Abstract
Our recent work has shown that activation of the Ras/Raf/ERK pathway extends the half-life of the Myc protein and thus enhances the accumulation of Myc activity. We have extended these observations by investigating two N-terminal phosphorylation sites in Myc, Thr 58 and Ser 62, which are known to be regulated by mitogen stimulation. We now show that the phosphorylation of these two residues is critical for determining the stability of Myc. Phosphorylation of Ser 62 is required for Ras-induced stabilization of Myc, likely mediated through the action of ERK. Conversely, phosphorylation of Thr 58, likely mediated by GSK-3 but dependent on the prior phosphorylation of Ser 62, is associated with degradation of Myc. Further analysis demonstrates that the Ras-dependent PI-3K pathway is also critical for controlling Myc protein accumulation, likely through the control of GSK-3 activity. These observations thus define a synergistic role for multiple Ras-mediated phosphorylation pathways in the control of Myc protein accumulation during the initial stage of cell proliferation.
Keywords: Myc, Ras, ERK, GSK-3, Ser 62, Thr 58, stability
Considerable effort devoted to elucidating the intracellular signaling pathways that control cellular proliferation has identified the Ras and Myc proteins as two critical components that are key for the control of normal cell growth. Moreover, various studies of oncogenic events that disrupt normal cell growth regulation have revealed the frequent alteration of Myc and Ras and have further suggested a synergistic relationship between these two activities. However, despite these efforts, the precise manner in which the pathways controlled by these two proteins might interact to synchronously regulate cell growth is still poorly understood.
The ras proto-oncogene encodes a small GTP-binding protein that plays a critical role in cell growth control as a central component of mitogenic signaling events (White et al. 1995). Ras activation initiates a complex array of signal transduction pathways including the Raf/MAPK (ERK) pathway, primarily involved in plasma membrane-to-nucleus signaling crucial for mitogen-induced cell proliferation (Seger and Krebs 1995; Lavoie et al. 1996), the PI3 kinase/AKT pathway, which is involved in cell survival signaling (Kauffmann-Zeh et al. 1997), the Rac/Rho pathway, involved in cytoskeletal remodeling (Lamarche et al. 1996), and the Rac/JNK and Rac/p38 pathways, both of which appear to be involved in cell stress responses, growth inhibition, and apoptotic signals (Coso et al. 1995; Minden et al. 1995; Xia et al. 1995). Activation of Ras-signaling pathways has been shown to be essential for cells both to leave a quiescent state and to pass through G1 phase of the cell cycle (Peeper et al. 1997).
c-Myc is the most ubiquitous and best studied member of a family of proteins that includes N-Myc, L-Myc, S-Myc, and B-Myc. The N terminus of Myc proteins contains the transcriptional activation domain, within which are two 20–amino acid segments termed Myc boxes 1 and 2 that are conserved in most Myc family proteins and appear in most cases to be crucial for all biological activities (Sakamuro and Prendergast 1999). The C terminus of Myc includes the basic/helix–loop–helix/leucine zipper (b/HLH/Z) motif that mediates oligomerization with the small b/HLH/Z partner protein Max and sequence-specific DNA recognition of E-box motifs (Luscher and Larsson 1999). The Max protein also acts as a heterodimeric partner for the Mad family of b/HLH/Z proteins that form transcriptional repressors on the same E-box sequence elements and that can antagonize Myc function (Foley and Eisenman 1999). While Max is ubiquitously and constitutively expressed, both Myc and Mad expression is tightly regulated in relation to cell growth; Myc levels are high in cycling cells but decrease as cells cease to proliferate and differentiate, and Mad expression follows the opposite pattern. Thus, precise regulation of the levels of Myc and Mad expression is critical to determine the formation of either Myc/Max or Mad/Max heterodimers and consequently cell growth or inhibition, respectively.
A variety of studies demonstrate that tight regulation of Myc protein levels is essential for normal cell function. Whereas homozygous deletion of myc genes results in embryonic lethality (Charron et al. 1992; Davis et al. 1993), constitutive overexpression of Myc proteins in cultured cells as well as in transgenic animals blocks differentiation, induces neoplastic transformation, and can initiate apoptosis (Coppola and Cole 1986; Evan et al. 1992). Moreover, a wide variety of naturally occurring tumors exhibit both chromosomal translocations and amplification of the c-myc locus that result in constitutive overexpression of Myc proteins (Cole 1986; Spencer and Groudine 1991). Perhaps some of the best evidence demonstrating the important role that fluctuations in Myc protein levels play in Myc function comes from studies in mice carrying inducible myc transgenes. It was observed that enforced expression of c-Myc in either skin or hematopoietic lineages in transgenic mice leads to neoplastic premalignant and malignant phenotypes, respectively, but when Myc expression is turned off in these systems, spontaneous regression of the neoplastic and malignant changes occurs (Felsher and Bishop 1999; Pelengaris et al. 1999).
Numerous studies have documented the growth-regulated accumulation of myc RNA (Kelly et al. 1983; Luscher and Eisenman 1990) resulting from increases in myc gene transcription and an increase in myc RNA stability (Jones and Cole 1987; Luscher and Eisenman 1990). However, posttranslational control of Myc protein levels has also recently been shown to contribute to the regulated accumulation of Myc activity. Myc protein exhibits an extremely short half-life, ∼30 min in growing cells (Hann and Eisenman 1984; Ramsay et al. 1986), because of its regulated destruction via the ubiquitin/26S proteasome pathway (Ciechanover et al. 1991; Flinn et al. 1998; Gross-Mesilaty et al. 1998; Salghetti et al. 1999). Moreover, Myc turnover by the ubiquitin/proteasome system appears to be regulated. It was shown that Myc half-life is decreased during erythroleukemia cell differentiation (Spotts and Hann 1990), and our recent work has demonstrated stabilization of Myc protein following stimulation of cell growth in vivo (Sears et al. 1999). In further experiments we demonstrated that the serum-induced increase in Myc half-life levels was mediated by Ras activation, involving the Raf/ERK kinase pathway and leading to inhibition of Myc ubiquitin-mediated degradation. Taken together, these results show that the increase in Myc protein normally observed following growth stimulation as cells enter the G1-phase of the cell cycle reflects both an increase in mRNA synthesis and a concomitant Ras-mediated stabilization of the newly translated Myc protein. Our results provide one mechanism to explain the synergistic activity of Ras and Myc for cell growth and transformation.
We now show that Myc protein stability is regulated by two adjacent N-terminal phosphorylation sites, Thr 58 and Ser 62, that exhibit opposing roles in the control of Myc protein stability. We find that phosphorylation of Ser 62 is involved in stabilizing Myc, while phosphorylation of Thr 58, which is dependent on prior Ser 62 phosphorylation, promotes Myc degradation through the ubiquitin/proteasome pathway. Moreover, we show that phosphorylation at these sites is regulated by mitogens and can be controlled by two Ras effector pathways, Raf/ERK and PI-3K/AKT. Our data points to a precise Ras-mediated regulatory mechanism controlling Myc protein stability and accumulation.
Results
N-terminal phosphorylation sites affect Myc protein stability
Our recent work has demonstrated a role for Ras-activated ERK kinases in mediating the stabilization of Myc protein (Sears et al. 1999). One mechanism for this stabilization could involve a direct phosphorylation of Myc by ERK. Indeed, many important cell cycle regulatory proteins turned over by the ubiquitin/proteasome pathway are targeted for degradation by specific growth-regulated phosphorylation events (King et al. 1996; Hoyt 1997). There are two phosphorylation sites in the N terminus of c-Myc, Thr 58 and Ser 62, that account for the majority of induced phosphorylation following stimulation of cell proliferation (Lutterbach and Hann 1994). These sites fall within the highly conserved Myc box I region and are absolutely conserved between c-Myc, N-Myc, B-Myc, and S-Myc (Fig. 1A). Both sites can be phosphorylated by the ERK/MAPKs in vitro, although Ser 62 is a more perfect consensus site (Seth et al. 1992; Pulverer et al. 1994). Several observations suggest a potential role for these phosphorylation sites in the control of Myc protein accumulation. First, the region including these sites is a mutational hot spot found in myc alleles isolated from Burkitt's lymphomas, AIDS-associated lymphomas, and mouse plasmacytomas, with the majority of mutations involving Thr 58 (Bhatia et al. 1993; Clark et al. 1994; Smith-Sorensen et al. 1996). Second, Thr 58 is mutant in all v-Myc proteins and restoration of the wild-type threonine severely inhibits the transformation properties of v-Myc (Palmieri et al. 1983; Papas and Lautenberger 1985). Third, mutagenesis of Thr 58 to alanine dramatically potentiates focus formation in a cotransformation assay of primary rat embryo fibroblasts with Ras, while mutation at Ser 62 severely inhibited transformation (Pulverer et al. 1994).
