G-CSF-R/gp130 chimera and sequenced. Episomal expression vectors pHPCAG, pBPCAG, and pHPPGK are described elsewhere (H. Niwa, I. Chambers, L. Forrester, M. Gassmann, and A.G. Smith, in prep.). The expression vector pPCAGIZ, which can be used as both an episomal and an integrated expression vector, was constructed by ligation of the encephalomyocarditis virus IRES (pCITE-1, Novagen) with the Streptoalloteichus bleomycin resistant gene (Sh ble:zeo) from pZeoSV (Invitrogen) and introduction into pPCAG (H. Niwa, I. Chambers, L. Forrester, M. Gassmann, and A.G. Smith, in prep.). cDNAs are inserted into a XhoI site 5′ to the IRES. The requirement for continuous relatively high-level expression of the zeo gene to confer antibiotic resistance allows direct selection for integrations into favorable expression sites. Consequently, using this vector, ES cell transfectants can readily be isolated that sustain stable transgene expression (H. Niwa, T. Burdon, I. Chambers, and A.G. Smith, unpubl.).
RNA and DNA hybridization analyses
Total RNA (Chomczynski and Sacchi 1987) was separated on a 0.66 m formaldehyde, 0.8% agarose gel and blotted onto nylon membranes (Hybond N, Amersham). Hybridization was performed with β-globin third exon, Rex-1, H19, and GAPDH cDNA probes labeled by random hexamer primed DNA synthesis in the presence of [α-32P]dCTP (3000 Ci/mmole).
For identification of targeted ES cell clones, genomic DNA was digested with SacI, separated on a 0.7% agarose gel, and analyzed by nonradioactive filter hybridization (Gene Image, Amersham) with an EcoRI–SacI fragment of the lacZ gene.
G-CSF-R binding assay
ES cells (1 × 106) were seeded in wells of a 24-well plate and grown for 24 hr. The cells were then cooled to 4°C and growth medium was replaced with 0.25 ml of ice-cold binding buffer (GMEM, 25 mm HEPES at pH 7.2, 0.2% BSA) containing 0.212 nm 125I-labeled G-CSF-R (Amersham) in the presence or absence of a 1000-fold molar excess of cold G-CSF-R. Binding reactions were incubated for 3 hr at 4°C and terminated by washing the cells three times with ice-cold binding buffer. Cells were then solubilized in 0.5% NP-40, and an aliquot was counted in a gamma counter. All treatments were performed in duplicate. No specific binding was detected to untransfected cells, and nondisplaceable binding was consistent between clones.
Self-renewal assays
To measure self-renewal of ES cells at cloning density, cells were plated at 1000 cells per well (∼100 cells/cm2) in 6-well dishes and cultured for 6 days. Cells were either grown in the absence of cytokines, in 100 U/ml recombinant LIF (Smith 1991), in 100 ng/ml IL-6 plus soluble IL-6R, or in 30 ng/ml G-CSF-R, as appropriate. On day 6, colonies were fixed and stained with Leishman’s reagent (Smith 1991) or for alkaline phosphatase activity (Sigma leukocyte alkaline phosphatase kit) (Bernstine et al. 1973). Numbers of stem cell and differentiated colonies were scored by microscopic examination, in some cases with computer-assisted image analysis. All assays were performed in duplicate or triplicate.
Stem cell-specific expression of β-galactosidase from the oct4 locus in D027 cells was quantified by ONPG assay on triplicate samples. Cells were plated at 5000 per well in 24-well dishes and cultured for 6 days in the presence or absence of cytokine as above. On day 6, cells were washed once with PBS and lysed in 0.4 ml of 0.25 m Tris (pH 7.5), 5 mm DTT, and 0.5% NP-40. Lysate (40 μl) was mixed with 100 μl of ONPG buffer (60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, 1 mm MgCl2, 50 mm 2-mercaptoethanol, 1.2 mm ONPG) in a microtiter plate and incubated at 37°C for 2–4 hr, and the absorbance was read at 420 nm.
