Abstract
To build complex organs, embryos have evolved mechanisms that integrate the development of cells unrelated to one another by cell type or ancestry. Here we show that the pha-4 locus establishes organ identity for the Caenorhabditis elegans pharynx. In pha-4 mutants, pharyngeal cells are transformed into ectoderm. Conversely, ectopic pha-4 expression produces excess pharyngeal cells. pha-4 encodes an HNF-3 homolog selectively expressed in the nascent digestive tract, including all pharynx precursors at the time they are restricted to a pharyngeal fate. We suggest that pha-4 is a key component of a transcription-based mechanism to endow cells with pharyngeal organ identity.
Keywords: C. elegans, fork head, HNF-3, pha-4, organogenesis, foregut development
The digestive tract consists of an epithelial tube that is spatially and functionally regionalized. In simple animals, such as Hydra, the gut contains only a few specialized cells and is divided into only two regions: the mouth and gut sac. In more complex animals, the digestive tract is composed of many different cell types and compartments. How is this complicated organ built? To tackle this question, we are studying foregut, or pharynx, development in the experimentally tractable organism Caenorhabditis elegans.
The nematode pharynx is a large neuromuscular organ used for feeding. Like the foregut compartments of more complex animals, the pharynx is composed of many different cell types such as muscles, glands, neurons, epithelia, and valves (Albertson and Thomson 1976). Two early blastomeres, ABa and MS, generate the pharynx (Sulston et al. 1983). Each blastomere gives rise to multiple cell types within the pharynx, and each uses a different genetic program to produce pharyngeal cells (for review, see Schnabel and Priess 1997). The ABa-derived pathway depends on intercellular signaling mediated by the GLP-1 receptor, whereas pharynx production from MS appears to be cell autonomous. Two predicted transcription factors, skn-1 and pop-1, are necessary for the MS developmental program, including the production of pharyngeal cells. Recent studies have begun to elucidate the gene networks controlling glp-1, skn-1, and pop-1 (for review, see Han 1997); however, the mechanisms that act downstream of these genes, to mediate pharyngeal organogenesis, are largely unknown.
The genetic pathways that generate the ABa and MS cell lineages converge on the pha-4 locus. Animals lacking zygotic pha-4 activity fail to produce pharynx cells from either ABa or MS (Mango et al. 1994a). In addition, these mutants lack a rectum. The rectum is descended from a third blastomere called ABp and does not derive from the pharyngeal cell lineages. Most other cells appear to be generated normally in pha-4 mutant embryos, including cells from ABa and MS that are not part of the pharynx. Thus, the pha-4 locus is critical to generate a group of cells related by function (the digestive tract) rather than by cell lineage (ABa, MS) or cell type (pharynx neuron, pharynx muscle, etc.). This phenotype reflects a transition during embryogenesis from maternal genes, like glp-1, skn-1, and pop-1, that control entire cell lineages and broad domains of axial patterning, to zygotic genes, like pha-4, that regulate the formation of specific organs, tissues, and cell types.
In the present study we address two potential mechanisms by which pha-4 could function. First, pha-4 could establish an organizing center that is critical for development of the entire pharynx. By analogy, loss of the anchor cell during C. elegans vulval development results in no vulva being formed (Greenwald 1997). Similarly, mutations in WT-1 or c-ret lead to the absence of the kidney in mouse embryos (Kreidberg et al. 1993; Schuchardt et al. 1994). Each of these phenotypes reflects a loss in cell signaling. For vulval development, signaling occurs between cells in two different organs, whereas in kidney formation WT-1 and c-ret are necessary for epitheliomesenchymal interactions among different tissues within the developing kidney primordium.
A second possible explanation for pha-4 function is that the protein could act within individual pharyngeal cells to establish pharynx identity. That is, pha-4 might be analogous to pax-6 and the network of transcription factors that initiate eye formation (Desplan 1997). Loss-of-function mutations in either eyeless, sine oculis, eyes absent, or dachshund leads to a complete absence of the eye in Drosophila. Conversely, each of these genes, either alone or in combination, is capable of inducing ectopic eyes. These phenotypes suggest that this group of factors synergize to define eye identity during development.
In this report we present evidence that pha-4 encodes an HNF-3 homolog that is expressed in all pharyngeal precursors and establishes their fate. We suggest that pha-4 and the Drosophila eye genes represent a new class of developmental regulator that specifies organ identity. Given that HNF-3 genes have been implicated in gut development in other organisms (Kaufmann and Knochel 1996), we propose that specification of organ identity within the digestive tract may be a conserved feature of HNF-3 proteins.
