Abstract
Although the role of miR-200s in regulating E-cadherin expression and epithelial-mesenchymal transition is well established, their influence on metastatic colonization remains controversial. Here, we use clinical and experimental models of breast cancer metastasis to discover a pro-metastatic role of miR-200s that goes beyond their regulation of E-cadherin and epithelial phenotype. Overexpression of miR-200s is associated with increased risk of metastasis in breast cancer and promotes metastatic colonization in mouse models, phenotypes that cannot be recapitulated by E-cadherin expression alone. Genomic and proteomic analyses revealed global shifts in gene expression upon miR-200 overexpression toward that of highly metastatic cells. MiR-200s promote metastatic colonization partly through direct targeting of Sec23a, which mediates secretion of metastasis suppressive proteins, including Igfbp4 and Tinagl1, as validated by functional and clinical correlation studies. Overall, these findings suggest a pleiotropic role of miR-200s in promoting metastatic colonization by influencing E-cadherin-dependent epithelial traits and Sec23a-mediated tumor cell secretome.
Keywords: miR-200, metastasis, Sec23a, secretome, epithelial-mesenchymal transition
Early events of metastatic dissemination are thought to be initiated by epithelial-mesenchymal transition (EMT) in carcinoma cells, promoting tumor cell migration and invasion1-3. While implications for EMT in cancer progression are widely recognized, the potential role of the reverse process, mesenchymal-epithelial transition (MET) is less clear. Histological analysis has revealed morphological similarities of primary tumors and metastatic lesions4,and it has been reported that E-cadherin levels are elevated in lymph node metastases relative to matched primary tumor samples, both suggesting that EMT in primary tumors may be followed by MET at distant sites5,6. Despite these correlative clinical findings, rigorous functional studies linking MET with metastatic colonization ability are scarce.
MicroRNAs (miRNAs) have been recognized as important regulators of normal and pathological processes in metazoan organisms7-9. Several miRNAs have also been shown recently to serve as promoters10-15 or suppressors16,17 of metastasis, particularly in the early step of tumor invasion. However, relatively little is known about the role of miRNAs in the late step — metastatic colonization of distant organs. The miR-200 family of miRNAs are important in neurogenesis18, regulation of embryonic and adult stem cells and cancer stem cells19-22, chemosensitivity and apoptosis23,24. Importantly, miR-200s have recently been shown to inhibit EMT and promote MET by direct targeting of E-cadherin transcriptional repressors Zeb1 and Zeb225-28. The fact that the miR-200 family has been shown to enforce the epithelial phenotype and inhibit EMT and invasion in vitro suggested that these miRNAs are likely to suppress metastasis. However, functional studies have yielded conflicting results in different models of metastasis29-31, casting doubt on the potential therapeutic utility of miR-200s. Furthermore, it remains unclear whether the metastasis-related functions of the miR-200s are mediated entirely or only partially through the Zeb-E-cadherin axis.
Here, we show that miR-200s promote metastatic colonization of breast cancer not only by influencing the cell-intrinsic epithelial traits through targeting the Zeb-E-cadherin axis but also by altering tumor cell-derived secretome through targeting of the Sec23a-mediated secretion of metastasis suppressive proteins, including Igfbp4 and Tinagl1. These findings provide novel insights into the molecular functions of miR-200s as well as the role of MET and tumor secretome in metastasis.
RESULTS
Correlation of miR-200 expression with metastatic colonization
To investigate the clinical significance of miR-200 expression (cluster 1: miR-200b/200a/429, cluster 2: miR-200c/141, Supplementary Fig. 1a), we performed a retrospective analysis on a series of breast tumor samples (n = 210, Oxford collection)32. A composite miR-200 family expression score calculated in each sample as either the median (Fig. 1a, P = 0.034) or the mean (P = 0.030) expression was significantly associated with poor distant relapse-fee survival (DRFS)33. In particular, miR-429 and -200a showed the most significant association with DRFS (Fig. 1a, P = 0.001 for miR-429 and P = 0.050 for miR-200a). The miR-200 family expression was associated with ER-positive status (P = 0.019, median expression) and correlated with poor DRFS only in the ER-positive tumors (P = 0.028, n = 122) but not ER-negative tumors (P = 0.48, n = 77). Furthermore, profiling miR-200 levels in ten human primary and lung-pleural metastasis samples, including six matched pairs (Meldola collection),revealed higher expression in metastases, reinforcing the potential role of miR-200s in metastatic colonization (Fig. 1b). These findings are consistent with a clinical correlation observed in serous ovarian carcinoma patients34 and an earlier observation in xenograft studies31.
Figure 1.
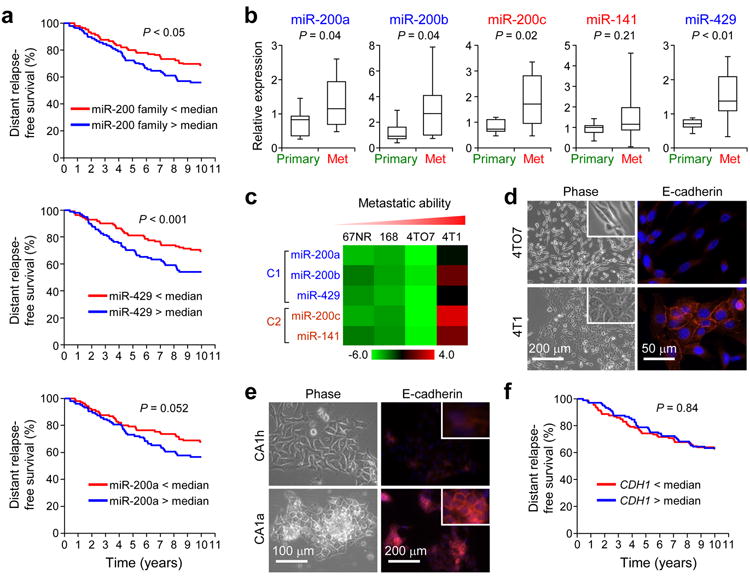
MiR-200s are associated with poor prognosis in breast cancer. (a) Kaplan-Meier curves showing the distant relapse-free survival of 210 patients with high or low expression of the entire miR-200 family (top panel), miR-429 (middle panel) and miR-200a (bottom panel) in breast tumors. P values were computed by a likelihood ratio test. (b) Box plots showing miR-200 expression levels in ten human primary and metastasis samples as assessed by qRT-PCR analysis. Data is normalized to U6 and P values were computed by Student’s t-test. (c) Heat map showing miRNA expression levels in 4T1 series. 168: 168FARN. (d) Phase contrast images (left panel) and immunofluorescence (IF) images of 4TO7 and 4T1 cells stained for E-cadherin (right panel). (e) Phase contrast images (left panel) and IF images for E-cadherin (right panel) of MCFCA1h and MCFCA1a cells. Inserts highlight the membrane localization of E-cadherin. (f) Kaplan-Meier curves showing the distant-relapse free survival of 210 patients with high or low CDH1 expression. P values were computed by a likelihood ratio test.
