Abstract
Background
Prostate cancer is the second leading cause of cancer-related deaths in men, accounting for over 30,000 deaths annually. The purpose of this study was to test whether variation in selected candidate genes in biological pathways of interest for prostate cancer progression could help distinguish patients at higher risk for fatal prostate cancer.
Methods
In this hypothesis-driven study, we genotyped 937 single nucleotide polymorphisms (SNPs) in 156 candidate genes in a population-based cohort of 1,309 prostate cancer patients. We identified 22 top-ranking SNPs (P ≤0.01, FDR ≤0.70) associated with prostate cancer-specific mortality (PCSM). A subsequent validation study was completed in an independent population-based cohort of 2,875 prostate cancer patients.
Results
Five SNPs were validated (P ≤0.05) as being significantly associated with PCSM, one each in the LEPR, CRY1, RNASEL, IL4, and ARVCF genes. Compared to patients with 0–2 of the at-risk genotypes those with 4–5 at-risk genotypes had a 50% (95% CI, 1.2–1.9) higher risk of PCSM and risk increased with the number of at-risk genotypes carried (Ptrend = 0.001), adjusting for clinicopathological factors known to influence prognosis.
Conclusion
Five genetic markers were validated to be associated with lethal prostate cancer.
Impact
This is the first population-based study to demonstrate that germline genetic variants provide prognostic information for prostate cancer-specific survival. The clinical utility of this five-SNP panel to stratify patients at higher risk for adverse outcomes should be evaluated.
Keywords: Prostate cancer-specific mortality, survival, genetic variants, single nucleotide polymorphisms, hazard ratio
Introduction
Prostate cancer accounts for over 200,000 cancer diagnoses each year in the U.S. Although many men at diagnosis have localized tumors that will remain indolent and slow-growing, a substantial number have tumors that will become aggressive, leading to over 30,000 prostate cancer-specific deaths each year in the U.S. (1) Disease features such as Gleason score, stage and serum prostate-specific antigen (PSA) may help distinguish patients at higher risk for adverse outcomes, (2–4) however these factors alone do not accurately stratify patients with indolent versus more aggressive tumors. Biomarkers that could predict which men are at higher risk for having life-threatening prostate cancer would substantially improve patient management, targeting aggressive therapy to those most likely to benefit and avoiding over-treatment of low-risk patients.
Genetic background is known to play a role in the development of prostate cancer, (5–7) and genetic variants have been associated with risk of more advanced disease (8) (9) or with biochemical recurrence, (10–12) although these particular SNPs have not been associated with PCSM. (13–17) Some earlier studies have correlated SNP genotypes with PCSM, (12, 18–24) but results have not been validated. Thus, while evidence suggests genetic background influences prostate cancer outcomes, validated genetic markers associated with lethal disease have yet to be characterized.
To search for genetic markers that distinguish high-risk patients for PCSM, we conducted a hypothesis-driven candidate gene study focused on biological pathways (e.g., steroid hormones, DNA repair, inflammation, circadian rhythm, vitamin D) for which there is evidence for a role in modulating prostate cancer progression. Two independent population-based prostate cancer patient cohorts, one from Seattle and one from Sweden, were studied. Top-ranking SNPs associated with fatal disease in the Seattle cohort (P ≤0.01) were subsequently genotyped in the Swedish cohort for validation. The results reported here are the first to validate a panel of five SNPs in five genes that provide prognostic information at diagnosis for risk stratification of patients with prostate cancer.
Materials and Methods
Study Subjects
Seattle Patient Cohort
The Seattle cohort was established from two population-based case-control studies of prostate cancer in King County, Washington. (25, 26) In the first study, cases were diagnosed between January 1, 1993, and December 31, 1996 and were 40–64 years of age at diagnosis. In the second study, cases were diagnosed between January 1, 2002, and December 31, 2005 and were 35–74 years of age at diagnosis. Overall, 2,244 eligible patients were identified and 1,754 (78%) were interviewed. Blood samples were drawn from 1,457 (83%) interviewed patients.
The current study includes 1,309 Caucasian patients with DNA available. These cases had confirmed adenocarcinoma of the prostate and were ascertained from the Seattle-Puget Sound SEER cancer registry, which provided information on Gleason score, (27) cancer stage at diagnosis, (28) diagnostic PSA level and primary therapy. (29) Vital status and underlying cause of death were also obtained through the cancer registry, which links quarterly to the Washington State Center for Health Statistics database. Underlying cause of death was coded according to the International Classification of Diseases (30). A total of 60 patients died of prostate cancer over an average follow-up period of 8.5 years (range 0.8 – 15.9 years); vital status for this analysis was determined as of January 2009.
The study was approved by the Institutional Review Board (IRB) of the Fred Hutchinson Cancer Research Center and written informed consent was obtained from all study participants. Genotyping was approved by the IRB of the National Human Genome Research Institute where genotyping was completed for the Seattle cohort.
Swedish Patient Cohort
The validation cohort is comprised of patients enrolled in Cancer of the Prostate in Sweden (CAPS), a population-based study. (31) Cases were recruited from four regions in Sweden through Regional Oncology Centers between July 2001 and October 2003. A total of 3,648 eligible patients were identified, 3,161 (87%) participated and blood samples were obtained from 2,893 (92%). For the current study, 2,875 patients of European descent had DNA available for genotyping. Information on clinicopathological factors was obtained from the National Prostate Cancer Register, (32) including Gleason score, stage, diagnostic PSA level and initial treatment. Follow-up for mortality as of June 2009 was based on record linkage to the Swedish Cause of Death Register. (33) Prostate cancer was confirmed as the underlying cause of death (30) in 501 patients.
Written informed consent was obtained from all study participants. The research ethics committees at the Karolinska Institutet and Umeå University Hospital approved the study.
Genotyping
A total of 937 SNPs primarily from candidate genes in biological pathways of interest for prostate cancer (e.g., steroid hormones, inflammation, growth factors, DNA repair, circadian clock, vitamin D) were genotyped in the Seattle cohort. Most SNPs were selected using the Genome Variation Server (34); a tagSNP approach based on the HapMap CEU population was utilized to select variants with a minor allele frequency (MAF) of ≥5% and an r2 ≥0.8. Priority was given to selection of nonsynonymous SNPs, followed by tagSNPs and SNPs previously reported to be associated with prostate cancer. For genotyping, the SNPlex Genotyping System (Applied Biosystems, Inc., Foster City, CA) was used with GeneMapper software to assign genotypes. Replicate samples (n = 140) were interspersed throughout all genotyping batches. Genotyping scores, including quality control data, were re-checked by different laboratory personnel to confirm the accuracy of each assay. Ninety-one SNPs were removed due to genotyping failure (n = 58), monomorphism (n = 27), or a low MAF (n = 6). The remaining 846 SNPs (Supplemental Table 1) were used for permutation testing to identify variants associated with PCSM.
