Abstract
The p53 tumor suppressor gene product interacts with the p300 transcriptional coactivator that regulates the transactivation of p53-inducible genes. The adenovirus E1A protein has been shown to bind to p300 and inhibit its function. E1A inhibits p53 transactivation and also promotes p53 accumulation by a p300-dependent mechanism. Murine double minute 2 (Mdm2) is a transcriptional target of p53 that binds to p53 and inhibits its transcriptional activity. E1A inhibited mdm2 transactivation without affecting the expression of p21WAF1 or Bax, which resulted in high levels of p53 accumulation and apoptosis. Ectopic expression of p300 restored Mdm2 levels and inhibited p53-dependent apoptosis, as did ectopic expression of Mdm2. Thus, p300 is required for mdm2 induction by p53 and the subsequent inhibition of p53 stabilization. Inhibition of p300 by E1A results in stabilization of p53 and causes apoptosis. Moreover, E1B 19K or Bcl-2 expression in E1A-transformed cells abrogated p53-dependent apoptosis by restoring mdm2 transactivation by p53. Hence, p300 regulation of mdm2 expression controls apoptotic activity of p53, and 19K or Bcl-2 bypass E1A inhibition of p300 transactivation of Mdm2.
Keywords: p53, Mdm2, p300, apoptosis, Bcl-2, E1B 19K, E1A, transcription
The tumor suppressor p53 gene product is a negative regulator of cellular growth and transformation (Vogelstein and Kinzler 1992; Ko and Prives 1996; Levine 1997; White 1996). Many studies have demonstrated that p53 functions as a transcriptional regulator by sequence-specific DNA binding (El-Deiry et al. 1992; Funk et al. 1992; Pietenpol et al. 1994). The p53 gene product regulates transcription by activation or repression (Ko and Prives 1996). Studies have shown that DNA damage increases p53 levels, which promotes cell cycle arrest allowing DNA repair to occur, or it induces apoptosis, presumably when damage is beyond repair (Ko and Prives 1996; White 1996; Levine 1997). The growth arrest function of p53 is implemented predominantly by transactivation of the cyclin-dependent kinase inhibitor p21/WAF1/CIP1 (El-Deiry et al. 1993) and p53 can induce apoptosis by up-regulating the death-promoting bax gene (Miyashita and Reed 1995). Other transcriptional targets for p53 include GADD45 (Kastan et al. 1992), murine double minute 2 (mdm2) (Barak et al. 1993; Wu et al. 1993), cyclin G (Okamoto and Beach 1994), and IGF-BP3 (Buckbinder et al. 1995). The DNA-binding ability of p53 appears to be important because the most frequently occurring p53 mutations in human tumors are found in this domain (Hollstein et al. 1991; Ko and Prives 1996). Hence, intact p53 transcriptional function is important to maintain genomic integrity.
The mdm2 gene was originally cloned because of its amplification in a spontaneously transformed murine BALB/c cell line (Fakharzadeh et al. 1991). The human homolog of Mdm2 protein was shown to be a negative regulator of p53. Mdm2 protein inhibits p53-mediated functions of G1 arrest and apoptosis (Chen et al. 1996a), most likely by binding to the amino-terminal transactivation domain of p53 (Momand et al. 1992; Oliner et al. 1993). Furthermore, Mdm2 appears to direct p53 degradation via the ubiquitin pathway (Haupt et al. 1997; Kubbutat et al. 1997). The mdm2 promoter contains p53 binding consensus sequences in which p53 binds and positively regulates its expression, creating a negative feedback loop for regulating the activity and levels of p53 (Barak et al. 1993; Haupt et al. 1996; Wu et al. 1993). The functional interdependence of Mdm2 and p53 was exemplified in studies with knockout mice. Loss of Mdm2 resulted in early embryonic lethality, which was rescued by deletion of p53 (Donehower et al. 1992; Montes de Oca Luna et al. 1995). Thus, Mdm2 is required in vivo for down-modulation of p53 function and perturbation of this regulation can be deleterious to embryonic development.
The CBP/p300 family members regulate transcription by functioning as transcriptional coactivators. Although the precise mechanism of transcriptional adaptor function is not known, CBP/p300 and an interacting protein, P/CAF, have been shown to have histone acetyltransferase activity (Bannister and Kouzarides 1996; Ogryzko et al. 1996; Yang et al. 1996), implicating a role for histone acetylation in transcriptional regulation. These proteins also interact with several transcription factors such as the TAFs (Thut et al. 1995), TBP (Abraham et al. 1993), CREB (Chrivia et al. 1993; Kwok et al. 1994), c-Jun/v-Jun (Bannister and Kouzarides 1995), c-Myb/v-Myb (Dai et al. 1996), c-Fos (Bannister and Kouzarides 1995), and others, which may determine the specificity of the regulation. The p300 family of proteins has been shown recently to bind to p53 and function as coactivators of p53-inducible genes (Avantaggiati et al. 1997; Gu et al. 1997; Lill et al. 1997; Scolnick et al. 1997). The amino-terminal activation domain of p53 interacts directly with the carboxy-terminal of p300 (Gu and Roeder 1997). It has also been shown that p300 can acetylate the carboxy-terminal domain of p53 and that this modification increases the sequence-specific DNA-binding ability of p53 (Gu and Roeder 1997). Thus, acetylation of specific transcription factors may reflect one level of p300 transcriptional regulation.
The adenoviral early region 1 (E1) genes encode for proteins that aid in cellular transformation by activating proliferation and suppressing apoptosis (White 1993; White and Gooding 1994). Expression of the adenoviral E1A gene stimulates cell cycle progression by interacting with and subverting the function of cellular proteins required for normal cell cycle and transcription regulation. E1A interacts with the retinobalstoma (Rb) gene product as well as its family members, p107 and p130 (Dyson and Harlow 1992; Moran 1993; Whyte et al. 1988). E1A also binds to and sequesters p300 (Moran 1993; Eckner et al. 1994; Yang et al. 1996). E1A interactions with these cellular proteins are important for transformation as suggested by the fact that E1A mutants that fail to interact with these proteins are incapable of promoting transformation. Expression of E1A alone, however, is insufficient to transform primary baby rat kidney (BRK) cells because cell cycle deregulation by E1A also stimulates p53-dependent apoptosis. Binding of p300 to E1A cosegregates with induction of p53 and stabilization (Sanchez-Prieto et al. 1995; Chiou and White 1997; Querido et al. 1997). Transcriptional activation of p53 target genes, such as bax (Han et al. 1996), followed by induction of caspase activation implements cell death (Rao and White 1997; Sabbatini et al. 1997). Transcriptional activation of the p53 target gene p21WAF1 is also induced, which contributes to implementation of cell cycle arrest by p53, which is not apparent because of cell death (Sabbatini et al. 1995a,b; Han et al. 1996).