Results and Discussion
pha-4 encodes the HNF-3 homolog Ce-fkh-1 and is expressed in the developing digestive tract
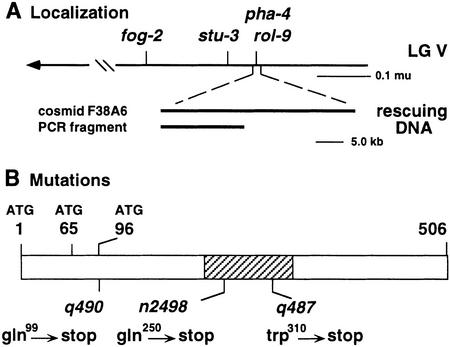
To understand the molecular mechanism of pha-4 function, we identified the gene by a combination of positional cloning and candidate gene approaches. Previous analyses showed that pha-4 is located within the YAC Y79B4 (S. Mango, unpubl.). YAC Y79B4 contains the predicted winged helix transcription factor Ce-fkh-1, which was an excellent candidate to encode pha-4 based on map position and mRNA expression (Azzaria et al. 1996; S. Mango, unpubl.). Three lines of evidence demonstrate that pha-4 corresponds to Ce-fkh-1. First, extrachromosomal arrays carrying either the cosmid F38A6, which carries Ce-fkh-1, or a 16.7-kb PCR fragment spanning the Ce-fkh-1 locus can rescue homozygous pha-4 mutants (Fig. 1A). Second, antisense Ce-fkh-1 RNA injected into wild-type hermaphrodites causes embryos and larvae to arrest with a weak pha-4 phenotype (see Materials and Methods). Third, three EMS-induced pha-4 alleles harbor mutations within Ce-fkh-1 (Fig. 1B). The Ce-fkh-1 locus generates three mRNAs, which are predicted to encode three proteins of 506, 441, and 410 amino acids (Azzaria et al. 1996). The allele pha-4(q490) carries a nonsense mutation at amino acid 99 relative to the largest pha-4 isoform and is predicted to truncate all three proteins severely. Therefore, this allele, which behaves like a null allele in genetic tests (Mango et al. 1994a), is likely to be a molecular null as well. The other two sequenced alleles, n2498 and q487, encode nonsense mutations at positions 250 and 310, respectively. These data show that pha-4 is encoded by Ce-fkh-1.
Figure 1.
pha-4 encodes the HNF-3 homolog Ce-fkh-1. (A) The genetic (top) and physical (bottom) location of pha-4. (B) Three pha-4 protein isoforms are indicated by ATG. The DBD is hatched. Each mutation depicted was induced by EMS (Mango et al. 1994a) and is the result of a C → T transition, as expected.
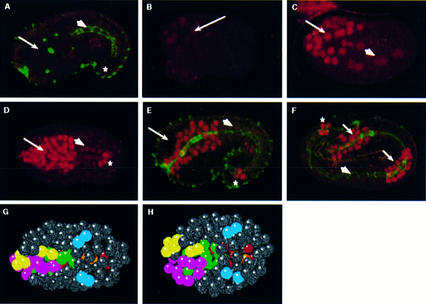
To determine when and where PHA-4 protein is expressed, we designed a polyclonal antibody that recognizes the PHA-4 carboxyl terminus. This antibody is specific, as no staining is observed in pha-4(q490) null embryos (Fig. 2A) or after preincubation with the immunizing peptide (Materials and Methods). We observe PHA-4 in the nuclei of cells destined to form the digestive tract. At the 4E stage, weak expression is detected in 8 MS great-granddaughters and 10 ABa descendants (ABaraaaa/p, ABaraapa/p, ABarapaa/p, ABalpaaa/p, ABalpapa/p) that each produce pharyngeal cells, as well as nonpharyngeal cells (Fig. 2B; we have staged embryos according to the number of E-derived midgut cells; see Fig. 3A for a diagram of the cell lineage). We also detect faint expression in the four midgut precursors. Brighter staining is observed in 6–8 MS-derived and 16–18 ABa-derived cells after the next cell division (MSaaaa/p, MSaapa/p, MSpaaa/p, MSpapa/p; Fig. 2C). Soon thereafter, at the 8–12E stage, PHA-4 is first observed in the rectal precursors. Expression is maintained in all pharyngeal, midgut, and rectal cells throughout embryogenesis. A similar pattern has been seen by J. Kalb, K. Lau, B. Goszczynski, T. Fukushige, D. Moons, P. Okkema, and J. McGhee (pers. comm.). In summary, PHA-4 expression defines the digestive tract during embryogenesis. The initiation of bright expression at the 8E stage corresponds to the time when the pharyngeal precursors are born. These cells are destined to produce only pharyngeal cells. However, they are not restricted to any particular cell type within the pharynx, consistent with pha-4’s proposed global role in specifying pharynx identity (Sulston et al. 1983; Mango et al. 1994a).
