Abstract
The epidermis of Arabidopsis wild-type primary roots, in which some cells grow hairs and others remain hairless in a position-dependent manner, has become an established model system to study cell differentiation. Here we present a molecular analysis of the RHL1 (ROOT HAIRLESS 1) gene that, if mutated, prevents the formation of hairs on primary roots and causes a seedling lethal phenotype. We have cloned the RHL1 gene by use of a T-DNA-tagged mutant and found that it encodes a protein that appears to be plant specific. The predicted RHL1 gene product is a small hydrophilic protein (38.9 kD) containing putative nuclear localization signals and shows no significant homology to any known amino acid sequence. We demonstrate that a 78-amino-acid sequence at its amino terminus is capable of directing an RHL1–GFP fusion protein to the nucleus. The RHL1 transcript is present throughout the wild-type plant and in suspension culture cells, but in very low amounts, suggesting a regulatory function for the RHL1 protein. Structural evidence suggests a role for the RHL1 gene product in the nucleolus. We have examined the genetic relationship between RHL1 and GL2, an inhibitor of root hair initiation in non-hair cells. Our molecular and genetic data with double mutants, together with the expression analysis of a GL2 promoter–GUS reporter gene construct, indicate that the RHL1 gene acts independently of GL2.
Keywords: Cell differentiation, root epidermal development, root hair initiation, Arabidopsis, RHL genes
During plant development, cells born in the meristem become progressively specified in relation to a particular cell fate, and then finally undergo a series of morphogenetic changes that result in a fully differentiated cell, such as a tracheid, a trichome, or a tapetal cell (Stewart 1978). An elegant model to study this sequence of events is the primary root epidermis of Arabidopsis. This consists only of two cell types, the trichoblasts, that go on to make a root hair, and the atrichoblasts, that develop into non-hair cells (Dolan et al. 1993, 1994). In the wild type, epidermal cells will only form a root hair if they overlie the cleft between two cortical cell files (Berger et al. 1998). As there is an invariant number of eight endodermal and eight cortical cell files in the Arabidopsis primary root, the number of hair cell files is fixed to eight as well. Sequential steps in time are spatially laid out in cell files along the root axis, an early initial cytoplasmic differentiation just above the meristematic zone, the visible onset of root hair formation at the end of the elongation zone and maturing root hairs up to the root–hypocotyl junction.
Several genes involved in the pathway leading to root hair initiation have been identified through their mutant phenotypes. Only very few mutants show root-specific phenotypes, and the erh 1 mutant that possesses ectopic root hairs, but appears to be unaffected in shoot development (Schneider et al. 1997) might be such a candidate. Most of the mutants with altered root hair patterning display pleiotropic phenotypes, indicating that the affected gene products might play a role in several developmental pathways.
One of the best characterized genes in this respect is GL2, which when mutated results in expanded leaf trichomes, ectopic root hairs, and a lack of proper seed coat mucilage (Koornneef 1981; Di Cristina et al. 1996; Masucci et al. 1996). The GL2 gene has been cloned and found to encode a homeodomain-containing protein (Rerie et al. 1994). In the primary root, the GL2 gene is preferentially expressed in atrichoblasts and therefore is thought to be either a negative regulator of root hair formation or a positive regulator of non-hair cell fate (Masucci et al. 1996). Another mutant, ttg, is characterized by ectopic root hairs, the lack of trichomes, and seed coat mucilage (Koornneef 1981, Galway et al. 1994). The TTG gene is potentially a positive regulator of GL2 in the wild-type (Di Cristina et al. 1996) and, therefore, is also itself a positive regulator of the non-hair cell fate.
A group of genes involved in plant growth factor pathways also confer alterations in root hair patterning when mutated. The auxin resistant mutant axr2 (Wilson et al. 1990) that displays a greatly reduced number of root hairs belongs to this class, as does the ethylene signal transduction mutant ctr1 (Kieber et al. 1993) that possesses ectopic root hairs (Dolan et al. 1994). The CTR1 gene has been cloned and the gene product shows homology to members of the Raf family of protein kinases (Kieber et al. 1993). Ethylene has been identified as a positive regulator of root hair formation in a dosage-dependent manner (Tanimoto et al. 1995). In all of these cases, ectopic root hairs are only produced on epidermal cells that have finished elongating, as in the wild-type case (Masucci and Schiefelbein 1996). Therefore, other events in cellular development like cell elongation, expansion, and morphogenesis may influence the morphogenetic process that is a consequence of progressive cell fate specification. The identification of an allele of pom1, a conditional root expansion mutant (Hauser et al. 1995), in a screen for ectopic root hair mutants (Schneider et al. 1997) provides further evidence for this assumption.
Recently, a new class of recessive mutants, the root hairless (rhl) mutants, have been identified in a visual screen for primary root phenotypes (Schneider et al. 1997). At the moment, three loci are known that cause similar phenotypes when mutated. A single T-DNA-tagged allele of rhl1 has been described earlier and the mutation mapped to the central part of chromosome I (Schneider et al. 1997). In this report we present the cloning of the RHL1 gene, its molecular characterization, the subcellular localization of the protein product, its expression pattern, its genetic relationship with GL2, and its possible role as a positive regulator of root hair initiation.