Twenty-two SNPs were found to be significantly associated with PCSM in the Seattle cohort. For these SNPs, call rates were >95% and there was >98% agreement between duplicate samples. In addition, all 22 SNPs were in Hardy-Weinberg equilibrium (P >0.05) in 1,266 genotyped Caucasian controls who were age-matched to the Seattle patient cohort.
The 22 top ranking SNPs discovered in the Seattle cohort were then genotyped in the validation cohort. The MassARRAY iPLEX system (Sequenom, Inc., San Diego, CA) was used to genotype samples at Wake Forest University. Duplicate samples (n = 106) and two negative controls that were blinded to the laboratory technician were included in each 96-well plate. The overall concordance level for 20 of the 22 SNPs was 99.9% among duplicated samples. Two SNPs (rs228697 and rs1029153) failed genotyping.
Statistical Analyses
SNP Selection by Permutation Testing
Six Cox regression models (three adjusting for age at diagnosis alone and three adjusting for age in addition to stage, Gleason score, diagnostic PSA, and primary treatment) were completed under dominant, recessive and log-additive (linear trend) genetic models. One-thousand permutation datasets were generated by randomly permuting all 846 SNPs together across subjects while keeping the clinical variables fixed, i.e., the clinical variables were retained with each subject and not permuted, and the same six Cox models were run for each permuted dataset to obtain the distribution of P-values under the null hypothesis of no SNP effect. False Discovery Rates (FDR, q value) (35) were calculated based on the distribution of permutation P-values and the observed P-values. This approach is the same as that of Morris and coworkers. (36) Using an a priori determined threshold of P ≤0.01 from the original data to select the top ranked SNPs, a total of 22 SNPs associated with PCSM were selected for validation. For these top ranking 22 SNPs, the FDR ranged from 0.26 to 0.70, which we deemed acceptable due to the affordable cost of genotyping in the validation cohort and the desire to not miss potentially informative SNPs for validation.
Cox Models
The hazard ratio (HR), 95% confidence interval (CI) and P-value were obtained for each of the 22 top ranked SNPs (Seattle cohort) under the best-fitting genetic model for each SNP. Cox models using the same underlying best-fitting genetic model from the Seattle cohort, but allowing for three sets of covariates (i.e., 1) age at diagnosis; 2) age at diagnosis, Gleason score, stage, and diagnostic PSA level; and 3) age at diagnosis, Gleason score, stage, diagnostic PSA level and initial treatment), were completed for each of the 20 SNPs genotyped in the Swedish cohort; a SNP was judged to be validated if the P-value from one of the three models was ≤0.05 (one-sided test) and the effect on mortality risk was in the same direction as in the Seattle dataset. HRs for the cumulative number of at-risk genotypes were calculated by Cox models adjusted for age alone and for age plus the four clinicopathological factors. The grouping by number of at-risk genotypes was done to ensure that each group had an expected number of at least five fatal events based on the Seattle cohort. The same grouping of at-risk genotypes (i.e., 0–2, 3, 4, or 5) was used to generate Kaplan-Meier (K–M) curves. A backward stepwise Cox model (adjusted for age and the four clinicopathological factors) was used to rank validated SNPs by level of statistical significance. SNP by SNP interactions were also examined, and an interaction was considered significant if the P-value associated with the HR was <0.001 (Bonferroni adjustment, P ≤0.05).
Results
Characteristics of the two patient cohorts are shown in Table 1. Patients in the Seattle cohort were younger at diagnosis than those in the Swedish cohort (mean age at diagnosis 59.9 versus 65.8 years, respectively, P <0.0001). A higher proportion of patients from Sweden (17.4%) had died of prostate cancer relative to those from Seattle (4.6%) during a median follow-up time of 6.5 years in each cohort. This is consistent with the higher prostate cancer mortality rate in Sweden relative to the U.S. (37) The Swedish population also had a greater proportion of cases with advanced clinicopathological features and who were treated with androgen deprivation therapy.
Table 1.
Clinicopathological Characteristics of Two Population-based Prostate Cancer Patient Cohorts
| Seattle Cohort (N=1,309) | Swedish Cohort (N=2,875) | |||
|---|---|---|---|---|
| N | % | N | % | |
| Age at diagnosis, years | ||||
| Mean | 59.9 | 65.8 | ||
| Median | 60.0 | 64.9 | ||
| Range | 35.0 – 74.0 | 44.6 – 80.4 | ||
| Follow-up time, years | ||||
| Mean | 8.5 | 6.0 | ||
| Median | 6.5 | 6.5 | ||
| Range | 0.8 – 15.9 | 0.3 – 8.6 | ||
| Prostate cancer-specific death | ||||
| No1 | 1,249 | 95.4 | 2,374 | 82.6 |
| Yes | 60 | 4.6 | 501 | 17.4 |
| Age at death, years | ||||
| Mean | 63.9 | 71.2 | ||
| Median | 65.2 | 71.2 | ||
| Range | 44.9–78.3 | 48.5–85.7 | ||
| Stage | ||||
| Local | 1,023 | 78.2 | 1,885 | 65.6 |
| Regional | 254 | 19.4 | 651 | 22.6 |
| Distant | 32 | 2.4 | 266 | 9.3 |
| Missing | 0 | 0.0 | 73 | 2.5 |
| Gleason score | ||||
| 2 – 4 | 67 | 5.1 | 106 | 3.7 |
| 5 – 6 | 680 | 51.9 | 1,269 | 44.1 |
| 7 | 432 | 33.0 | 782 | 27.2 |
| 8 – 10 | 126 | 9.6 | 467 | 16.2 |
| Missing | 4 | 0.3 | 251 | 8.7 |
| Diagnostic PSA level, ng/mL | ||||
| < 4 | 178 | 13.6 | 148 | 5.1 |
| 4 – 9.9 | 722 | 55.2 | 993 | 34.5 |
| 10 – 19.9 | 191 | 14.6 | 651 | 22.6 |
| ≥ 20 | 118 | 9.0 | 1,003 | 34.9 |
| Missing | 100 | 7.6 | 80 | 2.8 |
| Primary therapy | ||||
| Radical prostatectomy | 770 | 58.8 | 713 | 24.8 |
| Radiation therapy | 359 | 27.4 | 682 | 23.7 |
| Androgen deprivation | 61 | 4.7 | 927 | 32.2 |
| None | 115 | 8.8 | 488 | 17.0 |
| Other | 4 | 0.3 | 22 | 0.8 |
| Missing | 0 | 0.0 | 43 | 1.5 |
Includes men who died of other causes and were censored at time of death (Seattle, n=102; Sweden, n=258)
Permutation testing on 846 SNPs revealed 22 variants that were significantly (P ≤0.01) associated with PCSM in the Seattle cohort (Table 2). Genotyping data validated SNP rs1137100 minor allele G as being associated with a decrease in PCSM (HR = 0.82; 95% CI, 0.67–1.00; P =0.027) in the Swedish cohort under the same dominant genetic model adjusted for the same covariates as in the Seattle dataset. Also under the same dominant genetic model as used in the analysis of the Seattle dataset, but allowing for different covariates, four additional SNPs were validated as being significantly associated with PCSM in the Swedish cohort: rs627839 (P =0.024), rs2070874 (P =0.011), rs10778534 (P =0.022), and rs5993891 (P =0.024).
