Abstract
We explore a connection between the singular value decomposition (SVD) and functional principal component analysis (FPCA) models in high-dimensional brain imaging applications. We formally link right singular vectors to principal scores of FPCA. This, combined with the fact that left singular vectors estimate principal components, allows us to deploy the numerical efficiency of SVD to fully estimate the components of FPCA, even for extremely high-dimensional functional objects, such as brain images. As an example, a FPCA model is fit to high-resolution morphometric (RAVENS) images. The main directions of morphometric variation in brain volumes are identified and discussed.
Keywords: Voxel-based morphometry (VBM), MRI, FPCA, SVD, Brain imaging data
Introduction
Epidemiological studies of neuroimaging data are becoming increasingly common. Common features of these studies generally include large sample sizes and subtle effects under study. High-resolution three-dimensional brain images exponentially increase the volume of data, making many standard inferential tools computationally infeasible. This and other high dimensional data sets have motivated an intensive effort in the statistical community on methodological research for functional data analysis (FDA, Ramsay and Silverman, 2005). One group of FDA methods uses wavelets and splines to study and model curves as well as more general functional objects such as brain images (Guo, 2002; Mohamed and Davatzikos, 2004; Morris and Carroll, 2006; Reiss et al., 2005; Reiss and Ogden, 2008, 2010). Another group employs principal components as a basis for modeling functions and images (Crainiceanu et al., 2009, 2010; Di et al., 2008; Di and Crainiceanu, 2010; Greven et al., 2010; Staicu et al., 2010; Yao et al., 2005).
We follow the latter approach and put forward a generalization of principal components to understand major directions of variation in such large-scale neuroimaging studies. However, unlike most eigen-imaging approaches, we connect the methods to formal linear mixed models for imaging data. Our approach is based on FPCA (Ramsay and Silverman, 2005; Yao et al., 2005) which captures the principal directions of variation in the population. Subjects can be characterized in terms of their principal scores which are the coordinates in the space spanned by the principal components. Estimating both principal components and principal scores can be quite challenging in high-dimensional settings. We show how SVD can be efficiently adapted for the estimation problem. Zhang et al. (2007) explored SVD of individual functional objects which can be represented in a matrix form. In contrast, our approach employs SVD of the entire data matrix of vectorized neuroimages. It is well-known that left singular vectors of this SVD estimate principal components (Joliffe, 2002). To estimate principal scores for sparse FPCA models with measurement noise Yao et al. (2005) suggested to use estimated Best Linear Unbiased Predictors (BLUPs). We also estimate principal scores by BLUPs but our approach is different in two ways. Firstly, and most importantly, we consider high-dimensional FPCA settings where the covariance matrix of vectorized images is not invertible. Hence, the approach of Yao et al. (2005) cannot be applied directly. Secondly, we require neither specification of the distribution of the principal scores, nor that they be independent and show how right singular vectors can be used to estimate principal scores under minimal distributional assumptions. Once both principal components and principal scores are estimated hypothesis testing can be done using the standard tools of linear mixed model theory. Therefore, the approach yields a fully specified model and inferential framework. We further give a didactic explanation of easy methods for handling the necessary high dimensional calculations on even modest computing infrastructures.
Our proposed data-driven method applies generally, though in this manuscript we specifically apply it to morphometric images that would typically be used for voxel-based morphometry (Ashburner and Friston, 2000). In an imaging setting, the basic data requirement is a sample of spatially registered images, where the study of population variation in the registered intensities is of interest. Since the methods vectorize the imaging array information as a first step, whether the images are one, two, three or four (as in fMRI or PET studies) dimensional is irrelevant; though we stipulate that alternate methods that separate spatial and temporal variations (Beckmann and Smith, 2005; Caffo et al., 2010) are more relevant in the 4D cases. Regardless, the methods are generic and portable to a wide variety of imaging and non-imaging settings.
We also discuss the practical computing for the methods. We specifically demonstrate that model fitting can be performed via a SVD that can be applied iteratively, loading only components of the data at a time. Thereby, we demonstrate that the methods are scalable to large studies and can be executed on modest computing infrastructures.
The manuscript is laid out as follows. Motivating data section describes the motivating data, regional tissue volume maps (RAVENS maps) derived from structural brain MRI of former organolead manufacturing workers. Methods section explains why fitting FPCA model is identical to constructing SVD of the data matrix as well as provides necessary numerical adaptation to high-dimensional data. In Application to RAVENS images section, the method is applied to the RAVENS data. The last section concludes with a discussion.
Motivating data
The motivating data arise from a study of voxel-based morphom-etry (VBM) (Ashburner and Friston, 2000) in former organolead manufacturing workers. VBM is a common approach to analysis of structural MRI. The primary benefits of VBM are its lack of need for a priori specified regions of interest and its exploratory nature. VBM facilitates identification of complex, and perhaps previously unknown, patterns of brain structure via regression models of exposure or disease status on deformation maps.
However, VBM, as its name suggests, is applied at a voxel-wise level, resulting in tens or hundreds of thousands of tests considered independently. In contrast, regional analyses are primarily confirmatory, requiring both specified regional hypotheses as well as an anatomical parcellation. We instead analyze morphometric images to find principal directions of cross-sectional variation of brain image shapes. While this approach is useful for both analyzing deformation fields as an outcome (functional principal component analysis), it is also useful for regression models where morphometric deformation is a predictor (functional principal component regression) (Ramsay and Silverman, 2005).
The data were derived from an epidemiologic study of the central nervous system effects of organic and inorganic lead in former organolead manufacturing workers, described in detail elsewhere (Schwartz et al., 2000a,b; Stewart et al., 1999). Subject scans were from a GE 1.5 Tesla Signa scanner. RAVENS image processing (described further below) was performed on the T1-weighted volume acquisitions.
RAVENS stands for Regional Analysis of VolumE in Normalized Space, and represents a standard method for discovering localized changes in brain shape related to exposures (Goldszal et al., 1998; Shen and Davatzikos, 2003). It has been shown to be scalable and viable on large epidemiological cohort studies (Davatzikos et al., 2008; Resnick et al., 2009). The method analyzes smoothed deformation maps obtained when registering subjects to a standard template. Processing, and hence analysis, is performed separately for different tissue types (gray/white) and possibly for the analysis of cerebrospinal fluid (CSF), which may be informative for ventricular volume and shape. A complete description of RAVENS processing can be found in Goldszal et al. (1998) and Shen and Davatzikos (2003). In this study, we consider images collected over two visits roughly five years apart that were registered using a novel 4D generalization of RAVENS processing (Xue et al., 2006). Hence we investigate cross-sectional variation, separately at the first and second visits, as well as longitudinal variation as summarized by difference maps between the two time points.
We emphasize that our proposed modeling does not depend on imaging modality and processing. (Though, of course, processing and scientific context will dictate the utility of the models.) The necessary inputs for the procedure are images registered in a standardized space, where voxel-specific intensities are of interest. For example, the methods equally apply to PET images of a tracer or DTI summary (e.g. fractional anisotropy, mean diffusivity) maps.
Methods
In this section we discuss FPCA model. The relationship between FPCA and SVD will be highlighted. This link will allow us to address efficiently the computational issues arising for FPCA model in high-dimensional settings. Furthermore, the geometrical interpretation of left and right singular vectors within FPCA framework will be closely examined.
Single level FPCA
Suppose that we have a sample of images Xi, where Xi is a vectorized image of the ith subject, i=1,…,I. Every image is a 3-dimensional array structure of dimension p=p1×p2×p3. For example, in the RAVENS data described in Motivating data section, it has a dimension of p = 256×256×198 = 12,976,128. Of course, efficient masking of the data reduces this number drastically (to three million in the case of the RAVENS data). Hence, we represent the data Xi as a p×1 dimensional vector containing non-background voxels in a particular order, where the order is preserved across all voxels.
