Abstract
Cortical surface area measures appear to be functionally relevant and distinct in etiology, development, and behavioral correlates compared with other size characteristics, such as cortical thickness. Little is known about genetic and environmental influences on individual differences in regional surface area in humans. Using a large sample of adult twins, we determined relative contributions of genes and environment on variations in regional cortical surface area as measured by magnetic resonance imaging before and after adjustment for genetic and environmental influences shared with total cortical surface area. We found high heritability for total surface area and, before adjustment, moderate heritability for regional surface areas. Compared with other lobes, heritability was higher for frontal lobe and lower for medial temporal lobe. After adjustment for total surface area, regionally specific genetic influences were substantially reduced, although still significant in most regions. Unlike other lobes, left frontal heritability remained high after adjustment. Thus, global and regionally specific genetic factors both influence cortical surface areas. These findings are broadly consistent with results from animal studies regarding the evolution and development of cortical patterning and may guide future research into specific environmental and genetic determinants of variation among humans in the surface area of particular regions.
Keywords: cortex, cortical thickness, heritability
Introduction
Cortical surface area is a relatively understudied feature in neuroimaging studies of brain structure. To date, research on the behavioral correlates and biological underpinnings of brain size has focused predominantly on volumetric measures of brain structure, with a more recent emphasis on measures of cortical thickness. Despite limited study, findings have emerged to suggest that surface area is an important measure, distinct from cortical thickness in its contribution to volume.
The increased size of the human cortex, in comparison to that of other animals, appears to be driven primarily by expansion of the surface area, rather than an increase in thickness (Rakic 2009). Similarly, individual differences among humans in cortical volume are largely attributable to variability in surface area as opposed to thickness (Pakkenberg and Gundersen 1997; Im et al. 2008). In our recent twin study, we demonstrated genetic independence of surface area measures from measures of cortical thickness (Panizzon et al. 2009). The results of a family study (Winkler et al. 2010) and studies that have examined associations with particular genetic polymorphisms (Joyner et al. 2009; Rimol et al. 2010) have been consistent with such independence.
Examination of cortical surface area may prove useful in understanding normal brain development and brain aging, as well as structural effects of neuropathology. However, relationships such as the associations between regional measures of cortical surface area and age and cognition have only begun to be explored. Findings to date suggest that measures of cortical surface area, specifically total surface area and lobar surface area are negatively associated with age (Pakkenberg and Gundersen 1997; Ostby et al. 2009), even in samples exclusively of younger individuals. Recent findings also suggest that cortical surface area is related to cognitive performance and disease processes. For example, parietal lobe surface area has been shown to be positively associated with performance on a test of mental rotation ability in men (Koscik et al. 2009). Dickerson et al. (2009) compared cortical surface area, cortical thickness, and volume of medial temporal regions among patients with Alzheimer's disease and healthy younger and older adults and found that age-related differences (i.e., comparing healthy older with younger adults) were most prominent for volume and surface area measures, whereas disease-related differences (i.e., comparing patients with Alzheimer's disease to healthy older adults) were most prominent for volume and thickness measures. In adults with autism (Raznahan et al. 2010), older age was found to be associated with thicker cortical regions but not greater surface area, further emphasizing the dissociation between these measures.
Examination of the genetic and environmental influences on variations of cortical surface area between individuals is essential to understanding neural development, in regard to both normal processes and disease-related changes. Family and twin studies allow one to understand sources of individual differences in cortical surface area by partitioning the variance into genetic and nongenetic components, and population-based statistics, such as heritability, can be estimated. In a family study of baboons, total cerebral surface area was found to be highly heritable, such that, when controlling for age effects, 73% of the remaining variance was due to additive genetic effects (Rogers et al. 2007). Two small twin studies of humans found that familial effects were prominent in influencing total surface area (Tramo et al. 1998; White et al. 2002), hemispheric surface area, lobar surface area, and surface area in individual regions of interest (ROIs), especially in the left hemisphere (Tramo et al. 1998). Since both of these studies were small and only included monozygotic (MZ) twins, the conclusions that can be drawn from them are limited.
Our group recently reported a much larger-scale twin study of the genetic contributions to variations in global and lobar cortical surface area (Panizzon et al. 2009). We studied 474 individuals (including 110 MZ, or identical, twin pairs and 92 dizygotic [DZ], or fraternal, twin pairs) and found that 89% of the variance in total cortical surface area was attributable to genetic factors. Winkler et al. (2010) used family pedigrees to estimate heritability of global surface area and surface area of regional cortical parcellations in a sample of 486 participants and found similar genetic contributions. They found a heritability of 0.71 for global surface area; regional heritabilities ranged from 0.17 (frontal pole) to 0.68 (pericalcarine cortex) after correction for global size measures. Thus, the literature to date suggests that, while cortical surface area is highly heritable in general, the degree of heritability may vary by brain region.
In the current study, we examine in detail the genetic, shared environmental and unique environmental contributions to individual differences in regional surface area within the cortical parcellations of the Desikan–Killiany atlas (Desikan et al. 2006) using a large adult male twin sample. This work complements a previous report in which we detailed the genetic and environmental contributions to variations in cortical thickness within the same regions (Kremen et al. 2010) and expands on our previous study (Panizzon et al. 2009) by estimating within region heritabilities and by examining the impact of adjusting for the genetic and environmental effects shared between a region and total surface area. In contrast to previous reports, we also examine the degree to which apparent differences in the magnitude of heritability estimates are reliable (i.e., significant). Further, shared environmental contributions were not emphasized in our previous report, nor were they accounted for in the study of Winkler et al. (2010). Based on existing studies, we hypothesized that regional surface area measures would be generally quite heritable, with little contribution from shared environmental factors. We also expected, based on studies demonstrating high phenotypic correlations between regional and total surface area (Winkler et al. 2010), that the genetic contributions to individual differences in surface area of particular regional parcellations would be considerably smaller after accounting for genetic and environmental sources of variation in total surface area. To place the effect of adjustment for total surface area in context, we also examined the effect of adjusting for a global measure of cortical thickness on regional cortical thickness heritabilities. We hypothesized that the effects of global adjustment would be smaller for cortical thickness than for cortical surface area.
Materials and Methods
Participants
The Vietnam Era Twin Study of Aging (VETSA) project has been described in detail elsewhere (Kremen et al. 2006). Briefly, the VETSA sample of 1237 twins was drawn from an earlier study of over 3300 twin pairs from the Vietnam Era Twin (VET) Registry (Tsuang et al. 2001). The VET Registry is a sample of male–male twin pairs born between 1939 and 1957 who had both served in the United States military at some point between 1965 and 1975 (Goldberg et al. 2002). The study sample is not a Veterans Affairs (VA) or patient group, and the large majority of individuals were not exposed to combat. For this analysis, a subset of 474 individual VETSA participants with MRI data were included. Of those, 404 were paired (i.e., 202 twin pairs): 110 MZ and 92 DZ pairs. Twin zygosity was classified according to questionnaire and blood group information, and DNA verification has been made on a subset of 56% of the twins based on 25 microsatellite markers. As in the overall VETSA project, 95% of the questionnaire-based classifications agreed with the DNA-based classifications; when differences occurred, we used the DNA-based classifications.
