Abstract
Secreted proteins required for cellular movements along the circumference of the body wall in Caenorhabditis elegans include UNC-6/netrin and the novel TGF-β UNC-129. Expression of these proteins is graded along the dorsoventral (D/V) axis, providing polarity information to guide migrations. Here we show that the graded expression of UNC-129 in dorsal but not ventral body muscles depends on unc-130, which encodes a Forkhead transcription factor. The phenotype of unc-130 mutants closely mimics the reported effects of ectopically expressing unc-129 in both dorsal and ventral body muscles (Colavita et al. 1998). This fits our present finding that unc-130 cell autonomously represses unc-129 expression in the ventral body muscles. Thus the cell-specific effects of unc-130 on ventral, but not dorsal, body muscle expression of unc-129 accounts for the D/V polarity information required for UNC-129-mediated guidance. Genetic interactions between unc-130 and other guidance genes show that several molecular pathways function in parallel to guide the ventral to dorsal migration of distal tip cells (DTCs) and axonal growth cones in C. elegans. Genetic interactions confirm that UNC-129 does not require the only known type II TGF-β receptor in C. elegans (DAF-4) for its guidance functions. Also, unc-130 is partially required for male tail morphogenesis and for embryogenesis.
Keywords: Forkhead, TGF-β, cell migration, axon guidance, UNC-6/netrin
A fundamental question in development is how cellular movements are guided. Studying mutations that specifically affect guidance has proven to be an effective way to investigate this process. In the free-living nematode Caenorhabditis elegans, the secreted UNC-6/netrin guidance cue is expressed ventrally while UNC-5 and UNC-40/DCC (deleted in colorectal cancer), predicted receptors for UNC-6, are expressed in axons and cells that migrate along the dorsoventral (D/V) axis. UNC-5 and UNC-40 are required together for guidance away from UNC-6, while UNC-40 alone (or in combination with an undiscovered coreceptor) is required to guide migrations toward UNC-6 (Hedgecock et al. 1990; Ishii et al. 1992; Leung-Hagesteijn et al. 1992; Chan et al. 1996; Wadsworth et al. 1996). Studies with vertebrate homologs of these proteins suggest that guidance away from UNC-6/netrin is mediated by direct interaction with UNC-5 and UNC-40/DCC, which induces growth-cone repulsion, whereas guidance toward UNC-6/netrin is mediated by direct interaction with UNC-40/DCC, which induces growth-cone attraction (Keino-Masu et al. 1996; Leonardo et al. 1997; Hong et al. 1999).
In C. elegans, null mutations in unc-5 cause incompletely penetrant defects in dorsally oriented migrations, whereas null mutations in unc-40 or unc-6 cause incompletely penetrant defects in both dorsally and ventrally oriented migrations. Incomplete penetrance of single, double, and triple null mutants of these three genes suggest that they act in a single mechanism that guides migrations along the D/V axis and that other mechanisms act in parallel with UNC-6/netrin signaling to guide circumferential migrations (Hedgecock et al. 1990; Ishii et al. 1992; Leung-Hagesteijn et al. 1992; Chan et al. 1996). The incomplete effects of mutations in mouse netrin-1 and mouse DCC on circumferential axon guidance in the spinal cord similarly suggest that netrin-independent pathways also act to guide axons in the developing vertebrate spinal cord (Serafini et al. 1996; Fazeli et al. 1997). The nature of the parallel-acting D/V axon guidance mechanisms in C. elegans or in the mouse spinal cord has remained a mystery.
unc-129, which encodes a novel member of the TGF-β family of signaling ligands, also is required for circumferential migrations in C. elegans (Colavita and Culotti 1998; Colavita et al. 1998). Spatially restricted expression of UNC-129 in dorsal, but not ventral, body muscles is required for its guidance functions, suggesting that the resulting D/V-graded expression of UNC-129 conveys polarity information to cells and growth cones migrating along the D/V axis (Colavita et al. 1998). The mechanisms used by UNC-129 to guide cellular migrations remains unclear, as no other components of the UNC-129 signaling pathway have been identified. Mutants of the known TGF-β receptors in C. elegans do not appear to have unc-129-like phenotypes, suggesting unc-129 may act through a novel TGF-β receptor mechanism (Colavita et al. 1998; Krishna et al. 1999; see below).
How might other genes required to guide migrations along the D/V axis be identified? Null mutations in unc-5, unc-6, and unc-40 all lead to several visible phenotypes. In particular, as their names indicate, they are all uncoordinated (Unc). In addition, these mutations often affect the second (ventral to dorsal) phase of the triphasic distal tip cell (DTC) migration in the developing hermaphrodite (Hedgecock et al. 1990; Su et al. 2000). This creates ventral clear patches in the animal, which can be observed with a dissecting microscope (Hedgecock et al. 1990). We reasoned that other genes with these Unc and DTC phenotypes would be likely to participate either in the unc-5, unc-6, unc-40 circumferential guidance pathway or within a parallel-acting guidance pathway. Here, we report the characterization of a gene that identifies at least one such parallel-acting guidance mechanism.
Mutants of unc-130 share many phenotypes with unc-5, unc-6, and unc-40 mutants including motor axon guidance and DTC defects. The first unc-130 allele was identified in a screen for worms with this DTC phenotype (see Materials and Methods). Cloning and characterization of unc-130 show it encodes a Forkhead transcription factor that is required for the guidance of cellular migrations along the circumference of the body wall, both of pioneer axon growth cones and mesodermal cells (e.g., DTCs) in C. elegans. Analysis of genetic interactions between unc-130 and the known circumferential guidance genes suggests unc-130 acts in a pathway that parallels the unc-5, unc-6, unc-40 pathway. We report that the principle role of unc-130 in circumferential migrations is to initiate and maintain the dorsal body muscle specific expression pattern of UNC-129 TGF-β by repressing unc-129 expression in ventral body muscles. Consistent with this role, unc-129 mutations largely suppress the DTC migration defects of unc-130 mutant strains. This places unc-130 upstream of unc-129 as a negative regulator of UNC-129 expression in ventral muscle. This regulation is required to establish a graded distribution of UNC-129 along the D/V axis so UNC-129 can provide polarity information to guide circumferential migrations. Our results confirm that multiple mechanisms cooperate to promote guidance along the D/V axis in C. elegans and place unc-130 and possibly unc-129, in a pathway required for one such mechanism.
As unc-129 encodes a TGF-β ligand (Colavita et al. 1998) that is regulated by unc-130, we investigated interactions between unc-130 or unc-129 with genes known to function in TGF-β signaling. The nature of genetic interactions between daf-4 and unc-130 or unc-129 alleles reinforce the suggestion that UNC-129 function does not require DAF-4, the only known type II TGF-β receptor in C. elegans (Colavita et al. 1998). Our results indicate that eliminating the function of the TGF-β DBL-1, a regulator of body size with no previously reported migration defects, also alters migration along the D/V axis by affecting mechanisms that function in parallel to unc-130.
In addition to its role in circumferential guidance, unc-130 is required for the correct control of male tail morphogenesis and plays a role in embryonic development. The phenotype in the male tail and the embryonic phenotype are similar to those observed in mab-21 mutants (Chow et al. 1995), suggesting these genes affect similar developmental events. Unlike its role in circumferential guidance, unc-130 function in male ray morphogenesis and identity appears to be independent of unc-129. Thus, UNC-130 regulates distinct developmental processes in the body and in the tail by distinct mechanisms. Consistent with a role in cell fate determination in the male tail, UNC-130 also has been shown to regulate the fates of neurons in the head of C. elegans (Sarafi-Reinach and Sengupta, this issue).
Results
unc-130 is required for DTC and axonal growth-cone migrations along the D/V axis of the body wall
Mutations in unc-130 cause ventral clear patches indicative of a defect in the ventral to dorsal guidance (corresponding to the second migratory phase) of the DTCs (Fig. 1A). Normally, the two DTCs undergo a series of directed phases of migration during development, first migrating longitudinally along the ventral muscle band and away from their birthplace near the vulval precursor cells. They then turn and migrate dorsally along the epidermis, and finally, longitudinally along the dorsal muscle band back towards midbody (Hedgecock et al. 1990). In unc-130 mutants, as in mutants of unc-5, unc-6, and unc-40 (Hedgecock et al. 1990; Su et al. 2000), the DTCs often fail to undergo the second (ventral to dorsal) phase of their migration, but complete the third phase of migration along the ventral, rather than the dorsal, muscle band. The trajectory of the DTC determines the shape of the hermaphrodite gonad arm. In unc-130 mutants, when the second phase of DTC migration fails to occur, the distal arms of the gonad are misplaced ventrally, and cause a clear ventral patch in the body that is easily visualized with a dissecting microscope (Fig. 1A). These DTC migration defects are incomplete and temperature-sensitive for their penetrance in all alleles (Table 1A), including the null, unc-130(ev505) (see below). As for mutants of unc-5, unc-6, and unc-40, the anterior and posterior DTCs appear to act independently. Likewise, the posterior DTC is affected more often than the anterior DTC in all alleles, a phenomenon also observed in other DTC migration mutants (Hedgecock et al. 1990).
Figure 1.
