Abstract
Hedgehog (Hh) signaling is highly conserved in all metazoan animals and plays critical roles in many developmental processes. Dysregulation of the Hh signaling cascade has been implicated in many diseases, including cancer. Although key components of the Hh pathway have been identified, significant gaps remain in our understanding of the regulation of individual Hh signaling molecules. Here, we report the identification of novel regulators of the Hh pathway, obtained from an in vivo RNA interference (RNAi) screen in Drosophila. By selectively targeting critical genes functioning in post-translational modification systems utilizing ubiquitin (Ub) and Ub-like proteins, we identify two novel genes (dUba3 and dUbc12) that negatively regulate Hh signaling activity. We provide in vivo and in vitro evidence illustrating that dUba3 and dUbc12 are essential components of the neddylation pathway; they function in an enzyme cascade to conjugate the ubiquitin-like NEDD8 modifier to Cullin proteins. Neddylation activates the Cullin-containing ubiquitin ligase complex, which in turn promotes the degradation of Cubitus interruptus (Ci), the downstream transcription factor of the Hh pathway. Our study reveals a conserved molecular mechanism of the neddylation pathway in Drosophila and sheds light on the complex post-translational regulations in Hh signaling.
Introduction
Hedgehog (Hh) signaling is an evolutionarily conserved pathway that governs many crucial developmental events (reviewed in [1], [2]). Dysregulation of the Hh signaling pathway in humans often results in birth defects as well as tumorigenesis in adult organs (reviewed in [3], [4]). Key components of the Hh signaling cascade were initially identified through extensive genetic studies in Drosophila melanogaster, among which Hh (the ligand), Patched (Ptc, the receptor), Smoothened (Smo, the activator) and Cubitus interruptus (Ci, the transcription factor) are the most studied. The hh gene encodes a secreted protein that triggers a complex cascade of signaling events that are largely conserved from flies to mammals [1–4]. In the absence of Hh ligand, Ptc functions to suppress the activity of Smo. Due to this inhibition, Smo protein is retained in the cytoplasm, where it forms an inhibitory signaling complex with Costal2 (Cos2, a kinesin-like protein), Fused (Fu, a serine/threonine kinase) and Suppressor of Fused (SuFu, a novel regulator). This complex inhibits the activity of the transcription factor Ci by promoting its phosphorylation. Phosphorylated full-length Ci (CiFL, also known as Ci155) is subsequently processed into an N-terminal fragment (CiR, also known as Ci75) through partial degradation of the C-terminal portion of CiFL. CiR, lacking the co-activator binding domain, then moves into the nucleus to repress target gene transcription. In the presence of the Hh ligand, Hh signaling is initiated upon binding of Hh to Ptc, which releases Smo from Ptc inhibition. As a consequence, Smo protein is phosphorylated and relocalizes to the plasma membrane. This leads to dissociation of Ci from the inhibitory signaling complex, thus allowing CiFL to function as a transcription factor to activate the transcription of various Hh target genes ([1–4], and references therein).
Increasing evidence highlights a role of the ubiquitin-proteasome system (UPS) in the regulation of the stability and activity of Ci [5–13]. The majority of cellular protein degradation is subject to the UPS control, in which three different enzyme complexes, in a step-wise fashion, conjugate Ub to specific substrates. E1 (Ub-activating enzyme) and E2 (Ub-conjugating enzyme) are responsible for activating and conjugating Ub proteins, respectively. E3 functions as a Ub protein ligase to transfer Ub protein from the E2 enzyme onto specific substrates. Ubiquitinated substrates are subject to proteolysis in the 26S proteasome, and Ub proteins are recycled from the substrate by the deubiquitinating enzyme (DUB) (reviewed in [14–17]). It is well established that E3 ligases control the substrate specificity in the UPS [16], [17]. Genetic studies in Drosophila have identified two distinct E3 ligases for modulating Hh signaling, presumably targeting Ci for cleavage and/or degradation [5–8], [10–12]. Through a poorly understood mechanism, the Slimb (Supernumerary limbs)-Cul1 E3 complex is believed to regulate the activity of CiFL by promoting its partial degradation [5–8]. A second E3 complex, the Rdx (Roadkill)-Cul3 based E3 ligase, was shown to degrade Ci in Hh-responding cells [6], [10–12]. However, whether additional UPS components are involved in the regulation of Ci protein stability remains to be determined. Furthermore, the mechanism by which E3 ligases regulate Ci stability is not known.
Recent studies have revealed various ways in which the activity of these E3 ligase complexes is controlled. One such pathway relies on the covalent attachment of the Ub-like Neural precursor cell Expressed Developmentally Down-regulated protein 8 (NEDD8) to scaffolding Cullin proteins (reviewed in [18]). NEDD8 is conjugated to a conserved C-terminal lysine residue in Cullin proteins through the sequential action of a unique set of E1, E2, and E3 enzymes, a process known as neddylation [18–20]. Neddylated Cullins stimulate the ubiquitination activity of the E3 complex and prevents its association with the inhibitor CAND1 [21]. Neddylated Cullins are also subject to self-ubiquitination and degradation, thus providing a self-regulatory mechanism to maintain a proper level of ubiquitin ligase activity [22].
Drosophila wing morphogenesis is one of the most intensively investigated developmental processes for understanding Hh signaling. The stereotypical wing patterning and ample genetic tools make it a favorable system for genetic screens. Several genome-wide screens, using classical forward genetic strategies, have been reported and several novel regulators of the Hh signaling pathway were successfully identified [23–26]. Recently, large-scale in vitro RNAi screens have also been performed in cultured fly cells with promising outcomes [27], [28]. However, in vivo RNAi screens, aimed at identifying novel Hh signaling regulators, have not been reported. This lack of investigation is in contrast to what has been done for Notch signal transduction [29], [30].
Here, we report an in vivo RNAi screen to identify novel UPS regulators of Hh signaling. By assessing CiFL protein stabilization and dpp-lacZ reporter activity as simple but efficient readouts for Hh signaling, we identified two novel negative regulators of Hh signaling, each belonging to functionally distinct (E1 and E2) UPS complexes. Utilizing in vivo genetic and in vitro biochemical assays, we characterized these novel E1 and E2 genes as essential components of the neddylation pathway, which control the activity and stability of Cullin proteins and thereby regulate Ci protein stability and Hh signaling activity.
Results
The stability of endogenous Ci protein is regulated by the ubiquitin- proteasome system (UPS)
Ample genetic evidence highlights a role for UPS in the regulation of CiFL protein stability and activity, however, a previous report suggests that CiFL could also be degraded in the lysosome [31]. To distinguish the roles of these degradation pathways in the regulation of endogenous CiFL protein stability, we specifically prevented either UPS- or lysosome-mediated protein degradation, utilizing inhibitors in cultured fly cells as well as genetic manipulation in wing imaginal discs.
First, we examined the half-life of endogenous CiFL protein in cl-8 cells, a fly cell line that is responsive to Hh signaling [27], [28], [32], [33]. When treated with cychloheximide (CHX), an inhibitor of nascent protein synthesis, CiFL protein exhibited a rapid turnover with a half-life of approximately two hours (Figure 1A and B). Next, we tested whether CiFL protein degradation is regulated by a UPS- or lysosome-mediated process. cl-8 cells were incubated with specific UPS inhibitors (MG132, ALLN or lactacystin) or lysosomal inhibitors (E64, leupeptin or NH4Cl) for three hours followed by CHX treatment for an additional six hours. We found that UPS inhibitors, but not lysosomal inhibitors, were able to protect CiFL protein from CHX treatment-induced degradation in cl-8 cells (Figure 1C). To demonstrate physiological relevance of UPS-mediated CiFL degradation, we examined the effect of these inhibitors in cl-8 cells without CHX treatment. Significant accumulation of CiFL was observed when the UPS activity was attenuated (Figure 1D). Taken together, our results suggest that the UPS played a major role in regulating CiFL stability in vitro.
Figure 1. The stability of endogenous Ci is regulated by the UPS in vitro.
(A) Lysates extracted from cl-8 cells that were treated with 50 µg/ml cycloheximide (CHX) for the indicated hours (hrs) were immunoblotted (WB) with the 2A1 antibody, which specifically recognizes full-length Ci (CiFL) [87]. β-Tubulin detection served as the loading control in all figures. All immunoblotting data presented in the figures are representative of independent experiments that were performed at least three times. (B) Endogenous CiFL degraded rapidly with a half-life of approximately two hours (indicated by dashed lines) as determined by Image J densitometry. (C) CHX-induced CiFL degradation was rescued upon pre-incubation with UPS inhibitors (MG132, ALLN and Lactacystin), but not with DMSO or lysosomal inhibitors (E64, Leupeptin and NH4Cl). (D) In the absence of CHX treatment, incubation with UPS inhibitors alone resulted in significant accumulation of CiFL, while lysosomal inhibitors had no effect, thus suggesting a physiological relevance of the UPS-mediated Ci degradation in Hh signaling.
To investigate whether the stability of endogenous Ci is also subject to UPS control in vivo, we exposed Drosophila wing discs to inhibitors specific for either the UPS or the lysosome system. Hh protein is produced from cells present in the posterior compartment of the wing disc, and moves across the anterior/posterior (a/p) boundary to form a Hh morphogen gradient, thereby activating downstream target genes in anterior cells [1–4]. Those anterior cells abutting the a/p boundary receive the highest Hh signaling, while less signaling is transduced in the anterior-most cells. As a consequence, CiFL accumulates at a much higher level in anterior cells close to the a/p boundary (marked by the dashed line in Figure 2A), and levels sharply decline in more anterior cells. Consistent with our in vitro results, only the UPS inhibitors were sufficient to protect Ci protein from degradation in wing discs (Figure 2B and C; data not shown).
Figure 2. The stability of endogenous Ci is regulated by the UPS in vivo.
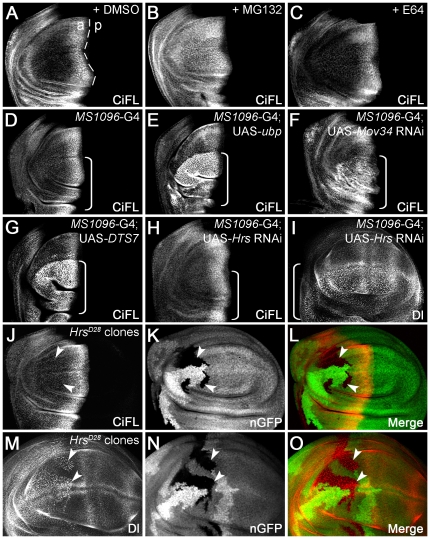
(A–C) UPS inhibition protected CiFL from degradation in the wing disc. CiFL, detected with the 2A1 antibody, accumulated abutting the anterior/posterior (a/p) boundary (marked by the dashed line) of a wildtype wing disc treated with DMSO (A). Incubation with the UPS inhibitor MG132 led to accumulation of CiFL in more anterior cells in the wing disc (B), while the lysosomal inhibitor E64 had no obvious effect (C). (D and E) Blockage of ubiquitination in the wing disc by overexpressing UAS-ubp resulted in accumulation of CiFL in more anterior cells (E). MS1096-Gal4 (G4), which was used in Figures 2, 3, 4 and 6 to drive transgene expression at a much higher level in the dorsal compartment of the wing disc (indicated by a box bracket), did not alter the stability of CiFL (D). (F–O) Genetic manipulation to disrupt UPS- or lysosome-mediated protein degradation in wing discs. Knockdown of the 19S proteasome subunit Mov34 by RNAi (F) or disrupting the function of the 20S proteasome core particle b2 subunit by expression of a dominant negative temperature-sensitive DTS7 transgene (G) in the dorsal compartment of wing discs (indicated by box brackets) resulted in significant accumulation of CiFL. In contrast, the expression pattern of CiFL was not altered when the lysosomal function was disrupted by Hrs RNAi (H, box bracket) or in HrsD28 loss-of-function somatic clones (J–L, arrowheads). As a control, accumulation of Delta protein (Dl), which normally undergoes endocytosis to the lysosome, was observed when Hrs function was disrupted (I, M–O). HrsD28 loss-of-function clones were negatively marked by nuclear GFP (nGFP; K and N). Note that the MS1096-Gal4 driver alone did not alter the expression patterns of CiFL or Dl in wing discs (see Figure S1).
