Abstract
The activities of several mRNA processing factors are coupled to transcription through binding to RNA polymerase II (Pol II). The largest subunit of Pol II contains a repetitive carboxy-terminal domain (CTD) that becomes highly phosphorylated during transcription. mRNA-capping enzyme binds only to phosphorylated CTD, whereas other processing factors may bind to both phosphorylated and unphosphorylated forms. Capping occurs soon after transcription initiation and before other processing events, raising the question of whether capping components remain associated with the transcription complex after they have modified the 5′ end of the mRNA. Chromatin immunoprecipitation in Saccharomyces cerevisiae shows that capping enzyme cross-links to promoters but not coding regions. In contrast, the mRNA cap methyltransferase and the Hrp1/CFIB polyadenylation factor cross-link to both promoter and coding regions. Remarkably, the phosphorylation pattern of the CTD changes during transcription. Ser 5 phosphorylation is detected primarily at promoter regions dependent on TFIIH. In contrast, Ser 2 phosphorylation is seen only in coding regions. These results suggest a dynamic association of mRNA processing factors with differently modified forms of the polymerase throughout the transcription cycle.
Keywords: RNA polymerase II, TFIIH, capping enzyme, Kin 28
Eukaryotic mRNAs undergo 5′ end capping, splicing of introns, and polyadenylation. Targeting of capping enzyme and other RNA processing factors is through binding to the carboxy-terminal domain (CTD) of the RNA polymerase II (Pol II) largest subunit (Cho et al. 1997; McCracken et al. 1997a,b; Yue et al. 1997; Hirose and Manley 1998; Pillutla et al. 1998; Hirose et al. 1999). Capping is the earliest modification, occurring when the transcript is 20 to 40 nucleotides long (Jove and Manley 1984; Rasmussen and Lis 1993). Phosphorylation of the CTD occurs soon after initiation and is necessary for capping enzyme recruitment. Other RNA-processing factors bind to both phosphorylated and unphosphorylated CTD and act much later during transcription. This raises the question of whether capping enzyme and other processing factors are simultaneously associated with RNA pol II throughout transcription or instead interact transiently at different stages. In vivo cross-linking is used here to show that capping enzyme is recruited to promoter regions dependent on TFIIH kinase activity, but does not remain associated with elongating polymerase. In contrast, the mRNA cap methyltransferase Abd1 and the polyadenylation factor CFIB/Hrp1 cross-link throughout transcribed regions. Surprisingly, Ser 5 phosphorylation of the CTD also localizes to promoters, suggesting dephosphorylation not long after escape into elongation phase. Ser 2 phosphorylation of the CTD shows a complementary pattern, with no cross-linking at the promoter and higher levels near the 3′ end of the gene. These results suggest a dynamic association of RNA processing factors with differently modified forms of the polymerase throughout the transcription cycle.
Results
Experimental design
To determine the in vivo distribution of capping enzyme relative to RNA polymerase along transcribed genes we used the chromatin IP method in which proteins are cross-linked in vivo to DNA using formaldehyde. Yeast cells are then lysed and the chromatin is isolated and sheared. The presence of individual proteins near specific DNA sequences is monitored by immunoprecipitating (IP) with the appropriate antibody, reversal of the cross-links, and PCR analysis of the coprecipitated DNA (Orlando et al. 1997; Orlando 2000).
Genes were chosen for study using several criteria: they are strongly and constitutively transcribed, the coding sequences are relatively long (to allow clear resolution of promoter-bound and elongating transcription complexes), and they do not obviously overlap neighboring genes. PMA1 encodes cytoplasmic H+–ATPase, PDR5 encodes a membrane protein identified as a multidrug resistance factor, ADH1 encodes alcohol dehydrogenase, PYK1 encodes pyruvate kinase, ACT1 encodes actin, and RPS5 encodes a ribosomal protein. Their estimated transcriptional rates are 80, 30 126, 101, 45, and 143 mRNAs per hour, respectively (Holstege et al. 1998). Several primer pairs were designed for each gene: one to amplify promoter regions and one or more further downstream within the coding sequences. Intergenic regions on chromosomes V or VII devoid of ORFs were used as controls for nontranscribed DNA. For each protein monitored, a single IP reaction was performed and the resulting DNA was used as template for the entire set of PCR reactions within each experiment.