Preparation of nuclear extracts and band-shift assays
One day after plating (1 × 106 cells per 60-mm dish), ES cells were washed with PBS and refed with medium lacking cytokines. The next day, cells were stimulated with IL-6 (100 ng/ml plus soluble receptor) or G-CSF-R (30 ng/ml) for 30 min, washed with ice-cold PBS, scraped off the plates, and collected by centrifugation. Nuclear extracts were prepared by the method described (Gobert et al. 1996) except that protease inhibitors (aprotinin, pepstatin, and leupeptin) were omitted from the cell lysis buffer. Protein concentrations of nuclear extracts were determined using a Bradford assay (Bio-Rad). Aliquots (2 μg) of nuclear extract were incubated with 0.25 ng of 32P-labeled double-stranded SIEm67 oligonucleotide probe (Sadowski et al. 1993) in binding buffer (20 mm HEPES at pH 7.5, 50 mm NaCl, 1 mm EDTA, 1 mm DTT, 0.05% NP-40, 10% glycerol, 2 μg/ml of poly[d(I-C)], and 1 mg/ml BSA) for 20 min at room temperature. Binding reactions were resolved by electrophoresis on a prerun 5% polyacrylamide gel in 0.25× TBE for 3 hr. Gels were fixed in 10% acetic acid, dried under vacuum, and subjected to autoradiography or quantitated on a Bio-Rad PhosphorImager.
Immunoblotting
One day after plating (1 × 106 cells per 60-mm dish), ES cells were refed with medium containing 1% FCS and lacking cytokines. Following overnight incubation, cells were transferred to serum-free medium for 4 hr prior to stimulation with IL-6 (100 ng/ml plus soluble receptor) or G-CSF-R (30 ng/ml) for 20 min. Cells were then washed once with ice-cold PBS and lysed on ice in 100 μl SDS sample buffer. Ten- microliter aliquots of the lysates were fractionated on a 10% SDS–polyacrylamide gel and electroblotted onto nitrocellulose. After overnight treatment in blocking buffer (25 mm Tris-HCl at pH 7.4, 2.7 mm KCl, 140 mm NaCl, 0.1% Tween 20, 5% nonfat dried milk), membranes were probed sequentially with the phospho-specific anti-ERK and anti-STAT3 antibodies according to the directions provided by the supplier (New England Biolabs). Blots were incubated with HRP-coupled anti-rabbit IgG and developed using ECL reagents (Amersham). Membranes were stripped between probings by incubation at 50°C for 30 min in 62.5 mm Tris-HCl (pH 6.8), 2% SDS, and 100 mm 2-mercaptoethanol.
Acknowledgments
We thank Alexander Medvinsky and John Bishop for comments on the manuscript. Craig Stracey is thanked for help with β-galactosidase assays, and Melany Jackson for assistance with image analysis. DO27 and LRKOh34 cells were generated by Christian Dani and Meng Li, respectively. We are grateful to Shizuo Akira and Daniel Nathans for the STAT3F and STAT3β constructs, respectively, James Darnell for STAT1 and STAT4 cDNAs, K. Akagi and Hermann Bujard for the tTA and hCMV*-1 constructs, and Tetsuya Taga for gp130 cDNA and recombinant IL-6 and sIL-6R. Recombinant G-CSF-R was a generous gift of Chugai Corporation. Photographic reproductions were by Graham Brown and colleagues. This work was supported by the Biotechnology and Biological Sciences Research Council of the United Kingdom, the Human Frontiers Science Program Organisation, and Stem Cell Sciences Pty.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL austin.smith@ed.ac.uk; FAX 44 131 667 0164.