E1A-induced cellular transformation is sustained by coexpression of the Bcl-2 adenoviral homolog, E1B 19K or Bcl-2 itself, which inhibit E1A-induced, p53-mediated apoptosis. E1B 19K and Bcl-2 inhibit apoptosis in part by binding to the death-promoting Bax protein (Han et al. 1996). Thus E1B 19K or Bcl-2 expression inhibits p53-mediated apoptosis but not growth arrest (Debbas and White 1993; Chiou et al. 1994b; Sabbatini et al. 1995a; Han et al. 1996). These co-operative functions between early adenoviral genes stimulating cell cycle progression and inhibiting apoptosis ensures efficient transformation.
We report that p300 is required specifically for transactivation of the mdm2 gene by p53 and for regulating p53-mediated apoptosis. Cells expressing E1A were unable to up-regulate Mdm2, causing stabilization of high levels of p53 that resulted in p53-dependent apoptosis. In contrast, BRK cells expressing c-Myc instead of E1A, up-regulated Mdm2, causing inhibition of p53 stabilization and inhibition of p53-dependent apoptosis. Thus, the inabilty to down-regulate p53 is associated with apoptosis and may explain the differences between the p53-dependent apoptotic response of E1A versus Myc transformed cells. Furthermore, E1B 19K or Bcl-2 expression restores mdm2 transactivation at the level of mRNA, bypassing the requirement for p300 cotransactivation. Thus the Bcl-2 family members may regulate the specificity of p53-dependent target gene activation and thereby contribute to inhibition of p53-dependent apoptosis.
Results
Mdm2 expression regulates p53-dependent apoptosis in BRK cells
BRK cells were transformed by a temperature-sensitive p53 (val135) mutant and either adenovirus E1A (p53A) or c-Myc (LTR.1A). As previously reported, both types of cell lines expressed the temperature-sensitive p53 (val135) mutant in which p53 is in the wild-type conformation at the permissive temperature of 32°C and in the mutant conformation at the nonpermissive temperature of 37.5°C (Debbas and White 1993; Sakamuro et al. 1995). There are two striking differences between E1A + tsp53 (val135)-transformed (p53A) and myc + tsp53 (val135)-transformed (LTR.1A) cell lines. First, BRK cells expressing E1A undergo massive apoptosis at the permissive temperature (Debbas and White 1993; Chiou et al. 1994a; Sabbatini et al. 1995a,b), whereas the Myc-expressing cells undergo a wave of apoptosis within the first 24 hr at the permissive temperature but are predominantly resistant to apoptosis (Sakamuro et al. 1995). Second, E1A-expressing cells maintain high levels of p53, whereas p53 levels are low at 32°C in Myc-expressing cells (Debbas and White 1993; Sakamuro et al. 1995). Thus, high levels of p53 correlate with augmentation of the apoptotic response in E1A-expressing p53A cells. Because E1A stabilizes p53 (Lowe and Ruley 1993; Chiou et al. 1994b) and this activity requires that E1A binds to p300 (Chiou and White 1997), this suggests a possible function for sequestration of p300 and induction of p53 levels. Furthermore, recent reports demonstrate the requirement for p300 in p53 transactivation (Gu et al. 1997; Lill et al. 1997; Scolnick et al. 1997) and a role for Mdm2 in promoting p53 degradation (Haupt et al. 1997; Kubbutat et al. 1997). We therefore investigated the possible role of p300 in the Mdm2-dependent negative-feedback loop and the control of p53-dependent apoptosis.
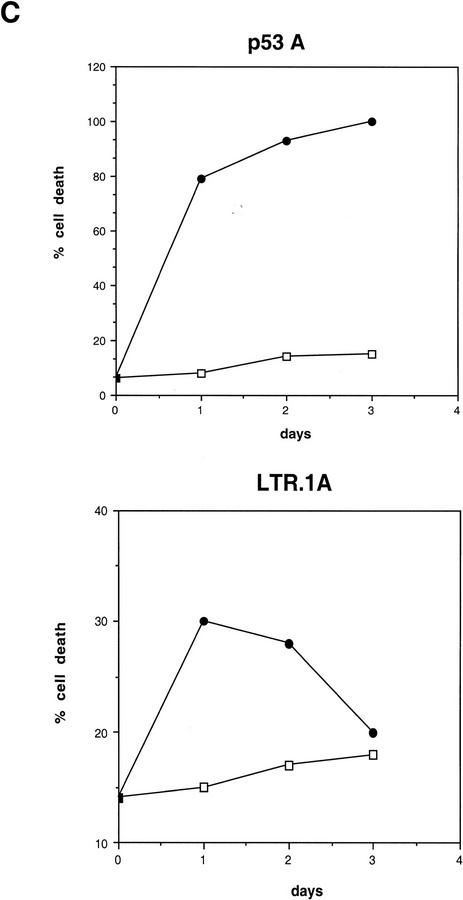
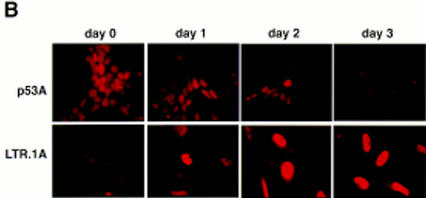
First, we determined whether the levels of p53-inducible gene products Mdm2, p21WAF1, and Bax were affected by the p300-binding protein E1A. Western blot analysis shows that high levels of p53 were stabilized in E1A-transformed cells and not stabilized in Myc-transformed cells at 32°C (Fig. 1A) as reported previously (Debbas and White 1993; Sakamuro et al. 1995). A lower band that may represent a proteolytic cleavage product of p53 was also observed in p53A cells as apoptosis progressed at the permissive temperature (Fig. 1A). High p53 levels correlated with apoptosis in p53A cells (Fig. 1, C, top, and D, top). Endogenous Mdm2 protein was initially present but was down-regulated at 32°C in p53A cells as shown by Western blot analysis (Fig. 1A) and immunofluorescence (Fig. 1B, top). Western blot analysis of Mdm2 in p53A cells at the permissive temperature showed a lower band at ∼50 kD, recognized specifically by the anti-Mdm2 monoclonal antibody (data not shown). This lower band may represent a cleavage product of Mdm2. However, this band disappeared as cells were incubated at the permissive temperature. Our studies did not investigate Mdm2 cleavage in apoptosis, however, we cannot rule out the possibility that Mdm2 cleavage may play a role in the regulation of apoptosis. Mdm2 down-regulation was surprising because mdm2 is a p53-regulated gene and suggested that p300 may be necessary for Mdm2 transactivation by p53.
Figure 1.
