Abstract
The POU transcription factor Oct-4 is expressed specifically in the germ line, pluripotent cells of the pregastrulation embryo and stem cell lines derived from the early embryo. Osteopontin (OPN) is a protein secreted by cells of the preimplantation embryo and contains a GRGDS motif that can bind to specific integrin subtypes and modulate cell adhesion/migration. We show that Oct-4 and OPN are coexpressed in the preimplantation mouse embryo and during differentiation of embryonal cell lines. Immunoprecipitation of the first intron of OPN (i-opn) from covalently fixed chromatin of embryonal stem cells by Oct-4-specific antibodies indicates that Oct-4 binds to this fragment in vivo. The i-opn fragment functions as an enhancer in cell lines that resemble cells of the preimplantation embryo. Furthermore, it contains a novel palindromic Oct factor recognition element (PORE) that is composed of an inverted pair of homeodomain-binding sites separated by exactly 5 bp (ATTTG +5 CAAAT). POU proteins can homo- and heterodimerize on the PORE in a configuration that has not been described previously. Strong transcriptional activation of the OPN element requires an intact PORE. In contrast, the canonical octamer overlapping with the downstream half of the PORE is not essential. Sox-2 is a transcription factor that contains an HMG box and is coexpressed with Oct-4 in the early mouse embryo. Sox-2 represses Oct-4 mediated activation of i-opn by way of a canonical Sox element that is located close to the PORE. Repression depends on a carboxy-terminal region of Sox-2 that is outside of the HMG box. Expression, DNA binding, and transactivation data are consistent with the hypothesis that OPN expression is regulated by Oct-4 and Sox-2 in preimplantation development.
Keywords: POU, Oct, Sox, osteopontin, preimplantation embryo
The fertilized oocyte undergoes cleavage until a uniform cluster of cells, the morula, is formed. The first apparent differentiation occurs as the inner cell mass (ICM) separates from the trophectoderm during blastocoel formation (Gardner 1983). Trophectoderm refers to the epithelial cell layer that encloses the ICM and blastocoel. Subsequently, cells dissociate from the ICM and cover its blastocoelic surface to form the hypoblast (also called primitive endoderm). These cells do not form a well-defined polarized epithelium, eventually loose cell contacts, and contain an extensive rough endoplasmatic reticulum, which is often swollen with secretory material (Nadijcka and Hillman 1974). The hypoblast differentiates into the parietal and visceral endoderms (Gardner 1983). Parietal endoderm cells form from hypoblast precursors that migrate and adhere to the thin basal lamina on the inner surface of the trophectoderm. In contrast, visceral endoderm cells do not migrate and consists of a columnar epithelial layer surrounding the late ICM or early epiblast (also called primitive endoderm). Relatively little is known about the molecular signals guiding cell proliferation, differentiation, and migration during establishment of these extraembryonic tissues that arise by the first differentiation events of the embryo.
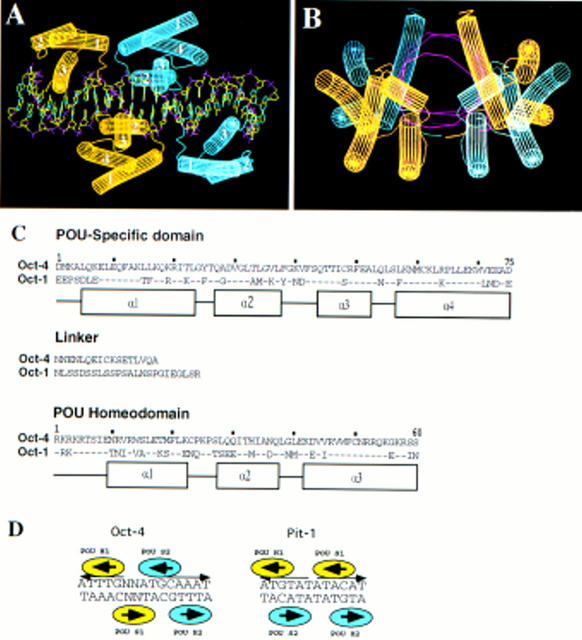
Oct-4 (also termed Oct-3 or Oct3/4) encodes a POU transcription factor (Okamoto et al. 1990; Rosner et al. 1990; Schöler et al. 1990a,b). DNA binding by POU factors is mediated by the 75-amino-acid POU-specific domain (POUS) and the 60-amino-acid carboxy-terminal POU homeodomain (POUHD) that is of the C-50 subtype (for review, see Herr and Cleary 1995). POUS and POUHD are connected by a linker that varies, in sequence and length, in the >20 metazoan POU factors that have been identified (Verrijzer and Van der Vliet 1993; Wegner et al. 1993; Herr and Cleary 1995). Regions outside the POU domain show no significant sequence homology.
