Abstract
The structural maintenance of chromosomes (SMC) family is a growing family of chromosomal ATPases. The founding class of SMC protein complexes, condensins, plays a central role in mitotic chromosome condensation. We report here a new class of SMC protein complexes containing XSMC1 and XSMC3, Xenopus homologs of yeast Smc1p and Smc3p, respectively. The protein complexes (termed cohesins) exist as two major forms with sedimentation coefficients of 9S and 14S. 9S cohesin is a heterodimer of XSMC1 and XSMC3, whereas 14S cohesin contains three additional subunits. One of them has been identified as a Xenopus homolog of the Schizosaccharomyces pombe Rad21p implicated in DNA repair and the Saccharomyces cerevisiae Scc1p/Mcd1p implicated in sister chromatid cohesion. 14S cohesin binds to interphase chromatin independently of DNA replication and dissociates from it at the onset of mitosis. Immunodepletion of cohesins during interphase causes defects in sister chromatid cohesion in subsequent mitosis, whereas condensation is unaffected. These results suggest that proper assembly of mitotic chromosomes is regulated by two distinct classes of SMC protein complexes, cohesins and condensins.
Keywords: Sister chromatid cohesion, chromosome condensation, the SMC family, X. laevis, cell-free system
Replication and segregation of the genetic information are two of the most fundamental events in cell reproduction. Following replication, chromosomal DNA undergoes three major structural transitions. First, the linkage of duplicated DNA molecules, termed sister chromatid cohesion, is established during or soon after S phase and is maintained throughout G2 phase of the cell cycle. Second, at the onset of mitosis, the DNA molecules start to condense, producing metaphase chromosomes consisting of paired sister chromatids. Sister chromatid cohesion at this stage is required for proper chromosome movements known as congression. Third, the linkage between the sister chromatids is dissolved highly synchronously at the metaphase–anaphase transition, allowing the two chromatids to segregate to opposite poles of the mitotic spindle. All of these steps are essential for faithful transmission of chromosomes and thereby must be regulated precisely. Despite recent progress in our understanding of the biochemical basis of cell cycle regulation, surprisingly little is known about the molecular mechanisms underlying the dynamic reorganization of chromosome architecture. Particularly, we have very limited information about structural protein components directly involved in these processes (for review, see Miyazaki and Orr-Weaver 1994; Yanagida 1995; Koshland and Strunnikov 1996).
The identification of the SMC (structural maintenance of chromosomes) family of ATPases has provided an important molecular clue to our understanding of higher-order chromosome dynamics (Hirano et al. 1995; Saitoh et al. 1995). SMC proteins are highly conserved from bacteria to humans. The completion of the yeast genome sequencing project has allowed reclassification of eukaryotic SMC proteins, which are now grouped into four types (SMC1–SMC4; Koshland and Strunnikov 1996). Among them, the functions of the SMC2- and SMC4-type proteins are best understood through biochemical and genetic analyses in multiple model organisms. In Xenopus laevis, XCAP-C (SMC4 type) and XCAP-E (SMC2 type) function as the core subunits of condensins, protein complexes that play a central role in mitotic chromosome condensation in vitro (Hirano and Mitchison 1994; Hirano et al. 1997; Kimura and Hirano 1997). In Saccharomyces cerevisiae, an smc2 mutant shows a defect in chromosome condensation as well as in chromosome segregation in mitosis (Strunnikov et al. 1995). In Schizosaccharomyces pombe, Cut3p (SMC4 type) and Cut14p (SMC2 type) are both essential for condensation and copurify in a complex (Saka et al. 1994; Sutani and Yanagida 1997). All of these results are consistent with the idea that the SMC2- and SMC4-type proteins are important components of the chromosome condensation machinery (condensins).
Previous genetic studies in S. cerevisiae showed that Smc1p is required for proper segregation of chromosomes (Strunnikov et al. 1993), but apparently not for condensation (Strunnikov et al. 1995). More recently, Smc1p and Smc3p were also identified in a genetic screen for mutants that show precocious separation of sister chromatids during mitosis (Michaelis et al. 1997). The same screen identified a third gene product, Scc1p/Mcd1p, that is homologous to the fission yeast Rad21p implicated in DNA repair (Birkenbihl and Subramani 1992). An independent study in yeast found physical and genetic interactions between Smc1p and Scc1p/Mcd1p (Guacci et al. 1997). On the basis of these observations, it has been proposed that Smc1p and Smc3p, along with Scc1p/Mcd1p, might be directly involved in sister chromatid cohesion (Michaelis et al. 1997). Despite all of the genetic data available in yeast, a functional and biochemical characterization of these proteins in vertebrate cells is lacking.
In this report, we have used Xenopus egg cell-free extracts to get insights into the biochemical basis for sister chromatid cohesion. We found that the Xenopus SMC1- and SMC3-type proteins associate with each other, forming large protein complexes (termed cohesins) with sedimentation coefficients of 9S and 14S. 14S cohesin contains additional subunits, one of which is structurally homologous to the yeast Scc1p/Mcd1p/Rad21p. 14S cohesin localizes to interphase chromatin, but, surprisingly, most of the complexes dissociate from the chromosomes at the onset of mitosis. Immunodepletion of cohesins during interphase results in cohesion defects that become apparent upon entry into mitosis. The current results, taken together with our previous results, provide a simple biochemical picture in which the two distinct classes of SMC protein complexes, cohesins and condensins, regulate sister chromatid cohesion and chromosome condensation, respectively, contributing to faithful segregation of chromosomes in eukaryotic cells.
Results
Molecular cloning of XSMC1 and XSMC3
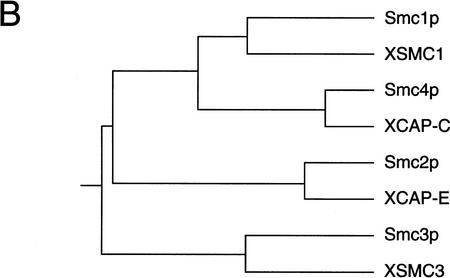
To amplify Xenopus cDNA sequences encoding SMC1- and SMC3-type proteins, we designed PCR primers on the basis of homologous sequences isolated from S. cerevisiae (Strunnikov et al 1993; Michaelis et al. 1997), Aspergillus nidulans (Holt and May 1996), Drosophila melanogaster (Hong and Ganetzky 1996), and human (Rocques et al. 1995). By use of the amplified fragments as hybridization probes, two groups of overlapping cDNAs were cloned from a Xenopus cDNA library. As expected, we found that the amino acid sequences encoded by the two groups of cDNAs corresponded to the SMC1- and SMC3-type proteins, and were not identical to either XCAP-C (SMC4 type) or XCAP-E (SMC2 type; Fig. 1A). We named these new polypeptides XSMC1 and XSMC3 (for Xenopus SMCs). Thus, Xenopus laevis has at least four members of the SMC family, each of which has an ortholog in S. cerevisiae (Fig. 1B).
Figure 1.
Four SMC subtypes in yeast and Xenopus. (A) Sequence alignment of four SMC subtypes within the DA-box region. Yeast sequences, Smc1p, Smc2p, Smc3p, and Smc4p; Xenopus sequences, XSMC1, XCAP-E, XSMC3, and XCAP-C. (B) A phylogenetic tree prepared on the basis of the DA-box sequences. The program GeneWorks (IntelliGenetics) was used for sequence analysis.
