Abstract
PIK3CA gain-of-function mutations are a common oncogenic event in human malignancy1–4, making PI3K an attractive target for cancer therapy. Despite the great promise of targeted therapy, resistance often develops, resulting in treatment failure. To elucidate mechanisms of resistance to PI3K-targeted therapy, we constructed a mouse model of breast cancer conditionally expressing human PIK3CAH1047R. Surprisingly, most PIK3CAH1047R-driven mammary tumors recurred following PIK3CAH1047R inactivation. Genomic analyses of recurrent tumors revealed multiple lesions, including focal amplification of c-Met or c-Myc. While c-Met amplification allowed tumor survival dependent on activation of endogenous PI3K, tumors with c-Myc amplification became independent of the PI3K pathway. Functional analyses demonstrated that c-Myc contributed to oncogene independence and resistance to PI3K inhibition. Importantly, PI3KCA mutations and increased c-MYC levels co-occur in a substantial fraction of human breast tumors. Together, these data suggest that c-MYC elevation represents a potential mechanism by which tumors develop resistance to current PI3K-targeted therapies.
More than 25% of breast cancers harbor somatic mutations in the PIK3CA-encoded p110α catalytic subunit of phosphatidylinositol 3-kinase (PI3K)1–4. These mutations usually occur in the helical region (E545K and E542K) or the kinase domain (H1047R) of p110α, with H1047R being the most common mutation (>50% of cases). Several experimental models have demonstrated that these tumor-associated PIK3CA mutations result in constitutive p110α activation and oncogenic transformation5–9, making the PIK3CA oncogene a major target for cancer therapy.
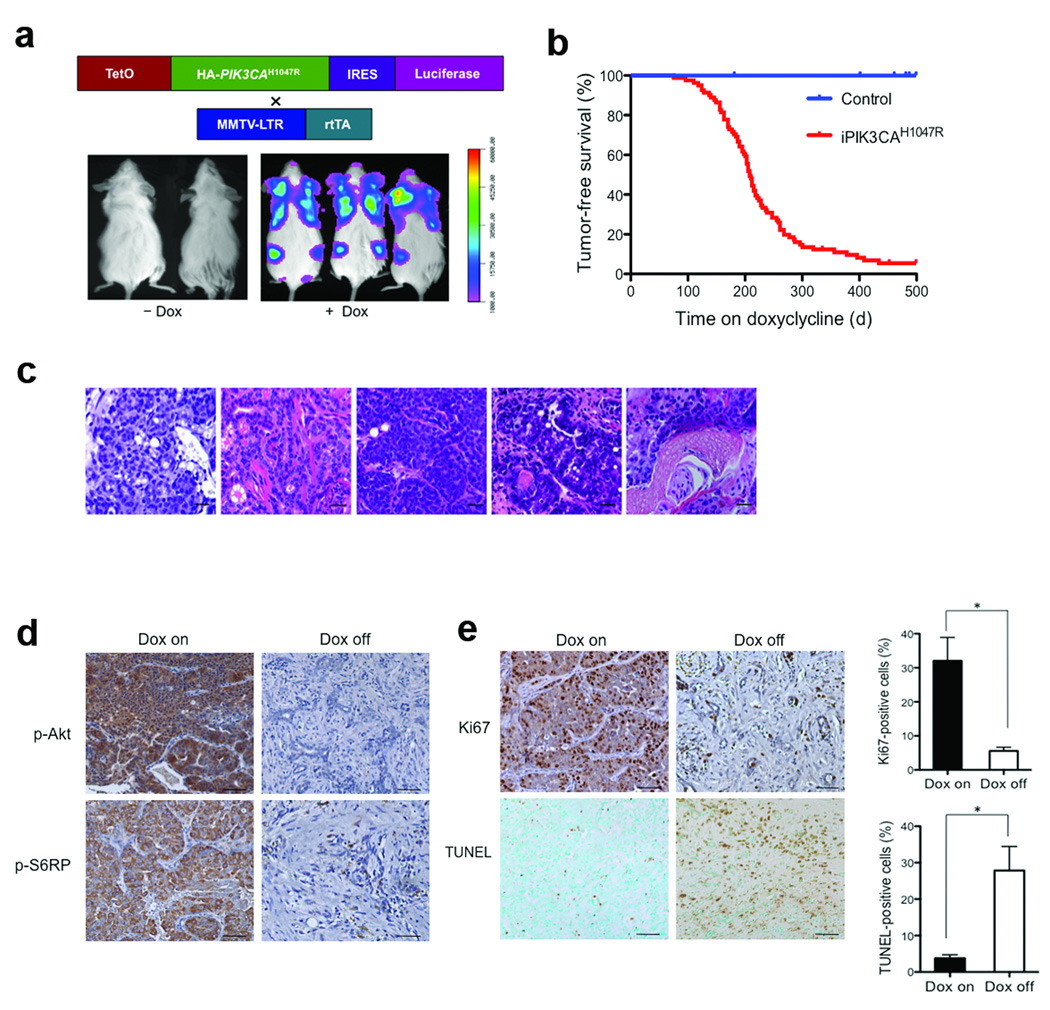
To study the effects of mutational activation of PI3K on breast tumorigenesis in vivo and to identify potential mechanisms of resistance to PI3K inhibition, we generated a transgenic mouse line expressing human PIK3CAH1047R in which transgene expression is under the control of a tetracycline-inducible promoter (TetO). PIK3CAH1047R expression is coupled with a luciferase reporter allowing transgene expression to be followed in vivo (Fig. 1a). To drive mammary-specific expression of PIK3CAH1047R, we crossed two tetO-PIK3CAH1047R founders (HR-2239 and HR-2251) to a previously described MMTV-rtTA (MTB) line10. The resulting bitransgenic MTB/tetO-PIK3CAH1047R mice were designated iPIK3CAH1047R. Quantitative RT-PCR analyses of mammary tissues isolated from bitransgenic females revealed that doxycycline treatment led to a substantial increase in PIK3CAH1047R expression as well as luciferase reporter activity, whereas endogenous mouse Pik3ca expression remained unaffected (Supplementary Fig. 1a,b). As mice derived from both iPIK3CAH1047R founder lines showed comparable mammary gland-specific and doxycycline-dependent transgene expression, the MTB/HR-2239 line was used for all subsequent experiments.
Figure 1.