To further validate the results obtained from inhibitor studies, we specifically disrupted UPS or lysosome function in wing discs by genetic manipulation. First, we overexpressed a UAS-ubp transgene, which encodes a yeast DUB enzyme that has been used in several Drosophila studies to efficiently remove Ub from ubiquitinated substrates [34–36]. As expected, CiFL was stabilized in the dorsal compartment of the wing disc (indicated by a box bracket) when ubp transgene expression was driven by the MS1096-Gal4 driver (Figure 2E; cf. Figure 2D); MS1096-Gal4 driver confers transgene expression at a much higher level in the dorsal compartment of the wing disc (Figure S1C). This experiment suggests that CiFL is ubiquitinated under normal physiological conditions and degraded in the cells away from the Hh signaling source.
Since the UPS and lysosome can both recognize ubiquitinated proteins as substrates for degradation, we further distinguished these two pathways by directly inhibiting either the proteasomal or lysosomal pathway. The most common form of the proteasome is the 26S complex, which is composed of two major subcomplexes: the 19S regulatory particle and the 20S proteolytic core particle (reviewed in [15]). RNAi-mediated knockdown of two well-conserved 19S proteasome regulatory particle subunit genes, Mov34 and Rpn6 [37], resulted in significant accumulation of CiFL protein (Figure 2F; data not shown). Similarly, when the function of 20S proteasome core particle b6 (Pros26) or b2 (Prosbeta2) subunit was disrupted in the dorsal compartment of the wing disc by the expression of dominant negative temperature-sensitive mutants DTS5 or DTS7 [38], CiFL protein was stabilized in more anterior cells (Figure 2G; data not shown). In contrast, when the essential lysosomal components, car (carnation) [39], dor (deep orange) [40], Hrs (Hepatocyte growth factor regulated tyrosine kinase substrate) or Stam (Signal transducer adaptor molecule) [41], [42], were specifically knocked down by RNAi, little if any effect on CiFL stability was observed (Figure 2H; data not shown). To rule out the possibility that the lack of Ci accumulation in the wing disc was due to the inefficiency of RNAi transgenes used, we generated loss-of-function somatic clones of lysosomal mutant alleles; clones were negatively marked by GFP (Figure 2K and N, arrowheads). Similar to our RNAi results, we found that the expression level of CiFL protein was un-altered in cells present in mutant clones (indicated by arrowheads) for dor, Hrs or Stam genes (Figure 2J–L; data not shown). As a control, Notch signaling ligand Delta (Dl) protein, which is normally endocytosed through lysosomal components [43], accumulated in the cells (i.e. dotted pattern, cf. Figure S1A) where lysosomal function was disrupted by Hrs RNAi or in loss-of-function Hrs mutant clones (Figure 2I and M–O). Taken together, the results from our in vitro and in vivo experiments strongly support a major role of UPS in regulating CiFL protein stability in Drosophila.
Targeted in vivo RNAi screen to identify novel UPS genes in regulating Hh signaling
To study the function of UPS regulators in Hh signaling, we searched the fly genome and identified a set of proteins that contain UPS-related domains [44]. A single E1 (Uba1, CG1782) [45], [46] and a single E2 (UbcD6, CG2013) [47], [48] enzyme are believed to function in UPS-mediated protein degradation in Drosophila. Seven additional E1s and 33 E2s have been identified based on the presence of signature protein domains [44], [49], [50]. Among those, Uba2 (CG7528) and Aos1 (CG12276) are believed to form a dimer and activate the Small Ub-like Modifier (SUMO) protein in Drosophila [51], [52], while Ubc9 (CG3018) functions as the E2 conjugating enzyme in the fly SUMO pathway [52], [53]. The function of other E1s or E2s are largely unknown in Drosophila. In contrast to limited numbers of E1s and E2s, there is a large array of E3 ligases that are responsible for targeting specific substrates for degradation. Based on sequence homology of their E2-binding domains, E3s can be generally classified into two major subfamilies: HECT (the homologous to E6-AP carboxyl terminus) domain- and RING (the really interesting new gene) finger domain-containing E3s [17], [44]. We identified 14 HECT- and 134 RING-containing proteins in the fly genome. We also found a set of proteins containing other domains that normally contribute to the formation of the E3 complex, including the F-box domain, Cullin domain, N-recognin domain, SKP1 domain and U-box domain. In total, we identified 8 E1, 34 E2 and 207 E3 genes in the fly genome (Table 1).
Table 1. UPS genes identified in the fly genome.
| Gene class | IPR domain | Number of genes | Number of RNAi lines |
| E1 | IPR000594 | 8 | 12 |
| E2 | IPR000608 | 34 | 61 |
| E3 (HECT) | IPR000569 | 14 | 25 |
| E3 (RING) | IPR001841 | 134 | 219 |
| E3 (Cullin) | IPR001373 | 7 | 14 |
| E3 (F-Box) | IPR001810 | 37 | 59 |
| E3 (SKP1) | IPR001232 | 8 | 13 |
| E3 (U-Box) | IPR003613 | 5 | 9 |
| E3 (N-recognin) | IPR003126 | 2 | 4 |
| In total | 248* | 414* |
*Note that CG15437 protein contains both an E2 domain and a F-Box domain.
The availability of two collections of transgenic RNAi libraries housed in the VDRC (Vienna Drosophila RNAi Center, Austria) [54] and the NIG-Fly Stock Center (National Institute of Genetics, Japan), makes it possible to screen nearly all UPS regulators that we identified for their effects on Hh signaling in vivo. We obtained 414 RNAi lines targeting 238 genes out of a total of 248 UPS genes (Table S1). Hh signaling functions as an important morphogen as well as a powerful mitogen during fly wing development. We therefore examined adult wing blade patterning and larval wing disc development as efficient and reliable readouts in our in vivo RNAi screen to identify Hh signaling-specific UPS genes.
We first knocked down the expression of individual UPS genes in wing discs by RNAi using the MS1096-Gal4 driver, and then examined the resulting adult wing blade phenotypes. We found that reducing the expression of 72 UPS genes altered adult wing morphology (Table S1). As several developmental signaling systems, including Hh, Wnt, TGF-β and Notch signaling, collaborate to control wing morphogenesis, a secondary screen was conducted to identify Hh signaling-specific UPS regulators. We evaluated the distribution patterns of CiFL in wing discs overexpressing UPS RNAi. To correlate Ci expression with Hh signaling activity, we also examined the expression of a decapentaplegic (dpp)-lacZ enhancer trap reporter; dpp is a direct transcriptional target of Hh signaling in the wing disc. To further validate our screen results, we investigated whether the effect of UPS regulation is direct by examining CiFL stability in UPS RNAi overexpressing clones (i.e. flip-out clones) [55], [56]. Only those UPS genes that cell-autonomously affect CiFL protein were chosen as true Ci regulators.
Our RNAi screen successfully identified E3 Ub ligase members which are known UPS regulators of CiFL stability [5–8], including slimb, Cul1 (lin 19), and Roc1a (Figure S2; data not shown). These genes encode Slimb-Cul1 complex components that have been shown to destabilize CiFL by promoting proteasomal degradation. In addition, we identified two novel genes (CG13343 and CG7375) that regulate CiFL stability and Hh signaling activity. To date, these genes are uncharacterized and their functions poorly understood. We therefore conducted genetic and biochemical studies to understand the molecular functions of these novel genes in Drosophila.
CG13343 functions as a neddylation E1-activating enzyme in Drosophila
The CG13343 transcript was uniformly expressed in the wing disc (Figure S3B). Reduced expression of CG13343 by RNAi in wing discs at 29°C resulted in lethality at the pupal stage. At 25°C, several escapers survived into adulthood, with their wing blades folded and damaged (data not shown). To accurately determine whether CG13343 played a role in Hh signaling, we investigated the effect of CG13343 knockdown in the wing disc, a primordium of the adult wing. During patterning in the developing wing, distinct target genes are activated in response to different strengths of Hh signaling activity (Figure 3A–C) [33]. Low-threshold Hh signaling stabilizes CiFL. The expression of one of these targets, dpp, is induced by intermediate-threshold Hh signaling, whereas two other targets, ptc and collier (Col), respond to high-threshold Hh signaling. Here, we examined the effects of reduced CG13343 activity on CiFL, dpp-lacZ reporter and Col protein in the wing disc.
Figure 3. CG13343 negatively regulates CiFL stability and Hh signaling.
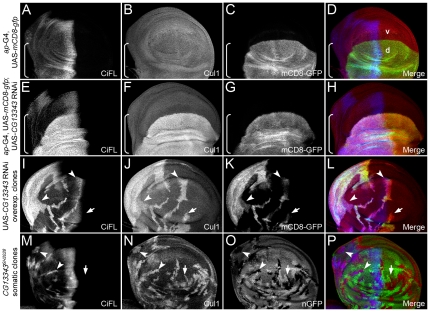
(A–C) Stabilization of CiFL (A), induction of Hh signaling reporter dpp-lacZ (B) and Col protein (C) abutting the a/p boundary (indicated by the dashed line in A) in wing discs correlate with low-, intermediate- and high-threshold Hh signaling activity, respectively. Box brackets mark the dorsal compartment of wing discs where MS1096-Gal4 exhibits a much higher activity (also see Figure S1C). (D–O) CG13343 negatively regulates Hh signaling. RNAi knockdown of CG13343 in the dorsal compartment of the wing disc (box bracket) led to accumulation of CiFL protein (D) and expansion of dpp-lacZ activity (E). Similarly, CiFL stabilization (G) and ectopic Col expression (J) were observed in CG13343 RNAi-overexpressing cells (positively marked by mCD8-GFP in H and K) in anterior clones (G–L, arrowheads), but not in posterior clones (arrow). Ectopic Col activation was also evident in CG13343SH2028 somatic clones (negatively marked by nGFP in N) located in the anterior compartment of the wing disc (M, arrowheads). Note that Ci and Col are not expressed in posterior cells.