Of the VETSA participants invited to undergo MRI scanning, only 6% declined to participate. Ultimately, 59% of those who initially agreed to participate were included. The remaining participants were not included for reasons such as possible metal in the body (7%), claustrophobia (3%), testing being conducted in the twins' hometown (5%), scanner problems (8%), cotwin being excluded (9%), and other reasons (9%).
Mean age of the MRI participants was 55.8 (2.6) years (range: 51–59), mean years of education was 13.9 (standard deviation = 2.1), and 85.2% were right handed. Most participants were employed full time (74.9%), 4.2% were employed part time, and 11.2% were retired. There were 88.3% non-Hispanic white participants, 5.3% African–American, 3.4% Hispanic, and 3.0% who were classified as “other.” Self-reported overall health status was as follows: excellent (14.8%); very good (36.5%); good (37.4%); fair (10.4%); and poor (0.9%). Demographic characteristics of the VETSA MRI sample did not differ from the larger sample and are comparable to US census data for similarly aged men (Centers for Disease Control and Prevention 2003; National Center for Disease Statistics 2003). There were no significant demographic differences between MZ and DZ twins.
All participants gave informed consent to participate in the research, and the study was approved by the Institutional Review Boards of the University of California, San Diego, Boston University and the Massachusetts General Hospital.
Image Acquisition
Images were acquired on Siemens 1.5 T scanners (241 at University of California, San Diego [UCSD]; 233 at Massachusetts General Hospital [MGH]). Sagittal T1-weighted magnetization prepared rapid gradient echo sequences were employed with a time to inversion = 1000 ms, time echo = 3.31 ms, time repetition = 2730 ms, flip angle = 7°, slice thickness = 1.33 mm, and voxel size 1.31.0 × 1.3 mm. Raw Digital Imaging and Communications in Medicine MRI scans (including 2 T1-weighted volumes per case) were downloaded to the MGH site. These data were reviewed for quality, registered, and averaged to improve signal to noise. Of the 493 scans available at the time of these analyses, quality control measures excluded 0.6% (3 cases) due to scanner artifact and 3% (16 cases) due to inadequate image processing results (e.g., poor contrast caused removal of nonbrain to fail).
Image Processing
The cortical surface was reconstructed using methods based on the publicly available FreeSurfer software package (Dale et al. 1999; Fischl et al. 1999; Fischl and Dale 2000; Fischl et al. 2004). Variation in image intensity due to magnetic field inhomogeneities was corrected, a normalized intensity image was created, and the skull (nonbrain) was removed from this image. A preliminary segmentation was then partitioned using a connected components algorithm, with connectivity not allowed across the established cutting planes. Interior holes in the components representing white matter were filled, resulting in a single filled volume for each cortical hemisphere. The resulting surface was covered with a polygonal tessellation and smoothed to reduce metric distortions. A refinement procedure was then applied to obtain a representation of the gray/white boundary, and the resulting surface was subsequently deformed outwards to obtain an explicit representation of the pial surface. Once generated, the cortical surface model was manually reviewed and edited for technical accuracy. Minimal manual editing was performed in alignment with standard, objective editing rules. Maps were placed into a common coordinate system using a nonrigid high-dimensional spherical averaging method to align cortical folding patterns. This procedure provides accurate matching of morphologically homologous cortical locations across subjects based on each individual's anatomy while minimizing metric distortion.
The surface was then divided into cortical regions of interest (Fischl et al. 2004). A label was given to each vertex based on 1) the prior probability of that label at that surface-based atlas location based on the manually parcellated training set, 2) local curvature information, and 3) contextual information, such as rules about spatial neighborhood relationships derived from the manual training set. Surface area was then calculated for the 66 ROIs (33 per hemisphere) in the parcellation scheme (Desikan et al. 2006) as the sum of the areas of each triangle falling within a given ROI. Calculations are made in each subjects' native space. We renamed the posterior cingulate as rostral posterior cingulate and isthmus of the cingulate as retrosplenial cortex for clarity of presentation in the tables. Total surface area was calculated as the sum of the areas of all ROIs.
Statistical Analysis
Models using twin data utilize MZ and DZ twin pair variances and covariance to estimate the proportion of total phenotypic variance due to additive genetic, shared environmental, and unique environmental influences. Additive genetic variance (A) refers to the additive genetic effects of alleles at every contributing locus. Shared environmental variance (C) is due to effects shared by a twin pair. Unique environmental variance (E) is due to effects not shared by a twin pair and also includes measurement error (Eaves et al. 1978; Neale and Cardon 1992).
Estimation of Genetic and Environmental Contributions
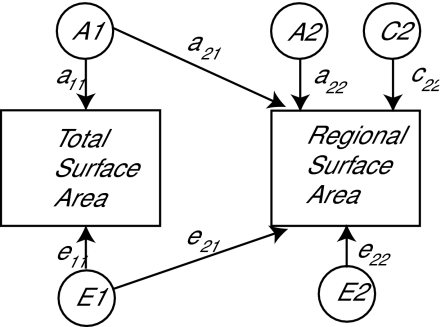
Bivariate twin models (using both regional and total surface area measures) were used to estimate the genetic and environmental contributions to the total phenotypic variance of a specific ROI. This approach allows for the estimation of “unadjusted” genetic and environmental effects on a specific ROI, which includes those genetic and environmental effects shared with total surface area. The bivariate model can also provide estimates of “adjusted” genetic and environmental contributions to regional surface area or the genetic and environmental effects specific to the ROI only (Fig. 1). The unadjusted heritability of a region of interest is calculated by summing the squared estimates of a21 and a22 and dividing by the total phenotypic variance. Adjusted heritability (i.e., heritability unique to the surface area of the particular region) is calculated by squaring the parameter labeled a22 in the figure and dividing by the total phenotypic variance. All bivariate models included the effects of site of data collection (MGH or UCSD) and age as fixed effects on the means.
Figure 1.