Identification of protein complexes containing XSMC1 and XSMC3
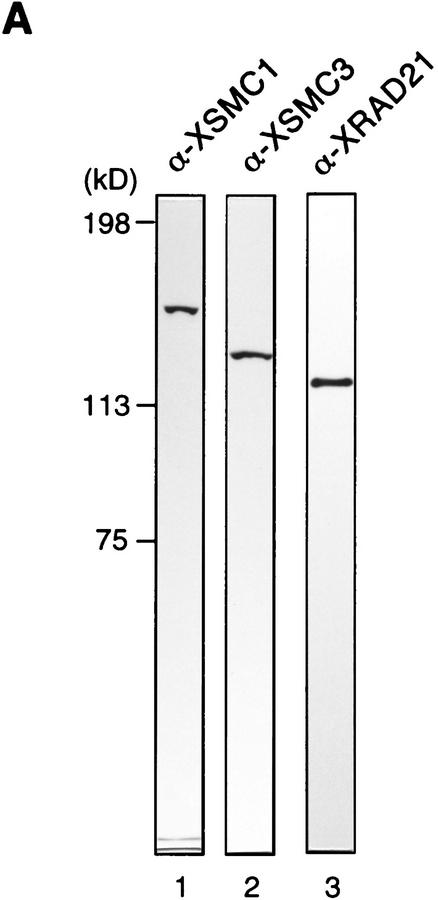
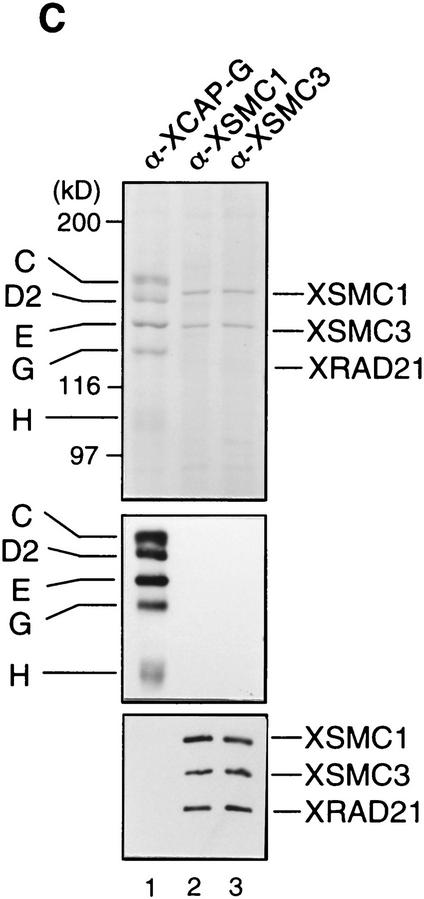
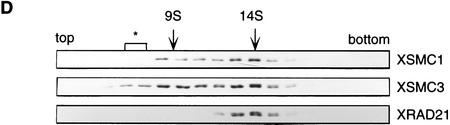
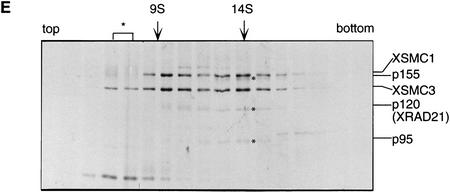
To characterize XSMC1 and XSMC3 biochemically, we raised antibodies against synthetic peptides corresponding to their carboxy-terminal sequences. These antibodies were very specific to XSMC1 and XSMC3 as judged by immunoblotting against total egg extracts (Fig. 2A). XSMC1 and XSMC3 coimmunoprecipitated with both antibodies, suggesting that the two polypeptides associated with each other in the extracts. The specificity of the immunoprecipitations was demonstrated by competition with the antigen peptides (Fig. 2B). Sucrose gradient fractionation of a total extract showed that XSMC1 and XSMC3 exist in two major forms with sedimentation coefficients of 9S and 14S (Fig. 2D). A minor peak of 4S–5S containing XSMC3 only was also detected. Next, the complexes were affinity-purified with an anti-XSMC3 peptide antibody and subjected to sucrose gradient centrifugation. We found that three other polypeptides cofractionated with XSMC1 and XSMC3 in the 14S complex. These three polypeptides (tentatively named p155, p120, and p95 according to their apparent molecular weights; Fig. 2E) also copurified with XSMC1 and XSMC3 on both cation- and anion-exchange column chromatography (data not shown). Immunoblot experiments identified p120 as a Xenopus homolog of the fission yeast Rad21p implicated in DNA repair (Birkenbihl and Subramani 1992) and of the budding yeast Scc1p/Mcd1p implicated in sister chromatid cohesion (Guacci et al. 1997; Michaelis et al. 1997). In this report, we refer to this protein as XRAD21 (for Xenopus RAD21; Fig. 2A,B,D). XRAD21 was immunoprecipitated from the extracts with both anti-XSMC1 and anti-XSMC3 (Fig. 2B) and, conversely, anti-XRAD21 immunoprecipitated XSMC1 and XSMC3 (data not shown), confirming their specific association. XRAD21 was exclusively found in the 14S form and no free population was detected in the extracts (Fig. 2D). Taking into account recent genetic studies in yeast (Birkenbihl and Subramani 1992; Guacci et al. 1997; Michaelis et al. 1997) and our biochemical studies in the Xenopus egg cell-free system (see below), we refer to the largest form of the complexes containing XSMC1 and XSMC3 as 14S cohesin and to the second largest as 9S cohesin. We also found that cytostatic factor (CSF)-arrested (metaphase II-arrested) and interphase extracts contain similar levels of cohesin subunits and a similar ratio of the 9S form to the 14S form (data not shown).
Figure 2.
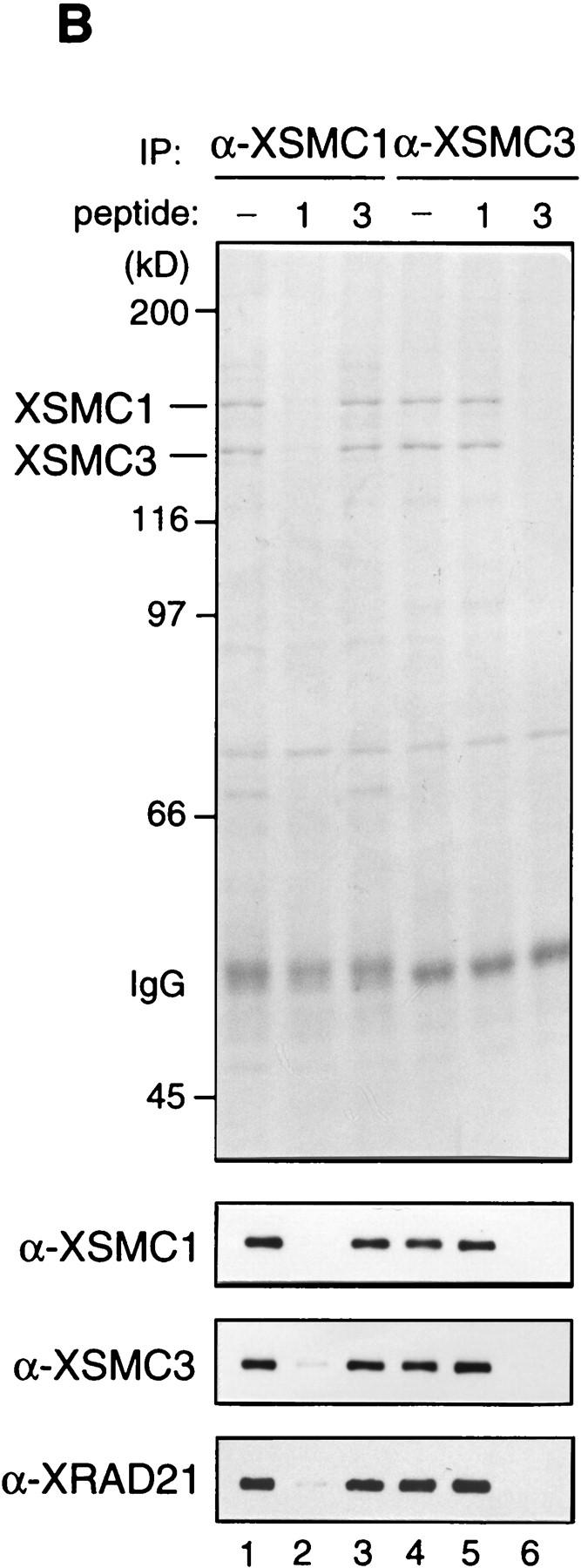
Biochemical characterization of protein complexes containing XSMC1 and XSMC3. (A) Immunoblot analysis of an egg extract with affinity-purified antibodies against XSMC1 (lane 1), XSMC3 (lane 2), or XRAD21 (lane 3). (B) Coimmunoprecipitation of XSMC1 and XSMC3. Aliquots of an interphase high-speed supernatant were incubated with anti-XSMC1 (lanes 1–3) or anti-XSMC3 (lanes 4–6) antibodies raised against the carboxy-terminal peptide sequences. The antigen peptide for XSMC1 (lanes 2,5) or XSMC3 (lanes 3,6) was added (0.4 mg/ml) to demonstrate the specificity of immunoprecipitation reactions. Immunoprecipitates were separated by SDS-PAGE followed by Coomassie blue stain (top) or by immunoblotting (bottom). (C) Independent immunoprecipitation of condensin and cohesin subunits. Immunoprecipitates obtained with anti-XCAP-G (lane 1), anti-XSMC1 (lane 2), or anti-XSMC3 (lane 3) were analyzed by Coomassie blue stain (top) or by immunoblotting (middle and bottom). (D) Sucrose gradient centrifugation of a total extract. An interphase high-speed supernatant was fractionated in a 5%–20% sucrose gradient, and fractions were analyzed by immunoblotting with anti-XSMC1, anti-XSMC3, and anti-XRAD21 antibodies. The two major peaks of 9S and 14S are indicated. A minor population of free XSMC3 is indicated by the asterisk. (E) Sucrose gradient centrifugation of an affinity-purified fraction. The cohesin complexes were affinity purified with anti-XSMC3 antibody and fractionated in a 5%–20% sucrose gradient. Fractions were TCA-precipitated, separated by SDS-PAGE, and stained with silver. Asterisks indicate positions of the bands corresponding to p155, p120, and p95 that cofractionate with XSMC1 and XSMC3 in the 14S peak.