Knockdown of CG13343 expression by RNAi in the wing disc resulted in the stabilization of CiFL (Figure 3D) and the activation of Hh signaling: dpp-lacZ (Figure 3E) and Col protein (data not shown) were ectopically expressed. Notably, CG13343 RNAi had little effect on the abundance of Ci75, the repressor form of Ci (Figure S4A). Further analyses of cells in CG13343 RNAi overexpressing clones, which were positively marked by mCD8-GFP (Figure 3H and K), confirmed that CG13343 functions cell autonomously to regulate CiFL stability (Figure 3G, arrowheads) and Col activation (Figure 3J, arrowheads). Three independent RNAi transgenic lines were tested and similar effects on CiFL stabilization and target gene activation were observed (Figure 3; data not shown). The efficiency and specificity of CG13343 RNAi was examined by semi-quantitative RT-PCR (Figure S5A).
To validate the CG13343 RNAi knockdown results, we generated loss-of-function somatic CG13343SH2028 mutant clones in the wing disc, which were negatively marked by nGFP (Figure 3N). The CG13343SH2028 is a recessive-lethal mutant that arose from a P-element insertion at the position 34 bps immediately after the ATG start codon in the CG13343 locus [57]. RT-PCR results indicated that CG13343SH2028 represented a null mutation for CG13343 (Figure S5C). Consistent with the CG13343 RNAi results, we found that CiFL was stabilized (Figure 4M) and Col was ectopically expressed in CG13343SH2028 clones (Figure 3M, arrowheads), indicating that CG13343 was required to control both low- and high-threshold Hh signaling.
Figure 4. The stability of Cul1 is regulated by CG13343.
(A–D) Uniform expression of Cul1 protein (B) in a wing disc expressing the ap-Gal4 driver. UAS-mCD8-gfp expression (C) reflects the ap-Gal4 activity in the dorsal (d) compartment (marked by the box bracket). (E–P) Regulation of Cul1 protein stability by CG13343. Knockdown of CG13343 expression by RNAi in the dorsal compartment of the wind disc resulted in significant accumulation of CiFL (E) and Cul1 (F). Analysis of CG13343 RNAi-overexpressing clones (positively marked by mCD8-GFP in K) in the anterior compartment (arrowheads) confirmed that the stability of CiFL (I) and Cul1 (J) was cell-autonomously regulated by CG13343. Similarly, stabilized CiFL (M, arrowheads) and Cul1 (N, arrowheads and arrow) were observed in CG13343SH2028 loss-of-function somatic clones (negatively marked by nGFP in O) in the wing disc.
Utilizing a protein domain search, we found that the CG13343 protein was highly conserved from yeast to human, and contain a Ub-activation domain normally found in Ub-activating E1 enzymes (Figure S6). Uba3 is the CG13343 ortholog in yeast and human, which functions as the NEDD8 activating E1 enzymes in the neddylation process [58]. The best-characterized substrates for neddylation are Cullin family proteins [18–22], [58]. Neddylation of Cullin proteins results in Cullin activation, but also leads to its own destabilization [18–22]. To investigate whether the function of CG13343 mimicked its yeast and vertebrate counterparts we examined the stability of Cullin proteins in wing discs. RNAi-mediated knockdown of CG13343 was carried out in the dorsal compartment of wing discs using an ap-Gal4 driver (expression pattern of ap-Gal4 is shown in Figure 4C). Notably, only one commercially available antibody raised against vertebrate Cullins (i.e. α-Cul1) worked for immunohistochemistry in wing discs. As expected, Cul1 accumulated in dorsal compartment cells (marked by box brackets) where CG13343 expression was down-regulated (Figure 4F, cf. Figure 4B). Consistent with this outcome, increased Cul1 protein expression (Figure 4J) was observed in clones overexpressing CG13343 RNAi (i.e. mCD8-GFP-positive cells in Figure 4K) as well as in loss-of-function somatic CG13343SH2028 clones (i.e. nGFP-negative cells in Figure 4N).
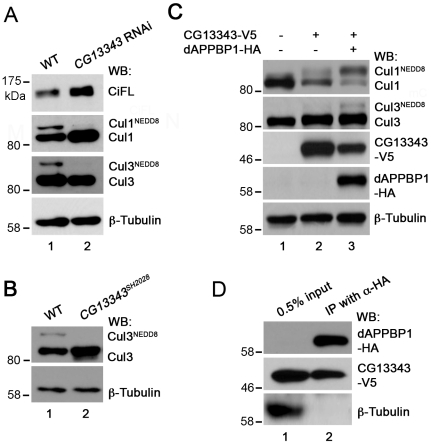
To investigate whether CG13343 protein was required for Cullin neddylation in vivo, we exploited the fact that neddylated Cullin migrates slower than its unmodified counterpart on SDS-PAGE [7], [22], [59]. To examine the extent of Cullin neddylation, antibodies specific for Cul1 and Cul3 were used. In wildtype wing disc lysates, both Cul1 and Cul3 proteins were neddylated (Figure 5A, lane 1). When CG13343 expression in wing discs was knocked down by RNAi, which stabilized CiFL, we found that neddylation of Cul1 or Cul3 was largely reduced (lane 2). These results are consistent with previous reports illustrating that neddylated Cullins are required for Ci degradation [22], [59]. As neddylated and activated Cullins are less stable, we observed that wing disc lysates overexpressing CG13343 RNAi had higher levels of Cul1, presumably resulting from stabilization of un-neddylated Cul1. However, it is interesting to note that the amount of un-neddylated Cul3 was not obviously changed in cells expressing CG13343 RNAi. The different sensitivity between Cul1 and Cul3 stabilization in response to CG13343 RNAi could be due to incomplete depletion of CG13343. To address this possibility, we examined the neddylation and stabilization of Cul3 in protein lysates extracted from first-instar larve homozygous of CG13343SH2028. As predicted, the accumulation of un-neddylated Cul3 protein was evident in the CG13343SH2028 mutant (Figure 5B, lane 2). These results confirmed that CG13343 played a role in the neddylation pathway. To definitively demonstrate that CG13343 protein functioned as a neddylation E1 enzyme, we examined Cullin neddylation using a cell-free neddylation assay. Minimal neddylation on Cul1 or Cul3 was detected in fly cl-8 cell lysates after one-hour incubation in the presence of purified E2 enzyme, NEDD8 and ATP (Figure 5C, lane 1). This approach provided a relatively clean system for testing the capacity of CG13343 as a neddylation E1 enzyme in vitro. When V5-tagged CG13343 (CG13343-V5) was overexpressed in cl-8 cells, we observed obvious, albeit weak, neddylation on Cul1, but not on Cul3 (lane 2).
Figure 5. CG13343 protein functions as the E1 enzyme for Cullin neddylation.
(A) Immunoblot analysis (WB) of lysates extracted from wildtype (WT) wing discs (lane 1) or wing discs overexpressing CG13343 RNAi driven by the MS1096-Gal4 driver (lane 2). CG13343 RNAi led to accumulation of both CiFL and Cul1 (lane 2). Furthermore, stabilized Cul1 was predominantly un-neddylated (lane 2). Neddylation of Cul3, another Cullin family protein, was also reduced. However, there was no accumulation of un-neddylated Cul3 (lane 2). Note that this Cul3 antibody does not work for immunohistochemistry in wing discs. (B) Immunoblot analysis of lysates extracted from wildtype (lane 1) or homozygous loss-of-function CG13343SH2028 first-instar larvae (lane 2). Neddylation of Cul3 protein was abolished and un-neddylated Cul3 was stabilized. (C) The E1 activity of CG13343 for Cullin neddylation. In an in vitro neddylation assay, purified human Ubc12 was used as E2 and cl-8 cell lysate provided the source for Cullin proteins. In the absence of added CG13343-V5, minimal neddylation activity was observed (lane 1). Overexpressed CG13343-V5 in cl-8 cells was sufficient to function as an E1 enzyme to neddylate Cul1, but not Cul3 (lane 2). The neddylation activity of CG13343-V5 was greatly enhanced when CG13343-V5 was co-expressed with dAPPBP1-HA in cl-8 cells (lane 3), resulting in the neddylation of both Cul1 and Cul3. Note that equal amounts of plasmid DNA were transfected in cl-8 cells, i.e. half amount of CG13343-V5 plasmid was transfected in lane 3 compared to that in lane 2. (D) CG13343 protein forms an E1 complex with dAPPBP1. cl-8 cells were transiently transfected with dAPPBP1-HA and CG13343-V5, and 0.5% of the cell lysate was loaded as input (lane 1). An anti-HA antibody was used for immunoprecipitation (IP) (lane 2).
Structural studies reveal that human Uba3 forms a heterodimer with β-Amyloid precursor protein binding protein 1 (APPBP1), and together they function as an active neddylation E1 complex [18–21]. Genetic evidence suggests that the Drosophila homolog of APPBP1 (dAPPBP1) may participate in the neddylation process [59]. We found that, when co-expressed in cl-8 cells, CG13343 protein was able to form a complex with dAPPBP1 (Figure 5D). Furthermore, this complex sufficiently acted as a potent neddylation E1 enzyme to neddylate both Cul1 and Cul3 (Figure 5C, lane 3; cf. lane 2). Taken together, our genetic and biochemical assays provide strong evidence that CG13343 protein was a functional homolog of Uba3 as it acted together with dAPPBP1 to function as a NEDD8 E1-activating enzyme. Hence, we propose the renaming of CG13343 to dUba3 (Drosophila Uba3).
CG7375 functions as a neddylation E2-conjugating enzyme in Drosophila
The second gene identified from our targeted RNAi screen was CG7375. Similar to CG13343, CG7375 is uniformly expressed in the wing disc (Figure S3E). knockdown of CG7375 expression by RNAi in the dorsal compartment of wing discs activated Hh signaling: elevated CiFL protein stabilization (Figure 6A and Figure S4B) as well as expanded dpp-lacZ (Figure 6B) and Col expression (data not shown) were observed. Analysis of cells in CG7375 RNAi overexpressing clones revealed that CG7375 acted cell autonomously to regulate CiFL stability (Figure 6D, arrowhead) and Col expression (Figure 6G, arrowhead).
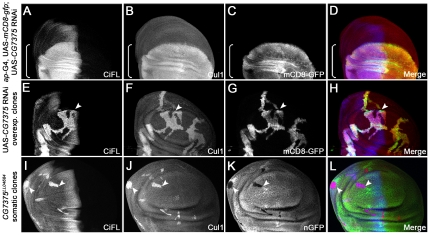
Figure 6. CG7375 negatively regulates CiFL stability and Hh signaling.
RNAi knockdown of CG7375 in the dorsal compartment of the wing disc (box bracket) led to accumulation of CiFL (A) and expansion of dpp-lacZ (B). Analysis of CG7375 RNAi-overexpressing clones (positively marked by mCD8-GFP in E and H) confirmed that CiFL stability (D, arrowhead) and Col expression (G, arrowhead) were cell-autonomously regulated by CG7375 in the anterior compartment of the wing disc. Similarly, ectopic Col expression (J, arrowheads) was observed in loss-of-function CG7375LL04684 somatic clones (negatively marked by nGFP in K) in the wing disc.