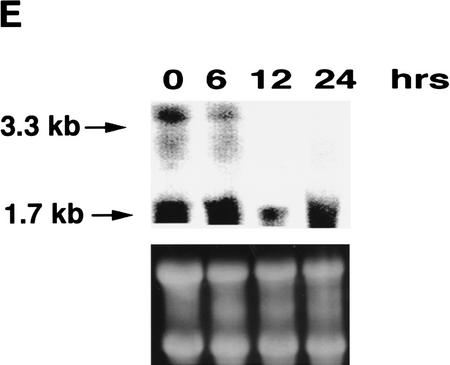
(A) Western blot analysis of p53, Mdm2, Bax, and p21WAF1 in p53A and LTR.1A cells after incubation at nonpermissive (37.5°C) and permissive (32°C) temperature at day 0 (lanes 1,6), day 1 (lanes 2,7), day 2 (lanes 3,8), day 3 (lanes 4,9), and day 4 (lanes 5,10). (B) Indirect immunofluorescence of Mdm2 in p53A (top) and LTR.1A (bottom) cells after incubation at 32°C from day 0 to day 3. (C) Cell viability by trypan blue staining of p53A and LTR.1A cells at the nonpermissive (37.5°C, □) and permissive (32°C, •) temperature from day 0 to day 3. (D) Analysis of p53A and LTR.1A cells incubated at 32°C from day 0 to day 4 by light microscopy to detect apoptotic cells. (E) Northern blot analysis of mdm2 expression of p53A cells incubated at 32°C for the indicated times.
In c-Myc-transformed LTR.1A cells, Mdm2 was up-regulated and p53 levels were low at 32°C (Fig. 1A,B, bottom). Therefore Myc- but not E1A-expressing BRK cells demonstrate p53-dependent induction of mdm2. Furthermore, high levels of Mdm2 in Myc-transformed BRK cells may have promoted p53 degradation (Fig. 1A). The LTR.1A cells initially undergo apoptosis but rapidly become resistant to p53-mediated cell death at 32°C (Fig. 1C, bottom, and 1D, bottom), which correlated with the low levels of p53 and up-regulation of Mdm2. Western blot analysis also showed an increase in the 50-kD lower band detected specifically by the anti-Mdm2 antibody (data not shown). Hence, the lower band that may represent Mdm2 cleavage product was also present in cells that were resistant to apoptosis. The decrease in Mdm2 levels observed in cells maintained at the restrictive temperature may be because of the confluency of the cells. At the restrictive temperature the BRK cell lines are proliferating and reach confluence in 2–3 days in this assay.
Mdm2 is only one of many p53-inducible genes that collectively act to modulate the physiological response to p53 induction. Therefore, p21WAF1 and Bax levels were also examined for transactivation in p53A and LTR.1A cells. In p53A cells, Bax and p21WAF1 levels were induced at 32°C even in the presence of E1A and inhibition of p300 (Fig. 1A) (Sabbatini et al. 1995b; Han et al. 1996). LTR.1A cells down-regulated Bax but up-regulated low levels of p21WAF1 at 32°C (Fig. 1A). We do not know why Bax is also present in LTR.1A cells at the restrictive temperature, but this suggests that Bax may require an activation step to induce cell death. Similar results (data not shown) were obtained with independent E1A + tsp53 (val135)-transformed and c-Myc + tsp53 (val135)-transformed clones (Debbas and White 1993; Sakamuro et al. 1995). These results indicate that E1A and Myc alter p53-dependent gene expression differentially.
To determine whether the absence of Mdm2 protein in p53A cells reflected a transcriptional event, we performed Northern blot analysis using cytoplasmic RNA extracted from E1A-transformed p53A cells incubated at the permissive temperature for the indicated time intervals (Fig. 1E). Two transcripts (3.3 and 1.7 kb) hybridized to an mdm2-specific probe. Both transcripts, particularly the 3.3 kb, were down-regulated to some extent as cells were incubated at 32°C. The detection of mdm2 mRNA in p53A cells was difficult because mdm2 expression was very low to begin with and was not induced by wild-type p53. Induction of mdm2 mRNA by p53, however, was observed in BRK cell lines expressing E1B 19K (see below). In contrast, we have previously reported that Bax and p21WAF1 mRNAs were up-regulated transiently in p53A cells incubated at the permissive temperature within the same time interval analyzed here for mdm2 mRNA expression (Sabbatini et al. 1995a,b; Han et al. 1996). Attempts to examine mdm2 mRNA levels at later times failed as the RNA in p53A cells became degraded as most of the cells became apoptotic. Therefore, E1A, although preventing mdm2 expression, did not affect p53’s ability to transactivate Bax and p21WAF1. These results suggest that p300 may be required for Mdm2 transactivation, but may be dispensable for Bax and p21WAF1 regulation.
p300 is required for the up-regulation of Mdm2 by p53 and inhibition of p53-mediated apoptosis
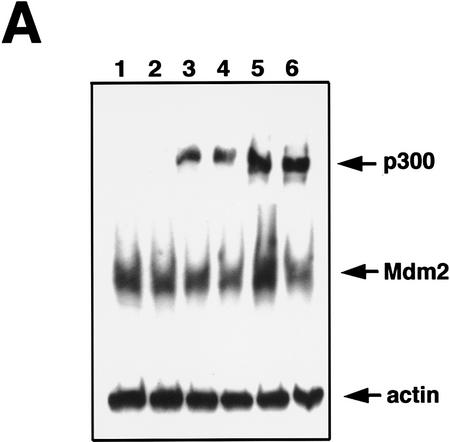
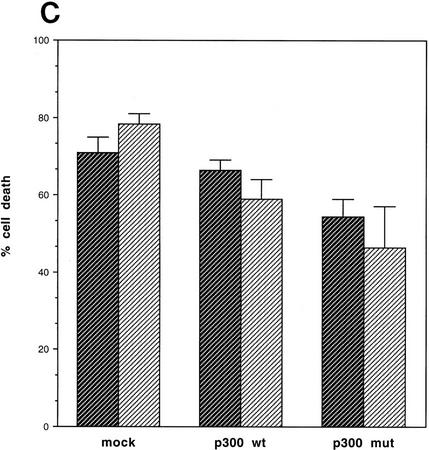
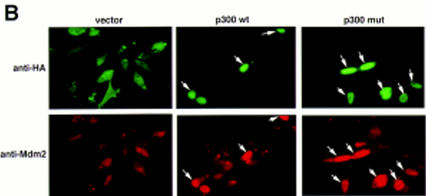
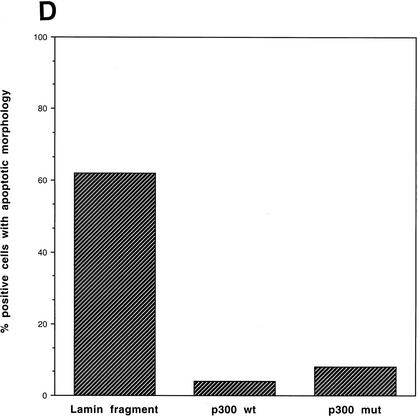
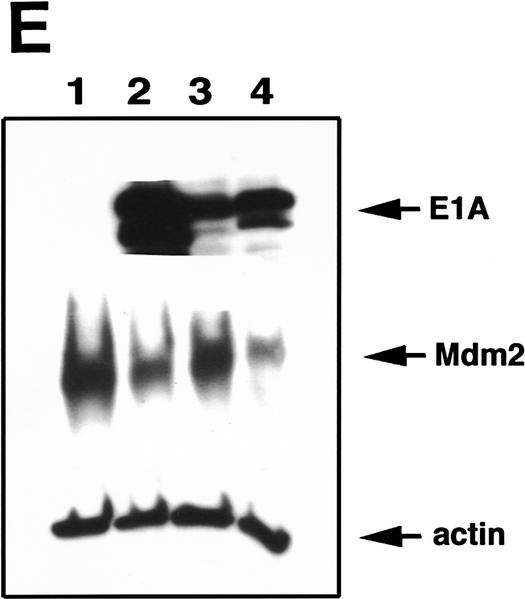
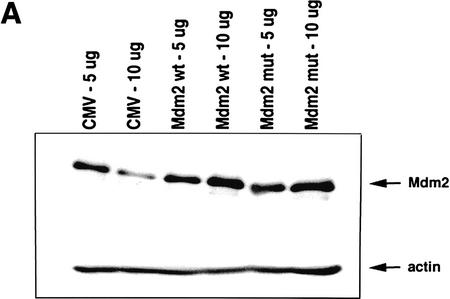