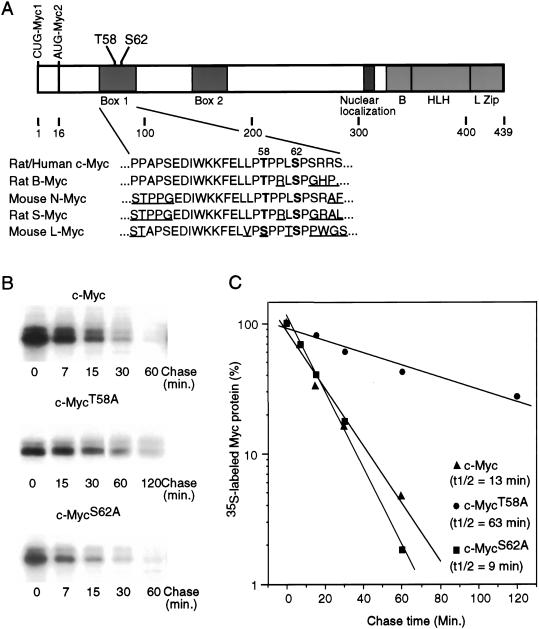
Figure 1.
Role of phosphorylation sites in determining the stability of Myc protein. (A) Schematic representation of the c-Myc protein. Translation start sites for Myc1 and Myc2 are shown. The helix–loop–helix (HLH)/leucine zipper (LZ) dimerization domains, the basic (B) DNA binding domain, the nuclear localization domain, and the two highly conserved Myc box 1 and 2 are shown. N-terminal phosphorylation sites Threonine 58 and Serine 62 are indicated. Depicted below is a comparison of the amino acid sequences surrounding the Thr 58 and Ser 62 residues in several related Myc proteins. Thr 58 and Ser 62 are in bold and divergent sequences are underlined. (B) Quiescent REF52 cells were infected with either Ad-Myc, Ad-MycT58A, or Ad-MycS62A, each at an MOI of 25. Infected cells were maintained in low serum (0.25%) medium. Eighteen hours postinfection, cells were labeled in vivo with 35S-methionine for 30 min and chased in low-serum medium containing excess unlabeled methionine for the indicated times. Labeled c-Myc was immunoprecipitated from equal cell numbers for each time point and analyzed by SDS-PAGE. (C) Immunoprecipitated 35S-labeled c-Myc from each sample shown in panel B was quantitated by PhosphorImager. Background was calculated from an equivalent area in each lane and subtracted from the value for labeled Myc in that lane. Time 0 was set at 100. Data is plotted on a semilog scale and best-fit lines were calculated using linear regression with the SigmaPlot program.
To directly examine a role for Thr 58 and Ser 62 phosphorylation in the regulation of Myc protein stability, we created recombinant adenoviruses that express mutant Myc proteins containing an alanine substitution at position 58 or 62. To measure the half-life of each of these mutant Myc proteins in quiescent cells, fibroblasts were infected with the recombinant Myc adenoviruses and incubated in low serum for an additional 16–18 h to allow for maximal expression of the adenoviral expressed proteins, and then the infected cells were pulse labeled with 35S-methionine and chased in media containing unlabeled methionine. As shown by the data in Figure 1B and C, both the wild-type c-Myc protein and the mutant MycS62A protein exhibit very short half-lives of 13 and 9 min, respectively. In contrast, the MycT58A mutant has a substantially longer half-life of 63 min, a result consistent with other recent data (Salghetti et al. 1999; Gregory and Hann 2000).
Opposing roles for Ser 62 and Thr 58 phosphorylation in determining Myc protein stability
To further explore the role of Ser 62 in the control of Myc protein stability, we have assayed the effect of growth stimulation, expression of Ras, or inhibition of proteasome function on the accumulation of the MycS62A mutant protein. Myc RNA levels expressed from the recombinant adenoviruses are not affected by these treatments (Sears et al. 1999). As shown in Figure 2A, wild-type c-Myc encoded by the Ad-Myc virus accumulates to a very low level in quiescent fibroblasts (lane 1), but is dramatically increased by either coexpression of activated Ras (lane 2), serum stimulation for 4 h (lane 4), or specific inhibition of the 26S proteasome with lactacystin (lane 3; Fenteany et al. 1995). In sharp contrast, while the accumulation of the MycS62A mutant protein is also very low in quiescent fibroblasts, it is not increased by either coexpression of activated Ras or serum stimulation (Fig. 2A, cf. lane 5 with lanes 6,8). The level of MycS62A is, however, enhanced in the presence of the proteasome inhibitor (Fig. 2A, lane 7), demonstrating that the observed low-level expression in quiescent cells is caused by rapid turnover by the ubiquitin/proteasome system.
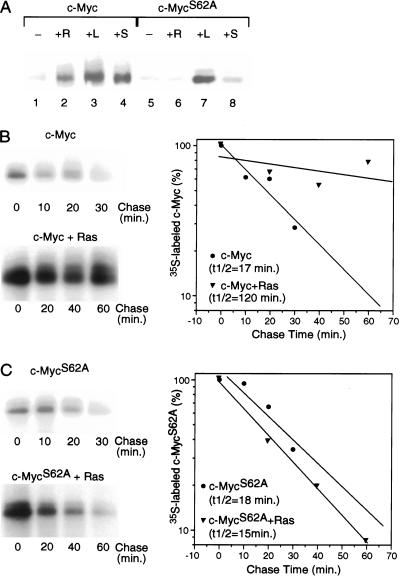
Figure 2.
Role of Serine 62 phosphorylation in Ras-mediated stabilization of Myc. (A) Quiescent REF52 fibroblasts were infected with either Ad-Myc or Ad-MycS62A (MOI = 25) together with either Ad-Ras61L (+R) or a control virus (Ad-Con; all other samples; MOI = 200). Infected cells were maintained in low-serum medium (0.25%) for an additional 18 h and then harvested for Western blot analysis. Ad-Con-infected cells were either untreated (−), treated with 20% FCS for 4 h before harvesting (+S), or treated with 10 mM lactacystin for 4 h before harvesting (+L). Western blot analysis used the C-33 monoclonal c-Myc antibody. (B) Quiescent REF52 cells were infected with Ad-Myc (MOI = 25) together with either Ad-Con (upper panel) or Ad-Ras61L (lower panel; MOI = 200). Infected cells were maintained in low-serum (0.25%) medium. Eighteen hours postinfection, cells were labeled in vivo with 35S-methionine for 30 min and chased in low-serum medium containing excess unlabeled methionine for the indicated times. Labeled c-Myc was immunoprecipitated from equal cell numbers for each time point and analyzed by SDS-PAGE. 35S-labeled c-Myc from each sample was quantitated by PhosphorImager. Background was calculated from an equivalent area in each lane and subtracted from the value for labeled Myc in that lane. Time zero was set at 100. Data is plotted on a semi-log scale and best-fit lines were calculated using linear regression with the SigmaPlot program. (C) Quiescent REF52 cells were infected with Ad-MycS62A (MOI = 25) together with either Ad-Con (upper panel) or Ad-Ras61L (lower panel). Pulse/chase analyses were performed on the infected cells and immunoprecipitated 35S-labeled Myc was quantitated as described for panel B.
We have also directly measured the half-life of the MycS62A mutant protein in the presence or absence of activated Ras in comparison with wild type. As shown in Figure 2B and C, wild-type c-Myc is stabilized by activated Ras, with a change in half-life from 17 min with control virus to 120 min with activated Ras virus. In sharp contrast, the half-life of the MycS62A protein is unchanged by the presence of activated Ras, yielding a half-life of 18 min with control virus and 15 min with Ad-Ras61L. These results strongly imply that phosphorylation at Ser 62 in c-Myc is responsible for stabilizing Myc following either Ras activation or serum stimulation and is consistent with a role for ERK in mediating this stabilization (Sears et al. 1999), as Ser 62 lies within a perfect consensus site for ERK phosphorylation.
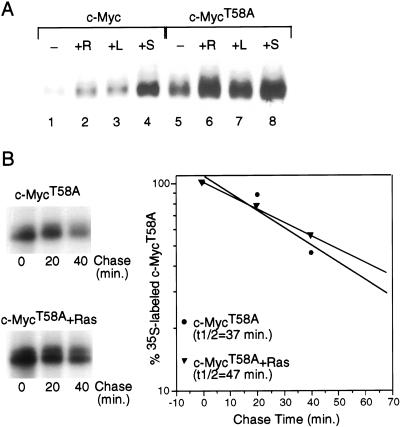
The role of Thr 58 phosphorylation in the control of Myc stability was studied in a similar manner. In contrast to results with the mutant c-MycS62A protein, we find that accumulation of the MycT58A protein is much higher than wild-type Myc under quiescent conditions (Fig. 3A, cf. lanes 1 and 5). This increased accumulation coincides with a four- to fivefold increase in the half-life of the MycT58A protein compared to wild type under serum-starved conditions (Fig. 1D). Moreover, while the accumulation of the mutant MycT58A protein is somewhat increased by coexpression of Ras, serum stimulation, or proteasome inhibition (Fig. 3A, cf. lane 5 with lanes 6–8), this effect is minor compared to the effect observed with wild-type Myc (Fig. 3A, cf. lane 1 with lanes 2–4). Likewise, pulse/chase experiments demonstrate only a small increase in the half-life of the mutant MycT58A protein in the presence of activated Ras, from 37 min in quiescence to 47 min with Ras (Fig. 3B). The half-life of wild-type Myc in this experiment was 11 min in quiescent cells and 45 min when coexpressed with Ras (data not shown). Thus, the MycT58A mutation appears to mimic Ras activation in that it results in an inhibition or substantial reduction in the normally rapid ubiquitin-mediated degradation of Myc.
Figure 3.