Schematic of statistical bivariate model used to estimate genetic and environmental variance components for regional surface area measurements with and without adjustment for total surface area. A1 represents additive genetic effects that influence both total and regional surface area and A2 are those effects unique to the particular region. Similarly, E1 represents unique environmental influences that affect both total and regional surface area and E2 are those only affecting the particular region. We only modeled shared or “common” environmental effects (C2) on regional surface area measures in this bivariate model because previous full ACE models for total surface area demonstrated that the contributions of shared environment to total surface area were very low. The unadjusted heritability of a region of interest is calculated by summing the squared values of a21 and a22 and dividing by the total phenotypic variance. Adjusted heritability (i.e., heritability unique to the surface area of the particular region) is calculated by squaring the parameter labeled a22 in the figure and dividing by the total phenotypic variance. Parameter estimates for C and E effects are also presented in the tables.
Optimization of model fit to these data was estimated using a maximum likelihood approach by calculating twice the log likelihood (−2LL) of the raw data for each twin pair and summing across all twin pairs. The use of the −2LL to estimate model fit allows for hypothesis testing between an original model (ACE) and its nested models (AE, CE, and E). The statistical significance of a genetic or environmental estimate was tested by calculating the difference in model fit between a full model with estimates of A, C, and E versus AE, CE, and E only submodels. This procedure produces nested submodels in which the difference in maximum likelihood asymptotically follows a 50:50 mixture distribution of zero and a χ2 with degrees of freedom equal to the difference in the number of free parameters (Eaves et al. 1978; Neale and Cardon 1992; Dominicus et al. 2006).
Two series of submodels of decreasing complexity were fitted. The first tested the significance of the shared environmental effects specific to total surface area and those common between total surface area and a specific ROI. This model therefore included A and E parameters related to total surface area and A, C, and E for each specific ROI (Fig. 1). This submodel was tested because a univariate ACE model of total surface area had determined that the parameter estimate for C was very small (c2 = 0.05 [0; 0.3]) and nonsignificant. This model was compared against a full bivariate model with A, C, and E for both total surface area and a specific ROI. This series of submodels was not found to significantly differ against their respective full bivariate model.
The second series of submodels tested for the effects of 1) additive genetic, 2) shared environmental, and 3) additive genetic and shared environmental effects specific to an ROI. This series of models were tested against models where there were A and E parameters related to total surface area and A, C, and E for each specific ROI.
We also wanted to determine whether the magnitudes of lobar heritability estimates were reliably different from one another. In order to place a significance level on the differences in heritability between pairs of lobar heritability estimates, we performed bootstrap analyses as follows: we randomly selected, with replacement, 110 MZ and 92 DZ twin pairs in each bootstrapped data set. The bivariate ACE–AE model was fit to each bootstrapped data set, and heritability estimates with and without adjustment for total surface area were extracted; this procedure was performed 20 000 times. For each iteration of the bootstrap, we computed all 12 × 11/2 = 66 differences among the 12 lobar regions, ordering the difference so that the region with smaller heritability (computed from the original data set) was subtracted from the larger. The resulting 20 000 bootstrap estimates were used to compute 2-sided bootstrapped P values for the difference of each pair of regions. These P values were then adjusted for multiple comparisons with a 0.05 false discovery rate using the procedure of Benjamini and Hochberg (1995).
In order to compare the effects of adjustment for a global covariate between regional surface area and regional thickness, we also fitted bivariate AE models for regional cortical thickness of each ROI and of mean cortical thickness. Cortical thickness of each ROI was measured as previously described (Kremen et al. 2010). The global metric was calculated using a weighted average: the mean cortical thickness of each region was multiplied by the proportion of total surface area occupied by that region and these values were summed in order to allow accurate representation of the mean thickness across the whole extent of the cortex (i.e., larger regions were weighted more and smaller regions less in the calculation of average thickness).
Data were passed from the statistical programming environment R (Ihaka and Gentleman 1996; R Development Core Team 2005) to Mx, a maximum likelihood–based structural equation modeling program (Neale et al. 2003).
Results
The heritability, or proportion of the total variance due to additive genetic effects, of total surface area was substantial, with near zero estimates of shared and unique environmental effects (MZ correlation = 0.94, DZ correlation = 0.52; A = 0.90 [95% confidence interval {CI} = 0.65; 0.96]; C = 0.05 [95% CI = 0; 0.3]; E = 0.05 [95% CI = 0.04; 0.08]). There were significant reductions in fit if either A or both A and C were dropped from the model (both Ps < 0.0001). There was no significant difference in model fit for a model without C compared with the full ACE model. Under an AE model, the heritability was estimated to be 0.95 (95% CI = 0.92; 0.96) and unique environmental contributions to individual differences in global surface area were again low (E = 0.05 [95% CI = 0.04; 0.08]).
Unadjusted Genetic and Environmental Contributions to Interindividual Variation in Regional Surface Area
MZ and DZ correlations as well as the proportions of variance accounted for by genetic and environmental effects for each lobar summary measure are presented in Table 1 and the variance components under an ACE model are presented graphically in Figure 2A. Values for each cortical parcellation region are presented in Supplementary Table 1. A greater difference in the correlations between MZ and DZ pairs suggests the presence of additive genetic effects on variation in regional surface area.
Table 1.