To further investigate the importance of miR-200s in metastasis, we profiled miR-200 expression levels in three cancer cell line series that model the progression of breast and bladder cancer. The 4T1 series, including 67NR, 168FARN, 4TO7 and 4T1, are near-isogenic mouse mammary tumor cell lines. Of these lines, only 4T1 cells are capable of spontaneously metastasizing and colonizing distant organs following orthotopic implantation35,36. We found that miR-200s showed greatest expression in the highly metastatic 4T1 cells (Fig. 1c and Supplementary Fig. 1b), which was consistent with acquisition of epithelial traits (Fig. 1d and Supplementary Fig. 2a,b) in 4T1 relative to the weakly metastatic 4TO7 cells37. Similar correlations between elevated expression of miR-200s with epithelial traits and increased metastatic ability were also made for the MCF10A human breast cancer38 (Fig. 1e and Supplementary Fig. 1b) and the TSU bladder carcinoma39 progression series (Supplementary Fig. 2c–e and Supplementary Table 1; see Supplementary Results). These observations collectively point to the possibility that MET induced by the miR-200 family may be crucial for successful completion of metastasis, particularly in the colonization step.
Since E-cadherin is an important mediator of MET, we investigated the clinical importance of E-cadherin expression in the Oxford cohort. As expected, miR-200 expression was positively correlated with E-cadherin expression (P < 0.001, Spearman correlation) and inversely correlated with Vimentin expression (P < 0.001, Spearman correlation). However, contrary to the clinical associations observed for miR-200s, E-cadherin expression alone did not have any prognostic power (Fig. 1f), suggesting that the influence of miR-200s on breast cancer metastasis goes beyond the regulation of E-cadherin and the epithelial phenotype and likely involves novel genetic targets.
MiR-200 overexpression enhances lung colonization
To directly test the functional role of miR-200s in metastasis, we stably overexpressed cluster 1 (C1 line), cluster 2 (C2 line) and clusters 1 and 2 simultaneously (C1+C2 line) in the mesenchymal-like, weakly metastatic 4TO7 cells (Supplementary Fig. 3a). We also generated E-cadherin (CDH1) overexpressing 4TO7 cells (CDH1 line) to test the importance of E-cadherin as a major downstream effector of miR-200s in metastasis. Interestingly, although ectopic expression of cluster 1 enforced expression of miRs-200b/200a/429, cluster 2 elevated expression of all five miRNAs from both clusters (Supplementary Fig. 3a), possibly due to a double-negative feedback mechanism involving Zeb factors25,40. As expected, the C1, C2 and C1+C2 lines expressed elevated levels of E-cadherin and lower levels of Zeb1 (Fig. 2a, b and Supplementary Fig. 3b,c) and adopted an epithelial-like phenotype (Fig. 2b). Interestingly, EMT inducers such as Snail and Twist, and mesenchymal markers such as N-cadherin and Vimentin, remained unaffected upon miR-200 expression (Fig. 2a,b). The CDH1 line also showed elevated expression of E-cadherin but maintained a mesenchymal morphology (Fig. 2a, b). All engineered lines mentioned possessed similar growth kinetics in vitro (data not shown) and in vivo (Supplementary Fig. 3d).
Figure 2.
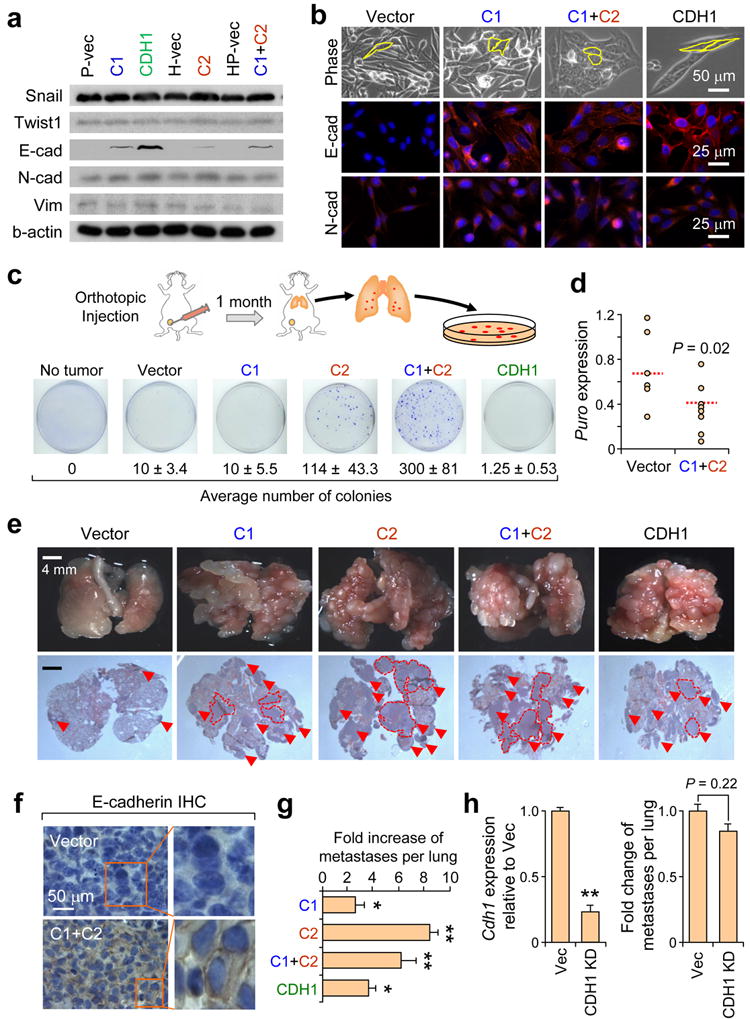
Ectopic miR-200 expression enhances spontaneous metastasis and colonization of distant organs. (a) Western blot showing expression of indicated proteins in various genetically modified 4TO7 cell lines. (b) Phase contrast and IF images of cell lines stained for E-cadherin and N-cadherin. Yellow outline emphasizes cell morphology. (c) Various cell lines were used to generate orthotopic mammary gland tumors. After one month, lungs were excised, dissociated and plated in selective media for colony formation. Average numbers of colonies were listed below representative plate images. Data represent mean ± s.e.m. from a single representative experiment out of three independent experiments. (n = 9–10). (d) Dot plot showing relative expression of puromycin resistance gene, an indicator of circulating tumor cells, by qRT-PCR analysis of genomic DNA from whole blood samples. Red lines represent median values. P = 0.02 (Student’s t-test). (e) Representative gross lung and H&E stained lung sections from animals intravenously injected with various 4TO7 cell lines. Red arrowheads and dotted lines highlight metastatic nodules. (f) Immunohistochemical staining for E-cadherin of lung nodules established from indicated cells. (g) Bar graph showing fold increase in number of pulmonary metastasis nodules for each group. Data represent mean fold increase ± s.e.m. from a single representative experiment out of three independent experiments. (n = 9–10). (h) Left: RT-PCR showing expression of Cdh1 in C1C2 cells with or without stable Cdh1knockdown. Right: Bar graph showing number of pulmonary lesions following intravenous inoculation of tumor cells. * P < 0.05, ** P < 0.01 (Student’s t-test).