Results
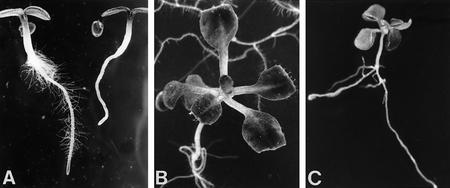
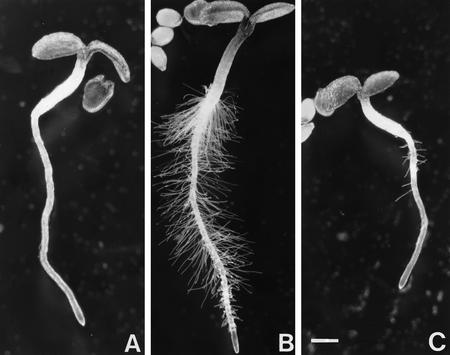
In a visual screen for root hair mutants, five rhl mutants that lack root hairs almost completely were identified (Schneider et al. 1997; Fig. 1). They fall into three complementation groups rhl1, rhl2, and rhl3, but their phenotypes are almost identical. Only occasionally are a few root hairs found around the emergence sites of the lateral roots. Ethylene treatment induces a very limited number of hairs on primary rhl roots (Schneider et al. 1997). As there is no obvious early cytoplasmic differentiation between hair and non-hair cell files in the root epidermis of the rhl mutants, the RHL genes are likely to act at early stages of cell specification as well as later in hair initiation. There are also severe defects in the shoots of rhl plants (Schneider et al. 1997; Fig. 1B,C). The rhl leaf epidermis has only a few abnormally shaped and spaced trichomes and rhl plants only form small rosettes and die after ∼3 weeks.
Figure 1.
Arabidopsis wild-type and rhl1 mutant phenotypes. (A) Three-day-old seedlings, with wild type at left and rhl1 mutant at right; (B) 2-week-old wild type; (C) 2-week-old rhl1 mutant seedling; photographs for B and C were taken at the same magnification.
Molecular cloning of the RHL1 gene
The rhl1 mutant was originally identified in a population of Arabidopsis thaliana (WS ecotype), transformed with the 3850:1003 T-DNA construct (Feldmann 1991). As the rhl1 mutant is a seedling lethal, the mutation was maintained in segregating populations. Seven lines with single T-DNA insertions were generated (see Materials and Methods).
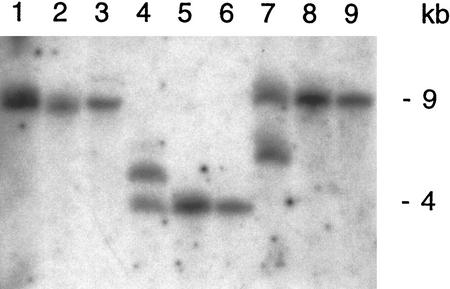
A plasmid rescue experiment identified 2 kb of plant genomic DNA adjacent to the left T-DNA border. In further genomic Southern blots, this 2-kb plant DNA fragment revealed a polymorphism between the DNA-banding pattern of heterozygous and wild-type plants (Fig. 2). Otherwise, under the stringent conditions chosen here, RHL1 appears to be a single copy gene in wild-type Arabidopsis.
Figure 2.
Genomic Southern analysis of T-DNA adjacent plant DNA. Genomic DNA from plants heterozygous for rhl1 (lanes 1,4,7), from WS wild-type plants (lanes 2,5,8), and from Columbia wild-type plants (lanes 3,6,9) was digested with SalI (lanes 1,2,3), HindIII (lanes 4,5,6), and PstI (lanes 7,8,9). The 2-kb insert obtained in the left border plasmid rescue experiment was used as a probe. A second band hybridizing in the DNA digests of the heterozygote indicates the presence of the T-DNA disrupted RHL1 allele.
The 2-kb plant genomic fragment from the plasmid rescue experiment served as a probe to isolate four genomic clones from a cosmid library prepared from Columbia wild-type DNA (C. Lister and C. Dean, unpubl.). A 4-kb BamHI fragment and an overlapping 2.4-kb EcoRI fragment, spanning 5.2 kb of genomic sequence around the putative RHL1 locus, were subcloned for sequence analysis. As the 2-kb plant genomic fragment identified a transcript on a Northern blot, it was directly used for the isolation of cDNA clones. Of four independent clones, one cDNA insert appeared to be slightly shorter than the other three, which all showed identical restriction patterns. One of the latter was therefore chosen for the nucleotide sequence analysis that allowed 1283 bp of the putative RHL1 transcript, followed by a poly(A)+ tail, to be determined (Fig. 3).
Figure 3.
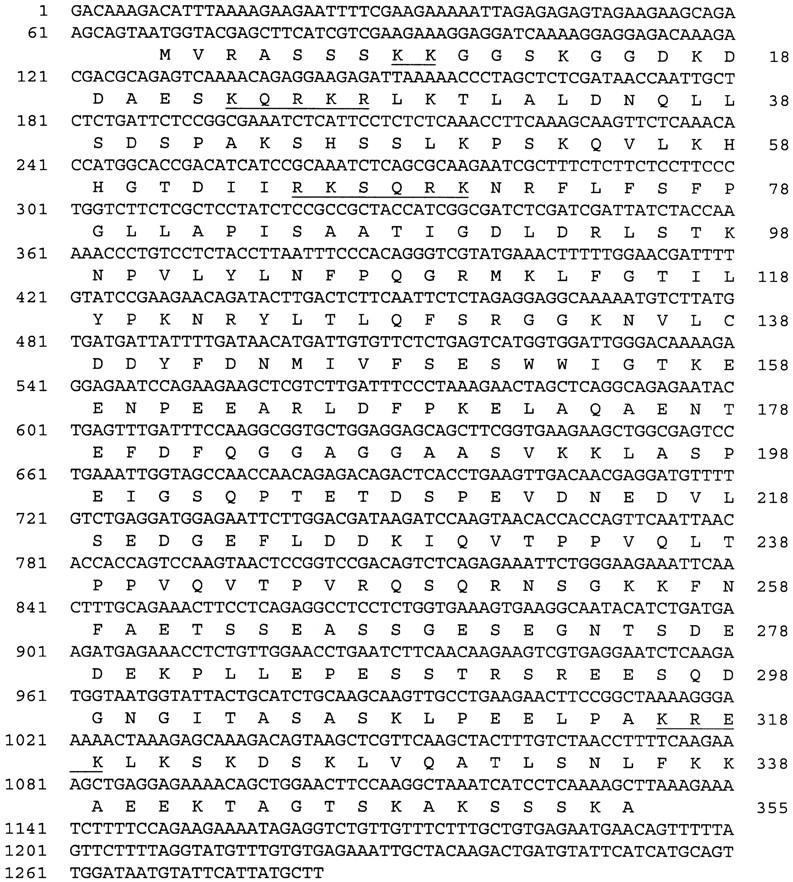
Nucleotide sequence of the RHL1 cDNA. The nucleotide sequence of the RHL1 cDNA is shown with the predicted amino acid sequence of the longest ORF, assumed to encode the RHL1 gene product (below). The nucleotide numbers are shown at the left; the amino acid numbers are given at the right. Putative nuclear localization signals are underlined.
