Abstract
The lipoprotein encoded by the Francisella tularensis subsp. tularensis locus FTT1103 is essential for virulence; an FTT1103 deletion mutant is defective in uptake and intracellular survival, and mice survive high dose challenges of greater than 108 bacteria. This protein has two conserved domains; one is found in a class of virulence proteins called macrophage infectivity potentiator (Mip) proteins, and the other in oxidoreductase Disulfide Bond formation protein A (DsbA)-related proteins. We have designated the protein encoded by FTT1103 as FipB for Francisella infectivity potentiator protein B. The locus FTT1102 (fipA), which is upstream of fipB, also has similarity to same conserved Mip domain. Deletion and site-specific mutants of fipA and fipB were constructed in the Schu S4 strain, and characterized with respect to intracellular replication and in vivo virulence. A nonpolar fipA mutant demonstrated reduced survival in host cells, but was only slightly attenuated in vivo. Although FipB protein was present in a fipA mutant, the abundance of the three isoforms of FipB was altered, suggesting that FipA has a role in post-translational modification of FipB. Similar to many DsbA homologues, FipB contains a cysteine-any amino acid-any amino acid-cysteine (CXXC) motif. This motif was found to be important for FipB's role in virulence; a deletion mutant complemented with a gene encoding a FipB protein in which the first cysteine was changed to an alanine residue (AXXC) failed to restore intracellular survival or in vivo virulence. Complementation with a gene that encoded a CXXA containing FipB protein was significantly defective in intracellular growth; however, only slightly attenuated in vivo.
Introduction
Francisella tularensis subspecies tularensis, also known as type A Francisella, causes a potentially life-threatening disease called tularemia. Tularemia can be contracted by the bite of an arthropod vector, or through inhalation of contaminated particles or bacteria. Concerns over use of F. tularensis as a biological weapon have arisen due to its documented use as a bioweapon in WWII, and reports of the development of weaponized strains that are resistant to antibiotics and vaccines [1], [2]. These concerns have led to increased interest in defining the mechanisms of virulence, and immunity as means towards identifying new targets for therapy and immune protection. There are several other less virulent subspecies and species of Francisella including the Live Vaccine Strain (LVS), an attenuated strain of F. tularensis subsp. holarctica that has been used as a protective vaccine in some parts of the world, but has never been licensed in the United States [3].
F. tularensis is a facultative intracellular bacterium that can invade a variety of cell types including macrophages, endothelial cells, and hepatocytes [4], [5], [6]. After phagocytosis F. tularensis resides in a phagosome for up to 4 hours before escaping to the cytoplasm [7], [8], [9]. Once in the cytoplasm bacteria replicate, induce apoptosis or pyroptosis, and are eventually released from cells. F. tularensis can also reenter the endocytic pathway and reside in a large membrane-bound compartment that has characteristics of an autophagocytic vacuole [10]. Thus far only a few loci have been directly implicated in phagosome survival or escape. Most of these loci are located on the Francisella pathogenicity island (FPI) [11], [12], [13].
Previously we identified a novel non- FPI encoded F. tularensis lipoprotein, encoded by locus FTT1103, that is defective in intracellular growth, and essential for virulence in vivo in the highly pathogenic F. tularensis subsp. tularensis strain Schu S4 [14]. We have designated the protein encoded by FTT1103 as FipB for Francisella infectivity potentiator protein B. FipB consists of a unique combination of conserved domains that are found in a class of virulence proteins called Mips (macrophage infectivity potentiator) [15], and DsbA oxidoreductases [16]. Mip is a homodimeric protein with peptidyl-prolyl cis/trans isomerase (PPIase) activity. Mip proteins are characterized by two conserved domains, the Forskolin-binding protein-N (FKBP-N), which is found at the amino-terminal end of Mip and also FipB, and FKBP-C, which encodes the PPIase activity. Mip protein was first identified in Legionella pneumophila as a virulence factor that was required for optimal intracellular survival and virulence in vivo [17]. Orthologs have been subsequently identified in several other Gram-negative bacteria in including Coxiella burnetii, Neisseria gonorrhoeae [18], and Chlamydia species [19]. A number of other Gram-negative bacteria have a Mip ortholog at least by bioinformatic annotation. F. tularensis has a Mip ortholog encoded by locus FTT1043.
Directly upstream of fipB in the Schu S4 genome is the FTT1102 locus, which we have designated as fipA. In the original annotation of the Schu S4 genome fipA was annotated as a pseudogene. However, proteomic analysis of LVS membrane fractions identified a peptide encoded by fipA [20]. The FipA protein is predicted to encode a 96 amino acid lipoprotein, and differs in only a single amino acid between Schu S4 and LVS. Like FipB, FipA shares some sequence and structural similarity to the FKBP-N domain. FipA and FipB are 28% identical to each other in a 54 amino acid overlap. FipA and FipB are both highly conserved (>98% identity) in all sequenced isolates and subspecies of F. tularensis.
Canonical DsbA proteins are periplasmic oxidoreductases that catalyze disulfide formation in nascent proteins in the periplasmic space [21]. The active site of DsbA is minimally defined by a Cysteine- any amino-acid-any amino acid- Cysteine (CXXC) motif embedded in a thioredoxin-like fold [22]. FipB contains a CXXC motif, and Straskova et al. have shown that recombinant protein of the LVS ortholog of FipB has oxidoreductase activity in vitro [20]. The Schu S4 and LVS FipB orthologs are highly similar; they differ in seven amino acid residues, which are scattered throughout the protein. LVS FipB also has 11 extra amino acids on its carboxyl terminus. There are at least eight families of DsbA-related proteins in the NCBI conserved domain database (www.ncbi.nlm.nih.gov/cdd). FipB is most similar to the DsbA_Com1_ like protein family (NCBI; conserved domain cd03023). Com1 is an outer membrane-associated protein of Coxiella burnetii [16]. Com1-like proteins are present in a number of gram-negative pathogens, but their roles as virulence factors or as oxidoreductases have not been fully explored.