Parental 4TO7 cells, when inoculated orthotopically in the mammary fat pad, spontaneously disseminate to lungs but have a very low efficiency of colonization35,36. C2 (P = 0.035) and C1+C2 (P = 0.01) lines, both of which overexpress all five members of the miR-200 family, formed 10–30 fold higher number of lung-derived tumor colonies suggesting that ectopic miR-200 expression can enhance lung colonization efficiency (Fig. 2c and Supplementary Fig. 4a,b; see Supplemental Methods), consistent with an earlier report31. Curiously, the C1 line, which expresses elevated levels of E-cadherin similar to the C2 and C1+C2 lines, was incapable of efficiently colonizing lungs (Fig. 2c, P = 0.38), implying that 1) E-cadherin is likely not the only functional downstream effector of miR-200s and 2) gene targeting mediated by all five members may be necessary to efficiently stimulate metastasis. In support of this, the CDH1 line also failed to phenocopy the C2 and C1+C2 lines (P = 0.08), confirming that miR-200-mediated E-cadherin regulation is not sufficient to promote spontaneous metastatic colonization but rather, other genetic pathways must also be involved (Fig. 2c and Supplementary Fig. 4a,b).
Since metastasis is a multi-step process, we aimed to elucidate the influence of miR-200s on early and late steps of metastasis. To determine whether invasion or intravasation was regulated by miR-200s, whole blood analysis for circulating tumor cells was performed. In contrast to the results obtained from the lung colony assays, miR-200 expression reduced tumor cell entry into circulation from primary tumors (Fig. 2d and Supplementary Fig. 5a), possibly through inhibition of EMT, migration and invasion as observed in vitro (Supplementary Fig. 5c). Similarly, there was also reduced dissemination of CDH1 cells from primary tumors (Supplementary Fig. 5b). In aggregate, these data suggested that although miR-200 expression can hinder entry of tumor cells into circulation, those that do intravasate may be more capable of colonizing distant organs.
To test this hypothesis, we inoculated cells directly into venous circulation and measured the incidence of pulmonary metastasis. Tail vein inoculation resulted in enhanced metastasis burden for all cell lines tested, with C2 (P < 10−7) and C1+C2 (P < 0.01) lines showed the greatest metastasis potential (Fig. 2e–g). MiR-200 overexpressing lines were only partly dependent on Cdh1 for colonization since stable knockdown of Cdh1 in C1+C2 lines modestly but insignificantly reduced colonization potential (Fig. 2h). In summary, results from both clinical and experimental analyses collectively show that miR-200s can promote distant colonization of breast cancer cells. However, the fact that E-cadherin overexpression alone cannot fully recapitulate the metastasis potential of miR-200 overexpressing lines, suggests that other genes or signaling pathways are also likely to be simultaneously targeted to enhance colonization efficiency.
miR-200 family induces global changes in gene expression
To identify such novel functional gene and pathway targets of miR-200s, we performed microarray analysis. Although we observed global changes in gene expression in C2 and C1+C2 lines, significantly less genetic changes were observed in C1 and CDH1 lines (Fig. 3a). To analyze the extent of the global changes in gene expression observed, we performed an unbiased hierarchical clustering based on a 1,218 gene-set signature that distinctively defines the parental 4TO7 and the highly metastatic 4T1 cell lines. Strikingly, we found that C2 and C1+C2 lines clustered with the highly metastatic 4T1 cells, whereas the C1 and CDH1 lines remained clustered with the parental 4TO7 cells (Fig. 3a), mirroring their spontaneous metastasis potentials (Fig. 2c). Rigorous gene set enrichment analyses (GSEA) were also performed to: 1) confirm the genome-wide shifts in gene expression, 2) reveal that PicTar-derived miR-200 targets were regulated at a global level, 3) highlight the global repression of the EMT-like genetic program upon miR-200 expression in 4TO7 cells, 4) show that Cdh1 overexpression alone causes only a modest expression shift, suggesting that E-cadherin is unlikely to be the only functional mediator of miR-200s, and 5) reveal that these genetic changes may influence many cellular functions, including epithelial traits and protein transport and secretion (Fig. 3b–c, Supplementary Fig. 6, Supplementary Table 2; see Supplementary Results). Furthermore, there was a significant negative association between miR-200 family expression and the miR-200 down-regulated gene signature in the Oxford cohort of tumor samples (P < 0.001), suggesting that the global changes in gene expression observed in vitro are likely to be pathologically relevant in breast tumors.