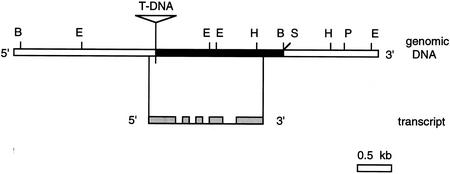
The alignment of cDNA and genomic sequence revealed that the RHL1 gene consists of five exons and four introns and covers only 2 kb of the Arabidopsis genome. The T-DNA integration disrupts the coding sequence in the first exon (Fig. 4).
Figure 4.
Molecular organization of the RHL1 gene. (Top) The genomic DNA at the RHL1 locus. A number of restriction sites are indicated (B, BamHI; E, EcoRI; H, HindIII; S, SalI; P, PstI). The integration site of the T-DNA is marked and the sequence isolated by the left border plasmid rescue experiment is shown in black. (Bottom) A schematic representation of the RHL1 transcript with its five exons shaded in gray.
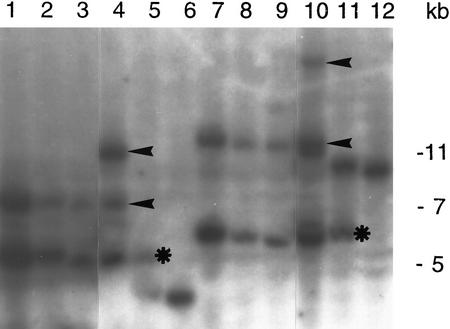
The formal proof for the cloning of the RHL1 gene requires the molecular complementation of the rhl1 mutant with the RHL1 wild-type gene. Therefore, the 4-kb BamHI fragment of genomic DNA—comprising ∼2 kb of the putative 5′ RHL1 promoter region, the entire assumed RHL1 coding region, and a small stretch of ∼200 bp of the 3′ genomic region of the RHL1 gene—was cloned into a plant transformation vector conferring hygromycin resistance. Difficulties were encountered with complementing the rhl1 mutant directly, so heterozygous plants were chosen for transformation instead. The progeny of the primary transformants were germinated on medium containing kanamycin and hygromycin. Twelve plants were recovered that were resistant to both antibiotics, and all had a fully restored wild-type phenotype. This result implies full functional complementation. Kanamycin- and hygromycin-resistant plants from the next generation of the complemented plants, that again exhibited the wild-type phenotype, were taken for genomic Southern analysis (Fig. 5). Those plants with only the T-DNA-tagged mutant copy of the RHL1 gene, together with one or more RHL1 transgenes that are different in restriction fragment size from the wild-type RHL1 copy, are proof of successful complementation.
Figure 5.
Genomic Southern analysis of the RHL1 gene in complemented plants. Genomic DNA from individual progeny of complemented plants harboring only the T-DNA-disrupted RHL1 allele and the RHL1 transgene (lanes 1–4,7–10), genomic DNA of the heterozygote (lanes 5,11), and the WS wild type (lanes 6,12) were digested with HindIII (lanes 1–6) and PstI (lanes 7–12). The RHL1 cDNA was used as a probe. (Asterisks) Bands representing the T-DNA-disrupted rhl1 allele; (arrowheads) bands resulting from the complementation construct.
Sequence analysis and subcellular localization of the RHL1 gene product
The longest ORF identified in the RHL1 cDNA encodes a polypeptide of 355 amino acids starting with the indicated methionine residue (Fig. 3). The nearest in-frame stop codon preceding the first ATG is 77 nucleotides upstream of the start of the cDNA in the genomic sequence and there is no other methionine in between, strongly suggesting that we have isolated the full coding sequence of the RHL1 gene. In the databases, we found a perfectly matching Arabidopsis EST (GenBank accession no. H37372) that is identical over the first 366 nucleotides in the 5′ region to the RHL1 cDNA. We performed RT–PCR with 5′ primers annealing to the RHL1 cDNA and to the genomic sequence ∼60 bp upstream of the start site of the RHL1 cDNA. Whereas we got the expected result with the internal 5′ primer, the 5′ primer annealing upstream of the RHL1 cDNA failed to generate a product (Fig. 6) supporting our determined RHL1 transcription start site.
Figure 6.