Before gastrulation Oct-4 RNA is expressed in all cells of the embryo proper (Rosner et al. 1990; Schöler et al. 1990b). The proportion of embryonic cells expressing Oct-4 mRNA gradually declines as the trophectodermal and somatic lineages are established, and Oct-4 transcripts are eventually confined to the male and female germ cell lineages (Rosner et al. 1990; Schöler et al. 1990b; Yeom et al. 1996, Pesce et al. 1998a; for review, see Pesce et al. 1998b). In contrast to the immediate down-regulation of Oct-4 in trophectodermal and somatic lineages, Oct-4 protein levels are increased initially in cells of another nongerm-line tissue, namely the premigratory hypoblast (Palmieri et al. 1994). Perhaps the initial steps of visceral and parietal endoderm formation depend on increased Oct-4 expression levels. Proliferation, differentiation, and migration are three processes in which Oct-4 might be involved during formation of these tissues.
Oct-4 is also expressed in undifferentiated embryonal cell lines, each of which represent cells of distinct developmental stages (Schöler et al. 1989a,b; Okamoto et al. 1990). Cultured embryonic stem (ES) and embryonal carcinoma (EC) cells exhibit features peculiar to specific cell types found in early embryos (Robertson 1987). On the basis of biochemical markers, F9 EC cells are a model system for embryonal cells that differentiate by way of a hypoblast-like cell type into visceral or parietal endoderm cells (Strickland and Mahdavi 1978; Strickland et al. 1980; Hogan et al. 1981).
High mobility group (HMG) box proteins are transcription factors that interact functionally with POU domain proteins (Leger et al. 1995; Zwilling et al. 1995; Ambrosetti et al. 1997). Sox-2 belongs to the Sox (Sry-related HMG box-containing) gene family and is expressed in preimplantation embryos and in ES and EC cells in a similar manner as Oct-4 (Yuan et al. 1995; Collignon et al. 1996; R. Lovell-Badge, pers. comm.). Later in development, Sox-2 is again coexpressed with Oct-4 in postmigratory primordial germ cells (Collignon et al. 1996). Sox-2 and Oct-4 are able to act synergistically on reporter genes in transient transfection studies (Yuan et al. 1995). The HMG box DNA-binding domain of Sry and other Sox proteins induces a strong bend on binding to the DNA (Ferrari et al. 1992; Giese et al. 1992). Thus, the role of Sry and Sry-related factors may be architectural, facilitating functional protein–protein interactions on enhancers (Ferrari et al. 1992; Giese et al. 1992; Werner et al. 1995).
Understanding the molecular and genetic framework in which Oct-4 operates during the first differentiation processes in development requires identification of its target genes. Several potential target genes of Oct-4 have been proposed (Rosfjord and Rizzino 1994; Kraft et al. 1996; Liu and Roberts 1996; Saijoh et al. 1996). However, the only conclusive candidate gene in early mouse development is fgf-4 (Schoorlemmer and Kruijer 1991; Dailey et al. 1994; Rizzino and Rosfjord 1994). The fgf-4 gene has an octamer-containing enhancer downstream of the coding region, which is activated synergistically by Oct-4 and Sox-2 in transient transfection assays (Yuan et al. 1995). Furthermore, fgf-4 is coexpressed with Oct-4 and Sox-2 in the ICM (Niswander and Martin 1992) and in EC and ES cells (Schoorlemmer and Kruijer 1991).