Figure 2.
PHA-4 is expressed in the developing digestive tract. (A–F) PHA-4 (red) and MH27 adherens junction (green) staining. (A) pha-4(q490) null embryos. (B) 4E stage embryo (expression in ABaraaaa/p, ABaraapa/p, ABarapaa/p, ABalpaaa/p, ABalpapa/p, all MS great-granddaughters, midgut). (C) 8E stage embryo (expression in MSaaaa/p, MSaapa, MSpaaa/p, MSpapa, ABa descendants, midgut). (D–E) Midstage embryos [97 ± 3 PHA-4+ cells in the head (expression in all pharyngeal cells, pharynx–intestinal valve cells, and arcade cells, based on cell position and double staining (n = 3)], rectal cells (vir, rep, KK‘), and midgut. (F) Terminal embryo (expression in entire pharynx, midgut, and rectum). (Arrow) Pharynx; (arrowhead) midgut; (*) rectum. Nuclear positions in pha-4(+) (G) and pha-4(−) (H) embryos at the 8E stage, ventral view. Pharyngeal cells are colored according to their lineal ancestry (yellow, ABalp; pink, ABara; green, MS). Midgut cells are red and rectal cells are blue. The embryos in B and C are rotated relative to the embryos shown here. Anterior is left (A–H); each embryo is ∼50 μm.
Figure 3.
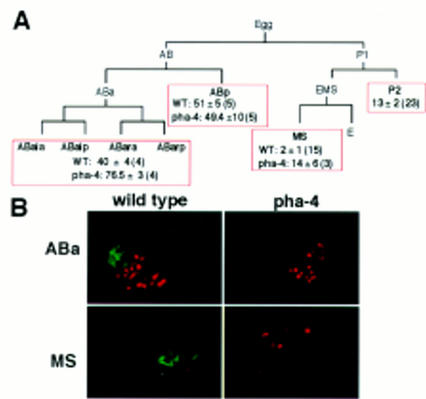
pha-4 embryos produce extra ectodermal cells. (A) Partial cell lineage depicting the number of LIN-26+ cells produced by isolated blastomeres. In parentheses is the number of ablated embryos analyzed. (B) Example of LIN-26 (red) and 3NB12 pharynx muscle (green) staining in operated embryos.
pha-4 is required to establish the pharyngeal precursors
We used lineage analysis to show that pha-4 function is required by the 8E stage in development. Previous studies demonstrated that pha-4 mutants lack a pharynx (Mango et al. 1994a). Here we investigate the pha-4 mutant phenotype during early developmental stages and show that pha-4 function is required for the earliest signs of pharyngeal development during embryogenesis. In the wild type, the pharyngeal precursors congregate at the ventral midline, where they ingress, beginning at the 4–8E stage (Fig. 2G). In pha-4 embryos, ingression is delayed or aborted so that most of these cells remain on or near the ventral surface. In addition, whereas wild-type pharyngeal precursors cluster into a column as they gastrulate, this process usually fails in pha-4 mutants (Fig. 2H). All other embryonic blastomeres behave normally at this time, including midgut cells (4/4 embryos lineaged completely to the 8E stage). Thus, pha-4 activity is required for the proper migration and possibly adhesion of the pharyngeal precursors by the 4–8E stage.
In the absence of pha-4 function, pharyngeal cells adopt an ectodermal fate. Staining with an antibody that recognizes LIN-26, a zinc-finger protein expressed in epidermis and neuronal support cells (Labouesse et al. 1994, 1996), shows that excess numbers of LIN-26+ cells are made in pha-4 mutants (M. Labouesse and S. Mango, unpubl.; see also Chanal and Labouesse 1997). To determine the source of these cells, we allowed individual blastomeres to develop after all other blastomeres were killed with a laser microbeam. This experiment demonstrated that excess LIN-26+ cells were made by isolated ABa and MS blastomeres, whereas their sister cells, ABp and P2, were unaffected (Fig. 3). Therefore, cells that would become part of the pharynx in a wild-type embryo, appeared to be transformed into LIN-26+ ectodermal cells in a pha-4 mutant. We confirmed this conclusion by following the late cell lineages of ABa and MS in pha-4 embryos. The terminal cell divisions and programmed cell deaths were altered in the cell lineages that would normally generate pharyngeal cells, suggesting that these cells no longer had pharynx identity. For example, in wild-type embryos, ABaraapapaa and ABaraapppaa divide to generate one daughter that dies and one pharyngeal neuron (Sulston et al. 1983). In pha-4 embryos, ABaraapapaa and ABaraapppaa arrested without undergoing the terminal cell division (2/2 embryos followed). In addition, no programmed cell deaths were observed for ABalpaaapap, ABarapaaapp, or MSaaappa (2/2 embryos examined for each cell).