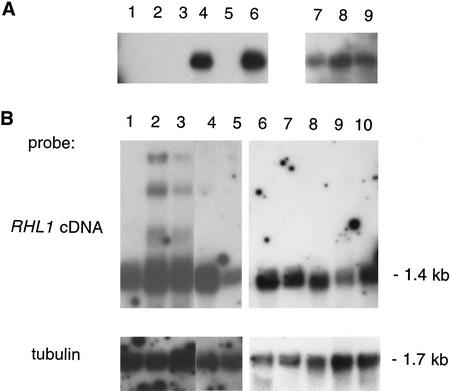
Analysis of RHL1 transcription. (A) RT–PCR. One microgram of total RNA of the following genotypes was primed with oligo(dT)–adaptor (B26/25); (lanes 1,2,7) rhl1 mutant; (lanes 3,4,8) rhl1 heterozygote; (lanes 5,6,9) wild type. For PCR amplification, gene-specific 5′ primers were used (see Materials and Methods) (lanes 1,3,5) C3204 priming ∼60 nucleotides upstream of the RHL1 cDNA (see Materials and Methods) and potentially generating a product of 1.35 kb; (lanes 2,4,6) C3004 priming the 5′ end of the RHL1 cDNA and generating a product of 1.2 kb; (lanes 7,8,9) control with a primer against the 5′ end of polyubiquitin cDNA generating a series of PCR products of which the largest, a 1.2-kb fragment, was taken as a standard. (B) Northern analysis. Five micrograms of poly(A)+ RNA of 5-week-old wild-type plants (lane 1) and two independent transformants possessing only the T-DNA-interrupted allele of the RHL1 gene and complemented with the RHL1 transgene (see text; lanes 2,3), Arabidopsis suspension culture cells (lane 4), and immature siliques (lane 5) was used for Northern blots and hybridized with either the RHL1 cDNA or a conserved part of the tubulin transcript as an internal loading standard. On a different blot, 4 μg of poly(A)+ RNA of 3-week-old wild-type seedlings (lane 6), shoots (lane 7), roots (lane 8), 5-day-old seedlings (lane 9), and flower buds (lane 10) was processed in the same way.
The sequence of the putative RHL1 gene product was scanned for homologies, at both the DNA and the protein levels. However, there was no significant homology to any other known gene product in the databases. Therefore, it is likely that the RHL1 gene is both novel and plant specific. In fact, a low stringency genomic Southern with DNA samples from a range of monocotyledonous and dicotyledonous plants indicated that even within the plant kingdom, RHL1-related sequences may rapidly diverge (data not shown).
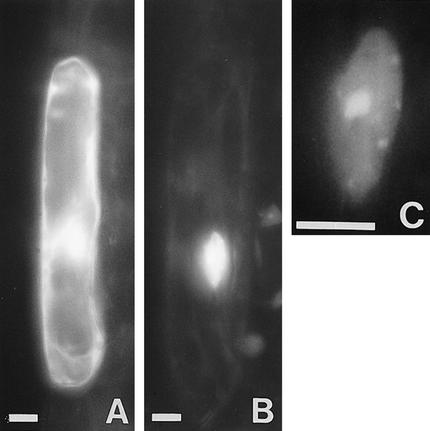
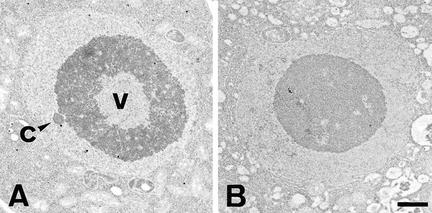
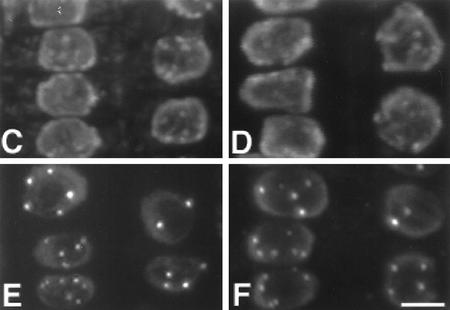
From the primary sequence, we predict a molecular weight of 38.9 kD and a pI of 7.5 for the putative RHL1 protein. Almost 31% of the amino acid residues of the putative RHL1 protein are charged and an additional 18% are hydroxylated, indicating that the potential RHL1 gene product is very hydrophilic. No transit or signal sequences were identified, nor was there any feature of a membrane-associated domain. However, the sequence reveals a number of potential nuclear localization signals (Raikhel 1992) as indicated in Figure 3. To test their functionality, we performed transient expression assays using engineered green fluorescent protein (GFP) as a vital marker (Chiu et al. 1996). We fused parts of the RHL1 cDNA sequence and the total coding region to the 5′ end of the GFP cDNA and expressed the constructs in the pRTL2 vector system (Carrington et al. 1991). Because the RHL1 protein might be specific to the Brassicaceae, it might require the presence of species-specific factors for its correct localization; therefore, we chose the homologous system, the Arabidopsis seedling, for our transformation experiments. Biolistic bombardment was used to deliver the DNA to the plant cells. Approximately 20 transformed cells, primarily located in the epidermal layer of the hypocotyl and in the cortical layer of the root, were identified per shot in three independent series of experiments. All cells transformed with the GFP control showed green fluorescence throughout the cytoplasm and the nucleus (Fig. 7). However, in cells transformed with a fusion between the 300-bp 5′ region of the RHL1 cDNA and GFP, the fluorescence was confined to the nucleus. Within the nucleus we even detected structures that were more strongly fluorescent, indicating an accumulation of the RHL1 protein. Among these structures, the nucleolus was prominent (Fig. 7). Constructs containing parts of the RHL1 3′ region and the total coding region fused to GFP failed to provide detectable fluorescence (data not shown). This negative result suggests that either high concentrations of the 3′ region of the RHL1 encoding sequence are unstable in the plant cell or the conformation of the fusion protein prevents fluorescence from GFP. In an attempt to link the molecular data of the RHL1 gene product with the phenotype of the rhl1 mutant at the subcellular level, we compared electron micrographs of wild-type and rhl1 nuclei in root epidermal cells (Fig. 8A,B). In the wild type, the nucleolus often contained large cavities typical of many plant cells (Shaw 1996). We identified nucleolar cavities in 16 of 30 wild-type trichoblasts and in 4 of 9 wild-type atrichoblasts. In the rhl1 root epidermal cells, there were no nucleolar cavities in 56 trichoblasts and only a single cavity was seen in 49 atrichoblasts. Two general aspects of nuclear functional architecture were examined by confocal microscopy of whole root preparations. The general level and distribution of heterochromatin were assessed by DAPI staining, and the number of coiled bodies (Lamond and Carmo-Fonseca 1993) in each nucleus was determined by immunofluorescence with an antibody against the splicing component U2B′′ snRNP (small nuclear ribonucleoprotein). The resulting images (Fig. 8C–F) show that for both nuclear features there is no difference between wild type and rhl1. In addition, the difference in coiled body number between hair and non-hair cell files (K. Boudonck, unpubl.) is preserved in rhl1. The images also show that the cell size difference between the two cell types, seen in wild type, remains unchanged in rhl1.
