Abstract
To learn about the acquisition of neural fate by ectodermal cells, we have analyzed a very early sign of neural commitment in Drosophila, namely the specific accumulation of achaete–scute complex (AS-C) proneural proteins in the cell that becomes a sensory organ mother cell (SMC). We have characterized an AS-C enhancer that directs expression specifically in SMCs. This enhancer promotes Scute protein accumulation in these cells, an event essential for sensory organ development in the absence of other AS-C genes. Interspecific sequence comparisons and site-directed mutagenesis show the presence of several conserved motifs necessary for enhancer action, some of them binding sites for proneural proteins. These and other data indicate that the enhancer mediates scute self-stimulation, although only in the presence of additional activating factors, which most likely interact with conserved motifs reminiscent of NF-κB-binding sites. Cells neighboring the SMC do not acquire the neural fate because the Notch signaling pathway effectors, the Enhancer of split bHLH proteins, block this proneural gene self-stimulatory loop, possibly by antagonizing the action on the enhancer of the NF-κB-like factors or the proneural proteins. These data suggest a mechanism for SMC committment.
Keywords: Neurogenesis, proneural gene, Notch, E(spl) complex, achaete–scute complex, Drosophila
A key process in development is the assignment of different fates to initially equivalent cells. In Drosophila, the analysis of the early development of the adult peripheral nervous system (PNS), which includes cuticular organs like the sensory bristles, has provided considerable insight into the genes that decide between epidermal and neural fates. Thus, the proneural genes, which are expressed by groups of cells known as proneural clusters, confer to these cells the potential to become neural precursors (for review, see Campuzano and Modolell 1992; Modolell 1997). Subsequently, cell–cell interactions mediated by the genes of the Notch (N) pathway allow only one or a few cells of each cluster to acquire the neural fate, whereas the remaining cells become epidermis (Artavanis-Tsakonas and Simpson 1991).
Among the best characterized proneural genes are achaete (ac) and scute (sc), two members of the ac–sc complex (AS-C) (Campuzano and Modolell 1992). They encode transcription factors of the basic helix–loop–helix (bHLH) family. The Ac and Sc proteins, which to a large extent are functionally redundant, appear to function in vivo as heterodimers with the bHLH protein encoded in the daughterless (da) gene (Murre et al. 1989). The expression of ac and sc in proneural clusters of the imaginal discs appears to be regulated by a combinatorial prepattern of transcription factors that interact with cis-controlling regions (enhancers) of the AS-C DNA and activate both ac and sc, so that they are coexpressed in all proneural clusters (Ghysen and Dambly-Chaudière 1988; Gómez-Skarmeta et al. 1995). A typical cluster of the imaginal wing disc that will give rise to a notum macrochaetae consists of 20–30 cells, but the sensory mother cell (SMC) appears to be selected among a smaller group of cells (the proneural field) that accumulates higher levels of Ac/Sc proteins than its neighbors (Cubas et al. 1991; Skeath and Carroll 1991; Cubas and Modolell 1992). Proneural field cells and the SMC, which accumulate still higher levels of Ac/Sc, always occupy the same position within the cluster. The SMC also accumulates Asense, another bHLH protein encoded in the AS-C (Brand et al. 1993; Domínguez and Campuzano 1993; Jarman et al. 1993). Together, these bHLH proteins are thought to be instrumental in implementing the neural differentiation pathway.
Cell–cell signaling among proneural cluster cells is mediated by the transmembrane proteins Delta (Dl) and Notch, acting as emitter and receptor of the signal, respectively (Heitzler and Simpson 1991). In the receptor cell, N activation promotes, by means of the Suppressor of Hairless [Su(H)] transcription factor, the expression of bHLH genes of the Enhancer of split complex [E(spl)-C] (Bailey and Posakony 1995; Lecourtois and Schweisguth 1995). Their products antagonize the function of the proneural proteins, probably by interfering with the activation of their target genes and by down-regulating proneural gene expression (Oellers et al. 1994; Ohsako et al. 1994; Van Doren et al. 1994; Heitzler et al. 1996). It is thought that the more proneural protein a cell accumulates, the stronger is its ability to signal and, consequently, the less inhibited it will be by its neighbors. Thus, in a proneural cluster, the cells that have the highest levels of proneural proteins tend to escape from the inhibition. When a cell does so, it becomes an SMC, signals maximally, and prevents its neighbors from acquiring the same fate (lateral inhibition) (Simpson 1990, 1997; Heitzler and Simpson 1991).
In this scenario, a little understood event is the acquisition of the neural fate by the cell that becomes the SMC (Modolell 1997). One of the earliest signs of its commitment is a large accumulation of proneural protein, which is independent from the controls that govern ac–sc expression in the proneural cluster (Cubas et al. 1991; Skeath and Carroll 1991). In the AS-C DNA and within 3.7 kb 5′ from the sc gene, there is an enhancer capable of directing lacZ expression specifically in SMCs (Martínez and Modolell 1991). Because this enhancer may be responsible for the increased expression of sc in these cells, we have further characterized it in the hope of learning about SMC commitment. We show that the increased Sc accumulation mediated by this enhancer in the SMC is essential, in the absence of other AS-C genes, for macrochaetae development. The enhancer mediates sc self-stimulation, although only in the presence of additional activating factors. In the remaining cells of the proneural cluster, the N signaling pathway effectors, the E(spl) bHLH proteins, block this sc self-stimulatory loop and prevent them from acquiring a neural fate.
Results
An SMC-specific enhancer
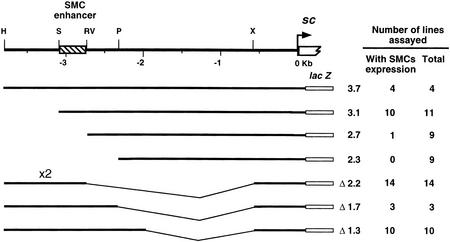
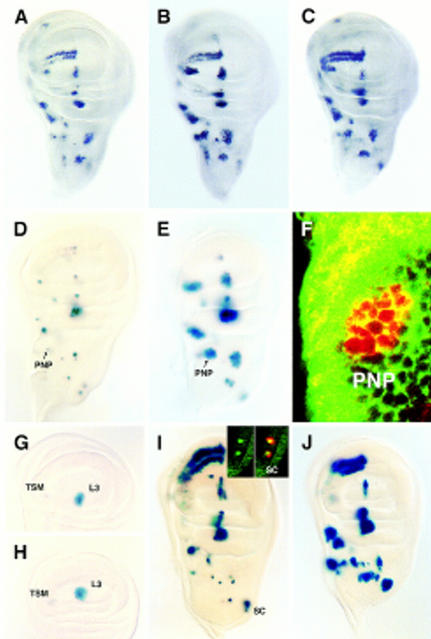
A 3.7-kb DNA fragment, extending upstream from and comprising the sc transcription initiation site, directs expression of a reporter lacZ gene in many single cells of the wing imaginal discs, which correspond to SMCs of notum macrochaetae and other sensilla of the wing (Martínez and Modolell 1991; Gómez-Skarmeta et al. 1995). To delimit the sequences responsible for expression in SMCs, subfragments of the 3.7-kb fragment were assayed for their ability to drive lacZ expression in wing discs. The necessary sequences were within a 356-bp SacII–EcoRV fragment (Fig. 1). This fragment linked to a heterologous hsp70 promoter and the lacZ gene (SRV–lacZ) specifically directed expression in SMCs (Fig. 2A). It also promoted expression in SMCs of other imaginal discs and of the embryonic PNS, but not in neuroectoderm neuroblasts (not shown).
Figure 1.
Localization of the DNA harboring an SMC enhancer. (Top line) A map of the 3.7-kb sc upstream region. Only the restriction sites used to prepare deletions of the 3.7 sc fragment are shown. Restriction site nomenclature: (H) HindIII; (P) PstI; (RV) EcoRV; (S) SacII; (X) XbaI. In the following lines, the extent of the sc promoter fragments used to prepare the transgenes, the nomenclature of each transgene, the number of lines that showed expression in SMCs, and the total number of stained lines are indicated. Construct Δ2.2 contains a direct tandem repeat of the HindIII–EcoRV distal DNA fragment.
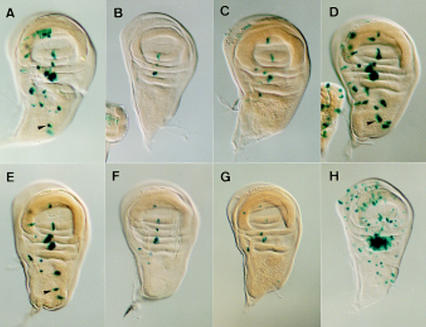
Figure 2.