Role of Threonine 58 in controlling Myc stability. (A) Quiescent REF52 fibroblasts were infected with either Ad-Myc or Ad-MycT58A (MOI = 25) together with either Ad-Ras61L (+R) or Ad-Con (all other samples; MOI = 200). Infected cells were maintained in low-serum medium (0.25%) for an additional 18 h and then harvested for Western blot analysis. Ad-Con-infected cells were either untreated (−), treated with 20% FCS for 4 h before harvesting (+S), or treated with 10 mM lactacystin for 4 h before harvesting ( +L). Western blot analysis used the C-33 monoclonal c-Myc antibody. (B) Quiescent REF52 cells were infected with Ad-MycT58A (MOI = 25), together with either Ad-Con (upper panel) or Ad-Ras61L (lower panel; MOI = 200). Infected cells were maintained in low-serum (0.25%) medium. Eighteen hours postinfection, cells were labeled in vivo with 35S-methionine for 30 min and chased in low-serum medium containing excess unlabeled methionine for the indicated times. Labeled c-MycT58A was immunoprecipitated from equal cell numbers for each time point and analyzed by SDS-PAGE. 35S-labeled c-MycT58A from each sample was quantitated by PhosphorImager. Background was calculated from an equivalent area in each lane and subtracted from the value for labeled Myc in that lane. Time 0 was set at 100. Data is plotted on a semi-log scale and best-fit lines were calculated using linear regression with the SigmaPlot program.
On the basis of these results, we conclude that Ser 62 is required for stabilization of Myc in response to Ras, whereas Thr 58 is required for the normal degradation of Myc. The opposing roles that phosphorylation at either Ser 62 or Thr 58 play in Myc protein stability could provide a molecular explanation for previous results in which mutation at Thr 58 resulted in a more oncogenic protein, while mutation of Ser 62 yielded a Myc protein with severely compromised transforming ability (Pulverer et al. 1994).
Changes in the phosphorylation status of Thr 58 and Ser 62 in c-Myc correlate with changes in Myc protein stability
To further elucidate the roles that phosphorylation at Thr 58 and Ser 62 might play in controlling Myc protein stability, we performed two-dimensional phosphopeptide mapping to examine the phosphorylation status of these two sites in wild-type c-Myc under various cell growth conditions where Myc protein stability is altered. These experiments relied heavily on previous experiments that have mapped these phosphorylation sites in 32P-labeled c-Myc (Lutterbach and Hann 1994). Quiescent REF52 cells were infected with the Ad-Myc recombinant adenovirus plus either the control virus Ad-Con or the Ad-Ras61L virus. Where indicated, cells infected with control virus were serum stimulated with 20% FCS for 4 h before harvesting. Cells were then in vivo labeled with 32P-orthophosphate, and equal counts of radio-labeled c-Myc was digested with thermolysin and subjected to two-dimensional separation.
Typical results from phosphopeptide mapping experiments are shown in Figure 4. The positions of thermolysin cleavage sites within the c-Myc sequence surrounding Thr 58 and Ser 62 are depicted in Figure 4A. The phosphopeptide profile of wild-type c-Myc expressed under various cell growth conditions is shown in Figure 4, panel B. The four spots, a, b, c, and e, are labeled according to corresponding spots in phosphopeptide maps described by Lutterbach and Hann (1994) and represent the phosphorylated thermolytic peptides diagrammed in panel A. The composition of each of the labeled spots in the phosphopeptide maps was determined by additional experiments including two-dimensional analysis of Myc proteins with mutations at Thr 58 or Ser 62, secondary digest of isolated peptides with proline endoprotease and subsequent two-dimensional analysis, and acid hydrolysis of specific spots to determine the phosphoserine and phosphothreonine content (data not shown). Spots c and b, which fall along a diagonal line characteristic of different phosphoisoforms of the same peptide, represent the thermolytic peptide from amino acids 56 to 68, containing either one phosphate at Ser 62 (spot c) or two phosphates at Ser 62 and Thr 58 (spot b). Spot e also lies along this diagonal and most likely represents the same thermolytic peptide with three phosphates at Ser 62, Ser 64, and Ser 67. Hann and colleagues reported that spot e is eliminated on mutation of Ser 64/67 (Lutterbach and Hann 1994), and we have been unable to detect any phosphothreonine in this spot. Spot a represents another thermolytic fragment from amino acids 69 to 74, with a single phosphorylation likely at Ser 71.
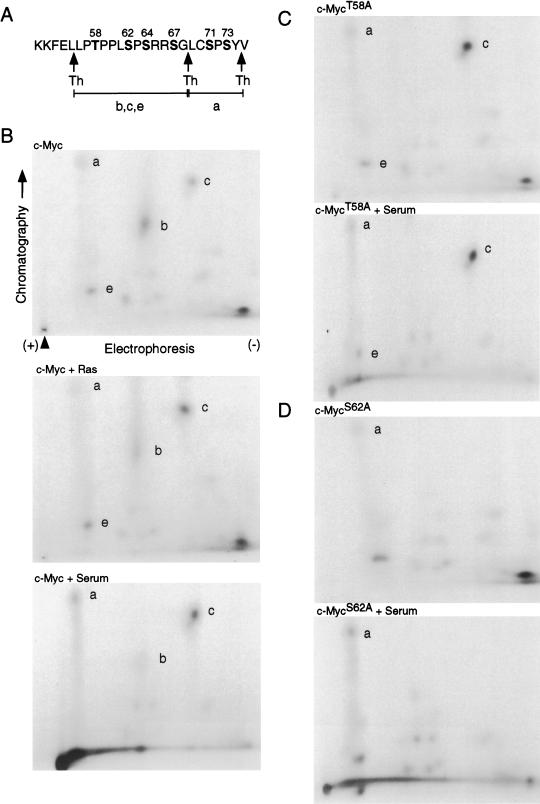
Figure 4.
Phosphopeptide mapping of c-Myc under various cell growth conditions. (A) Amino acid sequence of c-Myc between residues 51 and 75. Threonine and serine residues are in bold type and numbered. Thermolysin cleavage sites are indicated with arrows. Thermolytic peptides shown in the phosphopeptide maps are indicated (a,b,c,e). (B) Quiescent REF52 cells were infected with Ad-Myc (MOI = 25) together with either Ad-Ras61L (c-Myc + Ras) or Ad-Con (all other samples; MOI = 200). Infected cells were maintained in low-serum medium for 18 h and then in vivo labeled with 1 mCi/mL 32Pi for 2 h and harvested. Where indicated, Ad-Con-infected cells were stimulated with 20% FCS for 4 h before harvesting (c-Myc + Serum). 32P-labeled c-Myc was immunoprecipitated from cell lysates, fractionated by SDS-PAGE, transferred to Immobilon P membrane, and digested off the membrane with thermolysin. Thermolytic fragments were recovered and subject to two-dimensional separation. First-dimension electrophoresis was performed in buffer pH 1.9, and ascending chromatography was in phosphochromatography buffer. Chromatography and electrophoresis directions are shown, and the origin is indicated by an arrowhead. The same conditions were used for all two-dimensional maps shown. Thermolysin-digested phosphopeptides a, b, c, and e, described in panel A, are indicated. (C) Quiescent REF52 cells were infected and treated as described for panel B, except that Ad- Myc was replaced with Ad-MycT58A. Infected cells were in vivo labeled with 32Pi, and labeled MycT58A was subjected to thermolysin cleavage and phosphopeptide mapping as described for panel B. (D) Quiescent REF52 cells were infected and treated as described for panel B, except that Ad-Myc was replaced with Ad-MycS62A. Infected cells were in vivo labeled with 32Pi, and labeled MycS62A was subjected to thermolysin cleavage and phosphopeptide mapping as described for panel B.
By comparing the three different maps for wild-type Myc shown in Figure 4, panel B, quiescent cells (upper panel), coexpression of activated Ras (middle panel), and 4 h of serum stimulation (lower panel), it is clear that the phosphorylation status of the thermolytic fragment encompassing Thr 58 and Ser 62 changes with changing growth conditions that are known to alter Myc protein stability. Specifically, spot c, which represents a single phosphorylation at Ser 62, is enhanced in wild-type Myc with either coexpression of activated Ras or serum stimulation, both of which increase Myc stability, as compared to quiescent cells. In contrast, spot b, which represents double phosphorylation at Ser 62 and Thr 58, is markedly decreased in wild-type Myc in the presence of activated Ras or serum compared to quiescence, suggesting that secondary phosphorylation at Thr 58 is repressed under conditions that increase Myc stability. We also note that spot e is decreased with serum stimulation. It is not clear what the significance of this is except to suggest that a single phosphorylation at Ser 62 predominates under conditions where Myc protein is stable. We have not explored the importance of phosphorylations at Ser 64 or 67.