Osteopontin (OPN; also named bone sialo protein I, 2ar, Spp1, Eta-1, and pp69) is especially abundant in bone, kidney, decidua, and various epithelial cells (for review, see Denhardt and Guo 1993; Denhardt et al. 1995). OPN is an extracellular phosphoprotein containing a GRGDS motif. This peptide motif of OPN is capable of mediating adhesion to and migration along the surface of cell types expressing certain classes of integrins (for review, see Eble and Kühn 1997).
In this study we show that OPN is a candidate target gene of Oct-4 during the formation of the hypoblast of mouse embryos. EC cells were used as a cell culture model for the biochemical analysis of DNA–protein interactions that occur during hypoblast formation and differentiation. Pools of cross-linked F9 EC chromatin fragments bearing cis-acting elements that interact in vivo with Oct-4 were enriched by immunoprecipitation with Oct-4-specific antibodies. PCR analyses showed that the first intron of OPN (i-opn) was well represented in such a pool in comparison to other regions of OPN or to regulatory regions of other genes. The i-opn element contained an ES cell-specific enhancer composed of a cluster of high-affinity Oct-4-binding sites and sites for other transcription factors. In vitro, Oct-4 binds to i-opn both as a monomer and a dimer, but only the dimer confers transcriptional activation in transfection studies. Enhancer activity of i-opn is modulated in F9 EC cells through a Sox-binding site that is in close proximity to the Oct-4-binding sites. Sox-2 in vitro binds to i-opn and in cotransfection experiments interferes with Oct-4-mediated activity. This interference depends on a region outside the HMG domain. Oct-4, Sox-2, and OPN are coexpressed in the same cells of the early mouse embryo. In addition, OPN up- and down-regulation correlates with the pattern of Oct-4 and Sox-2 expression during differentiation of F9 EC cells. We suggest that genes such as OPN are tightly regulated by Oct-4 and Sox-2 in ICM and also in hypoblast cells, which will migrate along the trophectoderm to become parietal endoderm.
Results
Oct-4 can be isolated as part of the embryonal chromatin
Oct-4 is a transcription factor that binds to the octamer motif ATGCAAAT with high affinity in vitro (for review, see Schöler 1991). Statistically, a haploid mouse genome contains this motif ∼4 × 104 times. Hence, an approach to isolate target genes of Oct-4 on the basis of DNA sequence recognition alone is not feasible because of the high complexity of the mouse genome. One means to narrow down the pool of genes containing octamer motifs to those that may interact with Oct-4 in a physiologically relevant way is by isolating directly DNA sequences to which Oct-4 is bound within the context of a nucleus. The natural arrangement of proteins bound to DNA in the nucleus is preserved by cross-linking before lysis. Subsequent physical fragmentation yields a mixture of covalently linked aggregates of macromolecules that were in proximity of each other within the nucleus. Aggregates containing Oct-4 can be isolated by means of Oct-4-specific antibodies and decross-linked to yield a pool of Oct-4-associated macromolecules, from which DNA fragments can be subcloned. The ideal source of such a chromatin precipitate would be from cells that normally express Oct-4 in the animal. However, Oct-4 is only expressed in the toti- and pluripotent cells of the early mouse embryo. These provide insufficient starting material to perform the type of biochemical isolation outlined above. Hence EC cell lines that resemble cells of the early mouse embryo were used. EC cells express Oct-4 and can be grown in large enough quantities to perform chromatin precipitation experiments.