The goals of this study were to determine if fipA had roles in intracellular replication and in vivo virulence, and determine whether the active site of FipB was involved in this protein's essential role in virulence. FipB is a novel protein for several reasons; it contains the conserved amino-terminal domain of Mip proteins, and as we show in this paper, it consists of three isoforms, and it has an accessory protein, FipA, that may function in post-translational modification. Here we show that FipA is not essential for virulence, though this mutant does not appear to replicate intracellularly. We have also shown that in vivo virulence is dependent on the CXXC motif of FipB. However, only the first cysteine of the CXXC motif is essential for FipB activity.
Results
FipB is co-transcribed with fipA
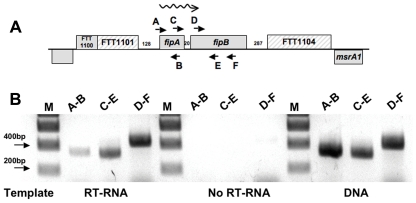
Although there are only 20 base pairs separating the open reading frames of fipA and fipB, it was important for later complementation studies to confirm that these two genes were transcribed from the same promoter. We verified that fipA and fipB are co-transcribed by reverse transcription PCR (RT-PCR) (Figure 1). As shown in Figure 1, PCR products using primer pair C/E amplified the intergenic region between fipA and fipB, and primer pairs A/B, and D/F amplified intragenic fragments of fipA and fipB, respectively. It is possible that additional loci are co-transcribed with fipAB. FipAB is flanked by FTT1100, and predicted pseudogenes, loci FTT1101 and FTT1104. If FTT1101 consisted of an intact open reading frame, translation would terminate about 120 base pairs (bps) from the translational start of fipA. The predicted start of FTT1104 is 287 bps from the end of fipB. For in-cis complementation of fipAB we included 262 bps upstream of fipA in the plasmid used for constructing the complemented strains (Table 1, pAQ162, pAQ163, pAQ164). Since this region of DNA was able to drive expression of fipB in the complemented strains (Figure 2), it suggests that this region contains a promoter element.
Figure 1. fipA and fipB are co-transcribed.
RNA was isolated from wild-type Schu S4, and then converted to cDNA. (A) Overview of chromosomal region containing fipA and fipB. Short arrows indicate the location of the PCR primers. The wavy arrow indicates the direction of transcription. The numbers between the genes indicate the size of the intergenic region. The hatched boxes (FTT1101 and FTT1104) indicate predicted pseudogenes. (B) PCR was performed using the indicated primers (RT-RNA). Controls were: No reverse transcriptase added to the cDNA synthesis reactions (No RT-RNA), and PCR using Schu S4 DNA as template and the indicated primers (DNA). Expected sizes of PCR products were as follows A–B: 314 bps, C–E: 311 bps, and D–F: 392 bps.
Table 1. Bacterial strains and plasmids used in this study.
| Name | Relevant characteristics | Source/Ref |
| Francisella strains | ||
| Schu S4 | F. tularensis tularensis, wild-type | CDC |
| BJM1031 | Schu S4 ΔfipB | Qin [14] |
| BJM1068 | Schu S4 ΔfipA | This study |
| BJM1069 | Schu S4 DfipA + B | This study |
| BJM1076 | fipA+B+ in cis complement of ΔfipAB | This study |
| BJM1077 | fipA+B C164A in cis complement of ΔfipAB | This study |
| BJM1078 | fipA+B C167A in cis complement of ΔfipAB | This study |
| Plasmids | ||
| pMP815 | Chromosomal integration system vector | LoVullo [46] |
| pGIR463 | sacB suicide vector | Sullivan [44] |
| pAQ136 | 5′flanking region of fipA in GIR463 (BM248/BM249) | This study |
| pAQ137 | 5′-and 3′flanking regions of fipA in GIR463 (BM250/BM251) | This study |
| pAQ138 | 5′-and 3′ flanking regions of fipAB in GIR463 (BM256/BM085) | This study |
| pAQ162 | fipA+fipB+ in pMP815 | This study |
| pAQ163 | fipA+fipB C164A in pMP815 | This study |
| pAQ164 | fipA+fipB C167A in pMP815 | This study |
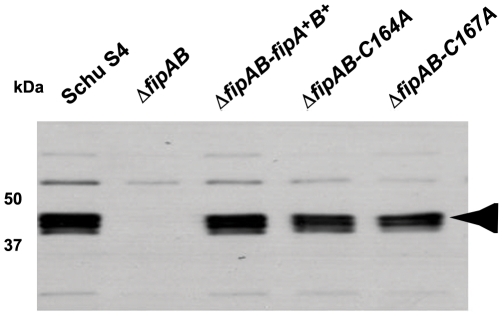
Figure 2. In cis complementation of ΔfipAB.
Wild-type and mutated copies of fipB were introduced into ΔfipAB and then selected for integration into the blaB locus as described in methods. Western blots of overnight cultures were prepared and incubated with anti-FipB specific antibody. Arrow indicates the location of the FipB triplet isoforms.
FipA affects post-translational processing of FipB
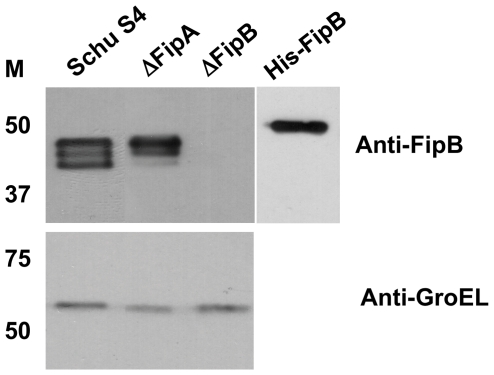
When Straskova et al. used various proteomic techniques to compare the protein profiles of wild-type LVS and an isogenic fipB (FTL_1096) mutant bacteria only two proteins were absent, FipB and FipA [20]. This suggested that FipB was required for FipA protein stability. To investigate the effects of FipA on FipB, and also the specific contributions of FipA to virulence, nonpolar mutants in ΔfipA, ΔfipB, and ΔfipAB were constructed. Western blots with anti-FipB antibody confirmed that the ΔfipA mutation was nonpolar (Figure 3). Anti-FipB specific antibody recognized three bands on Western blots. FipB has been identified as a glycosylated protein by carbohydrate detection and mass spectrometry techniques, so these three isoforms may reflect differences in glycosylation [23], [24]. All three bands disappeared in the ΔfipB mutant and also in the ΔfipAB mutant (data not shown). We noted that compared to the wild-type strain, the lowest migrating band of FipB was diminished in the fipA deletion mutant suggesting that FipA plays some role in FipB post-translation processing or modification such as glycosylation.