Lobar surface area measures adjusted for age and site: parameter estimates under bivariate models (AE influences on total surface area and either ACE or AE influences on regional surface area) and tests of submodels
| Region of interest | rMZ | rDZ | Parameter estimates with A, C, and E influences on region |
Model comparisons against model with A, C, and E influences on region |
Parameter estimates with A and E influences on region |
||||||||||
| A | 95% CI | C | 95% CI | E | 95% CI | No Aa | No Cb | No ACc | A | 95% CI | E | 95% CI | |||
| Left frontal | 0.94 | 0.52 | 0.94 | (0.91; 0.95) | 0.00 | (0; 0.01) | 0.06 | (0.05; 0.09) | <0.0001 | 1.00 | <0.0001 | 0.94 | (0.91; 0.95) | 0.06 | (0.05; 0.09) |
| Right frontal | 0.92 | 0.54 | 0.91 | (0.86; 0.94) | 0.02 | (0; 0.04) | 0.08 | (0.06; 0.11) | <0.0001 | 0.28 | <0.0001 | 0.92 | (0.9; 0.94) | 0.08 | (0.06; 0.1) |
| Left parietal | 0.88 | 0.45 | 0.89 | (0.82; 0.92) | 0.00 | (0; 0.05) | 0.10 | (0.08; 0.14) | <0.0001 | 0.89 | <0.0001 | 0.9 | (0.86; 0.92) | 0.1 | (0.08; 0.14) |
| Right parietal | 0.83 | 0.48 | 0.80 | (0.75; 0.87) | 0.05 | (0; 0.07) | 0.16 | (0.12; 0.21) | <0.0001 | 0.14 | <0.0001 | 0.85 | (0.8; 0.89) | 0.15 | (0.11; 0.2) |
| Left occipital | 0.79 | 0.50 | 0.75 | (0.57; 0.84) | 0.04 | (0; 0.18) | 0.21 | (0.16; 0.28) | <0.0001 | 0.57 | <0.0001 | 0.79 | (0.73; 0.84) | 0.21 | (0.16; 0.27) |
| Right occipitald | 0.65 | 0.53 | 0.56 | (0.48; 0.74) | 0.11 | (0; 0.17) | 0.33 | (0.25; 0.41) | <0.0001 | 0.21 | <0.0001 | 0.69 | (0.6; 0.76) | 0.31 | (0.24; 0.4) |
| Left lateral temporal | 0.86 | 0.36 | 0.86 | (0.80; 0.90) | 0.00 | (0; 0.04) | 0.14 | (0.1; 0.19) | <0.0001 | 1.00 | <0.0001 | 0.86 | (0.81; 0.9) | 0.14 | (0.1; 0.19) |
| Right lateral temporal | 0.79 | 0.38 | 0.75 | (0.68; 0.85) | 0.05 | (0; 0.09) | 0.20 | (0.15; 0.26) | <0.0001 | 0.30 | <0.0001 | 0.81 | (0.75; 0.86) | 0.19 | (0.14; 0.25) |
| Left medial temporal | 0.51 | 0.44 | 0.45 | (0.36; 0.64) | 0.09 | (0; 0.17) | 0.46 | (0.36; 0.57) | <0.0001 | 0.48 | <0.0001 | 0.55 | (0.44; 0.65) | 0.45 | (0.35; 0.56) |
| Right medial temporal | 0.54 | 0.21 | 0.55 | (0.42; 0.65) | 0.00 | (0; 0.1) | 0.45 | (0.35; 0.57) | <0.0001 | 1.00 | <0.0001 | 0.55 | (0.43; 0.65) | 0.45 | (0.35; 0.57) |
| Left cingulate cortex | 0.57 | 0.28 | 0.58 | (0.38; 0.68) | 0.00 | (0; 0.15) | 0.42 | (0.32; 0.54) | <0.0001 | 1.00 | <0.0001 | 0.58 | (0.46; 0.68) | 0.42 | (0.32; 0.54) |
| Right cingulate cortex | 0.63 | 0.32 | 0.67 | (0.39; 0.75) | 0.00 | (0; 0.21) | 0.33 | (0.25; 0.44) | <0.0001 | 1.00 | <0.0001 | 0.67 | (0.56; 0.75) | 0.33 | (0.25; 0.44) |
Note: rMZ = phenotypic correlation among MZ twins, rDZ = phenotypic correlation among DZ twins, A= additive genetic variance, C = shared environmental variance, E = unique environmental variance, Parameter estimates are listed in bold for greater readability.
Testing whether setting parameters a21 and a22 (see Fig. 1) to zero significantly reduces model fit. A significant change in fit indicates that a model without genetic influences on the region provides a worse representation of the data and provides a significance level for the heritability estimate.
Testing whether setting parameters c21 and c22 to zero significantly reduces model fit. A significant change in fit indicates that a model without shared environmental influences on the region fits significantly worse.
Testing whether setting parameters a21, a22, c21, and c22 to zero significantly reduces model fit. A significant change in fit indicates that a model without genetic and shared environmental influences on the region fits significantly worse.
Regions in which the shared environmental estimates are greater than 0.10, warranting caution in interpreting the A effects from an AE model as purely genetic in origin.
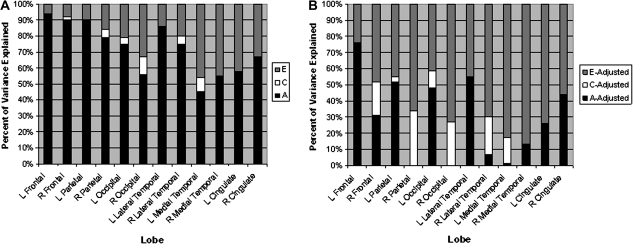
Figure 2.
Graphical representation of additive genetic (A), common environmental (C), and unique environmental (E) variance components for cortical lobes before (panel A) and after (panel B) adjustment for genetic and environmental effects shared with total surface area.
When estimated as part of a model containing A and E influences on total surface area and A, C, and E influences on each region, the average heritability across all individual regions was 0.38, noticeably lower than that of total surface area. Here, we are referring to the regional heritability estimates without adjustment for total surface area. Unadjusted heritability estimates were significant (as indicated by significance of the “no A” model comparison at P < 0.05 or a 95% confidence interval not containing zero) for 64 of 66 regions. Heritability estimates for bilateral frontal pole regions were not significantly greater than zero. Regionally, the estimates of shared environmental influences were very low, with 10 of 66 regions having C estimates greater than 0.10 and only one region (left parahippocampal gyrus) having an estimate that was significantly greater than zero (C = 0.23).
Because the majority of regions showed small contributions of C, heritabilities were also estimated as part of a model with only A and E influences on both total surface area and regional surface area. The mean unadjusted heritability across regions was 0.44 using this model and 63 of 66 regions were significantly heritable.
Under models with either ACE or AE influences on lobar surface area, bilateral frontal, parietal, and lateral temporal as well as left occipital heritabilities were the highest. Left frontal heritability was significantly greater than all other lobes (Ps < 0.003) with the exception of the right frontal lobe (P = 0.10). Bilateral medial temporal surface area heritabilities were the lowest and left medial temporal lobe heritability was significantly lower than most other lobes (Ps < 0.008) with the exception of right occipital (P = 0.14), right (P = 0.29) and left (P = 0.43) cingulate, and left medial temporal (P = 0.41). These lobar level findings were generally consistent with the findings for individual regions within the lobes. For example, within the medial aspect of the temporal lobe, none of the regions had heritabilities greater than 0.50, whereas all regions within the parietal lobe had heritabilities of this size or greater. Within each lobe, there were some regions that had much lower heritability estimates than others, and a few were not significantly heritable even under an AE model. Generally these regions were small in physical size and perhaps more prone to variability due to measurement error (which would tend to increase E and decrease A). We did not observe large differences between the heritability of left and right hemisphere structures for individual parcellations or at the lobar level, although there was a tendency at the lobar level for left hemisphere heritabilities to be nonsignificantly larger.
Adjusted Genetic and Environmental Effects on Regional Surface Area
Table 2 presents MZ and DZ correlations and variance component estimates for lobar surface areas adjusted for total surface area; Figure 2B shows the adjusted variance components graphically under an ACE model. Similar values for each cortical parcellation are shown in Supplementary Table 2. As we hypothesized, the heritability estimates for regional surface area decreased greatly after accounting for genetic variance associated with total surface area. Under a model with A and E effects estimated for both total surface area and ROI area, the average heritability across all regions after adjusting for total surface area was 0.22, a 50% reduction compared with the average estimate without adjustment for total surface area. Although reduced in magnitude, the majority of regions (45 of 66) still showed significant genetic influences under this model, however, only 4 regions had significant heritability under a model with A and E effects estimated for total surface area and A, C, and E effects estimated for ROI area.