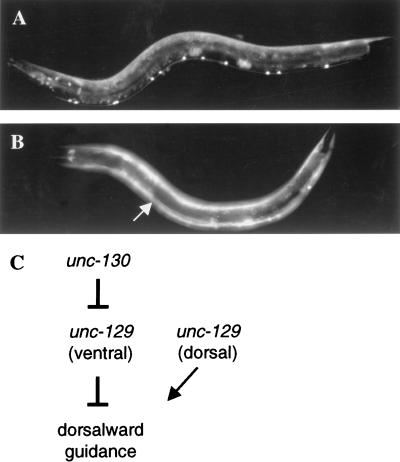
Phenotypes of unc-130 mutants. (A) The distal tip cell (DTC) migration (Mig) phenotype. In unc-130(ev505), L4 hermaphrodites grown at 25°C, the DTCs often fail to undergo the second, ventral-to-dorsal, phase of their migration leading to ventral clear patches (black arrows) flanking the vulva, caused by the ventral misplacement of the distal arm of the gonad. The DTCs act independently and show incomplete penetrance for the Mig phenotype, producing hermaphrodites with posterior (a), anterior (b), posterior and anterior (c), or no (not shown) ventral clear patches. (B) DA and DB motor neurons in wild-type adult animals grown at 25°C. DA and DB motor neurons visualized using evIs82 (neuronal-specific unc-129::gfp). These motor neurons extend growth cones from their cell bodies in the ventral nerve cord (white arrows) into the dorsal nerve cord (White et al. 1986). (C) unc-130(ev505); evIs82 adult hermaphrodite grown at 25°C. The DA and DB motor neurons often have axons with abnormal morphologies (white arrows). Instead of projecting directly from the ventral to dorsal nerve cord, axons often fail to reach the dorsal cord and run longitudinally at abnormal lateral positions. These aberrant axons indicate a defect in ventral-to-dorsal growth cone guidance. (D) DD and VD motor neurons visualized by staining an evIs113 (unc-5::lacZ; rol-6) adult hermaphrodite grown at 25°C for β-galactosidase activity (see Materials and Methods). As with the DA and DB motor neurons, the VD and DD motor neurons extend pioneer growth cones from the ventral nerve cord (black arrowhead) to the dorsal nerve cord (out of focus). (E) unc-130(ev505); evIs113 (unc-5::lacZ; rol-6) adult hermaphrodite grown at 25°C. DD and VD motor axons (white arrowheads) often fail to reach the dorsal nerve cord in unc-130 mutants. (F) Embryonic phenotype in unc-130(ev505). Embryos show variable defects. Cells often are found outside the anterior of the main embryo mass (black arrow) just before body elongation, which may be indicative of a defect in closure of the head epidermis. (G) unc-130(ev505) hatchling. L1 hatchlings sometimes have dorsal head bumps (black arrow). (H) him-5(e1490) male tail. Wild-type and control him-5(e1490) male tails have nine sensory rays on each side of the fan. Each ray (numbered 1–9) has a distinct position and morphology. (I) unc-130(ev505); him-5(e1490) male tail. unc-130 mutant males have variable fusions of sensory rays in the male tail. Shown here is a male tail with fusion of ray 3 with rays 4 and 6, and ray 7 with rays 8 and 9.
Table 1.
Penetrance of guidance defects in unc-130 mutant backgrounds
| A. DTC defects1
| ||||
|---|---|---|---|---|
| Genotype
|
Temperature (C)
|
% DTC migration defects
|
||
| anterior
|
posterior
|
N2
|
||
| unc-130(ev505) | 16 | 3 | 34 | 292 |
| 20 | 7 | 48 | 350 | |
| 25 | 21 | 85 | 330 | |
| unc-130(ev582) | 16 | 4 | 15 | 396 |
| 20 | 7 | 22 | 115 | |
| 25 | 4 | 52 | 180 | |
| unc-130(oy10) | 16 | 4 | 23 | 73 |
| 20 | 7 | 46 | 125 | |
| 25 | 12 | 57 | 152 | |
| unc-130(ev659) | 16 | 2 | 34 | 114 |
| 20 | 3 | 45 | 110 | |
| 25 | 9 | 64 | 116 | |
| N2 | 16 | 0 | 0 | 153 |
| 20 | 0 | 0 | 129 | |
| 25 | 0 | 0 | 200 | |
| B. Ventral to dorsal axon guidance defects1
| |||
|---|---|---|---|
| Genotype
|
Temperature (C)
|
% Defective axons
|
N2
|
| unc-130(ev505) | 16 | 18 | 107 |
| 20 | 23 | 104 | |
| 25 | 34 | 103 | |
| N2 | 16 | 0 | 150 |
| 20 | 0 | 165 | |
| 25 | 0 | 173 | |
| C. Dorsal to ventral axon guidance defects1
| ||||
|---|---|---|---|---|
| Genotype
|
Temperature (C)
|
% Defective axons
|
N2
|
|
| AVM
|
PVM
|
|||
| unc-130(ev505) | 25 | 14 | 14 | 100 |
| N2 | 25 | 1 | 0 | 100 |
Scored as described in the Results or in Materials and Methods.
N is the number of animals or embryos (section C) that were scored.
In order to determine whether unc-130 affects growth-cone migrations in addition to its effects on DTC migrations, the morphologies of the DA and DB motor neurons, and DD and VD motor neurons, visualized with unc-129::gfp and unc-5::lacZ reporters, respectively, were examined. The motor axons in unc-130(ev505) are disrupted in a manner consistent with a defect in ventral to dorsal guidance. Often, axons begin to extend dorsally but then turn longitudinally at variable lateral positions, frequently failing to reach the dorsal nerve cord (Fig. 1B–E; Table 1B). unc-130 does not appear to affect axon outgrowth, as axons appear to have normal lengths in the mutants. Motor axon guidance, like DTC migration, also appears to be temperature-sensitive in mutants of unc-130 (Table 1B), suggesting that unc-130 mutations may reveal an underlying temperature-sensitive process that affects both DTC and axon migration.
In order to examine the requirement for unc-130 in dorsal-to-ventral guidance, the morphology of lateral AVM and PVM touch neurons, which normally extend pioneer axons towards the ventral nerve cord, were examined in unc-130(ev505) and the wild type using a mec-7::gfp reporter. AVM and PVM axons are affected in a manner consistent with defects in ventral guidance. The initial phase of axon extension often is longitudinal or diagonal rather than ventrally oriented, with axons sometimes failing to reach the ventral nerve cord (Table 1C). Again, axon outgrowth does not seem to be affected. Consistent with a specific role for UNC-130 in D/V guidance, longitudinal ALM and PLM axons appear normal in unc-130(ev505).
unc-130 affects a ventral to dorsal guidance mechanism that acts in parallel to the UNC-6/netrin guidance pathway
Because the unc-130 mutant phenotype closely resembles the phenotypes of known UNC-6/netrin signaling mutants, we decided to examine genetically whether unc-130 might act within the UNC-6/netrin pathway. Double mutants of the unc-130(ev505) null allele with a unc-5, unc-6, or unc-40 null allele were compared with each single mutant. In each case, the penetrance of DTC defects was significantly higher in the double mutant than in the single null mutants (Table 2A; unc-40 data not shown). In addition, double mutants between unc-130(ev505) and weak alleles of unc-5, (e152) or (ev585), have more severe defects in dorsally oriented axon guidance, as scored by a failure of DA and DB motor neurons to reach the dorsal nerve cord, than the unc-130(ev505) single mutant (Table 2B). The double mutants also are more uncoordinated than any single mutant (data not shown). Taken together, these results indicate that unc-130 at least partially functions in parallel to the UNC-6/netrin pathway for dorsally oriented migrations of DTCs and axonal growth cones.
Table 2.
Penetrance of defects in single and double mutant backgrounds
| A. DTC defects1
| ||||
|---|---|---|---|---|
| Genotype
|
Temperature (C)
|
% DTC migration defects
|
||
| anterior
|
posterior
|
N2
|
||
| unc-130(ev505) | 25 | 13 | 84 | 292 |
| unc-130(ev505)/ mnDf77* | 25 | 12 | 83 | 275 |
| unc-5(ev585) | 25 | 30 | 53 | 330 |
| unc-130(ev505); unc-5(ev585) | 25 | 51 | 92 | 350 |
| unc-5(e152) | 25 | 18 | 57 | 180 |
| unc-130(ev505); unc-5(e152) | 25 | 50 | 87 | 396 |
| unc-5(e53) | 25 | 22 | 65 | 115 |
| unc-130(ev505); unc-5(e53) | 25 | 36 | 91 | 256 |
| unc-6(ev400) | 25 | 30 | 53 | 156 |
| unc-130(ev505); unc-6(ev400) | 25 | 51 | 92 | 220 |
| unc-130(ev505) | 25 | 12 | 85 | 143 |
| unc-129(ev554) | 25 | 0 | 0 | 225 |
| unc-130(ev505); unc-129(ev554) | 25 | 1 | 16 | 239 |
| unc-130(ev549) | 25 | 18 | 64 | 181 |
| unc-130(ev549); unc-129(ev554) | 25 | 0 | 8 | 220 |
| unc-130(ev505) | 16 | 3 | 34 | 120 |
| dbl-1(ev580) | 16 | 0 | 0 | 100 |
| unc-130(ev505); dbl-1(ev580) | 16 | 30 | 50 | 103 |
| daf-4(e1364) | 16 | 0 | 3 | 104 |
| unc-130(ev505); daf-4(e1364) | 16 | 38 | 64 | 396 |
| B. Ventral to dorsal axon guidance defects1
| |||
|---|---|---|---|
| Genotype
|
Temperature (C)
|
% Defective axons
|
N2
|
| unc-130(ev505) | 25 | 43 | 99 |
| unc-5(ev585) | 25 | 54 | 101 |
| unc-130(ev505); unc-5(ev585) | 25 | 94 | 99 |
| unc-5(e152) | 25 | 67 | 107 |
| unc-130(ev505); unc-5(e152) | 25 | 96 | 102 |
| C. Embryonic lethality1
| |||
|---|---|---|---|
| Genotype
|
Temperature (C)
|
% Lethality
|
N2
|
| unc-130(ev505) | 25 | 8 | 438 |
| dbl-1(ev580) | 25 | 8 | 230 |
| unc-130(ev505); dbl-1(ev580) | 25 | 89 | 113 |
| N2 | 25 | 1 | 218 |
Scored as described in Results or in Materials and Methods.