Figure 3. Detection of FipB in the ΔfipA bacteria.
On Western blots FipB migrates as three bands. In the ΔfipA mutant the lower band was diminished. Western blot of bacterial lysates of indicated strains with anti-FipB antibody; antibody to E. coli GroEL, which cross-reacts with the Francisella protein, was used as a loading control. Recombinant His-FipB was used to generate the anti-FipB antibody, and serves as a positive control.
ΔfipA mutant is defective in intracellular growth
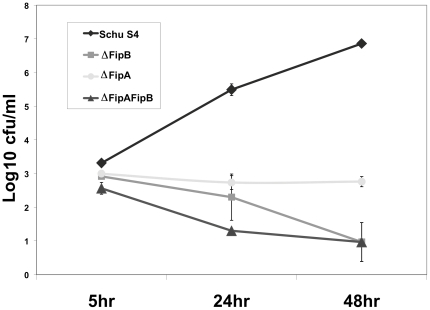
To explore and compare the roles of FipA and FipB in intracellular growth and virulence J774A.1 cells were infected with the fipA, fipB, or fipAB deletion strains, and then assayed for intracellular growth at several time points using gentamicin protection assays (Figure 4). All mutants exhibited statistically significant reduced growth when compared to wild-type Schu S4 at 5 hrs post-infection (p<0.001). The ΔfipB, and the double ΔfipAB mutants had similar phenotypes; in J774A.1 cells the number of CFUs recovered from these mutants at 24 hrs had decreased by about two logs from the 5 hr time point, while during this same time period Schu S4 CFUs had increased by almost 4 logs (Figure 4). In contrast, the ΔfipA mutant did not appear to replicate appreciably over this time period, and the number of recovered CFUs remained stable for up to 48 h. Similar patterns of defective growth for all of these mutant strains were also observed in A549 cells (data not shown). The difference in intracellular growth was not due to an inherent growth defect because all strains grew similarly in TSB/c and CDM media (data not shown). FipA and FipB may have independent functions in intracellular replication, but based on their sequence similarity, and altered levels of FipB isoforms in the fipA mutant, it seems likely that their roles in intracellular replication are linked.
Figure 4. ΔfipA bacteria are defective in intracellular growth.
J774A.1 cells were infected at an MOI of 50∶1 with the indicated strains of bacteria as described in materials and methods; cells were thoroughly washed, lysed at the indicated time points, and then diluted and plated to determine CFU/ml.
ΔfipA mutant is slightly attenuated in vivo
We have shown here and previously that fipB is essential for in vivo virulence in mice (Table 2) [14]. To test whether the intracellular growth defect of the fipA mutant would have any affect on virulence in vivo, C57BL/6 mice were challenged intranasally with 2800, 280, or 28 CFUs of ΔfipA bacteria. Despite a significant intracellular growth defect in vitro, the fipA mutant was only mildly attenuated in vivo; as few as 28 CFUs of the fipA mutant by an intranasal route was lethal, although there was 2–3 day delay in the time to death when compared to mice that received a challenge dose of 10 CFUs (Table 2).
Table 2. Survival of fipA, and ΔfipAB mice after intranasal inoculation.
| Strain | Relevant genotype | # of mice | Inoculation dose (CFU)1 | Days to Death2 |
| Schu S4 | 4 | 10 | 5,5,5,5 | |
| BJM1068 | ΔfipA | 3 | 28 | 7,7,8 |
| BJM1068 | ΔfipA | 3 | 280 | 6,7,8 |
| BJM1068 | ΔfipA | 2 | 2800 | 5,6 |
| BJM1069 | ΔfipAB | 4 | 7×107 | Survived >30 days |
| BJM1076 | ΔfipAB-fipA+B+ | 3 | 10 | 6,6,7 |
| BJM1076 | ΔfipAB-fipA+B+ | 3 | 100 | 6,6,6 |
| BJM1076 | ΔfipAB-fipA+B+ | 3 | 1000 | 5,5,5 |
| BJM1076 | ΔfipAB-fipA+B+ | 4 | 4.3×104 | 4,4,4,4 |
| BJM1077 | ΔfipAB-fipA+fipB C164A | 4 | 8.8×107 | Survived >30 days |
| BJM1078 | ΔfipAB-fipA+fipB C167A | 4 | 3.4×107 | 4,4,4,4 |
| BJM1078 | ΔfipAB-fipA+fipB C167A | 4 | 3.4×104 | 8,8,8,8 |
| BJM1078 | ΔfipAB-fipA+fipB C167A | 4 | 270 | 9,9,9,10 |
| BJM1078 | ΔfipAB-fipA+fipB C167A | 4 | 27 | 9,10,10, >20 days |
C57/BL6 mice were intranasally challenged with the indicated inoculum dose, which was confirmed by plating the inoculum.
Indicates the number of days after challenge that mice showed the first signs of irreversible mortality, and were euthanized. Mice were followed for a minimum of 20 days. Similar to the ΔfipAB mutant, mice similarly challenged with the ΔfipB mutant survive for more than 30 days without any signs of infection [14].