Expression of SRV–lacZ and several of its mutant forms in late third instar wing discs. (A) Expression of the unmodified SRV–lacZ transgene. (B–E) Expression of SRV–lacZ in which either the E1, E2, E3 or N box (see Fig. 4), respectively, have been mutated. (F,G) The α2 or the β2 box has been eliminated, respectively. Transgene mutated in the α3 was expressed as in F. Transgenes mutated in α1 and β3 had essentially wild-type expressions. Arrowheads in A, D, and E point at the posterior dorsocentral SMC. (H) Expression of SRV–lacZ in C-765; UAS–sc discs. The strong staining in the dorsal radius region is attributable to a large number of SMCs that differentiate in this region.
The SMC enhancer promotes macrochaetae development
One of the earliest known signs of SMC specification is an increased accumulation of Ac and Sc proteins in this cell (Cubas et al. 1991; Skeath and Carroll 1991). However, it is unknown whether this accumulation is necessary for sensory organ (SO) development. To test for its requirement, we prepared two transgenes that contained the sc structural gene and an AS-C DNA fragment that drives expression of ac and sc in the dorsocentral (DC) region of the prospective notum (Gómez-Skarmeta et al. 1995). In addition, one transgene (DC–SMC–sc), but not the other (DC–sc), contained the SMC enhancer (Fig. 3). As expected, in the absence of the endogenous ac and sc genes [In(1)sc10.1 background], both transgenes drove sc expression in the DC proneural cluster (Fig. 3A,B). However, single cells with high accumulation of Sc protein, presumably SMCs, were clearly distinguishable only within the cluster promoted by DC–SMC–sc (Fig. 3B). Still, both transgenes allowed formation of two DC macrochaetae per heminotum (Fig. 3C). Thus, under the conditions of our assay, the SMC enhancer was dispensable for DC macrochaetae formation.
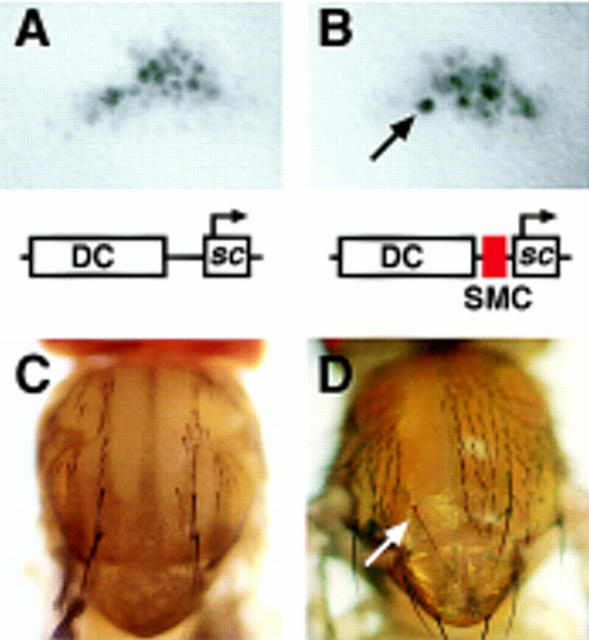
Figure 3.
Expression of sc minigenes and their rescuing ability of the DC macrochaetae. A 5.8-kb DNA fragment containing the DC enhancer was linked to the sc structural gene, with and without the 356-bp sequence comprising the SMC enhancer, as schematically represented. (A,B) sc expression in the DC region of late third instar wing discs promoted by these minigenes, in the absence of the endogenous ac and sc genes [In(1)sc10.1 background], as detected by an anti-Sc antibody. Arrow points to a cell with high levels of Sc, most likely an SMC (Cubas et al. 1991; Skeath and Carroll 1991). No single cells with preferential Sc accumulation were detected with the minigene lacking the SMC enhancer. (C) Notum of an In(1)sc10.1 fly carrying the DC–sc minigene. The four DC macrochaetae and several microchaetae were normally rescued. The same rescuing activity was observed with the DC–SMC–sc minigene (2.06 and 1.95 chaetae/heminotum in 36 and 42 heminota examined for DC–SMC–sc and DC–sc, respectively). (D) Rescue of a y DC macrochaeta within a clone of cells homozygous for the Df(1)260-1, which removes the y gene and the entire AS-C, and carrying the DC–SMC–sc minigene. In these flies, 14 DC chaetae positions were found within homozygous Df(1)260-1 territories and 13 DC chaetae developed. In DC–sc flies, only 1 DC chaetae developed in 13 positions within the Df(1)260-1 homozygous territories.
The In(1)sc10.1 chromosome contains a functional ase gene, which includes in its regulatory sequences a SMC-specific enhancer (Jarman et al. 1993) similar to that of sc (see below). Thus, ase and its associated enhancer might provide sufficient proneural protein to allow DC macrochaetae development and make the sc SMC enhancer dispensable in the above assay. To test this, mitotic recombination cell clones homozygous for the Df(1)260-1, which lack the complete AS-C (and therefore ase), were induced in larvae carrying either DC–sc or DC–SMC–sc. Df(1)260-1 cells regularly developed DC macrochaetae only if they carried the DC–SMC–sc transgene (Fig. 3D). This indicates that in the wild type the high accumulation of proneural proteins in SMCs is most likely a requisite for SO development. The sc SMC enhancer may help fulfill this requirement. The absence of chaetae outside the DC region in In(1)sc10.1; DC–SMC–sc flies further indicates that the SMC enhancer, without a proneural cluster-specific enhancer, is insufficient to promote SO development. This conclusion is reinforced by the fact that removal of ac–sc expression from specific proneural clusters, caused by chromosomal rearrangements that disconnect the corresponding enhancers, only eliminates the macrochaetae generated in the affected clusters, although the continuity between the endogenous sc–SMC enhancer and the sc gene is maintained (Campuzano et al. 1985; Gómez-Skarmeta et al. 1995).
Evolutionarily conserved sequences within the SMC enhancer
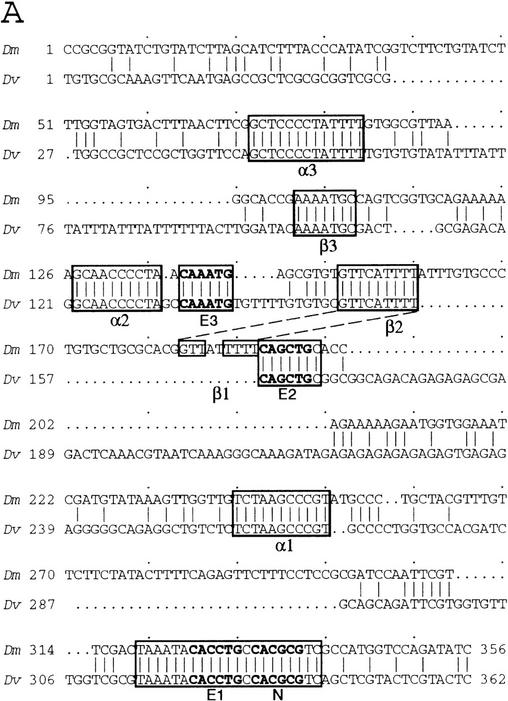
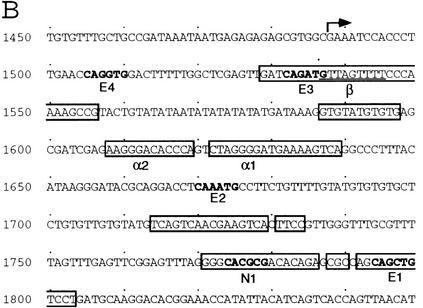
The sequence of the sc SMC enhancer was compared to the sequences upstream of the sc gene of Drosophila virilis. Significant similarities were found only within a 362-bp segment located 4.3 kb upstream of the D. virilis sc structural gene (Fig. 4A). The conserved sequences contained three E boxes, putative binding sites for bHLH proteins of the Ac, Sc, and Da type (for review, see Ohsako et al. 1994). The most proximal one was adjacent to an N box, a site that can be recognized by the E(spl)-C bHLH proteins (Tietze et al. 1992; Ohsako et al. 1994; Van Doren et al. 1994). In addition, there were three copies of a motif reminiscent of a consensus binding site for the NF-κB family of transcription factors (named α1, α2, and α3; Fig. 4C) (for review, see Lenardo and Baltimore 1989), and of a T-rich motif (β1, β2, β3), which does not fit with known protein-binding sequences (Fig. 4D).