The new cell lineage patterns observed in pha-4 embryos did not resemble a different branch of the wild-type lineage. Normally, individual sublineages can be identified by the total number of cell divisions within the lineage (e.g., 8, 9, or 10 for AB-derived blastomeres), the pattern of programmed cell deaths and the morphology of individual cells. In pha-4 embryos, AB-derived cells divided nine times and had few programmed cell deaths, unlike any specific sublineage in wild-type embryos. Thus, pha-4 mutant cells lost their pharyngeal lineage characteristics without adopting a pattern that mimicked another wild-type lineage. This result is consistent with pha-4’s involvement in establishing pharyngeal fate rather than a particular cell lineage.
pha-4 can specify pharyngeal fate
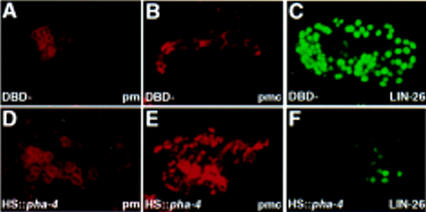
Expression of ectopic pha-4 is sufficient to confer pharynx identity to embryonic blastomeres. We used a heat shock promoter (Jones et al. 1986; A. Fire, pers. comm.) to express PHA-4 throughout the embryo. By Nomarski optics, all manipulated embryos arrested with a large pharynx-like organ, bounded by a basement membrane. In addition, these embryos failed to undergo normal morphogenesis and contained fewer epidermal cells or body wall muscles. We confirmed these observations by staining for pharyngeal muscles and marginal cells, which are generated by multiple branches of the lineage tree in wild-type animals (Sulston et al. 1983). Most heat-treated embryos contained at least 50% more pharyngeal muscle cells (Fig. 4A,D) and three times more pharyngeal marginal cells (Fig. 4B,E) compared to the wild type. Conversely, the number of LIN-26+ epidermal and neuronal support cells was strongly reduced after heat shock, as was the number of body wall muscles (Fig. 4C,F; Materials and Methods). These effects were only observed with an intact pha-4 gene and were not seen after heat shock of wild-type embryos or embryos carrying a control plasmid expressing a modified pha-4 construct that lacks the DNA binding domain (Fig. 4).
Figure 4.
Ectopic PHA-4 specifies pharyngeal identity. Embryos expressing full-length pha-4 (D–F) or pha-4 missing the DBD (A–C; DBD−) under control of the heat shock promoter, were stained with the pharyngeal muscle antibody 3NB12 (A,D; pm), the pharyngeal marginal cell antibody αIF (B,E; pmc), or anti-LIN-26 ectodermal antisera (C,F; Labouesse et al. 1996).
Not all embryonic cells were transformed into pharyngeal cells by ectopic pha-4 (Materials and Methods; Fig. 4). The incomplete transformation probably reflects two phenomena. First, cells may only be competent to respond to exogenous pha-4 for a brief period of time. For example, we observed the strongest effects when pha-4 was induced by the mid-4E stage. Induction after the 8E stage, on the other hand, gave rise to healthy L1 larvae with no obvious defects (n > 25 embryos for each stage; data not shown). A second reason for the partial transformation is that PHA-4 activity may be modulated by positive and negative cofactors that are each present in only a subset of embryonic blastomeres. Existence of cofactors could also explain the apparent paradox that ectopic pha-4 can confer pharyngeal identity, but the endogenous gene is normally expressed in the midgut and rectum. We propose that one or more cofactors cooperate with PHA-4 to promote pharynx development, another dampens PHA-4 activity in midgut precursors, and a third functions with PHA-4 to establish rectal cells. This hypothesis is consistent with our observation that midgut development is not inhibited by ectopic pha-4.