Figure 3.
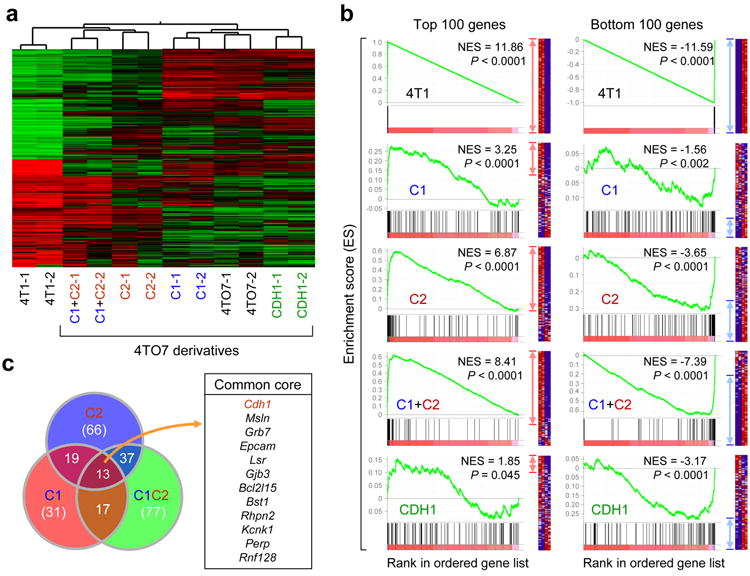
Ectopic miR-200 expression promotes global changes in gene expression. (a) Unsupervised clustering highlighting genome-wide changes in gene expression upon miR-200 expression in 4TO7 cells. Experiment was performed twice in duplicates. (b) GSEA showing influence of miR-200 overexpression on the overall gene expression profile of 4TO7 cells. Gene sets used are the top 100 (left panel) and bottom 100 (right panel) differentially expressed genes in the test (C1, C2, C1+C2 and CDH1) vs control lines. Gene list used include all mouse genes ranked by their differential expression between 4T1 and 4TO7. Enrichment of top and bottom 100 genes from 4T1 vs. 4TO7 ranked list is shown as an example of maximum possible enrichment. ES: enrichment score. NES: normalized enrichment score. Red and blue arrows denote the relative number of core genes for each analysis. (c) Venn diagram showing significant overlap of core genes from top 100 gene sets for C1 (red circle), C2 (blue circle) and C1+C2 (green circle) lines from (b). Common core genes between C1, C2 and C1+C2 lines are presented. Cdh1 is highlighted in red font to emphasize the positive influence of miR-200s on E-cadherin expression.
Genomic and proteomic identification of miR-200 targets
Recent studies suggest that in mammalian cells, miRNAs regulate most of their direct targets at both mRNA and protein levels41. As such, we combined microarray and mass spectrometry (MS) analysis to identify nine candidate genes down-regulated at both the RNA and protein level in C1+C2 cells versus control (Fig. 4a,b and Supplementary Table 3; see Supplementary Results). We confirmed reduced expression of the nine candidate MDA-MB-231 and TSU-PR1 human breast and bladder cancer cell lines, respectively (Fig. 4d). Furthermore, miR-200b/200c levels are negatively correlated with mean expression of the nine candidate genes in the NCI-60 panel of cell lines (Fig. 4e), implying conserved targeting.
Figure 4.
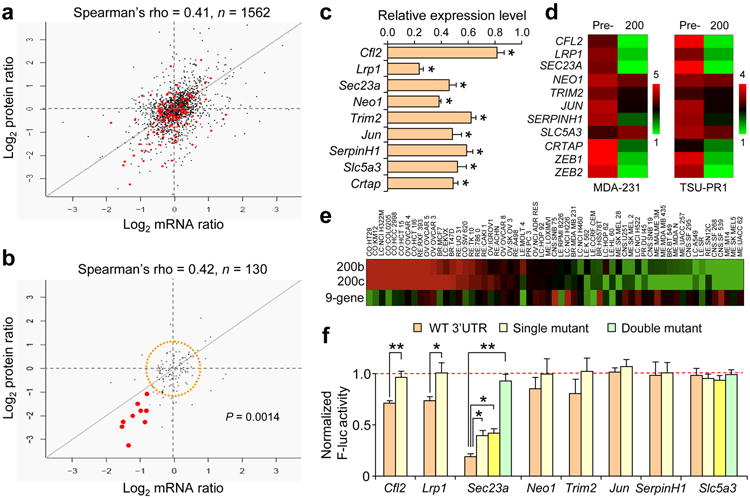
Identification of putative miR-200 targets using mass spectrometry. (a) Scatter plot shows the comparison between protein abundance and mRNA abundance for 1562 proteins in C1+C2 versus control cells. Red dots represent genes with miR-200 target sites. (b) Comparison between protein abundance and mRNA abundance for genes containing miR-200 target sites (n =130). Genes highlighted by red dots represent those that are significantly reduced in expression at both mRNA and protein level (n = 9). Dots confined within orange dashed circle represent genes harboring miR-200 target sites showing little or no change in gene expression. (c) qRT-PCR validation of reduced expression of the nine candidate genes highlighted by red dots in panel (b) in C1+C2 line compared to the control. Data represent mean ± s.e.m. * P < 0.05 (Student’s t-test). (d) Heat map showing expression of nine candidate genes in MDA-MB-231 (left) and TSU-PR1 (right) cells upon transient transfection of miR-200s. ZEB1 and ZEB2 were included as positive controls. (e) Heat map showing negative correlation between average expression of nine-candidate gene signature and miRs-200b and -200c in NCI-60 panel of cell lines. (f) Direct targeting of eight of nine candidate genes was tested by luciferase assays in HeLa cells. Data represents percent change in normalized luciferase activity after co-transfection of miR-200s relative to the negative control pre-miR ± s.e.m. * P < 0.05, ** P < 0.01 (Student’s t-test).