Table 2.
Hazard ratios (HR) for prostate cancer-specific mortality associated with a panel of 22 single nucleotide polymorphisms (SNPs) in candidate genes in a discovery cohort (Seattle) and a validation cohort (Sweden)
| Seattle Cohort | Swedish Cohort | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr. | Gene | Alleles1 | MAF2 | HR | 95% CI | P-value (q value) |
Model3 | MAF2 | HR | 95% CI | P-value4 | Model3 | P-value5 |
| rs1137100 | 1p31 | LEPR | A/G | 0.27 | 0.29 | 0.14–0.60 | 0.0001 (0.26) | Dom: ACP | 0.29 | 0.82 | 0.67–1.00 | 0.027 | Dom: ACP | 0.027 |
| rs228697† | 1p36 | PER3 | C/G | 0.11 | 0.25 | 0.10–0.60 | 0.0002 (0.26) | Dom: ACP | ||||||
| rs635261 | 1q25 | RNASEL | G/C | 0.36 | 0.22 | 0.07–0.65 | 0.0007 (0.56) | Rec: ACP | 0.35 | 0.98 | 0.74–1.29 | 0.44 | Rec: A | 0.48 |
| rs627839 | 1q25 | RNASEL | G/T | 0.47 | 3.98 | 1.64–9.65 | 0.0004 (0.26) | Dom: ACP | 0.47 | 1.22 | 1.00–1.50 | 0.024 | Dom: A | 0.22 |
| rs4583514 | 2p21 | MSH2 | G/A | 0.38 | 2.49 | 1.21–5.10 | 0.01 (0.70) | Dom: ACP | 0.43 | 0.98 | 0.81–1.18 | NS | Dom: A | NS |
| rs4608577 | 2p21 | MSH2 | T/G | 0.17 | 2.04 | 1.36–3.07 | 0.002 (0.57) | Tre: A | 0.21 | 0.93 | 0.80–1.09 | NS | Tre: A | NS |
| rs523349 | 2p23 | SRD5A2 | C/G | 0.29 | 0.49 | 0.28–0.86 | 0.01 (0.66) | Dom: A | 0.32 | 1.15 | 0.94–1.40 | NS | Dom: ACP6 | NS |
| rs12467911 | 2p23 | SRD5A2 | C/T | 0.28 | 0.45 | 0.24–0.81 | 0.005 (0.57) | Dom: A | 0.32 | 1.16 | 0.95–1.41 | NS | Dom: ACP6 | NS |
| rs11710277 | 3p21 | SEMA3F | A/G | 0.09 | 3.71 | 1.75–7.90 | 0.001 (0.53) | Dom: ACP | 0.08 | 0.86 | 0.66–1.11 | NS | Dom: A | NS |
| rs11205 | 5q23 | HSD17B4 | A/G | 0.39 | 0.21 | 0.06–0.70 | 0.001 (0.61) | Rec: ACP | 0.44 | 1.08 | 0.84–1.39 | NS | Rec: ACP6 | NS |
| rs2070874 | 5q31 | IL4 | C/T | 0.16 | 2.16 | 1.27–3.67 | 0.005 (0.57) | Dom: A | 0.19 | 1.27 | 1.04–1.56 | 0.011 | Dom: ACP6 | 0.047 |
| rs1799964 | 6p21 | TNF/LTA | T/C | 0.21 | 0.39 | 0.20–0.77 | 0.003 (0.57) | Dom: A | 0.20 | 0.91 | 0.74–1.13 | 0.20 | Dom: ACP | 0.44 |
| rs4645959 | 8q24 | C-MYC | A/G | 0.04 | 0 | 0.00-inf. | 0.003 (0.57) | Tre: ACP | 0.02 | 1.23 | 0.84–1.78 | NS | Tre: A | NS |
| rs1029153 † | 10q11 | CXCL12 | T/C | 0.31 | 0.22 | 0.07–0.75 | 0.005 (0.57) | Tre: A | ||||||
| rs2839685 | 10q11 | CXCL12 | C/T | 0.15 | 28.2 | 7.21–110.2 | 0.0003 (0.43) | Rec: ACP | 0.10 | 1.31 | 0.49–3.53 | 0.30 | Rec: ACP6 | 0.34 |
| rs2308327 | 10q26 | MGMT | A/G | 0.13 | 0.32 | 0.13–0.78 | 0.004 (0.57) | Tre: A | 0.13 | 0.92 | 0.76–1.12 | 0.20 | Tre: A | 0.20 |
| rs10778534 | 12q23 | CRY1 | T/C | 0.36 | 2.21 | 1.19–4.12 | 0.008 (0.57) | Dom: A | 0.35 | 1.23 | 1.00–1.51 | 0.022 | Dom: ACP | 0.036 |
| rs2494750 | 14q32 | AKT1 | C/G | 0.07 | 0.22 | 0.07–0.70 | 0.001 (0.50) | Tre: ACP | 0.06 | 0.93 | 0.70–1.24 | 0.31 | Tre: ACP | 0.31 |
| rs1799814 | 15q24 | CYP1A1 | C/A | 0.05 | 0.13 | 0.03–0.57 | 0.0003 (0.26) | Tre: ACP | 0.03 | 1.47 | 0.86–1.28 | NS | Tre: ACP6 | NS |
| rs25487 | 19q13 | XRCC1 | G/A | 0.36 | 0.49 | 0.31–0.77 | 0.003 (0.57) | Tre: A | 0.35 | 0.95 | 0.83–1.08 | 0.20 | Tre: A | 0.40 |
| rs915927 | 19q13 | XRCC1 | A/G | 0.43 | 2.54 | 1.24–5.18 | 0.005 (0.57) | Dom: A | 0.44 | 0.88 | 0.71–1.08 | NS | Dom: ACP | NS |
| rs5993891 | 22q11 | ARVCF | C/T | 0.05 | 0.21 | 0.07–0.61 | 0.0004 (0.26) | Dom: ACP | 0.05 | 0.72 | 0.52–1.01 | 0.024 | Dom: A | 0.21 |
These SNPs were not evaluated in the Swedish cohort due to genotyping failure.
Major/minor allele
MAF: minor allele frequency, calculated from cases that did not die of prostate cancer.
Genetic model of best fit (Dom=dominant, Rec=recessive, Tre=trend) adjusted for age (A) alone, age + clinicopathological (ACP) factors (Gleason score, stage, diagnostic PSA level and primary treatment).
NS= not significant as the HR in the validation (Swedish) dataset is in the opposite direction as in the discovery (Seattle) dataset.
P-value for the Swedish dataset using the same best fitting genetic model and the same covariates as for the Seattle dataset.
Adjusted for age + clinicopathological factors as in footnote 3 above, excluding primary treatment.