Exogenous pha-4 is sufficient to down-regulate LIN26 expression. During normal pharynx development, PHA-4 and LIN-26 are not found in the same cell or cell lineage (Labouesse et al. 1996; this work). Therefore, it is unlikely that PHA-4 initiates pharynx formation by repressing lin-26, which is essential for epidermal and glial cell development (Labouesse et al. 1994, 1996). To test this idea, we examined the phenotype of embryos lacking both pha-4 and lin-26. Pharynx development was not restored in mutants carrying null mutations in both genes, suggesting that repression of lin-26 by PHA-4 is not sufficient for pharynx development (see Materials and Methods). One facet of maintaining pharynx identity, however, may be the continual repression of lin-26. Because pha-4 encodes a DNA-binding protein, it is possible that PHA-4 represses lin-26 transcription directly. Interestingly, there are two consensus binding sites for HNF-3 within the lin-26 promoter, and these sites are conserved in lin-26 from Caenorhabditis briggsae (P. Chanal and M. Labouesse, unpubl.).
How might pha-4 govern pharynx fate? One idea is that pha-4 regulates the proper positioning and migration of cells, and the new location of cells in pha-4 mutants interferes with normal pharyngeal development. Alternatively, pha-4 might specify pharynx fate directly, and the loss of pharyngeal identity in pha-4 mutants lead to altered behaviors. Two lines of evidence support the latter hypothesis. First, most embryos with severe defects in morphogenesis and cell placement still form a pharynx (e.g., Ahnn and Fire 1994; Labouesse 1997; Schnabel and Priess 1997). Second, the ectopic pharyngeal cells seen in our heat shock experiments are often located in regions of the embryo that do not normally produce pharyngeal cells. Thus, normal patterns of cell migration, adhesion, and ingression depend on correct pharynx fate specification by pha-4.
Compartmentalization of the gut during development
Our studies suggest that the subdivision of the digestive tract into foregut, midgut, and hindgut is evolutionarily ancient and relies on conserved molecular mechanisms. We have shown that pha-4 encodes an HNF-3 homolog necessary to specify pharyngeal and rectal precursors. In other organisms, HNF-3 proteins have been implicated in foregut and hindgut development, the equivalent of the nematode pharynx and rectum. In Drosophila fork head mutants, for example, foregut and hindgut cells are transformed into head sclerite structures, whereas in vertebrates, the absence of HNF-3β leads to severe abnormalities in the foregut (for review, see Kaufmann and Knochel 1996). Moreover, the nematode nkx2.5 homolog ceh-22 is expressed in foregut muscle, similar to its murine and Drosophila counterparts (Okkema and Fire 1994; Harvey 1996 and references therein; Okkema et al. 1997), and end-1, a GATA factor involved in nematode midgut development, resembles the Drosophila midgut regulator serpent (Reuter 1994; Rehorn et al. 1996; Zhu et al. 1997). The conserved patterns of expression and phenotype for these three genes strongly support the notion that the mechanisms that compartmentalize the digestive tract have been phylogenetically conserved.
The anatomy of the foregut, midgut, and hindgut differs substantially from animal to animal. This observation implies that homologs of pha-4 and end-1 regulate different target genes in different animals, either directly or indirectly. Few targets have been identified, so it is unclear at what point the pathways diverge. Genes such as ceh-22 (Mango et al. 1994a; J. Kalb, K. Lau, B. Goszczynski, T. Fukushige, D. Moons, P. Okkema, and J. McGhee, pers. comm.) and K07C11.10, a paired-box homolog (A. Chisolm, pers. comm.; M. Domeier, and S. Mango, unpubl.), are good candidates to be regulated by pha-4/HNF-3 proteins in many animals. On the other hand, no obvious match to lin-26 has been identified in another species, nor to pha-1, another gene required for C. elegans pharynx formation and potential PHA-4 target (Schnabel and Schnabel 1990). In Drosophila, the HNF-3 homolog fork head represses trachealess to promote salivary gland development (Kuo et al. 1996). At least two trachealess homologs exist in C. elegans, one of which we have inactivated by RNA interference. No pharyngeal phenotype was observed (M. Domeier and S. Mango, unpubl.). From this short list of genes it is impossible to generalize about how HNF-3 proteins produce different specialized structures in different animals. However, it is interesting to note that both trachealess and lin-26 appear to be repressed by HNF-3 homologs. In rats, HNF-3β blocks activation of aldolase B transcription directly, by binding to the promoter (Gregori et al. 1993, 1994). These data suggest that PHA-4 and its homologs may function as transcriptional repressors, as well as activators, to control developmental fates.