We next attempted to rescue p53A cells from apoptosis at the permissive temperature by ectopically expressing p300. Vector alone, wild-type p300, and a functional but truncated form of p300 with a deletion of E1A-binding domain (Arany et al. 1994) were transiently transfected in p53A cells. p53-dependent apoptosis was examined at the permissive temperature 48 hr post-transfection. p53A cells transiently expressed the exogenous wild-type p300 and the E1A-binding mutant form of p300, neither of which were detected in mock transfected cells (Fig. 2A,B). The up-regulation of Mdm2 in p300-transfected cells was not detected by Western blot analysis above endogenous levels (Fig. 2A) because the transfection efficiency was low in transient assays. However, the up-regulation of Mdm2 in both wild-type and E1A-binding mutant of p300 was easily observed in 90% of transfected cells by indirect immunofluorescence when individual transfected cells were examined (Fig. 2B, arrows). Furthermore, ectopic expression of wild-type and mutant p300 rescued cells from apoptosis as cell viability was elevated as much as 20%–40% above mock-transfected cells (Fig. 2C). The mutant form of p300, which was unable to bind E1A, protected cells better than wild-type p300, presumably because it can evade E1A sequestration. In addition, we determined the percentage of cells with apoptotic morphology that were expressing wild-type p300 ectopically or the E1A-binding mutant of p300 following transient transfection with 24 μg of plasmid DNA. Apoptotic morphology was determined by counting cells that were rounded in shape and detaching from the dish. The percentage of cells with apoptotic morphology transiently expressing a control protein, a fragment of the lamin A protein (Rao et al. 1996), compared to p300 and mutant p300 transfected p53A cells at the permissive temperature was determined by indirect immunofluorescence (Fig. 2D). Only 4%–8% of the cells expressing wild-type p300 and mutant p300, respectively, have apoptotic morphology compared to 62% cells expressing the lamin fragment (Fig. 2D). These results reaffirm that expression of wild-type p300 or the E1A-binding p300 mutant is sufficient to rescue cells from p53-mediated apoptosis.
Figure 2.
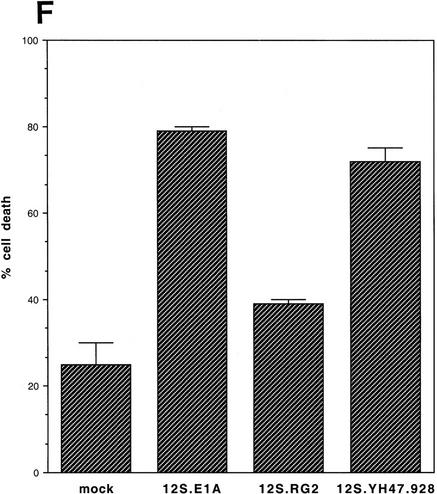
Transient expression of p300 and 12S E1A plasmids in p53A and LTR.1A cells, respectively, reversed its apoptotic phenotype at the permissive temperature. (A) Western blot analysis of ectopically expressed HA-tagged p300, endogenous Mdm2, and actin in p53A cells transfected transiently by electroporation with pCMVβ alone (lane 1, 12 μg; lane 2, 24 μg), pCMVβp300 (p300 wild-type) (lane 3, 12 μg; lane 4, 24 μg), or p300Δ30 (p300 mutant) (lane 5, 12 μg; lane 6, 24 μg). (B) Indirect immunofluorescence of HA-tagged p300 and endogenous Mdm2 in p53A cells transfected transiently with 24 μg of vector alone, wild-type p300, and mutant p300 (arrows indicate cells with ectopic p300 expression and up-regulation of Mdm2). (C) Cell viability as measured by trypan blue staining of p53A cells transfected transiently with vector alone, wild-type p300, or mutant p300. (Dark hatched bars) 12 μg; (light hatched bars) 24 μg. (D) Percentage of apoptotic cells that are positive for ectopic expression of a control lamin fragment, p300 wild-type and p300 mutant determined by cell morphology using indirect immunofluorescence in p53A cells transfected transiently with 24 μg of pCEP4-LA(1–406). pCMVβp300 or pCMVβp300Δ30 plasmid DNAs. (E) Western blot analysis of E1A, Mdm2, and actin in LTR.1A cells transfected transiently with 10 μg of vector alone (lane 1), 12S E1A (lane 2), 12S.RG2 (lane 3), or 12S.YH47.928 (lane 4). (F) Cell viability measured by trypan blue exclusion of LTR.1A cells transfected transiently with vector alone, 12S E1A, 12S.RG2, or 12S.YH47.928.
To determine whether LTR.1A cells can be made susceptible to p53-mediated death, we transiently transfected wild-type E1A (12S E1A) into these cells. Transient expression of 12S E1A in LTR.1A cells (Fig. 2E) promoted apoptosis at the permissive temperature (Fig. 2F). In addition, Mdm2 levels were down-regulated in cells transfected with 12S E1A as detected by Western blot analysis (Fig. 2E). Because E1A can also bind to Rb, we transiently transfected LTR.1A cells with 12S E1A mutant (12S.RG2) that is unable to bind to p300 but retains it ability to bind to Rb, and a 12S E1A mutant (12S.YH47.928) that binds to p300 but not to Rb (Wang et al. 1992), and examined p53-dependent apoptosis at the permissive temperature. Cells transfected with 12S.RG2 did not down-regulate Mdm2 (Fig. 2E) and were resistant to p53-dependent apoptosis (Fig. 2F). In contrast, cells transfected with 12S.YH47.928 down-regulated Mdm2 (Fig. 2E) and were susceptible to p53-dependent apoptosis at the permissive temperature (Fig. 2F). These results suggest that the ability of E1A to bind to p300 and inhibit Mdm2 induction correlates with E1A-mediated, p53-dependent apoptosis.