To assess which of these candidate genes are directly targeted by miR-200s, we cloned the 3’-UTRs of eight of the nine candidates for standard luciferase assays. We confirmed direct targeting of three candidates, including cofilin 2 (Cfl2), low-density lipoprotein receptor-related protein 1 (Lrp1), and Sec23 homolog A (Sec23a) (Fig. 4f). The 3’-UTRs of Cfl2 and Lrp1 each contain one functional miR-200 target sequence, while Sec23a contains two evolutionarily conserved miR-200 target sites that function cooperatively to suppress Sec23a expression (Fig. 4f and Supplementary Fig. 7).
Sec23a is a miR-200 target that suppresses metastasis
To test which of the three direct targets likely mediates miR-200 function in metastasis, we stably knocked-down (KD) each of the genes in 4TO7 cells (Supplementary Fig. 8a–c). Stable KD did not influence morphology (Supplementary Fig. 8a), cell proliferation or E-cadherin expression (data not shown). Of the three genes, Sec23a KD reduced transwell migration (Fig. 5a) and enhanced pulmonary colonization (Fig. 5b,c), phenocopying miR-200 overexpression (Supplementary Fig. 5c; Fig. 2e,f). In support of the functional data, endogenous Sec23a levels are less abundant in highly metastatic 4T1 and MCFCA1a lines relative to the weakly metastatic 4TO7 and MCFCA1h lines and is reduced in 4TO7 line upon miR-200 overexpression (Supplementary Fig. 8d–e). Furthermore, SEC23A levels are significantly lower in clinical metastases relative to primary tumors (Fig. 5d–e), consistent with its role as a suppressor of metastatic colonization.
Figure 5.
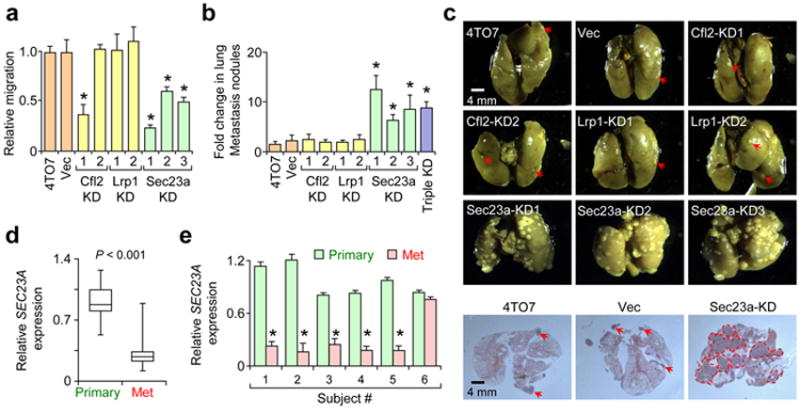
Sec23a knockdown phenocopies miR-200s in inhibiting migration and promoting metastatic colonization. (a) Transwell migration assays were performed in triplicate and is presented as the mean migration of experimental lines as a ratio relative to parental 4TO7 cells ± s.e.m. * P < 0.05 (Student’s t-test). (b) Bar graph showing fold change in number of pulmonary nodules relative to 4TO7 parental line. Data presented as mean ± s.e.m. * P < 0.05 (Student’s t-test). (c) Representative gross lung images and H&E stained lung sections (lower panel) from mice intravenously injected with various cell lines as indicated. Red arrows were used to highlight metastatic nodules, except in Sec23a-KD samples which contained large numbers of nodules. (d) Box plot showing relative expression of SEC23A in ten human primary tumors versus ten metastases. GAPDH was used to normalize expression. P value computed by Student’s t-test. (e) Bar graph showing relative expression of SEC23A in matched primary and metastasis samples collected from six individuals. GAPDH was used to normalize expression. * P < 0.05 (Student’s t-test).
Neither knockdown in 4TO7 cells nor overexpression of Sec23a in 4TO7-C1C2 cells significantly influenced spontaneous metastasis (Supplementary Fig. 8f–h). Since Sec23a inhibits migration but increases colonization, it is possible that these two effects may balance out each other. Indeed, evaluation of SEC23A expression as a primary tumor prognostic marker in a large public clinical database of breast cancer42 failed to show significant associations. Overall, both clinical data analysis and experimental animal models suggest that Sec23a plays an important role in suppressing metastasis specifically at the step of colonization. However, it should be noted that although Sec23a KD can enhance metastatic colonization, overexpression of Sec23a alone is not sufficient to significantly suppress metastasis (Supplementary Fig. 8i,j).
miR-200 overexpression suppresses Sec23a-mediated secretion
Sec23a is an essential component of COPII vesicles and is involved in anterograde transport of proteins from the ER to Golgi apparatus. Although several recent studies have shown that Sec23a is indispensible for the secretion of ECM components such as collagens and cartilage oligomeric matrix protein (Comp), and in craniofacial and chondrocyte development43-45, little else is known of other classes of proteins influenced by Sec23a-mediated secretion and the potential role they may play in metastasis. To this end, we performed mass spectrometry analysis on conditioned media (CM) from two Sec23a KD lines. There was a global reduction in protein secretion (Supplementary Fig. 9a), including those involved in wounding (P = 6.4e−6), cell adhesion (P = 1.6e−5), extracellular structure organization (P = 3.8e−3), inflammatory and immune responses (P = 1.0e−4 and P = 4.0e−4 respectively) (Supplementary Table 4) as defined by the Database for Annotation, Visualization and Integrated Discovery (DAVID ) Bioinformatics Resources46. Reduced secretion led to intracellular accumulation of proteins such as Comp44 (Supplementary Fig. 9b) which in turn resulted in modest ER distension as assessed by electron microscopy (EM) analysis (Supplementary Fig. 9c). There were strong correlations of Sec23-dependent secreted proteins between the two KD lines (Fig. 6a, R = 0.9, P < 0.0001) and between Sec23a KD#2 and the C1+C2 line (Fig. 6a, R = 0.43, P < 0.001). Similar to the Sec23a KD line, C1+C2 line also showed modest ER distension by EM analysis, suggesting functional disruption of protein transport (Supplementary Fig. 9c). Taken together, these data suggest that expression of Sec23a and the secretory pathway are inhibited by miR-200s.
Figure 6.