Hazard ratios were then calculated according to cumulative number of at-risk genotypes. As shown in Table 3 for the Swedish cohort, compared to men with 0–2 at-risk SNP genotypes, those with four (HR = 1.51; 95% CI, 1.16–1.97) or five (HR = 1.46; 95% CI, 0.97–2.19) at-risk genotypes had approximately a 50% higher risk of dying from prostate cancer, after adjustment for age and clinicopathological factors. In these analyses, the HRs increased directly with the cumulative number of at-risk genotypes (Ptrend =0.0005, adjusting for age only, and =0.001 adjusting also for clinicopathological factors). The proportion of all patients carrying four (28%) or five (6%) at-risk alleles was similar in both cohorts.
Table 3.
Hazard ratios (HR) for prostate cancer-specific mortality associated with the cumulative number of at-risk genotypes for a panel of five validated single nucleotide polymorphisms (SNPs)
| Seattle Cohort | Swedish Cohort | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of at-risk genotypes1 |
Number at-risk / Number of fatal events |
HR2 | 95% CI | HR3 | 95% CI | Number at-risk / Number of fatal events |
HR2 | 95% CI | HR3 | 95% CI |
| 0 – 2 | 314 / 4 | 1.00 | 1.00 | 803 / 113 | 1.00 | 1.00 | ||||
| 3 | 446 / 16 | 2.82 | 0.94–8.45 | 8.14 | 2.44–27.12 | 1047 / 181 | 1.24 | 0.98–1.57 | 1.05 | 0.81–1.37 |
| 4 | 322 / 16 | 3.92 | 1.31–11.74 | 9.08 | 2.73–30.15 | 797 / 154 | 1.44 | 1.13–1.83 | 1.51 | 1.16–1.97 |
| 5 | 69 / 13 | 15.80 | 5.14–48.52 | 15.12 | 4.44–51.56 | 176 / 39 | 1.69 | 1.17–2.43 | 1.46 | 0.97–2.19 |
Genotypes for SNPs rs1137100, rs627839, rs2070874, rs10778534, and rs5993891; patients missing data for any of the five SNP genotypes were excluded from the analysis (Seattle n=158; Sweden n=52).
Hazard ratio adjusted for age at diagnosis; p-value for trend = 0.0005 in the Swedish cohort.
Hazard ratio adjusted for age at diagnosis, stage, Gleason score, diagnostic PSA, and primary treatment; p-value for trend = 0.001 in the Swedish cohort.
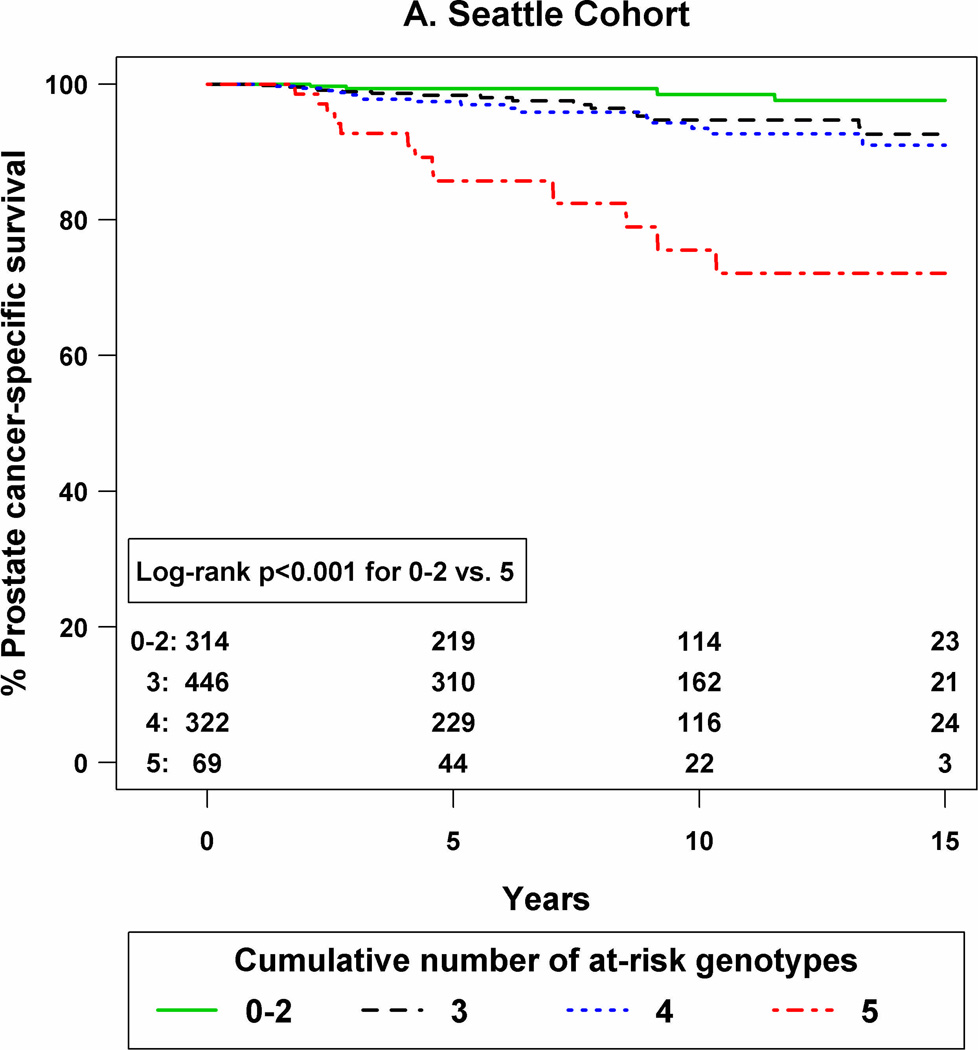
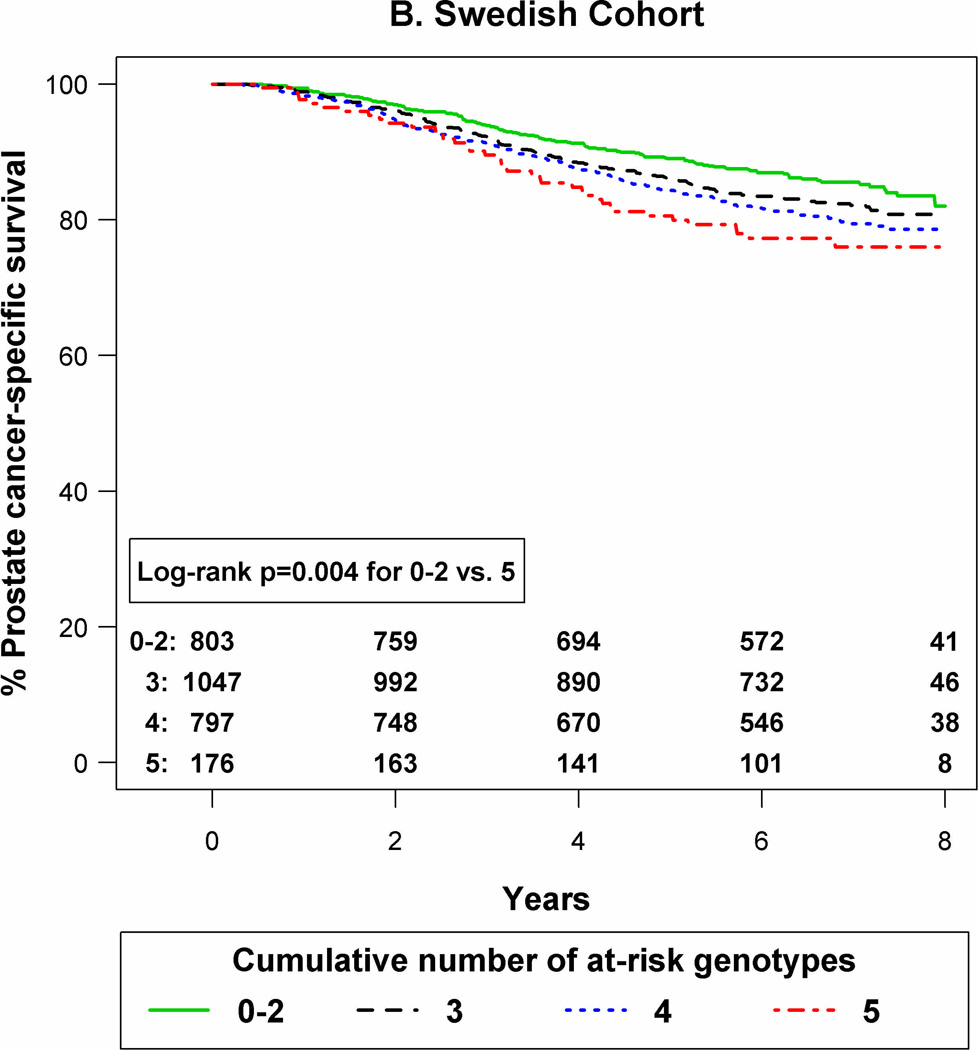
K–M curves were constructed according to the number of at-risk genotypes in each cohort (Figure 1). As shown, compared to patients with 0–2 at-risk genotypes, those with all five at-risk SNP genotypes had the lowest prostate cancer-specific survival in both the Seattle (P <0.001) and the Swedish (P =0.004) cohorts.
Figure 1.
Kaplan-Meier curves for prostate cancer-specific survival by number of at-risk genotypes for a 5-SNP panel in the Seattle cohort (panel A) and the Swedish cohort (panel B).
Stepwise backward selection Cox models were completed to evaluate the relative ranking of the five SNP genotypes in relation to PCSM. Based on models adjusted for age at diagnosis and the four clinicopathological variables, the most significant SNP in the Seattle dataset was rs1137100 (P =0.001) in the LEPR gene and in the Swedish dataset was rs10778534 (P =0.045) in the CRY1 gene.
Lastly, we tested the five genetic variants for SNP by SNP interactions. No evidence for significant interaction between these markers was found (all P-values >0.001).
Discussion
Prostate cancer is a heterogeneous disease and current clinical and pathological features are not reliably accurate for predicting individual patient outcomes. (38, 39) The ability to distinguish patients at elevated risk for having aggressive, life-threatening prostate cancer at the time of diagnosis could improve care for the subset of cases most likely to benefit from aggressive therapy and help avoid over-treatment of patients whose tumors are likely to remain indolent. Better biomarkers that can stratify patients according to tumor aggressiveness are urgently needed. Thus, we undertook a hypothesis-testing candidate gene approach to identify and validate genetic variants as prognostic markers for fatal prostate cancer.
The current study was developed based on the notion that the genetic background upon which cancer develops likely modulates tumor growth rate and propensity to metastasize as well as treatment response. This idea is consistent with results from animal models that demonstrate that genetic background influences cancer progression and metastatic potential. (40, 41) Further support comes from a recent study of squamous cell skin cancer in humans that showed somatic events in tumors can depend on an individual’s germline genotype. (42)
