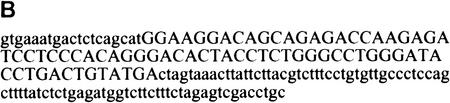Abstract
Using an in vitro randomization and functional selection procedure, we have identified three novel classes of exonic splicing enhancers (ESEs) recognized by human SF2/ASF, SRp40, and SRp55, respectively. These ESEs are functional in splicing and are highly specific. For SF2/ASF and SRp55, in most cases, only the cognate SR protein can efficiently recognize an ESE and activate splicing. In contrast, the SRp40-selected ESEs can function with either SRp40 or SRp55, but not with SF2/ASF. UV cross-linking/competition and immunoprecipitation experiments showed that SR proteins recognize their cognate ESEs in nuclear extract by direct and specific binding. A motif search algorithm was used to derive consensus sequences for ESEs recognized by these SR proteins. Each SR protein yielded a distinct 5- to 7-nucleotide degenerate consensus. These three consensus sequences occur at higher frequencies in exons than in introns and may thus help define exon–intron boundaries. They occur in clusters within regions corresponding to naturally occurring, mapped ESEs. We conclude that a remarkably diverse set of sequences can function as ESEs. The degeneracy of these motifs is consistent with the fact that exonic enhancers evolved within extremely diverse protein coding sequences and are recognized by a small number of SR proteins that bind RNA with limited sequence specificity.
Keywords: SR proteins, exonic splicing enhancers, SF2/ASF, RNA sequence motifs, SELEX
Pre-mRNA splicing consists of two trans-esterification reactions, which occur in a large RNA–protein complex termed the spliceosome. This high-fidelity process requires precise recognition of the intron–exon borders by the spliceosome. The poorly conserved metazoan splice sites and branch site do not provide sufficient information for this recognition. Additional intron and exon sequences are often necessary for efficient and/or accurate splicing of many higher eukaryotic pre-mRNAs. The positive exon cis-acting elements, known as exonic splicing enhancers (ESEs), are often, though not always, found in a purine-rich context. A well-studied example is the ESE in the alternative exon M2 of the mouse IgM gene. This 73-nucleotide ESE is essential for splicing of the preceding intron between exons M1 and M2. The M2 ESE can also stimulate splicing of a heterologous regulated intron of the Drosophila doublesex (dsx) gene. Enhancer activity in the context of the IgM pre-mRNA could also be obtained by insertion of certain natural or synthetic purine-rich sequences in place of the natural ESE. However, deletion of the purine-rich sequences within the M2 ESE did not abolish its activity completely (Watakabe et al. 1993; Tanaka et al. 1994). In agreement with this finding, SELEX experiments revealed that certain nonpurine-rich sequences can also function as enhancers (Tian and Kole 1995; Coulter et al. 1997). Most natural ESEs have been identified in tissue-specific or developmentally regulated exons, which typically have weak splice sites and require the ESE for exon inclusion. In some cases, ESEs are specifically recognized by one or more SR proteins (Lavigueur et al. 1993; Sun et al. 1993; Tian and Maniatis 1993, 1994; Ramchatesingh et al. 1995; Gontarek and Derse 1996). In turn, SR proteins are expressed at different levels in different tissues, and their expression also appears to be regulated by alternative splicing (Jumaa et al. 1997; for review, see Cáceres and Krainer 1997).
The SR proteins are a family of highly conserved serine/arginine-rich RNA-binding proteins. They are essential splicing factors (Krainer et al. 1990b, 1991; Ge et al. 1991; Zahler et al. 1992) and also regulate the selection of alternative splice sites in a concentration-dependent manner (Ge and Manley 1990; Krainer et al. 1990a; Zahler et al. 1993a), in part by antagonizing the activity of hnRNP A1 (Mayeda and Krainer 1992). The SR proteins act very early in spliceosome assembly (Krainer et al. 1990a; Fu and Maniatis 1992; Staknis and Reed 1994). They promote the binding of U1 snRNP to the 5′ splice site (Eperon et al. 1993; Wu and Maniatis 1993; Kohtz et al. 1994; Staknis and Reed 1994; Zahler and Roth 1995) and of U2AF65 to the 3′ splice site (Wu and Maniatis 1993), apparently by interacting with U1 70K and U2AF35, respectively. These observations have led to the hypothesis that SR proteins bound to ESEs recruit splicing factors to bind to the splice sites of adjacent introns (Wu and Maniatis 1993; Staknis and Reed 1994).
Nine human SR proteins are presently known: SF2/ASF, SC35, SRp20, SRp40, SRp75, SRp55, 9G8, SRp30c, and the somewhat more divergent p54. These proteins are closely related in primary structure and share the ability to complement splicing in a HeLa cell S100 extract (Ge et al. 1991; Krainer et al. 1991; Fu et al. 1992; Zahler et al. 1992, 1993b; Cavaloc et al. 1994; Screaton et al. 1995; Zhang and Wu 1996). SR proteins appear to have partially redundant functions, such that several different members of the family can complement an S100 extract to splice the same pre-mRNA, and/or stimulate use of the same alternative 5′ splice sites in vitro or in vivo. However, substrate-specific differences in general splicing, enhancer-dependent splicing, or alternative splicing mediated by different SR proteins have also been reported (Fu 1993; Sun et al. 1993; Zahler et al. 1993a; Cáceres et al. 1994; Wang and Manley 1995; Chandler et al. 1997). Drosophila SRp55/B52 has been shown to be essential for development (Ring and Lis 1994; Peng and Mount 1995), and at least one copy of the chicken SF2/ASF gene is required for survival of a B-lymphocyte cell line (Wang et al. 1996), demonstrating that at least some functions important for development or cell viability are uniquely carried out by single SR proteins in vivo. Individual SR proteins also differ in their subnuclear localization signals and in their ability to shuttle between the nucleus and the cytoplasm (Cáceres et al. 1997, 1998). Finally, individual SR proteins exhibit striking phylogenetic sequence conservation of all their constituent domains (Birney et al. 1993). Taken together, these observations demonstrate that individual SR proteins have some unique, specific functions.
Although SR proteins have been clearly implicated in ESE recognition and function, predictive rules for the recognition of ESEs by different SR proteins have not been derived. In this study, we sought to determine the specificity of individual SR proteins in ESE recognition by performing a randomization and selection procedure under splicing conditions.
Results
Identification of SR protein target sequences from a random pool under splicing conditions
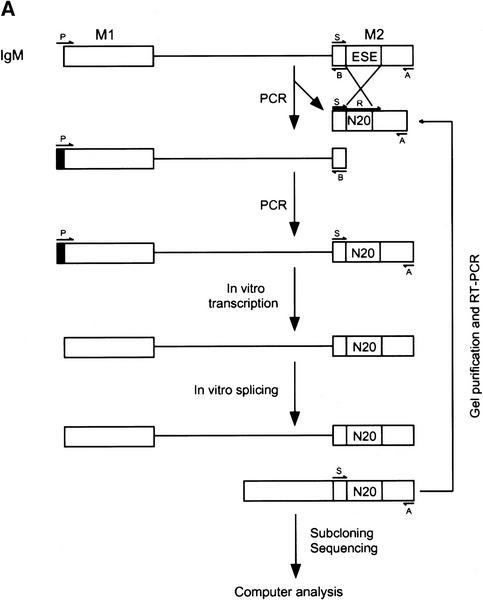
To find specific target sequences recognized by individual SR proteins under splicing conditions, a procedure based on SELEX (Tuerk and Gold 1990) was utilized imposing a selection for splicing (Tian and Kole 1995; Coulter et al. 1997), rather than for binding (Heinrichs and Baker 1995; Tacke and Manley 1995; Shi et al. 1997; Tacke et al. 1997). We modified the procedure further by carrying out the splicing reactions in the presence of a single, recombinant SR protein, which was used to complement HeLa extracts deficient in SR proteins— either an S100 extract or an SR protein-depleted nuclear extract. We performed the selection for ESEs in the context of a well-characterized IgM minigene transcript, comprising the last intron flanked by the M1 and M2 membrane isoform-specific exons. A prototypical ESE was previously mapped to a 73 nucleotide fragment of exon M2 (Watakabe et al. 1993). This ESE was found to be essential for IgM pre-mRNA splicing in nuclear or S100 extract (Watakabe et al. 1993; H.-X. Liu et al., unpubl.). The scheme for the randomization and selection procedure is outlined in Figure 1A (for details, see Materials and Methods). First, the natural ESE in the M2 exon (Fig. 1B) was replaced by 20 nucleotides of random sequence. A library of pre-mRNAs representing 1.2 × 1010 different sequences was spliced in S100 extract complemented by either recombinant SF2/ASF, SRp40, or SRp55 (Fig. 2). As a control, equivalent samples from the same library were spliced in nuclear extract, which contains all the SR proteins. A significant proportion of the pre-mRNAs from the initial pool contained sequences that functioned as enhancers in the nuclear extract, thus resulting in easily detectable levels of spliced mRNA (lanes 1,4,7). In contrast, no splicing could be detected by this direct assay in the S100 extract alone (lanes 2,5,8), or in reactions with S100 extract plus one of the SR proteins (lanes 3,6,9). Nevertheless, we assumed that a small proportion of the initial randomized sequences could function as enhancers in the presence of single SR proteins.