Figure 7.
Subcellular localization of the RHL1 protein by biolistic bombardment with GFP as a vital marker. Micrographs of cells transiently transformed with GFP (A), an RHL1::GFP fusion (B), and a magnification of the nucleus in B printed more lightly (C). Scale bar, 10 μm.
Figure 8.
Cellular and nuclear architecture of wild-type (A) and rhl1 (B) root epidermal cells. (A,C,E) Wild-type; (B,D,F) rhl1 mutant. (A,B) Transmission electron micrographs of thin sections of nuclei from cells in trichoblast positions. (c) Coiled body; (v) nuclear vacuole. Scale bar for A and B, 1 μm. (C,D) Projection of a confocal image series of DAPI-stained cells in trichoblast (left cell file) and atrichoblast position (right file). (E,F) Projection of a confocal image series of cells, labeled with 4G3 antibody, in trichoblast (left file) and atrichoblast position (right file). The labeling is restricted to the nucleus with strongly labeled coiled bodies. Scale bar for C–F, 5 μm.
Transcription of the RHL1 gene in Arabidopsis
To assess transcription of the RHL1 gene, we performed Northern analysis and RT–PCR. Steady-state RHL1 transcript levels throughout the plant were found to be extremely low. Routinely 4–7 μg of poly(A)+ RNA was used per sample in Northern analysis, and even then, long exposure times were required to obtain reasonable signals.
The RHL1 transcript is not detectable in rhl1 mutants by RT–PCR, but one product of the expected size is amplified in both heterozygous and wild-type plants (Fig. 6). By Northern analysis, the RHL1 transcript size was determined to be ∼1.4 kb. In the complemented plants, the RHL1 transcript level is restored to the wild-type amount. Higher molecular weight transcripts suggest some incomplete processing of the transcript from the transgene.
There appear to be equal levels of RHL1 transcript in shoots and roots of 3-week-old seedlings as well as in flower buds, but transcription is slightly lower in 5-day-old seedlings and in immature siliques. The RHL1 transcript was also detected in Arabidopsis cell cultures.
Genetic interaction between RHL1 and GL2
The GL2 gene is a well-characterized gene involved in root epidermal cell differentiation both at the genetic and the molecular level (see introductory section; Rerie et al. 1994; Masucci et al. 1996), and its possible interaction with the RHL1 gene was therefore investigated.
When the double mutant between gl2 and rhl1 was constructed, its roots had hardly any root hairs. It essentially looks like the rhl1 single mutant in all aspects of the plant, revealing formal epistasis of rhl1 to gl2 (Fig. 9).
Figure 9.
rhl1 and gl2 single and double mutant phenotypes. Three-day-old seedlings of rhl1 single mutant (A), gl2 single mutant (B), and rhl1 gl2 double mutant (C). Scale bar, 500 μm.
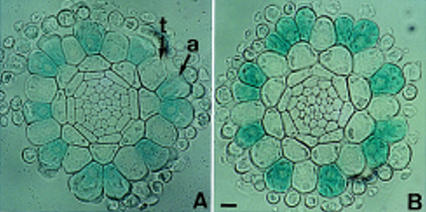
To determine GL2 transcription in rhl1 mutant roots, the expression of the GL2 promoter–GUS fusion was monitored. In the wild type, GUS staining was predominantly detected in the atrichoblasts (Masucci et al. 1996; Fig. 10A). In rhl1 mutants, the GL2 promoter induced GUS expression preferentially in atrichoblast cells (Fig. 10B) with ∼15% of stained cells in trichoblast cell files which is comparable with the wild type. Therefore, induction of GL2 transcription does not appear to require a functional RHL1 gene product in Arabidopsis roots.
Figure 10.
GL2 promoter–GUS expression in wild-type and rhl1 primary roots. Transverse sections (10 μm thick) of a GUS-stained primary root of the wild type (A) or rhl1 mutant (B). (a) atrichoblast; (t) trichoblast. Bar in B, 10 μm.
Discussion
In this report we show the cloning of the RHL1 gene that is required for hair initiation on Arabidopsis primary root epidermal cells, affects leaf epidermal patterning, and causes a seedling lethal phenotype when mutated (Schneider et al. 1997). It is the first member of the RHL gene class, comprising at least three loci, that has been characterized at the molecular level.
The RHL1 gene product is novel and may play a regulatory role in different plant organs
At present, the databases contain no similar sequences to the putative RHL1 gene product other than an EST and it also appears to be plant specific. Therefore, we are limited in our interpretation of possible functions to the primary sequence, the expression pattern of the RHL1 gene, and the mutant phenotype.
The steady-state level of RHL1 transcript is very low. Thus, a regulatory role of the RHL1 gene product is more likely than an enzymatic or a structural function. We detected the RHL1 transcript at all developmental stages and in all organs tested so far. The RHL1 gene is even transcribed in suspension culture cells. However, we have no data on post-transcriptional control. The RHL1 gene product is likely to play an important role in most, if not all, plant organs and the seedling lethal phenotype of the rhl1 mutant supports this assumption.