N is the number of animals that were scored.
unc-129::gfp is ectopically expressed in ventral body muscles in unc-130 mutants
unc-129 mutants have defects in D/V guidance of motor axons but do not affect DTC migrations. However, unc-129 has been shown to affect DTC migrations when ectopically expressed in both ventral and dorsal body muscles (Colavita et al. 1998). This mimics the unc-130 phenotype almost exactly, so we examined unc-129 expression in unc-130 mutant backgrounds. A full-length reporter for unc-129 promoter activity, evIs79, is normally expressed in dorsal body muscles and in DA and DB motor neurons, but not in ventral body muscles (Fig. 2A) (Colavita et al. 1998). When this reporter construct is used to assay unc-129 expression in unc-130 mutants, ectopic GFP expression is observed in ventral body muscles (Fig. 2B). Thus, unc-130 function is required to repress transcription from the unc-129 promoter in ventral muscles (Fig. 2C). However, unc-130 does not affect expression of unc-5, unc-6 or unc-40 (data not shown). This suggests that ectopic expression of endogenous UNC-129 is the cause of most of the ventral to dorsal guidance defects observed in unc-130 mutants.
Figure 2.
unc-130 is required to repress expression of UNC-129 in ventral body muscles. (A) evIs79 (full-length unc-129::gfp) in wild-type genetic background grown at 25°C. GFP is expressed in dorsal body wall muscles, DA and DB motor neurons, intestine, seam cells (not apparent), and some undefined cells in the head and tail (Colavita et al. 1998). (B) evIs79 (full-length unc-129::gfp) in unc-130(ev505) grown at 25°C. GFP is expressed ectopically in ventral body wall muscles (white arrow) in all mutant animals. (C) Model of unc-130 function in distal tip cell guidance: unc-130 is required to repress the expression of unc-129 in ventral muscle. This repression is required in order to maintain the dorsoventrally graded spatial pattern of UNC-129 expression, thereby allowing dorsally expressed UNC-129 to guide, directly or indirectly, dorsally oriented cellular movements.
To further examine this regulatory interaction, we constructed double mutants between the putative null unc-129 allele (ev554), and unc-130 alleles. The double mutants have far fewer DTC migration defects than the unc-130 alleles alone (Table 2A). Thus, loss of unc-129 function partially suppresses DTC migration defects caused by unc-130 mutations, placing unc-130 genetically upstream of, and inhibitory to, unc-129. However, at 25°C unc-129(ev554) only suppresses ∼80% of the defects in the migration of the posterior DTC caused by unc-130(ev505). This indicates that in addition to affecting DTC migrations by acting upstream of unc-129, unc-130 also affects these migrations by another mechanism. The unc-130; unc-129 double mutant is not any more uncoordinated than either single mutant. Furthermore, DA and DB motor axon guidance defects in the unc-130(ev505); unc-129(ev554) double mutant (43% misguided, n = 256) were not significantly more penetrant than in unc-129(ev554) (40% misguided, n = 320) but were more penetrant than in unc-130(ev505) (28% misguided, n = 256), suggesting that these two genes largely act in the same pathway for motor axon guidance.
unc-130 is required cell autonomously to repress unc-129::gfp expression in ventral muscle
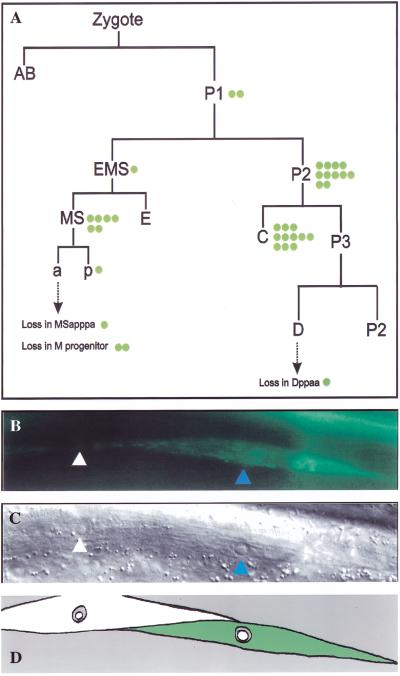
In principle, unc-130 could act within ventral muscle to regulate expression of unc-129 or it could act indirectly in another tissue. We tested the cell autonomy of unc-130 function with respect to unc-129 repression in ventral muscle by carrying out mosaic analysis. unc-130 (ev505); evIs79(full-length unc-129::gfp); ncl-1(e1865) L4 hermaphrodites were transformed by germ-line injection of unc-130(+) and ncl-1(+) rescuing DNAs (see Materials and Methods). ncl-1(e1865) causes the nuclei of mutant cells to become enlarged and is rescued cell-autonomously by ncl-1(+) DNA (Hedgecock and Herman 1995). A line carrying an extrachromosomal array, which partially rescued the DTC migration defects and frequently failed to ectopically express unc-129::gfp in individual ventral body muscles, was selected for further analysis. Animals mosaic for the rescuing array were identified by scoring for patches of ncl-1(−) or ncl-1(+) cells. The cell division where loss of the rescuing array occurred then was determined by scoring the Ncl phenotype in all body muscle cells and in cells descended from diverse lineages. In 36/36 mosaic losses, every ventral body muscle cell derived from the lineage that had lost the unc-130(+) rescuing array (as determined by the Ncl phenotype) concomitantly expressed unc-129::gfp (Fig. 3). Conversely, every ventral body muscle cell derived from lineages that retained the unc-130(+) array were rescued for the ectopic expression of unc-129::gfp. The range of mosaic animals confirmed that unc-130 is required cell autonomously for unc-129 repression in all lineages giving rise to ventral body muscles. In addition, loss of the rescuing array in a single body muscle cell and not in its sister cell also led to cell-autonomous ectopic expression of unc-129::gfp, eliminating a cell nonautonomous function for unc-130 in closely related muscle cells. Thus, unc-130 is required cell autonomously within ventral body muscle cells to repress unc-129 transcription.
Figure 3.
unc-130 acts cell autonomously to repress unc-129 expression in ventral body muscles. Mosaic analysis was carried out to determine the ability of unc-130(+)-containing extrachromosomal arrays to rescue cell autonomously the ectopic expression of unc-129::gfp in unc-130 mutants. unc-130(ev505); ncl-1(e1865); evIs79 (full-length unc-129::gfp) animals were transformed with a unc-130 rescuing construct (evEx114) and ncl-1(+) rescuing DNA. The resulting strains were used to identify mosaics that had lost the rescuing array in some ventral body muscles during development. The presence or absence of the rescuing array was determined by scoring the Ncl phenotype (enlarged nucleoli in mutant cells) and ventral muscle cells were scored for the presence or absence of ectopically expressed unc-129::gfp. (A) Summary of cells in which the rescuing extrachromosomal array was lost (green dot) as inferred from examining the Ncl phenotype (see panel C) in cells representative of most lineages. Thus, for a given mosaic loss, all descendants of the cell indicated by a green dot no longer contained unc-130 rescuing DNA. All ventral muscle cells that lost the rescuing array, as determined by the Ncl(−) phenotype, also expressed ectopic unc-129::gfp. Those ventral muscle cells that retained the array failed to express unc-129::gfp. (B) Example of a mosaic animal: failure of unc-130 rescue in a Ncl(−) ventral muscle cell (blue arrowheads in panels B and C) is indicated by ectopic unc-129::gfp expression in the cell. An adjacent Ncl(+) ventral muscle cell (white arrowheads in panels B and C) is rescued for ectopic unc-129::gfp expression. (C) The Ncl phenotype of the two ventral body muscles of panel B observed by D.I.C. microscopy. (D) Schematic summary of panel B plus panel C results.
UNC-130 acts in parallel to DBL-1 and DAF-4 mediated signaling
unc-129 encodes a TGF-β ligand (Colavita et al. 1998). TGF-β ligands normally signal through a conserved family of type I and type II TGF-β receptors. The phenotypes of mutations in the known C. elegans TGF-β receptors do not resemble unc-129 mutant phenotypes, suggesting unc-129 does not signal positively through a classical TGF-β receptor mechanism (Colavita et al. 1998). We therefore investigated genetic interactions between unc-130 and daf-4, which encodes the only type II TGF-β receptor in C. elegans (Estevez et al. 1993; Krishna et al. 1999), as a way to assess the interaction between ectopically expressed UNC-129 and DAF-4 mediated signaling. If UNC-129 acted positively through DAF-4, eliminating daf-4 function would be expected to suppress unc-130 DTC migration defects as do unc-129 mutations. However, daf-4 mutations enhance the DTC migration defects caused by unc-130(ev505) (Table 2A). This places daf-4 and unc-130 in parallel pathways for the ventral-to-dorsal guidance of DTCs. In addition, this strongly suggests that unc-129 does not act positively through the only known type II TGF-β receptor in C. elegans. Conversely, if UNC-129 acted to inhibit DAF-4 mediated signaling, daf-4 mutations should be epistatic to unc-129. This is not the case because daf-4(e1364); unc-129(ev554) double mutant animals have the same locomotion defects as unc-129 mutants and are small, like daf-4 mutants. Thus, unc-129 and daf-4 appear to act independently, suggesting that unc-129 does not positively or negatively regulate the known type I/type II TGF-β receptor-mediated signaling cascade to carry out its guidance functions.