Experiments performed half a century ago described the ability of dissociated organ rudiments to selectively recognize and adhere to cognate organ cells (Townes and Holtfreter 1955; Moscona 1957). This behavior constitutes a cellular description of organ identity. However, the underlying molecular mechanisms were unknown. We propose that pha-4 is an essential component of a transcription-based process that endows pharyngeal cells with organ identity. Regulators of organ identity may act combinatorially with factors that control positional, blastomere, or cell type identity to build complex organs and tissues.
Materials and methods
Genetics
C. elegans strains were maintained as described (Brenner 1974). Mutations and chromosomal balancers used were dpy-2(e489) lin-26(mc1), unc-4(e120), mnC1 LGII, unc-101(sy108) LGIV, fog-2(q71), stu-3(q265), rol-9(sc148), pha-4(q490), pha-4(q487), pha-4(q500), and pha-4(n2498) LGV.
For the lin-26;pha-4 double mutant analysis, embryos were collected from dpy-2 lin-26 unc-4/mnC1; fog-2 pha-4(q490)/stu-3 rol-9 mothers, aged, and stained for LIN-26, PHA-4, P granules, and 3NB12. About 15% of embryos produce one to three extra 3NB12+ cells in single pha-4 mutants (n = 120) as well as double pha-4;lin-26 embryos (n = 22).
Molecular biology
RNA interference was performed as described (Guo and Kemphues 1995) using pE38, which is a nearly full-length pha-4 cDNA. Fifty percent of injected mothers (n = 56) gave rise to arrested progeny, all of which lacked a pharynx by visual inspection. Fewer 3NB12+ pharynx muscles and excess LIN-26+ ectoderm were made by the progeny.
Microinjection rescue (Mello et al. 1991) was performed using YAC Y79B4, cosmids, or a PCR fragment amplified from genomic DNA and containing the equivalent of nucleotides 30–16,703 of F38A6.
To sequence pha-4 mutations, exons and flanking splice sites were amplified from genomic DNA obtained from unhatched pha-4 embryos. Purified PCR products were sequenced by the ABI method. To confirm the original mutation, the relevant fragment was reamplified and sequenced on both strands. Primer sequences are available upon request.
For ectopic PHA-4 expression, the cDNA corresponding to the longest pha-4 mRNA (pJM33; Azzaria et al. 1996) was inserted into pPD49.78 to generate plasmid pML422. The control plasmid pML423 was obtained by deleting the putative PHA-4 DNA-binding domain (DBD) from pML422. The resulting protein has the sequence IRRHGKVKEREPS across the DNA-binding region. N2 animals were injected (Mello et al. 1991), and integrated lines established. Embryos were placed at 33°C for 30 min and returned to normal growth conditions. The age of the embryos was determined by visual inspection. More than 95% of embryos carrying plasmid pML423 developed normally after heat shock, whereas >80% of the embryos carrying pML422 failed to undergo morphogenesis and displayed the phenotypes discussed in the text. Antibody staining confirmed that PHA-4 was expressed throughout the embryo for at least 10 hr after heat shock.
The average numbers of 3NB12+ pharyngeal muscles were 40 ± 16 after ectopic PHA-4 expression (n = 76), 18.3 ± 1.7 in wild-type heat shock-treated embryos (n = 16), and 18.2 ± 1.6 when the DBD was deleted (n = 21). There were 29 ± 10 pharyngeal marginal cells in the strain carrying HS::pha-4 (n = 21) compared to 9.4 ± 0.9 for the DBD mutant (n = 27). The numbers of LIN-26+ cells were 45 ± 42.0 after ectopic PHA-4 expression (n = 81), 131.2 ± 6.3 in wild-type heat shock-treated embryos (n = 16), and 126.4 ± 5.0 with the DBD deleted (n = 21). We obtained 28 ± 14 (n = 29) body wall muscles using HS::pha-4 as compared to 81 ± 6 (n = 14) with the DBD deleted control.
Antibody production and staining
A peptide consisting of the 18 carboxy-terminal amino acids [(C)VYQNTLYSSTNPNSAANL] was conjugated to KLH and used to generate affinity-purified rabbit polyclonal antibodies by Quality Controlled Biochemicals. In situ antibody staining was performed as described (Mango et al. 1994a,b). Antibodies used were α-LIN-26; pharyngeal muscle antibodies 3NB12 and 9.2.1; the intermediate filament antibody αIF, which recognizes pharyngeal marginal cells; the adherens junction antibody MH27; the body wall muscle antibody NE8-3C6; and J126, which detects pharyngeal glands, midgut, and rectal–intestinal valve cells (Mango et al. 1994b; Miller and Shakes 1995; Labouesse et al. 1996; S. Strome, pers. comm.).