To validate the CG7375 RNAi knockdown results, we generated loss-of-function somatic CG7375LL04684 mutant clones in the wing disc, which were negatively marked by nGFP (Figure 6K). The CG7375LL04684 is a recessive-lethal mutant that arose from a piggyBac insertion at the position 19 bps immediately after the ATG start codon in the CG7375 locus [60]. RT-PCR results indicated that CG7375LL04684 represented a null mutation for CG7375 (Figure S5D). Consistent with the CG7375 RNAi results, we found that CiFL was stabilized (Figure 7I) and Col was activated in CG7375LL04684 clones (Figure 6J, arrowheads). These results suggested that CG7375 behaved similarly to CG13343 to regulate Ci stability and Hh signaling activation.
Figure 7. CG7375 controls Cul1 protein stability.
(A–D) Knockdown of CG7375 expression by RNAi in the dorsal compartment of the wing disc (marked by mCD8-GFP expression in C) resulted in significant accumulation of CiFL (A) and Cul1 (B). (E–L) Cell-autonomous stabilization of CiFL (E and I) and Cul1 (F and J) was observed when CG7375 function was disrupted in CG7375 RNAi-overexpressing clones (arrowhead in E–H; positively marked by mCD8-GFP in G) or in CG7375LL04684 loss-of-function somatic clones in wing discs (arrowheads in I–L; negatively marked by nGFP in K).
CG7375 protein harbors two distinct functional motifs: the E1 binding motif and E2 activity core (Figure S7). Recently, CG7375 has been predicted to be involved in neddylation, most likely acting as an E2 NEDD8 conjugating enzyme [61]. This hypothesis partially relies on the fact that CG7375 contains a small N-terminal extension shared only by E2 Ubc12 family members specific for NEDD8 conjugation [62]. However, to date, no functional studies have been conducted to demonstrate that CG7375 acts as the Drosophila NEDD8 E2 enzymes [61], [63].
To determine whether CG7375 protein functions in fly neddylation, we first examined the expression pattern of Cul1 in wing discs where CG7375 expression was reduced. Both CiFL and Cul1 protein levels were significantly increased in the dorsal compartment of the disc where CG7375 RNAi was overexpressed by the ap-Gal4 driver (Figure 7A–D). Utilizing CG7375 RNAi overexpressing clones (Figure 7E–H, arrowhead) as well as loss-of-function somatic CG7375LL04684 clones (Figure 7I–L, arrowheads), we demonstrated that the effect of CG7375 on the stabilization of Ci and Cul1 was cell autonomous.
To examine whether the elevated Cullin expression in wing discs was due to reduced neddylation, we compared Cullin neddylation in protein lysates extracted from wing discs with or without overexpressed CG7375 RNAi. As expected, Cul1 neddylation was significantly reduced (Figure 8A, lane 2; cf. lane 1), which resulted in significant accumulation of Cul1 in wing discs expressing CG7375 RNAi (Figure 7). However, CG7375 RNAi had little effect on the stabilization of un-neddylated Cul3 (lane 2). Consistent with the result observed in dUba3 loss-of-function larvae (Figure 5B), we found that un-neddylated Cul3 accumulated in protein lysates extracted from the loss-of-function CG7375LL04684 mutant larvae (Figure 8B, lane 2).
Figure 8. CG7375 protein functions as the E2 enzyme for Cullin neddylation.
(A) Immunoblot analysis (WB) of lysates extracted from wildtype (WT) wing discs (lane 1) or wing discs overexpressing CG7375 RNAi driven by the MS1096-Gal4 driver (lane 2). CG7375 RNAi led to accumulation of both CiFL and un-neddylated Cul1 (lane 2). Similarly, neddylation of Cul3 was reduced, but the amount of un-neddylated Cul3 was not obviously changed (lane 2). (B) Immunoblot analysis of lysates extracted from wildtype (lane 1) or homozygous CG7375LL04684 first-instar larvae (lane 2). Neddylation of Cul3 protein was abolished and un-neddylated Cul3 was stabilized. (C and D) The E2 activity of CG7375 for Cullin neddylation. In an in vitro neddylation assay, purified human Uba3/APPBP1 complex was used as E1 and lysates extracted from wing discs expressing CG7375 RNAi provided the source for Cullin proteins. CG7375 RNAi wing disc lysates did not display neddylation activity (as 90% endogenous CG7375 was knocked down by CG7375 RNAi; Figure S5C), unless purified GST-CG7375 protein was added (lane 2 in C; lane 3 in D): both Cu1 and Cul3 were neddylated. Purified GST protein was used as a negative control (lane 1 in C and D). The neddylation activity of GST-CG7375 was dependent on the presence of purified E1 complex (lane 3 in C) and ATP (lane 4 in C). The N-terminus of human ortholog of CG7375 (Ubc12) is required to selectively recruit NEDD8's E1 to promote thioester formation between E2 and NEDD8 (Figure S7). Deletion of this conserved N terminal motif in GST-CG7375DN abolished its neddylation E2 activity (lane 2 in D). (E) In vitro reconstitution of Drosophila neddylation cascade. Cul1 and Cul3 were neddylated when both E1 complex (CG13343-V5 and dAPPBP1-HA produced in cl-8 cells) and E2 enzyme (GST-CG7375) were added to cl-8 lysates, which provided the source of Cullins (lane 4). Adding E1 (lane 2) or E2 (lane 3) alone did not result in neddylation of Cul1 or Cul3.
To demonstrate that CG7375 protein functions as a neddylation E2 enzyme, we tested whether CG7375 protein could transfer NEDD8 to Cullin proteins in an in vitro cell-free assay. Cullins were provided from lysates extracted from wing discs whose endogenous CG7375 mRNA was significantly reduced by CG7375 RNAi (Figure S5B). In this situation, neither Cul1 nor Cul3 was notably neddylated (Figure 8A, lane 2). Addition of GST protein alone to the lysates did not result in Cullin neddylation (Figure 8C, lane 1). In contrast, addition of purified GST-CG7375 fusion protein was sufficient to conjugate NEDD8 to almost all Cul1 protein present in wing disc lysates (lane 2). Furthermore, neddylation was dependent on the presence of a functional E1 enzyme complex (lane 3) and ATP (lane 4). The ability of CG7375 to conjugate NEDD8 was also true for Cul3, although the overall degree of Cul3 neddylation was weaker than that of Cul1 (lane 2).
Recent crystal structure studies on the human E2 neddylation enzyme, Ubc12, suggest that a unique motif present in the N terminus of the proteins (Figure S7) is crucial for recruiting NEDD8's E1 enzyme to promote thioester formation between Ubc12 and NEDD8 [62]. We therefore tested whether this motif plays a conserved role in Drosophila. We found that deletion of the characteristic N terminal motif (CG7375DN) completely abolished CG7375 protein's neddylation E2 activity (Figure 8D, lane 2). Together, our genetic and in vitro biochemical analyses demonstrate that CG7375 is a bona fide NEDD8 E2-conjugating enzyme. Thus, we propose the renaming of CG7375 to dUbc12.
In vertebrates, both Uba3-APPBP1 and Ubc12 are required for NEDD8 conjugation to Cullin proteins. Consistent with this, CG13343-dAPPBP1 E1 complex (Figure 8E, lane 2) or purified GST-CG7375 alone (lane 3) was unable to promote Cul1 or Cul3 neddylation in a cell-free neddylation assay, unless both enzymes were present (lane 4). These experiments indicate that dUba3 (i.e. CG13343) and dUbc12 (i.e. CG7375) function together in an enzyme cascade for neddylation in Drosophila.
Discussion
In this study, we utilized a targeted RNAi screen and identified several candidate UPS regulators in patterning of the Drosophila wing. Focused investigation on two candidate genes, CG13343 and CG7375, demonstrated that they played a critical role in Hh signal transduction by controlling the stability of Hh signaling transcription factor Ci to regulate both low- and high-threshold Hh signaling. Importantly, we provided genetic and biochemical evidence that protein products of these two genes participated in a conserved protein degradation process in Drosophila, functioning as the NEDD8 E1-activating and E2-conjugating enzymes in neddylation, respectively. Consistent with our biochemical analysis, reduction of dNEDD8 modifier was able to elicit a full spectrum of Hh pathway responses in the wing disc (Figure S8). Thus, we propose a model whereby the neddylation pathway negatively regulates Hh signaling at the level of Ci stability (Fig. S9). The activity of Cul1-based E3 ubiquitin ligase complex is activated by neddylation, which in turn promotes proteolytic cleavage of CiFL to Ci75, thereby antagonizing low-to-intermediate threshold Hh signaling. On the other hand, neddylation activates Cul3-based E3 ubiquitin ligase complex, which degrades CiFL to prevent high-threshold Hh signaling.
A general requirement of the UPS in the regulation of Ci protein stability
Hh signaling activates downstream target genes in a de-repression manner, thereby protecting the transcription factor Ci from degradation and/or processing. Two distinct ubiquitin ligase complexes, Slimb-Cul1 and Rdx-Cul3, have been identified as key regulators of Ci stability [5–13]. Both complexes recognize CiFL as the substrate, targeting it for either partial or complete degradation. Two subcellular compartments, the lysosome and proteasome, are important for regulated protein degradation. Although there is general consensus that Ci degradation takes place in the proteasome, in vivo evidence directly demonstrating the requirement of the proteasome for endogenous Ci degradation is lacking.
Here, we examined the stability of endogenous CiFL when the UPS or lysosome function was disrupted either by treating cultured fly cells with specific inhibitors or by genetically manipulating wing discs. Our data, consistent with previous studies on Ci degradation/processing [5–13], strongly support a major role of the UPS in controlling endogenous CiFL stability. However, this conclusion is in direct conflict with a previous study by Dai et al., suggesting that a multivesicular body-localizing protein Debra (Dbr) might direct CiFL degradation to the lysosome [31].
To solve this apparent discrepancy, we carefully compare the experimental conditions we employed to examine CiFL stability in cl-8 cells with those in Dai et al. Ectopically expressed HA-CiFL in Dai et al. exhibits a half-life of 15 hours, which is significantly longer than that of endogenous CiFL (approximately two hours, Figure 1B) in this study as well as that of the overexpressed Myc-CiFL (approximately three hours) demonstrated by Jia et al. [64]. Thus, it is not surprising that ectopic HA-CiFL was unable to respond to either UPS inhibitor MG132 or lysosomal inhibitor E64 treatment in cl-8 cells unless additional Dbr proteins were provided. In contrast, endogenous CiFL in cl-8 cells (Figure 1) and in wing discs (Figure 2), as well as over-expressed Myc-CiFL in cl-8 cells [7], [10], [64] can be readily protected from degradation by inhibiting the UPS function. Our conclusion is further supported by the fact that, overexpressed Gli1, one of the vertebrate orthologs of Ci, is also subject to proteasomal regulation [65]. We suspect that the Dbr-mediated lysosomal degradation of HA-CiFL may reflect a backup/alternative mechanism when UPS regulation is overwhelmed by highly overexpressed HA-CiFL in vitro. In the future, it will be interesting to investigate the molecular mechanism of Dbr-mediated degradation of endogenous Ci, and more importantly, the relationship with the UPS-regulated Ci degradation in vivo.