The RHL1 gene product may be localized in the nucleus
Using 300 bp of the RHL1 cDNA 5′ region in a transient expression assay with GFP as a visual tag, we showed that the RHL1 protein is located in the nucleus. Assuming that translation starts with the first methionine residue indicated in Figure 3, 78 amino acids of the RHL1 amino terminus corresponding to an 8.7-kD peptide are sufficient for targeting. That implies that the selected polypeptide contains a functional nuclear localization site for nuclear import; potential candidates have already been identified (Fig. 3). For the 8.7-kD RHL1 peptide, we calculated an isoelectric point of pI 11.4. The extremely basic nature of this protein fragment is in line with possible interactions with nucleic acids. By use of only this small part of the RHL1 protein in the construct, the RHL1–GFP fusion protein appears to accumulate in subnuclear structures like the nucleolus. The 8.7-kD RHL1 peptide contains numerous serine residues (16.7%), reminiscent of other proteins that accumulate in the nucleolus (Shaw and Jordan 1995). In those cases, however, the serine residues appeared more clustered than in the RHL1 peptide. Accumulation in the nucleolus is less likely to involve a targeting sequence, but it is possibly achieved by functional domains interacting with other proteins or nucleic acids that direct the complexes to the nucleolus, the site of ribosome assembly (Shaw and Jordan 1995). The subnuclear localization of the wild-type RHL1 protein could also be influenced by the other domains not present in our translational fusion with GFP. Functional assays with the entire RHL1 protein are needed to elucidate its mode of action and its interacting partners.
We have looked at various aspects of functional nuclear architecture, and from Figure 8 we can conclude that, at the structural level, chromatin condensation, cell cycle and size regulation, and the formation of coiled bodies appear to be unaffected in the rhl1 mutant. The only clear structural alteration in the nuclei of rhl1 root epidermal cells was the relative loss of nucleolar cavities. This loss suggests that the RHL1 protein may have a role in normal nucleolar processes that may include mRNA export functions in addition to ribosome production. The presence of nucleolar cavities has been correlated with active RNA metabolism (Shaw 1996).
RHL1 acts independently of GL2 in Arabidopsis roots
To assess the genetic relationship between RHL1 and GL2, we constructed the rhl1 gl2 double mutant that revealed epistasis of rhl1 over gl2. The roots of the double mutant had only very few root hairs.
This is consistent with the result from the analysis of the rhl-like rhl erh double mutants that root hair initiation requires the RHL genes (Schneider et al. 1997). However, this double-mutant result does not tell us whether RHL1 and GL2 are interacting in one linear pathway and if so, in which order they act. The possibility that RHL1 acts directly downstream of GL2 is unlikely, because phenotypic defects are visible in rhl1 roots much earlier than in gl2 roots (Masucci et al. 1996; Schneider et al. 1997), and RHL1 transcription appears to be unaffected in the gl2 background (data not shown).
To determine whether RHL1 acts upstream of GL2, we analyzed the transcription of the GL2 promoter at the cellular level in rhl1 mutant roots. However, we do not find any significant differences between the GL2 expression pattern in rhl1 and wild-type roots. Therefore, GL2 expression cannot depend on RHL1 gene activity. Thus, at present, it is most likely that RHL1 acts in a pathway required for root hair initiation and that GL2, induced by TTG, may act separately as a negative regulator in atrichoblasts.
In rhl1 mutant roots, cell specification events still result in cells in an appropriate epidermal position to make hairs, but that now lack some general nuclear function that helps to initiate a polar outgrowth or hair. The fact that an occasional hair can be initiated in response to excess ethylene suggests that the RHL1 protein is not essential for the machinery of hair growth itself, but is normally required to initiate it. Like the products of many other genes that cause pleiotropic phenotypes when mutated, the RHL1 gene product is a paradigm for components that are part of normal cellular activity, but can acquire a morphogenetic function within cells committed to a particular differentiation pathway.
Materials and methods
Plant material and growth conditions
The rhl1, erh1, pom1-12, and erh3 mutants have been described in detail recently (Schneider et al. 1997). The gl2 mutant and the line transformed with the GL2 promoter–GUS gene fusion have been presented elsewhere (Masucci et al. 1996). After surface sterilization, the seeds were vernalized and germinated on plates containing MS medium (pH 5.8), 3% sucrose, and 1% agarose. To select for transformed plants, the appropriate antibiotic was added to the autoclaved medium: 50 μg/ml of kanamycin and 15 μg/ml of hygromycin, respectively. The Arabidopsis cell suspension culture was grown as described before (May and Leaver 1993).
Genetic analysis
Potential heterozygotes that were selected for kanamycin resistance, a codominant marker of the T-DNA insertion, were outcrossed three times to the WS wild type. Thus, seven lines were generated that segregated in the 3:1 ratio of kanamycin-resistant to kanamycin-sensitive seedlings expected for a single T-DNA insertion (data not shown). All of the >400 rhl1 mutants analyzed were kanamycin resistant, indicating very tight linkage between the T-DNA insertion and the RHL1 gene. The fact that the T-DNA insert at the mutant locus was not rearranged was confirmed by genomic Southern analysis of heterozygous plant material by use of border and internal fragments of the T-DNA sequence as probes (data not shown). The rhl1 gl2 double mutant was constructed as described recently for other double mutants between root hairless and ectopic root hair mutants (Schneider et al. 1997). The GL2 promoter–GUS construct (Masucci et al. 1996) was introduced into the rhl1 mutant as follows. Plants heterozygous for rhl1 were fertilized with pollen from plants homozygous for the GL2 promoter–GUS construct. The F1 generation was selected for kanamycin resistant plants that were allowed to self-pollinate. Seedlings with the rhl1 phenotype in the F2 generation were submitted to the GUS-staining procedure.
Molecular biology techniques
Routine molecular biology work was performed as described by Sambrook et al. (1989).
Genomic plant DNA was extracted according to a slightly modified Dellaporta protocol (Dellaporta et al. 1983). For genomic Southern analysis, restriction fragments were separated in 0.7% agarose gels and transferred to nitrocellulose. Hybridization was carried out in 3× SSC, 0.1% SDS, and 1× Denhardt’s solution for 16 hr at 65°C. The filters were washed in 2× SSC, 0.1% SDS, and subsequently in 0.1× SSC, 1% SDS at 65°C for high stringency hybridizations. For low stringency hybridizations, only two washes with 2× SSC, 0.1% SDS were performed at 58°C.