References
- Argetsinger LS, Hsu GW, Myers MG, Billestrup N, White MF, Carter-Su C. Growth hormone, interferon-γ, and leukemia inhibitory factor promoted tyrosyl phosphorylation of insulin receptor substrate-1. J Biol Chem. 1995;270:14685–14692. doi: 10.1074/jbc.270.24.14685. [DOI] [PubMed] [Google Scholar]
- Baumann H, Wong GG. Hepatocyte-stimulating factor-lll shares structural and functional identity with leukemia-inhibitory factor. J Immunol. 1989;143:1163–1167. [PubMed] [Google Scholar]
- Baumann H, Gearing D, Ziegler S. Signaling by the cytoplasmic domain of hematopoietin receptors involves two distinguishable mechanisms in hepatic cells. J Biol Chem. 1994a;269:16297–16304. [PubMed] [Google Scholar]
- Baumann H, Symes AJ, Comeau MR, Morella KK, Wang Y, Friend D, Ziegler SF, Fink JS, Gearing DP. Multiple regions within the cytoplasmic domains of the leukemia inhibitory factor receptor and gp130 cooperate in signal transduction in hepatic and neuronal cells. Mol Cell Biol. 1994b;14:138–146. doi: 10.1128/mcb.14.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstine EG, Hooper ML, Grandchamp S, Ephrussi B. Alkaline phosphatase activity in mouse teratoma. Proc Natl Acad Sci. 1973;70:3899–3903. doi: 10.1073/pnas.70.12.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio P. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391:285–288. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- Boeuf H, Hauss H, De Graeve F, Baran N, Kedinger N. Leukemia inhibitory factor-dependent transcriptional activation in embryonic stem cells. J Cell Biol. 1997;138:1207–1217. doi: 10.1083/jcb.138.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Boulter CA, Aguzzi A, Williams RL, Wagner EF, Evans MJ, Beddington R. Expression of v-src induces aberrant development and twinning in chimaeric mice. Development. 1991;111:357–366. doi: 10.1242/dev.111.2.357. [DOI] [PubMed] [Google Scholar]
- Boulton TG, Stahl N, Yancopoulos GD. Ciliary neurotrophic factor/leukemia inhibitory factor/interleukin-6/oncostatin M family of cytokines induces tyrosine phosphorylation of a common set of proteins overlapping those induced by other cytokines and growth factors. J Biol Chem. 1994;269:11648–11655. [PubMed] [Google Scholar]
- Brook FA, Gardner RL. The origin and efficient derivation of embryonic stem cells in the mouse. Proc Natl Acad Sci. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Tay A, Guy GR, Tan YH. Activation and association of stat3 with src in v-src-transformed cell lines. Mol Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Cozens A, Broadbent J, Robertson M, Lee M, Li M, Smith A. Structure of the mouse leukaemia inhibitory factor receptor gene: Regulated expression of mRNA encoding a soluble receptor isoform from an alternative 5′ untranslated region. Biochem J. 1997;328:879–888. doi: 10.1042/bj3280879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Conover JC, Ip NY, Poueymirou WT, Bates B, Goldfarb MP, DeChiara TM, Yancopoulos GD. Ciliary neurotrophic factor maintains the pluripotentiality of embryonic stem cells. Development. 1993;119:559–565. doi: 10.1242/dev.119.3.559. [DOI] [PubMed] [Google Scholar]
- Davis S, Aldrich TH, Stahl N, Pan L, Taga T, Kishimoto T, Ip NY, Yancopoulos GD. LIFRβ and gp130 as heterodimerizing signal transducers of the tripartite CNTF receptor. Science. 1993;260:1805–1808. doi: 10.1126/science.8390097. [DOI] [PubMed] [Google Scholar]
- Dittrich E, Haft CH, Muys L, Heinrich PC, Graeve L. A di-leucine motif and an upstream serine in the interleukin-6 (IL-6) signal transducer gp130 mediate ligand-induced endocytosis and down-regulation of the IL-6 receptor. J Biol Chem. 1996;271:5487–5494. doi: 10.1074/jbc.271.10.5487. [DOI] [PubMed] [Google Scholar]