Differential neddylation of Cul1 and Cul3 in Hh signaling
Our work, together with other studies [22], [59], [66], [67], demonstrates that the activities of both Cul1 and Cul3 are controlled by neddylation in Drosophila. However, it should be noted that there are differences in their respective neddylation patterns in response to reduced neddylation. In hypomorphic dUba3 or dUbc12 RNAi-expressing wing discs, reduced neddylation led to high-level accumulation of un-neddylated Cul1, consistent with the notion that neddylated Cullin proteins are unstable [20]. However, the levels of un-neddylated Cul3 in this sensitized background seemed to be unaffected in wing disc lysates, although the reduction of neddylated Cul3 was evident (Figure 5A and Figure 8A). In contrast, when the neddylation process was compromised in dUba3 or dUbc12 mutant larvae, both Cul proteins were stabilized (Figure 5B and Figure 8B; data not shown). Similarly, much less neddylation of Cul3 is observed than that for Cul1 in our in vitro neddylation assays (Figure 5C and Figure 8C–E) as well as in vertebrate Cullins when tested in an in vitro assay [68]. This differential regulation of Cul1 and Cul3 is also observed in the fly mutants of CSN5 and Int6, two genes that are essential for de-neddylation [22], [66]. Although our results could simply reflect that Cul3 neddylation requires a much higher neddylation activity than Cul1, we believe that intrinsic differences may exist between Cul1 and Cul3 proteins. Neddylated Cul3 might degrade more rapidly than neddylated Cul1, which could explain distinct behaviors of Cul1 and Cul3 in our study. Indeed, the percentage of neddylated Cul3 in the total pool of Cul3 proteins in wildtype wing discs (i.e. 50% lower as determined by Image J densitometry in this study) and in brain lobes and eye discs [67] is significantly lower than that of Cul1, suggesting differential stability of neddylated Cullins. Further analyses are required to test this hypothesis and to elucidate the functional significance of differentially regulated Cullin proteins.
Cul1 and Cul3 are required for regulating Ci stability, but they function in very different manners (Figure S9). The Slimb-Cul1 complex destabilizes Ci in the absence of Hh signaling through direct binding between Slimb and phosphorylated Ci [6], [7]. Hh signaling prevents Ci phosphorylation and thus protects Ci from Slimb-Cul1 mediated degradation, as seen in the cells in the anterior compartment of wing and eye discs that receive Hh from the posterior compartment. The Rdx-Cul3 complex, on the other hand, constitutively degrades Ci independent of phosphorylation modifications even in the presence of Hh signaling [9], [10], [69]–[71]. Therefore, the activity of the Rdx-Cul3 complex has to be strictly controlled to ensure a proper Hh signaling outcome. One way to restrict Rdx-Cul3 activity is to utilize Rdx as a direct Hh signaling target [9], [10], [70]. In cells receiving low to intermediate levels of Hh signaling, Rdx is not expressed. In cells receiving the highest level of Hh signaling, Rdx expression is induced, thus allowing the formation of Rdx-Cul3 complex to degrade un-phosphorylated Ci. As the result, cells abutting the a/p boundary in the wing disc and posterior to the morphogenic furrow in the eye disc express much lower levels of Ci. Maintaining a low but steady level of Ci in these cells is crucial for transducing high-threshold Hh signaling, as further downregulation or abnormal accumulation of Ci proteins leads to patterning defects in the wing and eye [9], [10], [69], [70]. Our hypothesis that neddylated Cul3 is highly labile may, in part, provide a solution. We believe that neddylated Cul3 could act as an intrinsic brake to prevent Ci from complete degradation by the Rdx-Cul3 complex. Interestingly, a similar mechanism may also exist in the regulation of the cyclin E activity. Phosphorylated cyclin E is subject to Cul1-mediated degradation [72–75], whilst a Cul3-based complex targets cyclin E for ubiquitination independent of protein phosphorylation [76], [77]. Further studies will reveal the impact of such differential activity of neddylated Cul1 and Cul3 in Hh signaling as well as cell cycle control.
A conserved role of the neddylation process in regulating developmental signaling
NEDD8 was originally identified as one of a set of genes that is highly expressed in the embryonic mouse brain and was found to be down-regulated during development [78]. Subsequently, it was realized that NEDD8 is a Ub-like (UBL) protein, and is highly conserved in eukaryotes (reviewed in [79]). NEDD8 is ubiquitously expressed in most tissues and is essential for the viability of most model organisms (reviewed in [80]). Among the UBL family proteins, NEDD8 exhibits the highest protein sequence similarity with Ub and is conjugated to substrate proteins through a very similar enzyme cascade. However, the neddylation process utilizes its own set of enzymes to insure a specific conjugation pathway. Contrary to Ub, which is processed by a single E1 protein Uba1, a heterodimer of APPBP1 and Uba3 is required for NEDD8 activation. APPBP1 is homologous to the N-terminus of the Uba1 protein, whereas Uba3 is to the C-terminus [18–20], [49]. Studies in several organisms indicate that Ubc12 functions exclusively as the NEDD8 E2 enzyme [50], [78]–[80]. Much is known about the importance of the neddylation pathway in the regulation of developmental processes in Drosophila [6], [22], [59], [63], [66], [67], [69], but neither the identities nor the mechanisms of the fly NEDD8 E1 and E2 enzymes are known.
Our genetic and biochemical analyses demonstrate that CG13343 and CG7375 are functional orthologs of Uba3 and Ubc12 in Drosophila. Ubiquitous knockdown of either dUba3 or dUbc12 by driving RNAi transgenes with tub-Gal4 or act-Gal4 results in early larval lethality (data not shown). Similarly, homozygous mutants of the CG13343SH2028 or CG7375LL04684 allele die at early larval stages (data not shown). Our results are consistent with observations that null mutants of dNEDD8, dAPPBP1, or components of the de-neddylation CSN complex also die in early larval stages [6], [13], [22], [59], highlighting a critical role of neddylation in normal animal development.
The best-characterized neddylation substrates are the Cullin family proteins, which serve as the scaffold for the SCF ubiquitin E3 complexes. The SCF E3s regulate numerous developmentally important substrates, such as cell cycle regulator cyclin E [72–77] and signaling transduction effectors, including Ci [5–13] and Armadillo/β-catenin [81], [82]. NEDD8 has also been implicated in transcriptional regulation, by neddylating another substrate, the p53 tumor suppressor protein; neddylated p53 inhibits its own transcription activity [83], [84]. The number of identified NE88-target proteins is growing and interestingly a recent study in Drosophila indicates that many non-Cullin proteins can be neddylated in vivo [85]. The mechanisms regulating the neddylation pathway and the roles these processes in modulating animal development is more complicated than we previously anticipated. Further studies of dUba3 and dUbc12 in a highly amenable genetic model system, like Drosophila, will contribute substantially to our understanding of how neddylation functions in development.
Materials and Methods
Fly genetics
Act5C>yw>Gal4, ap-Gal4, MS1096-Gal4, and dpp-lacZ were described previously [33], [55]. Transgenic RNAi flies targeting predicted UPS genes (Table S1) were obtained from the Vienna Drosophila RNAi Center (VDRC) [54] and the Fly Stocks of National Institute of Genetics (NIG-Fly). Targeted RNAi screen was conducted by crossing individual RNAi lines with MS1096-Gal4 at 29°C for altered adult wing blade morphogenesis. For those lines displaying defective wing patterning, CiFL protein stabilization and dpp-lacZ induction were examined in third-instar larval wing discs for their effects on Hh signaling.
Specific fly strains and cross conditions as shown in the figures are listed below. For Figure 2E and F, UAS-ubp (gift of Liqun Luo) [34], UAS-Mov34 RNAi (V26183) or UAS-Rpn6 RNAi (V18021) was crossed with MS1096-Gal4 at 18°C. For Figure 2G, UAS-DTS5 or UAS-DTS7 [38] was crossed with MS1096-Gal4 at 29°C. For Figure 2H and I, UAS-Hrs RNAi (Bloomington 28964 and 28026), UAS-dor RNAi (V33733 and V107053), UAS-car RNAi (TRiP HMS00972) or UAS-STAM RNAi (V22497) was crossed with MS1096-Gal4 at 29°C. For Figure 2J–O, second-instar larvae from the crosses hs-flp;; ubi-gfp, FRT40A×HrsD28, FRT40A/Gla, Bc (gift of Hugo Bellen) [41], or hs-flp;; ubi-gfp, FRT40A×STAM2L3297, FRT40A/CyO (gift of Markus Affolter) [42], or ubi-gfp, hs-flp, FRT19A×dor8, FRT19A/FM7 (gift of Helmut Krämer) [39] were heat-shocked at 37°C for one hour to generate loss-of-function somatic clones in the wing disc. For Figure 3D–F, UAS-CG13343 RNAi lines (V17137, V17139 and V105141) were crossed with MS1096-Gal4; dpp-lacZ/CyO at 29°C. For Figure 3G–L and Figure 4 I–L, overexpressing (“flip-out”) clones were generated by heat-shocking second-instar larvae from the crosses of hs-flp; Act5C>yw>Gal4, UAS-mCD8-gfp×UAS-CG13343 RNAi lines at 37°C for one hour. For Figure 3M–O and 4M–P, late second-instar progeny of the cross hs-flp; FRT42D, ubi-gfp×FRT42D, CG13343SH2028/CyO (Drosophila Genetic Resource Center at Kyoto, 122114) were heat-shocked at 37°C for one hour. For Figure 4E–H, UAS-CG13343 RNAi lines were crossed with ap-Gal4, UAS-mCD8-gfp at 29°C. For Figure 6D–I and Figure 7E–H, the same heat-shocking condition was used as for Figure 4I–L. For Figure 6A–C, UAS-CG7375 RNAi (V35219, V35220 and V100761) was crossed with MS1096-Gal4; dpp-lacZ/CyO at 29°C. For Figure 7A–D, UAS-CG7375 RNAi lines were crossed with ap-Gal4, UAS-mCD8-gfp at 29°C. For Figure 6J–L and 7I–L, late second-instar progeny of the cross hs-flp;; ubi-gfp, FRT2A×CG7375LL04684, FRT2A / TM6B (Drosophila Genetic Resource Center at Kyoto, 141316) were heat-shocked at 37°C for one hour. For Figure S8, late second-instar progeny of the cross hs-flp; ubi-gfp, FRT40A×dNEDD8AN015, FRT40A / CyO (gift of Cheng-Ting Chien) [6] were heat-shocked at 37°C for one hour.