It is striking to note that the condensin complexes also exist in two major forms in Xenopus egg extracts: 8S condensin is a heterodimer of XCAP-C and XCAP-E, whereas 13S condensin contains three additional non-SMC subunits, XCAP-D2, XCAP-G, and XCAP-H (Hirano et al. 1997). None of the condensin subunits were detectable in the XSMC immunoprecipitates and, conversely, XSMC1, XSMC3, and XRAD21 were not associated with condensins (Fig. 2C). Thus, Xenopus egg extracts contain two distinct classes of SMC protein complexes and they share no common subunit.
Cell cycle-dependent chromosomal association and dissociation of cohesins
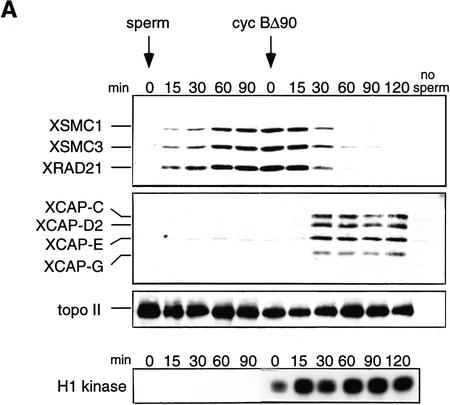
A cell-free system derived from Xenopus egg extracts is capable of reproducing cell cycle-specific chromosomal events in vitro, including DNA replication and chromosome condensation (Lohka and Masui 1983; Blow and Laskey 1986; Newport and Spann 1987). When sperm chromatin is incubated with an interphase extract, the nuclear envelope assembles around the chromatin in which DNA replication initiates. After completion of replication, the reaction mixture can be triggered to enter mitosis by addition of a CSF-arrested extract (or a nondegradable form of cyclin B) and the duplicated chromatin is converted into metaphase chromosomes with paired sister chromatids (Shamu and Murray 1992). We set up this in vitro reaction and took aliquots of the mixture at time intervals. Proteins bound to chromatin were isolated at each time point and analyzed by immunoblotting. Increasing amounts of XSMC1, XSMC3, and XRAD21 associated with chromatin as incubation proceeded, but most of these proteins (>95% as judged by quantitative immunoblotting) dissociated gradually upon mitotic activation (Fig. 3A, top). Chromosomal association and dissociation of the three polypeptides occurred with the same kinetics, providing additional evidence that they act as subunits of the same protein complex (14S cohesin). We could not test whether 9S cohesin also binds to chromatin as no polypeptide specific to this form has been identified. The behavior of 14S cohesin contrasted sharply with that of 13S condensin, which was undetectable in interphase chromatin but was associated specifically with mitotic chromosomes (Fig. 3A, middle). In contrast, chromosomal association of topoisomerase IIα was not cell cycle dependent (Fig. 3A, bottom).
Figure 3.
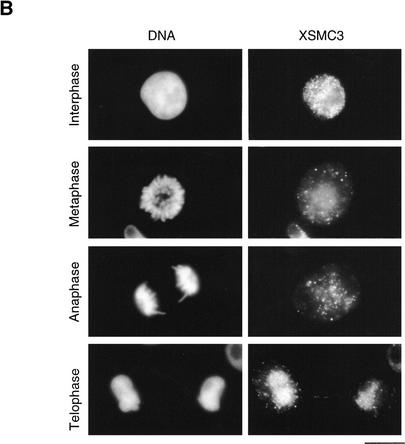
Cell cycle-dependent chromosomal targeting of cohesins in vivo and in vitro. (A) Cell cycle-dependent chromosomal targeting of cohesins and condensins. Sperm chromatin was incubated with an interphase low-speed supernatant for 90 min and then a non-degradable form of sea urchin cyclin B (cyc BΔ90) was added to drive the cell cycle into mitosis. Aliquots were taken at the indicated time points, and chromatin-bound protein fractions were isolated and analyzed by immunoblotting with antibodies against cohesin subunits (XSMC1, XSMC3, and XRAD21), condensin subunits (XCAP-C, XCAP-D2, XCAP-E, and XCAP-G) or topoisomerase IIα. The level of H1 kinase activity in the extract was also assayed. (B) Immunolocalization of XSMC3 in tissue culture cells. Xenopus XL177 tissue culture cells fixed with paraformaldehyde were stained with DAPI (left) and anti-XSMC3 (right). Bar, 10 μm.
Consistent with these in vitro results, immunolocalization experiments with Xenopus tissue culture cells showed that XSMC3 was localized in the nucleus during interphase, but dissociated from chromosomes in mitosis (Fig. 3B). Upon nuclear envelope breakdown, XSMC3 started to diffuse throughout the cytoplasm, and it re-entered the nucleus in telophase. Little, if any, staining was observed on condensed chromosomes under two different fixation conditions tested (see Materials and Methods). Similar results were obtained with anti-XSMC1. It is formally possible that a minor population of cohesins remains on condensed chromosomes and is enriched in a specific chromosomal region, for example, in centromeres. However, we have failed to detect such an enrichment on chromosomes either isolated from tissue culture cells or assembled in vitro (data not shown).
Chromatin association of 14S cohesin is not dependent on DNA replication
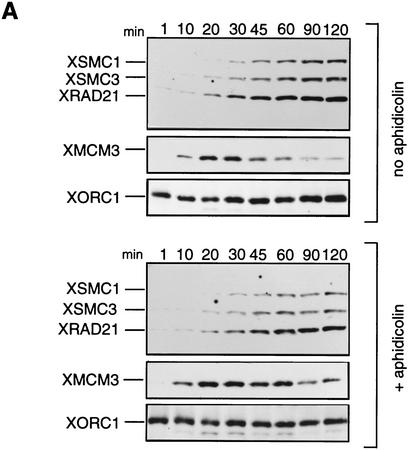
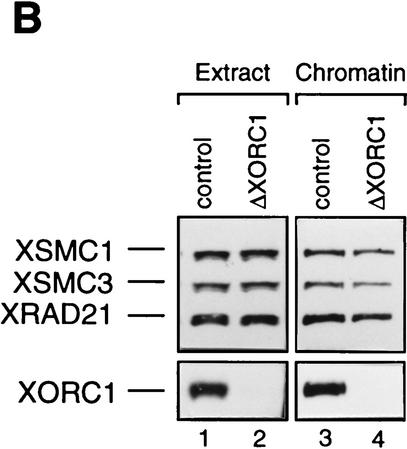
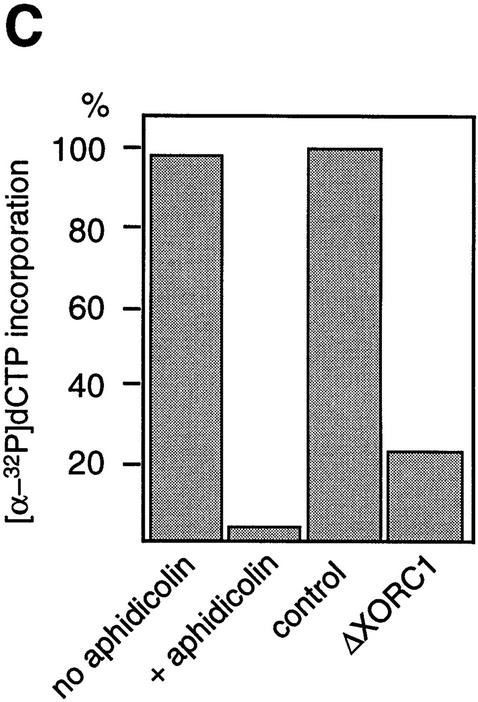
In several ways we tested whether the association of 14S cohesin with chromatin is dependent on DNA replication. Assembly of the nuclear envelope around sperm chromatin is a prerequisite for initiation of DNA replication in the Xenopus cell-free system (Sheehan et al. 1988). Removal of membrane vesicles from extracts by centrifugation prevents nuclear envelope formation, thereby impeding DNA replication. We found that this treatment had little effect on the kinetics of chromatin binding of 14S cohesin (data not shown). DNA replication in interphase extracts containing membrane vesicles starts ∼30 min after addition of sperm chromatin and is completed 1 hr later. The synthesis of DNA can be blocked by aphidicolin, a specific inhibitor of DNA polymerase α (Fig. 4C). We monitored the behavior of cohesin subunits as well as two replication factors, XMCM3 and XORC1, in the presence or absence of this drug (Fig. 4A). In both cases, XORC1 bound to chromatin within 1 min upon incubation of sperm nuclei with the extracts. Binding of XMCM3 occurred at 10–20 min, and was followed by binding of cohesin subunits at 20–30 min. No difference was apparent in the binding of cohesins to the replicating or nonreplicating chromatin. As reported previously, XMCM3 partially dissociated from chromatin as DNA replication proceeded in the extract without aphidicolin (Kubota et al. 1995). However, such a replication-dependent dissociation was not observed in any of the subunits of 14S cohesin. Because XORC provides the assembly site for other DNA replication factors including XMCM3 (Coleman et al. 1996; Romanowski et al. 1996; Rowles et al. 1996), we tested whether the association of cohesins with chromatin might be dependent on XORC. To this end, XORC was immunodepleted from an extract by use of anti-XORC1 antibodies (Fig. 4B, lane 2). We found that the cohesin subunits were still able to bind to chromatin in the XORC-depleted, replication-defective extract (Fig. 4B, lane 4, and C). These results suggest that the association of 14S cohesin with chromatin is not dependent on ongoing DNA replication in this cell-free system.