The conserved CXXC motif of FipB is required for intracellular replication
The CXXC motif has been shown to be critical for the enzymatic activity of Escherichia coli DsbA (EcDsbA), and also for substrate interactions [27], [28]. Therefore, we predicted that the CXXC motif of FipB would also be important for FipB's role in intracellular replication, and virulence. To investigate the importance of the CXXC motif in intracellular growth copies of fipB in which the cysteines in the CXXC motif had been replaced with alanines, (C164A and C167A), were integrated in to the blaB gene in a ΔfipAB strain along with the native promoter and wild-type fipA. Expression of the mutated genes was confirmed by Western Blots (Figure 2). The level of FipB in the in-cis complemented strains was similar to wild-type.
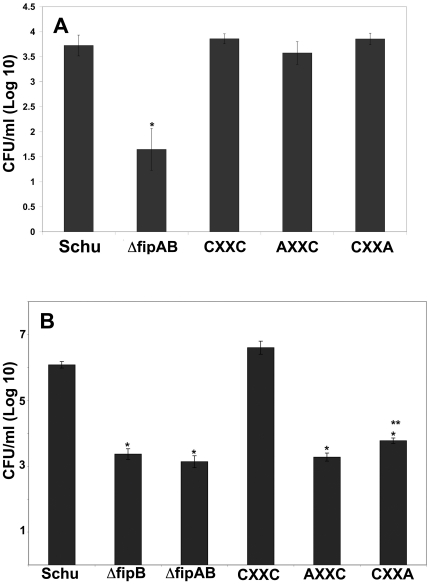
To examine uptake and the intracellular growth phenotype of the CXXC mutants, J774A.1 cells were incubated with the ΔfipAB or complemented strains, treated with gentamicin, and then assayed for growth at 2 and 24 hrs (Figure 5). At 2 hrs post-infection the ΔfipAB mutant had reduced uptake compared to the wild-type bacteria (Figure 5A). A ΔfipB mutant was also defective in uptake, but a ΔfipA mutant had uptake levels similar to wild-type bacteria (data not shown). Complementation with the C167A or C164A fipB genes (CXXA or AXXC, respectively) also restored uptake to wild-type levels. At 24 hrs post-infection complementation of the fipAB mutant with the wild-type fipB gene restored intracellular replication to wild-type levels. The numbers of bacteria recovered from fipAB mutant, or the strains complemented with CXXA or AXXC genes were significantly reduced compared to wild-type Schu S4 (p value<0.00001). However, when the complemented strains were compared to the ΔfipAB mutant, the number of CFUs recovered from the CXXA strain was statistically higher (p value<0.007) than the ΔfipAB mutant, while there was no statistical difference between the AXXC and ΔfipAB mutants. These results indicated that the CXXC motif was not required for uptake, but was important for FipB mediated intracellular replication, however, the C164 amino acid was more critical for function.
Figure 5. Effect of CXXC mutations on uptake and intracellular replication in J774A1.
Monolayers of J774A.1 cells were infected with indicated strains at an MOI of 100∶1. The number of intracellular bacteria per well after infection for 2 (A) or 24 (B) hrs was determined as described in methods; cells were thoroughly washed, lysed at the indicated time points, and then diluted and plated to determine CFU/ml. Bars represent the mean ±SD of a representative experiment performed in triplicate. This experiment was repeated two times with similar results. For panel A “*” indicates p<0. 003 compared to Schu S4. For panel B “*” indicates p<0.00001 compared to Schu S4 and “**”<0.007 compared to Δ fipAB.
The conserved CXXC motif of FipB is required for in vivo virulence
In the fipB and fipAB deletion mutants a defect in intracellular growth correlated with avirulence in mice ([14] & Table 2). However, this paradigm was not true of the ΔfipA mutant; despite a significant intracellular growth defect this mutant still retained virulence. One difference between ΔfipA, and ΔfipAB, was that between 24 hrs and 48 hrs post-infection the number of viable ΔfipA bacteria was stable, while viable ΔfipAB bacteria had decreased in number. To test whether the increased number of bacteria recovered from the CXXA complemented strain, compared to the ΔfipAB strain, translated to a difference in virulence in vivo, mice were challenged intranasally with decreasing doses of these various strains (Table 2). Similar to the parental deletion strain, the AXXC mutant appeared to be avirulent; mice survived challenges of 8.8×107 CFUs for more than 30 days. The CXXA mutant was also attenuated but still retained virulence; mice challenged with 3.4×104 CFUs died on day 8. Mice challenged with 10 CFUs of Schu S4 died on day 5 post-infection, but few as 27 CFUs of the CXXA, with one exception, also resulted in a fatal infection on day 9 or 10 post-infection. These results were consistent with the intracellular replication data (see Figure 4), and support a critical requirement for the first cysteine in the CXXC motif of FipB for virulence.
Discussion
FipB is a novel lipoprotein that is required for uptake, intracellular replication and in vivo virulence of F. tularensis. Although FipB is novel it has two conserved domains, DsbA_Com-1_like (cd03023), and FKBP-N, the amino-terminal domain of FKBP-type peptidyl prolyl isomerases (cl03173), which are also known as Mip proteins. To date, this combination of conserved domains is unique to FipB. FipB is also a lipoprotein, which is unusual, but not unique for DsbA proteins. Most DsbA are thought be periplasmic proteins, but a few lipoproteins have been identified; Neisseria sp., have three DsbA orthologs, and two of these are lipoproteins [29]. The other novel aspects of FipB include the presence of three isoforms, which are likely due to post-translational modification [24], and the presence of an accessory protein, FipA.
In the Schu S4 genome the FTT1102/fipA locus was annotated as a pseudogene. However, consistent with the proteomic data from Straskova et al. [20], which detected the 96 amino acid peptide corresponding to FipA, we have shown that fipA is indeed a functional locus. Deletion of fipA resulted in a significant intracellular growth defect in vitro, though the impact of this gene loss was not appreciably significant in vivo. It is possible that ΔfipA bacteria can replicate in other cell lineages that we have not tested. Horzempa et al. found that a Schu S4 pyrF mutant, which could not replicate in human primary macrophages, was able to replicate in HEK-293 and was virulent in mice [30]. Our results also suggest that replication in macrophages is not an essential requirement for virulence.