Molecular biology
Standard PCR method was used to amplify CG13343, CG7375 and dAPPBP1 coding sequences using cDNAs synthesized with mRNAs extracted from yw third-instar larvae. CG13343-V5 and dAPPBP1-HA were cloned into pIZ-V5 vector (Invitrogen) for overexpressing in cl-8 cells. CG7375 or CG7375ΔN (amino acids 2–23 were deleted) were cloned into pGST- parallel2 vector for generating GST-fusion proteins. Primers used are listed as follows: 5′-GTACGAATTCATGTCTGTCCACTCACCC-3′ and 5′-CTGATCTAGATAGACCATCTCCACCTCATT-3′ for CG13343; 5′-ATGCGAATTCTATGTCCTCGCCAGCCCCC-3′ and 5′-TCAGTCTAGATTAGAGGCTAGCGTAATCAGGAACGTCGTAAGGGTATAGCTTCAATGTGACACT-3′ for dAPPBP1; 5′- AGTCGAATTCAAATGATTAAACTATTCACG-3′ and 5′-CATGCTCGAGTCACTTGAGCAGACAGCACTC-3′ for CG7375; 5′-AGTCGAATTCAAATGGCGTCCGCCGCCCAGCTG-3′ and 5′- CATGCTCGAGTCACTTGAGCAGACAGCACTC-3′ for CG7375ΔN.
RT-PCR was used to measure the abundance of CG13343 and CG7375 mRNA after RNAi manipulation or in loss-of-function mutant alleles. To test RNAi efficiency, RNA were extracted from wing discs (100 pairs per sample) of third-instar larvae that expressed RNAi transgene under the control of the MS1096-Gal4 driver at 29°C. For characterization of loss-of-function alleles, RNA were extracted from GFP-negative first-instar larvae (40 larvae per sample) of CG13343SH2028/CyO, Kr-gfp or CG7375LL04684/TM3, twi-gfp flies. Total RNA was isolated with TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Contaminated DNA was digested using RNase-free DNase followed by a phenol/chloroform extraction to remove protein. First strand cDNA was synthesized from 1 µg of each sample using SuperScript III reverse transcriptase (Invitrogen). Semi-quantitative PCR was performed utilizing 20–35 cycles. The linear amplification stage for each primer set was determined by running the same volumes of amplified products on an agarose gel. α-tubulin primers were used for loading control. Primers used are listed as follows: 5′-GGCGTTGTCAAGCACATCATTC-3′ and 5′-TTTATCACATCCTCCAGCGTGG-3′ for CG13343 RNAi; 5′-GTACGAATTCATGTCTGTCCACTCACCC-3′ and 5′-CTGATCTAGATAGACCATCTCCACCTCAT-3′ to amplify full-length CG13343 cDNA in CG13343SH2028 mutant; 5′-GGAARCCAGTGCTGAACATCAACTC-3′ and 5′-ACGCATCGCCTTCTTTACATTG-3′ for CG7375 RNAi; 5′-AGTCGAATTCAAATGATTAAACTATTCACG-3′ and 5′-ATGCTCTAGACACTTGAGCAGACAGCACT-3′ to amplify full-length CG7375 cDNA in CG7375LL04684 mutant; 5′-GATCGTCGATCTGGTTCTGGACAG-3′ and 5′-CCAGTGGACGAAGGCACGCTT-3′ for α-tubulin.
cl-8 cells and wing disc cultures
Hh-responsive, Drosophila wing disc-derived clone-8 (cl-8) cells [86] were cultured at 25°C as described [33]. Effectene transfection reagent (Qiagen) was used for all transfection experiments. Cycloheximide (50 µg/ml; Sigma) was used to inhibit nascent protein synthesis in cl-8 cells. MG132 (50 mM; Sigma), ALLN (50 mM; Sigma) and lactacystin (20 mM; Boston Biochem) were used to inhibit the UPS activity. E64 (50 mM; Sigma), leupeptin (50 mM; Sigma) and NH4Cl (50 mM; Sigma) were used to inhibit lysosome function. In some experiments, cl-8 cells were pre-treated for 3 hours (9 hours in total) with lysosomal or UPS inhibitors prior to cycloheximide treatment for 6 hours. Third-instar larvae were dissected and incubated at 25°C for 4 hours in cl-8 cell medium supplemented with either lysosomal or UPS inhibitors before fixation.
In situ hybridization, immunofluorescence staining, immunoblotting and immunoprecipitation
The coding regions of CG13343 and CG7375 were used to generate RNA probes for in situ hybridization as described previously [33]. Wing discs from third-instar larvae were fixed in 4% paraformaldehyde and labeled with the following primary antibodies: rat anti-Ci (1∶20; 2A1; gift of Robert Holmgren) [87], mouse anti-Col (1∶100; gift of Alain Vincent) [88], rabbit anti-Cul1 (1∶100; Zymed) [6], mouse anti-Dl (1∶200; C594.9B; DSHB) and rabbit anti-β-galactosidase (1∶4000; Cappel). Alexa fluor-conjugated secondary antibodies (1∶400; Invitrogen) were used. The fluorescence images were acquired with a Zeiss Axio Imager2 equipped with an ApoTome.
cl-8 cells, first-instar larvae or wing discs dissected from third-instar larvae were lysed in NP-40 buffer (1% NP-40, 150 mM NaCl and 50 mM Tris-Cl, pH 8) supplemented with protease inhibitor cocktail (Roche). Protein concentrations of the cell lysates were measured using a BCA Protein Assay (Thermo). The following antibodies were used for immunoblotting: rat anti-Ci (1∶10; 2A1), rabbit anti-Ci (1∶20000; AbN; gift of Thomas Kornberg) [89], rabbit anti-Cul1 (1∶1000; Zymed), mouse anti-Cul3 (1∶1000; BD Transduction Lab.) [13], mouse-anti-GST (1∶20000; B-14; Santa Cruz), rabbit anti-HA (1∶1000; Y-11; Santa Cruz), mouse anti-β-Tubulin (1∶6000; Covance), and mouse anti-V5 (1∶2000; Invitrogen). Note that Cul1 and Cul3 antibodies for this study have been extensively used to reveal migratory differences between neddylated and un-neddylated Cullin proteins on immunoblots [7], [22], [59]. Anti-HA-conjugated agarose (Vector Lab.) was used to immunoprecipitate dAPPBP1-HA complexes in cl-8 cells.
In vitro neddylation assay
Cell-free in vitro neddylation assays were carried out with a NEDDylation kit according to the manufacturer's instructions (Enzo; UW0590). Typically, a 20 µl neddylation reaction includes human (supplied with the kit) or fly E1 and E2 enzymes, supplemented with 2 µl 10× NEDDylation buffer, 2 µl 10× NEDD8 (supplied with the kit), 1 µl 10× Mg-ATP (Sigma), 0.4 µl 50 mM DTT (Sigma) and 4 µl 100 U/ml IPP (NEB). For experiments shown in Figure 5C, 9.6 µl cl-8 cell lysates with or without transfected CG13343-V5 and dAPPBP1-HA, and 1 µl 20× human Ubc12 (supplied with the kit) were used as E1 and E2, respectively, to neddylate Cullin proteins present in cl-8 cell lysates. For experiments shown in Figure 8C and D, 2 µl 10× human NEDD8 E1 complex (supplied with the kit) and 2 µg GST or GST-CG7375 proteins bound on Glutathione Sepharose 4B beads (GE Healthcare) were used. For experiments shown in Figure 8E, 10.6 µl cl-8 cell lysates overexpressing the fly E1 complex, and 2 µg GST-CG7375 proteins bound on GST beads were used as E1 and E2, respectively, to constitute the fly neddylation cascade in vitro. All immunoblotting data presented in the figures are representative of independent experiments that were performed at least three times.
Supporting Information
Expression pattern of the MS1096 -Gal4 driver in the wing disc. MS1096-GAL4-driven mCD8-gfp was expressed at a much higher level in the dorsal (d) compartment of the wing disc (C). MS1096-Gal4 driver alone had no effect on the expression of Dl (A) or CiFL (B). Merged image is shown in (D).
(TIF)
Slimb as a negative regulator of CiFL stability. Inhibition of slimb function by RNAi in the dorsal compartment of the wing disc (indicated by a box bracket) led to accumulation of CiFL protein (A) and expansion of dpp-lacZ activity (B). Similarly, knockdown of slimb expression cell-autonomously stabilized CiFL in an anterior clone (D–F, arrowhead), but was incapable of inducing de novo Ci expression in a posterior clone (D–F, arrow). Note that ci transcript is not expressed in posterior cells.
(TIF)
Expression patterns of CG13343 and CG7375 in the wing disc. Endogenous CG13343 (B) and CG7375 transcripts (E) were detected by in situ hybridization in wildtype (WT) wing discs using antisense RNA probes specific to CG13343 and CG7375, respectively. Sense RNA probes (A and D) were used as the negative control. Ectopic expression of CG13343 (C) and CG7375 (F) was detected in the dorsal compartment of the wing disc (indicated by a box bracket) from ap-Gal4 driven EP[G8197] and EY[22840] flies, respectively. Note that the UAS-containing P-elements in EP[G8107] and EY[22804] are inserted on the 5′ UTR of CG13343 and CG7375, respectively. Elevated CG13343 or CG7375 expression in the wing dics was not sufficient to disrupt adult wing development (data not shown), presumably due to a limited amount of Cul proteins or NEDD8 modifier in the neddylation pathway.
(TIF)
The effect of CG13343 and CG7375 on the amounts of CiFL and Ci75 in wing discs. (A) Lysates extracted from wildtype (lane 1) or CG13343 RNAi overexpressing wing discs (lane 2) were immunoblotted (WB) with a Ci antibody (AbN), which recognizes both CiFL (ie. Ci155) and Ci75 [89]. Overexpression of CG13343 RNAi led to a significant accumulation of CiFL. However, the amount of Ci75 was not obviously changed. β-Tubulin was used as the loading control. (B) Lysates extracted from wildtype (lane 1) or CG7375 RNAi overexpressing βwing discs (lane 2) were immunoblotted with a Ci antibody (AbN). Overexpression of CG7375 RNAi resulted in a significant accumulation of CiFL. However, the amount of Ci75 was slightly reduced.
(TIF)
Reduced expression of CG13343 and CG7375 transcripts by RNAi and in loss-of-function alleles. (A and B) The levels of CG13343 (A) and CG7375 mRNAs (B) in wing discs overexpressing RNAi transgenes were evaluated by semi-quantitative RT-PCR. PCR products were quantified by Image J densitometry. RNAi overexpression resulted in significant reduction of the expression of CG13343 (70% reduction) and CG7375 (90% reduction) in wing discs. In contrast, the expression of CG11020, which is an off-target of the CG13343 RNAi transgene, did not change. α-tubulin was used as the internal control. (C and D) The levels of full-length transcripts of CG13343 (C) and CG7375 (D) in first-instar larvae were examined by RT-PCR. Full-length transcripts of CG13343 (C) and CG7375 (D) were not detected in CG13343SH2028 and CG7375LL04684 homozygous mutants, respectively. a-tubulin was used as the internal control.
(TIF)
ClustalX alignment of CG13343 protein and its Uba3 orthologs in Homo sapiens (Hs), Mus musculus (Mm) and Schizosaccharomyces pombe (Sp). Sequences used are Dm NP_610913.1, Hs NP_003959.3, Mm NP_ 035796.1 and Sp NP_ 592940.1. The UBA/THIF-type NAD/FAD binding domain (IPR000594) is shaded in yellow. The ubiquitin-activating enzyme repeat (IPR000127) is shaded in blue. The Nedd8 specificity determination residue is shaded in grey. The catalytic cysteine residue of E1-activating enzyme is shown in green. Purple shade marks the E2 binding domain (IPR014929).