Following Di et al. (2008) we consider a single level functional model: Xi(v) = μ(v) + Zi(v),i = 1,…,I and v denotes a voxel coordinate. The image μ(v) is the overall mean image and Zi(v) is a subject-specific image deviation from the overall mean. We assume that μ(v) is fixed and Zi(v) is a zero-mean second-order stationary stochastic process with continuous covariance function K(v1,v2) = EZi(v1)Zi(v2). Using Karhunen-Loeve expansions of the random processes (Karhunen, 1947) , where ϕk(v) are the eigenfunctions of the covariance function K(v1, v2) and ζik are uncorrelated eigenscores with non-increasing variances σk. For practical purposes, we consider a model projected on the first N components. We define vectors of eigenscores ζi = (ζi1,…,ζiN)′ which we assume to be independent and identically distributed random vectors with zero mean and diagonal covariance matrix Σ = diag{σk}. Note that from the above assumption it follows that for each i components ζ′iks are uncorrelated. In other words, we do not impose any additional assumptions on the distribution of eigenscores ζ′iks. Further, it will be more convenient to consider normalized scores . With these changes the FPCA model becomes a linear mixed effect model (McCulloch and Searle, 2001, Ch.6)
| (1) |
where denotes uncorrelated zero mean random variables with unit variance. Eq. (1) can be written in a convenient matrix form as , where columns of p × N matrix ΦN are principal components ϕk's, and matrix is diagonal with on the main diagonal. Statistical estimation of model (1) includes estimating eigenimages ϕk with eigenvalues σk and eigenscores ξik. After these parameters are estimated, inference, including hypothesis testing, can be done using standard linear mixed models techniques (Demidenko, 2004; McCulloch and Searle, 2001).
The natural estimate of μ, the vectorized version of μ(v), is the sample point-wise arithmetic average . The unexplained part of the image, X̃i = Xi − μ̂, is eigen-analyzed to obtain the eigenvectors ϕk and eigenvalues σk. Denote X̃ = (X̃1,…, X̃I) where X̃i is a centered p × 1 vector containing the unfolded image for subject i. Then covariance operator K is estimated as . Given rank (K̂) = r the estimated covariance operator K̂ can be decomposed as where p × r matrix Φ̂r has orthonormal columns, ϕ̂k, and r×r diagonal matrix Σ̂r has non-negative diagonal elements σ̂1≥σ̂2≥ .. ≥σ̂r > 0. A small number of principal components (or eigenimages), N, can usually explain most of the variation (Di et al., 2008). The number of principal components, N, is typically chosen to make the explained variability (σ̂1 + … + σ̂N) / (σ̂1 + … + σ̂r) large enough. Alternatively, restricted likelihood ratio tests within linear mixed model theory can be adapted to choose N by formally testing if the corresponding variance component is zero (Crainiceanu et al., 2009; Staicu et al., 2010).
The size of the covariance operator K̂ is p × p. For high-dimensional p the brute-force eigenanalysis requires O(p3) operations and as a result is infeasible. Calculating and storing K̂ becomes impossible when p reaches infeasible levels.
Nevertheless, it is still possible to get eigendecomposition of K̂ by using the fact that the number of subjects, I, is typically much smaller than p. Indeed, if I<p then matrix X̃ = (X̃1,…, X̃I) has at most rank I and the SVD of X̃
| (2) |
can be obtained with O(pI2 + I3) computational effort (Golub and Loan, 1996). Here, the matrix V is p × I with I orthonormal columns, S is a diagonal I × I matrix and U is a I × I orthogonal matrix. Full details on efficient SVD calculation for ultra high-dimensional p will be provided in the next section. Now we will show the relation between FPCA (1) and SVD (2).
Assume for a moment that we calculated Eq. (2). Then . Given all eigenvalues are different, the eigendecomposition of K̂ is unique. Thus,
| (3) |
where p × N matrix VN consists of the first N left singular vectors and SN is N × N diagonal matrix with squares of the first N singular values on the main diagonal. Identities in Eq. (3) determine the estimates of eigenimages ϕ̂k and eigenvalues σ̂k. Estimated eigenfunctions ϕ̂k and eigenvalues σ̂k are used to calculate the estimated best linear unbiased predictors (EBLUPs) of the scores ξik. For linear models with invertible covariance matrix Var (X̃i) and given μ BLUPs can be calculated as (McCulloch and Searle, 2001, Ch.9)
| (4) |
For instance, Yao et al. (2005) showed that for sparse FPCA models with measurement noise Eq. (4) is equivalent to E(ξi|X̃i) under a normality assumption on principal scores. Greven et al. (2010) did not impose a normality assumption and employed Eq. (4) to estimate principal scores in longitudinal FPCA models. Brute-force calculation of EBLUPs based on Eq. (4) requires the inversion of p × p matrices (see also Crainiceanu et al., 2009; Di and Crainiceanu, 2010) and becomes prohibitive for high-dimensional problems.
Our case is different in that Var (X̃i) is not invertible for models (1) and (4) and can not be applied directly. The derivation of the EBLUPs presented below requires neither specification of the distribution of the principal scores ξik, nor that they be independent, and is based on a projection argument in Harville (1976). For model (1) the BLUP is expressed via pseudo-inverse matrices (Harville, 1976) as
| (5) |
where (ΦNΣNΦ′N) is the unique generalized inverse of the matrix ΦNΣNΦ′N. Using results from Demidenko (2004, Appendix) we can write . SVD representation (2) allows us to express X̃i as X̃i = VS1/2U′(:,i), where U′(:,i) is the ith column of matrix U′. Combining this with the estimators for ΦN and ΣN in Eq. (3), we obtain the estimated BLUPs as . It is easy to see that where U′(1: N,i) denotes the first N coordinates of vector U′(:,i). Therefore, we get .
The result formally derived above can be informally and more intuitively seen as follows. For the data matrix X̃ = (X̃1,…,X̃I) we have two representations. The first one comes from the data generating representation of X̃ as X̃ = ΦNΣNξ, where ξ = (ξ1, …,ξI) is N × I matrix of principal scores. From the other side, there is SVD representation (2). Putting the two together we demonstrated that fitting FPCA model (1) to data X̃ is equivalent to finding the best rank N approximation of X̃.
To summarize, we demonstrated that: i) the eigenvectors ϕk are given by the left singular vectors vk; ii) the normalized principal scores ξik are given by the rows of matrix U truncated to the first N coordinates and scaled up by iii) the variances σk are estimated by the singular values sk scaled down I.
Implementation
Now we give details of a fast and efficient algorithm for calculating SVD with O(pI2 + I3) computational effort and sequential access to the memory. It was easily implemented on a regular PC and completed in minutes for the former lead workers RAVENS data. First step is to use I ×I symmetric matrix X̃′X̃ and its spectral decomposition X̃′X̃ = USU′ to get U and S1/2. For high-dimensional p the matrix X̃ can not be loaded into the memory. The solution we suggest is to partition it into M slices as X̃′ = [(X̃1)′|(X̃2)′|…|(X̃M)′], where the size of the mth slice, X̃m, is p/M × I which can be adapted to the available computer memory and optimized to reduce implementation time. The matrix X̃′X̃ is calculated as and requires O(pI2) operations. Spectral decomposition forX̃′X̃ requires O(I3) operations and calculates matrices U and S. The p × I matrix V can now be obtained as V =X̃US−1/2. Actual calculations can be performed on the slices of the partitioned matrix X̃ as Vm = X̃mUS−1/2, m = 1;M and can be done with O(pI2) operations. The concatenated slices [ (V1)′ | (V2)′ |…|(VM)′] form the matrix of the left singular vectors V′. Hence, all components of the SVD can be calculated without loading the entire data matrix into memory. The analysis scales to nearly arbitrary large parameter spaces on very modest computing infrastructures.