FipA also has similarity to the FKBP-N domain and is predicted to be a lipoprotein. One could speculate that fipA and fipB arose as gene duplications, and then fipA was truncated either in the process or subsequently. However, this event would have to have been an early event in the speciation of Francisella, because FipA is highly conserved among the various species and subspecies of Francisella, including the more distantly related Francisella philomiragia. There has also been considerable sequence divergence between FipA and FipB, which are only ∼28% identical at the amino acid level. The conserved domain FKBP-N, which is shared by the two proteins, has previously only been found in the amino-terminal region of Mip proteins. The crystal structure of LpMip has been determined [15]. Mip forms a dimer [31], and has its two conserved domains connected by a very long alpha-helix. The C-terminal domain of Mip (100–213 amino acids) has PPIase activity, and amino terminal FKBP-N domain contains the alpha helical N-term portion of the protein, which is required for dimerization [15]. The LpMip protein has a complex role in virulence. It is required for optimal replication in human macrophages and amoebae [17], migration through an epithelial barrier [32], and secretion of a phospholipase C-like activity in culture supernatants [33]. It has been reported that the FKBP-N or dimerization domain, but not the PPIase domain, is required for full virulence in Acanthamoeba castellanii [34], suggesting that dimerization and PPIase domains have separable functions. One model for FipA function is that it physically interacts or dimerizes with FipB, which then stabilizes a conformation state that facilitates post-translational modification or processing. This model is consistent with our observation that the level of one of the isoforms of FipB was diminished in a FipB mutant. FipA dimerization with FipB could also enable a different functional role for FipB. By proteomic analysis of membrane fractions Straskova et al. could not detect FipA in a FipB mutant [20], so FipA may be unstable unless it is able to associate with FipB. The presence of the FKBP-N dimerization domain in FipB also suggests that FipB could form a homodimer, which FipA could potentially regulate or influence. This will require further investigation. We have observed higher molecular sized complexes on nonreducing gels, but these complexes were sensitive to reducing agents, and are likely artifactual interactions between the cysteines in the active site.
The other conserved region of FipB, the DsbA-Com1-like domain, is one of several DsbA-related conserved domains. DsbA was first identified in E. coli, and is the best characterized DsbA protein both biochemically, and structurally [28]. The conserved CXXC motif, present in most all DsbA-related conserved domains, is critical for the oxidoreductase enzymatic activity of DsbA. In our studies we found that the CXXC motif was critical for FipB's role in intracellular survival and in vivo virulence, but does not appear to be essential for bacterial uptake. In the intracellular growth assays mutation of two cysteines individually did not produce identical results; the first cysteine residue was essential, while the second cysteine mutant had only reduced function. This finding is consistent with functional characterization of the CXXC motif of EcDsbA. Based on biochemical and crystallographic studies, the first cysteine is the nucleophilic residue that forms a mixed disulfide bond with its substrates. The second cysteine is hidden within the molecule and less accessible to solvent. One study that illustrates this difference examined the oxidation of beta-lactamase, a substrate of EcDsbA. When a CXXS mutant of EcDsbA was expressed in a wild-type strain it acted as a dominant negative mutant, which produced a decrease in the oxidation of beta-lactamase [35]. However, addition of oxidized glutathione to the media restored beta-lactamase folding. A SXXC mutant expressed in a wild-type strain did not exhibit a similar dominant negative phenotype. Based on this model, our in vivo experiments suggest that the in vivo environment must be sufficiently oxidizing so that the CXXA FipB mutant is able to carry its function at a level that is sufficient to promote virulence.
It is likely that at least part of FipB's role in virulence is through the folding of substrates that have critical roles in virulence. In other pathogenic bacteria DsbA is important for the structure or function of a number of virulence factors including the biogenesis of type IV pili in bacteria such as EPEC E. coli, and Pseudomonas aeruginosa [36], [37], the assembly and function of type III secretion systems in Salmonella typhimurium and Shigella flexneri [38], [39], [40], and the Dot/Icm type IV secretion system of L. pneumophila [41]. Identifying FipB substrates will help to define the essential elements of F. tularensis pathogenicity. However, proteins that contain a conserved DsbA pfam motif can be quite diverse [42]. With the exception of the active site and a few other conserved amino acids, many share very little additional sequence similarity. Therefore, it is likely that the function or structure of some proteins that contain the DsbA pfam is different, narrowed, or expanded. A number of bacteria have more than one DsbA-related protein, which also suggests more specialized functions [42]. Salmonella typhimurium, for example, contains plasmid encoded protein SrgA, which is a DsbA-related protein that has a restricted substrate specificity for the plasmid encoded fimbriae PefA [43]. The usual features of FipB, which include the FKBP-N domain and some interaction with FipA, suggest that FipB may also have specialized roles in virulence. The observation that complementation with CXXA and AXXC alleles was able to restore uptake supports a specialized role for FipB. FipB may act as a chaperone, or perhaps more directly mediate this activity. Indentifying these roles will help to define the essential aspects of F. tularensis subsp. tularensis virulence.
Materials and Methods
Ethics Statement
All experimental procedures and care of animals was approved by the University of Virginia's Institutional Animal Care and Use Committee. The University's Animal Welfare Assurance number is Animal Welfare Assurance #A3245-01, and the vivarium is accredited by the Association for Assessment Accreditation of Laboratory Animal Care International.
Bacterial strains, primers, plasmids and culture
Bacterial strains, plasmids, and primers used in these experiments are listed in Tables 1 and 3. Plasmids pGIR463 and pMP815 were kind gifts of Girija Ramakrishnan and Martin Pavelka, respectively. E. coli strains were grown in Luria-Bertani (LB) broth or on LB plates with kanamycin (50 µg/ml) or ampicillin (100 µg/ml) when required. F. tularensis subsp. tularensis (type A) Schu S4 was cultured on cysteine supplemented Muller-Hinton agar (MHA/c) or in cysteine supplemented Trypticase Soy broth (TSB/c) [44]. For F. tularensis strains 15 µg/ml of kanamycin was added when appropriate. Studies involving Schu S4 and derived strains were carried out in an approved Biosafety Level 3 laboratory.