We also performed phosphopeptide mapping on both the mutant MycT58A and MycS62A proteins under various cell growth conditions in parallel with the analysis of the wild-type protein. A comparison of the phosphopeptide map of the MycT58A protein with wild-type Myc under quiescent conditions (Fig. 4B,C, upper maps) reveals two specific differences. Most obvious is the fact that spot b is missing in the MycT58A phosphopeptide maps. This is not surprising as spot b appears to represent a peptide doubly phosphorylated at Thr 58 and Ser 62, which cannot exist when Thr 58 is mutated. The second difference is that spot c is enhanced in the MycT58A mutant compared to wild-type Myc under quiescent conditions. Hann and colleagues made a similar observation with the same Myc mutant (Lutterbach and Hann 1994). The increase in spot c is interesting as the mutant MycT58A protein is inherently stable, and spot c is enhanced in wild-type Myc under conditions where it is stable. In addition, an examination of the phosphopeptide map of MycT58A following serum stimulation (Fig. 4C, lower panel) reveals that in contrast to wild-type Myc, spot c from the MycT58A protein is not enhanced by serum stimulation or coexpression of Ras (data not shown), consistent with the fact that the MycT58A protein stability is not significantly changed with either of these cell growth conditions. Thus, the analysis of both wild-type c-Myc and the mutant MycT58A protein indicates that an increase in the proportion of Myc protein that is singly phosphorylated on Ser 62 correlates with stable Myc protein, and mutation of Thr 58 to alanine mimics the effects of mitogen stimulation on both Myc stability and the phosphorylation status of Ser 62.
Phosphopeptide mapping of the MycS62A protein shows a loss of both spots b and c under quiescent conditions, as well as in the presence of serum (Fig. 4D) or activated Ras (data not shown). This result suggests that in the absence of Ser 62 there is no singly or doubly phosphorylated form of the thermolytic fragment spanning amino acids 56–68, which would mean that phosphorylation of Thr 58 requires prior phosphorylation of Ser 62 and that phosphorylation of Ser 64/67 does not exist alone. A similar observation was reported by Hann and others, who first suggested a hierarchical ordering to phosphorylation at Ser 62 and Thr 58 (Lutterbach and Hann 1994). We also note that there is a spot in a similar position to spot e that is unlikely to be a triple phosphorylation of the thermolytic fragment aa 56–68, since in the absence of Ser 62 and Thr 58 phosphorylation there are not enough sites left to make a triple phosphorylated peptide. The significance of this spot is not known.
Myc degradation is controlled by the hierarchical phosphorylation of Ser 62 and Thr 58
The phosphopeptide analysis of the mutant MycS62A protein suggests a hierarchical ordering to phosphorylation of Ser 62 and Thr 58 whereby phosphorylation of Thr 58 requires prior phosphorylation at Ser 62. We have now used two phospho-specific peptide antibodies that recognize Myc protein phosphorylated at either Thr 58 or Ser 62 to more clearly demonstrate this hierarchy. An antibody raised against a Myc peptide doubly phosphorylated at Ser 62 and Thr 58 (P-S62/P-T58; New England Biolabs) fortuitously recognizes Myc that is singly phosphorylated at Thr 58 and doubly phosphorylated at Thr 58 and Ser 62 but does not recognize Myc that is singly phosphorylated at Ser 62 or that is unphosphorylated (data not shown); we refer to this as α-Myc(T58-P). Conversely, a second peptide-purified antibody was produced, directed specifically to a Myc peptide singly phosphorylated at Ser 62, that recognizes the doubly phosphorylated Myc as well as Myc singly phosphorylated at Ser 62 but does not recognize Myc either singly phosphorylated at Thr 58 or unphosphorylated Myc (data not shown); we refer to this as α-Myc(S62-P). Thus, while the specificity of these two antibodies overlaps in terms of their recognition of doubly P-T58/P-S62-phosphorylated Myc, they have the ability, when used together, to distinguish Myc protein that is singly phosphorylated at Thr 58 or Ser 62.
Using these antibodies, we performed Western blot analysis of wild-type Myc, MycT58A, or MycS62A under quiescent conditions and in the presence of the proteasome inhibitor lactacystin to allow Myc to accumulate. As shown in Figure 5A, wild-type Myc is phosphorylated on both Ser 62 (upper panel, lane 1) and Thr 58 (middle panel, lane 1) when expressed in quiescent cells. Interestingly, only the α-Myc (T58-P) antibody was able to recognize slower migrating bands of Myc that accumulate in the presence of lactacystin (middle panel, bracketed), likely representing ubiquinated forms of Myc. Analysis of the MycT58A protein shows that it is phosphorylated on Ser 62. Moreover, the enhanced phosphorylation of Ser 62 in the MycT58A mutant under quiescent conditions compared to wild-type Myc, observed in the phosphopeptide maps, is also evident in this analysis (Fig. 5, cf. lanes 1 and 2, upper panel). In contrast, the MycS62A protein is not phosphorylated on Thr 58 when expressed in quiescent conditions or in the presence of activated Ras (data not shown), clearly demonstrating the requirement of Ser 62 phosphorylation for Thr 58 to become phosphorylated.
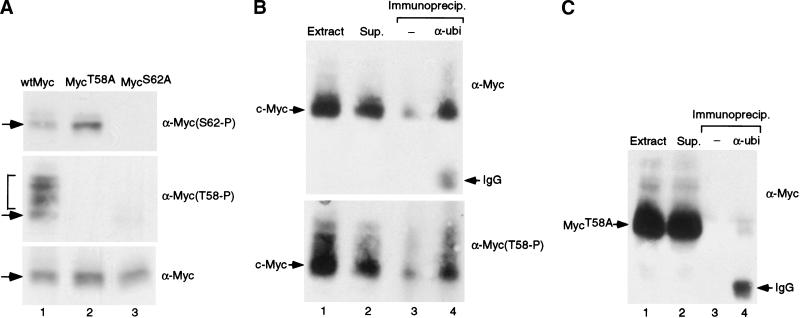
Figure 5.
Hierarchical phosphorylation of Serine 62 and Threonine 58 residues controls c-Myc ubiquitin-mediated degradation. (A) Quiescent REF52 cells were infected with either Ad-Myc, Ad-MycT58A, or Ad-MycS62A (MOI = 25). Infected cells were maintained in starvation medium for an additional 18 h. Four hours before harvesting, infected cells were treated with lactacystin. Each sample was analyzed by Western blotting with three separate antibodies: the monoclonal C-33 c-Myc antibody (lower panel), a polyclonal phosphospecific antibody that recognizes phosphorylated Thr 58 (middle panel), and a polyclonal phosphospecific antibody that recognizes Ser 62 (upper panel). Overlapping bands are indicated with arrows. Slower-mobility Myc bands are bracketed. (B) Quiescent REF52 cells were infected with Ad-Myc (MOI = 50). Infected cells were maintained in starvation medium for an additional 20 h. Six hours before harvesting, infected cells were treated with lactacystin. Ubiquitinated proteins were immunoprecipitated from harvested cell lysates with an anti-Ubiquitin antibody (Zymed; lane 4) or control extracts were mock treated and precipitated with proteinA/+G beads alone (lane 3). Immunoprecipitates along with one-tenth volume of the original extract (lane 1) or one-tenth volume of the supernatant following immunoprecipitation with anti-Ubi (lane 2) were all subjected to Western blot analysis with either the C-33 c-Myc antibody (upper panel) or the Thr 58 phosphospecific antibody (lower panel). C-Myc protein is indicated along with the IgG heavy chain from the anti-Ubi antibody. (C) Quiescent REF52 cells were infected with Ad-MycT58A (MOI = 50). Infected cells were treated as described in panel B. Ubiquitinated proteins were immunoprecipitated from harvested cell lysates, and Western blot analysis with the C-33 c-Myc antibody was performed on extract, supernatant, and immunoprecipitates as described for panel B.
Our data strongly indicate a role for the phosphorylation of Thr 58 in promoting Myc degradation. As Myc is degraded by the ubiquitin proteasome pathway, we have examined the relationship of Myc Thr 58 phosphorylation and ubiquitination. Quiescent REF52 cells were infected with Ad-Myc and incubated with lactacystin for 6 h before harvesting. Ubiquitinated Myc was then immunoprecipitated with an α-ubi antibody and visualized with either the monoclonal αMyc antibody or the αT58-P phosphospecific antibody. As shown in Figure 5B, wild-type Myc could be specifically immunoprecipitated with the α-ubi antibody (lane 4, upper panel). Importantly, the α-ubi-immunoprecipitated Myc is phosphorylated on Thr 58 (lower panel, lane 4). In contrast to wild-type Myc, the mutant MycT58A protein is not precipitated by the α-ubi antibody (Fig. 5C, lane 4), indicating that the targeting of Myc for ubiquitination is dependent on phosphorylation at Thr 58 at least when Ser 62 is wild type. Taken together, these data indicate that c-Myc degradation is controlled by a progression of phosphorylation events from phosphorylation at Ser 62 to Thr 58 phosphorylation leading to ubiquitin-mediated destruction.
Distinct Ras effector pathways, Raf/ERK and PI-3K/AKT, regulate Myc protein degradation
The phosphorylation of Ser 62, resulting in stabilization of the Myc protein, is likely mediated by ERK kinase. In particular, Ser 62 is within a perfect ERK consensus sequence and the ERK inhibitor PD098059 blocks Ras-mediated Myc stabilization (Sears et al. 1999). A candidate for the kinase responsible for Thr 58 phosphorylation is the glycogen synthase kinase, GSK-3. Previous work has shown that both isoforms of GSK-3 can phosphorylate c-Myc on Thr 58 and Ser 62 in vitro (Lutterbach and Hann 1994; Pulverer et al. 1994). However, it is likely that Thr 58 is the preferred in vivo target for GSK-3 as it lies within a better consensus site and GSK-3 appears to normally act in a processive fashion, phosphorylating sites adjacent to previously phosphorylated amino acids (DePaoli-Roach 1984). A role for GSK-3 in phosphorylation of Thr 58, and thus in promoting Myc degradation, is intriguing as GSK-3 activity is regulated by the PI-3K/AKT effector arm of the Ras pathway (Cross et al. 1995), thus providing a second mechanism for Ras to control Myc protein stability.