Simulations
In this section, we will illustrate the proposed methods in a simulation study. We generated 1000 data sets according to the following FPCA model
| (6) |
where eigenimages ϕk are displayed in Fig. 1, ν = [1,300] × [1,300], and denotes independent identically distributed random variables following normal distribution with zero mean and variance σk. The eigenimages can be thought of as 2D grayscale images with pixel intensities on the [0,1] scale. The black pixels are set to 1 and the white ones are set to zero. We set I=350 to replicate the sample size of the RAVENS data set. The eigenvalues were set to be σk = 0.5k−1, k = 1,…,5. Generated images Xi's were unfolded to obtain vectors of size p = 300·300 = 90,000. The simulation study took 32 min on a PC with a quad core i7-2.67 Ghz processor and 6 Gb of RAM memory.
Fig. 1.

True grayscale eigenimages.
Fig. 2 displays means of the estimated eigenimages in the top panel, and the pointwise 5th and 95th percentile images in the middle and the bottom rows, respectively. To obtain a grayscale image with pixel values in the [0,1] interval, each estimated eigenvector, ϕ̂ = (ϕ̂1, …,ϕ̂p), was normalized as ϕ̂→(ϕ̂ − minsϕ̂s) / maxsϕ̂s − minsϕ̂s). Top row of Fig. 2 displays how on average our method recovers the spatial configuration. The percentile images in the middle and bottom rows of Fig. 2 show a similar pattern as the average with small distortions from the true functions (please note the light gray areas). We conclude that the estimation of the 2D eigenimages is very good.
Fig. 2.
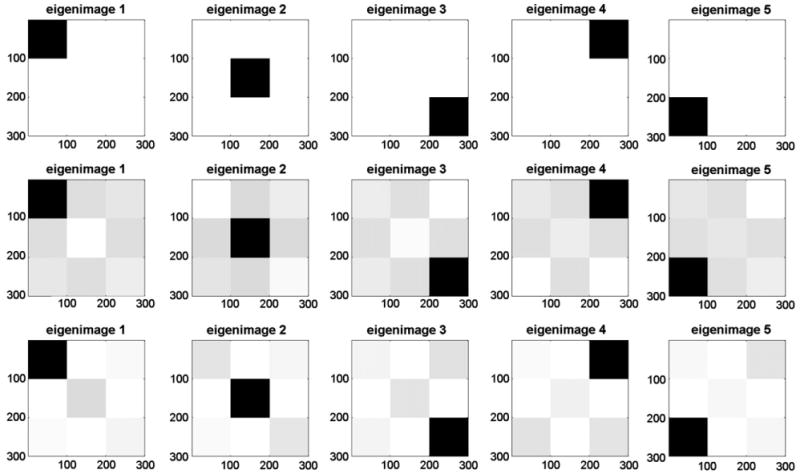
Grayscale images of the averages (top row), the 5th pointwise percentiles (middle row), and the 95th pointwise percentiles (bottom row) from the simulation study.
The total number of the normalized scores ξik in this simulation study was 350,000 for each k. The left column of Fig. 3 shows the distribution of the true (generated) scores at the top and the distribution of the corresponding estimated scores at the bottom. We see that the EBLUPs recover the form of the underlying distributions. The accuracy of EBLUPs can be seen in the right column of Fig. 3 which shows the distribution of the differences between true and estimated scores. The top graph displays the boxplots of the differences. The bottom one shows the medians, 0.5%, 5%, 90% and 99.5% quantiles of the distribution of the difference. One very noticeable pattern here is the scores corresponding to the eigenimages with lower variances have larger spread due to a reduced signal to noise ratio. Results show that the EBLUPs estimate true scores very well.
Fig. 3.

Left panel shows the distribution of the true scores (top) and the estimated scores (bottom). Right panel shows the boxplots of the difference (ξik−ξ̂ik).Boxplots are given at the top. The bottom picture shows the medians (black marker), 5% and 95% quantiles (blue markers), and 0.5% and 99.5% quantiles (red markers). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4 shows the boxplots of the estimated eigenvalues. We display the centered and standardized eigenvalues, (σ̂k−σk) / σk. The results indicate that eigenvalues are estimated with essentially no bias.
Fig. 4.

Boxplots of normalized estimated eigenvalues, (σ̂k−σk)/σk. The zero is shown by the solid black line.
Application to RAVENS images
In this section we apply our method to the RAVENS images described in Motivating data section. The RAVENS images are 256 × 256 × 198 dimensional for 352 subjects, each with two visits roughly five years apart. We analyze visit 1 and visit 2 separately. In addition, to identify the principal directions of the longitudinal change we consider a difference between images taken at visit 1 and visit 2. Although the data contains both white and gray matter as well as CSF, for illustration, the analysis is restricted only to the processed gray matter data. A small technical concern was of a few artifactual negative values in the data from the preprocessing. These voxels were removed from the analysis. After processing, the intersection of non-background voxels across images was collected. Such an intersection greatly reduced the dimension of the data matrix from ten billion numbers to two billion numbers divided as three million relevant voxels per subject per visit with seven hundred and four subject-visits.
Following Motivating data section all calculations were performed in such a way that only one of the manageable submatrices X̃m needs to be stored in memory at any given moment. The data matrix, of size 704 by 3 million, was divided into 100 submatrices of size 704 by 30 thousand (ten million numbers each). Note that on lower-resource computers the only change would be to reduce the size of submatrices. All calculations repeated for each of the three data sets were performed in Matlab 2010a and took around 15 min for each set on a PC with a quad core i7-2.67 Ghz processor and 6 Gb of RAM memory.
In the analysis, we first estimated the mean by the empirical voxel-specific arithmetic average. The visit specific mean images are uniform over the template and simply convey the message that localized changes in morphometry within subgroups get averaged over. The same is true for the mean of the longitudinal differences. In our eigenimage analysis we de-mean the data by subtracting out these vectors and work with de-meaned matrix X̃.
Fig. 5 shows the proportions of morphometric variation explained by the first thirty eigenimages for visit 1, visit 2, and the longitudinal difference. Cumulatively, the first thirty eigenimages explain 46.6%, 45.7%, and 52.5% of variation in data for visit 1, visit 2, and the longitudinal difference, respectively. The way eigenvalues decay on the most right graph of Fig. 5 is a clear indication that the longitudinal changes can be accurately described by the first thirty principal components explaining more than half of the longitudinal variation. Although the number of principal components, N, is usually chosen to explain enough variation (Di et al., 2008), our primary interest is the first few which identify the regions of brain exhibiting the most morphometric variation. The pattern of the percentage decrease on all three graphs of Fig. 5 flattens out after approximately the first ten principal components. Therefore, we concentrate our analysis on the first ten principal components.
Fig. 5.
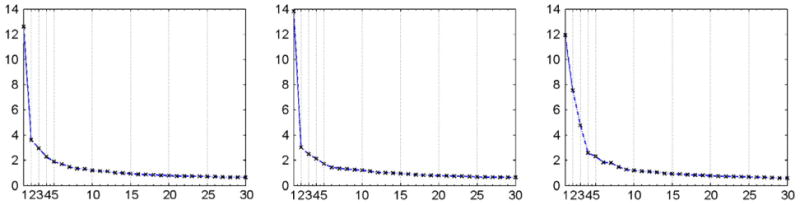
Proportions of morphometric variation explained by the first thirty eigenimages (from left to right: visit 1, visit 2, and the longitudinal difference).
Table 5 provides the cumulative percentages of variability explained by the first ten eigenimages. For visit 1 (top row) and visit 2 (middle row), they explain roughly the same amount of observed variation, 30%. For the longitudinal difference (bottom row), they explain 36.5% of the observed variability.
Table 5.