Figure 4.
Cohesin subunits associate with chromatin independently of DNA replication. (A) Effect of aphidicolin treatment on the association of cohesins with chromatin. Sperm chromatin was incubated with interphase low-speed supernatants in the absence (top) or presence of aphidicolin (bottom). At the indicated times after sperm addition, aliquots were removed from the extracts and chromatin-bound proteins were analyzed by immunoblotting with antibodies against the cohesin subunits, XMCM3 and XORC1. (B) Effect of XORC depletion on the association of cohesins with chromatin. Interphase extracts were immunodepleted with a control serum (lanes 1,3) or an anti-XORC1 serum (lanes 2,4). Extracts (lanes 1,2) or chromatin-bound fractions (lanes 3,4) were analyzed by immunoblotting with antibodies against the cohesin subunits or XORC1. (C) DNA replication in aphidicolin-treated or XORC-depleted extracts. DNA replication in the extracts was assayed by measuring the incorporation of [α-32P] dCTP into sperm chromatin. DNA synthesis is expressed as a percentage of the synthesis achieved in the control-depleted extract.
Normal nuclear envelope assembly and DNA replication in the absence of cohesins
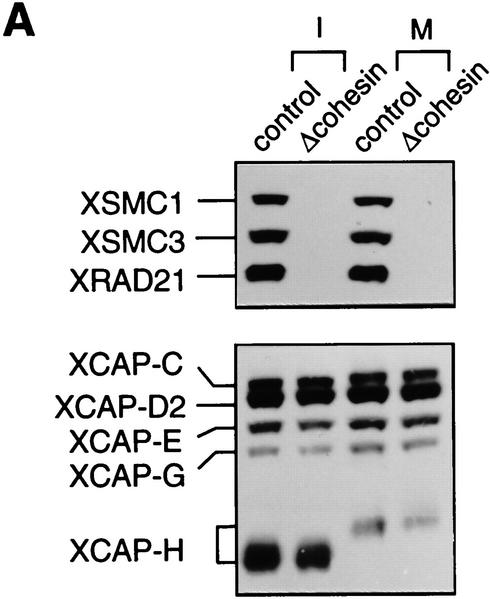
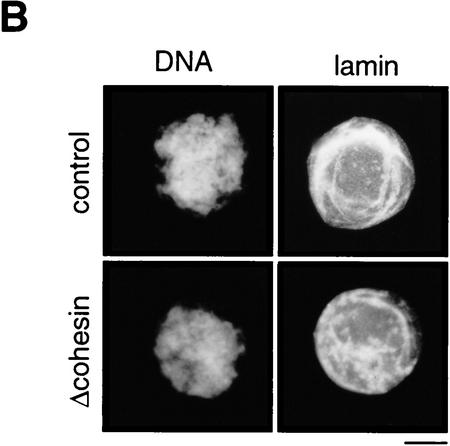
We tested a requirement for cohesins in interphase nuclear functions in vitro. Both 9S and 14S cohesins were immunodepleted from extracts with a combination of anti-XSMC1 and anti-XSMC3 antibodies (Fig. 5A). Quantitative immunoblotting indicated that depletion efficiency was >95%. In interphase extracts, nuclear assembly occurred normally in the absence of cohesins as judged by chromatin morphology and lamin staining (Fig. 5B). DNA replication in the assembled nuclei was assayed by measuring the incorporation of radiolabeled nucleotides into chromosomal DNA. We found very little difference in the kinetics and extent of DNA replication in nuclei assembled in the cohesin-depleted and control extracts (Fig. 5C). As a second assay for DNA replication, biotinylated nucleotides were used to visualize replicated DNA by fluorescent microscopy. Again, nuclei prepared from both cohesin-depleted and control extracts were uniformly labeled with biotinylated nucleotides (data not shown), suggesting that cohesins are not required for DNA replication in this system.
Figure 5.
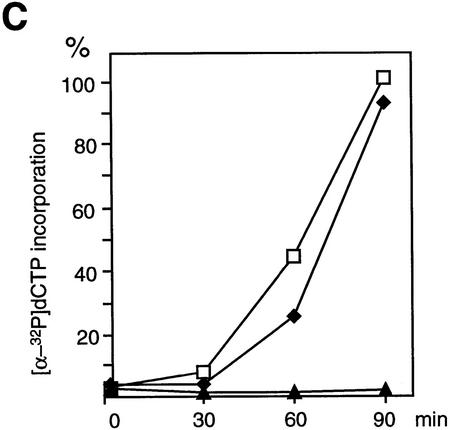
Normal interphase nuclear functions in the absence of cohesins. (A) Immunodepletion of cohesins. Interphase (I) or CSF-arrested (M) extracts were immunodepleted with control IgG (control; lanes 1,3) or a mixture of anti-XSMC1 and anti-XSMC3 (Δcohesin; lanes 2,4). The extracts were analyzed by immunoblotting with antibodies against the cohesin subunits (XSMC1, XSMC3, and XRAD21) or the condensin subunits (XCAP-C, XCAP-D2, XCAP-E, XCAP-G, and XCAP-H). The CSF-arrested extracts were used in the chromosome assembly assays described in Fig. 6. (B) Nuclear assembly in the absence of cohesins. Sperm chromatin was incubated in the cohesin-depleted (Δcohesin) or control extracts at 22°C for 2 hr, fixed, and stained with DAPI (DNA) or antilamin LIII antibody (lamin). Bar, 10 μm. (C) DNA replication in the absence of cohesins. DNA replication in the cohesin-depleted (♦) or control (□) extracts was assayed as in Fig. 4C. Incorporation of [α-32P]dCTP was measured after a 2 hr-incubation of the sperm chromatin with the corresponding extract. DNA replication in an aphidicolin-treated extract (▴) is also shown as a control.