Figure 4.
Comparison of the D. melanogaster sc SMC enhancer and ase gene leader sequences with the corresponding genomic regions of D. virilis. (A) sc SMC enhancer sequences. The longest conserved regions are boxed. Within these three E boxes and one N box are marked in bold. (B) Sequences of the ase gene of D. melanogaster. Transcription starts at the arrow. Regions conserved in the homologous DNA of D. virilis are boxed. Note the presence of E, N, α, and β boxes within the conserved sequences. (C) Comparison of the α-like motifs from several genes of D. melanogaster (Dm) and D. virilis (Dv) with the consensus for NF-κB binding sites (Lenardo and Baltimore 1989). Nucleotides that fit the consensus are highlighted. (D) Similar comparison of β-like boxes in the sc and ase SMC enhancers.
To investigate the functional significance of these motifs, each was mutated, except for β1, which has no clear counterpart in D. virilis, and the modified enhancers were assayed in vivo. Mutation of the E1 box and, to a slightly lower extent, the E2, α2, α3, and β2 boxes greatly reduced enhancer function. In contrast, mutation of the E3, α1, β3, or N boxes did not or only slightly modified it (see Fig. 2). Simultaneous mutation of α2 and α3 did not decrease further the residual enhancer activity observed with only one mutated motif.
Self-stimulation of sc in SMCs
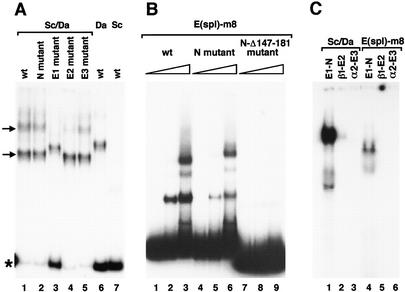
The fact that E boxes are required for the function of the SMC enhancer suggests that Sc and other proneural proteins bind to these sites and participate in sc activation. To examine this possibility, we analyzed in gel retardation assays the ability of the Sc protein to bind to wild-type and mutated enhancers. Binding occurred to the wild-type enhancer and, as shown previously (Murre et al. 1989; Cabrera and Alonso 1991; Van Doren et al. 1991), it required the bHLH protein Da (Fig. 5A, lanes 1,6,7). At least two DNA/protein complexes with different mobilities were detected, which suggests that the enhancer has at least two binding sites for Sc/Da heterodimers, consistent with the presence of two functional E boxes in the enhancer. In the absence of the E1 box, only one complex was detected (Fig. 5A, lane 3), which indicates that E1 is a binding site for the Sc/Da heterodimer. Removal of the E2 box preferentially depleted the slower migrating complex (Fig. 5A, lane 4). This suggests that E2 is also a binding site. However, the large amount of free probe in lane 3 compared with lanes 1 and 4 indicates that affinity for site E2 is lower than that for E1. Removal of the E3 and N boxes, which barely affects enhancer function (see above), did not modify the retardation profile (Fig. 5A, lanes 2,5). These data were verified by assaying the binding of Sc/Da to oligonucleotides bearing the different E boxes of the enhancer. Binding was strongest with E1 (plus N) box, weak with the E2 (plus β1) box, and undetectable with the E3 (plus α2) box (Fig. 5C). A DNase footprint assay indicated that Sc/Da bound to E1, but not to the N box (not shown). Taken together, these results indicate that Sc/Da heterodimers interact with the E boxes important for enhancer activity (see Fig. 2). As proneural proteins promote transcriptional activation (Cabrera and Alonso 1991; Van Doren et al. 1992, 1994; Ohsako et al. 1994), it is most likely that the high accumulation of Sc in SMCs is attributable to sc self-stimulation. Given that Ac/Da and Ase/Da dimers, also present in SMCs, recognize similar E boxes (Cabrera and Alonso 1991; Van Doren et al. 1991; Jarman et al. 1993; Ohsako et al. 1994; Singson et al. 1994), they may also participate in sc activation in these cells.
Figure 5.
Binding of Sc/Da heterodimers and E(spl)-m8 protein to SMC enhancer motifs. (A) Electrophoretic mobility-shift assays (EMSA) performed with a mixture of Sc and Da proteins (lanes 1–5) or with only Da (lane 6) or Sc (lane 7) proteins. DNA probe was the 356-bp enhancer in wild-type or mutant forms, as indicated. Arrows point at the complexes that probably contain one (lower band) and two Sc/Da heterodimers. Asterisk marks the free probe bands. (B) EMSA performed with 0, 1.3, and 6.6 μg of E(spl)-m8 protein and wild-type and mutant enhancer probes, as indicated. (C) EMSA carried out with mixtures of Sc and Da proteins (lanes 1–3) and with E(spl)-m8 protein (lanes 4–6). Oligonucleotide probes (see Materials and Methods) contained the boxes indicated.
We also examined whether high levels of Sc were sufficient to trigger the SMC enhancer and drive sc self-stimulation in cells other than SMCs. This was not apparently the case, as strong, generalized accumulation of Sc provided by a UAS–sc gene driven by Gal4 line C-765 (Gómez-Skarmeta et al. 1996) did not induce generalized expression of the SRV–lacZ transgene (see Fig. 2H), not even in those cells of the proneural clusters located near SMCs and that already contain elevated concentrations of endogenous Sc protein. β-Galactosidase accumulation only occurred in isolated cells (see Fig. 2H), which were most likely ectopic SMCs, as suggested by the many extra SOs that developed in adult flies (not shown). We conclude that sc self-stimulation, mediated by the SMC enhancer, is specific of SMCs and has requirements additional to a high level of Sc protein.
E and α boxes are sufficient to construct an SMC-specific enhancer
The above findings suggest that, to promote transcription, the SMC enhancer requires, besides proneural proteins, additional activating factors or the removal of inhibitors. Activating factors might interact with the α and β boxes necessary for efficient enhancer action (see Fig. 2F,G). To identify the minimum number of different motifs sufficient to constitute an SMC-specific enhancer, we assayed the enhancer activity of a synthetic oligonucleotide containing two E1 boxes and one α2 box and placed upstream of a basal hsp70 promoter linked to lacZ. It promoted β-galactosidase accumulation only in SMCs (not shown), although a weak one. A four tandem repeat of the same oligonucleotide drove much stronger lacZ activity and this also occurred exclusively in SMCs (Fig. 6I). In contrast, a four tandem repeat with E1 boxes, but without α2 boxes, drove strong expression in many cells of proneural clusters (Fig. 6J), revealing an inhibitory function of the α boxes in proneural cluster cells (but not in SMCs). Finally, a four tandem repeat of α2 boxes without E1 boxes failed to drive expression (not shown). Hence, both E and α boxes are sufficient, in the context of the minienhancer, to constitute an SMC-specific enhancer.
Figure 6.
Differential sensitivity of transcription directed by proneural cluster and SMC enhancers to N signaling. (A–C) Sc accumulation in wild-type, Nts, and da–Gal4/UAS–m8 imaginal wing discs, respectively. Note in B the presence of multiple cells with increased Sc accumulation, presumably SMCs, within proneural clusters. In C, overexpression of UAS–m8 did not modify appreciably Sc accumulation in proneural cluster cells, although it suppressed most SMCs. (D,E); SRV–lacZ expression in wild-type and Nts discs, respectively. Arrows point to postnotopleural (PNP) region. (F) Confocal section of a PNP cluster stained with anti-β-galactosidase antibody (red) and FITC–phalloidin (green) to visualize cell outlines. (G,H) lacZ expression driven by the L3/TSM enhancer in wild-type and Nts discs, respectively. Discs were understained (cf. with Fig. 5C of Gómez-Skarmeta et al. 1996) to better detect modifications in expression. (I,J) lacZ expression driven by minienhancers composed of four copies of an oligonucleotide containing E and α boxes or only E boxes, respectively. (Inset in I) Confocal image of the scutellar (SC) region of a disc carrying the complete minienhancer and showing that only single cells accumulate β-galactosidase (in cytoplasm, red). These cells are SMCs, as they also accumulate Ase protein (in nuclei, green).