Labeled regions of the brain template. Abbreviations: PLICICPL = posterior limb of internal capsule including cerebral peduncle left, PLICICPR = posterior limb of internal capsule including cerebral peduncle right.
| 1 | Medial front-orbital gyrus right |
| 2 | Middle frontal gyrus right |
| 3 | Lateral ventricle left |
| 4 | Insula right |
| 5 | Precentral gyrus right |
| 6 | Lateral front-orbital gyrus right |
| 7 | Cingulate region right |
| 8 | Lateral ventricle right |
| 9 | Medial frontal gyrus left |
| 10 | Superior frontal gyrus right |
| 11 | Globus pallidus right |
| 12 | Globus pallidus left |
| 14 | Putamen left |
| 15 | Inferior frontal gyrus left |
| 16 | Putamen right |
| 17 | Frontal lobe WM right |
| 19 | Angular gyrus right |
| 23 | Subthalamic nucleus right |
| 25 | Nucleus accumbens right |
| 26 | Uncus right |
| 27 | Cingulate region left |
| 29 | Fornix left |
| 30 | Frontal lobe WM left |
| 32 | Precuneus right |
| 33 | Subthalamic nucleus left |
| 34 | PLICICPL |
| 35 | PLICICPR |
| 36 | Hippocampal formation right |
| 37 | Inferior occipital gyrus left |
| 38 | Superior occipital gyrus right |
| 39 | Caudate nucleus left |
| 41 | Supramarginal gyrus left |
| 43 | Anterior limb of internal capsule left |
| 45 | Occipital lobe WM right |
| 50 | Middle frontal gyrus left |
| 52 | Superior parietal lobule left |
| 53 | Caudate nucleus right |
| 54 | Cuneus left |
| 56 | Precuneus left |
| 57 | Parietal lobe WM left |
| 59 | Temporal lobe WM right |
| 60 | Supramarginal gyrus right |
| 61 | Superior temporal gyrus left |
| 62 | Uncus left |
| 63 | Middle occipital gyrus right |
| 64 | Middle temporal gyrus left |
| 69 | Lingual gyrus left |
| 70 | Superior frontal gyrus left |
| 72 | Nucleus accumbens left |
| 73 | Occipital lobe WM left |
| 74 | Postcentral gyrus left |
| 75 | Inferior frontal gyrus right |
| 80 | Precentral gyrus left |
| 83 | Temporal lobe WM left |
| 85 | Medial front-orbital gyrus left |
| 86 | Perirhinal cortex right |
| 88 | Superior parietal lobule right |
| 90 | Lateral front-orbital gyrus left |
| 92 | Perirhinal cortex left |
| 94 | Inferior temporal gyrus left |
| 95 | Temporal pole left |
| 96 | Entorhinal cortex left |
| 97 | Inferior occipital gyrus right |
| 98 | Superior occipital gyrus left |
| 99 | Lateral occipitotemporal gyrus right |
| 100 | Entorhinal cortex right |
| 101 | Hippocampal formation left |
| 102 | Thalamus left |
| 105 | Parietal lobe WM right |
| 108 | Insula left |
| 110 | Postcentral gyrus right |
| 112 | Lingual gyrus right |
| 114 | Medial frontal gyrus right |
| 118 | Amygdala left |
| 119 | Medial occipitotemporal gyrus left |
| 128 | Anterior limb of internal capsule right |
| 130 | Middle temporal gyrus right |
| 132 | Occipital pole right |
| 133 | Corpus callosum |
| 139 | Amygdala right |
| 140 | Inferior temporal gyrus right |
| 145 | Superior temporal gyrus right |
| 154 | Middle occipital gyrus left |
| 159 | Angular gyrus left |
| 165 | Medial occipitotemporal gyrus right |
| 175 | Cuneus right |
| 196 | Lateral occipitotemporal gyrus left |
| 203 | Thalamus right |
| 243 | Background |
| 251 | Occipital pole left |
| 254 | Fornix right |
| 255 | Subarachnoid cerebro-spinal fluid |
Top panel of Fig. 6 provides the estimated actual eigenvalues for the eigenimages. Notice, however, that we are more interested in the relative size of the eigenvalues representing quantitative measure of variability of the related eigenscores. Bottom panel of Fig. 6 plots the distributions of the eigenscores corresponding to the first ten eigenimages. In Single level FPCA section we showed that the estimates of the normalized eigenscores are given by the right singular vectors of matrix X̃. Therefore, the estimates of unnormalized eigenscores can be obtained once we multiply them by the square root of the corresponding eigenvalues provided in the top panel of Fig. 6. The estimated eigenscores serve as (signed) quantifiers relating eigenimages to subjects and their RAVENS maps.
Fig. 6.

Normalized distributions of the eigenscores corresponding to the first ten eigenimages (from left to right: visit 1, visit 2, and the longitudinal difference).
As we can see, the distribution of eigenscores in visit 1 and visit 2 are close to each other. Comparisons of the principal scores versus age are shown in Fig. 7. The results show how the major directions of volumetric variation are associated with cross-sectional baseline, age adjusted for height. The scores for the first component, being related to total gray matter volume simply display the well known decrease in overall volume with age. The final row displays that, adjusting for height, brain aging is progressive, with greater total volume declines with age. The other plots display non-significant relationship after having accounted for baseline height. Thus principal directions of gray matter volumetric variation appear unrelated to age and aging. The EBLUPs of the scores allow to perform more rigorous hypothesis testing of a specific scientific conjecture using relevant covariates.
Fig. 7.
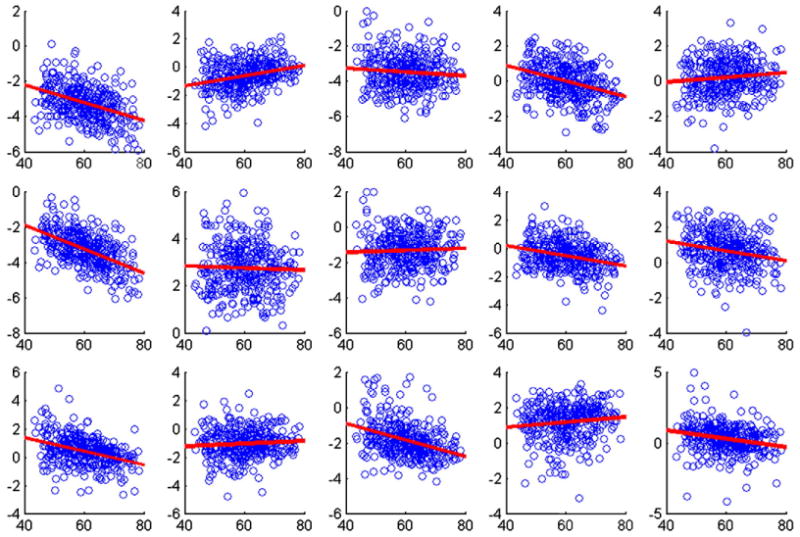
Plots of normalized eigenscores corresponding to the first 5 principal components against the age of the subjects adjusted for height. First row (visit 1), second row (visit 2), and third row (longitudinal difference). Red lines show fitted least squares lines adjusted for height. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
We now discuss overlap of the eigenimages with anatomical regions. Due to space limitations we discuss and depict only the first three eigenimages. We consider two independent separations of eigenimages. The first one quantifies the amount of variation of parcellated regions in template space. The kth eigenimage explains amount of variation. Recall, each coordinate of ϕk corresponds to a voxel in template space. Therefore, if the template is parcellated into R regions, then we can calculate the proportion of the variance explained by this particular region within eigenimage ϕk–on a scale from 0 to 1. Formally, all unfolded voxels {1,…,p} of template space can be regrouped as a union of R nonoverlapping subsets of voxels {Reg1,…,RegR}, where Regr consists of all region r voxels. Then eigenimage ϕk(v) can be represented as , where indicator I{v ε Regr} equals 1 if voxel v belongs to region r and equals 0 otherwise. Therefore, the variance explained by the kth eigenimage can be further decomposed as , where non-negative weights . Weights Wkr sum over the R regions to one and represent the proportions of variance σk explained by the regions. The second separation of eigenimages takes into account the sign of voxel values. Each eigenimage ϕk(v) can be split into two parts as , where is the positive loading and is the negative loading. Note that because of sign invariance of SVD, the separation between positive and negative loadings is comparable only within an eigenimage. Subject i is loaded on the kth eigenimage through score ξik. Therefore, subject i total loading can be split into the positive and negative parts as . In other words, for principal score ξik negative and positive voxel values correspond to the opposite directions (loadings) of variation. In our study, the template has been divided into R = 91 regions displayed in Table 5. However, the approach is general and applicable to any parcellation. In Table 2 we provide the variance explained by the labeled regions of the template for Visit 1. The twenty five regions with the highest loadings for each of the first three eigenimages are provided. Tables 1, 2, and 5 give now a way to determine a (signed) quantitative contribution of each particular region. For instance, the right middle temporal gyrus (130) explains 4.5% of the variance within eigenimage 1, which in turn explains 12.58% of the total variation. Hence, the right middle temporal gyrus explains 4.5% 12.58%= 0.57% of the total variation and has a mostly positive loading within eigenimage 1. Similarly, Tables 3 and 4 provide the regional quantifications of explained variation for Visit 2 and the longitudinal difference, respectively.