Cohesin depletion during interphase causes defects in sister chromatid cohesion in subsequent mitosis
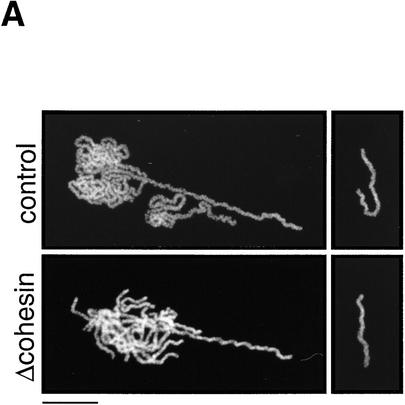
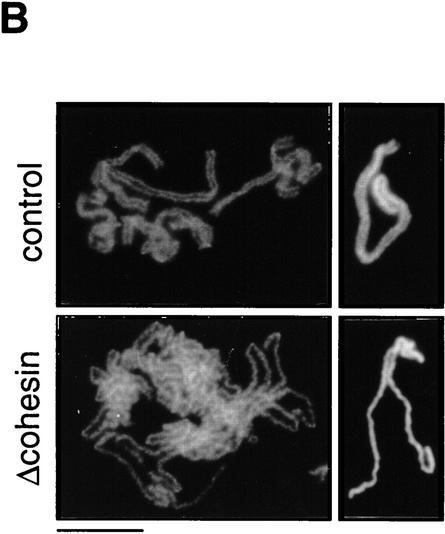
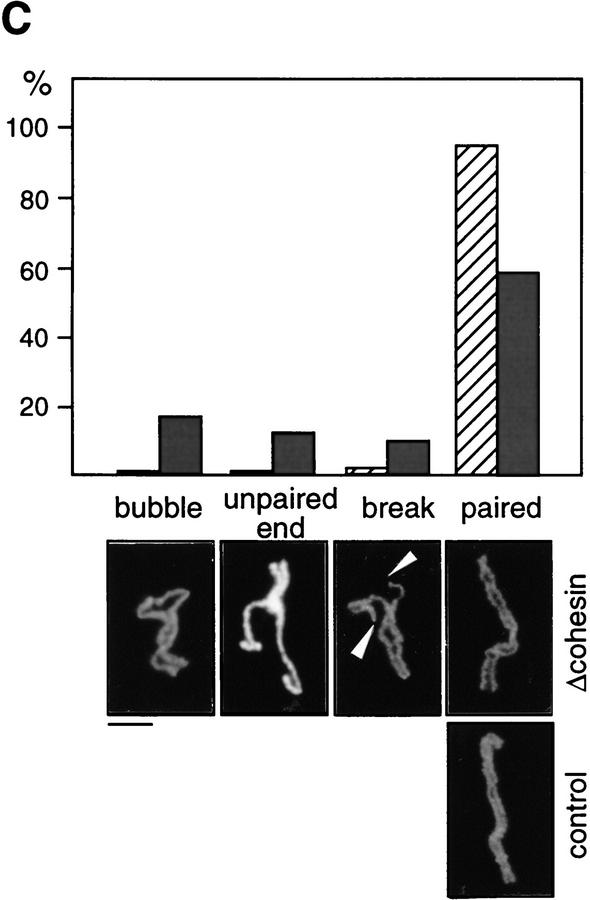
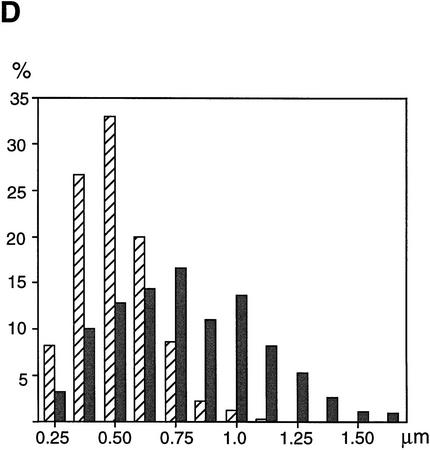
Although our results suggest that cohesins are not major structural components of mitotic chromosomes, it is possible that they contribute indirectly to mitotic chromosome assembly. To test this possibility, we used two types of in vitro assays. In the first assay, sperm chromatin was directly incubated in CSF-arrested extracts in which it was converted into mitotic chromosome-like structures consisting of single chromatids (single chromatid assembly assay; Hirano and Mitchison 1993). In this assay, no significant difference was detected in the morphology of chromosomes assembled in the presence or absence of cohesins (Fig. 6A). Second, nuclei were assembled in interphase extracts to allow DNA replication and then driven into mitosis by addition of CSF-arrested extracts (double chromatid assembly assay; Shamu and Murray 1992). The experiment was designed so that cohesins were present (control) or absent (cohesin depleted) throughout the two-step reactions. In the control extracts, duplicated chromatin was converted into metaphase chromosomes with sister chromatids that were tightly paired along their entire length (Fig. 6B, top). However, the pairing of sister chromatids formed in the cohesin-depleted extracts was largely impaired (Fig. 6B, bottom). For quantitative analysis, metaphase chromosome morphology was categorized as paired or unpaired. For chromosomes exhibiting unpaired morphology, pairing defects were further classified into three categories: bubble, unpaired end, and break, examples of which are shown in Figure 6C. More than 600 chromosomes from four independent experiments were counted and analyzed. The proportion of chromosomes with unpaired morphology was ∼40% in the depleted extracts, but they were rarely found in the control extracts (<4%). Although ∼60% of chromosomes in the depleted extracts were classified into the paired group, we found that the pairing of sister chromatids in these chromosomes was loose. The average distance between sister chromatids was significantly larger for chromosomes assembled in the cohesin-depleted extracts (0.79 ± 0.30 μm) compared with those assembled in the control extracts (0.51 ± 0.16 μm; Fig. 6D). The same level of cohesion defects was observed when interphase nuclei were first assembled in the absence of cohesins and then driven into mitosis by addition of a CSF-arrested extract containing cohesins (data not shown). These results suggest that cohesins’ function during interphase is essential for proper cohesion of sister chromatids in subsequent mitosis. Because no apparent difference was detected in the length/width of chromatids assembled in the depleted or control extracts, we conclude that cohesins do not play a major role in condensation.
Figure 6.
Cohesins are required for sister chromatid cohesion but not for condensation. (A) Single chromatid assembly assay. Sperm chromatin was incubated with CSF-arrested high-speed supernatants (cohesin-depleted or control), fixed, and stained with anti-XCAP-E. Individual chromosomes are shown on the right. Bar, 10 μm. (B) Double-chromatid assembly assay. Sperm chromatin was incubated with interphase extracts (cohesin-depleted or control) to allow nuclear assembly and DNA replication. The cohesin-depleted or control nuclei were converted into mitotic chromosomes by the addition of cohesin-depleted or control CSF-arrested extracts, respectively. The chromosomes were stained as described in A. Bar, 10 μm. (C) Classification of defective phenotypes. Chromosome morphology was classified into four groups: bubble (an unpaired region somewhere along the chromosome); unpaired end (an unpaired region at one end of the chromosome); break (a double-strand break in one of the chromatids); paired (chromatids paired along the length of the chromosome). (Hatched bars) Control; (solid bars) Δcohesin. Bar, 5 μm. (D) Distance between sister chromatids in paired chromosomes. Chromosomes formed in control (hatched bars) or cohesin-depleted extracts (solid bars) that were classified in the paired group were randomly selected, and the distance between the two sister chromatids measured at regular intervals along the entire length of the chromosomes (see Materials andMethods). The results were plotted as number of times (expressed as a percentage of the total number of measurements made, y-axis) in which the distance between the sister chromatids corresponded to a given value (distance in micrometers, x-axis). The mean values and standard deviations for each case are indicated in the legend.
Independent behavior of 14S cohesin and 13S condensin
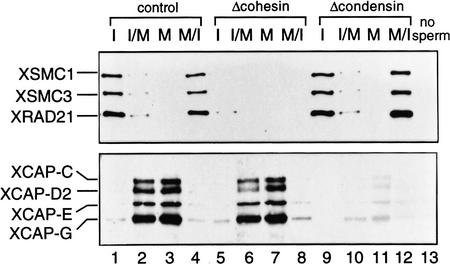
Even in the absence of cohesins, 13S condensin seems to bind to chromosomes and to promote normal chromosome condensation. Immunoblotting analysis of isolated chromatin fractions confirmed that this was the case: 13S condensin did not bind to interphase chromatin, but did bind to mitotic chromosomes upon mitotic activation in both cohesin-depleted and control extracts (Fig. 7, lanes 1,2,5,6). Conversely, when a mitotic extract was driven into interphase, 13S condensin prebound to mitotic chromosomes dissociated from them in the presence or absence of cohesins (Fig. 7, lane 3,4,7,8). We performed the same set of experiments using condensin-depleted extracts, and found that interphase-specific association of 14S cohesin with chromatin, as well as its dissociation on activation of mitosis, can be recapitulated in vitro in the absence of condensins in the extracts (Fig. 7, lanes 9–12). We conclude that chromosomal association and dissociation of 14S cohesin and 13S condensin can occur independently of each other in this cell-free system.
Figure 7.