- Dudley DT, Pang L, Decker SJ, Bridges AJ, Saltiel AR. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- Ernst M, Gearing DP, Dunn AR. Functional and biochemical association of Hck with the LIF/IL-6 receptor signal transducing subunit gp130 in embryonic stem cells. EMBO J. 1994;13:1574–1584. doi: 10.1002/j.1460-2075.1994.tb06420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst M, Oates A, Dunn AR. Gp130-mediated signal transduction in embryonic stem cells involves activation of Jak and Ras/mitogen activated protein kinase pathways. J Biol Chem. 1996;271:30136–30143. doi: 10.1074/jbc.271.47.30136. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman M. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: Involvement of STAT3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Ishizaka-Ikeda E, Seto Y, Nagata S. Expression cloning of a receptor for murine granulocyte colony-stimulating factor. Cell. 1990;61:341–350. doi: 10.1016/0092-8674(90)90814-u. [DOI] [PubMed] [Google Scholar]
- Gassmann M, Donoho G, Berg P. Maintenance of an extrachromosomal plasmid vector in mouse embryonic stem cells. Proc Natl Acad Sci. 1995;92:1292–1296. doi: 10.1073/pnas.92.5.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing DP, Bruce GA. Oncostatin M binds the high-affinity leukemia inhibitory factor receptor. New Biol. 1992;4:61–65. [PubMed] [Google Scholar]
- Gearing DP, Thut CJ, VandenBos T, Gimpel SD, Delaney PB, King J, Price V, Cosman D, Beckman MP. Leukemia inhibitory factor receptor is structurally related to the Il-6 signal transducer, gp130. EMBO J. 1991;10:2839–2848. doi: 10.1002/j.1460-2075.1991.tb07833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert S, Chretien S, Gouilleux F, Muller O, Pallard C, Dusanter-Fourt I, Groner B, Lacombe C, Gisselbrecht S, Mayeux P. Identification of tyrosine residues within the intracellular domain of the erythropoietin receptor crucial for STAT5 activation. EMBO J. 1996;15:2434–2441. [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi M, Murakami M, Saito M, Hirano T, Taga T, Kishimoto T. Molecular cloning and expression of an IL-6 signal transducer, gp130. Cell. 1990;63:1149–1157. doi: 10.1016/0092-8674(90)90411-7. [DOI] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988;15:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota H, Yoshida K, Kishimoto T, Taga T. Continuous activation of gp130, a signal-transducing receptor component for interleukin 6-related cytokines, causes myocardial hypertrophy in mice. Proc Natl Acad Sci. 1995;92:4862–4866. doi: 10.1073/pnas.92.11.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocke GM, Cui MZ, Fey GH. The LIF response element of the α2 macroglobulin gene confers LIF-induced transcriptional activation in embryonal stem cells. Cytokine. 1995;7:491–502. doi: 10.1006/cyto.1995.0067. [DOI] [PubMed] [Google Scholar]
- Ihara S, Nakajima K, Fukada T, Hibi M, Nagata S, Hirano T, Fukui Y. Dual control of neurite outgrowth by STAT3 and MAP kinase in PC12 cells stimulated with interleukin-6. EMBO J. 1997;16:5345–5352. doi: 10.1093/emboj/16.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle J. STATs: Signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- Kishimoto T, Taga T, Akira S. Cytokine signal transduction. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- Koblar SA, Turnley AM, Classon BJ, Reid KL, Ware CB, Cheema SS, Murphy M, Bartlett PF. Neural precursor differentiation into astrocytes requires signaling through the leukemia inhibitory factor receptor. Proc Natl Acad Sci. 1998;95:3178–3181. doi: 10.1073/pnas.95.6.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Sendtner M, Smith A. Essential function of LIF receptor in motor neurons. Nature. 1995;378:724–727. doi: 10.1038/378724a0. [DOI] [PubMed] [Google Scholar]
- Lutticken C, Wegenka UM, Yuan J, Buschmann J, Schindler C, Ziemiecki A, Harpur AG, Wilks AF, Yasukawa K, Taga T, Kishimoto T, Barbieri G, Pellegrini S, Sendtner M, Heinrich PC, Horn F. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science. 1994;263:89–92. doi: 10.1126/science.8272872. [DOI] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: A general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Fukada T, TakahashiTezuka M, Okuyama Y, Fujitani Y, Hanazono Y, Hirai H, Hirano T. Activation of Fes tyrosine kinase by gp130, an interleukin-6 family cytokine signal transducer, and their association. J Biol Chem. 1995a;270:11037–11039. doi: 10.1074/jbc.270.19.11037. [DOI] [PubMed] [Google Scholar]