Table 2.
Visit 1: Proportion of the variance explained by the regions of the template (see Table 5 for the template parcellation). The twenty five regions with the highest loadings are provided. Third column quantifies the positive loading (blue), and fourth column quantifies the negative loading (red).
| Eigenimage 1 | Eigenimage 2 | Eigenimage 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Label | Variance explained | Positive loading | Negative loading | Label | Variance explained | Positive loading | Negative loading | Label | Variance explained | Positive loading | Negative loading |
| 255 | 0.0508 | 0.0508 | 0.0000 | 27 | 0.0222 | 0.0222 | 0.0000 | 255 | 0.0719 | 0.0540 | 0.0179 |
| 130 | 0.0450 | 0.0450 | 0.0000 | 30 | 0.0179 | 0.0170 | 0.0009 | 83 | 0.0295 | 0.0009 | 0.0287 |
| 17 | 0.0410 | 0.0410 | 0.0000 | 255 | 0.0175 | 0.0124 | 0.0051 | 165 | 0.0285 | 0.0240 | 0.0045 |
| 30 | 0.0399 | 0.0399 | 0.0000 | 17 | 0.0143 | 0.0134 | 0.0008 | 64 | 0.0275 | 0.0011 | 0.0265 |
| 59 | 0.0381 | 0.0380 | 0.0001 | 7 | 0.0129 | 0.0129 | 0.0000 | 102 | 0.0255 | 0.0255 | 0.0000 |
| 145 | 0.0331 | 0.0331 | 0.0000 | 83 | 0.0124 | 0.0114 | 0.0010 | 95 | 0.0254 | 0.0000 | 0.0254 |
| 83 | 0.0298 | 0.0297 | 0.0000 | 59 | 0.0097 | 0.0085 | 0.0012 | 203 | 0.0235 | 0.0235 | 0.0000 |
| 61 | 0.0287 | 0.0287 | 0.0000 | 203 | 0.0073 | 0.0041 | 0.0032 | 30 | 0.0212 | 0.0180 | 0.0032 |
| 64 | 0.0268 | 0.0268 | 0.0000 | 6 | 0.0072 | 0.0072 | 0.0000 | 99 | 0.0202 | 0.0026 | 0.0177 |
| 27 | 0.0237 | 0.0237 | 0.0000 | 196 | 0.0066 | 0.0022 | 0.0044 | 108 | 0.0200 | 0.0199 | 0.0001 |
| 99 | 0.0221 | 0.0221 | 0.0000 | 105 | 0.0059 | 0.0055 | 0.0004 | 17 | 0.0185 | 0.0160 | 0.0025 |
| 2 | 0.0205 | 0.0205 | 0.0000 | 102 | 0.0059 | 0.0004 | 0.0054 | 94 | 0.0181 | 0.0025 | 0.0156 |
| 7 | 0.0201 | 0.0201 | 0.0000 | 3 | 0.0058 | 0.0058 | 0.0000 | 92 | 0.0176 | 0.0000 | 0.0176 |
| 75 | 0.0197 | 0.0197 | 0.0000 | 57 | 0.0057 | 0.0052 | 0.0005 | 21 | 0.0176 | 0.0000 | 0.0176 |
| 196 | 0.0187 | 0.0187 | 0.0000 | 90 | 0.0052 | 0.0052 | 0.0000 | 119 | 0.0172 | 0.0106 | 0.0066 |
| 119 | 0.0166 | 0.0166 | 0.0000 | 64 | 0.0051 | 0.0049 | 0.0001 | 196 | 0.0169 | 0.0071 | 0.0097 |
| 15 | 0.0158 | 0.0158 | 0.0000 | 119 | 0.0050 | 0.0017 | 0.0033 | 4 | 0.0158 | 0.0157 | 0.0001 |
| 105 | 0.0155 | 0.0154 | 0.0001 | 8 | 0.0050 | 0.0050 | 0.0000 | 59 | 0.0157 | 0.0048 | 0.0109 |
| 57 | 0.0150 | 0.0150 | 0.0000 | 75 | 0.0049 | 0.0047 | 0.0002 | 61 | 0.0118 | 0.0016 | 0.0101 |
| 165 | 0.0148 | 0.0148 | 0.0000 | 133 | 0.0047 | 0.0046 | 0.0001 | 88 | 0.0114 | 0.0103 | 0.0012 |
| 50 | 0.0147 | 0.0147 | 0.0000 | 61 | 0.0045 | 0.0031 | 0.0015 | 75 | 0.0114 | 0.0113 | 0.0001 |
| 4 | 0.0146 | 0.0146 | 0.0000 | 52 | 0.0045 | 0.0042 | 0.0003 | 114 | 0.0111 | 0.0107 | 0.0004 |
| 5 | 0.0144 | 0.0144 | 0.0000 | 99 | 0.0039 | 0.0025 | 0.0015 | 5 | 0.0108 | 0.0104 | 0.0004 |
| 108 | 0.0141 | 0.0141 | 0.0000 | 20 | 0.0039 | 0.0000 | 0.0039 | 145 | 0.0105 | 0.0100 | 0.0005 |
| 74 | 0.0116 | 0.0116 | 0.0000 | 32 | 0.0036 | 0.0036 | 0.0000 | 9 | 0.0100 | 0.0096 | 0.0004 |
Table 1.
Cumulative percentage of variation explained by first ten eigenimages for RAVENS data (visit 1 (top row), visit 2 (middle row), and the longitudinal difference (bottom row)).
| Visit 1 | ||||||||||
|
| ||||||||||
| Component | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Cum % var | 12.58 | 16.20 | 19.15 | 21.42 | 23.31 | 25.00 | 26.47 | 27.81 | 29.11 | 30.29 |
| Visit 2 | ||||||||||
|
| ||||||||||
| Component | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Cum % var | 13.81 | 16.82 | 19.30 | 21.43 | 23.15 | 24.57 | 25.92 | 27.22 | 28.48 | 29.68 |
| Longitudinal difference | ||||||||||
|
| ||||||||||
| Component | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Cum % var | 11.91 | 19.44 | 24.21 | 26.80 | 29.09 | 30.91 | 32.70 | 34.16 | 35.42 | 36.60 |
Table 3.