Figure 1.
Procedure for randomization and selection of ESEs. (A) The natural ESE in mouse IgM exon M2 was replaced by a 20-nucleotide segment of random sequence, and a library of pre-mRNAs was constructed by overlap–extension PCR and in vitro transcription. A sample of this pool, representing ∼1.2 × 1010 pre-mRNA molecules was then spliced in vitro by complementation of an S100 extract with individual recombinant SR proteins. The pool of spliced mRNA products was gel purified, and the sequences corresponding to the ESE region were rebuilt into pre-mRNA template molecules for a new round of selection, or subcloned and sequenced. The sequences were analyzed by a motif-search algorithm to identify common patterns. (B) Sequence of the M2 exon of the mouse IgM gene. The sequence of the previously mapped ESE is shown in uppercase.
Figure 2.
Splicing of the pre-mRNA pool prior to selection. Radiolabeled pre-mRNA (20 fmoles) from the unselected initial pool was incubated under splicing conditions in nuclear extract (lanes 1,4,7), in S100 extract only (lanes 2,5,8), or in S100 extract complemented with the indicated SR protein (lanes 3,6,9). The RNAs were analyzed by denaturing PAGE (12% polyacrylamide) and autoradiography. The structures and mobilities of the precursor, intermediates, and products of splicing (Watakabe et al. 1993) are shown next to each autoradiograph.
The presumptive spliced mRNAs, now carrying functional ESEs, were recovered from the expected region of denaturing polyacrylamide gels. For each S100 complementation reaction, the randomized region of exon M2 of the spliced mRNAs was then amplified by RT-PCR and reassembled into a new pool of pre-mRNAs for further selection. The amplification was carried out by overlap extension with two different upstream primers, to ensure that only spliced mRNAs were amplified (Fig. 1). PCR amplification confirmed the presence of spliced mRNAs after the initial round of selection. Two additional rounds of selection were carried out in SR protein-depleted nuclear extract (Blencowe et al. 1994) complemented with individual SR proteins, to mimic the conditions of nuclear extract, to minimize possible biases specific to the S100 extract, and to select the most efficient ESEs.
After three rounds of selection, the spliced mRNAs were amplified by RT-PCR, subcloned and sequenced. Twenty-four or more independent sequences obtained with each SR protein were analyzed to determine a consensus sequence, using the program Gibbs sampler (Lawrence et al. 1993). The defined motifs were used to generate a score matrix, according to the frequency of each nucleotide at each position. These score matrices were used to search the high-score motifs in each winner sequence. Small portions of the constant flanking regions (18 nucleotides of the 5′ region and 20 nucleotides of the 3′ region) were included during the search. The resulting alignments of sequences selected with SF2/ASF, SRp40, or SRp55 are shown in Figures 3A, 4, and 5, respectively. As a control, 30 sequences from the initial random RNA pool are shown in Figure 3B.
Figure 3.

Sequence alignment of SF2/ASF-specific ESEs (A) and sequences from the initial random pool (B). A consensus motif was identified as described in the text. The sequences are aligned on the basis of the best fit to the consensus within each sequence. Nucleotides in the boxed alignment that match the consensus position are shown white on black; mismatched nucleotides are not shaded. Underlined nucleotides are from the constant region flanking the randomized segment. The nucleotide composition of the randomized segment in the selected pool is provided in the lower left corner. S = G or C; R = A or G. Clones indicated in parentheses were selected for further characterization.
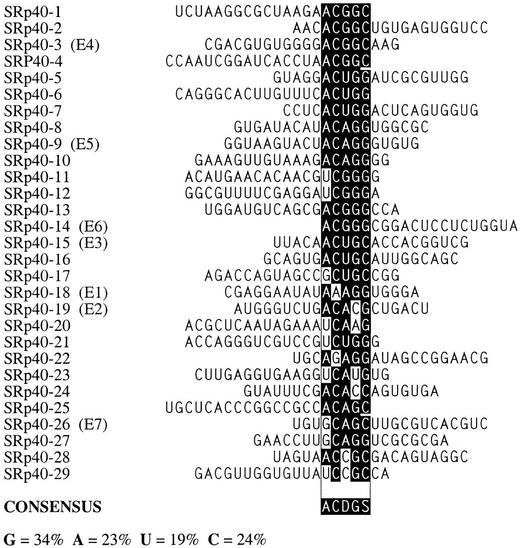
Figure 4.
Sequence alignment of SRp40-specific ESEs. D = A, G, or U. For details, see Fig. 3 legend.
Figure 5.
Sequence alignment of SRp55-specific ESEs. K = U or G; M = A or C. For details, see Fig. 3 legend.
The consensus sequences derived for each of the three SR proteins tested differed in both length and sequence. Each of the consensus sequences is relatively degenerate, and not all of the individual selected sequences match the consensus at every position. However, many of the individual sequences have more than one good match to the consensus, allowing for one or two mismatches.
The SF2/ASF winners gave the consensus sequence SRSASGA (S represents G or C, R represents purine), which only in some cases corresponds to a purine-rich motif. The content of U residues in the SF2/ASF winner pool was 16%, which represents a significant reduction from the 21% of U residues found in the initial random pool. This reduction can be accounted for by the absence of U residues from the consensus motif. The content of C residues increased by 4%. The frequency of A and G did not vary significantly upon selection (Fig. 3A,B). SF2/ASF was shown previously to recognize purine-rich sequences in SELEX procedures based on binding; the reported sequences, RGAAGAAC and AGGACRRAGC (Tacke and Manley 1995), are significantly different from, and simpler than, the consensus motif we found. Similar experiments, performed independently in our lab, revealed a different purine-rich consensus sequence, GARGAGC (A. Hanamura, I. Watakabe, and A.R. Krainer, unpubl.). In the present study, only 13 of 28 winners have uninterrupted purine-rich motifs longer than 5 nucleotides, indicating that SF2/ASF can productively recognize a far broader range of sequences. Indeed, the overall purine composition of the SF2/ASF-selected pool did not change significantly from that of the initial random pool.
The consensus for the SRp40-selected sequences is ACDGS (D represents residues other than C; S represents G or C). This consensus is also very different from that previously determined as an optimal RNA-binding site for SRp40, TGGGAGCRGTYRGCTCGY (Tacke et al. 1997). The content of G residues in the SRp40 winner RNA pool decreased from 39% in the initial random pool to 34%. The content of C residues increased by 5%. The frequency of A and U did not change significantly (Figs. 4 and 3B). The consensus motif for the SRp40 winners is relatively short but has a sufficient information content, such that, for example, it does not occur by chance in most of the RNAs sampled from the initial random pool. Winners SRp40-1, SRp40-2, SRp40-3, and SRp40-4, for example, are clearly not derived from a single founder sequence by accumulated mutations during PCR. However, they all share the sequence ACGGC, which matches the consensus, and is the only common sequence among these winners. Similar sequence relationships are seen among winners SRp40-5, SRp40-6, SRp40-7; SRp40-8, SRp40-9, SRp40-10; SRp40-11, SRp40-12; SRp40-13, SRp40-14; and SRp40-15, SRp40-16.
The SRp55 winners yielded the consensus sequence USCGKM (S represents G or C; K represents U or G; M represents A or C). The C residue content in the SRp55 winner pool increased significantly, from 19% in the starting pool to 26%. The content of G and U residues decreased by 5% and 4%, respectively (Figs. 5 and 3B). B52, which appears to be the Drosophila ortholog of human SRp55, was reported to have GRUCAACCNGGC GACNG as the optimal binding site (Shi et al. 1997). In that report, it was also suggested that a hairpin structure was required for efficient B52 binding. In contrast, we did not observe common secondary structure elements in our human SRp55 winner sequences.