Conservation of SMC—enhancer motifs in other genes
The ase sequences that direct expression in SMCs have been identified previously and contain several E boxes necessary for optimal expression in SMCs (Jarman et al. 1993). The corresponding DNA from D. virilis was sequenced and compared with that of D. melanogaster. Similarly to the sc SMC–enhancer, the stretches of conserved DNA contained E boxes, one N box, two α boxes, and one β box (see Fig. 4B–D), supporting the relevance of these boxes for SMC enhancer function. Moreover, the neurogenic gene Bearded, which is expressed in proneural clusters and SMCs (Singson et al. 1994), contains in its regulatory region one E box, necessary for its expression, and one motif identical to the α2 box (see Fig. 4C). An evolutionarily conserved α box is also found within the regulatory region of rough (see Fig. 4C), a homeobox gene important for restricting photoreceptor R8 specification (Tomlinson et al. 1988).
The SMC enhancer is controlled by N
N signaling prevents more than one or a few cells of a proneural cluster from becoming SMCs (Artavanis-Tsakonas and Simpson 1991). When the N pathway is not operative, for instance in Su(H) larvae or in larvae harboring a Nts allele raised at a nonpermissive temperature, Ac and Sc proteins accumulate in many cells of proneural clusters at levels higher than in the wild type (Schweisguth and Posakony 1994; Fig. 6A,B). The extra accumulation of Ac and Sc might be mediated by the cluster-specific enhancers, by the SMC enhancer, which under insufficient N signaling may promote expression in many cells of the proneural cluster as they become SMCs, or by both. To distinguish among these alternatives, we examined in wild-type and Nts discs the activity promoted by each type of enhancer. N inactivation allowed the SMC enhancer to drive expression in many cells of proneural clusters (Fig. 6D,E). Moreover, expression could occur in contiguous cells, indicating the failure of lateral inhibition (Fig. 6F). In contrast, N inactivation did not modify the activity of the enhancer (Gómez-Skarmeta et al. 1995) that drives expression in the vein L3 and TSM (twin sensilla of the wing margin) proneural clusters (Fig. 6G,H), although the accumulation of Sc in these clusters was increased (Fig. 6A,B). Hence, the SMC enhancer is responsible for most of the increased levels of proneural protein that occur in proneural clusters under insufficient N function.
N signaling, triggered by Ac–Sc in the emitter cell, promotes in the receptor cell the accumulation of E(spl)-C proteins, the main effectors of this signal (de la Concha et al. 1988; Jennings et al. 1994, 1995; de Celis et al. 1996a; Heitzler et al. 1996). E(spl)-C proteins are detectable in proneural cluster cells, except for the SMC (Jennings et al. 1995). This correlates with the SMC being the cells that signal maximally and inhibit their neighbors from acquiring the neural fate, while it is not itself inhibited. Ectopic accumulation of E(spl)-C protein prevents SMCs from emerging, as detected by a neuralized enhancer trap line (Huang et al. 1991) and the consequent absence of SOs in the adult fly (Tata and Hartley 1995; Nakao and Campos-Ortega 1996; J. Culí and J. Modolell, unpubl.). We have similarly found that overexpression of UAS–E(spl)–m8 or UAS–E(spl)–m7 transgenes driven by da–GAL4 or the C-253 GAL4 lines block the activity of the SMC enhancer and the development of the corresponding SOs (not shown). In contrast, either of these overexpressions allowed normal accumulation of Ac and Sc in proneural clusters (Fig. 6C) despite the high levels of ectopic E(spl)–m8 mRNA, which were severalfold higher than those in the wild type (not shown). However, in agreement with another report (de Celis et al. 1996a), overexpression with presumably stronger GAL4 drivers did interfere with ac–sc expression in proneural clusters (and SMC emergence; not shown). Taken together these results indicate that the function of the SMC enhancer is more sensitive to E(spl)-C inhibition than that of the proneural cluster enhancers and suggest that the first one is the main target of lateral inhibition mediated by the N pathway.
To analyze further the inhibition of SMC enhancer function by E(spl)-C, we examined whether E(spl)–m8 binds to this enhancer. EMSA and DNase footprint analyses showed that this protein binds to the N box and, unexpectedly, also protects a broad region of the enhancer (nucleotides 142–182), which does not contain sequences that fit the E(spl)-C consensus binding site (Fig. 5B and not shown). Binding to an enhancer with a mutated N box was weaker, and that to an enhancer without the N box and the second E(spl)–m8-binding site was undetectable (see Fig. 5B). Remarkably, the removal of one (Fig. 2E) or both (not shown) binding sites did not modify the SMC specificity of the enhancer, as might be expected if these binding sites mediated the repression of enhancer function in response to N signaling (Fig. 6B,E). Moreover, EMSA analyses showed that E(spl)–m8 was unable to bind to the synthetic SMC-specific minienhancer (not shown). These results were extended to other E(spl)-C proteins by verifying (not shown) that, similarly to E(spl)–m8 (Fig. 5C), E(spl)–m5 binds to an oligonucleotide with the E1–N sequence, but not to oligonucleotides containing only E2 or E3 boxes (not shown). Thus, it is concluded that the E(spl)-C proteins restrict enhancer function to SMCs by a mechanism that does not require direct interaction with enhancer DNA.
Discussion
To examine the early events that lead to the acquisition of neural fate by one or a few cells of the proneural clusters, we have characterized an enhancer that is found in the sc upstream regulatory region and that promotes expression exclusively in SMCs (Martínez and Modolell 1991), concomitantly with or shortly after their commitment to the neural fate. This enhancer is responsible, at least in part, for the high accumulation of Sc protein that occurs in SMCs (Cubas et al. 1991; Skeath and Carroll 1991). SMCs also accumulate Ac and Asense (Ase), two additional AS-C proneural proteins (Cubas et al. 1991; Skeath and Carroll 1991; Brand et al. 1993; Domínguez and Campuzano 1993; Jarman et al. 1993). Although the SMC enhancer found near sc might conceivably act also on ac and ase (Gómez-Skarmeta et al. 1995), this is probably not the case as additional SMC-specific enhancers are found in the vicinity of these genes. One, located in the transcribed sequences of ase (Jarman et al. 1993), has all the motifs that are functionally important in the sc SMC enhancer (E, α, and β boxes). The other one, whose presence is inferred by the observation that Ac accumulates in SMCs even when the ac region is separated from the rest of the AS-C by a chromosomal rearrangement (Gómez-Skarmeta et al. 1995), may comprise several E boxes located near the ac promoter (Van Doren et al. 1992, 1994; Martínez et al. 1993; Ohsako et al. 1994). We have failed, however, to find motifs similar to the α and β boxes in the ac available sequences.
SMC enhancers are necessary for SO development
We have examined the requirement of the sc SMC enhancer for DC macrochaetae development. In the absence of the endogenous ac and sc genes, but in the presence of ase, the sc SMC enhancer is dispensable. However, when ase is also removed, the SMC enhancer becomes essential for development of these chaetae. This indicates that SMC enhancers are necessary for macrochaetae development and that, at least to some extent, they can replace one another. Most likely they fulfill this function by providing SMCs with a high concentration of proneural proteins. Although SMCs for macrochaetae and for other types of sensilla accumulate Ac, Sc, and Ase, we have found that accumulation of only Sc allows macrochaetae development. This suggests that a high concentration of proneural protein, rather than its composition, is a requisite for neural development. However, the presence of Ac, Sc, and Ase in SMCs may still be important for correct development of SOs, as the bristles rescued by sc alone were abnormally small, whereas those generated in the presence of sc and ase were of normal size. Similarly, the specific proneural proteins expressed in at least some embryonic neuroblasts affect their identity, as measured by the combination of markers expressed in them and in their descendants (Parras et al. 1996).
sc self-stimulation in SMCs
Several data indicate that the SMC enhancer mediates sc self-stimulation in SMCs. Thus, the enhancer contains E boxes necessary for its efficient function and Sc/Da heterodimers bind to them in vitro. Moreover, in a yeast model system and in Drosophila cell cultures, Sc/Da or Ac/Da heterodimers activate transcription by binding to E boxes (Cabrera and Alonso 1991; Van Doren et al. 1992, 1994; Ohsako et al. 1994). We have also found that an oligonucleotide containing only several copies of one of the SMC enhancer E boxes drives transcription in most imaginal disc cells that contain Ac and Sc (Fig. 6J), and even in cells with transient ectopic Sc protein provided by an HSSC transgene (J. Culí and J. Modolell, unpubl.).