Visit 2: Proportion of the variance explained by the regions of the template (see Table 5 for the template parcellation). The twenty five regions with the highest loadings are provided. Third column quantifies the positive loading (blue), and fourth column quantifies the negative loading (red).
| Eigenimage 1 | Eigenimage 2 | Eigenimage 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Label | Variance explained | Positive loading | Negative loading | Label | Variance explained | Positive loading | Negative loading | Label | Variance explained | Positive loading | Negative loading |
| 255 | 0.0542 | 0.0542 | 0.0000 | 255 | 0.0425 | 0.0120 | 0.0306 | 255 | 0.0612 | 0.0500 | 0.0113 |
| 30 | 0.0438 | 0.0437 | 0.0001 | 99 | 0.0274 | 0.0258 | 0.0016 | 30 | 0.0342 | 0.0132 | 0.0211 |
| 17 | 0.0420 | 0.0419 | 0.0001 | 30 | 0.0165 | 0.0004 | 0.0161 | 17 | 0.0251 | 0.0061 | 0.0190 |
| 130 | 0.0390 | 0.0390 | 0.0000 | 165 | 0.0165 | 0.0066 | 0.0099 | 27 | 0.0222 | 0.0006 | 0.0215 |
| 59 | 0.0338 | 0.0336 | 0.0002 | 196 | 0.0153 | 0.0120 | 0.0033 | 83 | 0.0220 | 0.0015 | 0.0205 |
| 145 | 0.0307 | 0.0307 | 0.0000 | 17 | 0.0144 | 0.0005 | 0.0139 | 88 | 0.0218 | 0.0174 | 0.0043 |
| 61 | 0.0288 | 0.0288 | 0.0000 | 108 | 0.0137 | 0.0000 | 0.0137 | 105 | 0.0206 | 0.0059 | 0.0147 |
| 83 | 0.0275 | 0.0273 | 0.0002 | 119 | 0.0136 | 0.0107 | 0.0030 | 108 | 0.0189 | 0.0141 | 0.0047 |
| 64 | 0.0268 | 0.0268 | 0.0000 | 4 | 0.0129 | 0.0000 | 0.0129 | 64 | 0.0182 | 0.0005 | 0.0177 |
| 27 | 0.0214 | 0.0214 | 0.0000 | 203 | 0.0123 | 0.0000 | 0.0123 | 7 | 0.0178 | 0.0002 | 0.0176 |
| 2 | 0.0207 | 0.0207 | 0.0000 | 102 | 0.0115 | 0.0000 | 0.0115 | 61 | 0.0156 | 0.0061 | 0.0095 |
| 75 | 0.0187 | 0.0187 | 0.0000 | 15 | 0.0107 | 0.0000 | 0.0107 | 59 | 0.0141 | 0.0018 | 0.0123 |
| 7 | 0.0180 | 0.0180 | 0.0000 | 83 | 0.0099 | 0.0088 | 0.0011 | 165 | 0.0137 | 0.0126 | 0.0011 |
| 99 | 0.0174 | 0.0174 | 0.0000 | 75 | 0.0098 | 0.0000 | 0.0098 | 52 | 0.0125 | 0.0095 | 0.0030 |
| 50 | 0.0173 | 0.0173 | 0.0000 | 114 | 0.0084 | 0.0000 | 0.0084 | 57 | 0.0125 | 0.0025 | 0.0099 |
| 15 | 0.0170 | 0.0170 | 0.0000 | 59 | 0.0083 | 0.0049 | 0.0035 | 203 | 0.0119 | 0.0119 | 0.0000 |
| 105 | 0.0167 | 0.0165 | 0.0002 | 64 | 0.0082 | 0.0053 | 0.0029 | 102 | 0.0111 | 0.0111 | 0.0000 |
| 196 | 0.0166 | 0.0166 | 0.0000 | 95 | 0.0078 | 0.0078 | 0.0000 | 19 | 0.0110 | 0.0059 | 0.0050 |
| 57 | 0.0166 | 0.0165 | 0.0001 | 145 | 0.0076 | 0.0003 | 0.0073 | 130 | 0.0105 | 0.0023 | 0.0082 |
| 5 | 0.0148 | 0.0148 | 0.0000 | 9 | 0.0066 | 0.0000 | 0.0065 | 196 | 0.0091 | 0.0060 | 0.0031 |
| 165 | 0.0143 | 0.0143 | 0.0000 | 88 | 0.0065 | 0.0002 | 0.0063 | 4 | 0.0090 | 0.0051 | 0.0039 |
| 119 | 0.0133 | 0.0133 | 0.0000 | 94 | 0.0064 | 0.0050 | 0.0014 | 14 | 0.0089 | 0.0089 | 0.0000 |
| 74 | 0.0132 | 0.0132 | 0.0000 | 92 | 0.0062 | 0.0062 | 0.0000 | 9 | 0.0085 | 0.0067 | 0.0019 |
| 4 | 0.0130 | 0.0130 | 0.0000 | 130 | 0.0057 | 0.0020 | 0.0037 | 95 | 0.0082 | 0.0000 | 0.0082 |
| 108 | 0.0128 | 0.0128 | 0.0000 | 5 | 0.0054 | 0.0002 | 0.0052 | 92 | 0.0082 | 0.0000 | 0.0082 |
Table 4.
Longitudinal difference: Proportion of the variance explained by the regions of the template (see Table 5 for the template parcellation). The twenty five regions with the highest loadings are provided. Third column quantifies the positive loading (blue), and fourth column quantifies the negative loading (red).
| Eigenimage 1 | Eigenimage 2 | Eigenimage 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Label | Variance explained | Positive loading | Negative loading | Label | Variance explained | Positive loading | Negative loading | Label | Variance explained | Positive loading | Negative loading |
| 255 | 0.0620 | 0.0029 | 0.0591 | 64 | 0.0188 | 0.0000 | 0.0188 | 255 | 0.0687 | 0.0087 | 0.0599 |
| 30 | 0.0439 | 0.0003 | 0.0436 | 255 | 0.0179 | 0.0014 | 0.0165 | 64 | 0.0636 | 0.0000 | 0.0636 |
| 17 | 0.0419 | 0.0003 | 0.0416 | 130 | 0.0142 | 0.0000 | 0.0142 | 94 | 0.0344 | 0.0000 | 0.0344 |
| 27 | 0.0341 | 0.0000 | 0.0341 | 94 | 0.0133 | 0.0000 | 0.0133 | 83 | 0.0288 | 0.0023 | 0.0265 |
| 59 | 0.0320 | 0.0000 | 0.0320 | 83 | 0.0077 | 0.0008 | 0.0069 | 17 | 0.0259 | 0.0192 | 0.0067 |
| 145 | 0.0291 | 0.0000 | 0.0291 | 196 | 0.0070 | 0.0000 | 0.0070 | 30 | 0.0239 | 0.0058 | 0.0180 |
| 61 | 0.0283 | 0.0000 | 0.0283 | 102 | 0.0066 | 0.0066 | 0.0000 | 21 | 0.0227 | 0.0000 | 0.0227 |
| 83 | 0.0257 | 0.0000 | 0.0257 | 21 | 0.0060 | 0.0000 | 0.0060 | 130 | 0.0218 | 0.0021 | 0.0196 |
| 130 | 0.0235 | 0.0000 | 0.0235 | 30 | 0.0048 | 0.0008 | 0.0040 | 90 | 0.0190 | 0.0002 | 0.0189 |
| 7 | 0.0231 | 0.0000 | 0.0231 | 140 | 0.0047 | 0.0000 | 0.0047 | 95 | 0.0178 | 0.0000 | 0.0178 |
| 75 | 0.0211 | 0.0000 | 0.0211 | 59 | 0.0046 | 0.0011 | 0.0035 | 61 | 0.0171 | 0.0002 | 0.0169 |
| 4 | 0.0196 | 0.0000 | 0.0196 | 61 | 0.0044 | 0.0001 | 0.0043 | 140 | 0.0151 | 0.0004 | 0.0148 |
| 108 | 0.0179 | 0.0000 | 0.0179 | 50 | 0.0040 | 0.0000 | 0.0040 | 59 | 0.0145 | 0.0083 | 0.0062 |
| 64 | 0.0174 | 0.0000 | 0.0174 | 37 | 0.0040 | 0.0000 | 0.0040 | 4 | 0.0144 | 0.0144 | 0.0000 |
| 6 | 0.0169 | 0.0000 | 0.0169 | 17 | 0.0035 | 0.0012 | 0.0023 | 5 | 0.0118 | 0.0066 | 0.0052 |
| 99 | 0.0167 | 0.0000 | 0.0167 | 95 | 0.0031 | 0.0000 | 0.0031 | 6 | 0.0115 | 0.0004 | 0.0111 |
| 105 | 0.0160 | 0.0001 | 0.0159 | 52 | 0.0027 | 0.0002 | 0.0025 | 16 | 0.0098 | 0.0098 | 0.0000 |
| 57 | 0.0153 | 0.0001 | 0.0152 | 251 | 0.0025 | 0.0000 | 0.0025 | 15 | 0.0097 | 0.0009 | 0.0088 |
| 88 | 0.0149 | 0.0001 | 0.0148 | 145 | 0.0023 | 0.0003 | 0.0020 | 102 | 0.0097 | 0.0097 | 0.0000 |
| 90 | 0.0149 | 0.0000 | 0.0149 | 203 | 0.0023 | 0.0023 | 0.0000 | 75 | 0.0090 | 0.0076 | 0.0014 |
| 2 | 0.0145 | 0.0000 | 0.0145 | 90 | 0.0022 | 0.0003 | 0.0019 | 50 | 0.0088 | 0.0001 | 0.0088 |
| 52 | 0.0143 | 0.0000 | 0.0143 | 99 | 0.0021 | 0.0000 | 0.0021 | 154 | 0.0083 | 0.0000 | 0.0083 |
| 114 | 0.0141 | 0.0004 | 0.0137 | 15 | 0.0020 | 0.0000 | 0.0019 | 99 | 0.0080 | 0.0053 | 0.0027 |
| 196 | 0.0141 | 0.0001 | 0.0139 | 70 | 0.0019 | 0.0000 | 0.0019 | 145 | 0.0080 | 0.0073 | 0.0006 |
| 15 | 0.0137 | 0.0000 | 0.0137 | 6 | 0.0019 | 0.0005 | 0.0013 | 196 | 0.0079 | 0.0011 | 0.0069 |
As we showed in the Introduction section the estimated principal components (eigenimages) are left singular vectors of matrix X̃. Each left singular vector is of size p≈3·106 unfolded voxels. Therefore, each voxel is represented by a small value between negative and positive one and squares of the voxel values are summed to one. The distribution of the negative and positive voxel loadings are presented in Fig. 8 in red and blue, respectively. The voxel values of the estimated eigenimage ϕ̂ = (ϕ̂1,…,ϕ̂p) were transformed as ϕ̂→256·(ϕ̂−minsϕ̂s)/(maxsϕ̂s−minsϕ̂s) separately for voxels with positive and negative loadings. The transformed negative and positive loadings overlaid with the template are presented in Figs. 9, 10, and 11. Note that our approach does not incorporate spatial structure or smoothness. In part, this has been taken into account at the preprocessing step when a 3D Gaussian kernel had been used to smooth original brain images. However, the found eigenimages obey some regional boundaries despite the above-mentioned ignorance of spatial relationships. We believe that it highlights considerable potential of the methods within these settings.