The same sequence analysis programs were used to search the clones from the initial random pool, but no stable pattern was found. Each set of aligned winner sequences was used to create a score matrix that takes into account the overall nucleotide composition of the corresponding winner pool (see Materials and Methods). The scores of the SF2/ASF winner sequences ranged from 1.34 to 3.74, with a mean of 2.7; those of the SRp40 winner pool ranged from 0.33 to 1.68, with a mean of 1.2; those of the SRp55 winner pool ranged from 2.37 to 6.17, with a mean of 4.64. The sequence-scores for each SR protein correlated with the observed splicing efficiencies; however, sequence-scores for different SR proteins cannot be compared. The score matrices were then used to search the clones from all three of the winner pools and the initial random pool. The mean score of the corresponding SR protein-selected winner pool was always higher than that of the other three pools (data not shown). In particular, among the 30 sequences from the initial pool, only 3 had scores above the mean for the SF2/ASF-selected pool, 5 had scores above the mean for the SRp40-selected pool, and none had scores above the mean for the SRp55-selected pool.
The SELEX winner sequences function as bona fide ESEs
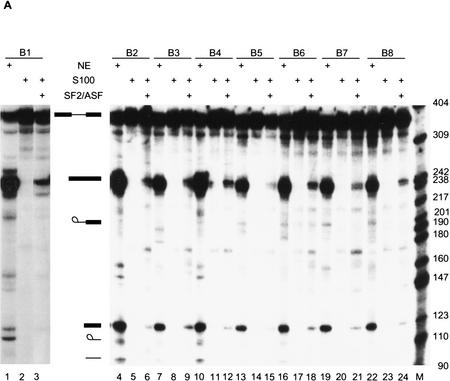
To investigate the functional importance of the winner sequences, several winners were randomly chosen from the winner pools of each SR protein. Their ability to function as enhancers was tested by splicing the corresponding pre-mRNAs in HeLa nuclear extract or in S100 extract plus specific SR proteins (Fig. 6).
Figure 6.
The selected sequences are functional SR protein-dependent ESEs. (A) In vitro splicing of pre-mRNAs containing the SF2/ASF-selected winner sequences in HeLa nuclear extract (lanes 1,4,7,10,13,16,19,22), S100 extract alone (lanes 2,5,8,11,14, 17,20,23), or S100 complemented by 20 pmoles of recombinant SF2/ASF (lanes 3,6,9,12,15,18,21,24). (B) Splicing of SRp40-selected winner sequences in nuclear extract (lanes 1,4,7,10, 13,16,19), S100 extract alone (lanes 2,5,8,11,14,17,20), or S100 extract complemented by 20 pmoles of recombinant SRp40 (lanes 3,6,9,12,15,18,21). (C) Splicing of SRp55-selected winner sequences in nuclear extract (lanes 1,5,9,13,17,21,25,29), S100 extract alone (lanes 2,6,10,14,18,22,26,30), or in S100 extract complemented by 10 pmoles SRp55 (lanes 3,7,11,15,19,23,27,31) or by 20 pmoles of SRp55 (lanes 4,8,12,16,20,24,28,32). The RNAs were analyzed by denaturing PAGE (5.5% polyacrylamide) and autoradiography. The structures and mobilities of the precursor, intermediates, and products of splicing (Watakabe et al. 1993) are shown next to each autoradiograph.
All the SF2/ASF-selected sequences promoted efficient splicing in nuclear extract (Fig. 6A, lanes 1,4,7,10, 13,16,19,22), compared to an enhancerless construct, which is essentially inactive (Watakabe et al. 1993; data not shown), indicating that the selected sequences could function as true ESEs. In each case, they promoted more efficient splicing in nuclear extract than any of several sampled round 0 sequences (data not shown). Furthermore, these ESE sequences promoted splicing in S100 extract plus recombinant SF2/ASF, albeit with variable efficiencies (Fig. 6A, lanes 3,6,9,12,15,18,21,24), but not in S100 extract only (Fig. 6A, lanes 2,5,8,11,14,17,20,23). The splicing efficiency in S100 extract plus SF2/ASF was lower than that of the nuclear extract. Winner sequences comprising either purine-rich (B1, B3, B4, B5, and B7) or nonpurine-rich motifs (B2, B6, and B8) resulted in a comparable range of splicing efficiencies.
Similar results were obtained in splicing assays with the SRp40 winners. All of the 7 winners tested spliced with somewhat variable efficiencies in nuclear extract (Fig. 6B, lanes 1,4,7,10,13,16,19) and in S100 extract plus recombinant SRp40 (Fig. 6B, lanes 3,6,9,12,15,18,21), but not in S100 extract only (Fig. 6B, lanes 2,5,8,11,14,17,20). The splicing efficiency of the SRp40 winners in nuclear extract was lower on average than that of the SF2/ASF winners (Fig. 6A,B). In S100 extract alone, several of the pre-mRNAs were extensively degraded (Fig. 6B, constructs E1, E2, E4, and E7). This observation is consistent with the notion that SR proteins are involved in the assembly of a commitment complex, such that substrates that are not productively assembled into commitment complexes and pre-spliceosomes are generally more susceptible to degradation by non-specific nucleases present in some batches of extract.
The eight tested SRp55 winners all spliced to variable extents in nuclear extract (Fig. 6C, lanes 1,5,9,13,17,21, 25,29) and in S100 extract plus SRp55 (Fig. 6C, lanes 4,8,12,16,20,24,28,32), but not in S100 extract only (Fig. 6C, lanes 2,6,10,14,18,22,26,30). Interestingly, four of the eight SRp55 winners tested spliced more efficiently in S100 extract plus SRp55 than in nuclear extract alone (Fig. 6C, C1, C4, C6, and C7), suggesting that SRp55 is the only SR protein required for effective recognition of these winner sequences. The higher splicing efficiency of C1, C4, C6, and C7 pre-mRNAs in S100 compared to nuclear extract cannot be accounted for by their increased stability, since the remaining winners, C2, C3, C5, and C7, were also greatly stabilized in S100 extract plus SRp55, but their splicing efficiencies were lower than in the nuclear extract.
We tested whether the short consensus motifs are sufficient to activate splicing. This was done by replacing sequences within template A13 by the short consensus motifs from the B1, B2, C4, or E7 winners (Figs. 3–5) and then testing the splicing activity of the corresponding pre-mRNAs. A13 was isolated from the initial random pool and had very low splicing activity in nuclear extract and no splicing activity in S100 extract complemented with any of the three SR proteins tested. Insertion of one copy of the consensus motifs was sufficient to activate splicing of the modified A13 pre-mRNA in S100 extract plus the cognate SR protein, although the splicing efficiencies were very low (data not shown). The low efficiency suggests that the sequence context surrounding the conserved motifs is also important for splicing, consistent with the observation that several of the winner sequences contain more than one match to the consensus. We also made and tested a number of clustered point mutations in several of the ESEs selected by each SR protein. We were unable to inactivate ESE function either by mutations in the best match to the consensus motif or by mutations on either side of the motif (data not shown). This unexpected observation suggests that each of the selected sequences has a high level of internal functional redundancy, which is probably necessary to allow efficient splicing.
SR protein specificity of the selected ESEs
The fact that each SR protein selected ESEs that fit a different consensus sequence suggests that SR proteins recognize the synthetic ESEs in a sequence-specific manner. On the other hand, the observation that most of the winner sequences promoted more efficient splicing in nuclear extract than in the S100 complementation reactions suggests that SR proteins may function cooperatively. We examined these possibilities by testing the effect of different SR proteins on splicing of pre-mRNAs with each type of winner. These experiments were performed in S100 extract complemented with individual SR proteins or with pairwise combinations thereof. Strikingly, the three kinds of SR protein winner sequences showed very different specificities (Table 1).
Table 1.