Table 3. Primers used in this study.
| Primer | Sequence 5′-3′ | Descript. | Restr. Enz. |
| BM063 | TCCATATGCAAGAAATGGCTGCTC | F fipB | NdeI |
| BM064 | GCGGCCGCTATAAGAAGGATAGGC | F fipA | NotI |
| BM066 | GGATCCTATCATCATCTTGGCTGAGC | R fipB | BamHI |
| BM150 | CTTTGATTATCAAGCTATGTACTGTTCTAAGCTTGC | F fipB C164A | |
| BM151 | GCAAGCTTAGAACAGTACATAGCTTGATAATCAAAG | R fipB C164A | |
| BM152 | CAATGTATGTACGCTTCTAAGCTTGCTTGCTCC | F fipB C167A | |
| BM153 | GGAGCAAGCAAGCTTAGAAGCTTACATACATTG | R fipB C167A | |
| BM245 | AGAAAATATGCGGCCGCGAAATAATAGGAG | F fipA | NotI |
| BM208 | TCCTCGAGCTTATTTCTTTTGAGCAGCC | R fipA | XhoI |
| BM248 | GAGCCCTAGGTAGAACAATGGCAACAGG | F fipA 5′deletion | AvrII |
| BM249 | AGGCGGCCGCATTATTTAGTTTCTCCTA | R fipA 5′deletion | NotI |
| BM250 | ATAGATCAGCGGCCGCATGCAATGATTGAATTCC | F fipA 3′deletion | NotI |
| BM251 | ATTAGAGCTCAACACTATCATCATCTTGGCTGAGC | R fipA 3′deletion | XhoI |
| BM256 | CTAAGTCTGCGGCCGCAACAACTAGTACTAGC | F fipAB 5′deletion | NotI |
| BM085 | CTCGAGATTACAGCATTACCAGCTGC | F fipAB 3′deletion | XhoI |
| BM297 | GAGGAATAATAAATGAAATTAACTAAAACTCT | F fipA | |
| BM298 | TTATTTCTTTTGAGCAGCCAT | R fipA |
DNA manipulation, cloning, and transformation
DNA was prepared and purified using a commercial kit (Qiagen, Valencia, CA). Oligonucleotides were synthesized by Integrated DNA Technologies Inc. (Coralville, IA). Restriction endonucleases and ligase were purchased from New England Biolabs (Ipswich, MA). HotStart® Taq (Qiagen) was used for routine PCR. FastStart® High fidelity PCR system (Roche, Indianapolis, IN) was used for construction of complementary and suicide plasmids. All cloning products were verified by DNA sequencing, which was performed at the University of Virginia Biomolecular Research Facility. Site direct mutagenesis was accomplished with a site-directed mutagenesis kit (QuikChange®, Agilent Technologies, Cedar Creek, TX) using primer pairs BM150/BM151 or BM152/153, and pAQ038 as template. Expression of fipB and mutant genes was verified by Western blot with rabbit anti-FipB antibody (1∶10,000) [14]. DNA transformation was performed as previously described [44].
Construction of deletion and integration plasmids
To confirm the non-polarity of our deletion mutants we first complemented these mutations using the Francisella shuttle vector, pFNLTP [25]. However, in-trans complementation only partially restored the intracellular growth defect, and these strains also grew poorly in liquid culture (data not shown). We hypothesized that over-expression of fipB was deleterious. To circumvent this problem we integrated the fipAB genes, along with the 262 bps upstream, into the blaB locus using the plasmid developed by Lovullo et al. [26]. To construct in-frame deletions of fipA and fipAB PCR products corresponding to regions upstream and downstream of fipA or fipAB were produced with primer pairs BM248/BM249 and BM250/BM251 for fipA and BM256/BM085 for fipAB (Table 3), and then cloned into the sacB suicide vector pGIR463 [45]. Plasmids for in cis complementation of fipA, and fipAB, and the CXXC mutants were produced by PCR amplification of the genes using the primers listed in Table 2. Each amplicon contained 262 bp of the sequence upstream of fipA. PCR products were ligated into the blaB region of pMP815 [26]. The resulting plasmids were introduced into the appropriate host strain, and integrants or subsequent gene deletion mutants were selected as previously described [14]. The nonpolarity of the fipA deletion was verified by the detection of FipB on Western blot with anti-FipB antibody.
Reverse Transcription PCR
Total RNA was isolated from overnight cultures using an RNeasy Protect mini kit (Qiagen) and treated with DNase I (Qiagen) to remove contaminating genomic DNA according to the manufacture's instructions. First strand cDNA was generated using SuperScript II Reverse transcriptase (Invitrogen) and random primers. A parallel transcription reaction without the reverse transcriptase enzyme was conducted to control for DNA contamination. Two µl of each reaction was used as a template for 50 µl PCR reaction.
Uptake and Intracellular growth assays
Assays were performed with murine macrophage J774A.1 (ATCC#TIB-67) cells propagated in high glucose DMEM supplemented with 10% fetal bovine serum. Cells (2.5×105/well) were seeded in 24 well plates, and incubated at 37°C, 5% CO2 for 18 h. Fresh cultures of F. tularensis were diluted in cell culture medium to reach the desired multiplicity of infection (MOI). Actual inoculum amounts of bacteria were determined by plating serial dilutions of the culture inoculum. The plates were centrifuged at 800×g for 8 min to start the infection, and then incubated at 37°C for l h. Cells were washed three times in PBS, and then extracellular bacteria were killed by gentamicin treatment (50 µg/ml). At the assay endpoint cells were washed, and then lysed with 0.1% sodium deoxycholate. Lysates were diluted and plated to determine the number of colony form units (CFU) in each well. Each experiment had triplicate wells and repeated a minimum of two times.