Overexpression of Mdm2 inhibits p53-mediated apoptosis
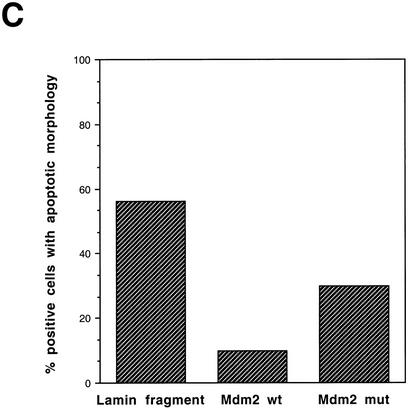
To determine whether Mdm2 expression can bypass the requirement for p300 cotransactivation and rescue p53A cells from p53-mediated death, we transiently transfected p53A cells with expression plasmids encoding wild-type Mdm2 and a truncated form of Mdm2 lacking the amino-terminal p53-binding domain (Haupt et al. 1997). Transient overexpression of wild-type and mutant Mdm2 were detected by Western blot analysis in p53A cells (Fig. 3A). Transient transfection of cells with 5 or 10 μg wild-type Mdm2 rescued cells from p53-dependent apoptosis at the permissive temperature (Fig. 3B). The truncated form of Mdm2 rescued cells from apoptosis less efficiently than the wild-type protein (Fig. 3B), presumably because it was not able to bind to p53 and promote its degradation. Furthermore, we examined the cell morphology of p53A cells expressing a wild-type and mutant Mdm2 plasmid DNA as compared to the morphology of cells expressing a control lamin fragment incubated at the permissive temperature using indirect immunofluorescence (Fig. 3C). The percentage of apoptotic cells expressing wild-type and mutant Mdm2 was only 10% and 30%, respectively, as compared to 56% apoptosis in cells expressing the control lamin fragment (Fig. 3C). The Mdm2 mutant, however, possessed greater apoptosis-suppressing activity than the control lamin fragment, which may indicate that Mdm2 can suppress apoptosis by a mechanism independent of p53 binding. These results demonstrate that overexpression of Mdm2 can rescue E1A-expressing cells from p53-mediated apoptosis by either promoting p53 degradation or by inhibiting its activity by direct interaction.
Figure 3.
Expression of Mdm2 rescues p53A cells from apoptosis at the permissive temperature. (A) Western blot analysis of p53A cells transfected transiently with 5 and 10 μg of pCMV vector alone, mdm2 wild-type (pCOC-X2), or mdm2 mutant (pCOC-ΔXM) plasmid DNAs. (B) Cell viability assessed by trypan blue staining of p53A cells transfected transiently with mdm2 expression plasmids incubated at 32°C. (Dark hatched bars) 5 μg; (light hatched bars) 10 μg. (C) Percentage of apoptotic cells that were positive for ectopic expression of a control lamin fragment, Mdm2 wild-type, and Mdm2 mutant determined by cell morphology using indirect immunofluorescence in p53A cells transfected transiently with 10 μg of pCEP4-LA (1–406), pCOC-X2, or pCOC-ΔXM plasmid DNAs.
E1B 19K or Bcl-2 can bypass E1A inhibition of p300 and restore mdm2 transactivation
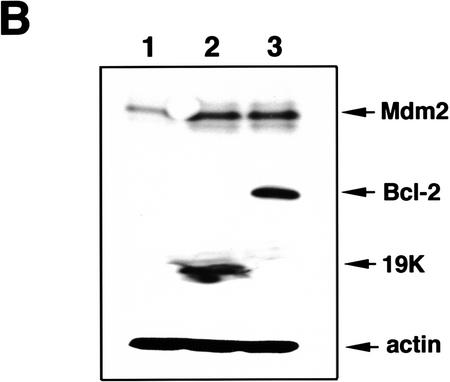
Another mechanism for suppressing p53-dependent apoptosis is through expression of Bcl-2 or E1B 19K, which function in part by binding to Bax and inactivating its pro-apoptotic function. To test if Bcl-2 or E1B 19K could also function to suppress apoptosis by influencing p53 target gene expression, we examined p53-inducible gene products, Mdm2, p21WAF1, and Bax, in p53A cells stably expressing adenovirus E1B 19K (19K1) or its human homolog Bcl-2 (4B).
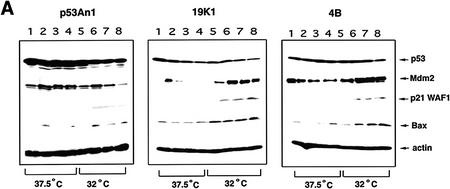
As reported previously, the expression of 19K or Bcl-2 abrogates p53-dependent apoptosis at 32°C (Chiou et al. 1994a; Sabbatini et al. 1995a; Han et al. 1996). Western blot analysis indicated that p21WAF1 and Bax levels increased at 32°C regardless of whether or not 19K or Bcl-2 were expressed and the levels of p53 remained high (Fig. 4A). However, the presence of 19K or Bcl-2 dramatically up-regulated Mdm2 levels, whereas the control E1A + tsp53 (val135)-transformed p53A cells transfected with vector alone (p53An1) down-regulated Mdm2 as expected (Fig. 4A). This suggested that E1B 19K or Bcl-2 could restore p53-dependent mdm2 expression in p53A cells.
Figure 4.
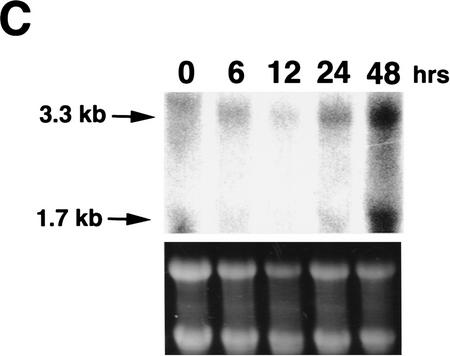
(A) Western blot analysis of p53, Mdm2, bax, and p21WAF1 in p53A cells stably expressing pSVneo (p53 An1), E1B 19K (19K1), or Bcl-2 (4B). p53An1, 19K1, and 4B cell lines were made as described previously (Sabbatini et al. 1995a). Cell lysates were made after incubating cells at 37.5 or 32°C during day 0 (lanes 1,5), day 1 (lanes 2,6), day 2 (lanes 3,7), and day 3 (lanes 4,8). (B) Western blot analysis of Mdm2, Bcl-2, E1B 19K, and actin in p53An1 cells transfected transiently with pCDNA3 alone (lane 1), pCMV194K (lane 2), and pCDNA3myc-tagged-bcl-2 (lane 3). (C) Northern blot analysis of mdm2 expression using 30 μg of cytoplasmic RNA of 19K1 cells incubated at 32°C for the indicated time intervals.
To determine whether the expression of E1B 19K or Bcl-2 was sufficient for up-regulation of Mdm2 levels, we transiently transfected 19K or Bcl-2 in p53An1 cells and examined Mdm2 levels at 32°C. Western blot analysis (Fig. 4B) and indirect immunofluorescence (data not shown) indicated that Mdm2 levels were up-regulated when either 19K or Bcl-2 was expressed transiently.