To test for the involvement of the PI-3K signaling pathway in the regulation of Myc stability, we made use of the chemical inhibitor Wortmannin, which specifically inactivates PI-3K but has no effect on MAPKs (Ui et al. 1995). Quiescent REF52 cells were infected with Ad-Myc together with either a control virus or the activated Ras virus, Ad-Ras61L. Eight hours postinfection, cells were either mock treated or treated with either the chemical inhibitor PD098059 to block activation of ERK kinases (Dudley et al. 1995) or Wortmannin to block PI-3K activity. Ten hours later cells were harvested and the amount of Myc accumulation was measured by Western blot analysis. As we have observed previously, activated Ras enhances Myc accumulation (Fig. 6A, cf. lanes 1and 2, and lanes 4 and 5) and treatment with the PD098059 inhibitor severely impairs Ras-mediated stabilization of Myc (Fig. 6, cf. lanes 2 and 3). Treatment with Wortmannin also specifically inhibits the ability of Ras to increase Myc accumulation at all doses tested. Wortmannin had no effect on the accumulation of Myc protein in the absence of Ras activation (data not shown). Thus, inhibition of PI-3K activity also impairs the ability of Ras to enhance Myc accumulation, indicating that both the ERK pathway and the PI-3K pathway are involved in controlling Myc protein levels.
Figure 6.
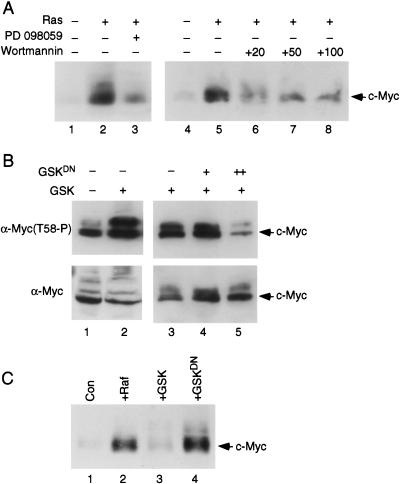
Role of ERK and GSK-3 in Myc phosphorylation and stability. (A) Quiescent REF52 cells were infected with Ad-Myc (MOI = 25) together with either Ad-Ras61L (Ras, +) or Ad-Con (Ras, −; MOI = 200). Infected cells were maintained in low-serum medium for an additional 18 h. Where indicated, the MEK inhibitor PD 098059 (lane 3, 10 μM), or the PI-3K inhibitor Wortmanin (lanes 6–8, at the indicated mM concentrations), were added for 10 h before harvesting. Western blot analysis was performed on cell lysates with the C-33 c-Myc antibody. (B) Quiescent REF52 cells were infected with Ad-Myc (MOI = 25) together with Ad-Con (lane 1, MOI = 20; lane 3, MOI = 500), Ad-GSK (lanes 2–5, MOI = 20), and Ad-GSKDN (lanes 4,5, MOI = 100[+] or 500[++]). Infected cells were maintained in starvation medium for 20 h and then harvested for Western blot analysis with either the C-33 c-Myc antibody (lower panel) or the phosphospecific Thr 58-P antibody (upper panel). (C) Quiescent REF52 cells were infected with Ad-Myc (MOI = 25), together with either Ad-Con (lane 1), Ad-RafCAAX (lane 2), Ad-GSK (lane 3), or Ad-GSKDN (lane 4; MOI = 200). Infected cells were maintained in low-serum medium and harvested for Western blot analysis 18 h later with the C-33 c-Myc antibody.
The role of PI-3K could be explained through an effect on GSK-3 activity. To more directly study a role for GSK-3 in phosphorylating Thr 58 and affecting Myc protein stability, we have made use of two recombinant adenovirus vectors, one that expresses wild-type GSK-3, which is constitutively active in the absence of inhibitory phosphorylation by AKT, and the other expressing a GSK-3DN mutant that is catalytically inactive and has been shown to function as a dominant negative mutant (Dominguez et al. 1995; Diehl et al. 1998). As shown by the data in Figure 6B, coinfection of Ad-Myc with the Ad-GSK-3 virus in quiescent REF52 cells leads to enhanced phosphorylation of Myc at Thr 58, as measured by Western blotting with the Thr 58 phosphospecific antibody (Fig. 6, cf. lanes 1 and 2, upper panel). Moreover, coexpression of increasing amounts of the dominant negative GSK-3 mutant protein together with GSK3 inhibits phosphorylation at Thr 58 (Fig. 6, cf. lane 3 with lanes 4 and 5, upper panel). The lower panel in Figure 6B shows the total amount of Myc, which does not significantly change in this experiment as cells were treated with the proteasome inhibitor Lactacystin to increase and normalize the expression level of Myc.
We also examined Myc protein accumulation following infection with Ad-Myc and either control viruses, Ad-RafCAAX (which expresses a mutant activated Raf kinase), Ad-GSK-3, or Ad-GSK-3DN. As we have reported previously, Myc accumulation in serum-starved cells is very low, whereas in the presence of activated Raf, Myc is stabilized and accumulates to high levels (Fig. 6C, cf. lanes 1 and 2; Sears et al. 1999). In contrast to activation of the Ras/Raf/ERK kinase pathway that stabilizes Myc, expression of active GSK-3 kinase does not significantly affect the level of Myc accumulation in quiescent fibroblasts (Fig. 6C, cf. lanes 1 and 3). However, inhibition of endogenous GSK-3 activity in serum-starved cells with the GSK-3DN mutant results in a dramatic increase in Myc accumulation (Fig. 6C, cf. lanes 1 and 4). This result suggests that the rapid turnover of Myc protein in quiescent cells is dependent on endogenous GSK-3 activity, which is constitutively active under these conditions. In summary, our data supports a dual mechanism for Ras-activated Myc stabilization. Ras activation, via the Raf/ERK kinases or via the p13K/AKT-mediated inactivation of GSK-3 kinase, can enhance Myc protein stability and allows high-level Myc accumulation (Fig. 6C, lanes 2,4).
GSK-3 activity is required for the normal down-regulation of Myc protein accumulation
To further explore the specific role of GSK-3 in regulating Myc protein levels following a growth response, we have analyzed the kinetics of endogenous Myc protein accumulation in the presence or absence of GSK-3 activity. After growth stimulation of normal fibroblasts, Myc protein rapidly accumulates because of both an increase in myc gene transcription and an increase in the stability of the newly synthesized Myc protein. Myc protein accumulation reaches maximal levels approximately 4 h after mitogen stimulation and then decreases to half-maximal levels in mid-G1; this level of Myc is then maintained as the cell continues to grow. The initial accumulation of Myc can be attributed in part to the stabilizing effect of ERK-mediated phosphorylation, as demonstrated by the fact that the PD098059 inhibitor dramatically reduces the accumulation of endogenous Myc protein 4 h after serum stimulation without affecting myc RNA levels (Sears et al. 1999). Conversely, the subsequent decline in Myc protein could be in part caused by GSK-3-mediated phosphorylation of Thr 58. GSK-3 activity is initially inhibited by the action of AKT but then becomes active as AKT activity subsides. At this time, GSK-3 could phosphorylate Myc at Thr 58 and trigger its degradation. If correct, we would expect that the inhibition of GSK-3 activity during a growth response would result in a sustained elevated level of Myc protein following the stimulation of cell growth. To test this possibility, we have made use of the GSK-3DN mutant.
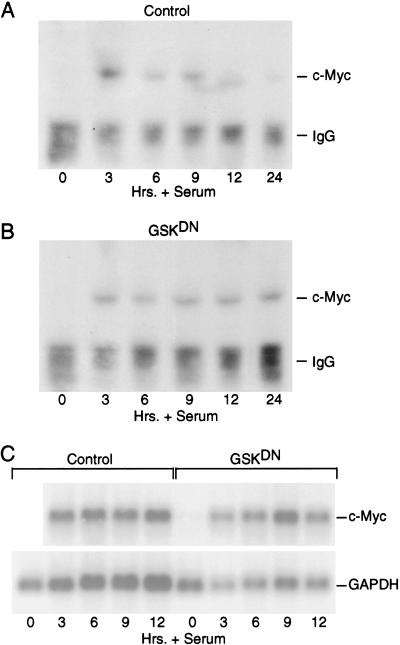
Briefly, quiescent REF52 fibroblasts were either infected with control virus or Ad-GSK-3DN. Eighteen hours later, cells were serum stimulated with 20% FCS and endogenous Myc was concentrated by immunoprecipitation and then visualized by Western blot analysis at various times following stimulation. As shown in Figure 7A, the accumulation of endogenous Myc protein follows the expected pattern in control cells. In contrast, while inhibition of GSK-3 activity did not alter the initial accumulation of Myc, it did prevent the subsequent decline, resulting in the maintenance of Myc at elevated levels through 24 h after growth stimulation (Fig. 7B). Myc RNA expression was not affected by the expression of the GSK-3DN protein compared to control cells (Fig. 7C).
Figure 7.