Differential association of mRNA processing factors during transcription
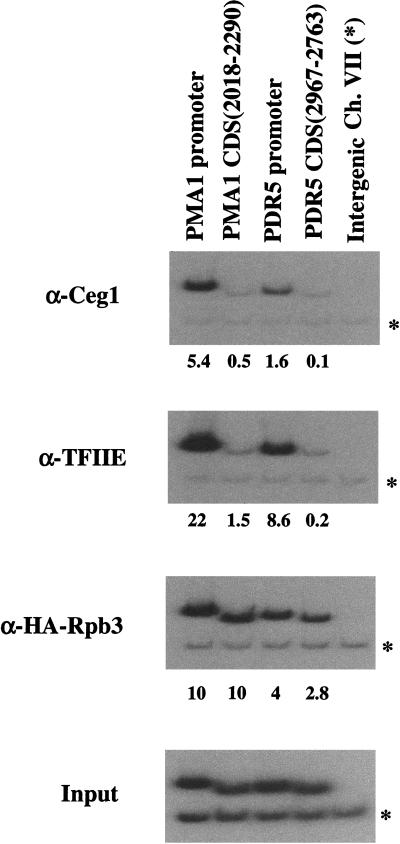
In Saccharomyces cerevisiae, the capping machinery consists of three polypeptides. Cet1 (capping enzyme triphosphatase) and Ceg1 (capping enzyme guanylyltransferase) form a complex referred to as capping enzyme. The third protein, Abd1, is an mRNA guanine-N7-methyltransferase that purifies independently of capping enzyme (for review, see Mizumoto and Kaziro 1987; Shuman 1995). In the chromatin IP assay, Cet1 and Ceg1 show strong cross-linking to the promoter regions of both PMA1 and PDR5. In contrast, little cross-linking is seen in the coding regions of these genes (Figs. 1 and 2). PhosphorImager quantitation indicates at least a 10-fold difference. Also, no cross-linking of capping enzyme was observed at a Pol III-transcribed promoter (data not shown). The cross-linking pattern of capping enzyme closely resembles that of transcription initiation factors TFIIE, TFIIB, TBP, and Kin28 (Figs. 1 and 2; see below). In contrast, an epitope-tagged Rpb3 subunit of RNA Pol II cross-links evenly in both the promoter and distal regions of the coding sequence (Figs. 1–4). This suggests that the capping enzyme does not remain associated with the elongating polymerase.
Figure 1.
Capping enzyme is localized to promoter regions. Cross-linked and sheared chromatin was immunoprecipitated with the indicated antibodies. After reversal of cross-linking and purification of the DNA, PCR was used to test for the presence of promoter or coding sequences (CDS) from the indicated genes. Each PCR reaction contained a second primer pair that amplifies a region of chromosome VII devoid of ORFs, thus providing an internal background control (indicated by *). Input (bottom) shows the signal from the chromatin before immunoprecipitation. HA–Rpb3 is an epitope-tagged subunit of RNA Pol II, Ceg1 is the guanylyltransferase subunit of capping enzyme, and TFIIE was immunoprecipitated with a polyclonal antibody directed against the small subunit Tfa2. Primer pairs are as described in Materials and Methods. The numbers immediately below each lane are quantitated PCR signals in arbitrary units after normalization for amplification efficiency and subtraction of background.
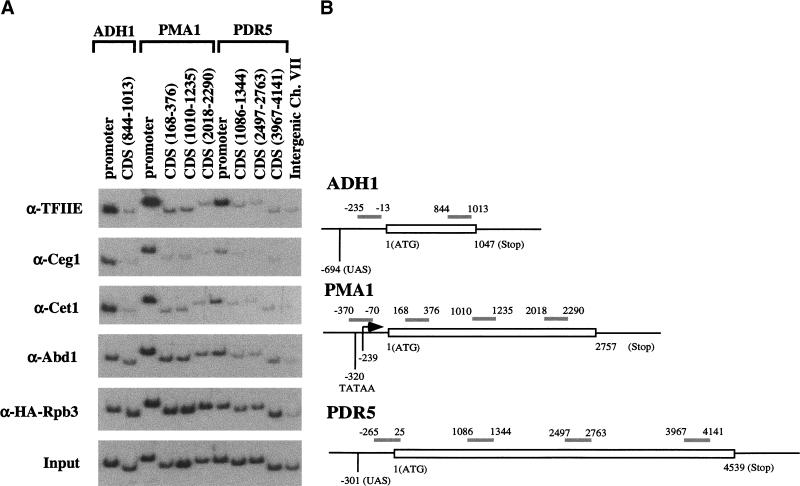
Figure 2.
Capping enzyme and cap methyltransferase show different patterns of association during transcription. (A) Chromatin IP/PCR was carried out as in Fig. 1 except that the Intergenic region control primers (last lane) were not included in the other PCR reactions. The immunoprecipitating antibodies are listed to the left of the autoradiographs. Cet1 is the RNA triphosphatase subunit of capping enzyme, and Abd1 is the mRNA guanine N7-methyltransferase. The small subunit of TFIIE (Tfa2) was used to monitor the transcription initiation complex and a triple HA-tagged polymerase subunit (HA–Rpb3) was used to monitor Pol II. (B) Schematic diagram of PCR primer pairs for PMA1, PDR5, and ADH1. Open bar represents the gene ORF. The gray bars represent PCR products with coordinates relative to the initiation codon of the ORF. Mapped features of PMA1, ADH1, and PDR5 promoters are also indicated.