As DAF-4 appears to have a role in the guidance of DTCs, but not as a receptor for UNC-129, we investigated whether another TGF-β ligand, DBL-1, which does not have any known role in guidance along the D/V axis, affects the ventral-to-dorsal guidance of DTCs. Introducing dbl-1(ev580) into a unc-130(ev505) background does indeed increase the penetrance of DTC migration defects, suggesting DBL-1 acts independently of UNC-130 to guide DTCs dorsally (Table 2A). In addition, at 25°C unc-130(ev505); dbl-1(ev580) double mutants have a high penetrance of embryonic lethality (Table 2C). Interestingly, dbl-1(ev580) appears to produce a small proportion of embryos that fail to hatch (Table 2C). These dying embryos have phenotypes similar to those observed in unc-130(ev505) (Fig. 1F) and mab-21(bx41) (data not shown).
Positional cloning of unc-130
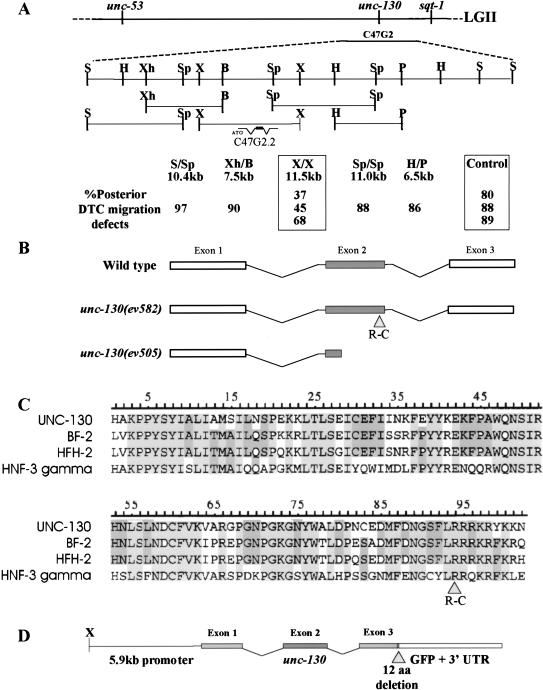
unc-130 maps to LGII, to the right of unc-53 and just left of sqt-1 (see Materials and Methods). One cosmid from this region, C47G2, was found to rescue the DTC migration defects in unc-130(ev505). An integrated array carrying C47G2 has no DTC migration defects. Furthermore, the integrated cosmid array rescues the morphology of the DD and VD motor axons visualized using a unc-5::lacZ reporter construct (see Materials and Methods). One subclone of C47G2, an 11.5-kb XbaI fragment, was capable of rescuing the DTC migration defects and axon guidance defects of unc-130(ev505) (Fig. 4A).
Figure 4.
Molecular characterization of unc-130. (A) unc-130 map location and rescue. (Top) unc-130 lies between unc-53 and sqt-1 on linkage group II (LGII). A single cosmid from the region, C47G2, rescues unc-130 phenotypes. (Enlarged region) Subclones of C47G2 aligned beneath its restriction map were tested for their ability to rescue the distal tip cell (DTC) migration defects of unc-130(ev505). Stable lines carrying extrachromosomal arrays containing the various constructs were scored for the penetrance of posterior DTC migration defects at 25°C. Only lines carrying arrays including an XbaI 11.5-kb fragment (X/X) consistently showed significant rescue when compared to control transgenic lines (boxes). Restriction site abbreviations (S) SalI; (H)HindIII; (Xh) XhoI; (Sp) SpeI; (X) XbaI; (B) BamHI; (P) PstI). (B) Molecular characterization of unc-130 alleles. unc-130 contains three exons (rectangles). The middle exon encodes the Forkhead DNA-binding (DB) domain (gray box). unc-130(ev582) has a missense mutation in a conserved arginine (codon 217) near the 3′ end of the Forkhead domain (gray triangle; see panel C). unc-130(ev505) contains a large deletion of ∼1.7 kb (see Materials and Methods). The left breakpoint of the deletion is within the Forkhead domain, whereas the right breakpoint lies ∼1 kb to the right of the 3′ end of the coding region. The next flanking gene in this direction is a pseudogene ∼5 kb away. (C) Alignment of UNC-130, BF-2, HFH-2 and HNF-3γ Forkhead domains. Amino acids that are conserved in all four proteins are highlighted in light gray; those conserved in three of four proteins are in dark gray. The R217C mutation in unc-130(ev582) is indicated by the gray triangle. (D) Structure of the unc-130::GFP translational fusion construct, pBN17. The fusion includes the entire 5′ end of pBN11 (the 11.5-kb XbaI-rescuing fragment) fused to the coding region of GFP with a 12 codon deletion at the 3′ end of exon 3 of unc-130 (see Materials and Methods). The construct is functional, as it is capable of rescuing both the DTC migration defects and the ventral-to-dorsal growth cone guidance defects of unc-130(ev505).
UNC-130 is a Forkhead transcription factor related to the vertebrate proteins BF-2 and HFH-2
The rescuing subclone was predicted to encode a putative Forkhead transcription factor. In order to confirm that disruption of this predicted gene was the cause of the phenotypes observed in unc-130 mutants, the coding region of two mutant alleles was analyzed. unc-130 (ev505) contains a deletion that eliminates the 3′ end of the predicted open reading frame including the Forkhead domain (Fig. 4B; see also Materials and Methods). We predict that unc-130(ev505) is a null. Consistent with this prediction, unc-130(ev505) homozygotes have the same penetrance of DTC defects as unc-130(ev505)/mnDf77 hemizygotes (Table 2A). unc-130(ev582), obtained in a noncomplementation screen (see Materials and Methods), has a missense mutation in a conserved arginine codon within the Forkhead domain converting it to a cysteine codon (Fig. 4B,C). Thus, unc-130 is C47G2.2, the putative Forkhead transcription factor-encoding gene that lies within the rescuing fragment. An unc-130::GFP translational fusion (see Materials and Methods) also rescues unc-130(ev505) defects, confirming the correct identification of the gene and suggesting the 3′ untranslated region of unc-130 plays little or no specific role in its function (Fig. 4D).
UNC-130 is most closely related to the vertebrate genes Brain Factor-2 (BF-2; Hatini et al. 1994) and Homolog of Forkhead-2 (HFH-2; Clevidence et al. 1993). The UNC-130 Forkhead domain is 86% identical to both vertebrate genes (Fig. 4C). BF-2 and HFH-2 are 95% identical within this region. For comparison, UNC-130 is 66% identical to the more distantly related HNF-3γ within the Forkhead domain.
unc-130 expression correlates with its function
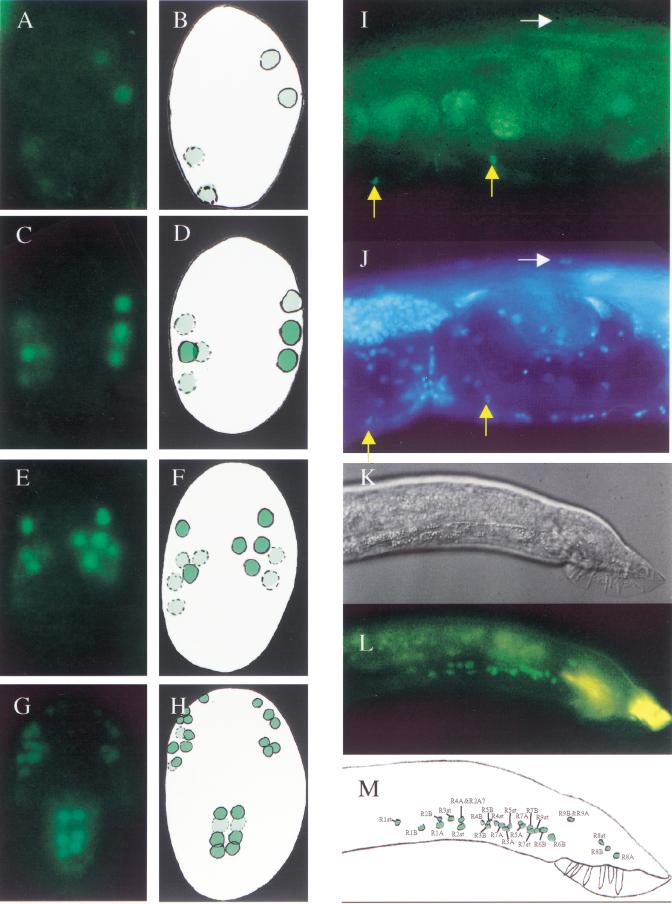
To investigate where unc-130 is expressed, animals transgenic for a chromosomally integrated version of the rescuing unc-130::GFP translational fusion were made and observed (Fig. 4D; see Materials and Methods). The unc-130::GFP construct is expressed in a dynamic pattern during embryogenesis in head hypodermal cells, as well as in muscle and intestinal precursor cells (Fig. 5A–H). In adults, unc-130::GFP is observed in ventral muscle (Fig. 5I,J), consistent with the cell autonomous requirement for unc-130 function in these cells (see above), as well as in intestine and other cells in the head and tail (not shown). In adult males, unc-130 is also expressed in the structural cells and two neurons of each ray (Fig. 5K–M), consistent with its role in male tail morphology. Thus, unc-130 expression and function appear to correlate well in the embryo, in ventral muscle, and in the male tail, as indicated by the mutant phenotypes and mosaic analysis, suggesting that the expression pattern observed using this GFP construct includes the functionally relevant portion of endogenous unc-130 gene expression. The expression pattern of UNC-130 in the head of C. elegans is described fully in the accompanying paper (Serafi-Reinach and Sengupta 2000).