To determine whether the expression of E1B 19K up-regulates Mdm2 at the transcriptional level, we performed Northern blot analysis using cytoplasmic RNA from cells expressing E1B 19K (19K1) incubated at 32°C for the indicated time intervals (Fig. 4C). As shown in Fig. 4C, mdm2 mRNA expression was up-regulated after 48 hr incubation at permissive temperature. This suggested that E1B 19K and Bcl-2 may restore mdm2 transactivation by p53, which would otherwise be inhibited by E1A binding to p300. In addition, previous studies have shown that Bax and p21WAF1 mRNA and protein were also up-regulated in 19K1 cells incubated at the permissive temperature (Sabbatini et al. 1995a,b; Han et al. 1996). Therefore the presence of 19K did not affect the expression of Bax or p21WAF1, however, E1B 19K can directly bind to Bax and inhibit its function (Han et al. 1996). E1B 19K and Bcl-2 have been shown to relieve transcriptional repression by E1A and p53, which may be responsible for inhibition of apoptosis (Shen and Shenk 1994; Sabbatini et al. 1995a; Murphy et al. 1996) and Mdm2 may be a physiological target of this activity.
Discussion
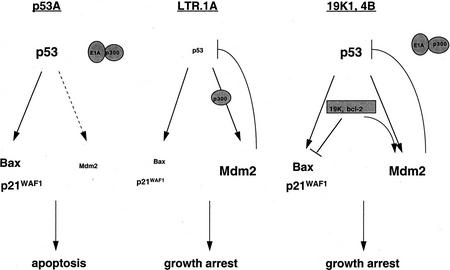
E1A + tsp53 (val135)-transformed p53A cells were unable to transactivate mdm2 as a result of E1A inhibition of p300, indicating that p53 requires p300 to transactivate mdm2 (Fig 5). Furthermore, low Mdm2 levels in p53A cells relieve the negative-feedback regulation of p53 resulting in high p53 accumulation leading to apoptosis. In contrast, p300 function is intact and able to transactivate mdm2 with p53 in c-Myc + tsp53 (val135)-transformed LTR.1A cells (Fig. 5). Up-regulation of mdm2 inhibits p53 function and promotes p53 degradation (Haupt et al. 1997; Kubbutat et al. 1997), which in turn inhibits p53-mediated cell death resulting in growth arrest in LTR.1A cells. The cellular levels of p53 thereby control the biological response to p53 such that lower levels of p53 induce growth arrest, whereas higher levels induce apoptosis (Chen et al. 1996b). Therefore, we can conclude that p300 regulation of Mdm2 levels determines whether the physiological response to p53 is growth arrest or apoptosis. Curiously, the expression of the 19K or Bcl-2 proteins in p53A cells (19K1, 4B) bypasses the requirement for p300 in mdm2 transactivation (Fig. 5). Thus, prosurvival members of the Bcl-2 family can inhibit p53-dependent apoptosis by restoring the capacity of p53 to transactivate mdm2.
Figure 5.
A model illustrating the regulation of cell death in p53A, LTR.1A, and 19K1/4B cells (for details, see text).
Interestingly, p300 cotransactivation is not required for the other p53-inducible genes, Bax and p21WAF1. It has been reported previously that E1A inhibits p53-mediated transactivation using promoter reporter assays (Steegenga et al. 1996). Our studies, however, demonstrate that E1A specifically inhibits endogenous mdm2 transactivation and not other p53-inducible genes such as bax and p21WAF1. p300/CBP functions by interacting not only with p53, but also with other factors such as the TAFs (Thut et al. 1995), TBP (Abraham et al. 1993), CREB (Chrivia et al. 1993; Kwok et al. 1994), c-Jun/v-Jun (Bannister and Kouzarides 1995), c-Myb/v-Myb (Dai et al. 1996), c-Fos (Bannister and Kouzarides 1995), and others. p300 binding to p53 alone may not regulate the specificity of cotransactivation. However, as numerous other protein interactions with p300 occur, this may determine the specificity of transactivation, accounting for differential regulation of p53-inducible genes by p300.
p300/CBP transcriptional coactivators have been shown to have histone acetyltransferase activity, which can modify chromatin structure and enhance gene expression (Bannister and Kouzarides 1996; Ogryzko et al. 1996). Recently, p300 has been shown to acetylate p53 itself (Gu and Roeder 1997). The acetylation of p53 increases the binding activity of p53 to specific consensus sequences. In vivo, this activity may be required for facilitating p53 tetramer interactions with the DNA template, thereby promoting mdm2 transactivation. However, acetylation of p53 by p300 may not be the only regulatory mechanism to control transactivation of p53-inducible genes. Gene transactivation may also depend on interactions with activators as well as coactivators at the promoter site to initiate transcription. Hence, different levels of regulation may play a role in transactivation of p53-inducible genes.
Mutations that inactivate p300 have been described in colorectal and gastric carcinomas (Muraoka et al. 1996), which may suggest that p300 functions as a negative regulator of cell growth. Misense mutations of p300, coupled to the deletion of the second allele of the gene, was observed in these carcinomas (Muraoka et al. 1996). Gene mutations were found in the Cys/His-rich regions of p300, which are known to play an important role in p300 function. These observations suggests that inactivation of p300 may play a role in the development of carcinomas. E1A inhibition of p300 transactivation may be one mechanism whereby this viral protein promotes cellular transformation. Interestingly, E1A binds Cys/His-rich region 3 thereby inactivating p300’s function (Eckner et al. 1994). Moreover, p300 inactivation may render cells susceptible to agents that induce p53-mediated cell death such as UV and ionizing radiation. It is intriguing to speculate that acetyltransferase inhibitors specific for p300 may be used in cells that do not respond to chemotherapy regardless of having wild-type p53.
Finally, E1B 19K and Bcl-2 can bypass E1A inhibition of p300 function and restore mdm2 transactivation, thereby inhibiting p53-dependent apoptosis. The up-regulation of mdm2 expression by E1B 19K and Bcl-2 may occur by alleviating the repressive function of adenovirus E1A protein caused by p300 inhibition. These results suggest that members of the anti-apoptotic family may promote survival by bypassing the requirement for p300 function. Both E1B 19K and Bcl-2 have been shown to relieve transcriptional repression by E1A and p53 (Yoshida et al. 1987; Shen and Shenk 1994; Sabbatini et al. 1995a). In addition, E1B 19K can block repression by E1A caused by p300 inhibition (Yoshida et al. 1987; Stein et al. 1991; Lee et al. 1995). We cannot rule out the possibility that E1B 19K and Bcl-2 might up-regulate a p300-like activity that can then co-operate with p53 to transactivate the mdm2 gene. Hence, E1B 19K and Bcl-2 may promote survival, not only by binding and inhibiting Bax, but also by relieving transcriptional repression or by enhancing transcription.