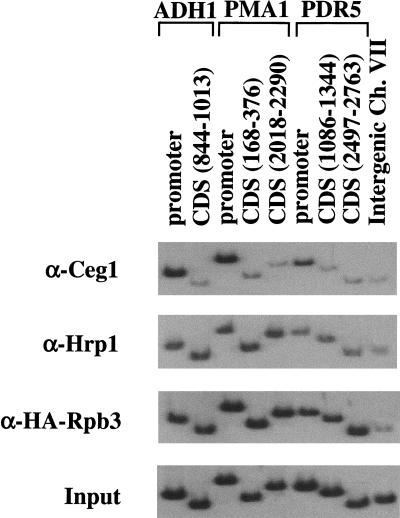
Figure 4.
mRNA cleavage factor IB (Hrp1) cross-links throughout the gene. Chromatin IPs were performed using antibodies indicated to the left of the panels. Hrp1 is an RNA-binding protein implicated in polyadenylation and mRNA degradation (Kessler et al. 1997; Chen and Hyman 1998; Minvielle-Sebastia et al. 1998). For comparison, cross-linking patterns of the capping enzyme guanylyltrasferase Ceg1, the Rbp3 subunit of RNA Pol II, and the Input chromatin are shown.
Additional intermediate primer pairs were used to increase the resolution of the analysis (Fig. 2). The reduction in capping enzyme cross-linking is seen with all primer pairs outside of the promoter region, even as close as 200 nucleotides downstream of the PMA1 or ADH1 promoters (Figs. 2 and 3; data not shown). Because the average length of chromatin fragments generated by our method is roughly 300 bp, we cannot determine more precisely when capping enzyme dissociates from Pol II. However, it is clear that dissociation occurs not long after the polymerase leaves the promoter.
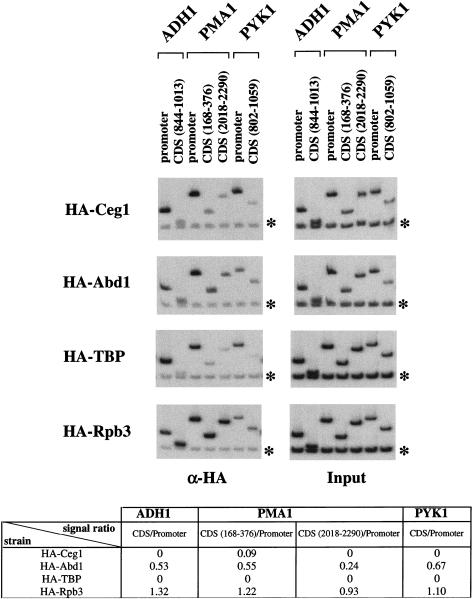
Figure 3.
Differences between capping enzyme and cap methyltransferase are not due to the specific antibodies. Chromatin IP/PCR reactions were carried out on strains containing HA-tagged Ceg1 (capping enzyme guanylyltransferase), Abd1 (methyltransferase), TATA-binding protein (TBP), or Rpb3 (Pol II). Signals were normalized to the input DNA signal (right panels) and background (Intergenic primer pair on chromosome V, denoted by asterisk) subtracted. The ratio of cross-linking in coding (CDS) to promoter regions was calculated for each factor (see table at bottom). A zero indicates that the ratio was <0.05.
We also tested the distribution of the mRNA guanine-N7-methyltransferase Abd1, which is not associated with the Ceg1/Cet1 complex but acts on the mRNA immediately after capping enzyme. The cross-linking pattern of Abd1 is somewhat different from that of Cet1 and Ceg1. Although Abd1 is enriched at the promoter region, cross-linking above background is clearly seen in the coding sequences of most genes (Fig. 2). To rule out that the differential cross-linking of Abd1 and Ceg1 was due to differences in the specific antibodies, this result was confirmed using strains containing epitope-tagged Abd1 or Ceg1 proteins recognized by the same monoclonal antibody (Fig. 3). Therefore, the mRNA methyltransferase appears to dissociate from the elongating polymerase at later times than the capping enzyme.
The mRNA polyadenylation machinery also appears to be targeted to RNA Pol II through the CTD (McCracken et al. 1997b; Hirose et al. 1999). We attempted to assay several polyadenylation factors (Rna15, Fip1, Cft1, Brr5, Pta1, and Pap1; antibodies provided by C. Moore, Tufts University, Boston, MA) by chromatin IP, but only obtained a signal for Hrp1. Hrp1 is an RNA-binding protein that was identified as cleavage factor IB in yeast (Kessler et al. 1997; Chen and Hyman 1998; Minvielle-Sebastia et al. 1998) and has also been implicated in mRNA turnover (Gonzalez et al. 2000). Hrp1 cross-links to promoter regions, but also throughout the coding sequences (Fig. 4). In fact, PhosphorImager quantitation indicates that Hrp1 cross-links approximately twofold better to coding sequences than to the promoter, suggesting that Hrp1 and perhaps other processing factors can load onto transcribing RNA Pol II even after escape into elongation.