Mammary gland-specific expression of PIK3CAH1047R induces mammary tumors. (a) Generation of a transgenic mouse model expressing HA (haemagglutinin)-tagged human PIK3CAH1047R under the control of a tetracycline-inducible promoter (TetO). The expression of PIK3CAH1047R is coupled through an IRES with downstream expression of luciferase. These mice were crossed with MMTV-rtTA (MTB) mice to generate bi-transgenic iPIK3CAH1047R animals to drive the expression of HAPIK3CAH1047R in mammary glands. Lower panels demonstrate bioluminescence imaging of iPIK3CAH1047R mice maintained in the presence or absence of doxycycline. (b) Tumor-free survival curve for iPIK3CAH1047R mice maintained on doxycycline (n = 81, median tumor free survival 208 days), and three groups of control mice: MTB (n = 12) and tetO-PIK3CAH1047R (n = 10) mice maintained with doxycycline, and iPIK3CAH1047R (n = 14) mice maintained in the absence of doxycycyline. All three types of control mice are represented by the blue line. (c) Representative haematoxylin and eosin (H&E)-stained sections of primary mammary tumors from iPIK3CAH1047R mice subjected to chronic doxycycline treatment. Scale bars, 25 µm. (d) Immunohistochemistry (IHC) for p-AKT(Ser473) and pS6RP(S235/236) performed on tumors isolated from iPIK3CAH1047R mice maintained on doxycycline (Dox on panels) or 6 days following doxycycline withdrawal (Dox off panels). Representative images are shown. Scale bar, 50 µm. (e) IHC for Ki67 or TUNEL performed on tumors isolated from iPIK3CAH1047R mice maintained on doxycycline (Dox on panels) or 3 days following doxycycline withdrawal (Dox off panels). Representative images are shown. Scale bar, 50 µm. n = 6 for each group. * P < 0.005 (Student’s t-test).
To determine whether expression of PIK3CAH1047R can initiate transformation of mammary epithelium, we analyzed mammary glands isolated from iPIK3CAH1047R females treated with doxycycline for 4 weeks. Histological examination showed increased mammary ductal side-branching and enlarged focal nodular structures filled with hyperproliferative cells characteristic of early neoplastic lesions (Supplementary Fig. 2a,b). Immunohistochemical (IHC) analyses demonstrated strong p-AKT signals in proliferating epithelial cells in the mammary glands from doxycycline-treated mice (Supplementary Fig. 2c), indicating activation of PI3K signaling in response to the induction of PIK3CAH1047R. Consistent with the phenotype noted above, chronic doxycycline induction of the PIK3CAH1047R transgene in bitransgenic mice resulted in mammary tumors with 95% penetrance and a mean latency of 7 months (Fig. 1b). These primary tumors displayed heterogeneous pathological phenotypes, including adenocarcinomas and adenosquamous carcinomas (Fig. 1c and Supplementary Table 1). In contrast, no tumors were observed in any of the control groups over the same time period (Fig. 1b). Thus, sustained induction of oncogenic PIK3CA expression leads to mammary tumor formation.