Table 2.
Lobar surface area measures adjusted for age and site and total surface area: residual parameter estimates from bivariate ACE model and tests of submodels
| Region of interest | rMZ | rDZ | Residual parameter estimates under model with A, C, and E influences on region | Model comparisons against model with A, C, and E influences on region | Residual parameter estimates under model with A and E influences on region | ||||||||||
| A | 95% CI | C | 95% CI | E | 95% CI | No Aa | No Cb | No ACc | A | 95% CI | E | 95% CI | |||
| Left frontal | 0.79 | 0.21 | 0.76 | (0.6; 0.82) | 0.00 | (0; 0.15) | 0.24 | (0.18; 0.32) | 0.01 | 0.00 | <0.0001 | 0.76 | (0.68; 0.82) | 0.24 | (0.18; 0.32) |
| Right frontald | 0.50 | 0.38 | 0.31 | (0; 0.63) | 0.20 | (0; 0.51) | 0.48 | (0.37; 0.64) | 0.49 | 0.00 | <0.0001 | 0.54 | (0.4; 0.64) | 0.46 | (0.36; 0.6) |
| Left parietal | 0.57 | 0.27 | 0.52 | (0.1; 0.65) | 0.03 | (0; 0.39) | 0.45 | (0.35; 0.59) | 0.42 | 0.00 | <0.0001 | 0.55 | (0.42; 0.65) | 0.45 | (0.35; 0.58) |
| Right parietald | 0.37 | 0.30 | 0.00 | (0; 0.46) | 0.34 | (0; 0.46) | 0.66 | (0.52; 0.78) | 1.00 | 0.00 | <0.0001 | 0.37 | (0.23; 0.5) | 0.63 | (0.5; 0.77) |
| Left occipitald | 0.59 | 0.35 | 0.48 | (0.09; 0.69) | 0.11 | (0; 0.43) | 0.41 | (0.31; 0.54) | 0.43 | 0.00 | <0.0001 | 0.59 | (0.47; 0.69) | 0.41 | (0.31; 0.53) |
| Right occipitald | 0.28 | 0.27 | 0.00 | (0; 0.42) | 0.27 | (0; 0.39) | 0.73 | (0.57; 0.86) | 1.00 | 0.00 | 0.000 | 0.31 | (0.15; 0.45) | 0.69 | (0.55; 0.85) |
| Left lateral temporal | 0.57 | 0.18 | 0.55 | (0.3; 0.67) | 0.00 | (0; 0.2) | 0.45 | (0.33; 0.59) | 0.30 | 0.00 | <0.0001 | 0.55 | (0.41; 0.67) | 0.45 | (0.33; 0.59) |
| Right lateral temporald | 0.32 | 0.25 | 0.07 | (0; 0.44) | 0.23 | (0; 0.41) | 0.70 | (0.55; 0.84) | 1.00 | 0.00 | <0.0001 | 0.33 | (0.17; 0.46) | 0.67 | (0.54; 0.83) |
| Left medial temporald | 0.19 | 0.19 | 0.01 | (0; 0.34) | 0.16 | (0; 0.3) | 0.82 | (0.66; 0.96) | 1.00 | 0.00 | 0.002 | 0.2 | (0.04; 0.35) | 0.8 | (0.65; 0.96) |
| Right medial temporal | 0.17 | −0.05 | 0.13 | (0; 0.28) | 0.00 | (0; 0.19) | 0.87 | (0.72; 1) | 1.00 | 0.00 | 0.021 | 0.13 | (0; 0.28) | 0.87 | (0.72; 1) |
| Left cingulate cortex | 0.26 | 0.10 | 0.26 | (0; 0.41) | 0.00 | (0; 0.29) | 0.74 | (0.59; 0.92) | 0.72 | 0.00 | 0.001 | 0.26 | (0.08; 0.41) | 0.74 | (0.59; 0.92) |
| Right cingulate cortex | 0.42 | 0.23 | 0.44 | (0; 0.57) | 0.00 | (0; 0.36) | 0.56 | (0.43; 0.73) | 0.92 | 0.00 | <0.0001 | 0.44 | (0.29; 0.57) | 0.56 | (0.43; 0.71) |
Note: rMZ = phenotypic correlation among MZ twins, rDZ = phenotypic correlation among DZ twins, A= additive genetic variance, C = shared environmental variance, E = unique environmental variance.
Testing whether setting the parameter a22 to zero significantly reduces model fit. A significant change in fit indicates that a model without residual genetic influences on the region provides a worse representation of the data and provides a significance level for the heritability estimate.
Testing whether setting parameter c22 to zero significantly reduces model fit. A significant change in fit indicates that a model without residual shared environmental influences on the region fits significantly worse.
Testing whether setting parameters a22 and c22 to zero significantly reduces model fit. A significant change in fit indicates that a model without residual genetic and shared environmental influences on the region fits significantly worse.
Regions in which the shared environmental effects are greater than 0.10, warranting caution in interpreting the A effects from an AE as purely genetic in origin.
It should be noted that some of these A estimates may also contain shared environmental (C) influences that are not separately estimated in the AE model. Heritability estimates derived from an AE model can be particularly biased for regions that have moderate shared environmental effects, even if the C estimates are not significantly greater than zero (e.g., those labeled with a d in Tables 1 and 2 or that are starred in Supplementary Table 2.). For example, under an ACE model, the estimated heritability of the right paracentral lobule when accounting for total surface area is 0.01, but since there appears to be some contribution of shared environment to this region (C = 0.24), the heritability estimate under the AE model is 0.29.
At the lobar level, adjustment for total surface area also reduced heritability estimates; however, there was evidence for remaining genetic influences on the surface area of left frontal lobe, left parietal lobe, left lateral temporal, and left occipital lobes under an ACE model. When an AE model was used for each lobe, there were also moderate to large (20–76%) reductions in the degree of heritability following adjustment for total surface area, but all lobar estimates of heritability remained significant after adjustment for total surface area with the exception of right medial temporal lobe. In terms of regional differences in heritability, we found that total surface area–adjusted left frontal heritability was the highest (0.76) and was significantly greater than right lateral temporal (P = 0.0006), right (P = 0.0003) and left (P = 0.0001) medial temporal, right parietal (P < 0.0003), right occipital (P < 0.0001), and right (P = 0.0036) and left (P = 0.0006) cingulate adjusted heritability estimates. In addition, right (P = 0.0032) and left (P = 0.0017) medial temporal and right occipital (P = 0.0048) adjusted heritabilities were significantly lower than left lateral temporal adjusted heritability.