We tested whether intact RNA was required for the cross-linking signal of Hrp1 as well as mRNA capping enzyme and methyltransferase. Extensive treatment of the cross-linked chromatin with RNase A did not appreciably diminish the signal (data not shown). It seems unlikely that all of these RNA-processing factors are in close contact with the DNA. It is more likely that the formaldehyde creates a network of protein–protein as well as protein–DNA cross-links. One cannot draw any conclusions about the lack of signal for the other polyadenylation factors, as this result could indicate that the factors are not present in the elongation complex, not in a position to be cross-linked to DNA, or that the antibodies do not precipitate under these conditions. Because we previously observed biochemical and genetic interactions between the polyadenylation factor Pta1 and the phosphorylated CTD (Rodriguez et al. 2000), we constructed HA-epitope-tagged Pta1 and Hrp1 strains. We then used a monoclonal antibody against the epitope tag for chromatin IPs. Under conditions that gave cross-linking of HA–Hrp1, we still did not observe cross-linking of HA–Pta1 (data not shown), suggesting that the lack of Pta1 signal is not simply due to technical problems with the polyclonal antibody.
Different phosphorylated forms of RNA Pol II during transcription
Because capping enzyme was associated only with promoter regions whereas other RNA-processing factors (Abd1 and Hrp1) cross-linked to both promoters and coding regions, it was important to monitor the status of the RNA polymerase itself. Using an antibody against an epitope-tagged Rpb3 subunit, roughly equal cross-linking of polymerase was seen to both promoters and coding regions, but not to a nontranscribed intergenic region (Figs. 1–4). A similar result was obtained using the monoclonal antibody 8WG16 (Thompson et al. 1989), which recognizes unphosphorylated CTD repeats (even if the CTD is partially phosphorylated) within the Rpb1 subunit (see below). Therefore, polymerase is present at both promoters and coding regions.
The CTD repeat sequence YSPTSPS is thought to be phosphorylated at several positions in vivo, predominantly at serines 2 and 5 (Patturajan et al. 1998, and references therein). In vitro, capping enzyme binds to phosphorylated GST–CTD and is recruited to an RNA Pol II initiation complex only upon CTD phosphorylation (Cho et al. 1997; McCracken et al. 1997a; Yue et al. 1997). Although there are probably multiple CTD kinases in vivo, genetic interactions between CEG1 and the TFIIH subunit KIN28 suggest that Kin28 is the kinase responsible for capping enzyme recruitment (Rodriguez et al. 2000). Kin28 phosphorylates Ser 5 (Hengartner et al. 1998) and mutations at this position, but not Ser 2, show genetic interactions with CEG1 (Rodriguez et al. 2000). Also, mammalian capping enzyme will bind in vitro to CTD peptides phosphorylated at either Ser 2 or Ser 5, but guanylyltransferase activity is only stimulated by the Ser 5 phosphopeptide (Ho and Shuman 1999).
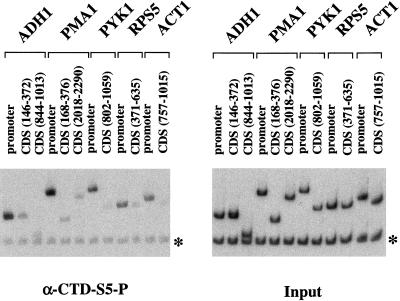
To determine the phosphorylation state of the CTD, chromatin IPs were performed with the monoclonal antibodies H5 and H14, which recognize CTD repeats phosphorylated at Ser 2 and Ser 5, respectively (Bregman et al. 1995; Patturajan et al. 1998). The H14 epitope was strongly cross-linked to promoter regions but not coding regions, exactly paralleling the distribution of capping enzyme (Figs. 5 and 6). Therefore, Ser 5 of the CTD becomes phosphorylated at or near the promoter. However, by the time polymerase has elongated to 200 nucleotides downstream of the promoter, the Ser 5 phosphate is either removed or the CTD is further modified in a way that blocks the H14 epitope (Fig. 5).
Figure 5.
Phosphorylation of CTD Ser 5 is localized to promoters. Chromatin IP/PCR was performed with monoclonal H14, an antibody that specifically recognizes Ser 5 phosphorylation of the CTD heptamer repeat (Bregman et al. 1995; Patturajan et al. 1998). Primer pairs for promoter or coding sequences (CDS) of the indicated genes were used. In addition, PCR reactions contained a primer pair recognizing an Intergenic region of chromosome V as an internal negative control.
Figure 6.
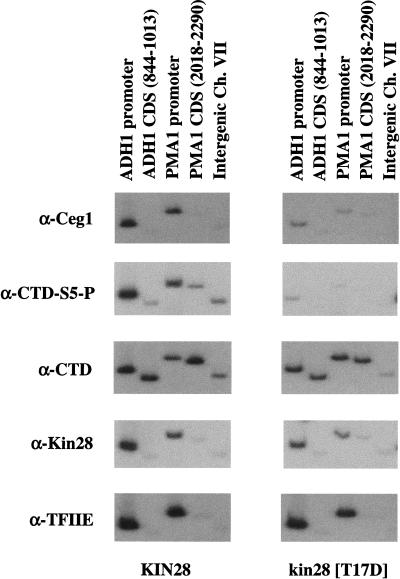
The TFIIH kinase Kin28 is required for CTD Ser 5 phosphorylation and capping enzyme recruitment to promoters in vivo. Chromatin IP/PCR reactions were carried out with wild-type and Kin28 mutant yeast strains. Kin28 is the catalytic kinase subunit of basal transcription factor TFIIH. The Kin28 (T17D) allele produces a stable protein with dramatically reduced kinase activity (M. Keogh and S. Buratowski, unpubl.). Immunoprecipitating antibodies are indicated to the left of the autoradiographs. The CTD signal is with monoclonal 8WG16, which recognizes the unphosphorylated CTD, whereas CTD phosphorylated at Ser 5 (CTD-S5-P) is recognized by the monoclonal H14. TFIIE was monitored using a polyclonal antiserum against the small subunit Tfa2.