Production of FipB antiserum
The DNA sequence of fipB gene was resynthesized with a his-tag and codons that were biased for E. coli expression (Accession #JN120022), and then cloned in pET Universal [46]. The expression of recombinant fipB was induced by the addition of 1 mM IPTG to log phase bacteria. The protein was purified from induced lysates using Talon beads (Clontech) and eluted with imidazole according to manufacturer's recommendations. Purified protein was dialyzed against PBS, and protein concentration was determined by BCA assay (Pierce). Purity was verified by SDS-PAGE and Western blots. Rabbit anti-his-tagged FipB serum was prepared by Covance Research products Inc (Emeryville, CA). Mouse anti-FipB serum was made in house.
Mouse virulence studies
For intranasal inoculation 8 to 10-week-old C57BL/6 mice (Jackson Laboratory) were anesthetized with ketamine-HCl-xylazine. Twenty microliters of bacteria or PBS was inoculated into the nares. The actual inoculation doses were confirmed by viable plate counting. The mice were monitored daily. Mice were humanely euthanized when death was considered to occur within 24 h. The University of Virginia's Animal Care and Use Committee approved all mouse studies.
Statistical analysis
All values were expressed as Mean ±SD and evaluated by using Student's unpaired Two-tailed t test with log transformed data, and assuming unequal variance.
Acknowledgments
The authors acknowledge Girija Ramakrishnan for helpful suggestions regarding this manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by NIH/NIAID grants R56AI79035 to BJM and Middle Atlantic Regional Center of Excellence for Biodefense U54 AI05716. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alibek K. Biohazard. New York: Random House; 1999. [Google Scholar]
- 2.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, et al. Tularemia as a biological weapon: medical and public health management. Jama. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 3.Oyston PC. Francisella tularensis vaccines. Vaccine. 2009;27(Suppl 4):D48–51. doi: 10.1016/j.vaccine.2009.07.090. [DOI] [PubMed] [Google Scholar]
- 4.Forestal CA, Benach JL, Carbonara C, Italo JK, Lisinski TJ, et al. Francisella tularensis selectively induces proinflammatory changes in endothelial cells. J Immunol. 2003;171:2563–2570. doi: 10.4049/jimmunol.171.5.2563. [DOI] [PubMed] [Google Scholar]
- 5.Conlan JW, North RJ. Early pathogenesis of infection in the liver with the facultative intracellular bacteria Listeria monocytogenes, Francisella tularensis, and Salmonella typhimurium involves lysis of infected hepatocytes by leukocytes. Infect Immun. 1992;60:5164–5171. doi: 10.1128/iai.60.12.5164-5171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anthony LD, Burke RD, Nano FE. Growth of Francisella spp. in rodent macrophages. Infect Immun. 1991;59:3291–3296. doi: 10.1128/iai.59.9.3291-3296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun. 2008;76:2671–2677. doi: 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clemens DL, Lee BY, Horwitz MA. Francisella tularensis phagosomal escape does not require acidification of the phagosome. Infect Immun. 2009;77:1757–1773. doi: 10.1128/IAI.01485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, et al. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Checroun C, Wehrly TD, Fischer ER, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–14583. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nano FE, Schmerk C. The Francisella pathogenicity island. Ann N Y Acad Sci. 2007;1105:122–137. doi: 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- 12.Santic M, Molmeret M, Barker JR, Klose KE, Dekanic A, et al. A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell Microbiol. 2007;9:2391–2403. doi: 10.1111/j.1462-5822.2007.00968.x. [DOI] [PubMed] [Google Scholar]
- 13.Barker JR, Chong A, Wehrly TD, Yu JJ, Rodriguez SA, et al. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol Microbiol. 2009;74:1459–1470. doi: 10.1111/j.1365-2958.2009.06947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin A, Scott DW, Thompson JA, Mann BJ. Identification of an essential Francisella tularensis subsp. tularensis virulence factor. Infect Immun. 2009;77:152–161. doi: 10.1128/IAI.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riboldi-Tunnicliffe A, Konig B, Jessen S, Weiss MS, Rahfeld J, et al. Crystal structure of Mip, a prolylisomerase from Legionella pneumophila. Nat Struct Biol. 2001;8:779–783. doi: 10.1038/nsb0901-779. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix LR, Mallavia LP, Samuel JE. Cloning and sequencing of Coxiella burnetii outer membrane protein gene com1. Infect Immun. 1993;61:470–477. doi: 10.1128/iai.61.2.470-477.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wintermeyer E, Ludwig B, Steinert M, Schmidt B, Fischer G, et al. Influence of site specifically altered Mip proteins on intracellular survival of Legionella pneumophila in eukaryotic cells. Infect Immun. 1995;63:4576–4583. doi: 10.1128/iai.63.12.4576-4583.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leuzzi R, Serino L, Scarselli M, Savino S, Fontana MR, et al. Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol Microbiol. 2005;58:669–681. doi: 10.1111/j.1365-2958.2005.04859.x. [DOI] [PubMed] [Google Scholar]
- 19.Neff L, Daher S, Muzzin P, Spenato U, Gulacar F, et al. Molecular characterization and subcellular localization of macrophage infectivity potentiator, a Chlamydia trachomatis lipoprotein. J Bacteriol. 2007;189:4739–4748. doi: 10.1128/JB.01889-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Straskova A, Pavkova I, Link M, Forslund AL, Kuoppa K, et al. Proteome analysis of an attenuated Francisella tularensis dsbA mutant: identification of potential DsbA substrate proteins. J Proteome Res. 2009;8:5336–5346. doi: 10.1021/pr900570b. [DOI] [PubMed] [Google Scholar]