To examine whether established tumors require continued PIK3CAH1047R expression to maintain their malignant state, we withdrew doxycycline from a cohort of tumor-bearing mice. All tumors exhibited regression during the first week following doxycycline removal. The suppression of PIK3CAH1047R expression following doxycycline withdrawal was confirmed by RT-PCR in primary tumors (Supplementary Fig. 3). IHC analyses revealed dramatically reduced levels of both p-Akt and p-S6RP in doxcycycline-off tumors as compared to those maintained on doxycycline (Fig. 1d). Moreover, while a robust Ki67 signal was detected in tumors maintained on doxycycline, the number of proliferating cells significantly decreased in tumors following doxycycline withdrawal (Fig. 1e). Conversely, while only a few apoptotic cells were detected in tumors on doxycycline, a markedly increased number of TUNEL-positive cells were observed in tumors after doxycycline removal (Fig. 1e). These data indicate that reduced cellular proliferation and increased apoptosis are responsible for the initial phase of tumor regression following downregulation of oncogenic PIK3CA.
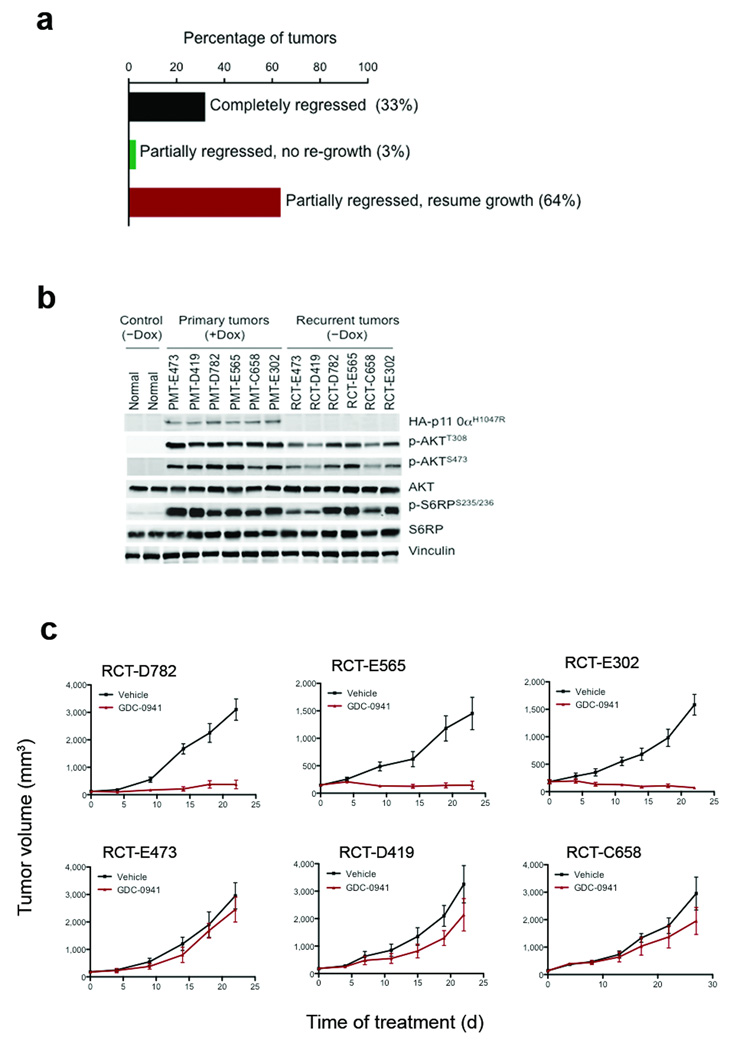
To determine whether the continued inactivation of oncogenic p110αH1047R resulted in sustained regression of mammary carcinomas initiated by the expression of PIK3CAH1047R, we followed a large cohort of tumors after doxycycline withdrawal for up to 6 months. We found that one-third of tumors rapidly and completely regressed to a non-palpable state within 1–2 months following doxycycline withdrawal with no re-growth (Fig. 2a and Supplementary Fig. 4a), indicating that these tumors remained dependent on p110αH1047R for their maintenance. While a small fraction of tumors regressed partially and did not resume growth following doxycycline removal, about two-thirds of tumors partially regressed but then resumed growth in the absence of doxycycline (Fig. 2a and Supplementary Fig. 4b). We confirmed that all recurrent tumors showed sustained downregulation of the PIK3CAH1047R transgene and its protein product (Fig. 2b). Thus, PIK3CAH1047R-initiated mammary tumors frequently failed to regress completely upon PIK3CAH1047R inactivation and recurred in a PIK3CAH1047R-independent manner.