Capping enzyme association with promoters was assayed in a strain carrying a Kin28 mutant (T17D) with reduced levels of kinase activity (Rodriguez et al. 2000; M. Keogh and S. Buratowski, unpubl.). Cross-linking of capping enzyme and CTD–Ser 5 phosphorylation were markedly reduced, providing strong in vivo evidence that Kin28 activity is essential for CTD phosphorylation and recruitment of capping enzyme to promoters (Fig. 6). We previously observed that total cellular levels of Ceg1 protein but not mRNA were reduced in the Kin28 (T17D) mutant strain and believe that this reflects preferential degradation of excess guanylyltransferase not bound to polymerase (Rodriguez et al. 2000). RNA polymerase and TFIIE were still present at the promoter, demonstrating that the effect on capping enzyme was not indirectly attributable to loss of initiation complexes. Surprisingly, Pol II (as assayed by 8WG16) is not reduced at either promoters or coding sequences, suggesting that Kin28-mediated phosphorylation of the CTD is not essential for escape into elongation in vivo. This agrees well with many in vitro studies that find no transcriptional requirement for CTD phosphorylation (Serizawa et al. 1993; Makela et al. 1995; Tirode et al. 1999 and references therein). However, it is important to note that the complete absence of the Kin28 subunit (as opposed to lack of catalytic activity) is likely to have a much more severe effect on transcription.
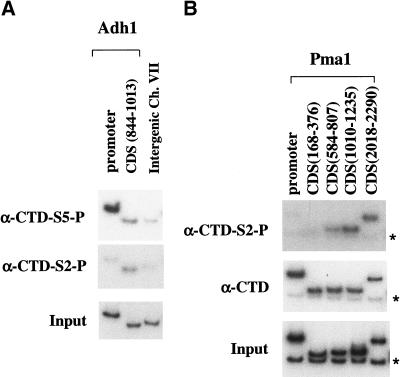
In vivo cross-linking of polymerase to mRNA suggests that the phosphorylated form Pol IIo is the major species associated with nascent transcripts, although some unphosphorylated Pol IIa is also observed (Cadena and Dahmus 1987). Staining of Drosophila polytene chromosomes with antibodies against different forms of Pol II shows that some bands contain Pol IIo, whereas others have Pol IIa or a mixture of the two (Weeks et al. 1993). Therefore, it was surprising that Ser 5 phosphorylation localized to promoters. We also tested for the presence of Ser 2 phosphorylation using the monoclonal antibody H5 (Fig. 7). Remarkably, phosphorylation of Ser 2 shows a pattern opposite to that of Ser 5. The H5 epitope was not seen at promoters, but was cross-linked to coding regions. On the Pma1 gene, the Ser 2 phosphorylation signal appeared to increase toward the 3′ end of the gene, suggesting that this phosphorylation occurs during elongation or that pol II phosphorylated at Ser 2 is more likely to reach the end of the gene. Although it is difficult to compare quantitatively different antibodies in this assay, the signal with H5 was generally lower than that seen with other antibodies. However, the location of the Ser 2 phosphorylation clearly differed from that of Ser 5.
Figure 7.
Ser 2 phosphorylation of the CTD is localized to coding regions of the gene. Chromatin IP/PCR reactions were carried out using the indicated antibodies. The presence of the CTD is probed with monoclonal antibody 8WG16, the phosphorylated Ser 5 (CTD-S5-P) is recognized by monoclonal H14, and the phosphorylated Ser 2 (CTD-S2-P) is recognized by monoclonal H5. Primer pairs are as in previous figures. Asterisks in B designate the product from the Intergenic primer pair (chromosome VII), which was used as an internal negative control in this experiment.