Materials and methods
Cells and culture conditions
Primary Fisher BRK cells were prepared from 6-day-old baby rats and cultured in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum as described previously (White et al. 1991). Plasmids were transfected by electroporation, and cells were maintained in DMEM with 10% fetal bovine serum. The transformed BRK cell line, p53A, was derived from transfection of pCMVE1A and pLTRcGval135 plasmids (Debbas and White 1993). The 19K1 and 4B cell lines were derived from transfecting pCMV19K and pSVBcl-2, respectively, and a neomycin-resistance plasmid, pSVneo, by electroporation (Debbas and White 1993; Chiou et al. 1994a; Sabbatini et al. 1995a). Control cell line p53An1 was derived from p53A cells containing only the neomycin-resistance plasmid (Debbas and White 1993). The LTR.1A cells were generated by transfecting primary BRK cells with 10 μg of LTR H-myc, which expresses human c-Myc, and pLTRcGval135 as described previously (Sakamuro et al. 1995).
Antibodies
SMP14, a mouse monoclonal anti-Mdm2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was used at 1:1000 (Western) or 1:100 (immunofluorescence) to detect Mdm2. PAb248, a murine-specific anti-p53 mouse monoclonal antibody which was kindly provided by A. Levine was used at 1:5000 to detect p53. Mouse monoclonal anti-p21WAF1 (Ab-4, Calbiochem, San Diego, CA) was used at 1:50 dilution to detect p21WAF1. A rabbit polyclonal anti-Bax (P-19, Santa Cruz) was used at 1:100 to detect Bax and a mouse monoclonal anti-actin antibody (Amersham) was used at 1:1000 dilution to detect actin. Anti-HA mouse monoclonal antibody (Babco, Richmond, CA) at 1:1000 dilution was used to detect ectopically expressed p300 and a mouse monoclonal anti-E1A antibody M73 (Calbiochem) was used at 1:1000 dilution to detect E1A. A monoclonal anti-myc antibody (Calbiochem) was used at 1:1000 dilution to detect myc-tagged proteins, Bcl-2, and lamin mutant. A polyclonal anti-E1B 19K antibody was used at 1:10,000 to detect transiently expressed E1B 19K.
Cell viability and morphology
Cell viability was measured by trypan blue staining. Cells were trypsinized, centrifuged, resuspended in PBS, and counted using a hemocytometer after diluting in trypan blue. Apoptotic morphology was assessed by scoring for cells that are positive by fluorescence microscopy and are rounded and nonadherent. Cell morphology was also documented by photography at 100× magnification on a Nikon phase-contrast microscope and camera using Kodak film.
Western blot analysis
Cells were incubated at 37.5°C and 32°C for various time intervals prior to lysing in buffer containing 4% SDS and 5% β-mercaptoethanol. Equal amounts of protein (20–30 μg) were electrophoresed on 7.5%–12% SDS-PAGE gels and transferred to PVDF membranes. Membranes were blocked for 15 min at room temperature in 5% nonfat Carnation dry milk in PBS containing 0.1% Tween 20 (PBST). Membranes were incubated with primary antibody followed by washes in PBST and then incubated with horseradish peroxidase (HRP)-conjugated sheep anti-mouse IgG or donkey anti-rabbit IgG monoclonal antibody at 1:2000 dilution. After several washes, blots were developed using the ECL chemiluminescence detection kit according to manufacturer’s recommendations (Amersham Life Sciences, Arlington Heights, IL).
Indirect immunofluorescence and microscopy
Cells were fixed in cold methanol prior to incubation with primary antibodies. Subsequent to washes in PBS, cells were incubated with rhodamine-conjugated goat anti-mouse antibody or fluorescein-conjugated goat anti-rabbit antibody. Cells were then washed, mounted, and analyzed by fluorescence microscopy using a Nikon FXA epifluorescence microscope.
Northern blot analysis
Cytoplasmic RNA was extracted from p53A and 19K1 cells using the NP-40 lysis protocol as described previously (Muraoka et al. 1996). Northern blots were done by loading 30 μg of cytoplasmic RNA on a formaldehyde gel and blotted as described previously (Muraoka et al. 1996) using Hybond-N membranes (Amersham) for transfer. The membrane was hybridized with a random-primed labeled murine mdm2-specific probe in ExpressHyb Hybridization solution (CloneTech, Palo Alto, CA) at 65°C. Blots were washed in 0.1× SSC and 0.1% SDS at 50°C and exposed to film for 2 days.
Transient transfections
The indicated amount of plasmid DNA was transfected by electroporation. After transfection, cells were incubated at 37.5°C for 24 hr to recover, and were then shifted to 32°C for 24 hr before analysis for viability by trypan blue staining and apoptotic morphology. For p300 transfections, p53A cells were electroporated with 12 or 24 μg of pCMVβ alone, pCMVβp300, or pCMVp300Δ30 mutant (deletion in E1A-interacting domain) (Arany et al. 1994). For transient expression of E1A proteins, LTR.1A cells were electroporated with 10 μg of pCMVβ alone, pCMV12SE1A, pCMV12S.RG2, or pCMV12S.YH47.928 (Wang et al. 1992). For Mdm2 transient expression, p53A cells were electroporated with 5–10 μg of pCMVβ alone, pCOC-X2 (wild-type Mdm2), and pCOC-ΔXM (mutant Mdm2 with deletion of the amino-terminal p53-binding domain) (Haupt et al. 1997). p53A cells were transfected transiently with 10 μg of pCEP4-myc–LA(1–406) by electroporation to detect transient expression of a lamin fragment as a control protein (Rao et al. 1996). Transient expression of Bcl-2 and E1B 19K was achieved by electroporating 10 μg of pCDNA3-Bcl-2 and pCMV19K plasmids into p53A cells.
Acknowledgments
We thank Moshe Oren for providing the mdm2 expression plasmids, Steve Grossman for the p300 expression plasmids, Elizabeth Moran for E1A mutant expression plasmids, and Arnold Levine for providing murine-specific anti-p53 antibody. This work was supported by National Institutes of Health grant CA-60088 to E.W.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ewhite@mbcl.rutgers.edu; FAX (732) 235-5795.