Heritability Estimates for Regional Cortical Thickness after Adjusting for Global Mean Thickness
For comparison purposes, we also calculated adjusted heritability estimates for regional measures of cortical thickness. These estimates have been previously reported with adjustment for estimated intracranial volume (Kremen et al. 2010). Intracranial volume adjustment had little effect on heritabilities, but we did not previously examine how regional heritabilities might be affected by adjustment for a global thickness measure. In the current analyses, we found that thickness heritability averaged across all the regions decreased from 0.49 to 0.36 after adjustment for mean cortical thickness; a reduction of 27%. After adjustment, all but 5 regions (left banks of the superior temporal sulcus, right inferior parietal, left supramarginal, and bilateral frontal pole) still showed significant heritability of cortical thickness. At the lobar level, there were significant genetic influences on cortical thickness in all lobes; none of the lobar heritabilities was greatly different from the others (including medial temporal lobe, which had adjusted thickness heritabilities of 0.49 [95% CI = 0.35; 0.61] on the left and 0.37 [95% CI = 0.22; 0.5] on the right).
Discussion
As hypothesized, we found that genetic factors contributed greatly to variation in surface area for almost all cortical parcellations, with heritabilities as high as 0.70 estimated from models with genetic and unique environmental variance components. Thus, genetic variation is an important determinant of individual differences in cortical surface area. These are likely to be genes related specifically to overall brain size, as opposed to body size more generally, because, although there were substantial reductions in regional heritabilities after adjusting for total surface area, we found a low correlation between height and total surface area in this sample (r = 0.24). Common environmental influences contributed to surface area measures in a small number of regions but generally accounted for less than 20% of the variance. Before any global adjustment, the heritabilities of regional surface areas (0.38 on average based on ACE models, 0.44 on average based on AE models) were significantly greater than zero but substantially smaller than that from an AE model of total surface area (0.95). Some of the difference may be due to greater measurement error for regional versus global measures, which would serve to increase unique environmental variance and decrease genetic variance estimates. This is supported by the fact that lobar heritabilities were intermediate in size between regional and global measures.
We found relatively little evidence for strong differences between individual parcellations in the degree of genetic and environmental contributions. Although heritabilities ranged from 0 to 0.70, there was generally considerable overlap in the confidence intervals. At the lobar level, we were able to test more directly for the reliability of differences between lobes in heritability estimates. One region in which unadjusted heritability was significantly lower for surface area was in the medial aspect of the temporal lobe. We did not observe such a large discrepancy for cortical thickness heritability in this region compared with other lobes, so to the extent that lower medial temporal lobe heritabilities for surface area are replicable and not due to greater measurement error in this region, this phenomenon may be related to environmental factors acting to expand or contract the numbers of neurons rather than their length and connections. In a previous family pedigree study using the same parcellation scheme to examine regional heritability of surface area (Winkler et al. 2010), genetic influences on medial temporal lobe surface areas were also slightly lower than those of other regions as determined by averaging their reported regional heritabilities (0.38 for medial temporal lobe compared with an average of other regions of 0.58).
It remains to be seen what sorts of environmental factors might be more strongly related to variation among middle-aged men in surface area of this medial temporal region, but those associated with normal aging processes are likely candidates since a recent study found reduction in mean surface area across age groups in these same regions (Dickerson et al. 2009) One might also speculate that environmental influences are more important in determining individual differences in cortical regions adjacent to subcortical structures such as the hippocampus which are particularly susceptible to toxic insults (e.g., hypoxia; Zola-Morgan et al. 1992). It should be noted that before adjustment for total surface area, there was still a meaningful contribution of genetic factors to the surface area of all regions within the medial aspect of the temporal lobe. This is consistent with the finding of an association between specific genetic polymorphisms (of the MECP2 gene known to be affected in Rett syndrome) and surface area of a region within the fusiform gyrus in a recent map-based study (Joyner et al. 2009). After adjustment, however, only 2 of the medial temporal regions (right entorhinal and left parahippocampal) had significant heritability estimates.
Extending our examination of variability among regions in genetic and environmental influences, we also examined whether regionally specific influences remained after controlling for genetic and environmental contributions to overall (i.e., total) surface area. We chose total surface area as our global covariate rather than intracranial or total brain volume since prior work suggests independent genetic influences on surface area and cortical thickness, both of which would contribute to intracranial volume and total brain volume (Panizzon et al. 2009). Consistent with our hypothesis, regional heritabilities were greatly reduced after controlling for genetic contributions to total surface area, although most remained significantly larger than zero under a model with only A and E effects. When region-specific C effects were also in the model, however, significant genetic influences could not be demonstrated in most regions. Winkler et al. (2010) also reported reductions in heritability estimates following adjustment for global variables, although their decreases were smaller (17–23% reductions). It is unclear what might account for the differences between these 2 studies, given that both used the same parcellation and image processing methods. The studies differed in being twin versus extended pedigree designs and in having a narrow versus a wide age range; however, it is not clear that these factors would account for the difference in the effect of adjusting for total surface area.
The effect of adjustment for total surface area on regional surface area heritability estimates was slightly greater than the effect of adjustment for mean cortical thickness on regional cortical thickness heritability estimates. After adjustment, there were no notable differences among lobar thickness heritabilities, whereas there were among the lobar surface area heritabilities. Specifically, adjusted left frontal surface area heritability was significantly greater than several other lobes. This suggests that there is a large amount of remaining genetic variance in left frontal surface area after accounting for genetic influences on total surface area. Since aging is known to have particular impact on the size of the frontal lobe (Raz et al. 2004; Fjell et al. 2009), it may be that aging-relevant genes are having strong regional influence.
Still, overall, we found that genetic effects were reduced after controlling for associations with global variables. Furthermore, almost all phenotypic correlations between total surface area and regional ROIs were strong, ranging from 0.12 to 0.96 (P < 0.0001). These phenotypic correlations were found to be driven by significant genetic covariances ranging from 0.22 (95% CI = 0.06; 0.33) to 0.84 (95% CI = 0.06; 0.92), meaning that many of the genes responsible for variation in the area of each individual region are expected to be the same as those responsible for variation in surface area as a whole. Our finding of greater environmental influences on regional surface area after controlling for genetic contributions to determination of total surface area could be meaningful, but it also could point to limitations of using traditional cortical parcellation schemes. To the extent that genes may act developmentally on areas that cross the regional boundaries we enforced with the Desikan–Killiany atlas (Rubenstein and Rakic 1999), we may have underestimated regional heritabilities and the extent to which there are important variations in this regional heritability across the cortex even after accounting for global effects. It also may be the case that regional variability will be more evident in the degree to which genetic or environmental influences change during aging. Our sample is characterized by a narrow age range and follow-up MRIs are being performed to detect particular regions in which genetic influences may increase or decrease with age.