However, in the context of the whole enhancer, the E boxes alone do not allow efficient sc self-activation. Other evolutionarily conserved motifs, among them sequences reminiscent of an NF-κB factor-binding site (α boxes) (Lenardo and Baltimore 1989), are also necessary. E and α boxes are clearly the most important motifs for enhancer function; an oligonucleotide with several copies of these boxes largely recapitulates the main characteristic of the enhancer, namely, its promoting expression exclusively in SMCs. Because Sc/Da dimers do not bind to α boxes, we postulate that additional factors, which we call Xα, interact with these boxes and allow sc self-stimulation. We have attempted to identify Xα among the several known Drosophila members of the NF-κB superfamily. These are the rel proteins encoded by the dorsal, dif (drosophila immunity factor), and relish genes (Steward 1987; Ip et al. 1993; Dushay et al. 1996). However, a null dorsal allele does not affect bristle development, not even in the presence of reduced proneural gene function [In(1)sc10.1/+ or scM6/Y genetic backgrounds; Gómez-Skarmeta et al. 1995; J. Culí, unpubl.]. Dif and Relish proteins accumulate homogenously, albeit weakly, in the nuclei of disc cells (R. Cantera and Y. Engstrom, pers. comm.) and remain candidates. Their roles in SMC development, however, are difficult to establish without mutations at their respective loci. Moreover, overexpression of UAS–dif or UAS–relish in proneural clusters (C253 GAL4 driver) does not affect the bristle pattern (J. Culí, S. Govind, and M. Dushay, unpubl.). Similarly, the mutation Toll10b, another form of overexpression as it enhances the expression of dif and the translocation of dorsal and dif products to nuclei, does not modify appreciably that pattern (J. Culí, unpubl.). Evidently, these gain-of-function experiments do not rule out that one of these proteins is Xα. The factor might be present in saturating amounts and its overexpression would cause little effect. The MBP (major histocompatibility complex binding protein) transcription factors also interact with NF-κB DNA-binding sites (Fan and Maniatis 1990). schnurri (shn) belongs to this class (Arora et al. 1995; Grieder et al. 1995), but chaetae arise normally within territories of cells homozygous for a null shn allele (J. Culí, unpubl.). Xα thus remains unidentified.
The SMC enhancer as a target for N signaling
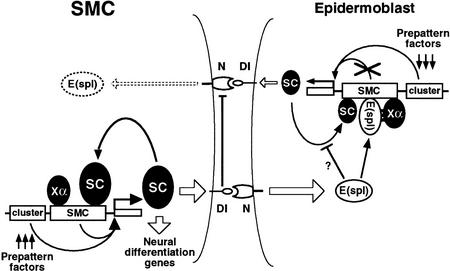
Cell–cell inhibitory interactions mediated by the N signaling pathway limit the number of SMCs that arise from a proneural cluster. According to a current model (for review, see Simpson 1997), in the signal-emitting cell, Ac and Sc proteins activate Dl and this relays the signal to the receiving cell by means of the N receptor (Kunisch et al. 1994). Activation of the N pathway promotes transcription of the final effectors, the bHLH proteins of the E(spl)-C, which somehow antagonize the function of the AS-C genes and prevent the receiving cell from becoming an SMC. Thus, mutual inhibitory interactions tending to magnify differences are established between neighboring cells of the proneural cluster. Within a proneural cluster, and probably because of the heterogeneous topographical distribution of activators and repressors—prepattern factors (Stern 1954; Gómez-Skarmeta et al. 1996)—that control ac–sc transcription in proneural clusters, a few cells accumulate more Ac/Sc proteins than their neighbors (Cubas et al. 1991; Skeath and Carroll 1991). Eventually, a cell of this group, which signals the most, will be released from the inhibitory loop, as its level of E(spl)-C bHLH protein becomes minimal (Jennings et al. 1995). According to our findings, this cell would then turn on ac–sc self-stimulation by means of the SMC enhancers and become an SMC (Fig. 7). The SMC signals maximally to its neighbors and prevents them from following the same fate.
Figure 7.
Model for the regulation of sc in SMCs and neighboring epidermoblasts. In all proneural cluster cells, including the emerging SMC, sc transcription is activated by prepattern genes acting on proneural cluster-specific enhancers (Gómez-Skarmeta et al. 1996). This activation is largely independent of the N signaling pathway. Additionally in the SMC (left), the sc SMC-specific enhancer, by means of E boxes that bind Sc protein (as heterodimers with Da), promotes sc self-activation. The self-stimulatory loop also requires the unidentified activating factor Xα, which should bind to α boxes (NF-κB-like binding sites). The resulting increased Sc accumulation promotes strong activation of Dl in the SMC, which signals to the neighboring proneural cluster cells (right) and promotes in them the transcription of E(spl)-C genes. The E(spl)-C proteins prevent the sc self-stimulatory loop in these cells, which become epidermoblasts, by interacting with the Xα factor and, possibly, by complexing with Sc and Da proteins (Gigliani et al. 1996). (DNA sites capable of binding E(spl) proteins, although dispensable, may facilitate or stabilize their interaction with the Xα factor.) As a consequence, the Sc concentration remains low and the inhibitory signaling through Dl to the SMC will be weak. The fact that E(spl)-C proteins in SMCs are undetectable (Jennings et al. 1995) suggests the presence of a mechanism that reduces E(spl)-C transcription in SMCs. As proposed for dorsoventral boundary formation at the wing margin (de Celis et al. 1996b), sequestering of N molecules by the high concentration of active Dl in the SMC may block reception of signaling from the surrounding cells. In addition, low levels of N activation in the SMC may be compatible with lack of E(spl)-C transcription, as small amounts of Su(H) activator can be titrated by the constitutive levels of the Hairless antagonist (Bang et al. 1995).
In this scenario, how do the E(spl)-C proteins antagonize neurogenesis? One possibility is by down-regulating the overall levels of ac–sc transcription in proneural clusters, as, under some conditions, overaccumulation of E(spl)-C proteins reduces these levels (de Celis et al. 1996a, J. Culí, unpubl.). However, we have found that ac–sc transcription in proneural clusters, which is regulated by cluster-specific enhancers, is much less sensitive to variation in levels of E(spl)-C protein than that governed by the sc SMC enhancer. Thus, extra E(spl)–m7 or E(spl)–m8 protein, supplied in amounts sufficient to block SMC emergence (as determined by the activity of the SRV–lacZ transgene or the neuralized A101 enhancer trap line) does not modify sc expression in proneural clusters. Conversely, an insufficency of E(spl)-C protein, induced by reduced N signaling, allows the SMC enhancer to drive transcription in many proneural cluster cells, but it does not modify that promoted by a proneural cluster enhancer. Interestingly, in the neuroectoderm, the expression of ac, sc, and l’sc in neuroblasts is also more sensitive to overexpression of E(spl)-C transgenes than that in proneural clusters (Nakao and Campos-Ortega 1996). These results point to SMC- and neuroblast-specific enhancers, rather than the proneural cluster enhancers, as preferential or main targets of N signaling.
The sc (and also the ase) SMC enhancer possesses an evolutionarily conserved N box capable of binding E(spl)-C proteins. This suggests that these proteins may bind to it and prevent enhancer function in all the proneural cluster cells except the SMC, which is essentially devoid of them (Jennings et al. 1994, 1995). However, removal of this box (and another nonconsensus binding site) does not provoke enhancer-promoted transcription in proneural cluster cells. This suggests that the E(spl)-C proteins block enhancer function by a different mechanism. Our observations that a synthetic minienhancer composed of only E boxes drives expression in most or all proneural cluster cells, whereas a minienhancer composed of both E and α boxes promotes expression almost exclusively in SMCs, suggests that E(spl)-C proteins somehow interfere with the function of the α box or of the factors binding to it. E(spl)-C m8 protein does not bind detectably in vitro to α boxes. However, the bHLH domain of these proteins is not strictly required for repressing neurogenesis (Giebel and Campos-Ortega 1997), which suggests that DNA binding is not essential for this repression. In fact, they appear to interact with other proteins by means of at least two other domains (Paroush et al. 1994; Dawson et al. 1995; Giebel and Campos-Ortega 1997). This should greatly enlarge the types of interacting partners and possible repression mechanisms. Therefore, we envision the Xα activator as a protein capable of interacting with E(spl)-C proteins and bringing them onto the enhancer. The bound Xα/E(spl)-C protein complex would prevent the enhancer-mediated self-stimulatory loop. Thus, the E(spl)-C proteins would convert Xα from an activator in the SMC to a repressor in the remaining proneural cluster cells (Fig. 7). According to this view, the N box and other E(spl)-C-binding sites in SMC enhancers might facilitate or stabilize the binding of the Xα/E(spl)-C protein complex. Still, E(spl)-C proteins might also function in other ways, like by sequestering proneural proteins in complexes not capable of binding to DNA, similarly to the Emc/Id proteins that antagonize proneural/myogenic protein function (Ellis et al. 1990; Garrell and Modolell 1990). Although proneural/E(spl)-C protein complexes have not been detected by EMSA assays (Van Doren et al. 1991), interactions between these proteins have been detected in λ repressor and yeast two-hybrid assay systems (Gigliani et al. 1996; Alifragis et al. 1997).