GSK-3 regulates c-Myc accumulation following a growth response. (A) Quiescent REF52 cells were infected with Ad-Con (MOI = 200). Infected cells were maintained in starvation medium for 15 h and then stimulated with 20% FCS. Cells were harvested at the indicated times after serum stimulation, and endogenous c-Myc from cell lysates was concentrated by immunoprecipitation with the C-33 c-Myc antibody. Immunoprecipitated c-Myc was then visualized by Western blot analysis with the same C-33 Myc antibody. C-Myc protein and the IgG heavy chain from the immunoprecipitation are indicated. (B) Quiescent REF52 cells were infected with Ad-GSK-3DN (MOI = 200). Infected cells were treated as described for panel A. Endogenous c-Myc from each time point was immunoprecipitated and visualized as described for panel A. (C) Quiescent REF52 cells were infected with Ad-Con (control) or AGSKDN (MOI = 200). Infected cells were treated as described for panel A. Poly A+ RNA was isolated from approximately 300 μg of total RNA from each time point and subjected to Northern blot analysis. The blot was probed for c-Myc and GAPDH (loading control) as indicated.
On the basis of these results, we propose that the normal accumulation of c-Myc protein following the stimulation of cell growth represents the concerted action of at least two Ras-dependent phosphorylation events—a role for ERK in augmenting the initial burst in Myc accumulation and a role for GSK-3 in targeting Myc degradation.
Discussion
Our previous work has pointed to a role for Ras-mediated control of Myc protein accumulation resulting from the action of the Raf/ERK pathway in promoting the stabilization of Myc (Sears et al. 1999). The data presented here now demonstrate that the stabilizing effect of Raf/ERK action coincides with the phosphorylation of Ser 62 in the N-terminal domain of Myc. In addition, our results show that there is a second N-terminal phosphorylation site, Thr 58, that has an opposing effect on Myc stability, targeting Myc for ubiquitin-mediated proteasome degradation. Moreover, our experiments also demonstrate that the role of Ras in controlling Myc protein accumulation is complex, with a second Ras effector pathway involving PI-3K/AKT also playing an important role in the control of Myc stability apparently by down-regulating the kinase GSK-3 that phosphorylates Thr 58 and promotes Myc degradation.
Distinct roles for two Ras pathways in the control of Myc stability
Our previous finding of a role for Ras activation of ERKs in mediating the stabilization of Myc can now be seen as the direct consequence of phosphorylation of Ser 62. The amino acid sequence surrounding Ser 62 represents a consensus ERK recognition sequence, and other work has provided evidence that ERK can mediate the phosphorylation of Myc at Ser 62 (Seth et al. 1992; Lutterbach and Hann 1994; Pulverer et al. 1994). The data we present here show that mutation of Ser 62 prevents mitogen- and Ras-induced stabilization of Myc. Moreover, phosphorylation at Ser 62 is enhanced under conditions where Myc is stabilized. The importance of Ser 62 in the control of Myc stability is seen not only from our experiments, which demonstrate a strict requirement for the stabilization of Myc by Ras, but also from other work that has demonstrated an impaired transforming function when Ser 62 is altered (Pulverer et al. 1994).
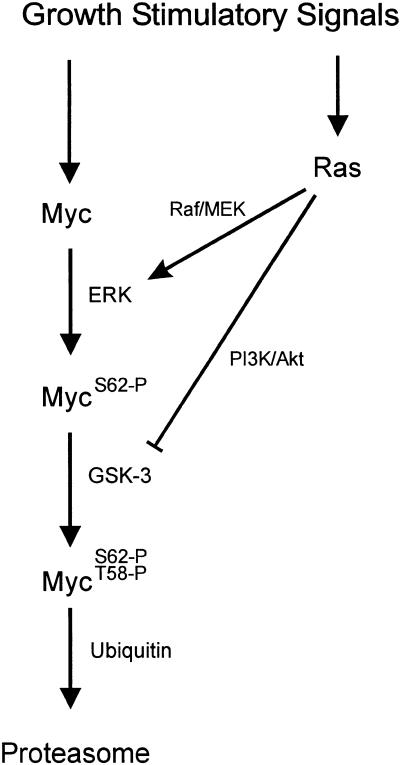
In contrast, phosphorylation at Thr 58 coincides with a decreased stability of Myc and mutations that prevent Thr 58 phosphorylation lead to stable Myc protein. Once again, this coincides with other work that has shown that alteration of Thr 58 enhances the transforming activity of Myc (Pulverer et al. 1994) and that mutations at this site are common in Myc proteins derived from tumors (Bhatia et al. 1993; Smith-Sorensen et al. 1996). Various lines of work suggest that the GSK-3 protein kinase is most likely responsible for the phosphorylation of Myc at Thr 58. Thr 58 lies within an established consensus, and GSK-3 has been shown to phosphorylate Thr 58 in Myc in vitro (Lutterbach and Hann 1994; Pulverer et al. 1994). However, unlike ERK, which is tightly regulated by cell growth, the level of GSK-3 protein is constant and does not fluctuate with cell growth. Nevertheless, despite the continual presence of GSK-3 protein, the activity of the kinase is regulated during the initial phase of cell proliferation (He et al. 1995). In particular, GSK-3 activity is inhibited through the action of PI-3K/AKT (Cross et al. 1995). Thus, as Ras initiates the PI-3K/AKT pathway, GSK-3 activity is held in check, preventing the phosphorylation of Thr 58. Only when AKT activity declines would GSK-3 then have the capacity to phosphorylate Thr 58 to induce the degradation of Myc. Thus, Ras activation elicits two responses within the cell that can cooperate to enhance Myc stability: a direct effect of ERK and an indirect effect of AKT (Fig. 8).
Figure 8.
Pathways controlling Myc phosphorylation and accumulation.
A mechanism to insure a transient accumulation of Myc
While activation of Ras results in stable Myc protein and an increase in Myc protein levels in early G1, proper control of Myc function requires this induction in Myc to be transient such that Myc protein levels decline as the cell continues to proliferate. The interplay between the two Ras effector pathways (Raf and PI-3K), coupled with the opposing effects of Thr 58 and Ser 62 phosphorylation on Myc stability and the hierarchical nature of the phosphorylation at Ser 62 and Thr 58, creates a mechanism that insures a transient accumulation of Myc following the stimulation of cell growth. In short, growth stimulation leads to a rapid activation of Ras and accumulation of both Raf/ERK and PI-3K/AKT kinase activities. Induction of Myc gene transcription follows shortly, such that the initial Myc product would be stabilized through the ERK-mediated phosphorylation of Ser 62. The phosphorylation of Ser 62 also primes Myc for degradation, as the ability of GSK-3 to phosphorylate Thr 58 is dependent on the prior phosphorylation of Ser 62. But GSK-3 activity would be held in check by the action of AKT. Then, as Ras activity subsides, resulting in a decline of AKT activity, GSK-3 would become active, leading to phosphorylation of Thr 58 and thus triggering Myc degradation.
The mechanistic basis for how these phosphorylation events actually control Myc degradation remains to be determined. Certainly, our data and that of others points to a mechanism involving ubiquitin-mediated degradation by the proteasome (Flinn et al. 1998; Gross-Mesilaty et al. 1998; Salghetti et al. 1999). Moreover, other work has clearly demonstrated a role for phosphorylation in triggering recognition of target proteins by the ubiquitination machinery, particularly those proteins regulated in the cell cycle (King et al. 1996; Hoyt 1997). Our data show that ubiquitinated Myc is phosphorylated on Thr 58. As such, one possibility is that phosphorylation of Thr 58 allows Myc to be recognized by a ubiquitin targeting enzyme, thus directing degradation by the proteasome. Nevertheless, it is likely that the actual mechanism is more complex, as mutation of Ser 62, which prevents phosphorylation at Thr 58, generates an unstable Myc protein that is degraded by the ubiquitin/proteasome pathway. In addition, Myc proteins that lack the first 100 amino acids (c-Mycs) have been shown to be degraded by the ubiquitin system (Gregory and Hann 2000). Thus, in the absence of Ser 62 phosphorylation, rapid degradation of Myc appears not to require Thr 58 phosphorylation.
Our data also indicate that high–molecular mass forms of Myc, which accumulate in the presence of a proteasome inhibitor, may be singly phosphorylated on Thr 58. This observation suggests the possibility that Myc degradation may not be triggered by Thr 58 phosphorylation per se, but rather by a subsequent dephosphorylation of Ser 62. Our data, and those of others, also show that mutation of Thr 58 results in an increase in Ser 62 phosphorylation, which could, in fact, be the mechanism behind the stability of Myc proteins that are mutant at Thr 58. The enhanced Ser 62 phosphorylation of Myc proteins mutant at Thr 58 also indicates that Thr 58 may in fact facilitate Ser 62 dephosphorylation and, in this way, play a positive role in promoting Myc degradation. In any case, our data demonstrate that at least two events appear to be associated with the control of Myc protein stability, phosphorylation of Ser 62, which protects Myc from degradation, and phosphorylation of Thr 58, which promotes Myc degradation. Our data demonstrate that at least two events appear to be associated with the control of Myc protein stability—phosphorylation of Ser 62, which protects Myc from degradation, and phosphorylation of Thr 58 that promotes Myc degradation. The Thr 58 phosphorylation appears to override the protective effects of Ser 62 phosphorylation, perhaps by triggering Ser 62 dephosphorylation, and subsequent Myc ubiquitination and degradation.