Discussion
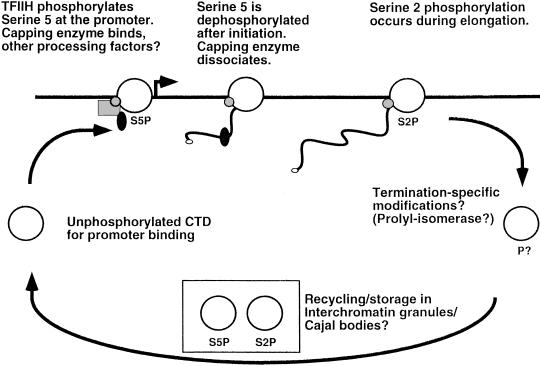
In summary, earlier studies combined with our chromatin IP results lead to the following model (Fig. 8). Unphosphorylated RNA Pol II assembles within the initiation complex at the promoter. At this stage, the CTD may interact with factors important for regulation of transcription initiation. The CTD is then phosphorylated at Ser 5 by the TFIIH kinase subunit. This may dissociate initiation-specific factors (Svejstrup et al. 1997) and acts as a signal for binding of capping enzyme and perhaps other RNA-processing factors. In vitro, CTD phosphorylation by TFIIH is neither dependent on (Cho et al. 1997) nor required for (Serizawa et al. 1993; Makela et al. 1995; Tirode et al. 1999 and references therein) transcription initiation. However, it is likely that these two events are somehow coordinated in vivo. Surprisingly, after escape into elongation (no later than 200 nucleotides) the CTD is dephosphorylated at Ser 5 or further modified in such a way that capping enzyme dissociates. The mRNA cap methyltransferase apparently dissociates at a slower rate, whereas at least one polyadenylation factor (Hrp1) remains associated with the elongation complex. As elongation proceeds, the level of CTD phosphorylation at Ser 2 is increased by an as yet unidentified kinase, and this could act as a recognition site for factors involved in elongation, termination, or mRNA 3′-end processing. Although capping enzyme absolutely requires CTD phosphorylation for binding, several splicing and polyadenylation factors can bind both phosphorylated and unphosphorylated forms of the CTD in vitro (Neugebauer and Roth 1997; Hirose and Manley 2000). This flexibility may allow these factors to recognize multiple forms of phosphorylated CTD or remain associated even after CTD dephosphorylation. Furthermore, both splicing and polyadenylation factors will recognize the appropriate sequences within the nascent mRNA, which is likely to contribute to association with the elongation complex at later times.
Figure 8.
Schematic diagram of the RNA Pol II CTD cycle. The white circle represents Pol II, the black line represents the gene with the arrow denoting the promoter, and the smaller shapes represent capping enzyme and other RNA-processing factors. See discussion for details.
Earlier models proposed a two-step transcription cycle in which unphosphorylated polymerase assembled at the promoter and phosphorylated Pol IIo carried out transcription elongation (Dahmus 1994). We have discovered that phosphorylation patterns and association of RNA-processing factors are dynamic during elongation. Our results suggest a more complex CTD cycle in which different modified forms predominate at different stages of transcription. We find that polymerase phosphorylated at Ser 5 (Pol II–S5P) is localized to promoters, whereas Ser 2 phosphorylation (Pol II–S2P) occurs primarily during transcription elongation or termination. Obviously, it will now be essential to identify all the relevant kinases and phosphatases of the transcription cycle, as well as which modified forms of the CTD are recognized by various mRNA processing, elongation, and termination factors.
It is also pertinent to note that cytological studies have identified a population of phosphorylated Pol II and processing factors in interchromatin nuclear bodies that are not sites of transcription (Bregman et al. 1995; Gall et al. 1999; Matera 1999 and references therein). It has been proposed that these may be important for assembly or recycling of multisubunit complexes involved in gene expression. Interestingly, the H5 and H14-reactive polymerases also localize to discrete locations within these bodies (Gall et al. 1999), hinting that this phase of the polymerase cycle may also include different modified forms.
Other known CTD modifications include phosphorylation at tyrosine, glycosylation, and perhaps isomerization of prolines (Dahmus 1994; Morris et al. 1999; Wu et al. 2000). It will be very interesting to determine when the other CTD modifications occur, which enzymes are responsible for adding and removing modifications, and whether they act to regulate transcription initiation, elongation, or RNA processing.
Materials and methods
Yeast strains used in this study: HA-Ceg1: MATa; ura3–52; leu2–2,112; his3Δ200; ceg1Δ1::HIS3; [pRS315–HA3–CEG1]. HA-Abd1: MATα; ura3–52; leu2Δ1; trp1Δ63; abd1Δ::TRP1; [pRS315– HA3–ABD1]. YSB675: MATa; ura3–52; leu2; trp1; his3Δ200; ade2; taf40Δ::LEU2; spt15Δ::(HA3)SPT15::URA3; [pRS313–TAF40]. YSB756: MATα; ura3–1; leu2–3,112; trp1–1; his3–11,15; kin28Δ::LEU2; ade2–1; ade3–22; can1–100; [pRS314–HA–Kin28]. YSB592: MATα; ura3–1; leu2–3,112; trp1–1; his3–11,15; kin28Δ::LEU2; ade2–1; ade3–22; can1–100; [pRS314–HA–kin28–T17D]. Z780: MATa; ura3–52; leu2–3,112; his3Δ200; rpb3–(HA3)::LEU2. YMK16α: MATα; ura3–1; leu2–3,112; trp1–1; his3–11,15; ade2–1; can1–100; fcp1Δ:LEU2; [pMK86 = FCP1, URA3, CEN/ARS].