Figure 2.
Tumor responses to doxycycline withdrawal. (a) Primary tumors (135 primary tumors were derived from 107 tumor bearing bi-transgenic mice; 81 mice carried one tumor, 21 mice bore two tumors and 4 mice had three tumors) had three types of response to doxycycline withdrawal. 45 of 135 (33%) regressed completely without re-growth (black bar), 4 of 135 (3%) regressed partially with no re-growth (green bar) and 86 of 135 (64%) regressed partially but then re-grew (red bar). (b) Western blot analyses of HA-p110αH1047R, p-Akt and p-S6RP in six recurrent tumors (RCT) in the absence of doxycycline and their matched primary tumors (PMT) maintained on doxycycline. Mammary gland tissues from uninduced iPIK3CAH1047R mice were used as controls. (c) Responses of recurrent tumor transplants to GDC-0941 or vehicle treatment. Data are shown as mean ± S.E.M (n = 6). * P < 0.001 (Student’s t test).
We next examined whether the PI3K pathway remained active in recurrent tumors, thus compensating for the loss of PIK3CAH1047R expression. Western blot analyses of six paired primary and recurrent tumors revealed that, while both p-AKT and p-S6RP signals were robust in all six primary tumors maintained on doxycycline, in three recurrent tumors these signals were maintained at comparably high levels, but were reduced substantially in the other three recurrent tumors (Fig. 2b). These six recurrent tumors were then transplanted into the mammary fat pads of athymic mice, and the tumor-bearing recipients treated with GDC-0941, a pan-Class I PI3K inhibitor currently in clinical trials11,12. Three recurrent tumors (RCT-D782, RCT-E565 and RCT-E302), all of which retained high levels of both p-AKT and p-S6RP, were sensitive to GDC-0941 treatment (Fig. 2c upper panels). In contrast, the three recurrent tumors (RCT-E473, RCT-D419 and RCT-C658), which showed reduced p-AKT and p-S6RP signals, were resistant to GDC-0941 (Fig. 2c lower panels). These data suggest that some recurrent tumors escaped addiction to the oncogenic PIK3CA but remained dependent on the PI3K pathway, while others acquired the ability to grow independently of both the PIK3CA oncogene and the PI3K pathway.
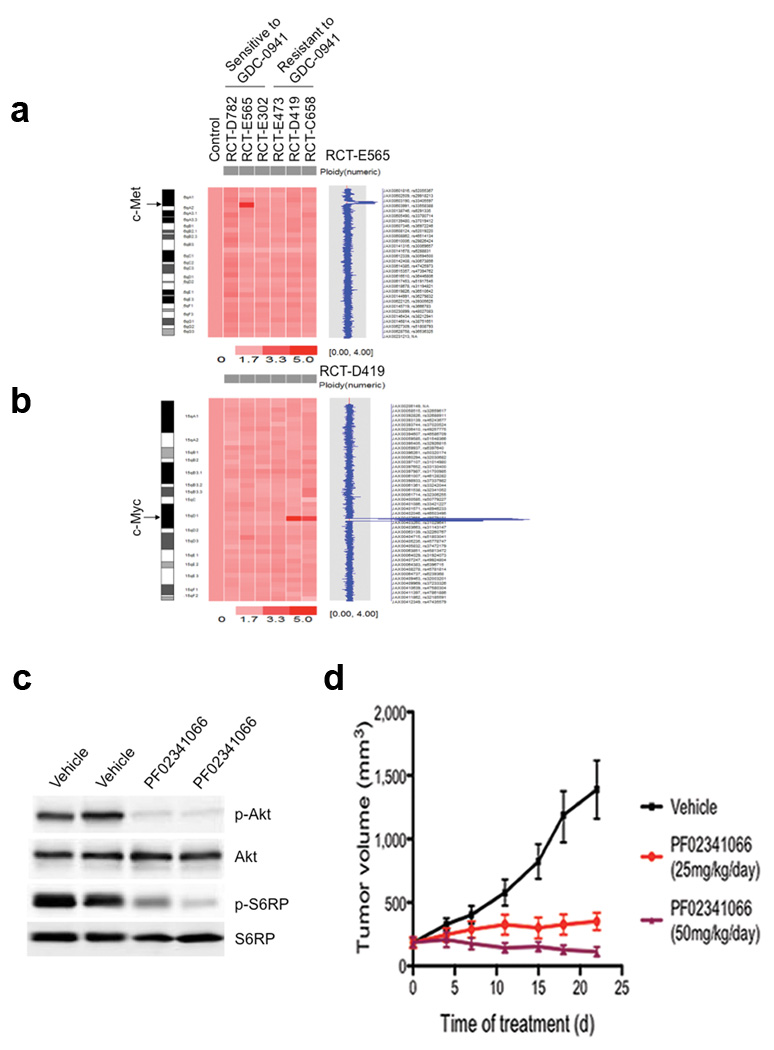
To search for genomic aberrations associated with this recurrence, we carried out mouse SNP array analyses of six recurrent tumors. A GDC-0941 sensitive tumor, RCT-E565, had a narrow amplification region encompassing c-Met (Fig. 3a) and also harbored a single copy loss of the tumor suppressor gene Cdkn2a (Supplementary Fig. 5a). Notably, two of three tumors that were resistant to GDC-0941 had a common amplification on chromosome 15 spanning 1.48 Mb (Chromosome 