Embryo manipulations
Time-lapse recordings were carried out as described (Schnabel et al. 1997). Wild-type or fog-2 pha-4(q490)/stu-3 rol-9 embryos, were recorded until the 1.5-fold stage. We followed all cells to the 350-cell stage in two N2 embryos, three Pha-4 embryos, and one non-Pha-4 embryo (i.e., pha-4/+ or +/+). For two Pha-4 embryos, we observed the complete cell lineage pattern for cells that would normally follow a pharyngeal fate. Pha-4 embryos were identified by the absence of a pharynx, which is easily scored by late embryogenesis. To generate the images in Figure 2, G and H, we noted the positions of all cells every 15–20 min using SIMI BioCell software, which can reconstitute the tridimensional aspect of the embryo (Schnabel et al. 1997).
Individual cells were destroyed with a laser microbeam as described (Sulston and White 1980; Avery and Horvitz 1987; Mango et al. 1991) except that the Micropoint laser system (Photonic Instruments) was used. After ablation, embryos were allowed to mature and then stained. The genotype of individual operated embryos was determined by staining for pharyngeal muscle (ABa or MS; 3NB12) or for rectal valve cells (ABp; J126). The genotype of P2-derived cells could not be determined; however, 25% of these embryos are predicted to be pha-4.
Acknowledgments
We thank J. Kalb and J. McGhee for their generosity with reagents and unpublished information and B. Bamber, E. Jorgensen, S. Prescott, H. Roehl, and J. Yost for thoughtful comments on the manuscript. M.L. is grateful to R. Schnabel and J. Vonesch for help setting up the 4D-microscope. We thank P. Reid, C. Thummel, and J. Vonesch for use of their confocal microscopes; A. Coulson, S. Chissoe, A. Fire, and Y. Kohara for numerous DNA clones; C. Mello for rol-9(zu156); the Caenorhabditis Genetics Center for nematode strains, and the Utah Health Sciences Sequencing Facility for DNA analysis. S.E.M. was supported by the Huntsman Cancer Institute Center for Children and the March of Dimes. M.L was funded by CNRS, INSERM, Hopital Universitaire de Strasbourg, and by grants from the European Economic Community (program HCM) and the Ministère de l’Education Nationale de la Recherche et de la Technologie (program ACC-SV4). J.K. was supported by the Howard Hughes Medical Institute and grants from the National Institutes of Health, National Science Foundation, and American Cancer Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL smango@genetics.utah.edu; FAX (801) 581-7796.
References
- Ahnn J, Fire A. A screen for genetic loci required for body-wall muscle development during embryogenesis in Caenorhabditis elegans. Genetics. 1994;137:483–498. doi: 10.1093/genetics/137.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell. 1987;51:1071–1078. doi: 10.1016/0092-8674(87)90593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzaria M, Goszczynski B, Chung MA, Kalb JM, McGhee JD. A fork head/HNF-3 homolog expressed in the pharynx and intestine of the Caenorhabditis elegans embryo. Dev Biol. 1996;178:289–303. doi: 10.1006/dbio.1996.0219. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanal P, Labouesse, M. M. A screen for genetic loci required for hypodermal cell and glial-like cell development during Caenorhabditis elegans embryogenesis. Genetics. 1997;146:207–226. doi: 10.1093/genetics/146.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplan C. Eye development: Governed by a dictator or a junta? Cell. 1997;91:861–864. doi: 10.1016/s0092-8674(00)80475-4. [DOI] [PubMed] [Google Scholar]
- Greenwald I. Development of the vulva. In: Riddle DL, et al., editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 519–542. [PubMed] [Google Scholar]
- Gregori C, Kahn A, Pichard AL. Competition between transcription factors HNF1 and HNF3, and alternative cell-specific activation by DBP and C/EBP contribute to the regulation of the liver-specific aldolase B promoter. Nucleic Acids Res. 1993;21:897–903. doi: 10.1093/nar/21.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Activity of the rat liver-specific aldolase B promoter is restrained by HNF3. Nucleic Acids Res. 1994;22:1242–1246. doi: 10.1093/nar/22.7.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Han M. Gut reaction to wnt signaling in worms. Cell. 1997;90:581–584. doi: 10.1016/s0092-8674(00)80517-6. [DOI] [PubMed] [Google Scholar]
- Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- Jones D, Russnak RH, Kay RJ, Candido EP. Structure, expression and evolution of a heat shock gene locus in Caenorhabditis elegans that is flanked by repetitive elements. J Biol Chem. 1986;261:12006–12015. [PubMed] [Google Scholar]