Summary of the activities and specificities of three types of in vitro-selected ESEs
|
|
No SR proteins
|
SF2/ASF
|
SRp40
|
SRp55
|
|---|---|---|---|---|
| B1 | − | +++ | ± | ± |
| B2 | − | ++ | ± | + |
| B3 | − | +++ | − | − |
| B4 | − | +++ | − | ± |
| B5 | − | + | ± | ± |
| B6 | − | +++ | ± | ± |
| B7 | − | ++ | ± | ± |
| B8 | − | +++ | N.D. | N.D. |
| E1 | − | − | +++ | +++ |
| E2 | − | − | ++ | +++ |
| E3 | − | ± | ++ | + |
| E4 | − | − | ++ | ++ |
| E5 | − | ± | + | ++ |
| E6 | − | − | +++ | +++ |
| E7 | − | − | ++ | ++ |
| C1 | − | ++ | ++ | +++ |
| C2 | − | − | − | + |
| C3 | − | ± | − | +++ |
| C4 | − | − | − | +++ |
| C5 | − | − | − | + |
| C6 | − | ± | − | ++ |
| C7 | − | ± | ± | +++ |
| C8 | − | N.D. | N.D. | ++ |
The SRp40-selected winner sequences failed to splice in S100 extract plus SF2/ASF (Fig. 7A, lanes 1,4,7,10,13, 16,19), even at higher concentrations of SF2/ASF (data not shown). However, they did splice in S100 extract plus SRp55 (Fig. 7B, lanes 1,4,7,10,13,16). Adding two SR proteins together did not significantly increase the splicing efficiency, although additive effects were observed with two of the winners, E4 and E6, using SRp40 and SRp55 (Fig. 7A,B).
Figure 7.
SR protein specificity of in vitro-selected ESEs. (A) SRp40-selected ESEs are inactive with SF2/ASF. Splicing was carried out in HeLa S100 extract complemented with 20 pmoles of SF2/ASF (lanes 1,4,7,10,13,16,19), 20 pmoles of SRp40 (lanes 2,5,8,11,14,17,20), or 20 pmoles of SF2/ASF plus 20 pmoles of SRp40 (lanes 3,6,9,12,15,18,21). (B) SRp40-selected ESEs can function in the presence of SRp55. Splicing was carried out in S100 extract complemented with 20 pmoles of SRp55 (lanes 1,4,7,10,13,16), 20 pmoles of SRp40 (lanes 2,5,8,11,14,17), or 20 pmoles of SRp55 plus 20 pmoles of SRp40 (lanes 3,6,9,12,15,18).
The SF2/ASF-selected winner sequences gave a different result. The B2 winner spliced poorly in the presence of S100 extract and SRp55, whereas the remaining winners, B1, B3, B4, B5, B6, and B7, failed to splice under these conditions (Table 1). SRp40 did not activate splicing of any of the SF2/ASF-selected winners tested. Moreover, SRp40 inhibited splicing of the SF2/ASF-selected winners even in the presence of SF2/ASF (data not shown).
The SRp55 winner C1 spliced in S100 extract plus any of the three SR proteins we examined. However, addition of two SR proteins did not increase its splicing efficiency. All the six other SRp55 winners tested failed to splice in S100 extract plus SF2/ASF or SRp40 (Table 1).
SR proteins bind specifically to the ESEs in nuclear extract
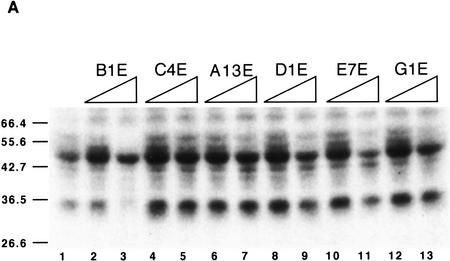
We have selected ESEs that respond specifically to individual SR proteins under splicing conditions. Because the selection was on the basis of function, it does not necessarily follow that the SR proteins bind directly to the cognate ESEs, although this is generally thought to be the case for at least some natural enhancers (see Discussion). To determine whether SR proteins directly contact the novel ESEs, we carried out UV cross-linking experiments under splicing conditions in nuclear extract. We used radiolabeled RNA fragments comprising the M2 exon with the different ESEs. Twenty femtomoles of an M2 exon RNA comprising the SF2/ASF-selected winner B1 (referred to as B1E) was incubated in the presence of excess cold exon M2 RNA competitors with different ESEs. The reaction mixtures were then irradiated with UV light on ice, digested with RNases A and T1 and analyzed by SDS-PAGE. B1E RNA cross-linked primarily to 34- and 50-kD polypeptides (Fig. 8A, lane 1). The 50-kD product is a nonspecific RNA-binding protein that cross-links to a wide variety of RNAs (data not shown), and is useful as an internal control for recovery and loading. Addition of cold RNA competitors showed that the 34-kD polypeptide bound to the B1E RNA with the greatest specificity (lanes 2,3). Other competitors, comprising an SRp55-selected ESE (lanes 4,5), an SRp40-selected ESE (lanes 10,11), ESEs selected by other SR proteins (lanes 8,9,12,13), or a sequence from the initial random pool (lanes 6,7) failed to compete specifically with B1E for binding to the 34-kD polypeptide (compare the reduction in intensity of the 34-kD band relative to that of the 50-kD band). A minor cross-linked product of 60 kD also decreased in intensity in the presence of the B1E competitor (lane 3), but not in the presence of the other competitors. This may represent an additional protein that interacts specifically with this ESE, or multimerization of the 34-kD product through additional RNA–protein or protein–protein cross-linking.
Figure 8.
Specific binding of SF2/ASF to an SF2/ASF-selected ESE. (A) UV cross-linking competition binding assay. Radiolabeled exon M2 RNA (20 fmoles) comprising the B1 winner sequence (B1E) was incubated under splicing conditions in HeLa nuclear extract. Subsequent UV cross-linking and RNase digestion resulted in label transfer predominantly to two proteins of 34 and 50 kD. The former, which binds specifically, is presumed to be SF2/ASF (see below). Cold competitor RNAs containing either the B1 winner insert, an SRp55-selected insert (C4E), an SRp40-selected insert (E7E), or other control sequence inserts, were present in excess, as indicated above the autoradiograph. (Lane 1) No competitor; in the remaining lanes, the indicated competitors were present in 5-fold excess (even lanes) or 50-fold excess (odd lanes) over the labeled B1E RNA. (B) Immunoprecipitation of SF2/ASF UV cross-linked to the B1E RNA. UV cross-linking was carried out as in A, lane 1. A 5% equivalent of the input was loaded directly (lane 1). Parallel reactions were incubated with a control antibody (lane 3), or with anti-SF2/ASF monoclonal antibody (lane 2), and the immunoprecipitates were recovered in SDS gel loading buffer. In A and B, the samples were analyzed by SDS-PAGE and autoradiography.
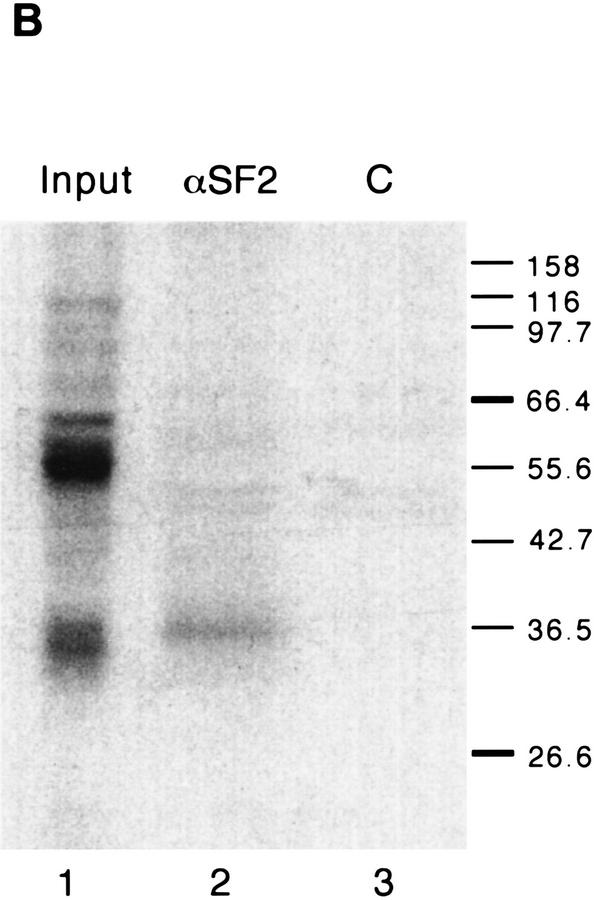
To confirm that the 34-kD polypeptide is SF2/ASF, we carried out immunoprecipitations after UV cross-linking and RNase digestion (Fig. 8B) (Sun et al. 1993). As expected, the 34-kD polypeptide was the major crosslinked product immunoprecipitated by a monoclonal antibody specific for SF2/ASF (lane 2) but not by a control monoclonal antibody (lane 3).