Figure 5.
unc-130 expression pattern. The unc-130::GFP expression pattern is dynamic during embryogenesis, first turning on in four cells at ∼315 min of development at 20°C (panel A schematized in B). Expression then turns on in progressively more cells, including muscle precursors and hypodermal cells in the head (panels C–F), along with intestinal precursors (two vertical rows of cells in panels G and H). In adults, weak expression of unc-130::GFP in ventral body muscle is observed (yellow arrows in panel I, DAPI staining in panel J). Weak expression in dorsal muscle is also occasionally observed (white arrows in I and J). Dorsal muscle expression of unc-130 apparently does not normally repress unc-129 expression in these cells; however, strains carrying extrachromosomal arrays that include unc-130(+) rescuing DNA have been observed to affect the expression of unc-129::gfp in dorsal muscle (data not shown), possibly because the levels of unc-130 expression are higher in these strains. Expression also is observed in all three cells that form each of the nine sensory rays in the male tail (panel K is a D.I.C. image of the male tail, panel L shows unc-130::GFP expression in the same male tail shown in K; panel M is a schematic of L). The cells observed here are cells that will form the male ray sensillae (Sulston et al. 1980). In panels A–H, anterior is up. In panels I–L, anterior is to the left and dorsal is up.
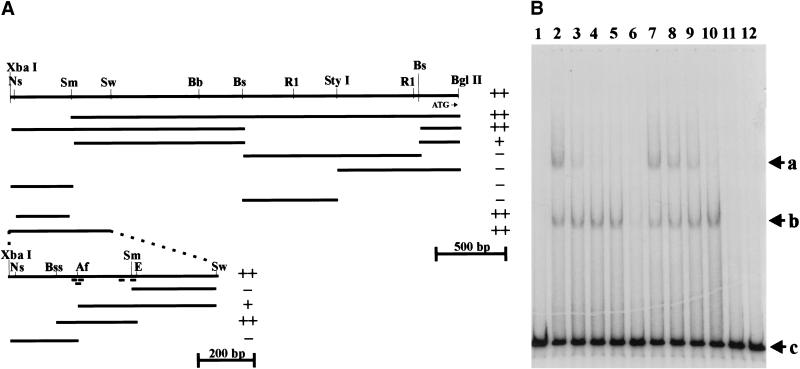
UNC-130 binds to predicted Forkhead transcription factor binding sites in a 277-bp region of the unc-129 promoter.
A purified maltose binding protein (MBP) fusion with UNC-130 (MBP–UNC-130) was made in Escherichia coli (see Materials and Methods) and examined for its ability to shift the electrophoretic mobility of individual DNA fragments (gel-shift assays) corresponding to subdomains of the putative unc-129 promoter (Colavita et al. 1998). The MBP–UNC-130 protein was found to bind at least two sites, one with high affinity and another with low affinity, contained within a 2-kb fragment previously identified as a regulatory region for muscle specific unc-129 expression (Colavita et al. 1998). The two MBP–UNC-130 binding sites were delimited to a 277-bp fragment from within this 2-kb region (Fig. 6). MBP alone or a mutant MBP–UNC-130 fusion protein carrying the same Arg to Cys alteration in the DNA binding site as the oy10 and ev582 protein failed to bind to this or any other fragments in the 3.1-kb unc-129 promoter region. The shifted 277-bp fragment contains five putative Forkhead transcription factor binding sites, all of which appear capable of binding wild-type but not mutant MBP–UNC-130 when present within 22-, 37-, and 23-bp double-strand oligonucleotides matching normal unc129 promoter sequence (Fig. 6; see also Materials and Methods). Oligonucleotides mutated in the consensus Forkhead binding sites were unable to bind wild-type or mutant MBT–UNC-130 proteins (data not shown). These results suggest that unc-129 is a direct target for transcriptional repression by UNC-130.
Figure 6.
Maltose binding protein (MBP)–UNC-130 binds to a 277-bp fragment in the muscle-specific regulatory region of the unc-129 promoter. (A) A restriction map of the entire previously characterized unc-129 5′ regulatory region (Colavita et al. 1998) and DNA fragments derived from this region are indicated. The ability of wild-type MBP–UNC-130 (but not MBP alone or R217C mutant MBP–UNC-130) to shift the electrophoretic mobility of these fragments (see Materials and Methods) is indicated to the right of each fragment: (++) strong specific binding, (+) weaker specific binding, (−) no detectable binding. These experiments identified a 277-bp BssHII–EcoRV fragment capable of specifically binding wild-type MBP–UNC-130. Restriction site abbreviations: (Ns) NsiI; (Sm) SmaI; (Sw) SwaI; (Bb) BbsI; (Bs) BstEII; (R1) EcoRI; (Bss) BssHII; (Af) AflII; (E) EcoRV. Short lines under the XbaI–SwaI fragment restriction map at the bottom of this panel represent consensus Forkhead transcription factor binding sites. Five potential target sites are located within the 277-bp BssH1–EcoRV fragment. (B) The specificity of binding to the 277-bp (BssHII–EcoRV) fragment is demonstrated. Lane 1 is unbound, radiolabeled, 277-bp DNA (b and c), lane 2 shows two gel-shifted bands (a and b) above the unbound DNA, while lanes 3–6 show binding in the presence of 10×, 50×, 100×, and 1000× molar excess of cold 277-bp competitor DNA. This competition experiment is repeated in lanes 7–10 with cold 245-bp XbaI–AflII fragment as competitor. Shifted band b represents an apparent high-affinity binding site that is not competed by 100×, but is largely competed by 1000×-specific, 277-bp competitor but not 1000×-nonspecific, 246-bp competitor. Shifted band a represents a low affinity site that is completely competed by 50×-specific, 277-bp competitor, but is not competed by 100×-nonspecific, 246-bp competitor. Lanes 11,12 show no binding of MBP alone or the R217C mutant form of MBP–UNC-130, respectively.
unc-130 is required for development of the male tail
Wild-type C. elegans males have nine sensory rays on each side of the fan in their tails that can be distinguished by morphology and position (Fig. 1H,I). In unc-130 mutants, rays frequently fail to maintain their distinct morphology, instead fusing with other rays in a variable way. The absence of rays 4 and 6 and their replacement by a large fused ray with morphological similarity to two fused ray 4s is the most common fusion (Fig. 1I). Ray 6, which normally has a cone-like shape, is rarely transformed into a longer, straighter, ray 4-like morphology when these two rays do not fuse (not shown). Other phenotypes observed in unc-130 mutant males include: (1) ectopic ray clusters, which probably result from ray lineage reiterations; (2) extra cells in ray precursor clusters, which probably result from defects in programmed cell deaths normally observed in certain ray lineages, and (3) fusion of R6.p with the tail seam (SET) instead of hyp-7.
Approximately 8% of unc-130(ev505) embryos grown at 25°C die before hatching with variable morphological abnormalities (Fig. 1F) and some hatchlings have variably abnormal morphologies (Fig. 1G). Some of the embryonic and the male tail defects (ectopic precursor clusters and R6.p fusions) of unc-130 are surprisingly similar to defects observed in mutants of mab-21, which encodes a nuclear localized protein that affects body size (Chow et al. 1995; our observations).
unc-130(ev505); unc-129(ev554); him-5(e1490) strains have male tail defects that are quantitatively indistinguishable from those observed in unc-130(ev505); him-5(e1490) (data not shown). The lack of suppression indicates that unlike its role in axon guidance, unc-130 does not act in male tail morphogenesis by negatively regulating unc-129 expression.