- Kaufmann E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Kuo YM, Jones N, Zhou B, Panzer S, Larson V, Beckendorf SK. Salivary duct determination in Drosophila: Roles of the EGF receptor signaling pathway and the transcription factors Fork head and Trachealess. Development. 1996;122:1909–1917. doi: 10.1242/dev.122.6.1909. [DOI] [PubMed] [Google Scholar]
- Labouesse M. Deficiency screen based on the monoclonal antibody MH27 to identify genetic loci required for morphogenesis of the Caenorhabditis elegans embryo. Dev Dyn. 1997;210:19–32. doi: 10.1002/(SICI)1097-0177(199709)210:1<19::AID-AJA3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Labouesse M, Sookhareea S, Horvitz HR. The Caenorhabditis elegans gene lin-26 is required to specify the fates of hypodermal cells and encodes a presumptive zinc-finger transcription factor. Development. 1994;120:2359–2368. doi: 10.1242/dev.120.9.2359. [DOI] [PubMed] [Google Scholar]
- Labouesse M, Hartwieg E, Horvitz HR. The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development. 1996;122:2579–2588. doi: 10.1242/dev.122.9.2579. [DOI] [PubMed] [Google Scholar]
- Mango SE, Maine EM, Kimble J. Carboxy-terminal truncation activates glp-1 protein to specify vulval fates in Caenorhabditis elegans. Nature. 1991;352:811–815. doi: 10.1038/352811a0. [DOI] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994a;120:3019–3031. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- Mango SE, Thorpe CJ, Martin PR, Chamberlain SH, Bowerman B. Two maternal genes, apx-1 and pie-1, are required to distinguish the fates of equivalent blastomeres in the early Caenorhabditis embryo. Development. 1994b;120:2305–2315. doi: 10.1242/dev.120.8.2305. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Shakes DC. Immunofluorescence microscopy. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern biological analysis of an organism. Vol. 48. San Diego, CA: Academic Press; 1995. pp. 365–394. [Google Scholar]
- Moscona A. The development in vitro of chimeric aggregates of dissociated embryonic chick and mouse cells. Proc Natl Acad Sci. 1957;43:184–194. doi: 10.1073/pnas.43.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Fire A. The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development. 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Ha E, Haun C, Chen W, Fire A. The Caenorhabditis elegans NK-2 homeobox gene ceh-22 activates pharyngeal muscle gene expression in combination with pha-1 and is required for normal pharyngeal development. Development. 1997;124:3965–3973. doi: 10.1242/dev.124.20.3965. [DOI] [PubMed] [Google Scholar]
- Rehorn K, Thelen H, Michelson AM, Reuter R. A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development. 1996;122:4023–4031. doi: 10.1242/dev.122.12.4023. [DOI] [PubMed] [Google Scholar]
- Reuter R. The gene serpent has homeotic properties and specifies endoderm versus ectoderm within the Drosophila gut. Development. 1994;120:1123–1135. doi: 10.1242/dev.120.5.1123. [DOI] [PubMed] [Google Scholar]
- Schnabel H, Schnabel R. An organ-specific differentiation gene, pha-1, from Caenorhabditis elegans. Science. 1990;250:686–688. doi: 10.1126/science.250.4981.686. [DOI] [PubMed] [Google Scholar]
- Schnabel R, Priess J. Specification of cell fates in the early embryo. In: Riddle DL, et al., editors. Caenorhabditis elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 361–382. [PubMed] [Google Scholar]
- Schnabel R, Hutter H, Moerman D, Schnabel H. Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: Variability of development and regional specification. Dev Biol. 1997;184:234–265. doi: 10.1006/dbio.1997.8509. [DOI] [PubMed] [Google Scholar]
- Schuchardt A, D’Agati AV, Larsson-Blomberg L, Costantini F, Pachnis V. Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature. 1994;367:380–383. doi: 10.1038/367380a0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, White JG. Regulation and cell autonomy during postembryonic development of Caenorhabiditis elegans. Dev Biol. 1980;78:577–597. doi: 10.1016/0012-1606(80)90353-x. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Townes PL, Holtfreter J. Directed movements and selective adhesion of embryonic amphibian cells. J Exp Zool. 1955;128:53–120. doi: 10.1002/jez.a.114. [DOI] [PubMed] [Google Scholar]
- Zhu J, Hill RJ, Heid PJ, Fukuyama M, Sugimoto A, Priess JR, Rothman JH. end-1 encodes an apparent GATA factor that specifies the endoderm precursors in Caenorhabditis elegans embryos. Genes & Dev. 1997;11:2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]