(TIF)
ClustalX alignment of CG7375 protein and its Ubc12 orthologs in Homo sapiens (Hs), Mus musculus (Mm) and Schizosaccharomyces pombe (Sp). Sequences used are Dm NP_648187.1, Hs NP_003960.1, Mm NP_663553.1 and Sp NP_588256.1. The ubiquitin-conjugating enzyme E2 activity core (IPR000608) is shaded in blue. The N-terminal E1 binding motif specific for neddylation [62] and the E2-conjugating enzyme catalytic cysteine residue are shaded in yellow and green, respectively. The N-terminal E1 binding motif was deleted in GST-CG7375DN (amino acids 2–23).
(TIF)
Reduced dNEDD8 expression regulates Cul stabilization to elicite a full spectrum of Hh signaling responses. Hypomorphic dNEDD8AN015 somatic clones (negatively marked by nGFP in B, E and H) were induced in wing discs. Cul1 protein (A) was stabilized in dNEDD8AN015 clones located at the anterior (arrowheads) and posterior (arrow) compartments of the wing disc. However, ectopic CiFL (D) and Col (G) were induced only in anterior clones (arrowheads).
(TIF)
A model illustrating that dUba3 and dUbc12 control the stability and activity of Cul1 and Cul3 to regulate a full spectrum of Hh signaling.
(TIF)
Targeted in vivo RNAi screen to identify the UPS regulators in Hh signaling.
(PDF)
Acknowledgments
We are grateful to Drs. Markus Affolter, Hugo Bellen, Cheng-Ting Chien, Robert Holmgren, Thomas Kornberg, Helmut Krämer, Liqun Luo, Alain Vincent, the Bloomington Stock Center, the Developmental Studies Hybridoma Bank (DSHB), the Drosophila Genetic Resource Center at Kyoto, Fly Stocks of National Institute of Genetics (NIG-Fly), the Transgenic RNAi Project at Harvard Medical School (TRiP), and the Vienna Drosophila RNAi center (VDRC) for fly stocks and antibodies.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Cleveland Clinic Startup fund, a March of Dimes Basil O1Connor Starter Scholar Award (5-FY07-41) and an NIH/NIGMS grant (R01GM085175) to AJZ, American Heart Association Postdoctoral Fellowship Awards (10POST4110011 and 0825591D) to JZ and YS, and an NIH/NICHD postdoctoral training grant (T32HD007104) to JKO. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hooper JE, Scott MP. Communicating with Hedgehogs. Nat Rev Mol Cell Biol. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- 2.Wilson CW, Chuang PT. Mechanism and evolution of cytosolic Hedgehog signal transduction. Development. 2010;137:2079–2094. doi: 10.1242/dev.045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barakat MT, Humke EW, Scott MP. Learning from Jekyll to control Hyde: Hedgehog signaling in development and cancer. Trends Mol Med. 2010;16:337–348. doi: 10.1016/j.molmed.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- 5.Noureddine MA, Donaldson TD, Thacker SA, Duronio RJ. Drosophila Roc1a encodes a RING-H2 protein with a unique function in processing the Hh signal transducer Ci by the SCF E3 ubiquitin ligase. Dev Cell. 2002;2:757–770. doi: 10.1016/s1534-5807(02)00164-8. [DOI] [PubMed] [Google Scholar]
- 6.Ou CY, Lin YF, Chen YJ, Chien CT. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16:2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jia J, Zhang L, Zhang Q, Tong C, Wang B, et al. Phosphorylation by double-time/CKIe and CKIa targets Cubitus interruptus for Slimb/β-TRCP mediated proteolytic processing. Dev Cell. 2005;9:819–830. doi: 10.1016/j.devcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Smelkinson MG, Kalderon D. Processing of the Drosophila Hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr Biol. 2006;16:110–116. doi: 10.1016/j.cub.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Kent D, Bush EW, Hooper JE. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development. 2006;133:2001–2010. doi: 10.1242/dev.02370. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, et al. A Hedgehog-induced BTB protein modulates Hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Price MA. A unique protection signal in Cubitus interruptus prevents its complete proteasomal degradation. Mol Cell Biol. 2008;28:5555–5568. doi: 10.1128/MCB.00524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Shi Q, Chen Y, Yue T, Li S, et al. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2009;106:21191–21196. doi: 10.1073/pnas.0912008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu JT, Lin WH, Chen WY, Huang YC, Tang CY, et al. CSN-mediated deneddylation differentially modulates Ci(155) proteolysis to promote Hedgehog signalling responses. Nat Commun. 2011;2:182. doi: 10.1038/ncomms1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–86. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 15.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat Rev Mol Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 16.Harper JW, Schulman BA. Structural complexity in ubiquitin recognition. Cell. 2006;124:1133–1136. doi: 10.1016/j.cell.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 17.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 18.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 19.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merlet J, Burger J, Gomes JE, Pintard L. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci. 2009;66:1924–1938. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu JT, Lin HC, Hu YC, Chien CT. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat Cell Biol. 2005;7:1014–1020. doi: 10.1038/ncb1301. [DOI] [PubMed] [Google Scholar]
- 23.Haines N, van den Heuvel M. A directed mutagenesis screen in Drosophila melanogaster reveals new mutants that influence Hedgehog signaling. Genetics. 2000;156:1777–1785. doi: 10.1093/genetics/156.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Végh M, Basler K. A genetic screen for Hedgehog targets involved in the maintenance of the Drosophila anteroposterior compartment boundary. Genetics. 2003;163:1427–1438. doi: 10.1093/genetics/163.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins RT, Cohen SM. A genetic screen in Drosophila for identifying novel components of the Hedgehog signaling pathway. Genetics. 2005;170:173–184. doi: 10.1534/genetics.104.039420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casso DJ, Liu S, Iwaki DD, Ogden SK, Kornberg TB. A screen for modifiers of Hedgehog signaling in Drosophila melanogaster identifies swm and mts. Genetics. 2008;178:1399–1413. doi: 10.1534/genetics.107.081638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, et al. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299:2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- 28.Nybakken K, Vokes SA, Lin TY, McMahon AP, Perrimon N. A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat Genet. 2005;37:1323–1332. doi: 10.1038/ng1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, et al. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature. 2009;458:987–992. doi: 10.1038/nature07936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saj A, Arziman Z, Stempfle D, van Belle W, Sauder U, et al. A combined ex vivo and in vivo RNAi screen for Notch regulators in Drosophila reveals an extensive Notch interaction network. Dev Cell. 2010;18:862–876. doi: 10.1016/j.devcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Dai P, Akimaru H, Ishii S. A Hedgehog-responsive region in the Drosophila wing disc is defined by Debra-mediated ubiquitination and lysosomal degradation of Ci. Dev Cell. 2003;4:917–928. doi: 10.1016/s1534-5807(03)00158-8. [DOI] [PubMed] [Google Scholar]
- 32.Zhu AJ, Zheng L, Suyama K, Scott MP. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev. 2003;17:1240–1252. doi: 10.1101/gad.1080803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su Y, Ospina J, Zhang J, Michelson AP, Schoen AM, et al. Sequential Phosphorylation of Smoothened Transduces Graded Hedgehog Signaling. Sci Signal. 2011;4:ra43. doi: 10.1126/scisignal.2001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, et al. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001;412:449–452. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- 35.Watts RJ, Hoopfer ED, Luo L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron. 2003;38:871–885. doi: 10.1016/s0896-6273(03)00295-2. [DOI] [PubMed] [Google Scholar]
- 36.Kuo CT, Jan LY, Jan YN. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci USA. 2005;102:15230–15235. doi: 10.1073/pnas.0507393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Udvardy A. Purification and characterization of a multiprotein component of the Drosophila 26S (1500 kDa) proteolytic complex. J Biol Chem. 1993;268:9055–9062. [PubMed] [Google Scholar]
- 38.Belote JM, Fortier E. Targeted expression of dominant negative proteasome mutants in Drosophila melanogaster. Genesis. 2002;34:80–82. doi: 10.1002/gene.10131. [DOI] [PubMed] [Google Scholar]
- 39.Sevrioukov EA, He JP, Moghrabi N, Sunio A, Krämer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell. 1999;4:479–86. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- 40.Sriram V, Krishnan KS, Mayor S. deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. J Cell Biol. 2003;161:593–607. doi: 10.1083/jcb.200210166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lloyd TE, Atkinson R, Wu MN, Zhou Y, Pennetta G, et al. Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell. 2002;108:261–269. doi: 10.1016/s0092-8674(02)00611-6. [DOI] [PubMed] [Google Scholar]
- 42.Chanut-Delalande H, Jung AC, Baer MM, Lin L, Payre F, et al. The Hrs/Stam complex acts as a positive and negative regulator of RTK signaling during Drosophila development. PLoS ONE. 2010;5:e10245. doi: 10.1371/journal.pone.0010245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fürthauer M, González-Gaitán M. Endocytic regulation of Notch signalling during development. Traffic. 2009;10:792–802. doi: 10.1111/j.1600-0854.2009.00914.x. [DOI] [PubMed] [Google Scholar]
- 44.Semple CA RIKEN GER Group, GSL Members. The comparative proteomics of ubiquitination in mouse. Genome Res. 2003;13:1389–1394. doi: 10.1101/gr.980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfleger CM, Harvey KF, Yan H, Hariharan IK. Mutation of the gene encoding the ubiquitin activating enzyme Uba1 causes tissue overgrowth in Drosophila. Fly. 2007;1:95–105. doi: 10.4161/fly.4285. [DOI] [PubMed] [Google Scholar]
- 46.Lee TV, Ding T, Chen Z, Rajendran V, Scherr H, et al. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development. 2008;135:43–52. doi: 10.1242/dev.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh CE, McMahon R, Benzer S, Tanouye MA. bendless, a Drosophila gene affecting neuronal connectivity, encodes a ubiquitin-conjugating enzyme homolog. J Neurosci. 1994;14:3166–3179. doi: 10.1523/JNEUROSCI.14-05-03166.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, et al. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci. 2007;120:1689–1700. doi: 10.1242/jcs.004663. [DOI] [PubMed] [Google Scholar]
- 49.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex of downstream signaling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long X, Griffith LC. Identification and characterization of a SUMO-1 conjugation system that modifies neuronal calcium/calmodulin-dependent protein kinase II in Drosophila melanogaster. J Biol Chem. 2000;275:40765–40776. doi: 10.1074/jbc.M003949200. [DOI] [PubMed] [Google Scholar]
- 52.Shih HP, Hales KG, Pringle JR, Peifer M. Identification of septin-interacting proteins and characterization of the Smt3/SUMO-conjugation system in Drosophila. J Cell Sci. 2002;115:1259–1271. doi: 10.1242/jcs.115.6.1259. [DOI] [PubMed] [Google Scholar]