SMC specification
Our results indicate that the SMC-specific enhancers of the AS-C are essential for macrochaetae development and that they function at the level of the specification of a single cell of a proneural cluster as an SMC. The resulting large accumulation of proneural proteins most likely activates neural differentiation genes. Is this accumulation of proneural protein the initial step of SMC specification, or is it the culmination of other earlier steps specific for this cell? Although there is as yet no solution to this alternative, the self-activation of proneural genes seems the earliest known event that is exclusive to the SMC. According to the model depicted in Figure 7, this activation would be concomitant with the depletion of the E(spl)-C antagonizing proteins. However, this depletion would probably occur in several of the neighboring cells that accumulate most Ac–Sc under the stimulus of the proneural cluster enhancers. The turning on of the self-stimulatory loop in one of these cells would increase its signaling and prevent its neighbors from triggering it. The activation of the neuralized A101 enhancer trap is also an early marker of SMC commitment (Huang et al. 1991). However, before strong expression occurs in the SMC, weak expression has been detected in two or three cells at the position where the SMC will emerge (Huang et al. 1991). Thus, a pre-SMC state has been invoked, a state that would not be exclusive of the cell that becomes the SMC and be shared by a few of the cells with maximal ac–sc levels (Cubas et al. 1991; Skeath and Carroll 1991; Cubas and Modolell 1992).
It is of interest that the cells with strong accumulation of Ac–Sc and the SMC appear in reproducible positions within proneural clusters giving rise to notum macrochaetae (Cubas et al. 1991). This suggests that the landscape of prepattern factors, by means of the control of ac–sc expression and the participation of antagonists of the proneural function like emc, predetermines with the uncertainty of a few cells the one that becomes the SMC (Cubas and Modolell 1992). As the N signaling pathway, acting on lateral inhibition, does not seem to affect substantially the distribution of proneural protein contributed by means of the proneural cluster enhancers, we suggest that, at least for macrochaetae, this pathway has no major function in determining which cell becomes the SMC.
Materials and methods
Drosophila stocks
Stock descriptions are in In(1)sc10.1, Df(1) 260-1, and Nts (Lindsley and Zimm 1992); da–Gal4 (daG32, Wodarz et al. 1995). GAL4 drivers C-765 and C-253 were provided by A. Brand (University of Cambridge, UK; see also Gómez-Skarmeta et al. 1996). UAS-m8 and UAS–sc stocks were provided by S. Sotillos and I. Rodríguez (both at Centro Biología Molecular Severo Ochoa) and UAS–m7 was from J.F. de Celis. To generate mitotic recombination clones homozygous for the Df(1) 260-1, M(1)osp/FM6; P[w+;DC–sc] or M(1)osp/FM6; P[w+;DC–SMC–sc] females were mated with Df(1)260-1;y−/Dp(1;Y)y2-67g males, and larvae were X-ray treated (1000 rads) at 60 hr after egg laying. Females Df(1) 260-1;y−/M(1)osp; P[w+,DC–sc or DC–SMC–sc]/+ were examined for clones homozygous for M+ and y. SRV–lacZ and LIII/TSM–lacZ transgene expression in hypomorphic N condition was analyzed in male larvae, hemizygous for the Nts allele and heterozygous for the transgenes, kept at 30°C for 12 hr in a water bath.
Construction of transgenes
Distal deletion series of the 3.7-kb sc SMC enhancer fragment were prepared with four DNA genomic fragments extending from an HpaII restriction site (AS-C coordinate 33.5; Campuzano et al. 1985), located within the sc leader sequences, to the PstI (35.9), EcoRV (36.3), SacII (36.7), and HindIII (37.4) sites, respectively. Fragments were subcloned into the transformation plasmid plac20 (Martínez and Modolell 1991) by fusing the HpaII cut end to the leader of the vector lacZ gene. The DNA constructs used for the intercalary deletion series shared a proximal XbaI–HpaII (34.2–33.5) 0.67-kb fragment. The distal parts of the constructs were composed of the HindIII–EcoRV (37.4–36.3) fragment duplicated in a direct tandem repeat (Δ2.2); the HindIII–PstI (37.4–35.9) fragment (Δ1.7); and the same fragment as in Δ1.7 plus a fragment obtained by PCR amplification and extending from the PstI (35.9) site to the map coordinate 35.5 (construct Δ1.3). Amplification from this coordinate was performed with oligonucleotide CCTCTAGAAAGCAAGTTAAG that introduced an XbaI site to aid in the construction. In all cases, the proximal and distal DNA fragments were ligated in Bluescript (Stratagene) and then excised and fused to the lacZ gene in plac20.
Constructs containing the sc SMC enhancer (0.36-kb SacII–EcoRV 36.7–36.3 fragment), the hsp70 heterologous promoter, and the lacZ gene were prepared in the transforming plasmid pHZ50PL (Gómez-Skarmeta et al. 1996). The sc SMC enhancer fragment was mutagenized by inverse PCR mutagenesis, essentially as described (Hemsley et al. 1989), except that Taq polymerase was supplemented with Taq extender (Stratagene). Oligonucleotides (Isogen) used were N box mutagenesis, TCCTC GCCATGGTCCAGA and TCCGCAGGTGTATTTAGTCG; E1 box mutagenesis, TCCCCACGCGTCGCCATG and TCCTATTTAGTCGAACGAATT; E2 box mutagenesis, TCCCACCAGAAAAAGAATGG and TCCAAAAATAACCGTGCGCAGCACAGGG; E3 box mutagenesis, TCCAGCGTGTGTTCATTTTAT and TCCTTAGGGGTTGCTTTTTTC; α1 box mutagenesis, TCCTTATGCCCTGCTACGTTT and TCCTCCCAACCAACTTTATAC; α2 box mutagenesis, GATCCACAAATGAGCGTGT and CGACGTTTTTTCTGCACCGA; α3 box mutagenesis, TCCGAAAGTGGCGTTAAGGCA and TCCGCACGAAGTTAAAGTCAC; β2 box mutagenesis, TCCCAATTTGTGCCCTGTG and TCCGACACGCTCATTTGTT; and for β3 box mutagenesis, TCCTCAGTCGGTGCAGAAA and TCCCGGTGCCTTAACGCCA. The wild-type motifs were replaced by the sequences shown in bold, without changing their length. In all cases a BamHI site was reconstituted. A double mutant was prepared by mutating the α3 box in an α2 mutated plasmid using the oligonucleotides described above. Another double mutant was prepared by deleting the 147–181 nucleotides of an N box-mutated enhancer by means of oligonucleotides GGTTATTTTTCAGCTGC and GCTCATTTGTTAGGGGT. All mu- tated enhancer fragments were sequenced and subcloned into pHZ50PL.
To construct the DC–sc minigene, a 1.9-kb BamHI genomic fragment containing the sc basal promoter and the complete transcribed region (sc sequence nucleotides 122–2015, GenBank accession no. M17119) and a 5.7-kb EcoRI (68.6–62.9 coordinates) fragment bearing the dorsocentral AS-C enhancer (Gómez-Skarmeta et al. 1995) were subcloned in transformation plasmid pW8 (Klemenz et al. 1987). To prepare the DC–SMC–sc minigene, the 0.36-kb SMC enhancer fragment was introduced between the DC and sc sequences in pW8 by using polylinker restriction sites. The relative orientations and order of the elements were the same as in the Drosophila genome.