Independent chromosomal association and dissociation of cohesins and condensins in Xenopus egg extracts. Sperm nuclei were incubated with interphase extracts (lanes 1,5,9), or with interphase extract followed by addition of a half volume of CSF-arrested extract to enter mitosis (lanes 2,6,10). Alternatively, sperm nuclei were incubated with CSF-arrested extracts (lanes 3,7,11), or with CSF-arrested extracts followed by addition of 0.4 mm CaCl2, which promotes exit from mitosis and entry into interphase (lanes 4,8,12). Chromatin fractions were isolated and analyzed by immunoblotting. Lanes 1–4, control; lanes 5–8, cohesin depletion; lanes 9–12, condensin depletion; lane 13, a control without sperm.
Discussion
Cohesins, a second class of SMC protein complexes in Xenopus
Our previous studies identified the founding class of SMC protein complexes, condensins, that contain the SMC2- and SMC4-type subunits (Hirano and Mitchison 1994; Hirano et al. 1997). The present study reports the biochemical identification and functional characterization of a second class of Xenopus SMC protein complexes, termed cohesins. Xenopus egg extracts contain two major forms of cohesins (9S and 14S), which share the SMC1- and SMC3-type polypeptides as their core subunits. The 14S form contains three additional subunits, one of which is a homolog of S. cerevisiae Scc1p/Mcd1p implicated in sister chromatid cohesion (Guacci et al. 1997; Michaelis et al. 1997) and S. pombe Rad21p implicated in DNA repair (Birkenbihl and Subramani 1992). Although the identity of the p155 and p95 subunits of 14S cohesin is currently unknown, they are immunologically distinct from the condensin subunits, suggesting that cohesins and condensins share no common subunit. Nevertheless, it should be noted that the two classes of SMC protein complexes share striking similarities in their structural organization: (1) Each class contains a heterodimer of SMC proteins that serve as core subunits; (2) each class exists in two major forms with different sedimentation coefficients; and (3) the larger form of each class contains three (or more) additional subunits (Hirano et al. 1997).
Previous genetic studies in yeast suggested that distinct SMC proteins might have nonoverlapping functions. For example, in budding yeast, although both smc1 and smc2 mutants showed defects in proper segregation of chromosomes, a condensation defect was detectable only in smc2, but not in smc1 mutants (Strunnikov et al. 1993, 1995). Moreover, a recent genetic screen for mutants exhibiting precocious sister chromatid separation identified smc1 and smc3, but not smc2 or smc4 (Michaelis et al. 1997). Our current study, taken together with the previous studies, reveals a clear biochemical picture of the division of labors among the different SMC proteins, providing a simple explanation for the genetic observations in yeast: XCAP-C/Smc4p and XCAP-E/Smc2p function together as the core subunits of condensins (Hirano and Mitchison 1994; Hirano et al. 1997; Sutani and Yanagida 1997), whereas XSMC1/Smc1p and XSMC3/Smc3p constitute the core subunits of cohesins. Our identification of XRAD21, a Xenopus homolog of yeast Scc1p/Mcd1p, as an intrinsic subunit of 14S cohesin also complements the genetic studies in yeast (Guacci et al. 1997; Michaelis et al. 1997). In yeast cells, it remains to be determined whether the three gene products (Smc1p, Smc3p, and Scc1p/Mcd1p) identified genetically also form a large complex equivalent to Xenopus 14S cohesin. So far, only a physical interaction between Smc1p and Mcd1p has been demonstrated in coimmunoprecipitation experiments (Guacci et al. 1997). Although stoichiometric association of SMC1- and SMC3-type proteins has also been found in recombination complex-1 (RC-1) purified from calf thymus, this complex is distinct from 14S cohesin as judged from its size and the identity of the associated polypeptides (Jessberger et al. 1996; R. Jessberger, pers. comm.).
Are cohesins involved in interphase-specific cohesion?
The association of 14S cohesin with chromatin is regulated tightly during the cell cycle. Cohesin subunits bind to interphase chromatin in vitro (the density of XSMC1 in isolated chromatin is estimated to be one molecule per ∼20 kb of DNA by quantitative immunoblotting), and, on mitotic activation, >95% of the subunits dissociate from the chromatin. This behavior of cohesins, which was also observed in vivo by immunofluorescent staining of tissue culture cells, stands in striking contrast to the mitosis-specific chromosomal binding of condensins (Hirano and Mitchison 1994; Hirano et al. 1997). Interphase nuclei assembled in extracts depleted of cohesins are apparently normal, at least as judged by nuclear lamina assembly and DNA replication. When these nuclei enter mitosis, however, the resulting sister chromatids fail to remain tightly associated with each other. The cohesion defect cannot be rescued when cohesins are supplemented on mitotic activation. From these data, it is reasonable to conclude that cohesins must bind to chromatin during interphase to execute their main function, probably constituting (or being part of) the cohesion machinery that holds sister chromatids together after DNA replication. We speculate that the cohesin-mediated sister chromatid cohesion during interphase has at least two functions. First, it is a prerequisite for proper assembly of mitotic chromosomes in subsequent mitosis: A loss of the linkage between sister DNA molecules during or after DNA replication would cause premature separation of interphase chromatids, leading to abnormal assembly of mitotic chromosomes with cohesion defects. Second, it would be important for other interphase nuclear functions such as recombinational DNA repair. Mutations in the S. pombe rad21 gene result in an increased sensitivity to γ-irradiation (Birkenbihl and Subramani 1992). In this regard, it is interesting to note that double-stranded DNA breaks are among the major chromosomal defects we observed in chromosomes assembled in cohesin-depleted extracts.
It is unclear how the cohesin-mediated sister chromatid linkage is established and maintained. Whereas chromatin association of 14S cohesin occurs in parallel with DNA replication in vitro, the two events are not functionally coupled: Blocking DNA replication by three different methods (membrane depletion, aphidicolin treatment, and XORC depletion) did not affect binding of cohesin subunits to chromatin. One possible explanation is that sister chromatid cohesion is established via a two-step mechanism: First, 14S cohesin binds to chromatin and then recruits additional cohesion factors in a DNA replication-dependent manner. Candidates for such factors include Scc2p (Michaelis et al. 1997), Trf4p (Castaño et al. 1996), and BimDp (Holt and May 1996), because these proteins have been shown to interact with some of the cohesin subunits, either genetically or physically.
In yeast, the abundance of Scc1p/Mcd1p fluctuates, being highest in S phase and reduced to a lower level by G2/M (Guacci et al. 1997), whereas the level of Smc1p (and perhaps Smc3p) is constant throughout the cell cycle (Strunnikov et al. 1993). Assuming that the level of Scc1p/Mcd1p reflects the level of functional cohesin complexes on chromatin, this result is consistent with their requirement during interphase. In the Xenopus system, CSF-arrested (arrested in metaphase II) and interphase extracts contain the same level of cohesin subunits and the same proportion of the two complex forms (9S and 14S). The cell cycle-dependent association of cohesins with chromatin might then involve post-translational modifications of some of the subunits or, alternatively, changes in the availability of their chromosomal target sites. Whereas condensin function appears to be regulated by mitosis-specific phosphorylation (Hirano et al. 1997), no cell cycle-specific phosphorylation has been observed for the cohesin subunits (A. Losada, unpubl.).