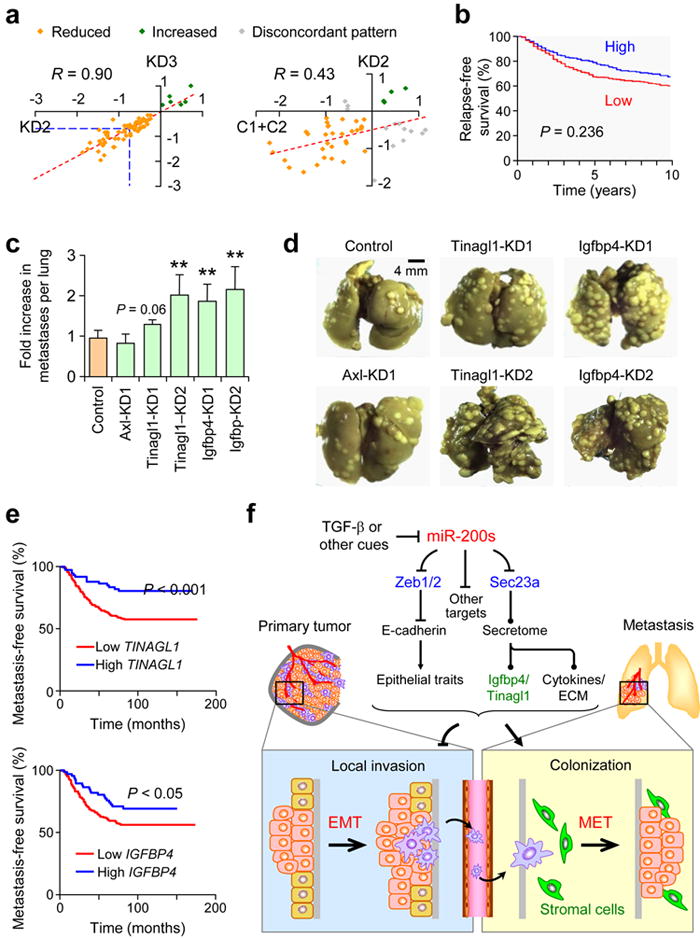
Sec23a knockdown disrupts secretion of proteins that are correlated with suppression of clinical metastasis. (a) Scatter plot showing correlation of secretome profiles between two different Sec23a KD lines and between Sec23a KD and C1+C2 lines. Proteins in common between different lines were used to generate the plots. Orange dots represent proteins reduced in abundance in both conditions; green dots represent proteins increased in abundance in both conditions whereas gray dots represent those proteins that show discordant expression patterns. (b) Kaplan-Meier curves showing relapse-free survival of patients with high and low median expression level of 35 genes reduced in secretion in Sec23a KD lines. (c) Bar graph showing fold increase in number of pulmonary metastases in 4TO7-derived lines stably knocking down Axl, Tinagl1 or Igfbp4 relative to vector control. (d) Representative gross lung images from animals injected via lateral tail vein with various knockdown lines from (c) along with vector control. ** P < 0.01 (Student’s t-test). (e) Kaplan-Meier plots of distant metastasis-free survival of patients in the EMC286 dataset stratified by expression of TINAGL1 (top) or IGFBP4 (bottom). P values were computed by log-rank test. (f) Schematic model of miR-200 function during metastasis. MiR-200s simultaneously target several genes including Zeb1, ZEB2 and Sec23a to inhibit local invasion but promote metastatic colonization. Targeting of Zeb1/2 influences cell-intrinsic epithelial traits whereas targeting of Sec23a modulates tumor-derived secretion of factors such as Igfbp4 and Tinagl1, which influence metastatic colonization by altering tumor-stromal interactions.
Igfbp4 and Tinagl1 are Sec23a-dependent metastasis suppressors
To evaluate the clinical importance of the secretome in cancer progression in human breast cancer patients, we analyzed the expression of 35 of 38 genes that were significantly reduced in secretion in both Sec23a KD lines (secretome gene signature) (Fig. 6a), in a large public microarray database42. Analysis revealed that low expression is significantly correlated with reduced RFS (Fig. 6b, P = 0.0236), reinforcing the role of these genes as metastasis suppressors. In contrast, average expression of eight of nine candidate miR-200 target genes identified through combined genomic and proteomic analysis (Fig. 4a,b) showed correlation with good RFS but failed to reach significance (P = 0.13, data not shown), highlighting the prominent influence of the Sec23a-mediated secretome in metastasis-free survival.
To uncover the central mediators of the secretome that possessed metastasis-related functions, we functionally tested three of six genes — Axl receptor tyrosine kinase (Axl), tubulointerstitial nephritis antigen-like 1 (Tinagl1) and insulin-like growth factor binding protein 4 (Igfbp4) — from the secretome signature that were significantly associated with RFS (Supplementary Fig. 9d). Stable knockdown of Tinagl1 and Igfbp4 in 4TO7 cells (Supplementary Fig. 9e) enhanced colonization after intravenous injection (Fig. 6c,d), phenocopying miR-200 overexpression and Sec23a KD. Importantly, both TINAGL1 and IGFBP4 were also significantly associated with better distant metastasis-free survival in an independent EMC286 clinical dataset47 (Fig. 6e), again supporting their role in reducing metastasis. Furthermore, IGFBP4 expression showed a significant lung-tropic association (Supplementary Fig. 9f) with metastases in the MSK82 clinical dataset48. In aggregate, our data implicates a significant role for the secretome as an important downstream mediator of miR-200s in metastasis, and uncovers Tinagl1 and Igfbp4 as secreted suppressors of lung metastasis with potential therapeutic applications.
Discussion
In this study, we show that miR-200s promote metastastic colonization through mechanisms that go beyond cell-intrinsic regulation of epithelial traits through the Zeb1/2-E-cadherin axis (Fig. 6f). MiR-200s also influence tumor cell secretome by direct targeting of Sec23a-mediated transport pathway, which affects cell-extrinsic tumor-stromal interactions. We further identified Tinagl1 and Igfbp4 as important Sec23a-mediated secretory proteins that significantly reduce metastatic colonization. Our findings support the dynamic roles of EMT and MET during different stages of metastasis — while EMT and low levels of miR-200s promote invasion and intravasation, MET and high miR-200 expression is required for efficient colonization of secondary organs.