We and others (43) have hypothesized that inherited predisposition influences prostate cancer progression. Variants in several candidate genes, including MIC-1, (18) SEP15, (21) VDR, CYP27B1,and CYP24A1, (22) TLR-9, (19) Megalin, (20) CYP17, (23) and FASN, (24) and two risk-loci (19q13 and 11q13) (12) have previously been associated with PCSM in individual studies, although results have not been validated in independent cohorts. More recently, a GWAS of 196 patients with either metastasis or PCSM found no genome-wide significant results, but one intergenic SNP was replicated (P =0.05) in a highly selected case series. (44) In the present study we investigated a subset of these same candidate gene variants, though none was among our top ranked 22 SNPs.
A nonsynonymous coding SNP (rs1137100) in exon 4 of the leptin receptor (LEPR) gene was validated as the strongest marker associated with PCSM in our study. LEPR is a cytokine receptor that is highly expressed in normal and malignant prostate tissue. (45) The binding of leptin to its receptor, LEPR, leads to several downstream effects that may affect prostate carcinogenesis, including stimulation of tissue growth, (46) inflammation, (47) angiogenesis, (48) and bone mass regulation. (49) The latter effect makes LEPR an interesting candidate for disease progression, (50) since the primary metastatic site for prostate cancer is the bone and bony metastases are predictive of fatal prostate cancer. (51–53)
The other SNPs significantly associated with survival in our study include rs627839, which tags the RNASEL gene within the hereditary prostate cancer 1 (HPC1) locus. (54) A role in prostate cancer has been suggested through the protein’s ability to increase apoptosis and inhibit inflammation, cell proliferation and adhesion. (55, 56) Of the five genes highlighted here, this is the only one previously evaluated in a study of prostate cancer that found no mortality association. (57) Variant rs2070874 is in the promoter region of Interleukin 4 (IL4), which plays a role in cancer via activation of the Stat6 transcription factor. (58) Studies have shown that IL4 inhibits tumor growth (59–62) and angiogenesis, (63) and prevents invasion and migration of colon cancer cells. (64) Of note, rs2070874 is in perfect LD with rs2243250, a promoter variant for which the minor allele confers diminished IL4 expression. (65) SNP rs10778354 tags the Cryptochrome 1 (CRY1) gene in the circadian rhythm pathway. (66–68) Circadian clock genes regulate androgen levels, (69) which are known to affect prostate cancer progression, (70) and may also function as tumor suppressors through regulation of cell proliferation, apoptosis and response to DNA damage. (71) Finally, rs5993891 is located in the ARVCF gene, a member of the p120catenin family of proteins. Increased expression of ARVCF has been shown to disrupt cell adhesion, (72) which may facilitate cancer progression. This SNP also tags the COMT gene, which encodes a protein that neutralizes the genotoxic effects of catechol estrogens. (73, 74)
The five validated SNPs highlighted above represent the first evidence for this panel of genetic variants being associated with prostate cancer mortality. Two variants (rs228697 in PER3 and rs1029153 in CXCL12) associated with PCSM in the Seattle cohort were not evaluated in the Swedish cohort due to genotyping failure. Thus, additional investigation of these variants is warranted. Strengths of this project include the population-based patient cohorts, the discovery-validation study design, the large number of patients and outcomes in the validation cohort, and the hypothesis-driven approach focused on genes in biological pathways of interest. One potential concern relates to the sample size in the discovery cohort, which may have missed additional SNP associations due to limited power. The smaller size of the Seattle cohort and regression to the mean likely explain the reduced HRs associated with the five SNP genotypes observed in the Swedish cohort relative to the Seattle cohort. Another potential issue is the difference in clinicopathological factors between the two cohorts. To accommodate these dissimilar features, adjustment covariates were allowed to vary in the Cox models, although this does potentially lead to multiplicity and false positives. It would be important to confirm our findings in a U.S. cohort with similar clinical features to those of the Seattle cohort so that covariate models could be fixed for validation. The focus on fatal events reduces potential bias related to different screening practices between the two cohorts that likely account for the differing clinical characteristics. Interestingly, the proportion of patients carrying 4 or 5 of the at-risk genotypes was similar in the two cohorts. Because Gleason score and stage are strong predictors of PCSM, adjustment for these factors in multivariate models is important even though it may diminish the magnitude of the SNP-PCSM associations.