For plasmid rescue experiments, genomic DNA from plants heterozygous for rhl1 was digested with either EcoRI or SalI, submitted to religation and transformed into Escherichia coli bacteria by electroporation (Feldmann 1992).
The λPRL-2 cDNA library, constructed from an amplified pool of cDNA samples from different Arabidopsis thaliana tissues, was prepared by T. Newman and associates (Michigan State University, Lansing) and obtained under accession number CD4-7 from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University.
DNA sequencing was performed with the Prism Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Perkin Elmer) on an ABI model 373A DNA sequencer (Applied Biosystems). DNA sequence analyses and searches were done with the software package of the University of Wisconsin Genetics Computer Group, version 8 (Devereux et al. 1984). The RHL1 cDNA sequence including the deduced amino acid sequence is available under GenBank accession number AF062371, and the DNA sequence of the RHL1 genomic clone has been published under GenBank accession number AF062372 in the databases.
To characterize transcription, Northern analysis was done according to Bartels et al. (1990) and RT–PCR was performed as described in Metzlaff et al. (1997). For RT–PCR all RNA samples were primed with the oligo(dT) adaptor (B26-B25). The following sequence specific 5′ primers were used: C3004, 5′-TCGTCGAAGAAAGGAGG-3′, annealing from nucleotides 83 to 99 of the RHL1 cDNA just one nucleotide upstream of the T-DNA insertion in the rhl1 mutant; C3204, 5′-TCCGTTGTTCTTCTTCC-3′, spanning nucleotides −70 to −54 on the genomic sequence just upstream of the start site of the RHL1 cDNA, and a primer annealing to the polyubiquitin cDNA (K.M. Davies et al., unpubl., GenBank accession no. X67957).
Complementation
For genetic complementation of the rhl1 mutant, a 4-kb BamHI fragment of genomic DNA comprising 2 kb upstream of the predicted RHL1 transcription start site, the entire coding region of the RHL1 gene, and 200 bp of 3′ genomic sequence was cloned into the pGDW31 plant expression vector (Wing et al. 1989) conferring hygromycin resistance to transformed plants. The construct was conjugated from E. coli to Agrobacterium and transformed into root explants from seedlings heterozygous for rhl1 by Agrobacterium-mediated Arabidopsis transformation as described (Koncz et al. 1994). Seeds of primary transformants were germinated on medium containing both kanamycin and hygromycin as selectable markers.
GUS staining of Arabidopsis thaliana roots, fixation, and embedding
Three-day-old seedlings were stained for 5 hr at 37°C in 0.2 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 1 mm potassium ferricyanide, 1 mm potassium ferrocyanide, 10 mm EDTA, and 0.1 m sodium phosphate buffer (pH 7). After three washes in the phosphate buffer, seedlings were prefixed in 1% glutaraldehyde for 1 hr and then embedded in a very thin layer of 1% agarose. A further fixation, the infiltration with Historesin (Leica), and the sectioning were essentially performed as described before (Schneider et al. 1997).
Plasmid constructs for transient assays and biolistic bombardment
A 300-bp fragment of the 5′ region of the RHL1 cDNA was PCR-amplified with specific primers introducing a HindIII, an XhoI, and an NcoI site at the termini for further cloning steps. 5′ primer, 5′-AAAAAACTCGAGCCATTGACAAAGACATTTAAAAG-3′ and 3′ primer, 5′-AAAAAAAGCTTAGGGAAGGAGAAGAGAAAGCG-3′. The 300-bp fragment was fused in-frame to the amino terminus of sGFPS65T (Chiu et al. 1996) by use of the HindIII site. The RHL1—GFP cassette was cut out and inserted into the pRTL2 vector (Carrington et al. 1991) creating the pRTL2::RHL1::GFP construct. As a control, the construct pRTL2::GFP was used.
Ten micrograms of Qiagen-purified plasmid DNA was precipitated onto 2.5 mg of gold containing a mixture of 0.7- and 0.95-μm big particles. An equal volume of 0.1 m spermidine was added. The volume was doubled by adding 2.5 m CaCl2. After vigorous mixing, the coating assay was incubated for 10 min at room temperature. The coated gold particles were washed with ice-cold ethanol and resuspended in a final volume of 2.5 ml ethanol by sonification. For each shot, 163 μl of the gold suspension was delivered to 3-day-old Arabidopsis seedlings by electric discharge particle acceleration (Christou et al. 1991). After 20–30 hr, the seedlings were viewed under an epifluorescence microscope (Nikon) at a wavelength of 488 nm.
Electron microscopy
Three-day-old seedlings were fixed in 2.5% glutaraldehyde in 0.05 m sodium cacodylate (pH 7.2) and treated with 1% osmium prior to dehydration and infiltration with LR White resin (Agar Scientific, Stansted, Essex, UK). Sections (0.1 μm thick) were collected onto carbon-coated copper grids and stained with uranyl acetate and lead citrate. The sections were examined in a JEOL 1200 EX electron microscope (JEOL, Tokyo, Japan).
Whole-mount immunofluorescence
Whole-mount immunofluorescence labeling was performed as described by Wymer et al. (1998). The 4G3 antibody (Habets et al. 1989) against the U2B′′ snRNP was used to visualize splicing components, and the cells were counterstained for total DNA content with DAPI.
Acknowledgments
We thank Sue Bunnewell for preparing medium for plates and for excellent photographic work. We are grateful to Ken Feldmann and the DuPont company for making the rhl1 mutant available. We acknowledge the ABRC at the Ohio State University for the λPRL-2 cDNA library. Our thanks also go to Clare Lister and Caroline Dean for making the cosmid library of genomic DNA from the Columbia ecotype available for screening. We are indebted to Csaba Koncz at the Max-Planck-Institut für Züchtungsforschung who hosted the complementation work in his laboratory and critically read the manuscript. Furthermore, we thank Michael Metzlaff for his advice on RT–PCR experiments and Eva Stöger for a successful introduction to biolistic bombardment techniques. We thank John Schiefelbein for providing seeds of the GL2 promoter–GUS line and the gl2 mutant and for sharing useful conversations. K.S. was supported by fellowships from the European Molecular Biology Organization (EMBO) (ALTF 568–1992) and the Human Frontiers Science Program (HFSP) (LT-135/93), and by a grant from the Gatsby Charitable Foundation. B.W., L.D., and K.R. acknowledge funding from the Biotechnology and Biological Sciences Research Council (BBSRC).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kschneid@mpiz-koeln.mpg.de; FAX (0221) 50 62 413.