Our results suggest high heritability of total cortical surface area and an apparent role of both genes and environment in the determination of individual differences in regional surface area measures. Total genetic influences on individual variation in surface area for any given region were generally high since there were both influences shared with total surface area as well as smaller, but generally significant, unique genetic influences on the area of the particular region. Although heritability is a population-based statistic having to do with variations among individuals, our findings in middle-aged men are broadly consistent with neurodevelopmental evidence of a protomap that establishes, very early on, relative position and numbers of cortical columns in human-specific cytoarchitectonic regions (Rubenstein and Rakic 1999; Rakic 2009). Genes that impact cell cycling in the first phase of symmetric divisions of neural stem cells could have a large effect on total surface area, and evolutionary effects on cortical surface area are thought to have acted during this phase (Rakic 1995). Similarly, genes that affect the organization of the protomap and regulate gradients of transcription factors and signaling molecules clearly have effects on the relative size of cortical regions (O'Leary et al. 2007). Regional cortical surface area in the mature adult human is likely a product of both these early determinants of numbers of neurons and subsequent effects (both growth and shrinkage) on synaptogenesis, dendritic arborization, intracortical myelination, and connectivity. Our data suggest that these subsequent effects are both genetic and environmental, perhaps related to genes involved in synaptogenesis and programmed cell death and to life experiences that may serve to increase connections within functional regions. Stochastic processes may also contribute to nongenetic variation in neural structure between individuals (Macagno et al. 1973). Relative expansion of surface area from macaque to human and from human infants to human adults is not uniform across cerebral cortex (Hill et al. 2010), with particularly large expansion in left dorsal frontal cortex and relatively less expansion in other regions, such as medial temporal cortex. Perhaps consistent with these differences in rates of expansion during development, which are thought to reflect differential maturity at birth, we found differences between these same regions in the relative contributions of genetic versus environmental influences.
There are several limitations to our study, which guide future work. First, our sample only included male twins, so the generalizability of our findings to women is unknown. Second, despite our very large sample size, we were underpowered to make inferences about shared environmental effects (Visscher et al. 2008). Most of the estimates of shared environmental effects were quite low. However, in a small number of cases the estimates were high enough (even if nonsignificant) to suggest that heritability estimates based on AE models might be biased for those regions. These regions were specifically noted in Supplementary Table 1. and 2. This highlights the necessity of beginning with full models that include C effects so that one can most accurately model the full range of genetic and environmental sources of variation and then make valid inferences about the likely contributions of purely genetic effects (Kendler and Neale 2009). Third, although we chose a widely used cortical parcellation scheme, these boundaries may not be optimal for examining genetic contributions. Future studies will address this limitation using continuous maps of the heritability of area expansion or contraction relative to a standard template at each point on the cortex. Fourth, although we identified some regions (e.g., within the medial temporal lobe) that had relatively low heritability of surface area compared with other regions, we did not undertake an examination of whether the genes that influence surface area in one region (or set of regions) are distinct from those that influence the surface area of other regions. This is an important future direction that will help to identify independent structural phenotypes for gene association studies. Fifth, although our results are consistent with neurodevelopmental models, we can only definitively conclude from our cross-sectional data that these are the patterns of genetic influences on cortical surface area in middle age.
In this large-scale twin study, we found high heritability for global cortical surface area and moderate genetic contributions to variations in regional surface area. We found some evidence for stronger environmental contributions to medial temporal lobe surface area and stronger genetic contributions to frontal lobe surface area compared with several other lobes. Due to the substantial genetic covariance between total surface area and the area of specific regions, the influence of genetic factors on individual differences was reduced after controlling for global measures, although most regions had some unique genetic contributions and substantial unique genetic effects on surface area were still observed in a few regions, such as left frontal lobe. Even if unique genetic effects were not found for some specific regions, that does not mean that there are no genetic influences on the surface area of those regions; rather, it indicates that the genetic variance is shared with that of global surface area. The results highlight the importance of examining genes that have widespread effects in order to understand individual variation in surface area but also suggest that future work examining environmental influences on medial temporal lobe surface area and the effect of particular genes on the relative area of the left frontal cortex could be fruitful.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/
Funding
National Institutes of Health (NIH) National Institute on Aging (U24 RR021382, R01 AG18386, R01 AG18384, R01 AG22381, and R01 AG 22982); National Institute for Mental Health (T32 MH20030); National Institute for Drug Abuse (R01 DA18673); and VA Desert Pacific Mental Illness Research Education and Clinical Center. Additional support was provided in part by the National Center for Research Resources (P41-RR14075 and the NCRR BIRN Morphometric Project BIRN002, U24 RR021382), the National Institute for Biomedical Imaging and Bioengineering (R01 EB006758), the National Institute for Neurological Disorders and Stroke (R01 NS052585-01), as well The Autism and Dyslexia Project funded by the Ellison Medical Foundation. The US Department of Veterans Affairs has provided support for the development and maintenance of the VET Registry.
Supplementary Material
Acknowledgments
Numerous organizations have provided invaluable assistance in developing and maintaining the VET Registry, including VA Cooperative Studies Program; Department of Defense; National Personnel Records Center, National Archives and Records Administration; the Internal Revenue Service; NIH; National Opinion Research Center; National Research Council, National Academy of Sciences; the Institute for Survey Research, Temple University; Schulman, Ronca, and Bucuvalas, Inc. Most importantly, we gratefully acknowledge the cooperation and participation of the members of the VET Registry and their families. Without their contribution this research would not have been possible. Conflict of Interest: None declared.