15:61,271,320–62,750,432), which contains the coding sequence for a single gene, c-Myc (Fig. 3b and Supplementary Fig. 6). In addition to c-Myc amplification, RCT-C658 also carried an amplification encompassing the Mdm2 oncogene (Supplementary Fig. 5b). Further analyses of c-Met, c-Myc and Mdm2 in a large collection of recurrent tumors showed that these oncogenes were upregulated in various fractions of recurrent tumors (Supplementary Fig. 7–12 and Supplementary Table 2). These data demonstrate that several of the most common gain- or loss-of-function genetic events in human cancers were recapitulated in this mouse tumor model.
Figure 3.
Genetic alterations associated with PIK3CAH1047R-independent tumor recurrence. Mouse SNP6.0 array analyses of six recurrent tumors identified an amplification region encompassing c-Met in RCT-E565 (a), and a common focal amplification at the c-Myc locus in RCT-D419 and RCT-C658 tumors (b). (c) Western blot analyses of p-Akt (Ser473) and pS6RP(S235/236) in two RCT-E565 xenograft tumors treated with vehicle or PF02341066. Samples were isolated 4 hours after the last dose from mice treated with PF02341066 for 3 days. (d) Responses of RCT-E565 xenograft tumors in NcrNu mice to PF02341066 or vehicle. Data are shown as mean ± S.E.M (each group, n = 6). *P < 0.005, ** P < 0.001 (Student’s t-test).
Since c-Met is a receptor tyrosine kinase known to activate the PI3K pathway via ERBB3 and GAB113,14, we tested whether c-Met amplification contributes to increased PI3K activity and tumor growth in the absence of PIK3CAH1047R expression. We confirmed that the RCT-E565 tumor, but not its parental primary tumor PMT-E565, had elevated c-Met mRNA and protein levels (Supplementary Fig. 13). We then treated mice bearing RCT-E565 tumor transplants with PF02341066, a c-Met inhibitor currently in clinical development15. PF02341066 abrogated both p-Akt and p-S6RP signals as well as tumor growth (Fig. 3c,d). These results suggest that c-Met elevation is one mechanism underlying the growth of recurrent tumors that have escaped oncogenic PIK3CA addiction but remain dependent on the PI3K pathway.
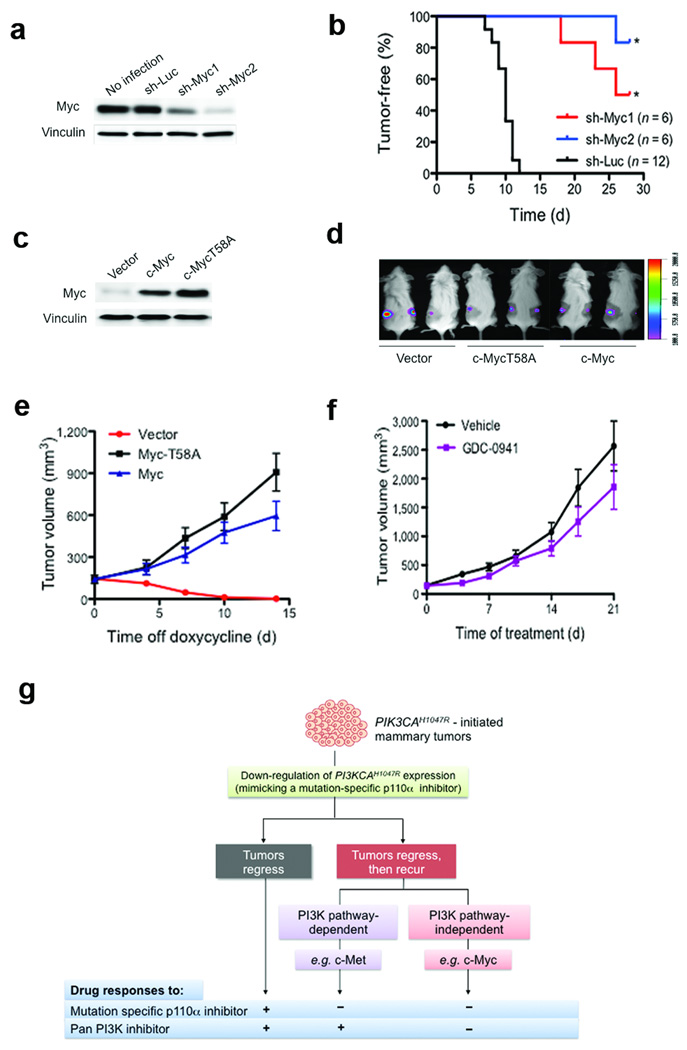
Because two of three GDC-0941-insensitive recurrent tumors featured c-Myc amplification (Fig. 3b) and overexpression (Supplementary Fig. 14), and given the known role of c-Myc functioning downstream of the PI3K pathway16, we hypothesized that c-Myc elevation might contribute to the recurrence of tumors that were resistant to PI3K inhibition. Further analyses of c-Myc for DNA copy number as well as both mRNA and protein levels in a large cohort of recurrent tumors (Supplementary Fig. 7–10) demonstrated that c-Myc