References
- Bartels D, Schneider K, Terstappen G, Piatkowski D, Salamini F. Molecular cloning of abscisic acid-modulated genes which are induced during desiccation of the resurrection plant Craterostigma plantagineum. Planta. 1990;181:27–34. doi: 10.1007/BF00202321. [DOI] [PubMed] [Google Scholar]
- Berger F, Haselhoff J, Schiefelbein J, Dolan L. Positional information in root epidermis is defined during embryogenesis and acts in domains with strict boundaries. Curr Biol. 1998;8:421–430. doi: 10.1016/s0960-9822(98)70176-9. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Freed DD, Leinicke AJ. Bipartite signal sequence mediates nuclear translocation of the plant potyviral Nla protein. Plant Cell. 1991;3:953–962. doi: 10.1105/tpc.3.9.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu W-I, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Christou P, Ford TL, Kofron M. Production of transgenic rice (Oryza sativa L.) plants from agronomically important indica and japonica varieties via electric discharge particle acceleration of exogenous DNA into immature zygotic embryos. BioTechnology. 1991;9:957–962. [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: Version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Devereux J, Haeberly P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristina M, Sessa G, Dolan L, Linstead P, Baima S, Ruberti I, Morelli G. The Arabidopsis Athb-10 (GLABRA) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313x.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. Cellular organization of the Arabidopsis thaliana root. Development. 1993;119:71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development. 1994;120:2465–2474. [Google Scholar]
- Feldmann KA. T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1991;1:71–82. [Google Scholar]
- ————— . T-DNA insertion mutagenesis in Arabidopsis: Seed infection/transformation. In: Koncz C, Chua N-H, Schell J, editors. Methods in Arabidopsis research. London, UK: World Scientific; 1992. pp. 274–289. [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol. 1994;166:740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- Habets WJ, Hoet MH, De Jong BAW, Van der Kemp A, Van Venrooij WJ. Mapping of B cell epitopes on small nuclear ribonucleoproteins that react with human autoantibodies as well as with experimentally-induced mouse monoclonal antibodies. J Immunol. 1989;143:2560–2566. [PubMed] [Google Scholar]
- Hauser M-T, Morikami A, Benfey PN. Conditional root expansion mutants of Arabidopsis. Development. 1995;121:1237–1252. doi: 10.1242/dev.121.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Koncz C, Martini N, Szabados L, Hrouda M, Bachmair A, Schell J. Plant molecular biology manual B2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. Specialized vectors for gene tagging and expression studies; pp. 1–22. [Google Scholar]
- Koornneef M. The complex syndrome of ttg mutants. Arabidopsis Inf Serv. 1981;18:45–51. [Google Scholar]
- Lamond AI, Carmo-Fonseca M. The coiled body. Trends Cell Biol. 1993;3:198–204. doi: 10.1016/0962-8924(93)90214-l. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell. 1996;8:1505–1517. doi: 10.1105/tpc.8.9.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW. The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development. 1996;122:1253–1260. doi: 10.1242/dev.122.4.1253. [DOI] [PubMed] [Google Scholar]
- May MJ, Leaver CJ. Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 1993;103:621–627. doi: 10.1104/pp.103.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzlaff M, O’Dell M, Cluster PD, Flavell RB. RNA-mediated RNA degradation and chalcone synthase A silencing in petunia. Cell. 1997;88:845–854. doi: 10.1016/s0092-8674(00)81930-3. [DOI] [PubMed] [Google Scholar]
- Raikhel N. Nuclear targetting in plants. Plant Physiol. 1992;100:1627–1632. doi: 10.1104/pp.100.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes & Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schneider K, Wells B, Dolan L, Roberts K. Structural and genetic analysis of epidermal cell differentiation in Arabidopsis primary roots. Development. 1997;124:1789–1798. doi: 10.1242/dev.124.9.1789. [DOI] [PubMed] [Google Scholar]
- Shaw PJ. Nuclear organization in plants. Essays Biochem. 1996;31:77–89. [PubMed] [Google Scholar]
- Shaw PJ, Jordan EG. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Stewart RN. Ontogeny of the primary body in chimeral forms of higher plants. In: Subtelney S, Sussex IM, editors. The clonal basis of development. New York, NY: Academic Press; 1978. pp. 131–159. [Google Scholar]
- Tanimoto M, Roberts K, Dolan L. Ethylene is a positive regulator of root hair development in Arabidopsis thaliana. Plant J. 1995;8:943–948. doi: 10.1046/j.1365-313x.1995.8060943.x. [DOI] [PubMed] [Google Scholar]
- Wilson A, Pickett FB, Turner JC, Estelle M. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol & Gen Genet. 1990;222:377–383. doi: 10.1007/BF00633843. [DOI] [PubMed] [Google Scholar]
- Wing D, Koncz C, Schell J. Conserved function in Nicotiana tabacum of a single Drosophila hsp70 promoter heat shock element when fused to a minimal T-DNA promoter. Mol & Gen Genet. 1989;219:9–16. doi: 10.1007/BF00261151. [DOI] [PubMed] [Google Scholar]
- Wymer CL, Beven AL, Boudonck K, Lloyd CW. Confocal microscopy of plant cells. In: Paddock S, editor. Protocols in confocal microscopy. Totowa, NJ: Humana Press; 1998. . (In press.) [Google Scholar]