Discussion
unc-130 was identified in a screen for mutants with defects in coordinated locomotion and in dorsally oriented guidance of DTCs. In addition to defects in both dorsally and ventrally oriented cell and axon growth cone migrations, unc-130 mutant worms are slightly uncoordinated, slightly dumpy, and have a range of male tail defects that suggest alterations in cell lineages and cell fates. The apparent cell fate changes in the male tail are consistent with the recent finding that unc-130 mutants affect the fates of several neurons in the head of C. elegans (Serafi-Reinach and Sengupta 2000). Taken together, these results suggest that unc-130, which encodes a putative Forkhead transcription factor, has multiple functions in the development of C. elegans.
unc-130 ensures normal D/V guidance in the body of C. elegans by establishing the correct spatial expression of unc-129
To guide migrating cells and axonal growth cones, molecules that function as guidance cues must be expressed in the correct spatial pattern in order to impart directional information. unc-130 is required for the correct spatial expression pattern and distribution of UNC-129, a novel TGF-β ligand, which may be acting as a guidance cue or patterning the formation of an unknown guidance cue (Colavita et al. 1998). unc-130 is expressed and required cell autonomously in ventral body wall muscle cells in order to repress unc-129 expression in these cells. Ectopic UNC-129 expression in ventral body muscles is known to induce DTC and motor axon migration defects (Colavita et al. 1998), therefore, in principle, the same ectopic UNC-129 expression observed in unc-130 mutants is sufficient to explain the unc-130 mutant DTC and Unc phenotypes. This is consistent with the ability of unc-129 mutations to suppress unc-130 DTC defects in double mutants. As unc-129 single mutants do not have DTC migration defects, the suppression of this phenotype in the double mutant means that in the absence of functional unc-129, unc-130 is no longer necessary to keep unc-129 from being expressed ventrally. Thus, the normal function of unc-130 appears to be to establish and possibly maintain the correct spatial pattern of unc-129 expression in dorsal, but not ventral, body muscles.
By genetic criteria, unc-130 acts in a pathway parallel to unc-5, unc-6, and unc-40 because double mutants carrying the unc-130(ev505) null allele, and null alleles of these three genes are more penetrant than the null phenotypes of each single mutant. There are two complications this interpretation. The first arises from the fact that unc-130 mutations appear to affect guided migrations by causing ectopic expression of UNC-129 in ventral body muscles. It is therefore possible that the genetic interactions between unc-130 and unc-5, unc-6 or unc-40 are due to neomorphic unc-129 function attributable to this ectopic expression. In this case, the genetic interactions between unc-130 and these three genes might not address whether unc-129 normally functions in parallel to unc-5, unc-6, and unc-40, only that it does so in this context. However, we have found that UNC-129 does normally affect DTC migrations. Normally recessive unc-6 null mutations can act as dominant enhancers of unc-129 DTC defects; for example, animals homozygous for unc-129(ev554) and heterozygous for unc-6(ev400) have 22% posterior DTC defects at 20°C (n = 100). Thus, animals lacking unc-129 function may be sensitized to the dose of unc-6(+) required for normal DTC migration. This indicates that UNC-129 does normally affect DTC migration, however, unc-129 mutants presumably do not have DTC migration defects because this role of UNC-129 is completely redundant. This result argues against a neomorphic effect of UNC-129 on guided migrations in a unc-130 mutant. In addition, it shows that at least some unc-6 function is required independently of unc-129 for normal DTC migration.
The second complication to the interpretation of the double mutant results is that unc-129 null mutations only partially suppress unc-130 DTC defects. This means that unc-130 mutations also affect DTC migrations by a unc-129-independent mechanism. Therefore, enhancement of unc-6 DTC defects by unc-130 may result from the loss of this other mechanism and not from ectopic expression of UNC-129. For this reason, we can not be absolutely certain that unc-129 functions in parallel to unc-6 even though unc-6 can at least partially function in parallel to unc-129. However, given that there is no apparent regulation of unc-5, unc-6, or unc-40 expression by unc-129 or unc-130 (Colavita et al. 1998; A. Colavita and B. Nash, data not shown), it is more likely that UNC-130 (and by extrapolation, UNC-129) act in parallel to UNC-6/netrin signaling to guide migrations along the D/V axis of C. elegans.
Taken together, our genetic results confirm that cells are guided in their movements along the D/V axis of the C. elegans body wall by at least two separate mechanisms. One involves the D/V-graded expression of the UNC-6 guidance cue (Wadsworth et al. 1996; Kim et al. 1999; Ren et al. 1999). The other mechanism likely involves the D/V-graded expression of a guidance cue whose function requires the corresponding graded expression of UNC-129 (but does not require classical TGF-β signaling through type I and type II receptors). These two mechanisms cooperate to ensure the exquisite level of guidance required for invariant development. Either pathway is partially capable of guiding cell movements, as revealed when the parallel pathway is compromised by mutation. When both pathways are eliminated by mutation, guidance is further compromised. Indeed, the observation that the two DTCs still sometimes migrate dorsally, but at different frequencies, in double mutants between unc-130(ev505) and unc-6, unc-5, or unc-40 null alleles suggests that at least one additional mechanism acts in parallel to UNC-6/netrin and UNC-129 signaling to guide migrations along the dorsoventral axis.
DBL-1 acts in parallel to UNC-130 to affect DTC migrations
The observation that DBL-1 signaling appears to act in parallel to UNC-130, as indicated by the enhanced percentage of DTC defects and embryonic lethality in unc-130; dbl-1 double mutants, reveals new roles for dbl-1 in development. The role in D/V patterning in the body may be analogous to dbl-1 function in male tail patterning, where dbl-1 is proposed to act as a dorsalizing factor. However, as dbl-1 is expressed in the ventral nerve cord, it seems likely that here dbl-1 may act as a ventralizing signal (Suzuki et al. 1999). This role may be conserved between dpp in Drosophila and BMP-2 and BMP-4 in vertebrates, the closest relatives to dbl-1, which also affect D/V patterning (for review, see Holley and Ferguson 1997).
UNC-130 is a Forkhead transcription factor family member
UNC-130 contains a highly conserved Forkhead DNA-binding domain and is able to bind three putative consensus binding sites in a gel-shifted 277-bp fragment of the muscle-specific regulatory region of unc-129. Among identified proteins, UNC-130 is most similar to vertebrates HFH-2 and BF-2. BF-2 helps pattern the forebrain, optic vesicle and kidney (Hatini et al. 1994, 1996; Yuasa et al. 1996; Gomez-Skarmeta et al. 1999).
The Xenopus homolog of BF-2 (XBF-2) is expressed in and helps maintain dorsolateral mesoderm during gastrulation by down-regulating the TGF-β family member BMP-4. XBF-2 also affects neural crest cell migration ; its expression must be down-regulated in order to allow migration to occur (Gomez-Skarmeta et al. 1999). XBF-2 lies downstream of the BMP antagonists noggin, cerberus, and gremlin and has neuralizing activity, probably as a result of its effects on BMP-4 expression (Mariani and Harland 1998; Gomez-Skarmeta et al. 1999). XBF-2 is a transcriptional repressor that converts ectoderm into neural tissue. HFH-2 (formerly Genesis) is expressed in premigratory and migrating neural crest cells in the early mouse embryo and in motor neuron progenitors in the developing spinal cord, and might therefore have some functions analogous to BF-2 (Hromas et al. 1999).
It is unclear whether BF-2 repression of BMP-4 is analogous to UNC-130 repression of unc-129 in ventral body muscles in C. elegans. However, the similarities are striking. BF-2 and UNC-130 are both required for the correct D/V-graded expression of a TGF-β superfamily member and both genes appear to have multiple functions in development, including effects on mesodermal and ectodermal cells. In the future, it will be interesting to determine the similarity between the molecular mechanisms used by these proteins to regulate TGF-β and BMP expression.
unc-130 affects multiple TGF-β dependent processes
unc-130 is required for the correct D/V spatial expression of the TGF-β ligand UNC-129. unc-130 thereby contributes to the correct guidance of circumferential cellular migrations during development in C. elegans. As mentioned above, it is unclear how UNC-129 guides circumferential migrations, although recent studies in vertebrates suggest that TGF-β ligands can act as guidance cues (Augsburger et al. 1999). The suppression of unc-130 DTC defects by unc-129 mutations is the basis for a screen for genes that act in the unc-129 signaling pathway. Identification of such genes should help define the molecular mechanisms by which UNC-129 functions to guide migrations.
In addition to its effect on unc-129, unc-130 interacts genetically with daf-4, which encodes the type II TGF-β receptor in C. elegans, with dbl-1, which encodes a TGF-β ligand required for male tail morphogenesis and the control of body size, and with mab-21, which has been shown to act in the dbl-1 TGF-β pathway in the male tail (Morita et al. 1999; B. Nash, data not shown). Thus, unc-130 appears to be intimately connected to several TGF-β-dependent processes. In other systems, Forkhead transcription factors have been shown previously to mediate TGF-β signaling, acting as transcriptional partners with Smad2, to effect expression of TGF-β ligands such as BMP-4 and to be targets of activin induction. Our results suggest that individual Forkhead transcription factors may interact both upstream and in concert with distinct TGF-β signaling pathways, further increasing the potential for complexity in the interactions between Forkheads and TGF-β-signaling pathways. Further investigation into the function of unc-130 and its relationship to other developmentally important genes, most notably TGF-β pathway components, should reveal the molecular mechanisms by which these interactions are regulated.
Materials and methods
Genetics
unc-130(ev505) was induced by irradiation with a cesium 137 (137Cs) source (2000 Rads) and isolated in an F2 screen for mutants with ventral clear patches indicative of defects in ventral to dorsal DTC migration. unc-130(ev582) was isolated in a noncomplementation screen with a unc-53(e404); unc-130(ev505); sqt-1(e1350) triple mutant chromosome. unc-130(ev549) was isolated in an F2 EMS (ethylmethansulfonate) screen. unc-130(oy10) was kindly provided by T. Sarafi-Reinach and P. Sengupta (Brandeis University, Waltham, MA). Other strains not newly isolated in our laboratory were obtained from the C. elegans Genetics Stock Center, care of T. Stiernagle (University of Minnesota) and/or from C. Kenyon (UCSF; mec-7::GFP).