Fig. 8.

Distributions of the intensities of the first three eigenimages (visit 1 (top row), visit 2 (middle row), and the longitudinal difference (bottom row)). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 9.
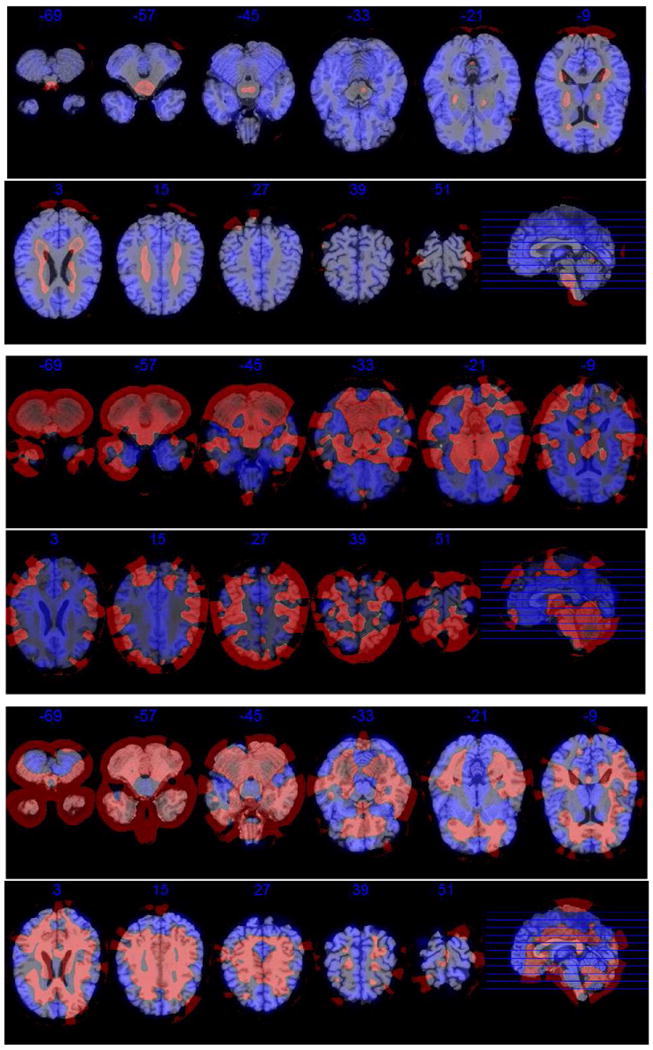
The first three estimated eigenimages for visit 1. Each eigenimage is represented by eleven equidistant axial slices. Negative loadings are depicted in red, and positive ones are in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 10.
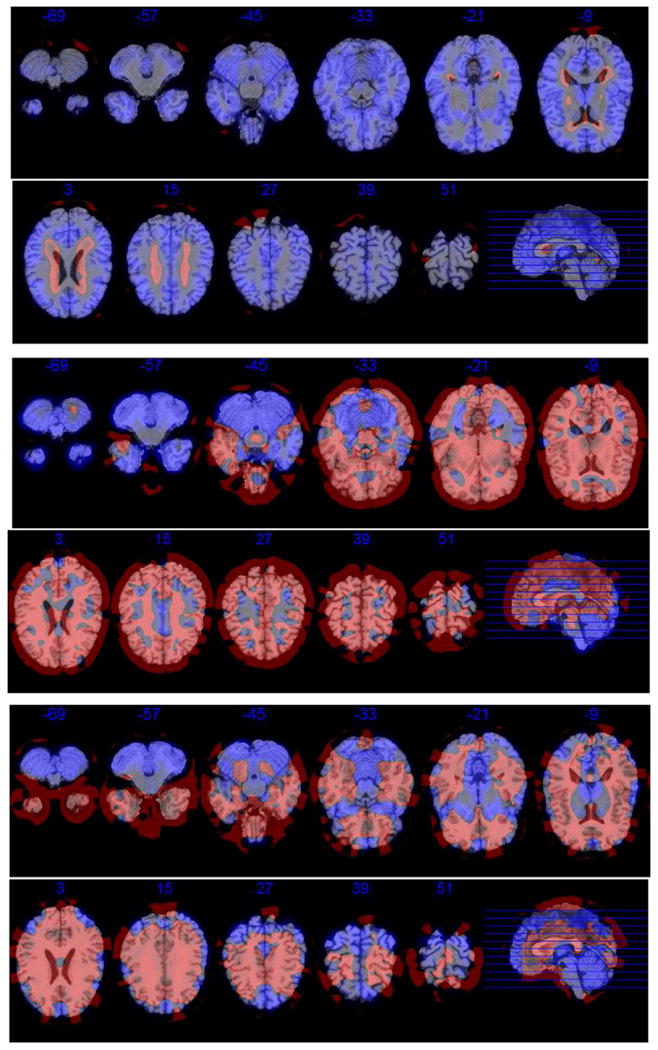
The first three estimated eigenimages for visit 2. Each eigenimage is represented by eleven equidistant axial slices. Negative loadings are depicted in red, and positive ones are in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 11.

The first three estimated eigenimages for the longitudinal difference. Each eigenimage is represented by eleven equidistant axial slices. Negative loadings are depicted in red, and positive ones are in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
In this paper we proved a connection between SVD and FPCA models. This coupling allowed us to develop efficient model-based computing techniques. The developed approach was applied to a novel morphometric data set with 704 RAVENS images. Principal components of morphometric variation were identified and studied. An alternative to our analysis would be a more formal separation of cross-sectional and longitudinal morphometric variation within multilevel functional principal component analysis framework suggested in Di et al. (2008).