Tight regulation of Myc protein turnover is essential for normal cell function
Elevated Myc expression is observed in a wide variety of human tumors (Nesbit et al. 1999). In many cases this is caused, at least in part, by gene amplification and, in the case of Burkhitt's lymphoma and AIDS-associated lymphomas, involves a translocation of c-myc gene to the immunoglobulin loci. In addition, point mutations are frequently found in the coding region of c-myc both in primary tumors and in the cell lines derived from these lymphomas. Hot spots for these mutations are found clustered around Thr 58, and many of these mutant Myc proteins have increased transforming potential associated with an inability to phosphorylate Thr 58 (Bhatia et al. 1993; Henriksson et al. 1993). Data presented here now provide an explanation for the increased oncogenic nature of Myc that cannot be phosphorylated on Thr 58, as we and others have shown that mutation of Thr 58 creates a stable Myc protein (Salghetti et al. 1999; Gregory and Hann 2000). Moreover, recent reports have demonstrated that many mutant Myc proteins derived from Burkitt's cell lines with mutations at or around Thr 58 are stable (Gregory and Hann 2000).
The complex nature of the ordered phosphorylation of Ser 62 and Thr 58 clearly allows for very tight regulation of Myc protein levels. Given the important role that Myc plays in controlling cell growth and the oncogenic nature of Myc overexpression, it is not surprising that such a tightly regulated mechanism has evolved. It is also interesting to note that c-Jun, another important cell growth regulator that has oncogenic potential when overexpressed and has similar kinetics of protein accumulation following cell growth stimulation, contains a sequence of six amino acids that is an exact match to those found in Myc surrounding Thr 58 and Ser 62. Moreover, c-Jun has been shown to be stabilized by MAPK/ERK phosphorylation (Musti et al. 1997). Thus, the ordered phosphorylation of two adjacent phosphorylated sites controlled by ERK and GSK-3 may be a more common mechanism for the control of immediate early protein accumulation following the stimulation of cell growth.
Materials and methods
Antibodies
The C-33 c-Myc monoclonal antibody is from Santa Cruz. The ubiquitin antibody is from Zymed. The Thr 58 phospho-specific antibody is from New England Biolabs. The phospho-Ser 62–specific antisera was raised against chemically synthesized, KLH-conjugated phosphopeptide PLS (PO3) PSRRSGC (amino acids 60–68) as previously described (Kitagawa et al. 1996; Shieh et al. 1997). The antisera was further affinity purified through a phosphopeptide-conjugated Sepharose CL-4B column. To deplete antibodies that recognize unphosphorylated c-Myc, the affinity-purified antibodies were then passed through a column conjugated with corresponding unphosphorylated peptide. Purified antibodies thus obtained specifically recognized phosphorylated but not unphosphorylated peptides in ELISA test.
Cells and viruses
REF52 cells were grown in D-MEM containing 5% fetal calf serum (FBS) and 5% calf serum (CS). To bring cells to quiescence, REF52 cells were plated at ∼3500 cells/cm2 and incubated overnight. The next day the culture medium was replaced with D-MEM containing 0.25% serum (FCS/CS) and cells were incubated for an additional 48 h. Viral stocks were created and virus purified as described previously (Nevins et al. 1997). Viral titers were determined by an indirect immunofluorescent assay specific for the viral 72K E2 gene product as described (DeGregori et al. 1995). Quiescent REF52 cells were infected with virus by incubation in D-MEM with 20 mM HEPES (pH 7.2) for 75 min at 37°C at a cell to volume ratio of 6 × 105 cells/mL. After infection, four volumes of D-MEM/0.25% serum was added to each plate and the cells were further incubated at 37°C for 18–20 h before harvest and analysis. Where indicated, cells were serum stimulated by the addition of 20% FCS directly to the D-MEM/0.25% serum medium. The construction of Ad-Con, Ad-Ras61L, and Ad RafCAAX have been previously described (DeGregori et al. 1997; Leone et al. 1997; Sears et al. 1999). Ad-GSK3β was constructed by digesting a pcDNA3 plasmid containing the cDNA for GSK3β (a gift from Dr. Woodgett, Ontario Cancer Institute) with ApaI, creating a blunt end with a Klenow fragment, and then finally digesting with KpnI. The KpnI/blunt end fragment of GSK3β was then ligated into the KpnI/EcorRV sites in the AdTrack–CMV plasmid used to construct recombinant adenoviruses as described in He et al. (1995). Ad-GSKDN was similarly constructed by cloning the cDNA encoding a catalytically inactive mutant GSK-3β protein, GSK3β K85R, (a gift from Dr. Sokol, Harvard Medical School, Boston, MA; Dominguez et al. 1995) into KpnI/EcorRV-digested AdTrack–CMV. Ad-Myc was similarly constructed by ligation of a HindIII–XbaI (blunted with Klenow) fragment from the murine c-Myc cDNA from the CMV–Myc plasmid (Sears et al. 1997) into the HindIII–EcoRV sites of the AdTrack–CMV plasmid. Ad-MycT58A and Ad-MycS62A were constructed by first making point mutations in the murine c-Myc sequence in the CMV–Myc plasmid using the GeneEditor kit (Promega) per manufacturer's instructions. The oligonucleotides used in the site-directed mutagenesis are as follows: MycT58A, 5′-CTGCTTCCCGCCCCGCCCCTGTC-3′; MycS62A, 5′-CACCCCGCCCCTGGCCCCGAGC-3′; selection oligo, 5′-CGCCGGTGATATCGATCACGATGCG-3′. The mutated c-Myc cDNAs were then cloned into AdTrack–CMV and recombinant adenoviruses were created as described for Ad-Myc.
Measurement of protein stability
Infected cells were prestarved by replacing the culture media with D-MEM without L-methionine and L-cysteine for 15 min. Cells were labeled in vivo with [35S]methionine/cysteine EXPRESS protein labeling mix from Dupont-NEN using 300 μCi/mL at a cell to volume ratio of 8.7 × 105 cells/mL for 30 min. After labeling, cells were immediately washed one time with D-MEM containing 5 mM L-methionine, 3mM L-Cysteine, and 0.25% serum and then incubated in this same media for the indicated chase times. Cells were harvested and labeled; Myc proteins were immunoprecipitated as described below. Labeled Myc was visualized by autoradiography and quantitated using a PhosphorImager (Molecular Dynamics).
Immunoprecipitations
Cells were harvested by washing 1× in cold (4°C) PBS, followed by lysis in cold Myc Ab-lysis buffer (Hann et al. 1985) containing protease and phosphatase inhibitors. Myc proteins were immunoprecipitated from equal numbers of cells for each sample at a cell to volume ratio of 1.3 × 106 cells/mL lysis buffer as described previously (Sears et al. 1999) using a Myc monoclonal antibody C-33 (Santa Cruz Biotechnology). Immunoprecipitated Myc was separated by SDS-PAGE.
Western analysis
Cells were harvested by scraping in hot 1× SDS sample buffer at a cell to volume ratio of 3.25 × 106 cells/mL. Protein concentrations were determined for normalization purposes using the ABC solutions (Pierce) per manufacturer's instructions. Equal protein for each sample was separated by SDS-PAGE, blotted to Immobilon-P membrane (Millipore), and detected with HRP-conjugated secondary antibodies and immunofluorescence using the ECL reagents from Amersham per the manufacturer's instructions.
Phosphopeptide mapping
Infected cells were labeled in vivo with [32P]orthophosphate for 2 h. 32P-labeled c-Myc was immunoprecipitated with the C-33 monoclonal Myc antibody (Santa Cruz Biotechnology), fractionated by SDS-PAGE, and transferred to immobilon-P membrane. Labeled Myc protein was then digested off the membrane with 10 μg Thermolysin and lyophilized as previously described (Boyle et al. 1991; Lutterbach and Hann 1994; van der Geer and Hunter 1994). Digested Myc was oxidized with performic acid (1 h at 4°C) followed by repeated lyophilizations. Yield was estimated by Cerenkov counting. Myc peptides were resuspended in pH 1.9 buffer (Boyle et al. 1991) and equal counts were spotted onto 0.1-mm cellulose TLC plastic sheets without fluorescent indicator (EM Science). Electrophoresis in the first dimension was with a Hunter thin-layer electrophoresis chamber in pH 1.9 buffer (1.5 kV, 25 min) and ascending chromatography for the second dimension in phosphochromatography buffer for 6.5 h (Boyle et al. 1991).
Detection of ubiquitinated Myc
Cells were washed 1× with cold PBS and lysed with Ripa buffer plus the following inhibitors: 5 mM N-ethylmaleimide (NEM; Sigma), 8.6 mM iodoacetamide (IAA; Sigma), and protease and phosphatase inhibitors at a cell to volume ratio of 1.25 × 107 cells/mL. Ubiquitinated proteins were immunoprecipitated with anti-Ubiquitin antibody (Zymed). Following precipitation with the protein A/+G-agarose beads, immunoprecipitates were only washed 1× with low-stringency buffer containing PBS and all inhibitors used with the Ripa buffer as well as 0.1% NP40 (Sigma). Immunoprecipitated proteins were then separated by 8% SDS-PAGE, blotted to immobilonP membrane, and detected with the indicated Myc antibodies.