Molecular biology
Standard methods were used unless other methods are mentioned. A. Coulson and J. Sulston (Sanger Centre, Cambridge, UK) kindly provided cosmids.
Microscopy
Male tail, embryonic and larval phenotypes were determined by examining animals mounted in 1mM levamisole or water (embryos) using differential interference contrast optics or fluorescence microsopy with a Leica DMR photomicroscope.
Statistical methods
All comparisons of significance were tested using a student's T-test, P = 0.05.
Scoring embryonic lethality
Adult hermaphrodites were allowed to lay ∼100 embryos to unseeded plates. The number of dead embryos then was compared to the total number of embryos laid.
Mapping and rescue
unc-130 was mapped by standard methods to LGII, within a region flanked by unc-53 and sqt-1. Cosmids in the region spanning the unc-130 physical map location were tested for their ability to complement unc-130(ev505) DTC defects by microinjecting hermaphrodite gonads of unc-130(ev505); dpy-20(e1282) with candidate clones plus dpy-20(+) DNA (pMH86, Clark et al. 1995) as a cotransformation marker (Mello and Fire 1995). dpy-20 rescued animals were compared to their nonrescued siblings for the penetrance of DTC defects. unc-130 mutant strains transgenic for the cosmid C47G2 were significantly rescued for DTC defects (25%–45% posterior DTC defects in dpy-20 rescued worms as opposed to 85%–90% defects in nonrescued siblings at 25°C). One array carrying C47G2 and pMH86 was integrated into a random chromosomal location in unc-130(ev505); dpy-20(e1282) by irradiation from a 137Cs source (1800 rads). The resulting strain has no DTC defects and is no longer Unc or Dpy and rescues the morphology of DD and VD motor neurons, as assessed by β-galactosidase staining of an integrated unc-5::lacZ reporter (Su et al. 2000) introduced into the strain. Fragments of cosmid C47G2 were subcloned into pBCKS (Stratagene) and tested for rescuing ability in a similar fashion. Strains transgenic for extrachromosomal arrays carrying an 11.5-kb XbaI fragment of C47G2 subcloned into pBCKS (pBN11) were the only lines that significantly rescued DTC (Fig. 4A), Dpy and Unc phenotypes. The best of these rescuing extrachromosomal arrays rescues the morphology of DA and DB axons as assessed by introduction of an integrated unc-129::GFP neuron-specific reporter construct evIs82 (Colavita et al. 1998) into the transgenic lines. [0% of axons failed to reach the dorsal cord in evEx118(pBN11, pMH86); evIs82; unc-130(ev505); dpy-20(e1282) hermaphrodites (n = 196) as opposed to 34% in unc-130(ev505); dpy-20(e1282) hermaphrodites (n = 99)].
The unc-130::GFP translational fusion construct (pBN17) was assembled as follows. pBN11 was cut with SacI and treated with Mung Bean nuclease to produce blunt ends and subsequently digested with XbaI. The resulting 7.6-kb fragment was cloned into pPD95.79 digested with XbaI and SmaI, thereby fusing the 5.9-kb promoter and unc-130 coding region lacking codons for the last 12 amino acids, to the GFP coding region and 3′ UTR in pPD95.79. The construct was coinjected with rol-6(su1006) DNA (pRF4) (Mello and Fire 1995) into unc-130(ev505) to create extrachromosomal arrays. Rollers from these lines were partially rescued for the Unc phenotype, suggesting the construct expresses functional UNC-130::GFP in a pattern sufficient to rescue the mutant phenotype. The unc-130::GFP extrachromosomal array (evEx114) was integrated into a random chromosomal location as described above. The integrated unc-130::GFP (evIs120) line then was tested for its ability to rescue the axon guidance defects observed in unc-130(ev505) by introducing evIs82(unc-129::gfp) into the unc-130(ev505); evIs120(pBN17, pRF4) background. The resulting strain had no motoraxon guidance defects (n = 170) as opposed to evIs82; unc-130(ev505), which exhibited 35% motoraxon defects (n = 103), confirming that this construct is capable of rescuing unc-130 guidance functions.
Sequencing
Cosmid C47G2 was sequenced by the Genome Sequencing Project. The only predicted coding sequence in the 11.5-kb XbaI rescuing fragment contained in pBN11 (C47G2.2) was sequenced in unc-130(ev582) following amplification of the three exons by PCR, identifying a point mutation (C to T transition) that causes a missense alteration (Arg to Cys) of codon 217 of C47G2.2 (Fig. 4B,C). Both PCR and southern analysis of unc-130(ev505) genomic DNA revealed an ∼1.7-kb deletion eliminating the 3′ region of C47G2.2 (Fig. 4B). Subsequent sequencing confirmed the nature of this deletion. The left breakpoint of the deletion is within the Forkhead domain of the coding sequence. The lesion results in an altered coding sequence, beginning at codon 156, and is predicted to encode 13 altered amino acids followed by a stop codon. If any translation of unc-130(ev505) transcript occurs, it is predicted to encode a truncated protein without most of the Forkhead DNA-binding domain. Primer sequences are available upon request.
Expression and purification of MBP–UNC-130 proteins and gel-shift assays
A wild-type unc-130 cDNA missing the first four codons was inserted into the pMal-c2 MBP vector (New England Biolabs) such that the remaining unc-130 coding region was in frame with MBP. A mutant UNC-130 (R217C) was made using a primer (containing the mutant sequence and a neighboring BamH1 restriction site) to synthesize a PCR product that was used to replace a corresponding fragment of the wild-type gene. The presence of only the desired mutation in the resulting cDNA was verified by sequencing the entire PCR-derived fragment plus flanking DNA. Expression and affinity purification of the MBP and MBP–UNC-130 fusion proteins were as described in Ring et al. (2000), except bacterial strain CAG621 (New England Biolabs) was transformed (by electroporation) for expression and protease inhibitors (1 mg/mL leupeptin, antipain, and pepstatin; 5 mg/mL aprotinin; 10 mg/mL benzamidine hydrochloride and soybean trypsin inhibitor; and 100mM PMSF) were added at 100-fold dilution before and after bacterial sonication, during gel elution, and again before freezing. Affinity purification was verified by Coomassie staining and western blotting of SDS-PAGE gels using anti-MBP antibodies (New England Biolabs) for immunodetection of fusion proteins. Although some degradation of both fusion proteins (but not MBP) was apparent, much of the affinity-purified protein was full length. The proteins therefore were tested for their ability to slow the rate of electrophoresis of radiolabeled DNA fragments (Ring et al. 2000) comprising the putative muscle regulatory region of the unc-129 promoter (Colavita et al. 1998; see Fig. 6, legend ). Protein concentrations were estimated by Bradford assays (Pierce Chemicals) and DNA by OD280. Equal molar amounts of protein and radiolabeled DNA were used in all binding assays. Radiolabeled double-strand oligonucleotides (ds oligos) also were examined for binding to MBP–UNC-130 wild-type and mutant proteins. These oligos were designed to include five consensus Forkhead transcription factor target sequences (WTRTTNN NKY and WWTRTTNNNNY) found in the 277-bp binding fragment of the unc-129 promoter. Three of these target sequences are clustered around the AflII site, a fourth is close to the Sma I site present in this fragment, and a fifth is between these (see Fig. 6 for positions). The AfiII-containing ds oligo (AGC AAATTATGTTAAGCTTAAGATAACATTGTGGAG), which included three potential target sequences, tightly bound wild-type MBP–UNC-130, but did not bind the R217C mutant form of this fusion protein. This was also true for the potential Forkhead target sequence included in the SmaI oligo (TCGATAT CATGTTTCCCCGGGATT) and the target site between SmaI and AflII (TTCTAAAGAAAACAATGAAAG), however, binding to the latter two ds oligos was greatly reduced relative to the AfiIII-containing oligo (data not shown).
Acknowledgments
We thank Piali Sengupta for the oy10 allele of unc-130 and for sharing unpublished data; the Cynthia Kenyon lab for the gift of a mec-7::gfp transgenic line; T. Stiernagle of the Caenorhabditis elegans Genetics Center, which is funded by the National Institutes of Health, Center for Research Resources, for providing strains; Rob van Weeghel and Katie McGhee for initial unc-130 genetic studies; Alan Coulson for cosmids; and the C. elegans Genome Sequencing Center for invaluable sequence data. We also owe many thanks to Sabine Cordes and Takehiro Kawano for guidance on gel shift assays and members of the Culotti lab for valuable discussions. This work was supported by a grant from the Canadian Medical Research Council to J.G.C., and University of Toronto Open and Ontario Graduate Fellowships to B.N.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL culotti@mshri.on.ca; FAX (416) 586-8588.
Article and publication date are at www.genesdev.org/cgi/doi/10.1101/gad.831500.