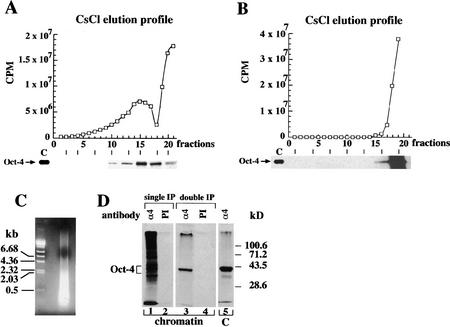
A stringent Oct-4 immunoprecipitation procedure for covalently fixed EC cell chromatin was established based on a procedure used to investigate target genes in Drosophila (Orlando and Paro 1993). P19 EC cells were labeled with [35S]methionine and fixed with formaldehyde in vivo to preserve native protein–DNA complexes by covalent cross-linking. Covalent bonds formed between endogenous Oct-4 and nearby DNA allows the use of stringent conditions during the subsequent purification of Oct-4-specific chromatin fragments (see Materials and Methods). Cross-linked chromatin was physically fragmented to an average DNA size of 400 bp (Fig. 1C) by an empirically calibrated sonication step and was fractionated on equilibrium cesium chloride (CsCl) gradients. The 35S-label was used to determine protein content of each gradient fraction showing a distribution in two peaks at different CsCl densities (Fig. 1A, top). One peak centered at fraction 15 had a density of ∼1.38 gram/cm3, whereas the second was at the top of the gradient (fraction 21). A parallel sedimentation profile of material that had been decross-linked before centrifugation showed only one 35S peak at the top of the gradient, indicating that this material consists of uncross-linked material (Fig. 1B, top). Consequently, the 1.38 gram/cm3 peak was considered as the cross-linked chromatin fraction. Its density is similar to those previously reported for fixed protein–DNA complexes (Solomon et al. 1988).
Figure 1.
Immunoprecipitation of Oct-4-containing chromatin of embryonal cells. (A) Protein elution profiles of isopyknic CsCl gradients of sheared cross-linked chromatin of P19 EC cells. (B) In parallel to equilibrium ultracentrifugation in CsCl the chromatin that was decross-linked before fractionation. In A and B CsCl density gradients were fractionated and the 35S levels (□) determined in each fraction. Fraction 1 is bottom of the gradient. The Oct-4 protein elution profile was obtained from aliquots of gradient fractions by hydrolyzing cross-links, and detected by 35S on SDS-PAGE. Lane C contains a control Oct-4 immunoprecipitate of 35S-labeled P19 EC protein extracts. Exposure of the Oct-4 profile in B is three times longer than in A to show that Oct-4 was only detected at the top of the gradient. (C) Cross-linked chromatin was physically fragmented to an average DNA size of 400 bp as determined by an agarose gel stained with ethidium bromide. (D) Immunoprecipitation of cross-linked chromatin with Oct-4-specific antibodies. All immunoprecipitations were performed with aliquots of fraction 15. 35S-Labeled proteins in precipitates were analyzed on 10% SDS-PAGE. (Lane 1) Immunoprecipitation with Oct-4 antibodies (α4); (lane 2) immunoprecipitation with preimmune (PI) antibodies; (lane 3) Oct-4 immunoprecipitated chromatin was hydrolyzed and subjected to a second round of immunoprecipitation with α4 antibodies; (lane 4) reverse cross-linked PI immunoprecipitated chromatin was hydrolyzed and subjected to a second round of immunoprecipitation with PI antibodies; (lane 5) α4-immunoprecipitates of 35S-labeled P19 EC protein extract. All immunoprecipitations were done with affinity-purified Oct-4 polyclonal antibodies.
Immunoprecipitation with an αOct-4 antibody (α4) was used to identify Oct-4 in gradient fractions of chromatin fragments decross-linked after or before fractionation. In the gradient of cross-linked chromatin fragments, Oct-4 was detected in the fraction 15 peak but not at the top of the gradient (Fig. 1A, bottom). In contrast, Oct-4 was only detected at the top of the gradient in chromatin that was decross-linked before fractionation (Fig. 1B, bottom). Therefore, Oct-4 was cross-linked efficiently to EC cell chromatin with formaldehyde.