- Matsuda T, TakahashiTezuka M, Fukada T, Okuyama Y, Fujitani Y, Tsukada S, Mano H, Hirai H, Witte ON, Hirano T. Association and activation of Btk and Tec tyrosine kinases by gp130, a signal transducer of the interleukin-6 family of cytokines. Blood. 1995b;85:627–633. [PubMed] [Google Scholar]
- Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira A. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc Natl Acad Sci. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountford P, Zevnik B, Duwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Dicistronic targeting constructs: Reporters and modifiers of mammalian gene expression. Proc Natl Acad Sci. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Hibi M, Nakagawa N, Nakagawa T, Yasukawa K, Yamanishi K, Taga T, Kishimoto T. IL-6-induced homodimerization of gp130 and associated activation of a tyrosine kinase. Science. 1993;260:1808–1810. doi: 10.1126/science.8511589. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, Kitaoka T, Fukada T, Hibi M, Hirano T. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–3658. [PMC free article] [PubMed] [Google Scholar]
- Narazaki M, Witthuhn BA, Yoshida K, Silvennoinen O, Yasukawa K, Ihle JN, Kishimoto T, Taga T. Activation of JAK2 kinase mediated by the interleukin 6 signal transducer gp130. Proc Natl Acad Sci. 1994;91:2285–2289. doi: 10.1073/pnas.91.6.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Chambers I, Smith A. Derivation of germline competent embryonic stem cells with combination of interleukin-6 and soluble interleukin-6 receptor. Exp Cell Res. 1994;215:237–239. doi: 10.1006/excr.1994.1338. [DOI] [PubMed] [Google Scholar]
- Okazawa H, Okamoto K, Ishino F, IshinoKaneko T, Takeda S, Toyoda YMM, Hamada H. The oct3 gene, a gene for an embryonic transcription factor, is controlled by a retinoic acid repressible enhancer. EMBO J. 1991;10:2997–3006. doi: 10.1002/j.1460-2075.1991.tb07850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennica D, King KL, Shaw KJ, Luis E, Rullamas J, Luoh S-M, Darbinne WC, Knutzon DS, Yen R, Chien KR, Baker JB, Wood WL. Expression cloning of cardiotrophin 1, a cytokine that induces cardiac myocyte hypertrophy. Proc Natl Acad Sci. 1995a;92:1142–1146. doi: 10.1073/pnas.92.4.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennica D, Shaw KJ, Swanson TA, Moore MW, Shelton DL, Zioncheck KA, Rosenthal A, Taga T, Paoni NF, Wood WI. Cardiotrophin-1. Biological activities and binding to the leukemia inhibitory factor receptor/gp130 signaling complex. J Biol Chem. 1995b;270:10915–10922. doi: 10.1074/jbc.270.18.10915. [DOI] [PubMed] [Google Scholar]
- Rathjen PD, Nichols J, Toth S, Edwards DR, Heath JK, Smith AG. Developmentally programmed induction of differentiation inhibiting activity and the control of stem cell populations. Genes & Dev. 1990;4:2308–2318. doi: 10.1101/gad.4.12b.2308. [DOI] [PubMed] [Google Scholar]
- Rose TM, Weiford DM, Gunderson NL, Bruce AG. Oncostatin M (OSM) inhibits the differentiation of pluripotent embryonic stem cells in vitro. Cytokine. 1994;6:48–54. doi: 10.1016/1043-4666(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Sadowski HB, Shuai K, Darnell JE, Gilman MZ. A common nuclear signal transduction pathway activated by growth factor and cytokine receptors. Science. 1993;261:1739–1744. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaefer TS, Sanders LK, Nathans D. Cooperative transcriptional activity of jun and Stat3β, a short form of Stat3. Proc Natl Acad Sci. 1995;92:9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Z, Knowlton K, Chen J, Hoshijima M, Brown JH, Chien KR. Cardiotrophin 1 (CT-1) inhibition of cardiac myocyte apoptosis via a mitogen activated protein kinase-dependent pathway. J Biol Chem. 1997;272:5783–5791. doi: 10.1074/jbc.272.9.5783. [DOI] [PubMed] [Google Scholar]