Similar UV cross-linking experiments were attempted with SRp40- and SRp55-selected ESEs. Cross-linked proteins of ∼40 and 55 kD were detected by use of radiolabeled exon M2 RNAs corresponding to the SRp40 winner E7 (E7E), and the 55-kD protein also cross-linked to the SRp55 winner C4 (C4E), although the background was high (data not shown). Neither of these RNAs cross-linked to proteins with the mobility of SF2/ASF. Cross-linking to the 55-kD protein was competed by an excess of cold C4E RNA, but not by B1E or E7E. Immunoprecipitation with a polyclonal antiserum that recognizes both SRp40 and SRp55 (Du et al. 1997) selectively precipitated cross-linked proteins of the expected size (data not shown). These results suggest that, similar to SF2/ASF, SRp55 and SRp40 also interact directly with their cognate in vitro-selected ESEs.
Sequences that fit the consensus for in vitro-selected ESEs are present in natural exons and known ESEs
Sequences identified by SELEX procedures do not necessarily correspond to functional elements that have evolved in nature (Irvine et al. 1991). To evaluate the biological significance of the novel ESE consensus sequences we identified, we analyzed their distribution in known sequences of natural genes. We reasoned that if the short consensus motifs derived from the in vitro-selected ESEs are akin to natural ESEs, they should be present with higher probability in regions corresponding to known ESEs than elsewhere in the exons or in intron sequences. The score matrices derived for each of the three SR proteins tested were used to search genes or exons with previously characterized ESEs. The resulting scores were then plotted against the position along the exons or genes (Fig. 9).
Figure 9.
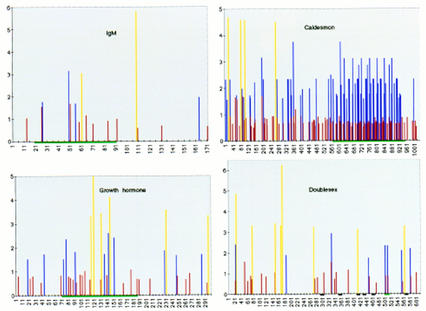
Distribution of in vitro-selected ESE consensus sequences within exons comprising natural ESEs. Score matrices were built for each class of in vitro-selected ESE, as described in Materials and Methods. The indicated natural exon sequences were searched with each score matrix, and the resulting scores (y-axis) were plotted against the nucleotide positions for each exon (x axis). Note that the x-axis scales are different in each case, because of the different exon sizes. Graphs are shown for mouse IgM exon M2, bovine growth hormone 3′ exon, Drosophila dsx female-specific exon, and chicken caldesmon exon 5. High score motif matches are shown by blue (SF2/ASF), red (SRp40), and yellow (SRp55) vertical bars. For each SR protein, only the sequence matches with a score greater than that of the lowest scoring winner sequence in Figs. 3–5 are shown. Note that there is no relation between the height of bars of different colors. The green horizontal bars under the x axis indicate previously mapped ESEs or the dsx PRE. The black horizontal bars denote the dsx repeat elements (dsxREs).
The natural sequence of the mouse IgM exon M2 was analyzed first, since our ESEs were selected in the context of this exon, after deletion of its natural ESE. Remarkably, a high density of motifs with high-score matches to the SF2/ASF and SRp40 consensus was found within the 73-nucleotide natural ESE mapped previously (Watakabe et al. 1993). In contrast, few matching sequences were found in the flanking regions of the exon, and most of these had lower scores, correlating with the lack of splicing upon deletion of the natural ESE. The distribution of motifs with high-score matches to the SRp55 ESE consensus did not correlate with the location of the natural ESE. The SR protein specificity of the natural M2 ESE was not known from previous work, but we have determined that IgM minigene transcripts comprising the natural ESE can function in S100 extract complemented with SF2/ASF, SRp40 or SRp55 (data not shown). The high-score SF2/ASF and SRp40 motifs are present in clusters, suggesting that multiple copies of these motifs are particularly effective as ESEs, or provide an optimal context. Indeed, multimerization of short repeats often results in increased ESE activity, both in natural and synthetic enhancer elements (Tian and Maniatis 1993; Tanaka et al. 1994; Tacke and Manley 1995).
We next analyzed the sequence of the last exon of the bovine growth hormone (bGH) gene, which contains a natural ESE previously mapped to a 115-nucleotide fragment, that is required for splicing of the preceding intron (Sun et al. 1993). The highest density of sequences matching the SF2/ASF, SRp40, and SRp55 consensus ESEs was found within the 115-nucleotide fragment corresponding to the natural ESE, compared to the rest of the 302-nucleotide exon. The highest scores for each of the three SR protein motifs were all found within the fragment with natural enhancer activity. Although the last intron of the bGH pre-mRNA does not splice in S100 extract in the presence of SR proteins, as it apparently requires additional factors, the ESE in the last exon was previously shown to bind SF2/ASF specifically, and this SR protein also stimulated bGH splicing in nuclear extract (Sun et al. 1993).
The caldesmon pre-mRNA is alternatively spliced in a tissue-specific manner (Humphrey et al. 1995). An alternative 5′ splice site within the large exon 5 is used in nonmuscle cells, which also exclude exon 6. In smooth muscle, the entire exon 5 is included and spliced to exon 6. A 32-nucleotide repeat present in multiple copies within the 3′ portion of exon 5 functions as an ESE to enhance usage of the upstream non-muscle-specific 5′ splice site (Humphrey et al. 1995). Our sequence analysis showed that SF2/ASF and SRp40 ESE consensus sequences are highly enriched within the 3′ portion of exon 5, whereas SRp55 consensus sequences are found much more frequently upstream of the non-muscle-specific 5′ splice site.
Female-specific alternative splicing of the Drosophila dsx pre-mRNA involves six 13-nucleotide repeat elements (dsxRE) and a purine-rich element (PRE) (Tian and Maniatis 1993; Lynch and Maniatis 1995). These cis-acting elements are essential for splicing of a dsx pre-mRNA in HeLa cell nuclear extract. UV cross-linking analysis showed that the fourth and fifth dsxREs bind specifically to the human SR protein 9G8, whereas the PRE binds preferentially to SF2/ASF and probably other SR proteins of similar size in HeLa nuclear extracts (Lynch and Maniatis 1996). Consistent with these results, our sequence analysis did not reveal any high-score motifs matching the SF2/ASF, SRp40, and SRp55 ESE consensus sequences within the fourth and fifth dsxREs, whereas high-score matches to the SF2/ASF ESE were found within the PRE.
Finally, we also analyzed the sequences of characterized ESEs present in exon 5 of chicken cardiac troponin T (Xu et al. 1993), in exon 3 of the Tat gene of equine infectious anemia virus (Gontarek and Derse 1996), in late pre-mRNAs of bovine papilloma virus type 1 (Zheng et al. 1996), in exon ED-A of human fibronectin (Lavigueur et al. 1993; Caputi et al. 1994), and in the exon downstream of the tat–rev intron of HIV-1 (Amendt et al. 1995; Staffa and Cochrane 1995). In all cases, the sequence analysis was consistent with the available data on these natural ESEs and the binding of SR proteins, when known (data not shown).
Next, we used the same score matrices to analyze the distribution of high-score motifs in human exons versus introns. A total of 570 intron-containing genes, corresponding to 2634 exons (431 kb) and 2079 introns (1300 kb), were extracted from the ALLSEQ data (Burset and Guigo 1996) and analyzed. We searched all sequences with a score equal to or greater than the mean score of the selected winner pool for each SR protein. Remarkably, high-score motifs matching each of the three SR protein ESE consensus sequences were found more frequently in exons than in introns. For SF2/ASF, the density of high-score motifs was 4.3 per kilobase of exon and 2.9 per kilobase of intron; for SRp40, the corresponding numbers were 7.9 per kilobase of exon and 6.8 per kilobase of intron; and for SRp55, they were 5.5 per kilobase of exon and 4.9 per kilobase of intron. The higher density of high-score motifs in exons than in introns is statistically significant because of the large database size, and the P-values for these pairwise comparisons were all <10−10.
Discussion
We have developed a method to identify ESE elements that can function specifically with individual SR proteins. This goal was accomplished by use of SR protein-deficient HeLa extracts complemented with individual SR proteins, and a pool of pre-mRNAs derived from mouse IgM, whose natural ESE in exon M2 was replaced by a 20-nucleotide segment of random sequence. Sequence analysis revealed that the motifs identified by the selection for function with SF2/ASF, SRp40, or SRp55 tend to be clustered in regions corresponding to known, natural ESEs, compared to other exon regions.