Both proteins and DNA that associate closely with Oct-4 in EC cell chromatin should be cross-linked to Oct-4 in the procedure described above. Analysis of the cross-linked proteins was investigated to define the specificity of the immunoprecipitation procedure. The set of proteins cross-linked to Oct-4 was ascertained by comparing the profiles of proteins precipitated from the fraction 15 peak by either the α4 antibody or total antibody from preimmune (PI) serum (Fig. 1D). The α4 immunoprecipitate (decross-linked just before SDS-PAGE) yielded a large number of proteins with molecular masses >30 kD (Fig. 1D, lane 1). The largest histone (H1) has a size of 22.5 kD, suggesting that these proteins are nonhistone. In addition, the different protein bands may represent associated proteins that bind either directly to Oct-4 or indirectly through DNA. A second precipitation of the decross-linked material with the Oct-4 antibody (Fig. 1D, double IP) gave only one band with the mobility of Oct-4, ruling out the possibility that the high apparent molecular mass of the cross-linked proteins was attributable to incomplete hydrolysis of cross-links. In contrast, PI immunoprecipitates (lane 2) contained no detectable protein, indicating that nonspecific precipitation was extremely low.
The α4 polyclonal antibody preparation used had been affinity purified by sequential passage over Oct-6– and Oct-4–Sepharose and was without detectable cross-reactivity in a variety of assays (Palmieri et al. 1994; data not shown). However, to ensure that the proteins immunoprecipitated by α4 from cross-linked chromatin were brought down by virtue of their association with Oct-4, an aliquot of the first α4 immunoprecipitate was decross-linked and immunoprecipitated again with α4. As expected, only a single band was detected that had exactly the size expected for Oct-4 (Fig. 1D, lane 3). The α4 antibody precipitates the same Oct-4 band from P19 EC extracts that were not cross-linked (lane 5). Thus, the α4 immunoprecipitations of cross-linked chromatin can be used to isolate pools of DNA fragments that lie close to Oct-4 in chromatin in vivo.
Enrichment of OPN in Oct-4 chromatin immunoprecipitates
A second approach used to identify target genes of Oct-4 was to screen the available sequence databases for genes that have octamer motifs together with other cis-acting elements that are active in early embryonal cells like Sox-2 recognition sites (Dailey et al. 1994; Yuan et al. 1995). The canonical octamer motif containing one possible A/T variation at position 5 (ATGCA/TAAT) and the Sox-binding site (TCTTTGTT) (Yuan et al. 1995) were used in one search. More than 2000 rodent sequences containing octamer motifs were found in the GenBank-EMBL sequence database. Only 17 of these contained the Sox element within 40 bp of the octamer motif.
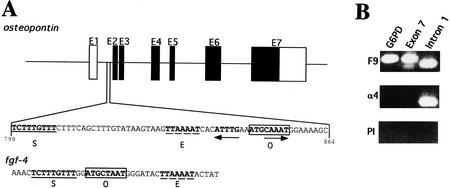
Oct-4 levels are increased transiently in the forming hypoblast of the developing blastocyst (Palmieri et al. 1994). Changes in cell adhesion and migration are two hallmarks of the forming hypoblast and occur during blastocyst development as the hypoblast cells delaminate from the ICM, migrate, and differentiate into visceral and parietal endoderm (Gardner 1983). Of the 17 candidate elements revealed in the sequence search, OPN was the only interesting candidate target gene of Oct-4 with respect to adhesion and migration processes in the preimplantation embryo. OPN had been proposed to have a role in the adhesion and migration of various vertebrate cell types (Denhardt and Guo 1993; Denhardt et al. 1995). The closely spaced octamer and Sox elements of OPN were located in the first intron (i-opn) (Fig. 2A). An engrailed-like (TTAAAAT) sequence, which is also active in early embryonal cells (Okamoto et al. 1990; Fig. 2A), was found in close proximity to the other two elements. An enhancer in the 3′ nontranslated region of fgf-4 also contains these three elements in close proximity (Curatola and Basilico 1990). The combination of Oct-4 and Sox-2 results in cooperative activation of the fgf-4 enhancer in transient transfection assays (Yuan et al. 1995). Therefore, fgf-4 is a candidate target gene of Oct-4 and is likely involved in controlling cell proliferation in the early embryo (Schoorlemmer and Kruijer 1991; Dailey et al. 1994; Rizzino and Rosfjord 1994; Feldman et al. 1995).