A minienhancer containing E1 and α2 boxes was prepared by annealing the two complementary oligonucleotides GATC-CAAAGCAACCCCTAAGAACTAAATACACCTGCGAGCT-AAATACACCTGCA and GATCTGCAGGTGTATTTAGCTCGCAGGTGTATTTAGTTCTTAGGGGTTGCTTTG and subcloning the resulting double-stranded oligonucleotide into Bluescript, cut in the BamHI site. The insert was sequenced and was transferred to the transforming plasmid pHZ50PL. The underlined bases form BamHI–BglII restriction sites. The bases shown in bold correspond to the E1 and α2 boxes and the flanking sequences found in the original SMC enhancer. A minienhancer containing four direct tandem repeat copies of the above double-stranded oligonucleotide was constructed by annealing, ligation, and digestion with BamHI and BglII, selection of the tandem of four copies in an agarose gel, and subcloning into Bluescript. Remaining procedures were as before. Tetramers of the minienhancer with disrupted E1 boxes were similarly prepared, by using the oligonucleotides GATCCAAAGCAACCCCTAAGAACTAAATAGACCTGCGAGCTAAATAGACCTGCA and GATCTGCAGGTCTATTTAGCTCGCAGGTCTATTTAGTTCTTAGGGGTTGCTTTG, and to make the tetramer with disrupted α2 boxes: GATCCAAATCCAGCCCAAAGAACTAAATACACCTGCGAGCTAAATACACCTGCA and GATCTGCAGGTGTATTTAGCTCGCAGGTGTATTTAGTTCTTTGGGCTGGATTTG. Italics show differences with the wild-type boxes.
Germ-line transformation and histochemistry
P element-mediated transformation (Rubin and Spradling 1982) was performed with 0.3 mg/ml DNA of the transforming plasmid and 0.15 mg/ml pUChsπΔ2-3 as transposase source. The recipient stocks were ry506 for vectors plac20 and pHZ50PL and y w1118 for vector pW8. Expression of transgenes was analyzed in wing imaginal discs by X-gal staining (Gómez-Skarmeta et al. 1995). Anti-sc antibody staining was performed as in Cubas et al. (1991). Labeling of cell outlines and β-galactosidase-containing cells was performed by incubating discs overnight with mouse anti-β-galactosidase antibody (1:200), followed by incubation for 2 hr with (1:200) biotin-conjugated anti-mouse IgG (Amersham) and for 1 hr in (1:200) lissamine rhodamine-conjugated streptavidin (Jackson). Discs were postfixed for 30 min in PBS-formaldehyde containing a 1:10 dilution of FITC-phalloidin 3.3 mm stock. Fluorescent double staining with anti-Ase-antibody (1:500; Brand et al. 1993) and anti-β-galactosidase antibody was performed as described (Gómez-Skarmeta et al. 1996). Images were acquired with a Zeiss LSM310 confocal microscope.
Gel mobility-shift assays
Sc, Da, and E(spl)-C m5 proteins were produced in bacteria with the expression plasmids pET14b–T4, pET14b–da, and pET14b–m5, respectively (Ohsako et al. 1994), using the conditions therein, except that the His tag was not cleaved from the fusion proteins. E(spl)-C m8 full coding sequence was PCR amplified using the oligonucleotides CGCGGATCCATGGAATACACCACCAAG and GCGGAATTCTTTACCAGGGGCGCCACA, which introduce BamHI and EcoRI sites, and fused in-frame to the glutathione S-transferase in vector pGEX-2T. The fusion protein was prepared and purified as described (Gómez-Skarmeta et al. 1996). Probes for EMSA assays were the wild-type 0.36-kb SMC enhancer and its mutated E1, E2, E3, N, and N-Δ147-181 forms 32P-labeled by Klenow filling, and the double-stranded oligonucleotides NE1, TAAATACACCTGCCACGCGTCGC; β1E2, GTTATTTTTCAGCTGCACC; and α2E3, GCAACCCCTAACAAATGAGCG 32P-end-labeled with T4 polynucleotide kinase. Probe and proteins were mixed in 20 mm HEPES (pH 7.6), 50 mm KCl, 10 mm DTT, 1 mm EDTA, 5% glycerol, 0.07 mg/ml double-stranded poly[d(I-C)], and 0.3 mg/ml bovine serum albumin (Ohsako et al. 1994). After 30 min at room temperature, 10 μl of the 15-μl reaction volume were loaded on a 4% (0.36-kb probes) or a 5% (oligonucleotides probes) polyacrylamide (30:1, acrylamide/bisacrylamide ratio) gel. In assays with Sc/Da heterodimers, 600 ng of Da and 500 ng of Sc were preincubated at 4°C during 30 min before probe addition.
DNA sequencing
DNA sequencing was carried out with the Promega fmole or ABI Taq cycle sequencing systems. Assembling and analysis of sequences were performed with the University of Wisconsin GCG software package (Devereux et al. 1984).
Acknowledgments
We are very grateful to J.A. Campos-Ortega, S. Campuzano, J.F. de Celis, F. Jiménez, M. Ruiz-Gómez, and colleagues of our laboratory for constructive comments on the manuscript; S. González-Crespo for advice; J.F. de Celis, I. Rodríguez, and S. Sotillos for allowing us to use the UAS-m7 and UAS-m8 before publication; R. Cantera, M. Dushay, Y. Engstrom, and S. Govind for communication and sharing of results and materials before publication; A. Brand and S. González-Crespo for fly stocks and antibodies; M. Caudy for expression vectors; and E. Madueño and M. Dermitzakis for help in DNA sequencing. A predoctoral fellowship from Comunidad Autónoma de Madrid to J.C. is acknowledged. This work was supported by grants from Dirección General de Investigación Científica y Técnica (PB93-0181), Comunidad Autónoma de Madrid, EC (contract CHRX-CT94-0692), and an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
The DNA sequences of the SMC enhancers reported in this paper have been submitted to the GenBank/EMBL library under accession nos. AF060506 (for D. melanogaster) and AF060507 (for D. virilis).
Footnotes
E-MAIL jmodol@cbm.uam.es; FAX 34-91-397-4799.
References
- Alifragis P, Poortinga G, Parkhurst SM, Delidakis C. A network of interacting transcriptional regulators involved in Drosophila neural fate specification revealed by the yeast two-hybrid system. Proc Natl Acad Sci. 1997;94:13099–13104. doi: 10.1073/pnas.94.24.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora K, Dai H, Kazuko SG, Jamal J, O’Connor MB, Letsou A, Warrior R. The Drosophila schnurri gene acts in the Dpp/TGFβ signalling pathway and encodes a transcription factor homologous to the human MBP family. Cell. 1995;81:781–790. doi: 10.1016/0092-8674(95)90539-1. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Simpson P. Choosing a cell fate: A view from the Notch locus. Trends Genet. 1991;7:403–408. doi: 10.1016/0168-9525(91)90264-q. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of Hairless directly activates transcription of Enhancer of split complex genes in response to Notch receptor activity. Genes & Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Bang AG, Bailey AM, Posakony JW. Hairless promotes stable commitment to the sensory organ precursor cell fate by negatively regulating the activity of the Notch signaling pathway. Dev Biol. 1995;172:479–494. doi: 10.1006/dbio.1995.8033. [DOI] [PubMed] [Google Scholar]
- Brand M, Jarman AP, Jan LY, Jan YN. asense is a Drosophila neural precursor gene and is capable of initiating sense organ formation. Development. 1993;119:1–17. doi: 10.1242/dev.119.Supplement.1. [DOI] [PubMed] [Google Scholar]
- Cabrera CV, Alonso MC. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano S, Modolell J. Patterning of the Drosophila nervous system: The achaete-scute gene complex. Trends Genet. 1992;8:202–207. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]
- Campuzano S, Carramolino L, Cabrera CV, Ruiz-Gómez M, Villares R, Boronat A, Modolell J. Molecular genetics of the achaete-scute gene complex of D. melanogaster. Cell. 1985;40:327–338. doi: 10.1016/0092-8674(85)90147-3. [DOI] [PubMed] [Google Scholar]
- Cubas P, Modolell J. The extramacrochaetae gene provides information for sensory organ patterning. EMBO J. 1992;11:3385–3393. doi: 10.1002/j.1460-2075.1992.tb05417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, de Celis JF, Campuzano S, Modolell J. Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing disc. Genes & Dev. 1991;5:996–1008. doi: 10.1101/gad.5.6.996. [DOI] [PubMed] [Google Scholar]
- Dawson SR, Turner DL, Weintraub H, Parkhurst S. Specificity for the Hairy/Enhancer of split basic helix-loop-helix (bHLH) proteins map outside the bHLH domain and suggests two separable models of transcriptional repression. Mol Cell Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, de Celis J, Ligoxygakis P, Preiss A, Delidakis C, Bray S. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: The E(spl) genes mediate only a subset of Notch activities during imaginal development. Development. 1996a;122:2719–2728. doi: 10.1242/dev.122.9.2719. [DOI] [PubMed] [Google Scholar]
- de Celis JF, García-Bellido A, Bray SJ. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development. 1996b;122:359–369. doi: 10.1242/dev.122.1.359. [DOI] [PubMed] [Google Scholar]