elevation is a frequent event selected in recurrent tumors following sustained PIK3CAH1047R inactivation. To test whether c-Myc elevation contributes to tumor recurrence in a PI3K pathway-independent manner, we examined the effects of c-Myc knockdown by short hairpin RNAs (sh-Myc1 and sh-Myc2) on the growth of recurrent tumors transplanted in the mammary fat pads of immunodeficient mice. Knockdown of c-Myc dramatically reduced tumor incidence and extended the time to tumor onset (Fig. 4a,b). Conversely, enforced expression of c-Myc or c-MycT58A, a more stable version of c-Myc17, rendered otherwise PIK3CAH1047R-dependent tumors able to grow in the absence of doxycycline (Fig. 4c). Moreover, these c-Myc- or c-MycT58A-expressing tumors were resistant to GDC-0941 treatment (Fig. 4f and Supplementary Fig. 15). Together, these data suggest that c-Myc elevation is a mechanism that renders tumors free of addiction to PIK3CAH1047R and provides resistance to PI3K inhibition.
Figure 4.
Elevation of c-Myc drives mammary tumors to become independent of PIK3CAH1047R and resistant to PI3K inhibition. (a) shRNA knockdown of c-Myc in primary tumor cells isolated from RCT-D419. Western blot analysis of c-Myc in RCT-D419 parental cells or cells infected with the indicated lentiviral shRNAs. Vinculin was used as a loading control. (b) RCT-D419 cells expressing sh-Luc, sh-Myc1 or sh-Myc2 were transplanted into NOD-SCID mice and tumor formation monitored. Downregulation of c-Myc suppressed tumor formation. * P < 0.001 (log-rank test) (c) Western blot analysis of ectopically expressed c-Myc or c-MycT58A in D777 tumor cells isolated from a PIK3CAH1047R-dependent primary tumor that had been maintained on doxycycline. (d) Bioluminescence imaging showing tumor establishment in NOD-SCID mice transplanted with D777 cells expressing vector, c-MycT58A or c-Myc. These mice were maintained on doxycycline to sustain PIK3CAH1047R expression. (e) Tumors established by D777 cells expressing a control vector in the presence of doxycycline regressed upon doxycycline withdrawal. Tumors established by D777 cells expressing c-Myc or c-MycT58A continued to grow in the absence of doxycycline. Data are shown as mean ± S.E.M (n = 6). (f) Mice bearing D777-MycT58A tumors were treated with either GDC-0941 or vehicle and tumor growth followed. Data are shown as mean ± S.E.M (n = 6). (g) A schematic diagram summarizing three outcomes of PIK3CAH1047R-initiated tumors following inactivation of PIK3CAH1047R expression; tumors either regress (gray box) or recur (red box) via a PI3K pathway-dependent or -independent mechanism. As illustrated, these tumor outcomes affect tumor responses to drug treatment.