In conclusion, our study provides initial validation for five germline genetic variants that are associated with lethal prostate cancer outcomes. Three of these polymorphisms (rs1137100, rs2070874, rs10778534) were significantly associated with PCSM in multivariate models that included the traditional clinical factors (i.e., Gleason score, stage) used to predict outcomes, suggesting that these variants contribute independent data beyond the standard prognostic variables. Two other SNPs (rs627839, rs5993891) were validated to be associated with PCSM in models that adjusted for age at diagnosis alone. There was also preliminary evidence for a dose-response effect according to the number of at-risk genotypes carried. A validation study of this five-SNP panel for stratification of patients at the time of diagnosis into those at higher risk for adverse outcomes is urgently needed. Understanding the individual prostate cancer patient’s risk for progression to lethal disease will allow more informed counseling of patients regarding therapy options, follow-up plans, and approaches for secondary prevention. Such high-risk patients should benefit most from early aggressive therapy and be ideal candidates for novel adjuvant treatment trials aimed at improving patient survival.
Supplementary Material
Acknowledgments
Funding
This work was supported by grants RO1 CA056678 (J.L.S.), R01 CA092579 (J.L.S.), R03 CA121871 (J.L.S.), RO3 CA137799 (J.L.S.), RO1 CA133009 (S.L.Z.), RO1 CA129684 (J.X.) and P50 CA097186 from the National Cancer Institute at the National Institutes of Health. Additional support was provided by the Fred Hutchinson Cancer Research Center, the Intramural Program of the National Human Genome Research Institute, the Cancer Risk Prediction Center (CRisP; www.crispcenter.org), a Linneus Centre (contract 70867902) financed by the Swedish Research Council, the Swedish Research Council (grant K2010-70X-20430-04-3), the Swedish Cancer Foundation (grant 09-0677), the Hedlund Foundation, the Söderberg Foundation, the Enqvist Foundation, and ALF funds from the Stockholm County Council.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010;60:1–24. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Partin AW, Kattan MW, Subong EN, Walsh PC, Wojno KJ, Oesterling JE, et al. Combination of prostate-specific antigen, clinical stage, and Gleason score to predict pathological stage of localized prostate cancer. A multi-institutional update. JAMA. 1997;277:1445–1451. [PubMed] [Google Scholar]
- 3.Ross PL, Scardino PT, Kattan MW. A catalog of prostate cancer nomograms. J Urol. 2001;165:1562–1568. [PubMed] [Google Scholar]
- 4.Stephenson AJ, Scardino PT, Eastham JA, Bianco FJ, Jr., Dotan ZA, Fearn PA, et al. Preoperative nomogram predicting the 10-year probability of prostate cancer recurrence after radical prostatectomy. J Natl Cancer Inst. 2006;98:715–717. doi: 10.1093/jnci/djj190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 6.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 7.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 8.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Zheng SL, Isaacs SD, Wiley KE, Wiklund F, Sun J, et al. Inherited genetic variant predisposes to aggressive but not indolent prostate cancer. Proc Natl Acad Sci USA. 2010;107:2136–2140. doi: 10.1073/pnas.0914061107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin HC, Liu CC, Kang WY, Yu CC, Wu TT, Wang JS, et al. Influence of cytokine gene polymorphisms on prostate-specific antigen recurrence in prostate cancer after radical prostatectomy. Urologia Internationalis. 2009;83:463–470. doi: 10.1159/000251189. [DOI] [PubMed] [Google Scholar]
- 11.Cheng I, Plummer SJ, Neslund-Dudas C, Klein EA, Casey G, Rybicki BA, et al. Prostate cancer susceptibility variants confer increased risk of disease progression. Cancer Epidemiol Biomarkers Prev. 2010;19:2124–2132. doi: 10.1158/1055-9965.EPI-10-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher DJ, Vijai J, Cronin AM, Bhatia J, Vickers AJ, Gaudet MM, et al. Susceptibility loci associated with prostate cancer progression and mortality. Clin Cancer Res. 2010;16:2819–2832. doi: 10.1158/1078-0432.CCR-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kote-Jarai Z, Easton DF, Stanford JL, Ostrander EA, Schleutker J, Ingles SA, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomarkers Prev. 2008;17:2052–2061. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FitzGerald LM, Kwon EM, Koopmeiners JS, Salinas CA, Stanford JL. Analysis of recently identified prostate cancer susceptibility loci in a population-based study: Associations with family history and clinical features. Clin Cancer Res. 2009;15:3231–3237. doi: 10.1158/1078-0432.CCR-08-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiklund FE, Adami HO, Zheng SL, Stattin P, Isaacs WB, Gronberg H, et al. Established prostate cancer susceptibility variants are not associated with disease outcome. Cancer Epidemiol Biomarkers Prev. 2009;18:1659–1662. doi: 10.1158/1055-9965.EPI-08-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penney KL, Salinas CA, Pomerantz M, Schumacher FR, Beckwith CA, Lee GS, et al. Evaluation of 8q24 and 17q risk loci and prostate cancer mortality. Clin Cancer Res. 2009;15:3223–3230. doi: 10.1158/1078-0432.CCR-08-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salinas CA, Koopmeiners JS, Kwon E, FitzGerald LM, Lin DW. Clinical utility of five genetic variants for predicting prostate cancer risk and mortality. Prostate. 2009;69:363–372. doi: 10.1002/pros.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes VM, Severi G, Southey MC, Padilla EJ, English DR, Hopper JL, et al. Macrophage inhibitory cytokine-1 H6D polymorphism, prostate cancer risk, and survival. Cancer Epidemiol Biomarkers Prev. 2006;15:1223–1225. doi: 10.1158/1055-9965.EPI-06-0063. [DOI] [PubMed] [Google Scholar]