References
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Centers for Disease Control and Prevention. Public health and aging: trends in aging United States and worldwide. MMWR CDC Surveill Summ. 2003;52:101–106. [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Feczko E, Augustinack JC, Pacheco J, Morris JC, Fischl B, Buckner RL. Differential effects of aging and Alzheimer's disease on medial temporal lobe cortical thickness and surface area. Neurobiol Aging. 2009;30:432–440. doi: 10.1016/j.neurobiolaging.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominicus A, Skrondal A, Gjessing HK, Pedersen NL, Palmgren J. Likelihood ratio tests in behavioral genetics: problems and solutions. Behav Genet. 2006;36:331–340. doi: 10.1007/s10519-005-9034-7. [DOI] [PubMed] [Google Scholar]
- Eaves LJ, Last KA, Young PA, Martin NG. Model-fitting approaches to the analysis of human behaviour. Heredity. 1978;41:249–320. doi: 10.1038/hdy.1978.101. [DOI] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, et al. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Fjell AM, Westlye LT, Amlien I, Espeseth T, Reinvang I, Raz N, Agartz I, Salat DH, Greve DN, Fischl B, et al. High consistency of regional cortical thinning in aging across multiple samples. Cereb Cortex. 2009;19:2001–2012. doi: 10.1093/cercor/bhn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J, Curran B, Vitek ME, Henderson WG, Boyko EJ. The Vietnam Era Twin Registry. Twin Res. 2002;5:476–481. doi: 10.1375/136905202320906318. [DOI] [PubMed] [Google Scholar]
- Hill J, Inder T, Neil J, Dierker D, Harwell J, Van Essen D. Similar patterns of cortical expansion during human development and evolution. Proc Natl Acad Sci U S A. 2010;107:13135–13140. doi: 10.1073/pnas.1001229107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Gr Stat. 1996;5:299–314. [Google Scholar]
- Im K, Lee JM, Lyttelton O, Kim SH, Evans AC, Kim SI. Brain size and cortical structure in the adult human brain. Cereb Cortex. 2008;18:2181–2191. doi: 10.1093/cercor/bhm244. [DOI] [PubMed] [Google Scholar]
- Joyner AH, J CR, Bloss CS, Bakken TE, Rimol LM, Melle I, Agartz I, Djurovic S, Topol EJ, Schork NJ, et al. A common MECP2 haplotype associates with reduced cortical surface area in humans in two independent populations. Proc Natl Acad Sci U S A. 2009;106:15483–15488. doi: 10.1073/pnas.0901866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Neale MC. “Familiality” or heritability. Arch Gen Psychiatry. 2009;66:452–453. doi: 10.1001/archgenpsychiatry.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscik T, O'Leary D, Moser DJ, Andreasen NC, Nopoulos P. Sex differences in parietal lobe morphology: relationship to mental rotation performance. Brain Cogn. 2009;69:451–459. doi: 10.1016/j.bandc.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen WS, Prom-Wormley E, Panizzon MS, Eyler LT, Fischl B, Neale MC, Franz CE, Lyons MJ, Pacheco J, Perry ME, et al. Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. Neuroimage. 2010;49:1213–1223. doi: 10.1016/j.neuroimage.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen WS, Thompson-Brenner H, Leung YM, Grant MD, Franz CE, Eisen SA, Jacobson KC, Boake C, Lyons MJ. Genes, environment, and time: the Vietnam Era Twin Study of Aging (VETSA) Twin Res Hum Genet. 2006;9:1009–1022. doi: 10.1375/183242706779462750. [DOI] [PubMed] [Google Scholar]
- Macagno ER, Lopresti V, Levinthal C. Structure and development of neuronal connections in isogenic organisms: variations and similarities in the optic system of Daphnia magna. Proc Natl Acad Sci U S A. 1973;70:57–61. doi: 10.1073/pnas.70.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Disease Statistics. Health, United States. Hyattsville (MD): U.S. Department of Health and Human Services, Centers for Disease Control and Prevention; 2003. [Google Scholar]
- Neale MC, Boker SM, Xie G, Maes HH. Mz: statistical modeling. Richmond (VA): Department of Psychiatry, Medical College of Virginia; 2003. [Google Scholar]
- Neale MC, Cardon LR. Methodology for genetic studies of twins and families. Dordrecht (The Netherlands): Kluwer Academic Publishers; 1992. [Google Scholar]
- O'Leary DD, Chou SJ, Sahara S. Area patterning of the mammalian cortex. Neuron. 2007;56:252–269. doi: 10.1016/j.neuron.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Ostby Y, Tamnes CK, Fjell AM, Westlye LT, Due-Tonnessen P, Walhovd KB. Heterogeneity in subcortical brain development: a structural magnetic resonance imaging study of brain maturation from 8 to 30 years. J Neurosci. 2009;29:11772–11782. doi: 10.1523/JNEUROSCI.1242-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakkenberg B, Gundersen HJ. Neocortical neuron number in humans: effect of sex and age. J Comp Neurol. 1997;384:312–320. [PubMed] [Google Scholar]
- Panizzon MS, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, Neale M, Jacobson K, Lyons MJ, Grant MD, Franz CE, et al. Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex. 2009;19:2728–2735. doi: 10.1093/cercor/bhp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2005. [Google Scholar]
- Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Nerosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz N, Gunning-Dixon F, Head D, Rodrigue KM, Williamson A, Acker JD. Aging, sexual dimorphism, and hemispheric asymmetry of the cerebral cortex: replicability of regional differences in volume. Neurobiol Aging. 2004;25:377–396. doi: 10.1016/S0197-4580(03)00118-0. [DOI] [PubMed] [Google Scholar]
- Raznahan A, Toro R, Daly E, Robertson D, Murphy C, Deeley Q, Bolton PF, Paus T, Murphy DG. Cortical anatomy in autism spectrum disorder: an in vivo MRI study on the effect of age. Cereb Cortex. 2010;20:1332–1340. doi: 10.1093/cercor/bhp198. [DOI] [PubMed] [Google Scholar]
- Rimol LM, Agartz I, Djurovic S, Brown AA, Roddey JC, Kahler AK, Mattingsdal M, Athanasiu L, Joyner AH, Schork NJ, et al. Sex-dependent association of common variants of microcephaly genes with brain structure. Proc Natl Acad Sci U S A. 2010;107:384–388. doi: 10.1073/pnas.0908454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Kochunov P, Lancaster J, Shelledy W, Glahn D, Blangero J, Fox P. Heritability of brain volume, surface area and shape: an MRI study in an extended pedigree of baboons. Hum Brain Mapp. 2007;28:576–583. doi: 10.1002/hbm.20407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Rakic P. Genetic control of cortical development. Cereb Cortex. 1999;9:521–523. doi: 10.1093/cercor/9.6.521. [DOI] [PubMed] [Google Scholar]
- Tramo MJ, Loftus WC, Stukel TA, Green RL, Weaver JB, Gazzaniga MS. Brain size, head size, and intelligence quotient in monozygotic twins. Neurology. 1998;50:1246–1252. doi: 10.1212/wnl.50.5.1246. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Bar JL, Harley RM, Lyons MJ. The Harvard Twin Study of Substance Abuse: what we have learned. Harv Rev Psychiatry. 2001;9:267–279. [PubMed] [Google Scholar]
- Visscher PM, Gordon S, Neale MC. Power of the classical twin design revisited: II detection of common environmental variance. Twin Res Hum Genet. 2008;11:48–54. doi: 10.1375/twin.11.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Andreasen NC, Nopoulos P. Brain volumes and surface morphology in monozygotic twins. Cereb Cortex. 2002;12:486–493. doi: 10.1093/cercor/12.5.486. [DOI] [PubMed] [Google Scholar]
- Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, Duggirala R, Glahn DC. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53:1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zola-Morgan S, Squire LR, Rempel NL, Clower RP, Amaral DG. Enduring memory impairment in monkeys after ischemic damage to the hippocampus. J Neurosci. 1992;12:2582–2596. doi: 10.1523/JNEUROSCI.12-07-02582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.