The initial randomized pool of IgM-derived substrates consisted of 20 fmoles of pre-mRNA (∼1.2 × 1010 molecules), which is large enough to include all possible 16-mers (∼4.3 × 109). The longest motif we identified was the 7-nucleotide consensus selected by SF2/ASF, indicating that the initial random pool had sufficient complexity. In parallel SELEX experiments, we also employed a different RNA pool, in which only 14 positions within the IgM M2 exon were randomized. Functional ESEs were also selected out of that library (data not shown), suggesting that the 20-mer library can potentially encode most, if not all, natural ESEs, and that the library size we used was adequate. The functional SELEX procedure was performed for only three rounds. This was deemed sufficient, as all the winner sequences tested proved to be functional. Additional rounds of selection would be expected to result in loss of consensus sequence information, as only the most efficiently spliced RNAs would be recovered.
Our experiments confirm and extend two previous studies that used functional selection from random pools to identify novel ESEs. Tian and Kole (1995) randomized a 20-nucleotide region within the context of a duplicated exon in a model β-globin pre-mRNA. They selected sequences that promoted inclusion of the middle duplicated exon in HeLa nuclear extract. The resulting ESEs after five or seven selection cycles included both purine-rich and nonpurine-rich motifs (Tian and Kole 1995). A related approach was used by Coulter et al. (1997) to identify ESEs that promote inclusion of the internal alternative exon 5 of chicken cardiac troponin T. In this case, the natural ESE was replaced by a 13-nucleotide randomized segment, and the selection for splicing was carried out by three rounds of transient transfection into QT35 quail cells. The resulting ESEs included both purine-rich elements and a novel class of AC-rich elements (ACEs; Coulter et al. 1997). These pioneering studies could not readily identify the factors responsible for ESE recognition—although SR proteins were obvious candidates—because they relied on crude nuclear extracts or cultured cells. In addition, the novel ESEs did not fall into obvious consensus sequences, most likely because they represent a complex collection of elements recognized by several distinct factors. We improved this general approach by performing the selections in extracts dependent upon addition of individual SR proteins, which allowed us to identify functional ESEs recognized and activated by each SR protein, and therefore to derive a corresponding consensus sequence. Our selections were carried out in a different context from those in the above two studies, which focused on inclusion of an optional exon; we used the last exon of the IgM pre-mRNA, which requires an enhancer for splicing of the last intron (Watakabe et al. 1993).
Previous work also attempted to address the specificity of SR proteins in ESE recognition by use of conventional SELEX procedures based on selection for high-affinity binding (Tacke and Manley 1995; Shi et al. 1997; Tacke et al. 1997). These studies gave very different results from our current results with some of the same SR proteins but with selection cycles based on function. First, the consensus sequences obtained by these two approaches are very different. The SF2/ASF motifs defined by binding (Tacke and Manley 1995; A. Hanamura, I. Watakabe, and A.R. Krainer, unpubl.), which are purine-rich, appear to be a subgroup of those defined by function, although they yield relatively low scores when analyzed with our SF2/ASF score matrix. It should be noted that a purine-rich composition is not sufficient for function, but rather, specific sequences are required (Tanaka et al. 1994; Ramchatesingh et al. 1995). Second, many of the winner sequences obtained by binding protocols were not functional as ESEs. In contrast, among the winner sequences obtained by our functional selection protocol, many were tested, and all of these were functional ESEs. Third, the complexity of the winner sequences identified by binding SELEX is much lower than that of the ESEs identified by functional SELEX. As a result, the consensus sequences obtained from the binding selection are less degenerate than those we obtained through functional selection. This may be attributable in part to the use of more selection/amplification cycles in some of the binding SELEX experiments. In the natural situation, exon sequences are obviously very diverse. Degenerate sequence specificity is probably essential for a limited number of SR proteins to be able to recognize a very large number of ESE-containing exons in different genes.
The different results obtained by binding selection and functional selection protocols shed light on the mechanisms of ESE function. The binding selection is based on the affinity of RNA–protein interactions, and the iterative protocol is designed to yield the binding sites with the highest affinity for the protein of interest. However, it appears that the best binding sites are not necessarily the best functional sites, and, in some cases, a high affinity may preclude function. In addition, optimal interactions between an SR protein and its cognate ESEs may require other splicing components, as opposed to just the purified protein. The binding protocol is carried out with the purified protein, whereas the functional selection protocol is carried out in the presence of all components required for splicing. There are also technical reasons why iterative binding protocols may not yield optimal functional sites. The binding affinity and/or the specificity of the binding may be significantly affected by the idiosyncrasies of the binding assay employed (Irvine et al. 1991). Indeed, several of the SF2/ASF and SRp40 winners identified by iterative binding failed to bind to the cognate SR protein when analyzed by a different binding assay (Tacke and Manley 1995; Tacke et al. 1997). Another contributing factor to the discrepancy between the results obtained in binding and functional assays may be that in at least some applications of the former, truncated proteins lacking the RS domain were used (Tacke and Manley 1995). Although the precise functions of the RS domains are not completely understood, they appear to be important for protein–protein and/or RNA–protein interactions (Wu and Maniatis 1993; Tacke et al. 1997; Xiao and Manley 1997). Thus, deletion of the RS domain may affect the binding specificity, as may an incorrect or incomplete phosphorylation state of the domain. In addition, the use of oligo-histidine or other tags in some studies may also affect the binding specificity. In the present study, we used unagged proteins expressed in Escherichia coli. Although these proteins are not phosphorylated when isolated, they are very rapidly phosphorylated upon addition to nuclear or S100 extracts (Hanamura et al. 1998). Finally, in the case of the different consensus sequences obtained previously for Drosophila B52 (Shi et al. 1997) and in the present study for human SRp55, the binding specificity may have diverged considerably between arthropods and vertebrates. For example, the enhancer complex formed on the PRE of the Drosophila dsx pre-mRNA binds SRp55/B52 in Drosophila Kc cell extracts, but does not appear to bind human SRp55 in HeLa cell extracts (Lynch and Maniatis 1996).
The specific recognition of ESEs by SR proteins is well documented. Some examples include recognition of purine-rich ESEs in bovine growth hormone pre-mRNA by SF2/ASF (Sun et al. 1993), in cardiac troponin T by SRp40 and SRp55 (Ramchatesing et al. 1995), and in the dsx pre-mRNA by one of the Drosophila SRp30 proteins (Lynch and Maniatis 1996). Our data address the molecular basis of the redundancy and specificity of SR proteins. The different consensus sequences of the three types of in vitro-selected ESEs, and their different responses to individual SR proteins provide an indication of the specificity of SR proteins in ESE recognition and function. The consensus sequence of SF2/ASF-selected ESEs, SRSASGA, matches the sequence of most purine-rich ESEs characterized to date. It is worthwhile to note that this sequence is devoid of U residues. In the HPRT and IgM genes, the presence of C residues within the purine-rich ESEs was compatible with enhancer function, whereas changing the C residues to U residues abolished their enhancer activity (Tanaka et al. 1994). The SRp55-selected winners also had a reduced U content, and we suspect that a low U composition contributes to the information content that defines ESEs recognized by these SR proteins.
The SRp40-selected ESEs share a relatively short consensus sequence, ACDGS. They could be activated by SRp55, but not by SF2/ASF. The fact that SRp55 could activate SRp40-selected ESEs suggests that these two SR proteins, which are closely related in domain structure, unmodified molecular mass (31.2 kD for SRp40; 39.6 kD for SRp55), and sequence (65% identity; Screaton et al. 1995), also have some functional overlap. It is unlikely that the sequences selected by SRp40 fortuitously comprise a distinct SRp55 recognition site, but not an SF2/ASF site, as all of the seven independent SRp40 ESEs tested had similar properties. When the SF2/ASF score matrix was used to search the seven examined SRp40-selected ESEs, most of them had a score lower than the minimum score of the SF2/ASF-selected ESEs, which could explain why SRp40-selected ESEs were not activated by SF2/ASF. We do not know why E4, which had a score higher than the average score of SF2/ASF-selected ESEs, was not activated by SF2/ASF. However, it is likely that the sequence context, for example, in the form of negative elements or silencers, somehow prevents activation of this motif by SF2/ASF. A related observation is the fact that SRp40 inhibited splicing of some SF2/ASF-selected ESEs even in the presence of SF2/ASF. This may be attributable to formation of inhibitory complexes with SRp40, such that SR proteins may also participate in exonic silencer function, depending on the sequence context. An interesting implication is that the variable expression levels of these antagonistic SR proteins may determine the cell type-specific function of certain ESEs.