Figure 2.
OPN first intron is enriched in EC chromatin αOct-4 immunoprecipitates. (A) Two elements identified by a computer search for genes containing combination of potential binding sites for Oct-4 (O, boxed) and Sox-2 (S, underlined). An engrailed-like factor-binding site is represented by E, dashed line. Schematic organization of OPN gene and localization of the first intron. Solid boxes are coding regions; open boxes are untranslated regions. PORE sequence is represented by inverted arrows. (B) Enrichment of OPN first intron in αOct-4 EC chromatin immunoprecipitates. PCR was performed on F9 chromatin immunoprecipitated with αOct-4 antibodies (α4) or with preimmune antibodies (PI). PCR on purified F9 genomic DNA (F9) serves as a control for the PCR reactions. Primer pairs were selected to amplify a promoter region of G6PD and exon 7 and intron 1 fragments of OPN.
To determine whether a physiologically relevant interaction takes place between Oct-4 and the i-opn fragment, the relative abundance of this fragment in α4 and PI immunoprecipitates of F9 EC cell chromatin was compared (Fig. 2B). F9 cells were used instead of P19 cells because several studies had indicated that they were more similar to hypoblast cells and could be used to model hypoblast differentiation in culture (Strickland and Mahdavi 1978; Strickland et al. 1980; Hogan et al. 1981). PCR analysis revealed that the i-opn fragment containing the octamer motifs was clearly present in the α4 chromatin immunoprecipitates, whereas it was not detected in corresponding immunoprecipitates with PI. In contrast, the promoter and exon 7 of OPN, and a fragment containing the glucose-6-phosphate-dehydrogenase (G6PDH) promoter were not enriched in either immunoprecipitate but could be detected in F9 genomic DNA (OPN promoter not shown). Amplification of the i-opn was verified by cloning and sequencing of the 214-bp band (data not shown). These data indicate that Oct-4 is closely associated with the first intron of OPN in the nuclei of undifferentiated F9 EC cells. Enrichment of the enhancer element of fgf-4 was analyzed in the same pair of immunoprecipitates and revealed that enrichment of the fgf-4 enhancer element is 8- to 16-fold lower than i-opn (data not shown).
Expression of OPN in embryonal cell lines and preimplantation embryos
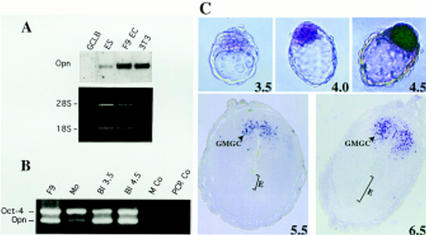
The close association of Oct-4 with the OPN cis-element in the nuclei of cultured cells and the presence of Oct-4-binding sites in this element suggests that Oct-4 regulates physiologically the level of OPN transcription. To test this hypothesis, OPN and Oct-4 expression levels were compared by Northern blot analysis of poly(A)+ RNA from four different embryonal cell lines representing different stages of embryonic development (Fig. 3A; for P19 see Fig. 7A, lane 0, below). MBL-1 ES cells and F9 EC cells resemble cells of the preimplantation embryo, namely ICM and early hypoblast cells, respectively (Strickland and Mahdavi 1978; Strickland et al. 1980; Hogan et al. 1981; Yeom et al. 1996). GCLB cells are derived from P19 EC cells and have many characteristics of early mesodermal cells (Pruitt 1994). All four cell lines express Oct-4 mRNA at similar levels (data not shown). Expression of OPN was strongest in F9 EC, intermediate in MBL-1 ES, weak in P19 EC, and not detected in GCLB cells (Figs. 3A and 7A, below). These results show that OPN expression varies significantly in Oct-4-positive cell lines. The highest levels are observed in cell lines resembling the preimplantation embryo, namely in F9 EC and ES, consistent with the idea that Oct-4 up-regulates OPN expression in the ICM or hypoblast. 3T3 cells were included in this comparison as a cell line that lacks Oct-4. The expression of OPN in the absence of Oct-4 (Fig. 3A) indicates that Oct-4 is not necessary for OPN expression in all cell types. It also suggests that the i-opn element bearing the Oct-4-binding sites may only be active during preimplantation development.