References
- Abraham SE, Lobo S, Yaciuk P, Wang HG, Moran E. p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- Arany Z, Sellers W, Livingston D, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300-CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger BR, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- Chen J, Wu X, Lin J, Levine AJ. mdm-2 inhibits the G1 arrest and apoptosis functions of the p53 tumor suppressor protein. Mol Cell Biol. 1996a;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes & Dev. 1996b;10:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- Chiou S-K, White E. p300 binding by E1A cosegregates with p53 induction but is dispensable for apoptosis. J Virol. 1997;71:3515–3525. doi: 10.1128/jvi.71.5.3515-3525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou S-K, Rao L, White E. Bcl-2 blocks p53-dependent apoptosis. Mol Cell Biol. 1994a;14:2556–2563. doi: 10.1128/mcb.14.4.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou S-K, Tseng CC, Rao L, White E. Functional complementation of the adenovirus E1B 19K protein with Bcl-2 in the inhibition of apoptosis in infected cells. J Virol. 1994b;68:6553–6566. doi: 10.1128/jvi.68.10.6553-6566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Hou DX, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes & Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- Debbas M, White E. Wild-type p53 mediates apoptosis by E1A which is inhibited by E1B. Genes & Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumors. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Dyson N, Harlow E. Adenovirus E1A targets key regulators of cell proliferation. Cancer Surv. 1992;12:161–195. [PubMed] [Google Scholar]
- Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB, Livingston DM. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcription adaptor. Genes & Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- El-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nature Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer E, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WD, Pak DT, Karas RH, Wright WE, Shay JW. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992;12:2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Gu W, Shi X-L, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- Han J, Sabbatini P, Perez D, Rao L, Modha D, White E. The E1B 19K protein blocks apoptosis by interacting with and inhibiting the p53-inducible and death-promoting Bax protein. Genes & Dev. 1996;10:461–477. doi: 10.1101/gad.10.4.461. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Barak Y, Oren M. Cell type-specific inhibition of p53-mediated apoptosis by mdm2. EMBO J. 1996;15:1596–1606. [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, El-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace AJ. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;13:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: Puzzle and paradigm. Genes & Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Lee JS, Galvin KM, See RH, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes & Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- Lowe S, Ruley HE. Stabilization of the p53 tumor suppressor is induced by adenovirus-5 E1A and accompanies apoptosis. Genes & Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Moran E. DNA tumor virus transforming proteins and the cell cycle. Curr Opin Genet Dev. 1993;3:63–70. doi: 10.1016/s0959-437x(05)80342-9. [DOI] [PubMed] [Google Scholar]
- Muraoka M, Konishi M, Yanoshita R, Tanaka K, Shitara N, Chong JM, Iwama T, Miyaki M. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12:1565–1569. [PubMed] [Google Scholar]
- Murphy M, Hinman A, Levine AJ. Wild-type p53 negatively regulates the expression of a microtubule-associated protein. Genes & Dev. 1996;10:2971–2980. doi: 10.1101/gad.10.23.2971. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Beach D. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. EMBO J. 1994;13:4816–4822. doi: 10.1002/j.1460-2075.1994.tb06807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol J, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumor suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- Pietenpol JA, Tokino T, Thiagalingam S, El-Deiry WS, Kinzler KW, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido EJ, Teodoro JG, Branton PE. Accumulation of p53 induced by the adenovirus E1A protein requires regions involved in the stimulation of DNA synthesis. J Virol. 1997;71:3526–3533. doi: 10.1128/jvi.71.5.3526-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L, White E. Bcl-2 and ICE family of apoptotic regulators: Making a connection. Curr Opin Genet Dev. 1997;7:52–58. doi: 10.1016/s0959-437x(97)80109-8. [DOI] [PubMed] [Google Scholar]
- Rao L, Perez D, White E. Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol. 1996;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbatini P, Chiou S-K, Rao L, White E. Modulation of p53-mediated transcription and apoptosis by the adenovirus E1B 19K protein. Mol Cell Biol. 1995a;15:1060–1070. doi: 10.1128/mcb.15.2.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbatini P, Lin J, Levine AJ, White E. Essential role for p53-mediated transcription in apoptosis but not growth suppression. Genes & Dev. 1995b;9:2184–2192. doi: 10.1101/gad.9.17.2184. [DOI] [PubMed] [Google Scholar]
- Sabbatini P, Han JH, Chiou S-K, Nicholson D, White E. Interleukin 1β converting enzyme-like proteases are essential for p53-mediated transcriptionally dependent apoptosis. Cell Growth Differ. 1997;8:643–653. [PubMed] [Google Scholar]
- Sakamuro D, Eviner V, Elliot KJ, Showe L, White E, Prendergast GC. c-Myc induces apoptosis in epithelial cells by both p53-dependent and p53-independent mechanisms. Oncogene. 1995;11:2411–2418. [PubMed] [Google Scholar]
- Sanchez-Prieto R, Lleonart M, Cajal S. Lack of correlation between p53 protein level and sensitivity to DNA-damaging agents in keratinocytes carrying adenovirus E1A mutants. Oncogene. 1995;11:675–682. [PubMed] [Google Scholar]
- Scolnick DM, Chehab NH, Stavridi ES, Lien MC, Caruso L, Moran E, Berger SL, Halazonetis TD. CREB-binding protein and p300-CBP-associated factor are transcriptional coactivators of the p53-tumor suppressor protein. Cancer Res. 1997;57:3693–3696. [PubMed] [Google Scholar]
- Shen Y, Shenk T. Relief of p53 mediated transcriptional repression by the adenovirus E1B 19-kDa protein or the cellular Bcl-2 protein. Proc Natl Acad Sci. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegenga WT, van Laar T, Riteco N, Mandarino A, Shvarts A, van der Eb AJ, Jochemsen AG. Adenovirus E1A proteins inhibit activation of transcription by p53. Mol Cell Biol. 1996;16:2101–2109. doi: 10.1128/mcb.16.5.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RW, Corrigan M, Yaciuk P, Whelan J, Moran E. Analysis of E1A-mediated growth regulation functions: Binding of the 300-kilodalton cellular product correlates with E1A repression function and DNA synthesis-inducing activity. J Virol. 1991;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thut CJ, Chen J-L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII400 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. p53 Function and dysfunction. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- Wang H-GH, Rikitake Y, Carter MC, Yaciuk P, Abraham SE, Zerler B, Moran E. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and the control of cell growth. J Virol. 1992;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Regulation of apoptosis by the transforming genes of the DNA tumor virus adenovirus. Proc Soc Exp Biol Med. 1993;204:30–39. doi: 10.3181/00379727-204-43631. [DOI] [PubMed] [Google Scholar]
- ————— Life, death, and the pursuit of apoptosis. Genes & Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- White E, Gooding LR. Regulation of apoptosis by human adenoviruses. In: Tomei LD, Cope FO, editors. Apoptosis: The molecular basis for cell death II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 111–141. [Google Scholar]
- White E, Cipriani R, Sabbatini P, Denton A. The adenovirus E1B 19-Kilodalton protein overcomes the cytotoxicity of E1A proteins. J Virol. 1991;65:2968–2978. doi: 10.1128/jvi.65.6.2968-2978.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte P, Buchkovich K, Horowitz JM, Friend SH, Raybuck M, Weinbert RA, Harlow E. Association between an oncogene and an anti-oncogene: The adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes & Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani YA. p300/CBP-associated factor that competes with the adenoviral oncoprotein E1a. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Venkatesh L, Kuppuswamy M, Chinnadurai G. Adenovirus transforming 19-kD T antigen has an enhancer dependent trans-activation function and relieves enchancer repression mediated by viral and cellular genes. Genes & Dev. 1987;1:645–658. doi: 10.1101/gad.1.7.645. [DOI] [PubMed] [Google Scholar]