In our model, PIK3CAH1047R-induced tumors have three potential outcomes in response to PI3K inhibition (Fig. 4g). For those tumors that escape oncogene addiction and recur, c-Myc elevation represents a potential resistance mechanism with respect to current PI3K-targeted therapies in clinical trials. To explore whether PIK3CA mutations and c-MYC elevation coexist in human breast cancer, we analyzed several breast cancer datasets containing both PIK3CA mutation status and c-MYC copy number or expression data18–21. Among these cohorts, substantial fractions of PIK3CA mutation positive tumors have increased c-MYC copy number as well as mRNA and c-MYC protein levels22,23 (Supplementary Fig. 16 and Supplementary Table 3). Taken together, our findings suggest that aberrant elevation of c-MYC represents a potential mechanism by which tumors develop resistance to PI3K inhibition, and thus combination therapies targeting both PI3K and c-MYC may be necessary to circumvent resistance to PI3K-targeted therapy.
METHODS
Transgenic mice
We cloned human PIK3CAH1047R into the BamHI site of pTRE2 (Clontech) and inserted an IRES-firefly luciferase sequence downstream of PIK3CAH1047R to generate the TetO-PIK3CAH1047R-IRES-luciferase plasmid. We linearized the plasmid and gel-purified the released fragment for injection into fertilized oocytes from superovulated FVB mice at the transgenic core facility at the Brigham & Women’s Hospital, Boston. We crossed TetO-PIK3CAH1047R mice with MMTV-rtTA (MTB) mice (generously provided by L. Chodosh) to produce mice with inducible PIK3CAH1047R transgene expression in mammary glands (iPIK3CAH1047R). We administered iPIK3CAH1047R mice with doxycycline in their drinking water (2mg/ml). We performed all mouse experiments in accordance with protocols approved by the Institutional Animal Care and Use Committees of Dana-Farber Cancer Institute and Harvard Medical School.
Bioluminescence imaging
We anesthetized mice with ketamine and xylazine, and administeredmice with D-luciferin (Promega) intraperitoneally to monitored luciferase gene expression in vivo. We analyzed images using KODAK Molecular Imaging Software (version 4.5.0b6 SE).
Western blotting
We prepared lysates for mammary glands, mammary tumors or tumor cells in ice-cold RIPA buffer (Sigma-Aldrich) containing protease inhibitor cocktail (Roche). We cleared lysates by centrifugation before subjecting them to separation on SDS-PAGE gels and performed western blot assays as described previously5 with antibodies against phospho-AKT (Ser473 or Thr 308), AKT, phospho-S6 ribosomal protein (Ser235/Ser236), S6 ribosomal protein, and c-Met (Cell Signaling Technology), c-Myc (Santa Cruz Biotechnology) and vinculin (Sigma-Aldrich). We used immunofluorescently labelled anti-mouse IgG (Rockland Immunochemicals) and anti-rabbit IgG (Molecular Probes) to visualize western blots on an Odyssey scanner (Li-Cor, Lincoln, NE).
Histology and immunohistochemistry
We fixed tumors in formalin overnight before paraffin embedding. Paraffin blocks were sectioned, and stained with hematoxylin and eosin at the DF/HCC Rodent Histopathology Core. We performed immunohistochemistry using the antibodies: Ki67 (Vector), phospho-AktSer473 (Invitrogen) and phospho-S6 Ribosomal Protein (Cell Signaling). We performed TUNEL assay using the ApoTag Plus Peroxidase in situ TUNEL Apoptosis Kit (Millipore) according to the manufacturer’s instructions.
Mouse SNP analyses
We isolated genomic DNAs from mammary tissues or tumors using the Allprep DNA/RNA Kit (Qiagen). SNP array analyses with Mouse Diversity Genotyping Arrays (Affymetrix) were performed at the Microarray Core at Dana-Farber Cancer Institute. The SNP data (GEO accession number, GSE27691) were analyzed using a SNP microarray copy number application24 in the software suite, dChip (http://biosun1.harvard.edu/complab/dchip/), to compare positions of copy difference between a normal tissue sample from the inbred strain of mouse used in this study, and each of the tumor samples from the same inbred strain.
Tumor cell culture and viral transduction
We isolated tumors and dissociated them into single cells as described25 with the exception that the cells were cultured in DMEM/F12 supplemented with 0.5% FBS and 10ng/ml EGF and doxycycline (2µg/ml). We produced retrovirus or lentivirus and infected cells according to the methods previously described26,27. Infected cells were selected in culture medium plus puromycin (0.5 µg/ml) for 2 days. Cells were passaged no more than twice before being used for injection or further analysis. The retroviral vectors used in this study were MSCV-PIG (Puro IRES GFP) (used as a control vector, Addgene plasmid 18751), MSCV-MYC-T58A (Addgene plasmid 20076) and MSCV-MYC (derived from MSCV-MYC-T58A by site-directed mutagenesis (Stratagene)). The lentiviral shRNA constructs, sh-Luc, shMyc-1 (ID TRCN 42513) and shMyc-2 (ID TRCN 42517) were obtained from the RNAi consortium (Broad Institute, Cambridge, MA).