There are a few important limitations in the presented methodology. First, we have not assumed noise in the model. RAVENS data represent preprocessed and smoothed images. However, there are a considerable number of studies collecting functional observations measured with non-ignorable noise. The ideas proposed in Huang et al. (2008) and Eilers et al. (2006) can be explored to develop a feasible smoothing procedure for spatial principal components. Another related development could be a rigorous incorporation of 3D spatial structure.
Our model assumes that the functional data are densely, rather than sparsely, observed. The issue of sparsity was addressed in Di et al. (2008) and Di and Crainiceanu (2010) for multilevel models. The proposed efficient solutions were based on smoothing of the covariance operator which is infeasible for high-dimensional data. Therefore, there is a great demand in computationally efficient procedures of covariance smoothing in the high dimensional context.
Acknowledgments
The authors would like to thank the reviewers and the editor for their helpful comments and suggestions which led to an improved version of the manuscript. The research of Vadim Zipunnikov, Brian Caffo and Ciprian Crainiceanu was supported by award number R01NS060910 from the National Institute of Neurological Disorders and Stroke. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke or the National Institutes of Health.
References
- Ashburner J, Friston K. Voxel-based morphometry–the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Beckmann C, Smith S. Tensorial extensions of independent component analysis for multisubject fMRI analysis. Neuroimage. 2005;25:294–311. doi: 10.1016/j.neuroimage.2004.10.043. [DOI] [PubMed] [Google Scholar]
- Caffo B, Crainiceanu C, Verduzco G, Joel S, Mostofsky SH, Bassett SS, Pekar JJ. Two-stage decompositions for the analysis of functional connectivity for fMRI with application to Alzheimer's disease risk. Neuroimage. 2010;51:1140–1149. doi: 10.1016/j.neuroimage.2010.02.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crainiceanu CM, Caffo BS, Luo S, Zipunnikov VV, Punjabi NM. Population Value Decomposition, a framework for the analysis of image populations. Johns Hopkins University, Dept of Biostatistics Working Paper 220. 2010 doi: 10.1198/jasa.2011.ap10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crainiceanu CM, Staicu AM, Di CZ. Generalized multilevel functional regression. J Am Stat Assoc. 2009;104(488):1550–1561. doi: 10.1198/jasa.2009.tm08564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davatzikos C, Fan Y, Wu X, Shen D, Resnick SM. Detection of prodromal Alzheimer's disease via pattern classification of magnetic resonance imaging. Neurobiol Aging. 2008;29:514–523. doi: 10.1016/j.neurobiolaging.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenko . Mixed Models: Theory and Applications. John Wiley and Sons; 2004. [Google Scholar]
- Di CZ, Crainiceanu CM. Multilevel sparse functional principal component analysis. Technical Report. 2010 doi: 10.1002/sta4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di CZ, Crainiceanu CM, Caffo BS, Punjabi NM. Multilevel functional principal component analysis. Ann Appl Stat. 2008;3(1):458–488. doi: 10.1214/08-AOAS206SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers PH, Currie ID, Durban M. Fast and compact smoothing on large multidimensional grids. Comput Stat Data Anal. 2006;50:61–76. [Google Scholar]
- Goldszal A, Davatzikos C, Pham D, Yan M, Bryan R, Resnick SM. An image processing protocol for the analysis of MR images from an elderly population. J Comput Assist Tomogr. 1998;22:827–837. doi: 10.1097/00004728-199809000-00030. [DOI] [PubMed] [Google Scholar]
- Golub GH, Loan CV. Matrix Computations. The Johns Hopkins University Press; 1996. [Google Scholar]
- Greven S, Crainiceanu CM, Caffo BS, Reich D. Longitudinal functional principal component analysis. Electron J Stat. 2010;4:1022–1054. doi: 10.1214/10-EJS575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W. Functional mixed effects models. Biometrics. 2002;58:121–128. doi: 10.1111/j.0006-341x.2002.00121.x. [DOI] [PubMed] [Google Scholar]
- Harville D. Extension of the Gauss–Markov theorem to include the estimation of random effects. Ann Stat. 1976;4(2):384–395. [Google Scholar]
- Huang JZ, Shen H, Buja A. Functional principal components analysis via penalized rank one approximation. Electron J Stat. 2008;2:678–695. [Google Scholar]
- Joliffe I. Principal Component Analysis. 2nd. Springer-Verlag; New-York: 2002. [Google Scholar]
- Karhunen K. Uber Lineare Methoden in der Wahrscheinlichkeitsrechnung. Ann Acad Sci Fenn. 1947;37:1–79. [Google Scholar]
- McCulloch C, Searle S. Generalized, Linear, and Mixed Models. Wiley Interscience; 2001. [Google Scholar]
- Mohamed A, Davatzikos C. Shape representation via best orthogonal basis selection. Med Image Comput Comput Assist Interv. 2004;3216:225–233. [Google Scholar]
- Morris JS, Carroll RJ. Wavelet-based functional mixed models. J R Stat Soc B. 2006;68:179–199. doi: 10.1111/j.1467-9868.2006.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay J, Silverman B. Functional Data Analysis. 2nd. Springer; 2005. [Google Scholar]
- Reiss P, Ogden RT. Functional generalized linear models with applications to neuroimaging. Poster Presentation Workshop on Contemporary Frontiers in High-Dimensional Statistical Data Analysis 2008 [Google Scholar]
- Reiss P, Ogden RT. Functional generalized linear models with images as predictors. Biometrics. 2010;1:61–69. doi: 10.1111/j.1541-0420.2009.01233.x. [DOI] [PubMed] [Google Scholar]
- Reiss PT, Ogden RT, Mann J, Parsey RV. Functional logistic regression with PET imaging data: a voxel-level clinical diagnostic tool. J Cereb Blood Flow Metab. 2005;25:s635. [Google Scholar]
- Resnick S, Espeland MA, Jaramillo SA, Hirsch C, Stefanick ML, Murray AM, Ockene J, Davatzikos C. Postmenopausal hormone therapy and regional brain volumes: the WHIMS-MRI Study. Neurology. 2009;72:135–142. doi: 10.1212/01.wnl.0000339037.76336.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B, Stewart W, Bolla K, Simon D, Bandeen-Roche K, Gordon B, Links J, Todd A. Past adult lead exposure is associated with longitudinal decline in cognitive function. Neurology. 2000a;55:1144. doi: 10.1212/wnl.55.8.1144. [DOI] [PubMed] [Google Scholar]
- Schwartz B, Stewart W, Kelsey K, Simon D, Park S, Links J, Todd A. Associations of tibial lead levels with BsmI polymorphisms in the vitamin D receptor in former organolead manufacturing workers. Environ Health Perspect. 2000b;108:199. doi: 10.1289/ehp.00108199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen DG, Davatzikos C. Very high resolution morphometry using mass-preserving deformations and HAMMER elastic registration. Neuroimage. 2003;18:28–41. doi: 10.1006/nimg.2002.1301. [DOI] [PubMed] [Google Scholar]
- Staicu AM, Crainiceanu CM, Carroll RJ. Fast analysis of spatially correlated multilevel functional data. Biostatistics. 2010;11(2):177–194. doi: 10.1093/biostatistics/kxp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart W, Schwartz B, Simon D, Bolla K, Todd A, Links J. Neurobehavioral function and tibial and chelatable lead levels in 543 former organolead workers. Neurology. 1999;52:1610. doi: 10.1212/wnl.52.8.1610. [DOI] [PubMed] [Google Scholar]
- Xue Z, Shen D, Davatzikos C. CLASSIC: consistent longitudinal alignment and segmentation for serial image computing. Neuroimage. 2006;30:388–399. doi: 10.1016/j.neuroimage.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Yao F, Muller HG, Wang JL. Functional data analysis for sparse longitudinal data. J Am Stat Assoc. 2005;100:577–590. [Google Scholar]
- Zhang L, Marron J, Shen H, Zhu Z. Singular value decomposition and its visualization. J Comput Graphical Stat. 2007;16:833–854. [Google Scholar]