- 19.Stark JR, Wiklund F, Gronberg H, Schumacher F, Sinnott JA, Stampfer MJ, et al. Toll-like receptor signaling pathway variants and prostate cancer mortality. Cancer Epidemiol Biomarkers Prev. 2009;18:1859–1863. doi: 10.1158/1055-9965.EPI-08-0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt SK, Karyadi DM, Kwon EM, Stanford JL, Nelson PS, Ostrander EA. Association of megalin genetic polymorphisms with prostate cancer risk and prognosis. Clin Cancer Res. 2008;14:3823–3831. doi: 10.1158/1078-0432.CCR-07-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penney KL, Schumacher FR, Li H, Kraft P, Morris JS, Kurth T, et al. A large prospective study of SEP15 genetic variation, interaction with plasma selenium levels, and prostate cancer risk and survival. Cancer Prev Res. 2010;3:604–610. doi: 10.1158/1940-6207.CAPR-09-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holt SK, Kwon EM, Koopmeiners JS, Lin DW, Feng Z, Ostrander EA, et al. Vitamin D pathway gene variants and prostate cancer prognosis. Prostate. 2010;70:1448–1460. doi: 10.1002/pros.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright JL, Kwon EM, Lin DW, Kolb S, Koopmeiners JS, Feng Z, et al. CYP17 polymorphisms and prostate cancer outcomes. Prostate. 2010;70:1094–1101. doi: 10.1002/pros.21143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen PL, Ma J, Chavarro JE, Freedman ML, Lis R, Fedele G, et al. Fatty acid synthase polymorphisms, tumor expression, body mass index, prostate cancer risk, and survival. J Clin Oncol. 2010;28:3958–3964. doi: 10.1200/JCO.2009.27.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanford JL, Wicklund KG, McKnight B, Daling JR, Brawer MK. Vasectomy and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1999;8:881–886. [PubMed] [Google Scholar]
- 26.Agalliu I, Salinas CA, Hansten PD, Ostrander EA, Stanford JL. Statin use and risk of prostate cancer: results from a population-based epidemiologic study. Am J Epidemiol. 2008;168:250–260. doi: 10.1093/aje/kwn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gleason DF. Histologic grading and clinical staging of prostatic carcinoma. Urologic pathology. In: Tannenbaum M, editor. The Prostate. Philadelphia: Lea and Febiger; 1977. pp. 171–197. [Google Scholar]
- 28.American Joint Committee on Cancer. Past Editions of the AJCC Cancer Staging Manual. 2010 [cited; Available from: http://www.cancerstaging.org/products/pasteditions.html.
- 29.Young JLJ, Roffers SD, Ries LAG, Fritz AG, Hurlbut AA. SEER Summary Staging Manual - 2000: Codes and Coding Instructions. Bethesda: National Cancer Institute; 2001. [Google Scholar]
- 30.Manual of the International Statistical Classification of Diseases, Injuries, and Causes of Death. Geneva: World Health Organization; 1977. [Google Scholar]
- 31.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 32.Adolfsson J, Garmo H, Varenhorst E, Ahlgren G, Ahlstrand C, Andren O, et al. Clinical characteristics and primary treatment of prostate cancer in Sweden between 1996 and 2005. Scand J Urol Nephrol. 2007;41:456–477. doi: 10.1080/00365590701673625. [DOI] [PubMed] [Google Scholar]
- 33.Swedish Cause of Death Register. [cited; Available from: www.socialstyrelsen.se.
- 34.Genome Variation Server. [cited; Available from: http://pga.gs.washington.edu/
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 36.Morris JS, Yin G, Baggerly KA, Wu C, Zhang L. Pooling information across different studies and oligonucleotide microarray chip types to identify prognostic genes for lung cancer. In: Shoemaker JS, Lin SM, editors. Methods of Microarray Data Analysis IV. New York: Springer-Verlog; 2005. pp. 51–66. [Google Scholar]
- 37.GLOBOCAN 2008 (IARC) Section of Cancer Information. [cited; Available from: http://globocan.iarc.fr/factsheets/populations/
- 38.Kattan MW, Eastham JA, Stapleton AM, Wheeler TM, Scardino PT. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90:766–771. doi: 10.1093/jnci/90.10.766. [DOI] [PubMed] [Google Scholar]
- 39.Andren O, Fall K, Franzen L, Andersson SO, Johansson JE, Rubin MA. How well does the Gleason score predict prostate cancer death? A 20-year followup of a population based cohort in Sweden. J Urol. 2006;175:1337–1340. doi: 10.1016/S0022-5347(05)00734-2. [DOI] [PubMed] [Google Scholar]
- 40.Lifsted T, Le Voyer T, Williams M, Muller W, Klein-Szanto A, Buetow KH, et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer. 1998;77:640–644. doi: 10.1002/(sici)1097-0215(19980812)77:4<640::aid-ijc26>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 41.Hunter KW, Crawford NP. Germ line polymorphism in metastatic progression. Cancer Res. 2006;66:1251–1254. doi: 10.1158/0008-5472.CAN-05-3705. [DOI] [PubMed] [Google Scholar]
- 42.Dworkin AM, Ridd K, Bautista D, Allain DC, Iwenofu OH, Roy R, et al. Germline variation controls the architecture of somatic alterations in tumors. PLoS Genet. 2010;6:1–9. doi: 10.1371/journal.pgen.1001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rebbeck TR. Inherited genotype and prostate cancer outcomes. Cancer Epidemiol Biomarkers Prev. 2002;11:945–952. [PubMed] [Google Scholar]
- 44.Penney KL, Pyne S, Schumacher FR, Sinnott JA, Mucci LA, Kraft PL, et al. Genome-wide association study of prostate cancer mortality. Cancer Epidemiol Biomarkers Prev. 2010;19:2869–2876. doi: 10.1158/1055-9965.EPI-10-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cioffi JA, Shafer AW, Zupancic TJ, Smith-Gbur J, Mikhail A, Platika D, et al. Novel B219/OB receptor isoforms: possible role of leptin in hematopoiesis and reproduction. Nat Med. 1996;2:585–589. doi: 10.1038/nm0596-585. [DOI] [PubMed] [Google Scholar]
- 46.Mantzoros CS, Flier JS, Rogol AD. A longitudinal assessment of hormonal and physical alterations during normal puberty in boys. V. Rising leptin levels may signal the onset of puberty. J Clin Endocrinol Metab. 1997;82:1066–1070. doi: 10.1210/jcem.82.4.3878. [DOI] [PubMed] [Google Scholar]
- 47.Loffreda S, Yang SQ, Lin HZ, Karp CL, Brengman ML, Wang DJ, et al. Leptin regulates proinflammatory immune responses. FASEB J. 1998;12:57–65. [PubMed] [Google Scholar]