The biphasic role of miR-200s in metastasis
Here, we present the first large scale breast cancer clinical study showing a significant clinical association of miR-200 family expression with poor DRFS, particularly in ER-positive breast cancers. Furthermore, miR-200 expression was increased in lung metastases compared to primary tumors. In support of the clinical associations, a survey across several different isogenic series of cancer cell lines revealed a strong correlation between miR-200 expression and metastatic ability, which was further supported by functional analysis in vivo.
Several recent studies in animal metastasis models have reported conflicting roles of miR-200s in metastatic progression29-31. These contradictory findings may in part due to the different models applied in these studies, where miR-200s may promote metastasis of breast cancer cells31 and hinder metastasis of lung adenocarcinoma29 and pancreatic neuroendocrine cells30. However, it is more likely that the final outcome of miR-200s in metastasis progression of various cancers depends on several variables such as the difference in the rate-limiting step of the metastatic cascade and the importance of MET in colonization in different models. In the model system we applied, the 4TO7 tumor cells are inherently highly migratory and invasive in vitro and in vivo, and although ectopic miR-200 expression reduces dissemination, it has a net effect of enhancing the rate-limiting colonization step. In contrast, this outcome is less likely in model systems where earlier steps of metastasis, such as early dissemination through acquisition of EMT-like properties, is the rate-limiting process, as may be the case for the model systems showing metastasis-suppressor functions of miR-200s29,30 (see Supplementary Discussion for further discussion on the biphasic role of miR-200s).
miR-200s influence cell-intrinsic and -extrinsic pathways
In our present study, both clinical and functional data revealed that E-cadherin alone is insufficient to recapitulate all miR-200 phenotypes, suggesting that miR-200s likely influence other genes or pathways during metastasis. We used integrated genomic and proteomic analysis to obtain a comprehensive understanding of the impact of miR-200s on gene expression in cancer cells. MiR-200 overexpression led to sweeping changes in gene expression toward that of the most metastatic cells. In addition to genes and pathways controlling epithelial characteristics, additional cellular processes such as protein transport and secretion were also implicated in miR-200-dependent regulation of metastatic ability. Importantly, we showed that the expression of Sec23a is directly suppressed by miR-200s through two evolutionarily conserved target sequences in its 3’-UTR (see Supplementary Discussion for summary of evidence supporting Sec23a as a functional target miR-200s).
Thus, in addition to the well established function of miR-200s in regulating intrinsic cellular properties through Zeb1/2 and E-cadherin, miR-200s may also possess the potential to influence the microenvironment by directed targeting of Sec23a-dependent secretome. Through regulation of this pathway, miR-200s may extend their reach to manipulate a neighboring population of tumor and stromal cells, influencing their collective behavior during metastasis. The ability of miRNAs to influence cell-intrinsic and –extrinsic properties of tumor cells has also recently been uncovered for the miR-17/20 cluster49-51. The miR-200 family and the miR-17/20 cluster may represent a growing number of miRNAs with the ability to influence both intracellular regulatory machineries and intercellular communication of tumor and stromal cells (see Supplementary Discussion for further comments on the role of secreted components in metastasis).
In summary, our study combined clinical and experimental studies to establish a biphasic role miR-200s in metastasis. We showed that miR-200s promote metastatic colonization by enhancing cell-intrinsic epithelial traits via the Zeb-E-cadherin axis and by inhibiting Sec23a-dependent regulation of tumor secretome. The fact that miR-200 targeting of Sec23a appears to have dichotomous roles in metastasis, hindering early steps of migration and invasion while promoting late step of metastatic colonization, may explain the contradictory roles of miR-200s in various models of metastasis. Although miR-200s and the Sec23a pathway may represent new therapeutic opportunities for metastatic cancer, their dichotomous functions warrant careful assessment of potential therapeutic benefits and adverse side effects when treatments are applied at different stages of the disease.
METHODS
Tumor xenografts
All procedures involving mice, such as housing and care, and all experimental protocols were approved by Institutional Animal Care and Use Committee (IACUC) of Princeton University. Cells were harvested from subconfluent cell culture plates, washed with PBS twice, and re-suspended at the appropriate concentration in PBS. 2×105 cells in 0.1 ml PBS were injected on day 0 into the lateral tail vein of 4-week-old, female BALB/c mice (NCI) using 26 G needles to generate pulmonary metastasis. For orthotopic primary tumor formation, female BALB/c mice at 6 weeks old were anaesthetized and a small incision was made to reveal the mammary gland. 106 cells resuspended in 10 μl PBS were injected directly into the mammary fat pad. The primary tumor growth was monitored weekly by measurement of the tumor size.
Statistical analysis
Results were reported as mean ± s.e.m. (standard error of the mean). Two-sided independent Student’s t-test without equal variance assumption or the Wilcoxon signed-rank test was performed to analyze gene and miRNA expression levels, differences in number of tumor colonies and nodules, end-points of in vitro luciferase assays and histology data. For clinical associations, Spearman rank correlation coefficients were used for studying the association between continuous variables. Tests of hypotheses on the location parameter (median) were performed using Mann-Whitney and Kruskall-Wallis. The log-rank test was used to test for differences in survival in univariate analysis. Expression values were introduced into survival analysis as continuous variables using a non-parametric approach in which samples were ranked by expression values, and ranks were normalized between 0 and 1. Statistical analyses were performed using R (www.r-project.org). DRFS and RFS were calculated as described by the STEEP criteria33. A standard chi-square goodness of fit test was used to determine significance of overlap in number of enriched GO categories between samples.
Additional experimental procedures are listed in the Supplementary Methods.
Supplementary Material
Acknowledgments
We thank S. Kyne at the mass spectrometry core facility, D. Storton and J. Buckles at the microarray core facility, E. Williams at the electron microscopy facility of Princeton University for expert technical advice and support, N. Sethi for insightful discussions and blinded verification of tumor counts, S.J. Parkinson for technical advice, and K. Socha for colony counting. We also thank E. Williams and F. Miller for the TSU-PR1 and 4T1 cell line series, respectively. Y.K. is a Champalimaud Investigator and a Department of Defense Era of Hope Scholar Award recipient. This research was supported by grants from the US National Institutes of Health (1R01-CA141062) and the Brewster Foundation to Y.K., and from Cancer Research UK, Oxford NHS Biomedical Research Centre and Friends of Kennington Cancer Fund to A.L.H. M.K. and Y.H. are recipients of Department of Defense pre-doctoral fellowships.