We did not observe any cooperative effects among the SR proteins tested. However, the fact that most of the ESEs we identified gave higher splicing efficiencies in nuclear extract than in S100 extract complemented with SR proteins suggests that other SR proteins and/or additional splicing factors may be required for optimal ESE recognition or function. With other substrates, such as β-globin or certain natural ESE-dependent pre-mRNAs, comparable splicing efficiencies can be obtained in the two systems. A few natural or synthetic ESE-dependent pre-mRNAs can only splice in the nuclear extract, apparently because they require one or more unknown factors, distinct from SR proteins, that are also absent in the S100 extract (Sun et al. 1993; Tacke and Manley 1995; Tacke et al. 1997). If this is a property of the ESE, rather than its context, this class of ESEs may not be well represented in the consensus sequences we derived. However, we did find high-score motif matches within one such natural ESE, that of the bGH last exon (Fig. 9). The ESEs we obtained were selected to function in S100 extract plus an SR protein, and hence, it is not surprising that at least basal function could be observed in this complementation system. However, maximal activity appears to require one or more additional factors that may be limiting in the S100 extract.
Many natural ESEs have been found in the last several years. Most of these well-defined ESEs are purine rich, although this nucleotide composition may reflect an experimental bias. First, many of the biochemical studies were carried out in HeLa cell nuclear extract, in which SF2/ASF, which prefers purine-rich sequences, may be the most abundant SR protein. Second, purine-rich motifs may be easier to find by visual inspection which, together with the precedent of known purine-rich ESEs, makes them more likely to be studied further. We have identified three new degenerate motifs, which are not necessarily purine rich. Significantly, the SF2/ASF and SRp40 motifs we defined occur more frequently (and with higher scores) within exon segments corresponding to known ESEs than elsewhere in the exons. All of the motifs also occur more often in exons than in introns and may thus contribute to defining exon–intron boundaries. These consensus sequences may be useful for the prediction of natural ESEs in uncharacterized exons. Our data also suggest that target sites for multiple SR proteins are clustered within natural ESEs. This may explain why large deletions are often required to inactivate natural ESEs. The SRp55 ESE consensus motif did not always correlate with the location of natural ESEs. Interestingly, in the caldesmon gene, SF2/ASF and SRp40 sites are enriched in the 3′ portion of exon 5 that is included in smooth muscle cells, whereas SRp55 sites occur more frequently in the 5′ portion of exon 5 that is included in all cell types. Inclusion of the constitutively spliced upstream segment of exon 5 may also require a functional ESE, which we would predict, on the basis of the present data, to be SRp55-dependent. The differential recognition of the alternative 5′ splice sites associated with the caldesmon exon 5 by different SR proteins may be responsible for the proper developmental and tissue-specific expression of caldesmon by alternative splicing.
Our results with just three of the nine known SR proteins indicate that a highly diverse set of sequences can function as SR protein-specific ESEs. In fact, of the more than 1010 20-mers we tested in the context of the IgM exon M2, a significant proportion contained functional enhancers. Thus, of the RNA molecules from the unselected library that remained at the completion of a splicing reaction in nuclear extract, ∼20% were spliced (Fig. 2). Likewise, in the present study and in two previous studies, about 10%–20% of random 20-mers or 13-mers comprised sequences that could enhance splicing significantly above background (Tian and Kole 1995; Coulter et al. 1997; and data not shown). This level of degeneracy in sequence-specific RNA recognition may be essential to allow evolution of effective ESEs interspersed with open reading frames that are constrained by the structure and function of the encoded proteins. These findings imply that many exons are likely to have multiple ESEs that are to some extent redundant, but which may also function additively or allow fine tuning of tissue-specific or developmentally regulated expression. On the other hand, we suspect that splicing silencer elements, which are less well understood, will prove to have similar complexity. As a result, it is likely that many natural sequences that match the simple motifs identified in this study will fail to function as ESEs unless they are placed in an appropriate context.
Materials and methods
Preparation of HeLa cell extracts and recombinant SR proteins
Nuclear and cytosolic S100 extracts were prepared from fresh 12-liter suspension cultures of HeLa cells, as described (Mayeda and Krainer 1998a).
Expression and purification of the authentic form of the recombinant SR proteins SF2/ASF, SRp40 and SRp55, by use of the expression vector pET9c (Novagen), were carried out as described previously (Krainer et al. 1991; Screaton et al. 1995). The integrity and purity of these recombinant SR proteins were checked by SDS-PAGE and their specific activities were determined by in vitro splicing of β-globin pre-mRNA in S100 extract (data not shown).
Randomization and selection
The SELEX procedure is outlined in Figure 1A. The sequence of the wild-type IgM exon M2 is shown in Figure 1B. The plasmids μM1-2 and μMΔ, which bear a mouse IgM minigene with or without the natural enhancer, respectively (Watakabe et al. 1993), were a generous gift from Prof. Y. Shimura. The randomized substrate pool was constructed by overlap-extension PCR (Horton et al. 1989; Tian and Kole 1995). Two sets of PCRs were performed with μMΔ as template. The first PCR was carried out with primers R and A. The second PCR used primers P and B. The products from the two reactions were then combined and further amplified with primers P and A. The resulting PCR product was then used for in vitro transcription with SP6 RNA polymerase to generate a radiolabeled pre-mRNA substrate pool. The pool of spliced mRNAs generated by in vitro splicing was excised from a urea–polyacrylamide gel, eluted in 0.5 m ammonium acetate plus 0.1% SDS, reverse transcribed by use of Superscript II RT (GIBCO-BRL), and amplified by PCR with primers P and A. The amplified product was further amplified with primers S and A. The PCR product was purified on a 2% agarose gel and reassembled into the pre-mRNA template by overlap-extension PCR for the next round of selection. The reverse transcription and PCR reactions were performed as suggested by the vendors (GIBCO-BRL and Stratagene, respectively). All of the PCRs were carried out with the high-fidelity Pfu DNA polymerase. The primers were purchased from Operon and were used at a concentration of 1 μm. After three rounds of selection, the amplified spliced products were subcloned into the vector PCR-Blunt (Stratagene) and sequenced by use of a Dye Terminator Cycle Sequencing kit (Perkin-Elmer) and an automated ABI 377 sequencer. The second and third rounds of SELEX were performed in nuclear extract depleted of total SR proteins by Mg2+ precipitation (Blencowe et al. 1994). The sequences of the primers were as follows: Primer P, 5′-ATTTAGGTGACACTATAGAATAC-3′; primer A, 5′-GCAGGTCGACTCTAGAAAGAAG-3′; primer S, 5′-GTGAAATGACTCTCAGCAT-3′; primer B, 5′-ATGCTGAGAGTCATTTCAC-3′; primer R, 5′-GTGAAATGACTCTCAGCAT(N)20CTAGTAAACTTATTCTTACGTC-3′.
Identification of consensus motifs among the selected sequences
Functional selected sequences for each SR protein were aligned by use of Gibbs sampler (Lawrence et al. 1993), with the assumption that there is a common sequence motif of length L present at least once in all of the sequences. Because Gibbs sampler is a stochastic algorithm, for each fixed L, at least 10 different runs (with different random seeds) were carried out for times sufficient to achieve convergence. A conservative value for L was determined empirically by observing a sharp drop in information per parameter (Lawrence et al. 1993) as L was increased. To exclude the possibility that the predicted consensus motif arose by chance, the information per parameter was also compared to alignments of random sequences obtained by shuffling the nucleotides within each sequence. The final alignment was manually adjusted in a few cases, when better matches to the consensus could be obtained by including a few flanking nucleotides in the alignment.
Construction of a scoring matrix
First, a frequency matrix fi (a) was calculated from the alignment (i is the position of nucleotide a). Given a background frequency for the set of sequences, p(a), the scoring matrix is defined by the following formula:
 |
where i = (1, 2, ..., L), a = (A, C, G, U), and ε = 0.5 is the Bayesian prior parameter (Lawrence et al. 1993).
A motif score is equal to the sum of the scores at each position. Motifs may be ranked by their scores. The top three scores in each sequence using all three different scoring matrices (SF2/ASF, SRp40, and SRp55) were calculated and tabulated (data not shown).
The sequence scores were consistent semiquantitatively with the gel intensity data when the sequence-scores for a given SR protein were defined as follows: (1) The maximum score for each selected sequence was calculated using the scoring matrix, and the threshold was defined as the minimum of these scores; and (2) The sequence score was defined as the number of nonoverlapping motifs that have a score greater than or equal to the threshold. This integer score correlated well with the corresponding gel intensity.
It should be noted that a motif scoring matrix may depend on the pre-mRNA substrate and on the experimental parameters, such as the concentration of SR protein.