- de la Concha A, Dietrich U, Weigel D, Campos-Ortega JA. Functional interactions of neurogenic genes of Drosophila melanogaster. Genetics. 1988;118:499–508. doi: 10.1093/genetics/118.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeverli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez M, Campuzano S. asense, a member of the Drosophila achaete-scute complex, is a proneural and neural differentiation gene. EMBO J. 1993;12:2049–2060. doi: 10.1002/j.1460-2075.1993.tb05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dushay MS, Asling B, Hultmark D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci. 1996;93:10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis HM, Spann DR, Posakony JW. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990;61:27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- Fan CM, Maniatis T. A DNA-binding protein containing two widely separated zinc finger motifs that recognize the same DNA sequence. Genes & Dev. 1990;4:29–42. doi: 10.1101/gad.4.1.29. [DOI] [PubMed] [Google Scholar]
- Garrell J, Modolell J. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- Ghysen A, Dambly-Chaudière C. From DNA to form: The achaete-scute complex. Genes & Dev. 1988;2:495–501. doi: 10.1101/gad.2.5.495. [DOI] [PubMed] [Google Scholar]
- Giebel B, Campos-Ortega JA. Functional dissection of the Drosophila Enhancer of split protein, a suppressor of neurogenesis. Proc Natl Acad Sci. 1997;94:6250–6254. doi: 10.1073/pnas.94.12.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigliani F, Longo F, Gaddini L, Battaglia PA. Interactions among the bHLH domains of the proteins encoded by the Enhancer of split and achaete-scute gene complexes of Drosophila. Mol & Gen Genet. 1996;251:628–634. doi: 10.1007/BF02174111. [DOI] [PubMed] [Google Scholar]
- Gómez-Skarmeta JL, Rodríguez I, Martínez C, Culí J, Ferrés-Marcó MD, Beamonte D, Modolell J. Cis-regulation of achaete and scute: Shared enhancer-like elements drive their coexpression in proneural clusters of the imaginal discs. Genes & Dev. 1995;9:1869–1882. doi: 10.1101/gad.9.15.1869. [DOI] [PubMed] [Google Scholar]
- Gómez-Skarmeta JL, Diez del Corral R, de la Calle-Mustienes E, Ferrés-Marcó D, Modolell J. araucan and caupolican, two members of the novel Iroquois complex, encode homeoproteins that control proneural and vein forming genes. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- Grieder NC, Nellen D, Burke R, Basler K, Affolter M. schnurri is required for Drosophila Dpp signalling and encodes a zinc finger protein similar to the mammalian transcription factor PRDII-BF1. Cell. 1995;81:791–800. doi: 10.1016/0092-8674(95)90540-5. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- Hemsley A, Arnheim N, Toney M, Cortopassi G, Galas D. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989;17:6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Dambly-Chaudière C, Ghysen A. The emergence of sense organs in the wing disc of Drosophila. Development. 1991;111:1087–1095. doi: 10.1242/dev.111.4.1087. [DOI] [PubMed] [Google Scholar]
- Ip YT, Reach M, Engstrom Y, Kadalayil L, Cai H, González-Crespo S, Tatei K, Levine M. Dif, a Dorsal related gene that mediates an immune response in Drosophila. Cell. 1993;75:753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Brand M, Jan LY, Jan YN. The regulation and function of the helix-loop-helix gene, asense, in Drosophila neural precursors. Development. 1993;119:19–29. doi: 10.1242/dev.119.Supplement.19. [DOI] [PubMed] [Google Scholar]
- Jennings B, Preiss A, Delidakis C, Bray S. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994;120:3537–3548. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- Jennings B, de Celis J, Delidakis C, Preiss A, Bray S. Role of Notch and achaete-scute complex in the expression of Enhancer of split bHLH proteins. Development. 1995;121:3745–3752. [Google Scholar]
- Klemenz R, Weber U, Gehring W. The white gene as a marker in a new P-element vector for gene transfer in Drosophila. Nucleic Acids Res. 1987;15:3947–3959. doi: 10.1093/nar/15.10.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisch M, Haenlin M, Campos-Ortega JA. Lateral inhibition mediated by the Drosophila neurogenic gene Delta is enhanced by proneural proteins. Proc Natl Acad Sci. 1994;91:10139–10143. doi: 10.1073/pnas.91.21.10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates transcriptional activation of the Enhancer of split complex genes triggered by Notch signaling. Genes & Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Lenardo MJ, Baltimore D. NF-κB: A pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989;58:227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Martínez C, Modolell J. Cross-regulatory interactions between the proneural achaete and scute genes of Drosophila. Science. 1991;251:1485–1487. doi: 10.1126/science.1900954. [DOI] [PubMed] [Google Scholar]
- Martínez C, Modolell J, Garrell J. Regulation of the proneural gene achaete by helix-loop-helix proteins. Mol Cell Biol. 1993;13:3514–3521. doi: 10.1128/mcb.13.6.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolell J. Patterning of adult peripheral nervous system of Drosophila. Persp Dev Neurobiol. 1997;4:285–304. [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nakao K, Campos-Ortega JA. Persistent expression of genes of the Enhancer of Split complex suppresses neural development in Drosophila. Neuron. 1996;16:275–286. doi: 10.1016/s0896-6273(00)80046-x. [DOI] [PubMed] [Google Scholar]
- Oellers N, Dehio M, Knust E. bHLH proteins encoded by the Enhancer of split complex of Drosophila negatively interfere with transcriptional activation mediated by proneural genes. Mol Gen Genet. 1994;244:465–473. doi: 10.1007/BF00583897. [DOI] [PubMed] [Google Scholar]
- Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes & Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with Hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Parras C, García-Alonso LA, Rodríguez I, Jiménez F. Control of neural precursor specification by proneural proteins in the CNS of Drosophila. EMBO J. 1996;15:6394–6399. [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Schweisguth F, Posakony JW. Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development. 1994;120:1433–1441. doi: 10.1242/dev.120.6.1433. [DOI] [PubMed] [Google Scholar]
- Simpson P. Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development. 1990;109:509–519. doi: 10.1242/dev.109.3.509. [DOI] [PubMed] [Google Scholar]
- ————— Notch signalling in development: On equivalence groups and asymmetric developmental potential. Curr Opin Genet Dev. 1997;7:537–542. doi: 10.1016/s0959-437x(97)80083-4. [DOI] [PubMed] [Google Scholar]
- Singson A, Leviten MW, Bang AG, Hua XH, Posakony JW. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibition signaling. Genes & Dev. 1994;8:2058–2071. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Carroll SB. Regulation of achaete-scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes & Dev. 1991;5:984–995. doi: 10.1101/gad.5.6.984. [DOI] [PubMed] [Google Scholar]
- Stern C. Two or three bristles. Am Scientist. 1954;42:213–247. [Google Scholar]
- Steward R. Dorsal, an embryonic polarity gene in Drosophila, is homologous to the vertebrate proto-oncogene c-rel. Science. 1987;238:692–694. doi: 10.1126/science.3118464. [DOI] [PubMed] [Google Scholar]
- Tata F, Hartley DA. Inhibition of cell fate in Drosophila by Enhancer of split genes. Mech Dev. 1995;51:305–315. doi: 10.1016/0925-4773(95)00377-0. [DOI] [PubMed] [Google Scholar]
- Tietze K, Oellers N, Knust E. Enhancer of splitD, a dominant mutation of Drosophila, and its use in the study of functional domains of a helix-loop-helix protein. Proc Nat Acad Sci. 1992;89:6152–6156. doi: 10.1073/pnas.89.13.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, Kimmel BE, Rubin GM. Rough, a Drosophila homeobox gene required in photoreceptors R2 and R5 for inductive interactions in the developing eye. Cell. 1988;55:771–784. doi: 10.1016/0092-8674(88)90133-x. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Ellis HM, Posakony JW. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Powell PA, Pasternak D, Singson A, Posakony JW. Spatial regulation of proneural gene activity: Auto- and cross-activation of achaete is antagonized by extramacrochaetae. Genes & Dev. 1992;6:2592–2605. doi: 10.1101/gad.6.12b.2592. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Bailey AM, Esnayra J, Ede K, Posakony JW. Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes & Dev. 1994;8:2729–2742. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- Wodarz A, Hinz U, Engelbert M, Knust E. Expression of crumbs confers apical character on plasma membrane domains of ectodermal epithelia in Drosophila. Cell. 1995;82:67–76. doi: 10.1016/0092-8674(95)90053-5. [DOI] [PubMed] [Google Scholar]