- 53.Bhaskar V, Valentine SA, Courey AJ. A functional interaction between dorsal and components of the Smt3 conjugation machinery. J Biol Chem. 2000;275:4033–4040. doi: 10.1074/jbc.275.6.4033. [DOI] [PubMed] [Google Scholar]
- 54.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 55.Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- 56.Pignoni F, Zipursky SL. Induction of Drosophila eye development by decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- 57.Oh S-W, Kingsley T, Shin H-h, Zheng Z, Chen H-W, et al. A P-element insertion screen identified mutations in 455 novel essential genes in Drosophila. Genetics. 2003;163:195–201. doi: 10.1093/genetics/163.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, et al. A new NEDD8-ligating system for cullin-4A. Genes Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim HJ, Kim SH, Shim SO, Park E, Kim C, et al. Drosophila homolog of APP-BP1 (dAPP-BP1) interacts antagonistically with APPL during Drosophila development. Cell Death Differ. 2007;14:103–115. doi: 10.1038/sj.cdd.4401935. [DOI] [PubMed] [Google Scholar]
- 60.Schuldiner O, Berdnik D, Levy JM, Chang AS, Moley KH, et al. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yi CH, Sogah DK, Boyce M, Degterev A, Christofferson DE, et al. A genome-wide RNAi screen reveals multiple regulators of caspase activation. J Cell Biol. 2007;179:619–626. doi: 10.1083/jcb.200708090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang DT, Miller DW, Mathew R, Cassell R, Holton JM, et al. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat Struct Mol Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Broemer M, Tenev T, Rigbolt KT, Hempel S, Blagoev B, et al. Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Mol Cell. 2010;40:810–822. doi: 10.1016/j.molcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 64.Jia H, Liu Y, Xia R, Tong C, Yue T, et al. Casein kinase 2 promotes Hedgehog signaling by regulating both smoothened and Cubitus interruptus. J Biol Chem. 2010;285:37218–37226. doi: 10.1074/jbc.M110.174565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huntzicker EG, Estay IS, Zhen H, Lokteva LA, Jackson PK, et al. Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 2006;20:276–281. doi: 10.1101/gad.1380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rencus-Lazar S, Amir Y, Wu J, Chien CT, Chamovitz DA, et al. The proto-oncogene Int6 is essential for neddylation of Cul1 and Cul3 in Drosophila. PLoS ONE. 2008;3:e2239. doi: 10.1371/journal.pone.0002239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim SH, Kim HJ, Kim S, Yim J. Drosophila Cand1 regulates Cullin3-dependent E3 ligases by affecting the neddylation of Cullin3 and by controlling the stability of Cullin3 and adaptor protein. Dev Biol. 2010;346:247–257. doi: 10.1016/j.ydbio.2010.07.031. [DOI] [PubMed] [Google Scholar]
- 68.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ou CY, Wang CH, Jiang J, Chien CT. Suppression of Hedgehog signaling by Cul3 ligases in proliferation control of retinal precursors. Dev Biol. 2007;308:106–119. doi: 10.1016/j.ydbio.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 70.Baker NE, Bhattacharya A, Firth LC. Regulation of Hh signal transduction as Drosophila eye differentiation progresses. Dev Biol. 2009;335:356–366. doi: 10.1016/j.ydbio.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seong KH, Akimaru H, Dai P, Nomura T, Okada M, et al. Inhibition of the nuclear import of Cubitus interruptus by Roadkill in the presence of strong Hedgehog signal. PLoS ONE. 2010;5:e15365. doi: 10.1371/journal.pone.0015365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dealy MJ, Nguyen KV, Lo J, Gstaiger M, Krek W, et al. Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat Genet. 1999;23:245–248. doi: 10.1038/13886. [DOI] [PubMed] [Google Scholar]
- 73.Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, et al. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 74.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 75.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, et al. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 76.Singer JD, Gurian-West M, Clurman B, Roberts JM. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 1999;13:2375–2387. doi: 10.1101/gad.13.18.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wimuttisuk W, Singer JD. The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Mol Biol Cell. 2007;18:899–909. doi: 10.1091/mbc.E06-06-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 79.Rabut G, Peter M. Function and regulation of protein neddylation. EMBO Rep. 2008;9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xirodimas DP. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 81.Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F- box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- 82.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, et al. The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 1999;13:270–83. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 84.Abida WM, Nikolaev A, Zhao W, Zhang W, Gu W. FBXO11 promotes the Neddylation of p53 and inhibits its transcriptional activity. J Biol Chem. 2007;282:1797–1804. doi: 10.1074/jbc.M609001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chan Y, Yoon J, Wu JT, Kim HJ, Pan KT, et al. DEN1 deneddylates non-cullin proteins in vivo. J Cell Sci. 2008;121:3218–3223. doi: 10.1242/jcs.030445. [DOI] [PubMed] [Google Scholar]
- 86.Van Leeuwen F, Samos CF, Nusse R. Biological activity of soluble wingless protein in cultured Drosophila imaginal disc cells. Nature. 1994;368:342–344. doi: 10.1038/368342a0. [DOI] [PubMed] [Google Scholar]
- 87.Motzny CK, Holmgren RA. The Drosophila Cubitus interruptus protein and its role in the wingless and hedgehog signal transduction pathways. Mech Dev. 1995;52:137–150. doi: 10.1016/0925-4773(95)00397-j. [DOI] [PubMed] [Google Scholar]
- 88.Crozatier M, Vincent A. Requirement for the Drosophila COE transcription factor Collier in formation of an embryonic muscle: Transcriptional response to Notch signalling. Development. 1999;126:1495–1504. doi: 10.1242/dev.126.7.1495. [DOI] [PubMed] [Google Scholar]
- 89.Aza-Blanc P, Ramírez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by Hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression pattern of the MS1096 -Gal4 driver in the wing disc. MS1096-GAL4-driven mCD8-gfp was expressed at a much higher level in the dorsal (d) compartment of the wing disc (C). MS1096-Gal4 driver alone had no effect on the expression of Dl (A) or CiFL (B). Merged image is shown in (D).
(TIF)
Slimb as a negative regulator of CiFL stability. Inhibition of slimb function by RNAi in the dorsal compartment of the wing disc (indicated by a box bracket) led to accumulation of CiFL protein (A) and expansion of dpp-lacZ activity (B). Similarly, knockdown of slimb expression cell-autonomously stabilized CiFL in an anterior clone (D–F, arrowhead), but was incapable of inducing de novo Ci expression in a posterior clone (D–F, arrow). Note that ci transcript is not expressed in posterior cells.
(TIF)
Expression patterns of CG13343 and CG7375 in the wing disc. Endogenous CG13343 (B) and CG7375 transcripts (E) were detected by in situ hybridization in wildtype (WT) wing discs using antisense RNA probes specific to CG13343 and CG7375, respectively. Sense RNA probes (A and D) were used as the negative control. Ectopic expression of CG13343 (C) and CG7375 (F) was detected in the dorsal compartment of the wing disc (indicated by a box bracket) from ap-Gal4 driven EP[G8197] and EY[22840] flies, respectively. Note that the UAS-containing P-elements in EP[G8107] and EY[22804] are inserted on the 5′ UTR of CG13343 and CG7375, respectively. Elevated CG13343 or CG7375 expression in the wing dics was not sufficient to disrupt adult wing development (data not shown), presumably due to a limited amount of Cul proteins or NEDD8 modifier in the neddylation pathway.
(TIF)
The effect of CG13343 and CG7375 on the amounts of CiFL and Ci75 in wing discs. (A) Lysates extracted from wildtype (lane 1) or CG13343 RNAi overexpressing wing discs (lane 2) were immunoblotted (WB) with a Ci antibody (AbN), which recognizes both CiFL (ie. Ci155) and Ci75 [89]. Overexpression of CG13343 RNAi led to a significant accumulation of CiFL. However, the amount of Ci75 was not obviously changed. β-Tubulin was used as the loading control. (B) Lysates extracted from wildtype (lane 1) or CG7375 RNAi overexpressing βwing discs (lane 2) were immunoblotted with a Ci antibody (AbN). Overexpression of CG7375 RNAi resulted in a significant accumulation of CiFL. However, the amount of Ci75 was slightly reduced.
(TIF)
Reduced expression of CG13343 and CG7375 transcripts by RNAi and in loss-of-function alleles. (A and B) The levels of CG13343 (A) and CG7375 mRNAs (B) in wing discs overexpressing RNAi transgenes were evaluated by semi-quantitative RT-PCR. PCR products were quantified by Image J densitometry. RNAi overexpression resulted in significant reduction of the expression of CG13343 (70% reduction) and CG7375 (90% reduction) in wing discs. In contrast, the expression of CG11020, which is an off-target of the CG13343 RNAi transgene, did not change. α-tubulin was used as the internal control. (C and D) The levels of full-length transcripts of CG13343 (C) and CG7375 (D) in first-instar larvae were examined by RT-PCR. Full-length transcripts of CG13343 (C) and CG7375 (D) were not detected in CG13343SH2028 and CG7375LL04684 homozygous mutants, respectively. a-tubulin was used as the internal control.
(TIF)
ClustalX alignment of CG13343 protein and its Uba3 orthologs in Homo sapiens (Hs), Mus musculus (Mm) and Schizosaccharomyces pombe (Sp). Sequences used are Dm NP_610913.1, Hs NP_003959.3, Mm NP_ 035796.1 and Sp NP_ 592940.1. The UBA/THIF-type NAD/FAD binding domain (IPR000594) is shaded in yellow. The ubiquitin-activating enzyme repeat (IPR000127) is shaded in blue. The Nedd8 specificity determination residue is shaded in grey. The catalytic cysteine residue of E1-activating enzyme is shown in green. Purple shade marks the E2 binding domain (IPR014929).
(TIF)
ClustalX alignment of CG7375 protein and its Ubc12 orthologs in Homo sapiens (Hs), Mus musculus (Mm) and Schizosaccharomyces pombe (Sp). Sequences used are Dm NP_648187.1, Hs NP_003960.1, Mm NP_663553.1 and Sp NP_588256.1. The ubiquitin-conjugating enzyme E2 activity core (IPR000608) is shaded in blue. The N-terminal E1 binding motif specific for neddylation [62] and the E2-conjugating enzyme catalytic cysteine residue are shaded in yellow and green, respectively. The N-terminal E1 binding motif was deleted in GST-CG7375DN (amino acids 2–23).
(TIF)
Reduced dNEDD8 expression regulates Cul stabilization to elicite a full spectrum of Hh signaling responses. Hypomorphic dNEDD8AN015 somatic clones (negatively marked by nGFP in B, E and H) were induced in wing discs. Cul1 protein (A) was stabilized in dNEDD8AN015 clones located at the anterior (arrowheads) and posterior (arrow) compartments of the wing disc. However, ectopic CiFL (D) and Col (G) were induced only in anterior clones (arrowheads).
(TIF)
A model illustrating that dUba3 and dUbc12 control the stability and activity of Cul1 and Cul3 to regulate a full spectrum of Hh signaling.
(TIF)
Targeted in vivo RNAi screen to identify the UPS regulators in Hh signaling.
(PDF)