- Shimozaki K, Nakajima K, Hirano T, Nagata S. Involvement of STAT3 in the granulocyte colony-stimulating factor-induced differentiation of myeloid cells. J Biol Chem. 1997;272:25184–25189. doi: 10.1074/jbc.272.40.25184. [DOI] [PubMed] [Google Scholar]
- Smith AG. Culture and differentiation of embryonic stem cells. J Tiss Cult Meth. 1991;13:89–94. [Google Scholar]
- Smith AG, Hooper ML. Buffalo rat liver cells produce a diffusible activity which inhibits the differentiation of murine embryonal carcinoma and embryonic stem cells. Dev Biol. 1987;121:1–9. doi: 10.1016/0012-1606(87)90132-1. [DOI] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Stahl N, Boulton TG, Farruggella T, Ip NY, Davis S, Witthuhn BA, Quelle FW, Silvennoinen O, Barbieri G, Pellegrini S, Ihle JN, Yancopoulos GD. Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science. 1994;263:92–95. doi: 10.1126/science.8272873. [DOI] [PubMed] [Google Scholar]
- Stahl N, Farrugella TJ, Boulton TG, Zhong Z, Darnell JE, Yancopoulos GD. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–1353. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- Starr R, Novak U, Willson TA, Inglese M, Murphy V, Alexander WS, Metcalf D, Nicola NA, Hilton DJ, Ernst M. Distinct roles for leukemia inhibitory factor receptor α-chain and gp130 in cell type-specific signal transduction. J Biol Chem. 1997;272:19982–19986. doi: 10.1074/jbc.272.32.19982. [DOI] [PubMed] [Google Scholar]
- Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida M, Yamamoto-Yamaguchi Y, Hozumi M. Purification of a factor inducing differentiation of mouse myeloid leukemic M1 cells from conditioned medium of mouse fibroblast L929 cells. J Biol Chem. 1984;259:10978–10982. [PubMed] [Google Scholar]
- Wen Z, Zhong Z, Darnell JE. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- Wolf E, Kramer R, Polejaeva I, Thoenen H, Brem G. Efficient generation of chimeric mice using embryonic stem cells after long term culture in the presence of ciliary neurotrophic factor. Transgenic Res. 1994;3:152–158. doi: 10.1007/BF01973982. [DOI] [PubMed] [Google Scholar]
- Yamamori T, Fukada K, Aebersold R, Korsching S, Fann M-J, Patterson PH. The cholinergic neuronal differentiation factor from heart cells is identical to leukemia inhibitory factor. Science. 1989;246:1412–1416. doi: 10.1126/science.2512641. [DOI] [PubMed] [Google Scholar]
- Yamanaka Y, Nakajima K, Fukada T, Hibi M, Hirano T. Differentiation and growth arrest signals are generated through the cytoplasmic region of gp130 that is essential for STAT3 activation. EMBO J. 1996;15:1557–1565. [PMC free article] [PubMed] [Google Scholar]
- Yasukawa K, Saito T, Futatsugi K, Saito T, Sekimori Y, Koishihara Y, Fukui H, Ohsugi Y, Matsuda T, Yawata H, Hirano T, Taga T, Kishimoto T. Purification and characterization of soluble human IL-6 receptor expressed in CHO cells. J Biochem. 1990;108:673–676. doi: 10.1093/oxfordjournals.jbchem.a123261. [DOI] [PubMed] [Google Scholar]
- Yin T, Yang Y-C. Mitogen-activated protein kinases and ribosomal S6 protein kinases are involved in signaling pathways shared by interleukin-11, interleukin-6, leukemia inhibitory factor, and oncostatin M in mouse 3T3-L1 cells. J Biol Chem. 1994;269:3731–3738. [PubMed] [Google Scholar]
- Yoshida K, Chambers I, Nichols J, Smith A, Saito M, Yasukawa K, Shoyab M, Taga T, Kishimoto T. Maintenance of the pluripotential phenotype of embryonic stem cells through direct activation of gp130 signalling pathways. Mech Dev. 1994;45:163–171. doi: 10.1016/0925-4773(94)90030-2. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Taga T, Saito M, Suematsu S, Kumanogoh A, Tanaka T, Fujiwara H, Hirata M, Yamagami T, Nakahata T, Hirabayashi T, Yoneda Y, Tanaka K, Wang W-Z, Mori C, Shiota K, Yoshida N, Kishimoto T. Targeted disruption of gp130, a common signal transducer for the interleukin 6 family of cytokines, leads to myocardial and hematological disorders. Proc Natl Acad Sci. 1996;93:407–411. doi: 10.1073/pnas.93.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]