Tumor transplantation and in vivo treatment studies
For tumor grafting, we injected 2–5 × 105 tumor cells into the inguinal mammary glands of recipient mice (NcrNu or NOD-SCID females, 10–12 week old, Taconic). GDC-0941 was purchased from commercial sources (Sai Advantium Pharma) and was reconstituted in 0.5% methylcellulose (Sigma) and 0.2% Tween 80 (Sigma) and administered by oral gavage (120 mg/kg/day). PF02341066 (Selleck Chemicals) was administered via oral gavage at doses of 25 or 50mg/kg/day in water. Tumor volumes were measured twice a week with calipers and calculated by the following formula: Tumor volume = (length × width2)/228.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Roberts, L. Cantley and W. Sellers for scientific discussions and suggestions. We thank L. Clayton and D. Silver for critical review of this manuscript. We thank R. Bronson for pathological analyses of tumor samples. We thank C. Li and E. Allgood for technical assistance. This work was supported by US National Institute of Health (NIH) grants CA134502 (JJZ), CA148164-01 (JJZ, NG) and K08CA122833 (RB). The Stand Up To Cancer (JJZ, GBM), a Dana Farber Harvard Cancer Center breast cancer SPORE grants P50 CA089393-08S1 (JJZ), the Department of Defense (BC051565 to JJZ.), the V Foundation (JJZ, RB) and the Claudia Barr Program (JJZ). In compliance with Harvard Medical School guidelines, we disclose that JJZ and RB are consultants for Novartis Pharmaceuticals, Inc.
Footnotes
Note: Supplementary information is available on the Nature Medicine website.
AUTHOR CONTRIBUTIONS
P.L., H.C. and J.J.Z. designed the experiments, interpreted the data and wrote the paper. P.L. and H.C. performed most of the experiments. S.S, A.I and D.J.S. assisted with biochemical analyses and mouse work. J.Y., C.C, E.A.F., J.M. and R.S. performed genome-wide DNA copy number profiling. N.S.G. provided GDC-0941 inhibitor. M.R. and R.B. analyzed co-occurrence of PIK3CA mutation with c-MYC amplification and overexpression in human breast tumors. F.Z. and G.B.M. provided the RPPA data on the co-occurrence of PIK3CA mutation with increased c-MYC protein levels in human breast tumors.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 2.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogt PK, Kang S, Elsliger MA, Gymnopoulos M. Cancer-specific mutations in phosphatidylinositol 3-kinase. Trends Biochem Sci. 2007;32:342–349. doi: 10.1016/j.tibs.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao JJ, et al. The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A. 2005;102:18443–18448. doi: 10.1073/pnas.0508988102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isakoff SJ, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 8.Samuels Y, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunther EJ, et al. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. Faseb J. 2002;16:283–292. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- 11.Raynaud FI, et al. Biological properties of potent inhibitors of class I phosphatidylinositide 3-kinases: from PI-103 through PI-540, PI-620 to the oral agent GDC-0941. Mol Cancer Ther. 2009;8:1725–1738. doi: 10.1158/1535-7163.MCT-08-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Brien C, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3' kinase inhibitor GDC-0941 in breast cancer preclinical models. Clin Cancer Res. 2010;16:3670–3683. doi: 10.1158/1078-0432.CCR-09-2828. [DOI] [PubMed] [Google Scholar]
- 13.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 14.Turke AB, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou HY, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 16.Sears R, et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeh E, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6:308–318. doi: 10.1038/ncb1110. [DOI] [PubMed] [Google Scholar]
- 18.Cizkova M, et al. Gene expression profiling reveals new aspects of PIK3CA mutation in ERalpha-positive breast cancer: major implication of the Wnt signaling pathway. PLoS One. 2010;5:e15647. doi: 10.1371/journal.pone.0015647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 20.Haverty PM, et al. High-resolution genomic and expression analyses of copy number alterations in breast tumors. Genes Chromosomes Cancer. 2008;47:530–542. doi: 10.1002/gcc.20558. [DOI] [PubMed] [Google Scholar]
- 21.Saal LH, et al. Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc Natl Acad Sci U S A. 2007;104:7564–7569. doi: 10.1073/pnas.0702507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stemke-Hale K, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tibes R, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol Cancer Ther. 2006;5:2512–2521. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 25.Moody SE, et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell 2, Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- 26.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 27.Moody SE, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.