Detecting endogenous c-Myc
Quiescent REF52 cells were infected as described above (see Cells and Viruses) except that after the infection four volumes of the previously used DMEM/0.25% medium was added back to each plate. Approximately 15 h later, the media was changed to fresh DMEM/20%FCS and samples were collected at the indicated times. Endogenous c-Myc was immunoprecipitated as described under Immunoprecipitations, except cells were lysed in Ab-lysis buffer at 7.5 × 106 cells/mL and precipitates were not washed but were loaded directly onto 8% SDS-PAGE.
Northern blot analysis
Total RNA was isolated using the TRIzol Reagent (GIBCO BRL). Poly(A)+ RNA was isolated from an equal amount of total RNA (∼300 μg) with a PolyATtract mRNA isolation system (Promega) as described previously (DeGregori et al. 1995). RNAs were electrophoresed, blotted to nylon membrane, and probed with [32P]-labeled random-primed cDNAs as described (Sears et al. 1997). cDNA probes include the mouse c-Myc cDNA from the CMV–Myc plasmid (Sears et al. 1997) and the GAPDH cDNA obtained from the American Type Culture Collection.
Acknowledgments
We wish to thank Kaye Culler for assistance in the preparation of the manuscript, Laszlo Jakoi for ELIZA analysis of phospho-specific antibodies, and Dr. Bob Abraham for helpful discussion and advice. J.R.N. is an Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL j.nevins@duke.edu; FAX (919) 681-8973.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.836800.
References
- Bhatia K, Huppi K, Spangler G, Siwarski D, Iyer R, Magrath I. Point mutations in the c-Myc transactivation domain are common in Burkitt's lymphoma and mouse plasmacytomas. Nat Genet. 1993;5:56–61. doi: 10.1038/ng0993-56. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, Alt FW. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes & Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- Ciechanover A, DiGiuseppe JA, Bercovich B, Orian A, Richter JD, Schwartz AL, Brodeur GM. Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proc Natl Acad Sci. 1991;88:139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark HM, Yano T, Otsuki T, Jaffe ES, Shibata D, Raffeld M. Mutations in the coding region of c-MYC in AIDS-associated and other aggressive lymphomas. Cancer Res. 1994;54:3383–3386. [PubMed] [Google Scholar]
- Cole MD. The myc oncogene: Its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- Coppola JA, Cole MD. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986;320:760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- Coso OA, Chiariello M, Yu JC, Termamoto H, Crespo P, Xu N, Miki T, Gutkind JS. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- Cross DAE, Alessi DR, Cohen P, Andejelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes & Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis and G1/S regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaoli-Roach AA. Synergistic phosphorylation and activation of ATP–Mg-dependent phosphoprotein phosphatase by F A/GSK-3 and casein kinase II (PC0.7) J Biol Chem. 1984;259:12144–12152. [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3b regulates cyclin D1 proteolysis and subcellular localization. Genes & Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez I, Itoh K, Sokol SY. Role of glycogen synthase kinase 3b as a negative regulator of dorsoventral axis formation in Xenopus embryos. Proc Natl Acad Sci. 1995;92:8498–8502. doi: 10.1073/pnas.92.18.8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DT, Pang L, Decker SJ, Bridges AJ, Saltiel AR. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GL, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematiopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber RD. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- Flinn EM, Busch CMC, Wright APH. myc Boxes, which are conserved in myc family proteins, are signals for protein degradation via the proteasome. Mol Cell Biol. 1998;18:5961–5969. doi: 10.1128/mcb.18.10.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley KP, Eisenman RN. Two MAD tails: What the recent knockouts of Mad1 and Mxi1 tell us about the MYC/MAX/MAD network. Biochim Biophys Acta. 1999;31:1423. doi: 10.1016/s0304-419x(99)00012-8. [DOI] [PubMed] [Google Scholar]
- Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: Stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol. 2000;20:2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Mesilaty S, Reinstein E, Bercovich B, Tobias KE, Schwartz AL, Kahana C, Ciechanover A. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc Natl Acad Sci. 1998;95:8058–8063. doi: 10.1073/pnas.95.14.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann SR, Eisenman RN. Proteins encoded by the human c-myc oncogene: Differential expression in neoplastic cells. Mol Cell Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann SR, Thompson CB, Eisenman RN. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. Nature. 1985;314:366–369. doi: 10.1038/314366a0. [DOI] [PubMed] [Google Scholar]
- He X, Saint-Jeannet J-P, Woodgett JR, Varmus HE, Dawid IB. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature. 1995;374:617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- Henriksson M, Bakardjiev A, Klein G, Luscher B. Phosphorylation sites mapping in the N-terminal domain of c-Myc modulate its transforming potential. Oncogene. 1993;8:3199–3209. [PubMed] [Google Scholar]
- Hoyt MA. Eliminating all obstacles: Regulated proteolysis in the eukaryotic cell cycle. Cell. 1997;91:149–151. doi: 10.1016/s0092-8674(00)80396-7. [DOI] [PubMed] [Google Scholar]
- Jones TR, Cole MD. Rapid cytoplasmic turnover of c-myc mRNA: Requirement of the 3′ untranslated sequences. Mol Cell Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann-Zeh A, Rodriquez-Viciana P, Elrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- Kelly K, Cochran BH, Stiles CD, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35:603–609. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- King RW, Deshaies RJ, Peters JM, Kirschner MW. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Higashi H, Jung H-K, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J-Y, Segawa K, Yoshida E, Nishimura S, et al. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- Lamarche N, Tapon N, Stowers L, Burbelo PD, Aspenstrom P, Bridges T, Chant J, Hall A. Rac andn Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- Lavoie JN, L'Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOG MAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- Leone G, DeGregori J, Sears R, Jakoi L, Nevins JR. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature. 1997;387:422–426. doi: 10.1038/387422a0. [DOI] [PubMed] [Google Scholar]
- Luscher B, Eisenman RN. New light on Myc and Myb. Part I. Myc. Genes & Dev. 1990;4:2025–2035. doi: 10.1101/gad.4.12a.2025. [DOI] [PubMed] [Google Scholar]
- Luscher B, Larsson L. The basic region/helix–loop–helix/leucine zipper domain of Myc proto-oncoproteins: Function and regulation. Oncogene. 1999;18:2955–2966. doi: 10.1038/sj.onc.1202750. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Hann SR. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol. 1994;14:5510–5522. doi: 10.1128/mcb.14.8.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minden A, Lin A, Claret FX, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- Musti AM, Treier M, Borhmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- Nevins JR, DeGregori J, Jakoi L, Leone G. Functional analysis of E2F. Methods Enzymol. 1997;283:205–219. doi: 10.1016/s0076-6879(97)83017-0. [DOI] [PubMed] [Google Scholar]
- Palmieri S, Kahn P, Graf T. Quail embryo fibroblasts transformed by four v-myc isolates show enhanced proliferation but are non-tumorigenic. EMBO J. 1983;2:2385–2389. doi: 10.1002/j.1460-2075.1983.tb01750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papas TS, Lautenberger JA. Sequence curiosity in v-myc oncogenes. Nature. 1985;318:237. doi: 10.1038/318237a0. [DOI] [PubMed] [Google Scholar]
- Peeper DS, Upton TM, Ladha MH, Neuman E, Zalvide J, Bernards R, DeCaprio JA, Ewen ME. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997;386:177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: Induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- Pulverer BJ, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett JR. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- Ramsay G, Stanton L, Schwab M, Bishop JM. The human proto-oncogene N-myc encodes nuclear proteins that bind DNA. Mol Cell Biol. 1986;6:4450–4457. doi: 10.1128/mcb.6.12.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamuro D, Prendergast GC. New Myc-interacting proteins: A second Myc network emerges. Oncogene. 1999;18:2942–2954. doi: 10.1038/sj.onc.1202725. [DOI] [PubMed] [Google Scholar]
- Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: Cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Ohtani K, Nevins JR. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–179. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- Seger R, Krebs EG. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- Seth A, Gonzalez FA, Gupta S, Raden DL, Davis RJ. Signal transduction within the nucleus by mitogen-activated protein kinase. J Biol Chem. 1992;267:24796–24804. [PubMed] [Google Scholar]
- Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Smith-Sorensen B, Hijmans EM, Beijersbergen RL, Bernards R. Functional analysis of Burkitt's lymphoma mutant c-Myc proteins. J Biol Chem. 1996;271:5513–5518. doi: 10.1074/jbc.271.10.5513. [DOI] [PubMed] [Google Scholar]
- Spencer CA, Groudine M. Control of c-myc regulation in normal and neoplastic cells. Adv Cancer Res. 1991;56:1–48. doi: 10.1016/s0065-230x(08)60476-5. [DOI] [PubMed] [Google Scholar]
- Spotts GD, Hann SR. Enhanced translation and increased turnover of c-myc proteins occur during differentiation of murine erythroleukemia cells. Mol Cell Biol. 1990;10:3952–3964. doi: 10.1128/mcb.10.8.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui M, Okada T, Hazeki K, Hazeki O. Wortmannin as a unique probe for an intracellular signalling protein, phosphoinositide 3-kinase. Trends Biochem Sci. 1995;20:303–307. doi: 10.1016/s0968-0004(00)89056-8. [DOI] [PubMed] [Google Scholar]
- van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by electrophoresis and chromatography on thin-layer cellulose plates. Electrophoresis. 1994;15:544–554. doi: 10.1002/elps.1150150173. [DOI] [PubMed] [Google Scholar]
- White MA, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler MH. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]