References
- Augsburger A, Schuchardt A, Hoskins S, Dodd J, Butler S. BMPs as mediators of roof plate repulsion of commissural neurons. Neuron. 1999;24:127–141. doi: 10.1016/s0896-6273(00)80827-2. [DOI] [PubMed] [Google Scholar]
- Chan SS, Zheng H, Su MW, Wilk R, Killeen MT, Hedgecock EM, Culotti JG. UNC-40, a C. elegans homolog of DCC (deleted in colorectal cancer), is required in motile cells responding to UNC-6 netrin cues. Cell. 1996;87:187–195. doi: 10.1016/s0092-8674(00)81337-9. [DOI] [PubMed] [Google Scholar]
- Chow KL, Hall DH, Emmons SW. The mab-21 gene of Caenorhabditis elegans encodes a novel protein required for choice of alternate cell fates. Development. 1995;121:3615–3626. doi: 10.1242/dev.121.11.3615. [DOI] [PubMed] [Google Scholar]
- Clark DV, Suleman DS, Beckenbach KA, Gilchrist EJ, Baillie DL. Molecular cloning and characterization of the dpy-20 gene of Caenorhabditis elegans. Mol Gen Genet. 1995;247:367–378. doi: 10.1007/BF00293205. [DOI] [PubMed] [Google Scholar]
- Clevidence DE, Overdier DG, Tao W, Qian X, Pani L, Lai E, Costa RH. Identification of nine tissue-specific transcription factors of the hepatocyte nuclear factor 3/forkhead DNA-binding-domain family. Proc Nat Acad Sci. 1993;90:3948–3952. doi: 10.1073/pnas.90.9.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavita A, Culotti JG. Suppressors of ectopic UNC-5 growth cone steering identify eight genes involved in axon guidance in Caenorhabditis elegans. Dev Biol. 1998;194:72–85. doi: 10.1006/dbio.1997.8790. [DOI] [PubMed] [Google Scholar]
- Colavita A, Krishna S, Zheng H, Padgett RW, Culotti JG. Pioneer axon guidance by UNC-129, a C. elegans TGF-β. Science. 1998;281:706–709. doi: 10.1126/science.281.5377.706. [DOI] [PubMed] [Google Scholar]
- Culotti JG, Merz DC. DCC and netrins. Curr Opin Cell Biol. 1998;10:609–613. doi: 10.1016/s0955-0674(98)80036-7. [DOI] [PubMed] [Google Scholar]
- Estevez M, Attisano L, Wrana JL, Albert PS, Massague J, Riddle DL. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature. 1993;365:644–649. doi: 10.1038/365644a0. [DOI] [PubMed] [Google Scholar]
- Fazeli A, Dickinson SL, Hermiston ML, Tighe RV, Steen RG, Small CG, Stoeckli ET, Keino-Masu K, Masu M, Rayburn H, et al. Phenotype of mice lacking functional deleted in colorectal cancer (DCC) gene. Nature. 1997;386:796–804. doi: 10.1038/386796a0. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, de la Calle-Mustienes E, Modolell J, Mayor R. Xenopus brain factor-2 controls mesoderm, forebrain and neural crest development. Mech Dev. 1999;80:15–27. doi: 10.1016/s0925-4773(98)00190-7. [DOI] [PubMed] [Google Scholar]
- Hamelin M, Zhou Y, Su MW, Scott IM, Culotti JG. Expression of the UNC-5 guidance receptor in the touch neurons of C. elegans steers their axons dorsally. Nature. 1993;364:327–330. doi: 10.1038/364327a0. [DOI] [PubMed] [Google Scholar]
- Hatini V, Tao W, Lai E. Expression of winged helix genes, BF-1 and BF-2, define adjacent domains within the developing forebrain and retina. J Neurobiol. 1994;25:1293–1309. doi: 10.1002/neu.480251010. [DOI] [PubMed] [Google Scholar]
- Hatini V, Huh SO, Herzlinger D, Soares VC, Lai E. Essential role of stromal mesenchyme in kidney morphogenesis revealed by targeted disruption of winged helix transcription factor BF-2. Genes & Dev. 1996;10:1467–1478. doi: 10.1101/gad.10.12.1467. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Hall DH. The unc-5, unc-6, and unc-40 genes guide circumferential migrations of pioneer axons and mesodermal cells on the epidermis in C. elegans. Neuron. 1990;4:61–85. doi: 10.1016/0896-6273(90)90444-k. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Herman RK. The ncl-1 gene and genetic mosaics of Caenorhabditis elegans. Genetics. 1995;141:989–1006. doi: 10.1093/genetics/141.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley SA, Ferguson EL. Fish are like flies are like frogs: Conservation of dorsal–ventral patterning mechanisms. Bioessays. 1997;19:281–284. doi: 10.1002/bies.950190404. [DOI] [PubMed] [Google Scholar]
- Hong K, Hinck L, Nishiyama M, Poo MM, Tessier-Lavigne M, Stein E. A ligand-gated association between cytoplasmic domains of UNC5 and DCC family receptors converts netrin-induced growth cone attraction to repulsion. Cell. 1999;97:927–941. doi: 10.1016/s0092-8674(00)80804-1. [DOI] [PubMed] [Google Scholar]
- Hromas R, Ye H, Spinella M, Dmitrovsky E, Xu D, Costa RH. Genesis, a winged helix transcriptional repressor, has embryonic expression limited to the neural crest, and stimulates proliferation in vitro in a neural development model. Cell Tissue Res. 1999;297:371–382. doi: 10.1007/s004410051365. [DOI] [PubMed] [Google Scholar]
- Ishii N, Wadsworth WG, Stern BD, Culotti JG, Hedgecock EM. UNC-6, a laminin-related protein, guides cell and pioneer axon migrations in C. elegans. Neuron. 1992;9:873–881. doi: 10.1016/0896-6273(92)90240-e. [DOI] [PubMed] [Google Scholar]
- Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, Tessier-Lavigne M. Deleted in colorectal cancer (DCC) encodes a netrin receptor. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- Kim S, Ren XC, Fox E, Wadsworth WG. SDQR migrations in Caenorhabditis elegans are controlled by multiple guidance cues and changing responses to netrin UNC-6. Development. 1999;126:3881–3890. doi: 10.1242/dev.126.17.3881. [DOI] [PubMed] [Google Scholar]
- Krishna S, Maduzia LL, Padgett RW. Specificity of TGF-β signaling is conferred by distinct type I receptors and their associated SMAD proteins in Caenorhabditis elegans. Development. 1999;126:251–260. doi: 10.1242/dev.126.2.251. [DOI] [PubMed] [Google Scholar]
- Leonardo ED, Hinck L, Masu M, Keino-Masu K, Ackerman SL, Tessier-Lavigne M. Vertebrate homologues of C. elegans UNC-5 are candidate netrin receptors. Nature. 1997;386:833–838. doi: 10.1038/386833a0. [DOI] [PubMed] [Google Scholar]
- Leonardo ED, Hinck L, Masu M, Keino-Masu K, Fazeli A, Stoeckli ET, Ackerman SL, Weinberg RA, Tessier-Lavigne M. Guidance of developing axons by netrin-1 and its receptors. Cold Spring Harb Symp Quant Biol. 1997;62:467–478. [PubMed] [Google Scholar]
- Leung-Hagesteijn C, Spence AM, Stern BD, Zhou Y, Su MW, Hedgecock EM, Culotti JG. UNC-5, a transmembrane protein with immunoglobulin and thrombospondin type 1 domains, guides cell and pioneer axon migrations in C. elegans. Cell. 1992;71:289–299. doi: 10.1016/0092-8674(92)90357-i. [DOI] [PubMed] [Google Scholar]
- Mariani FV, Harland RM. XBF-2 is a transcriptional repressor that converts ectoderm into neural tissue. Development. 1998;125:5019–5031. doi: 10.1242/dev.125.24.5019. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. . Review. [PubMed] [Google Scholar]
- Morita K, Chow KL, Ueno N. Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-β family. Development. 1999;126:1337–1347. doi: 10.1242/dev.126.6.1337. [DOI] [PubMed] [Google Scholar]
- Ren XC, Kim S, Fox E, Hedgecock EM, Wadsworth WG. Role of netrin UNC-6 in patterning the longitudinal nerves of Caenorhabditis elegans. J Neurobiol. 1999;39:107–118. [PubMed] [Google Scholar]
- Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127:307–317. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- Sarafi-Reinach, T.R. and Sengupta, P. 2000.The forkhead domain gene unc-130 generates chemosensory neuron diversity in C. elegans. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Serafini T, Colamarino SA, Leonardo ED, Wang H, Beddington R, Skarnes WC, Tessier-Lavigne M. Netrin-1 is required for commissural axon guidance in the developing vertebrate nervous system. Cell. 1996;87:1001–1014. doi: 10.1016/s0092-8674(00)81795-x. [DOI] [PubMed] [Google Scholar]
- Su M, Merz DC, Killeen MT, Zhou Y, Zheng H, Kramer JM, Hedgecock EM, Culotti JG. Regulation of the UNC-5 netrin receptor initiates the first reorientation of migrating distal tip cells in Caenorhabditis elegans. Development. 2000;127:585–594. doi: 10.1242/dev.127.3.585. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Albertson DG, Thomson JN. The Caenorhabditis elegans male: Postembryonic development of nongonadal structures. Dev Biol. 1980;78:542–576. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, Ross RM, Padgett RW, Wood WB. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development. 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- Wadsworth WG, Bhatt H, Hedgecock EM. Neuroglia and pioneer neurons express UNC-6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron. 1996;16:35–46. doi: 10.1016/s0896-6273(00)80021-5. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate JN, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil Trans R Soc of Lond. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Yuasa J, Hirano S, Yamagata M, Noda M. Visual projection map specified by topographic expression of transcription factors in the retina. Nature. 1996;382:632–635. doi: 10.1038/382632a0. [DOI] [PubMed] [Google Scholar]