- 21.Guilhot C, Jander G, Martin NL, Beckwith J. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc Natl Acad Sci U S A. 1995;92:9895–9899. doi: 10.1073/pnas.92.21.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin JL, Bardwell JC, Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature. 1993;365:464–468. doi: 10.1038/365464a0. [DOI] [PubMed] [Google Scholar]
- 23.Thomas RM, Twine SM, Fulton KM, Tessier L, Kilmury SL, et al. Glycosylation of DsbA in Francisella tularensis subspecies tularensis. J Bacteriol. 2011 doi: 10.1128/JB.00438-11. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balonova L, Hernychova L, Mann BF, Link M, Bilkova Z, et al. A multimethodological approach to identification of glycoproteins from the proteome of Francisella tularensis, an intracellular microorganism. J Proteome Res. 2010;9:1935–2005. doi: 10.1021/pr9011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier TM, Havig A, Casey M, Nano FE, Frank DW, et al. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl Environ Microbiol. 2004;70:7511–7519. doi: 10.1128/AEM.70.12.7511-7519.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LoVullo ED, Molins-Schneekloth CR, Schweizer HP, Pavelka MS., Jr Single-copy chromosomal integration systems for Francisella tularensis. Microbiology. 2009;155:1152–1163. doi: 10.1099/mic.0.022491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quan S, Schneider I, Pan J, Von Hacht A, Bardwell JC. The CXXC motif is more than a redox rheostat. J Biol Chem. 2007;282:28823–28833. doi: 10.1074/jbc.M705291200. [DOI] [PubMed] [Google Scholar]
- 28.Kadokura H, Beckwith J. Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid Redox Signal. 2010;13:1231–1246. doi: 10.1089/ars.2010.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sinha S, Langford PR, Kroll JS. Functional diversity of three different DsbA proteins from Neisseria meningitidis. Microbiology. 2004;150:2993–3000. doi: 10.1099/mic.0.27216-0. [DOI] [PubMed] [Google Scholar]
- 30.Horzempa J, O'Dee DM, Shanks RM, Nau GJ. Francisella tularensis DeltapyrF mutants show that replication in nonmacrophages is sufficient for pathogenesis in vivo. Infect Immun. 2010;78:2607–2619. doi: 10.1128/IAI.00134-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt B, Rahfeld J, Schierhorn A, Ludwig B, Hacker J, et al. A homodimer represents an active species of the peptidyl-prolyl cis/trans isomerase FKBP25mem from Legionella pneumophila. FEBS Lett. 1994;352:185–190. doi: 10.1016/0014-5793(94)00970-8. [DOI] [PubMed] [Google Scholar]
- 32.Wagner C, Khan AS, Kamphausen T, Schmausser B, Unal C, et al. Collagen binding protein Mip enables Legionella pneumophila to transmigrate through a barrier of NCI-H292 lung epithelial cells and extracellular matrix. Cell Microbiol. 2007;9:450–462. doi: 10.1111/j.1462-5822.2006.00802.x. [DOI] [PubMed] [Google Scholar]
- 33.Debroy S, Aragon V, Kurtz S, Cianciotto NP. Legionella pneumophila Mip, a surface-exposed peptidylproline cis-trans-isomerase, promotes the presence of phospholipase C-like activity in culture supernatants. Infect Immun. 2006;74:5152–5160. doi: 10.1128/IAI.00484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohler R, Fanghanel J, Konig B, Luneberg E, Frosch M, et al. Biochemical and functional analyses of the Mip protein: influence of the N-terminal half and of peptidylprolyl isomerase activity on the virulence of Legionella pneumophila. Infect Immun. 2003;71:4389–4397. doi: 10.1128/IAI.71.8.4389-4397.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kishigami S, Kanaya E, Kikuchi M, Ito K. DsbA-DsbB interaction through their active site cysteines. Evidence from an odd cysteine mutant of DsbA. J Biol Chem. 1995;270:17072–17074. doi: 10.1074/jbc.270.29.17072. [DOI] [PubMed] [Google Scholar]
- 36.Ha UH, Wang Y, Jin S. DsbA of Pseudomonas aeruginosa is essential for multiple virulence factors. Infect Immun. 2003;71:1590–1595. doi: 10.1128/IAI.71.3.1590-1595.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J. Inactivation of DsbA, but not DsbC and DsbD, affects the intracellular survival and virulence of Shigella flexneri. Infect Immun. 1998;66:3909–3917. doi: 10.1128/iai.66.8.3909-3917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miki T, Okada N, Danbara H. Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J Biol Chem. 2004;279:34631–34642. doi: 10.1074/jbc.M402760200. [DOI] [PubMed] [Google Scholar]
- 39.Ellermeier CD, Slauch JM. RtsA coordinately regulates DsbA and the Salmonella pathogenicity island 1 type III secretion system. J Bacteriol. 2004;186:68–79. doi: 10.1128/JB.186.1.68-79.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J, Kroll JS. DsbA: a protein-folding catalyst contributing to bacterial virulence. Microbes Infect. 1999;1:1221–1228. doi: 10.1016/s1286-4579(99)00239-7. [DOI] [PubMed] [Google Scholar]
- 41.Jameson-Lee M, Garduno RA, Hoffman PS. DsbA2 (27 kDa Com1-like protein) of Legionella pneumophila catalyses extracytoplasmic disulphide-bond formation in proteins including the Dot/Icm type IV secretion system. Mol Microbiol. 2011;80:835–852. doi: 10.1111/j.1365-2958.2011.07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heras B, Shouldice SR, Totsika M, Scanlon MJ, Schembri MA, et al. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol. 2009;7:215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- 43.Bouwman CW, Kohli M, Killoran A, Touchie GA, Kadner RJ, et al. Characterization of SrgA, a Salmonella enterica serovar typhimurium virulence plasmid-encoded paralogue of the disulfide oxidoreductase DsbA, essential for biogenesis of plasmid-encoded fimbriae. J Bacteriol. 2003;185:991–1000. doi: 10.1128/JB.185.3.991-1000.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin A, Mann BJ. Identification of transposon insertion mutants of Francisella tularensis tularensis strain Schu S4 deficient in intracellular replication in the hepatic cell line HepG2. BMC Microbiol. 2006;6:69. doi: 10.1186/1471-2180-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sullivan JT, Jeffery EF, Shannon JD, Ramakrishnan G. Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J Bacteriol. 2006;188:3785–3795. doi: 10.1128/JB.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]