- 48.Sierra-Honigmann MR, Nath AK, Murakami C, Garcia-Cardena G, Papapetropoulos A, Sessa WC, et al. Biological action of leptin as an angiogenic factor. Science. 1998;281:1683–1686. doi: 10.1126/science.281.5383.1683. [DOI] [PubMed] [Google Scholar]
- 49.Ducy P, Amling M, Takeda S, Priemel M, Schilling AF, Beil FT, et al. Leptin inhibits bone formation through a hypothalamic relay: a central control of bone mass. Cell. 2000;100:197–207. doi: 10.1016/s0092-8674(00)81558-5. [DOI] [PubMed] [Google Scholar]
- 50.Ribeiro R, Lopes C, Medeiros R. Leptin and prostate: implications for cancer prevention--overview of genetics and molecular interactions. Eur J Cancer Prev. 2004;13:359–368. doi: 10.1097/00008469-200410000-00002. [DOI] [PubMed] [Google Scholar]
- 51.Norgaard M, Jensen AO, Jacobsen JB, Cetin K, Fryzek JP, Sorensen HT. Skeletal related events, bone metastasis and survival of prostate cancer: a population based cohort study in Denmark (1999 to 2007) J Urol. 2010;184:162–167. doi: 10.1016/j.juro.2010.03.034. [DOI] [PubMed] [Google Scholar]
- 52.Hsiao W, Moses KA, Goodman M, Jani AB, Rossi PJ, Master VA. Stage IV prostate cancer: survival differences in clinical T4, nodal and metastatic disease. J Urol. 2010;184:512–518. doi: 10.1016/j.juro.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 53.Rana A, Chisholm GD, Khan M, Sekharjit SS, Merrick MV, Elton RA. Patterns of bone metastasis and their prognostic significance in patients with carcinoma of the prostate. Br J Urol. 1993;72:933–936. doi: 10.1111/j.1464-410x.1993.tb16301.x. [DOI] [PubMed] [Google Scholar]
- 54.Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet. 2002;30:181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 55.Malathi K, Paranjape JM, Bulanova E, Shim M, Guenther-Johnson JM, Faber PW, et al. A transcriptional signaling pathway in the IFN system mediated by 2'-5'-oligoadenylate activation of RNase L. Proc Natl Acad Sci U S A. 2005;102:14533–14538. doi: 10.1073/pnas.0507551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiang Y, Wang Z, Murakami J, Plummer S, Klein EA, Carpten JD, et al. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2',5'-oligoadenylates. Cancer Res. 2003;63:6795–6801. [PubMed] [Google Scholar]
- 57.Meyer MS, Penney KL, Stark JR, Schumacher FR, Sesso HD, Loda M, et al. Genetic variation in RNASEL associated with prostate cancer risk and progression. Carcinogenesis. 2010;31:1597–1603. doi: 10.1093/carcin/bgq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Witthuhn BA, Silvennoinen O, Miura O, Lai KS, Cwik C, Liu ET, et al. Involvement of the Jak-3 Janus kinase in signalling by interleukins 2 and 4 in lymphoid and myeloid cells. Nature. 1994;370:153–157. doi: 10.1038/370153a0. [DOI] [PubMed] [Google Scholar]
- 59.Toi M, Bicknell R, Harris AL. Inhibition of colon and breast carcinoma cell growth by interleukin-4. Cancer Res. 1992;52:275–279. [PubMed] [Google Scholar]
- 60.Obiri NI, Hillman GG, Haas GP, Sud S, Puri RK. Expression of high affinity interleukin-4 receptors on human renal cell carcinoma cells and inhibition of tumor cell growth in vitro by interleukin-4. J Clin Invest. 1993;91:88–93. doi: 10.1172/JCI116205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Topp MS, Koenigsmann M, Mire-Sluis A, Oberberg D, Eitelbach F, von Marschall Z, et al. Recombinant human interleukin-4 inhibits growth of some human lung tumor cell lines in vitro and in vivo. Blood. 1993;82:2837–2844. [PubMed] [Google Scholar]
- 62.Lahm H, Schnyder B, Wyniger J, Borbenyi Z, Yilmaz A, Car BD, et al. Growth inhibition of human colorectal-carcinoma cells by interleukin-4 and expression of functional interleukin-4 receptors. Int J Cancer. 1994;59:440–447. doi: 10.1002/ijc.2910590325. [DOI] [PubMed] [Google Scholar]
- 63.Volpert OV, Fong T, Koch AE, Peterson JD, Waltenbaugh C, Tepper RI, et al. Inhibition of angiogenesis by interleukin 4. J Exp Med. 1998;188:1039–1046. doi: 10.1084/jem.188.6.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uchiyama A, Essner R, Doi F, Nguyen T, Ramming KP, Nakamura T, et al. Interleukin 4 inhibits hepatocyte growth factor-induced invasion and migration of colon carcinomas. J Cell Biochem. 1996;62:443–453. doi: 10.1002/(SICI)1097-4644(19960915)62:4%3C443::AID-JCB2%3E3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 65.Rosenwasser LJ, Borish L. Genetics of atopy and asthma: the rationale behind promoter-based candidate gene studies (IL-4 and IL-10) Am J Respir Crit Care Med. 1997;156:S152–S155. doi: 10.1164/ajrccm.156.4.12tac-14. [DOI] [PubMed] [Google Scholar]
- 66.Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- 67.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 68.Kondratov RV, Kondratova AA, Lee C, Gorbacheva VY, Chernov MV, Antoch MP. Post-translational regulation of circadian transcriptional CLOCK(NPAS2)/BMAL1 complex by CRYPTOCHROMES. Cell Cycle. 2006;5:890–895. doi: 10.4161/cc.5.8.2684. [DOI] [PubMed] [Google Scholar]
- 69.Plymate SR, Tenover JS, Bremner WJ. Circadian variation in testosterone, sex hormone-binding globulin, and calculated non-sex hormone-binding globulin bound testosterone in healthy young and elderly men. J Androl. 1989;10:366–371. doi: 10.1002/j.1939-4640.1989.tb00120.x. [DOI] [PubMed] [Google Scholar]
- 70.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nature Reviews. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 71.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- 72.Reintsch WE, Mandato CA, McCrea PD, Fagotto F. Inhibition of cell adhesion by xARVCF indicates a regulatory function at the plasma membrane. Dev Dyn. 2008;237:2328–2341. doi: 10.1002/dvdy.21651. [DOI] [PubMed] [Google Scholar]
- 73.Guldberg HC, Marsden CA. Catechol-O-methyl transferase: pharmacological aspects and physiological role. Pharmacol Rev. 1975;27:135–206. [PubMed] [Google Scholar]
- 74.Fotsis T, Zhang Y, Pepper MS, Adlercreutz H, Montesano R, Nawroth PP, et al. The endogenous oestrogen metabolite 2-methoxyoestradiol inhibits angiogenesis and suppresses tumour growth. Nature. 1994;368:237–239. doi: 10.1038/368237a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.