Footnotes
AUTHOR CONTRIBUTIONS M.K. and Y.K. designed experiments. M.K., B.J.E., T.C.-T. and Y.H. performed the experiments. F.M.B., T.I, L.M., J.R., D.A. and A.L.H. provided clinical samples and associated analyses, M.A.B., Z.K., H.G., Y.W., G.H. and B.A.G. contributed genomic and proteomic analyses. M.K. and Y.K. wrote the manuscript. All authors discussed the results and commented on the manuscript.
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
Accession number Gene expression microarray data used to analyze global changes in gene expression induced by miR-200 family or Cdh1 alone has been deposited at the NCBI Gene Expression Omnibus with the accession GSE19631.
References
- 1.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Thompson EW, Williams ED. EMT and MET in carcinoma--clinical observations, regulatory pathways and new models. Clin Exp Metastasis. 2008;25:591–592. doi: 10.1007/s10585-008-9189-8. [DOI] [PubMed] [Google Scholar]
- 3.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Chaffer CL, Thompson EW, Williams ED. Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs. 2007;185:7–19. doi: 10.1159/000101298. [DOI] [PubMed] [Google Scholar]
- 5.Jeschke U, Mylonas I, Kuhn C, Shabani N, Kunert-Keil C, Schindlbeck C, Gerber B, Friese K. Expression of E-cadherin in human ductal breast cancer carcinoma in situ, invasive carcinomas, their lymph node metastases, their distant metastases, carcinomas with recurrence and in recurrence. Anticancer Res. 2007;27:1969–1974. [PubMed] [Google Scholar]
- 6.Park D, Karesen R, Axcrona U, Noren T, Sauer T. Expression pattern of adhesion molecules (E-cadherin, alpha-, beta-, gamma-catenin and claudin-7), their influence on survival in primary breast carcinoma, and their corresponding axillary lymph node metastasis. Apmis. 2007;115:52–65. doi: 10.1111/j.1600-0463.2007.apm_524.x. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 9.Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington’s disease. Neurobiol Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, Gimotty PA, Katsaros D, Coukos G, Zhang L, Pure E, Agami R. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–210. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 11.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 14.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) The Journal of biological chemistry. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 15.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell research. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 16.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 19.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell stem cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 20.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, Brunton VG, Morton J, Sansom O, Schuler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 21.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RA, Lao K, Clarke MF. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 Inhibition of Suz12 Leads to Polycomb-Mediated Repression Required for the Formation and Maintenance of Cancer Stem Cells. Mol Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schickel R, Park SM, Murmann AE, Peter ME. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Molecular cell. 2010;38:908–915. doi: 10.1016/j.molcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cochrane DR, Howe EN, Spoelstra NS, Richer JK. Loss of miR-200c: A Marker of Aggressiveness and Chemoresistance in Female Reproductive Cancers. Journal of oncology. 2010;2010:821717. doi: 10.1155/2010/821717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO reports. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 27.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, Kurie JM. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–2151. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, Libutti SK, Nakakura EK, Golub TR, Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4:e7181. doi: 10.1371/journal.pone.0007181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 33.Hudis CA, Barlow WE, Costantino JP, Gray RJ, Pritchard KI, Chapman JA, Sparano JA, Hunsberger S, Enos RA, Gelber RD, Zujewski JA. Proposal for standardized definitions for efficacy end points in adjuvant breast cancer trials: the STEEP system. J Clin Oncol. 2007;25:2127–2132. doi: 10.1200/JCO.2006.10.3523. [DOI] [PubMed] [Google Scholar]
- 34.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 35.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 36.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Lou Y, Preobrazhenska O, auf dem Keller U, Sutcliffe M, Barclay L, McDonald PC, Roskelley C, Overall CM, Dedhar S. Epithelial-mesenchymal transition (EMT) is not sufficient for spontaneous murine breast cancer metastasis. Dev Dyn. 2008;237:2755–2768. doi: 10.1002/dvdy.21658. [DOI] [PubMed] [Google Scholar]
- 38.Santner SJ, Dawson PJ, Tait L, Soule HD, Eliason J, Mohamed AN, Wolman SR, Heppner GH, Miller FR. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res Treat. 2001;65:101–110. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- 39.Chaffer CL, Brennan JP, Slavin JL, Blick T, Thompson EW, Williams ED. Mesenchymal-to-epithelial transition facilitates bladder cancer metastasis: role of fibroblast growth factor receptor-2. Cancer research. 2006;66:11271–11278. doi: 10.1158/0008-5472.CAN-06-2044. [DOI] [PubMed] [Google Scholar]
- 40.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 41.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 43.Lang MR, Lapierre LA, Frotscher M, Goldenring JR, Knapik EW. Secretory COPII coat component Sec23a is essential for craniofacial chondrocyte maturation. Nat Genet. 2006;38:1198–1203. doi: 10.1038/ng1880. [DOI] [PubMed] [Google Scholar]
- 44.Saito A, Hino S, Murakami T, Kanemoto S, Kondo S, Saitoh M, Nishimura R, Yoneda T, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Imaizumi K. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nature cell biology. 2009;11:1197–1204. doi: 10.1038/ncb1962. [DOI] [PubMed] [Google Scholar]
- 45.Townley AK, Feng Y, Schmidt K, Carter DA, Porter R, Verkade P, Stephens DJ. Efficient coupling of Sec23-Sec24 to Sec13-Sec31 drives COPII-dependent collagen secretion and is essential for normal craniofacial development. J Cell Sci. 2008;121:3025–3034. doi: 10.1242/jcs.031070. [DOI] [PubMed] [Google Scholar]
- 46.Huang DW, Sherman BT, Lempicki RA. integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 48.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 50.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, Wu K, Whittle J, Ju X, Hyslop T, McCue P, Pestell RG. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. The Journal of cell biology. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, Liu Y, McCue PA, Quong AA, Lisanti MP, Pestell RG. microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci U S A. 2010;107:8231–8236. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