Chromatin IPs were performed essentially as described in Kuras and Struhl (1999). Briefly, all yeast strains were grown to OD600 ∼0.6 in synthetic complete medium supplemented with 2% glucose. Formaldehyde was added to a final concentration of 1% for 20 min. Cross-linking was quenched by addition of glycine to 240 mM. Cells were collected by centrifugation, washed in 1× TBS, and lysed with glass beads in FA lysis buffer (50 mM HEPES-KOH at pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na deoxycholate, 0.1% SDS, 1 mM PMSF). Chromatin was sheared by sonication, therefore the average fragment size was between 200 and 700 bp (as determined by agarose gel electrophoresis). Sheared chromatin was stored in aliquots at −70°C.
Rabbit polyclonal antiserum recognizing Ceg1, Cet1, Abd1, Tfa2 (the small subunit of yeast TFIIE), and TBP were generated using standard methods by S. Buratowski, L. Fresco, T. Takagi, and N. Kuldell (Harvard Medical School, Boston, MA). Anti-Hrp1 serum was the gift of C. Moore (Tufts University, Boston, MA). Anti-Kin28 polyclonal serum and monoclonal antibodies H14 and H5 were purchased from Covance. For IPs, all antibodies except H14 and H5 were preincubated with protein A–Sepharose CL-4B beads (Amersham/Pharmacia) and washed once with FA lysis buffer. Chromatin solution was then added and reactions incubated for 90 min at room temperature. For the H5 immunoprecipitation, anti-mouse IgM antibodies coupled to agarose beads (Sigma) were bound to the H5 antibody and incubated with chromatin overnight at 4°C. For H14 IPs, protein A–Sepharose CL-4B beads were precoated with anti-IgM/IgG (Sigma), and together with H14 antibody were added directly to the chromatin solution before the 90-min incubation step. Immunoprecipitated chromatin was subsequently washed under stringent conditions, and subjected to protease treatment and reversal of cross-links as described (Kuras and Struhl 1999). A single preparation of immunoprecipitated DNA was used as template for all the PCR reactions within a given panel.
Conditions for PCR reactions were as follows: 0.25 μM each primer, 0.1 mM each dNTP, 1× PCR buffer (no Mg2+; GIBCO BRL), 1.5 mM MgCl2, 0.5 units Platinum Taq polymerase (GIBCO BRL), 0.06 mCi/ml [α-32P]dATP in 10-μL reaction volume. PCR cycle was once 90 sec at 94°C, followed by 25 cycles of 30 sec at 94°C, 30 sec at 55°C, and 1 min at 72°C, and then once 10 min at 72°C. PCR products were resolved in 8% polyacrylamide–1× TBE gels. For the Input controls, 0.005% of the amount of chromatin (decross-linked as above) used in the IPs was added as template to the PCR reaction. Where noted, PCR signals were quantitated by PhosphorImager (Fujix BAS 2040) scanning and normalization to the input DNA reaction and the internal Intergenic control primer pair (to correct for PCR efficiency and background signal). Error due to variability in multiple reactions was found to be approximately ±10% of the signal.
Primers used in this study are designated by the name of the gene followed by the position of its 5′ end relative to the translation initiation codon: ADH1−235: TTCCTTCCTTCATTCAC GCACACT; ADH1−13: GTTGATTGTATGCTTGGTATAGCT TG; ADH1146: ACGCTTGGCACGGTGACTG; ADH1372: ACCGTCGTGGGTGTAACCAGA; ADH1844: TTCAACCAAGT CGTCAAGTCCATCTCTA; ADH11013: ATTTGACCCTTTTCCATCTTTTCGTAA; ACT1−376: TACCCGCCACGCGTTTTTT TCTTT; ACT1−120: GGTTTGAGTAGAAAGGGGAAGGAAGA; ACT1757: GTATTGTTTTGGATTCCGGTGATGGTGTTA; ACT11015: ATTGAAGAAGATTGAGCAGCGGTTTG; PDR5−265: CTGAGCAATACAAACAAGGCCTCTCCTA; PDR525: TATT GTTAAGCTTGGCCTCGGGCATTTT; PDR51086: CACAGTG GCCATCTATCAATGTTC; PDR51344: GTTCATTTCCTTCGGG GTCTGTGGTAT; PDR52497: GTTGGGGAACGTAGTGACT TATCCAG; PDR52763: CCTTTCGGCCAAACAATCCAGAAGT GTG; PDR53967: AGGGGTGCTTTATTTTGGTTGTTC; PDR54141: TAGGCATGGCACTTGGGGTAG; PMA1−370: GG TACCGCTTATGCTCCCCTCCAT; PMA1−70: ATTTTTTTTCT TTCTTTTGAATGTGTG; PMA1168: CGACGACGAAGACAGT GATAACG; PMA1376: ATTGAATTGGACCGACGAAAAACAT AAC; PMA1584: AAGTCGTCCCAGGTGATATTTTGCA; PMA1807: AACGAAAGTGTTGTCACCGGTAGC; PMA11010: GTTTGCCAGCTGTCGTTACCACCAC; PMA11235: GCAGC CAAACAAGCAGTCAACATCAAG; PMA12018: CTATTATTGA TGCTTTGAAGACCTCCAG; PMA12290: TGCCCAAAATAATA GACATACCCCATAA; PYK1−327: GAATGCTTGTGATGTCT TCCAAGT; PYK1−23: TGATTGGTGTCTTGTAAATAGAAACA; PYK1802: GGTATTGAAATCCCAGCCCCAGAAG; PYK11059: GACAGCGGTTTCAGCCATAGTG; RPS5−236: CCTACCTTC GCCGCAGGCTTAGTG; RPS516: CTTCGGTGTCAGACATCT TTGGAATGG; RPS5371: TGACTGACCAAAACCCAATCC; RPS5635: TTGATAGCGTAAGAAGTAGAGGAACC; Intergenic VII-1: CCCACCACCGATAACGACAAG; Intergenic VII-2: CCAA CAAATGAGGCGGAACC; Intergenic V-1: GGCTGTCAGA ATATGGGGCCGTAGTA; Intergenic V-2: CACCCCGAAGCT GCTTTCACAATAC.