In vitro splicing
In vitro splicing was performed as described previously (Mayeda and Krainer 1998b). Briefly, 20 fmoles of 32P-labeled, 7CH3GpppG-capped SP6 or T7 transcripts generated from PCR products were incubated in 25-μl splicing reactions. The reactions contained 4 μl of HeLa nuclear extract or 7 μl of S100 extract in buffer D. The MgCl2 concentration was 4.8 mm. Twenty picomoles of the appropriate SR protein was used in S100 complementation assays. After incubation at 30°C for 4 hr, the RNA was extracted and analyzed on 5.5% or 12% polyacrylamide denaturing gels, followed by autoradiography.
UV cross-linking
UV cross-linking experiments were carried out under splicing conditions with or without a 5- to 50-fold molar excess of unlabeled RNA competitor. Polyvinyl alcohol was omitted from the cross-linking reactions. After a 30-min incubation at 30°C the reactions were exposed to 254 nm UV light by use of a Spectronics XL-1000 UV cross-linker at a setting of 1.8 J/cm2 on ice. RNase A (10 μg) and RNase T1 (100 units) were added and the reactions were incubated for 15 min at 37°C. The cross-linked proteins were analyzed by SDS-PAGE on a 12% gel, followed by autoradiography.
Immunoprecipitations
Immunoprecipitations were performed as described (Sun et al. 1993). The anti-SF2/ASF monoclonal antibody recognizes the amino terminus of SF2/ASF and does not cross-react with other human SR proteins (Hanamura et al. 1998). Polyclonal antiserum against SRp40 (anti-HRS/SRp40) was a generous gift from Drs. K. Du and R. Taub (Du et al. 1997). The cross-linking reactions were precleared after incubation with control antibodies and 50 μl of protein A–agarose (1:1 suspension) in 500 μl of IP buffer (50 mm Tris-HCl at pH 8.0, 150 mm NaCl, 0.05% NP-40) for 2 hr at 4°C. An unrelated monoclonal antibody of the same isotype was used for the SF2/ASF preclearing step and rabbit preimmune serum was used for the SRp40 preclearing step. After spinning in a microcentrifuge for 30 min at 4°C, the supernatants were transferred to tubes containing the appropriate antibody immobilized on protein A–agarose and rocked overnight at 4°C. The bound material was recovered by centrifugation, washed twice with 1 ml of IP buffer, eluted in 30 μl of sample buffer [62.5 mm Tris-HCl at pH 6.8, 2% (wt/vol) SDS, 10% (vol/vol) glycerol, 5% (vol/vol) 2-mercaptoethanol], and analyzed by SDS-PAGE and autoradiography.
Acknowledgments
We thank Dr. Y. Shimura for the gift of plasmids μM1-2 and μMΔ, Dr. A. Mayeda for recombinant SR proteins, and other members of our laboratory for sharing reagents and valuable ideas. We are grateful to Drs. A. Mayeda, M. Murray, L. Cartegni, and D. Horowitz for comments on the manuscript. This work was supported by the National Institutes of Health (NIH) grants GM42699 to A.R.K. and HG00010 to M.Z., and by fellowships from the Cold Spring Harbor Laboratory Association and from the U.S. Army Medical Research and Materiel Command under DAMD 17-96-1-6172 to H.-X.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL krainer@cshl.org; FAX (516) 367-8453.
References
- Amendt BA, Si Z-H, Stoltzfus CM. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: Evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Nickerson JA, Issner R, Pennman S, Sharp PA. Association of nuclear antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burset M, Guigo R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Krainer AR. Mammalian pre-mRNA splicing factors. In: Krainer AR, editor. Eukaryotic mRNA processing. Oxford, UK: IRL Press; 1997. pp. 174–212. [Google Scholar]
- Cáceres JF, Stamm S, Helfman DM, Krainer AR. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- Cáceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes & Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli R, Melo CA, Baralle FE. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Popielarz M, Fuchs JP, Gattoni R, Stévenin J. Characterization and cloning of the human splicing factor 9G8: A novel 35 kD factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler SD, Mayeda A, Yeakley JM, Krainer AR, Fu X-D. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci. 1997;94:3596–3601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter L, Landree M, Cooper T. Identification of a new class of exonic splicing enhancers by in vivo selection. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du K, Peng Y, Greenbaum LE, Haber BA, Taub R. HRS/SRp40-mediated inclusion of the fibronectin EIIIB exon, a possible cause of increased EIIIB expression in proliferating liver. Mol Cell Biol. 1997;17:4096–4104. doi: 10.1128/mcb.17.7.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon IC, Ireland DC, Smith RA, Mayeda A, Krainer AR. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. The 35-kD mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc Natl Acad Sci. 1992;89:1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Mayeda A, Maniatis T, Krainer AR. General splicing factors SF2 and SC35 have equivalent activities in vitro and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci. 1992;89:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Ge H, Zuo P, Manley JL. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Gontarek RR, Derse D. Interactions among SR proteins, an exonic splicing enhancer, and a lentivirus Rev protein regulate alternative splicing. Mol Cell Biol. 1996;16:2325–2331. doi: 10.1128/mcb.16.5.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamura A, Cáceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- Heinrichs V, Baker BS. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Humphrey MB, Bryan J, Cooper TA, Berget SM. A 32-nucleotide exon-splicing enhancer regulates usage of competing 5′ splice sites in a differential internal exon. Mol Cell Biol. 1995;15:3979–3988. doi: 10.1128/mcb.15.8.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine D, Tuerk C, Gold L. SELEXION. Systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. J Mol Biol. 1991;222:739–761. doi: 10.1016/0022-2836(91)90509-5. [DOI] [PubMed] [Google Scholar]
- Jumaa H, Guenet JL, Nielsen PJ. Regulated expression and RNA processing of transcripts from the SRp20 splicing factor gene during the cell cycle. Mol Cell Biol. 1997;17:3116–3124. doi: 10.1128/mcb.17.6.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990a;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes & Dev. 1990b;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Lavigueur A, LaBranche H, Kornblihlt AR, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes & Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, Wootto JC. Detecting subtle sequence signals: A Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- Lynch KW, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes & Dev. 1995;9:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- ————— Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes & Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Preparation of Hela cell nuclear and cytosolic S100 extracts for in vitro splicing. In: Haynes SR, editor. Methods in molecular biology, RNA–protein interaction protocols. Totowa, NJ: Humana Press; 1998a. . (In press.) [DOI] [PubMed] [Google Scholar]
- ————— . Mammalian in vitro splicing assays. In: Haynes SR, editor. Methods in molecular biology, RNA-protein interaction protocols. Totowa, NJ: Humana Press; 1998b. . (In press.) [Google Scholar]
- Peng X, Mount SM. Genetic enhancement of RNA-processing defects by a dominant mutation in B52, the Drosophila gene for an SR protein splicing factor. Mol Cell Biol. 1995;15:6273–6282. doi: 10.1128/mcb.15.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchatesingh J, Zahler AM, Neugebauer KM, Roth MB, Cooper TA. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring HZ, Lis JT. The SR protein B52/SRp55 is essential for Drosophila development. Mol Cell Biol. 1994;14:7499–7506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton GR, Cáceres JF, Mayeda A, Bell MV, Plebanski M, Jackson DG, Bell JI, Krainer AR. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Hoffman BE, Lis JT. A specific RNA hairpin loop structure binds the recognition motifs of the Drosophila SR protein B52. Mol Cell Biol. 1997;17:2649–2657. doi: 10.1128/mcb.17.5.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Mayeda A, Hampson RK, Krainer AR, Rottman FM. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes & Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Kole R. Selection of novel exon recognition elements from a pool of random sequences. Mol Cell Biol. 1995;15:6291–6298. doi: 10.1128/mcb.15.11.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- ————— A splicing enhancer exhibits both constitutive and regulated activities. Genes & Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Wang J, Manley JL. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- Wang J, Takagaki Y, Manley JL. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes & Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes & Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes & Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Xu R, Teng J, Cooper TA. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Roth MB. Distinct functions of SR proteins in recruitment of VI small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: A conserved family of pre-mRNA splicing factors. Genes & Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Neugebauer KM, Lane WS, Roth MB. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993a;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Neugebauer KM, Stolk JA, Roth MB. Human SR proteins and isolation of a cDNA encoding SRp75. Mol Cell Biol. 1993b;13:4023–4028. doi: 10.1128/mcb.13.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WJ, Wu JY. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z-M, He P, Baker CC. Selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site is regulated through an exonic splicing enhancer and its juxtaposed exonic splicing suppressor. J Virol. 1996;70:4691–4699. doi: 10.1128/jvi.70.7.4691-4699.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]