Differences between the yeast and Xenopus systems
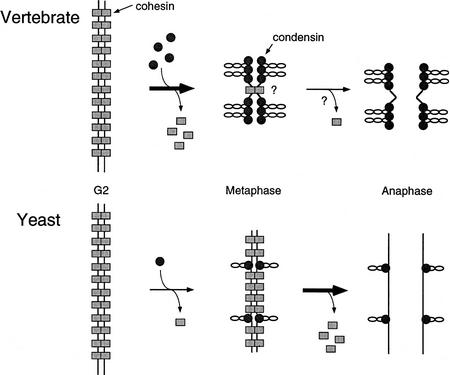
Our current results provide a first line of biochemical evidence for a role of SMC1- and SMC3-type proteins in sister chromatid cohesion, and are in general agreement with genetic studies performed in yeast (Guacci et al. 1997; Michaelis et al. 1997). There are, however, several notable differences between the two systems. For example, two observations in yeast raised the possibility that Scc1p/Mcd1p-mediated cohesion may persist until the metaphase–anaphase transition: Scc1p/Mcd1p is required for maintenance of cohesion in mitotically arrested cells (Guacci et al. 1997), and it dissociates from chromatin at the metaphase–anaphase transition and is degraded possibly by the anaphase promoting complex (Michaelis et al. 1997). In contrast, we find that >95% of cohesin subunits dissociate from chromatin at the onset of mitosis in our system. The possibility cannot be excluded, however, that the residual level of cohesins associated with metaphase chromosomes might be functionally significant (see below). To explain this apparent discrepancy between the two systems, we propose a model in which the balance between cohesion and condensation acts as a determinant for chromosome architecture (Fig. 8). We speculate that, in vertebrate cells, the major chromosomal reorganization (both morphological and biochemical) occurs at the onset of mitosis when most cohesins dissociate from chromosomes and are replaced by condensins (Fig. 8, top). Dissociation of cohesins reorganizes and loosens the linkage between sister chromatids, thereby relieving steric obstruction that could otherwise block condensin-mediated condensation. Cytological studies support the idea of a reorganization of cohesion at this stage of the cell cycle (e.g., Gimenez-Abian et al. 1995). The residual level of cohesins bound to metaphase chromosomes might be sufficient to hold sister chromatids together until the metaphase–anaphase transition. Alternatively, cohesion at this stage might be supported by a different mitosisspecific cohesion machinery, the identity of which is currently unknown. In yeast, the structural changes of chromosomes are less prominent at the onset of mitosis, resulting in the formation of less condensed, transcriptionally active chromosomes (Fig. 8, bottom). This different chromosomal architecture might be imposed by the unique cell cycle of S. cerevisiae in which the onset of mitosis cannot be clearly defined (Lew et al. 1997). In this organism, the major structural reorganization of chromosomes takes place at the metaphase–anaphase transition when the majority of cohesins (or at least the Scc1p/Mcd1p subunit) dissociate from chromosomes leading, directly or indirectly, to sister chromatid separation (Fig. 8, bottom).
Figure 8.
Sister chromatid cohesion and condensation in vertebrate and yeast chromosomes. Hypothetical chromosome architecture of vertebrate (top) and yeast (bottom) chromosome is shown. In vertebrates, the major transition occurs at the onset of mitosis (G2 to metaphase) when cohesion is largely reorganized and chromatids fully condense (top). In yeast, the structural reorganization of chromosomes takes place primarily at the metaphase–anaphase transition (bottom). Cohesins and condensins are shown by rectangles and circles, respectively. The single cohesion site drawn on the vertebrate metaphase chromosome by no means imply that cohesion is restricted to a centromeric region.
In yeast, the function of Mcd1p/Scc1p is required for cohesion near the centromeres as well as along the chromosome arms (Guacci et al. 1997; Michaelis et al. 1997). In higher eukaryotes, several observations indicate that there is a difference in the cohesion mechanisms acting along the chromosome arms and at the centromeric regions. This is especially apparent during meiosis in which cohesion of these chromosomal regions is dissolved at two different stages (Miyazaki and Orr-Weaver 1994). In our system, defects in centromeric cohesion (as judged by increased distance between sister kinetochores) were less prominent than defects in arm cohesion in the absence of cohesins (A. Losada, unpubl.). We hypothesize that cohesins are involved primarily in arm cohesion and that additional factors might contribute to centromeric cohesion in vertebrates. Such factors could be enriched in the large blocks of heterochromatin in which centromeres are embedded in vertebrates, but not in S. cerevisiae.
Guacci et al. (1997) have proposed that Mcd1p is a mitotic chromosome determinant, functioning as a linker molecule that connects the cohesion and condensation machineries on mitotic chromosomes. We provide three lines of evidence to suggest that this might not be the case in the Xenopus system. First, XRAD21, the Xenopus homolog of yeast Mcd1p/Scc1p, acts as a subunit of the putative cohesion machinery (14S cohesin), but does not associate with the condensation machinery (13S condensin), at least before it is targeted to chromosomes. Second, condensins bind normally to chromatin in the cohesin-depleted extracts. Third, immunodepletion of cohesins has little, if any, effect on the level of chromosome condensation in both the single and double chromatid assembly assays. We conclude that cohesion and condensation are largely separable processes in this cell-free system. The apparently conflicting observations in the two systems could be explained by the differential contribution of the condensation and cohesion machineries to metaphase chromosome organization (Fig. 8).
Two SMC-dependent processes in the eukaryotic chromosome cycle
In summary, we propose that proper assembly of mitotic chromosomes is regulated by two different SMC-dependent processes: cohesins (containing the SMC1- and SMC3-type proteins) participate in the establishment and maintenance of interphase sister chromatid cohesion after DNA replication, whereas condensins (containing the SMC2- and SMC4-type proteins) mediate chromosome condensation during mitosis. The two processes are functionally separable in Xenopus. However, given the structural similarities between cohesins and condensins, it is tempting to speculate that cohesion and condensation might have evolved from a common molecular mechanism. It will be interesting to test whether cohesins have biochemical activities similar to those associated with condensins (Kimura and Hirano 1997; Sutani and Yanagida 1997). It will also be important to determine how the two SMC-dependent processes are regulated precisely during the cell cycle. The current study provides a powerful biochemical system to address these questions in a vertebrate system.
Materials and methods
Cloning of XSMC1, XSMC3, and XRAD21
Poly(A)+ RNA was isolated from Xenopus eggs with a QuickPrep mRNA purification kit (Pharmacia Biotech) according to the manufacturer’s instructions. cDNA was synthesized from the RNA by use of a first-strand cDNA synthesis kit with the NotI–d(T)18 bifunctional primer (5′-AACTGGAAGAATTCGCGGCCGCAGGAAT18-3′; Pharmacia Biotech). The resulting single-stranded cDNA was used as a template for PCRs. For XSMC1, we designed a degenerate primer, OA2, that encodes the amino acid sequence PFFVLDE (amino acids 1152–1158 of Smc1p; Strunnikov et al. 1993): 5′-GCGGGATCCCC(TCAG)TT(TC)TT(TC)GT(TCAG)(TC)T(TCAG)GA(TC) GA-3′ (BamHI tag sequence is underlined). A PCR containing OA2 amplified a fuzzy band of ∼750 bp even without the addition of a second primer. Sequence analysis of the cloned fragment revealed that the NotI–d(T)18 primer carried over from the cDNA synthesis reaction functioned as the second primer, producing a band containing a short coding sequence followed by the 3′-end untranslated region of the cDNA. The 3′-end sequences of cDNAs for XSMC3 and XRAD21 were also amplified in the same way. The nucleotide sequences of the primers used were for XSMC3, 5′-GCGGGATCCTT(TC)TA(TC)(TC)T(TCAG) TT(TC)GA(TC)GA(AG)AT-3′, which encodes FYLFDEI (amino acids 1150-1156 of Smc3p; Michaelis et al. 1997); for XRAD21, 5′-GCGGGATCCCA(AG)GC(TCAG)GC(TCAG)GC(TCAG)AA (AG)TT(TC)TA-3′, which encodes QAAAKFY (amino acids 592–598 of human Rad21; McKay et al. 1996). The amplified PCR fragments were used as hybridization probes to screen a Xenopus oocyte cDNA library (Stratagene). To obtain full-length cDNAs, a second-round screening using different probes and nested PCRs were performed. We determined the full-length sequences for XSMC1 and XRAD21, and a 3′-end partial sequence for XSMC3.
Preparation of antibodies
Rabbit polyclonal antisera were raised against synthetic peptides corresponding to the carboxy-terminal amino acid sequences of XSMC1 (DLTKYPDANPNPND), XSMC3 (EQAKDFVEDDTTHG), and XRAD21 (SDIVATPGPRFHTV). Immunization and affinity purification of antibodies were performed as described previously (Hirano et al. 1997). We also raised antisera against recombinant XSMC1 and XSMC3 polypeptides and obtained results similar to those with the peptide antibodies.
Preparation of Xenopus egg extracts
CSF-arrested and interphase extracts were prepared in XBE2 buffer as described (Murray 1991) with the following modifications: (1) The crushing spin was performed at 10,000g for 15 min rather than 10 min; (2) unfertilized eggs were activated with Ca++ ionophore rather than an electric shock.
Immunoprecipitation, immunoaffinity purification, and immunodepletion
Immunoprecipitation and immunoaffinity purification were performed as described previously (Hirano et al. 1997). For immunodepletions, 50 μl of extract were incubated for 1 hr at 4°C with 25 μl of Affi-Prep Protein A Support (Bio-Rad) precoated with 5 μg each of anti-XSMC1 and anti-XSMC3 (cohesin depletion), 5 μg each of anti-XCAP-C, anti-XCAP-E, and anti-XCAP-G (condensin depletion), or 10 μg of control rabbit IgG (control depletion). XORC1 depletion was performed as described (Rowles et al. 1996).