Acknowledgments
We are extremely grateful to Laurent Kuras of the Struhl laboratory for help with the chromatin IP protocol and some PCR primers, M. Kobor and R. Young for yeast strains and plasmids, C. Moore and P. Silver for antibodies against polyadenylation factors, T. Takagi for capping enzyme antibodies, M. Keogh for the Kin28 mutant, and P. Sharp for helpful discussions. We thank the members of the Buratowski laboratory for helpful discussions and comments on the manuscript. This work is supported by grants GM46498 and GM56663 from the NIH to S.B. S.B. is a Scholar of the Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
We recently performed similar experiments using a new lot of monoclonal antibody H14 from Covance. The signal for serine 5 phosphorylation is still much stronger (5–10-fold) at promoters relative to coding regions. However, this batch of antibody detects a very low level of H14 epitope in coding regions that is not seen in intergenic regions. Although this result does not significantly change our conclusions, it may suggest a small fraction of CTD repeats remain phosphorylated at serine 5 during elongation or that a few polymerases escape dephosphorylation altogether.
Footnotes
E-MAIL steveb@hms.harvard.edu; FAX (617) 738-0516.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.824700.
References
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena DL, Dahmus ME. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J Biol Chem. 1987;262:12468–12474. [PubMed] [Google Scholar]
- Chen S, Hyman LE. A specific RNA–protein interaction at yeast polyadenylation efficiency elements. Nucleic Acids Res. 1998;26:4965–4974. doi: 10.1093/nar/26.21.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus M. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog Nucleic Acids Res Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu A, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez CI, Ruiz-Echevarria MJ, Vasudevan S, Henry MF, Peltz SW. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol Cell. 2000;5:489–499. doi: 10.1016/s1097-2765(00)80443-8. [DOI] [PubMed] [Google Scholar]
- Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- ————— RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes & Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Jove R, Manley JL. In vitro transcription from the adenovirus 2 Major Late Promoter utilizing templates truncated at promoter-proximal sites. J Biol Chem. 1984;259:8513–8521. [PubMed] [Google Scholar]
- Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes & Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Makela TP, Parvin JD, Kim J, Huber LJ, Sharp PA, Weinberg RA. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc Natl Acad Sci. 1995;92:5174–5178. doi: 10.1073/pnas.92.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: Multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 1997a;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997b;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Beyer K, Krecic AM, Hector RE, Swanson MS, Keller W. Control of cleavage site selection during mRNA 3′ end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K, Kaziro Y. Messenger RNA capping enzymes from eukaryotic cells. Prog Nucleic Acid Res. 1987;34:1–28. doi: 10.1016/s0079-6603(08)60491-2. [DOI] [PubMed] [Google Scholar]
- Morris DP, Phatnani HP, Greenleaf AL. Phospho-carboxyl-terminal domain binding and the role of a prolyl isomerase in pre-mRNA 3′-end formation. J Biol Chem. 1999;274:31583–31587. doi: 10.1074/jbc.274.44.31583. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM, Roth MB. Transcription units as RNA processing units. Genes & Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Pillutla RC, Yue Z, Maldonado E, Shatkin AJ. Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J Biol Chem. 1998;273:21443–21446. doi: 10.1074/jbc.273.34.21443. [DOI] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol Cell Biol. 2000;20:104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa H, Conaway JW, Conaway RC. Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature. 1993;363:371–374. doi: 10.1038/363371a0. [DOI] [PubMed] [Google Scholar]
- Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ, Li Y, Fellows J, Gnatt A, Bjorklund S, Kornberg RD. Evidence for a mediator cycle at the initiation of transcription. Proc Natl Acad Sci. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson NE, Steinberg TH, Aronson DB, Burgess RR. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- Tirode F, Busso D, Coin F, Egly JM. Reconstitution of the transcription factor TFIIH: Assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: Correlations with gene activity and transcript processing. Genes & Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- Wu X, Wilcox CB, Devasahayam G, Hackett RL, Arevalo-Rodriguez M, Cardenas ME, Heitman J, Hanes SD. The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J. 2000;19:3727–3738. doi: 10.1093/emboj/19.14.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]