In vitro assay for chromatin binding
Interphase extracts were incubated with sperm nuclei (1600 nuclei/μl) at 22°C in the presence or absence of 100 μg/ml aphidicolin and 20-μl aliquots were removed at different time points and placed on ice for 10 min. Chromatin fractions were then prepared as described (Kubota et al. 1995) except that XBE2 was used instead of EB. For mitotic activation, a half volume of a CSF-arrested extract or a nondegradable form of sea urchin cyclin B (cyclin BΔ90; Glotzer et al. 1991) was added to interphase extracts. H1 kinase activity was assayed as described (Murray 1991).
Immunofluorescent staining
Xenopus XL177 tissue culture cells were grown on polylysine-coated coverslips, and fixed with either cold methanol (−20°C, 10 min) or 2% paraformaldehyde in PBS (room temperature, 15 min) followed by treatment with 0.2% Triton X-100 in PBS (4°C, 5 min). Samples were first blocked for 30 min with 3% BSA/3% normal goat serum in PBS, and then incubated for 1 hr with primary antibodies diluted to 2 μg/ml in blocking solution. Rhodamine-labeled goat anti-rabbit IgG (Jackson) was used at 1:50 dilution. Samples were counterstained with 1 μg/ml DAPI and mounted in FITC-Guard (Testog). For lamin staining of interphase nuclei assembled in vitro, sperm nuclei (500 nuclei/μl) were incubated in interphase extracts for 90 min at 22°C, fixed for 15 min at room temperature with 10 volumes of 2% paraformaldehyde in XBE2, and sedimented onto coverslips through a cushion of 30% glycerol in XBE2 (Hirano and Mitchison 1993). Lamin monoclonal antibody L(0)46F7 (Benavente et al. 1985) was used at 1:100 dilution followed by rhodamine-labeled anti-mouse IgG (Jackson).
Assay for DNA replication
Demembranated sperm nuclei (500 nuclei/μl) and 5 μCi of [α-32P]dCTP were added to 50 μl of XSMC-depleted, control-depleted, or untreated extract in the presence or absence of 100 μg/ml aphidicolin. The reactions were incubated at 22°C, and 10-μl aliquots were removed at 30-min intervals and mixed with 10 μl of termination buffer [10 mm Tris-Cl (pH 8.0), 10 mm EDTA, 2% SDS, and 20 μg/ml proteinase K]. Samples were incubated at 37°C for 1 hr and analyzed on a 0.7% agarose gel followed by autoradiography. Incorporation of the labeled nucleotide was quantitated with an image analyzer (BAS 2000 Fuji Photofilm).
Condensation and cohesion assays
For the single-chromatid assembly assay, sperm nuclei (500 nuclei/μl) were incubated at 22°C in CSF-arrested high-speed supernatants (cohesin-depleted or control). After 2 hr, the mixtures were diluted, fixed, and spun onto coverslips. Chromosomes were stained with anti-XCAP-E followed by rhodamine-labeled goat anti-rabbit antibody (Jackson) and counterstained with DAPI. Identical results were obtained when crude extracts (low speed supernatants) were used. For the double-chromatid assembly assay, interphase (cohesin-depleted or control) extracts containing 100 μg/ml cycloheximide and 10 μm biotin–16-dUTP (Boehringer Mannheim) were incubated with sperm nuclei for 2 hr at 22°C. The extracts were driven into mitosis by the addition of a half volume of the corresponding CSF-arrested (cohesin-depleted or control) extracts containing cycloheximide, and incubated for another 90 min. Chromosomes were isolated and stained as described above, except that an additional incubation with 10 μg/ml fluorescein-conjugated avidin D was used to visualize the incorporation of biotin-16-dUTP. The average distance between sister chromatids was measured in chromosomes with paired morphology. Twenty-seven chromosomes assembled in control extracts (total length, 458 μm) and 31 chromosomes assembled in cohesin-depleted extracts (total length, 450 μm) were randomly selected from four independent experiments. Distance between the chromatids was measured at regular intervals of 0.6 μm along the entire length of the chromosomes.
Acknowledgments
We thank Alison Rowles and Julian Blow for anti-XORC1 and anti-XMCM3; Georg Krohne for anti-lamin LIII; and Michael Glotzer and Sandra Holloway for the cyclin BΔ90 expression plasmid. We are grateful to Doug Koshland, Kim Nasmyth, Rolf Jessberger, Hironori Funabiki, and Andrew Murray for discussions. We also thank Alain Verreault and Juan Mendez for critically reading the manuscript. This work was supported in part by grants from the National Institutes of Health (GM53926) and the Pew Scholars Program in the Biomedical Sciences to T.H. A.L. is the recipient of a fellowship from Ministerio de Educacion y Cultura, Spain, and is also supported by the Robertson Research Fund.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note
The GenBank accession numbers for XSMC1, XSMC3, and XRAD21 are AF051784, AF051785, and AF051786, respectively.
Footnotes
E-MAIL hirano@cshl.org; FAX (516) 367-8815.
References
- Benavente R, Krohne G, Franke WW. Cell type-specific expression of nuclear lamina proteins during development of Xenopus laevis. Cell. 1985;41:177–790. doi: 10.1016/0092-8674(85)90072-8. [DOI] [PubMed] [Google Scholar]
- Birkenbihl RP, Subramani S. Cloning and characterization of rad21, an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Castaño IB, Brzoska PM, Sadoff BU, Chen H, Christman MF. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae. Genes & Dev. 1996;10:2564–2576. doi: 10.1101/gad.10.20.2564. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Gimenez-Abian JF, Clarke DJ, Mullinger AM, Downes CS, Johnson RT. A postprophase topoisomerase II-dependent chromatid core separation step in the formation of metaphase chromosomes. J Cell Biol. 1995;131:7–17. doi: 10.1083/jcb.131.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993;120:601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ, Swedlow JR. The SMC family: From chromosome condensation to dosage compensation. Curr Opin Cell Biol. 1995;7:329–336. doi: 10.1016/0955-0674(95)80087-5. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Holt CL, May GS. An extragenic suppressor of the mitosis-defective bimD6 mutation of Aspergillus nidulans codes for a chromosome scaffold protein. Genetics. 1996;142:777–787. doi: 10.1093/genetics/142.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C-S, Ganetzky B. Molecular characterization of neurally expressing genes in the para sodium channel gene cluster of Drosophila. Genetics. 1996;142:879–892. doi: 10.1093/genetics/142.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger R, Riwar B, Baechtold H, Akhmedov AT. SMC proteins constitute two subunits of the mammalian recombination complex RC-1. EMBO J. 1996;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: A biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Weinert T, Pringle JR. Cell cycle control in Saccharomyces cerevisiae. In: Pringle JR, Broach JR, Jones EW, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 607–695. [Google Scholar]
- Lohka MJ, Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- McKay MJ, Troelstra C, van der Spek P, Kanaar R, Smit B, Hagemeijer A, Bootsma D, Hoeijmakers JHJ. Sequence conservation of the rad21 Schizosaccharomyces pombe DNA double-strand break repair gene in human and mouse. Genomics. 1996;36:305–315. doi: 10.1006/geno.1996.0466. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Miyazaki WY, Orr-Weaver TL. Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Newport J, Spann T. Disassembly of the nucleus in mitotic extracts: Membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell. 1987;48:219–230. doi: 10.1016/0092-8674(87)90425-9. [DOI] [PubMed] [Google Scholar]
- Rocques PJ, Clark J, Ball S, Crew J, Gill S, Christodoulou Z, Borts RH, Louis EJ, Davies K, Cooper CS. The human SB1.8 gene (DXS423) encodes a putative chromosome segregation protein conserved in lower eukaryotes and prokaryotes. Mol Human Genet. 1995;4:243–249. doi: 10.1093/hmg/4.2.243. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JPJ, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Goldberg I, Earnshaw WC. The SMC proteins and the coming age of the chromosome scaffold hypothesis. BioEssays. 1995;17:759–766. doi: 10.1002/bies.950170905. [DOI] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu CE, Murray AW. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Larionov VL, Koshland D. SMC1: An essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous family. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes & Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yanagida M. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature. 1997;388:798–801. doi: 10.1038/42062. [DOI] [PubMed] [Google Scholar]
- Yanagida M. Frontier questions about sister chromatid separation in anaphase. BioEssays. 1995;17:519–526. doi: 10.1002/bies.950170608. [DOI] [PubMed] [Google Scholar]