Figure 3.
OPN expression in embryonal cell lines and pregastrulation embryos. (A) Northern blot analyses of OPN mRNA (Opn) expression in F9 EC, GCLB, MBL-1 ES, and fibroblast (3T3) cell lines. Each lane contains 1 μg of poly(A)+ RNA. mRNA levels were normalized by comparison of corresponding ethidium bromide-stained 28S and 18S rRNA bands. (B) RT–PCR of OPN and Oct-4 mRNA in different stages of preimplantation embryos. Multiplex PCR of Oct-4 and Opn was performed on reverse transcripts obtained from F9 cells, morulae (Mo), blastocysts of 3.5 dpc (Bl 3.5) and 4.5 dpc (Bl 4.5). PCR negative controls were done with H2O (PCR Co) and M2 medium treated with RNA extraction buffers and reverse transcription buffers (M Co). (C) In situ hybridization of 3.5–6.5 dpc embryos with OPN DIG-labeled mRNA. Blastocyst staining is observed in ICM and forming hypoblast, but not in trophectoderm and 4.5 dpc hypoblast. In sections of 5.5 and 6.5 dpc embryos only granulated metrial gland cells (GMGC) are stained. (E) embryo. Hybridization with sense probes did not generate staining (not shown).
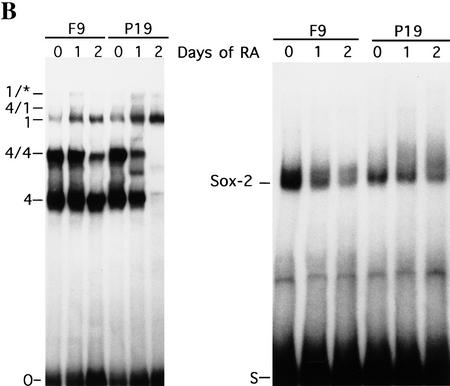
Figure 7.
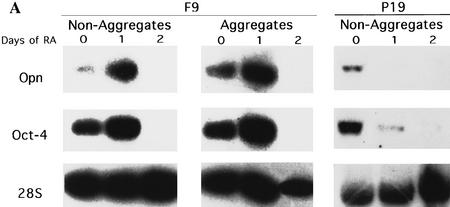
Correlation of OPN, Oct-4, and Sox-2 expression during F9 and P19 differentiation. (A) F9 EC cells were induced to differentiate into parietal endoderm by exposing adherent cells to 10−1 μm all-trans retinoic acid (RA) (nonaggregates) or into visceral endoderm by exposing cells grown in suspension to 10−1 μm RA (aggregates). P19 were treated with 1 μm all-trans RA. Total RNA was extracted at day 0 (untreated F9 and P19 cells), day 1 and 2 of RA treatment. Northern blot analyses were performed for OPN, Oct-4, and 28S rRNA probes. Each lane contains 10 and 30 μg of total mRNA in F9 and P19 EC cells, respectively. The same filter was sequentially hybridized to OPN, Oct-4, and 28S rRNA probes. (B) EMSA of differentiating F9 and P19 EC cell extracts. Whole cell extracts at day 0 (untreated F9 and P19 EC cells), day 1 and 2 of RA treatment were incubated with the O and S oligonucleotide used as probe. Complexes are denoted as follows: (4) Oct-4 monomer; (4/4) Oct-4 homodimer; (4/1) heterodimer of Oct-1 and Oct-4; (1) Oct-1 monomer; (1/*) Oct-1 homo- or heterodimer. PIO-1 complex in P19 EC cells could be observed only after longer exposure.
Expression of OPN was then analyzed in mouse embryos during the morula → blastocyst transition, when Oct-4 is expressed strongly and specifically in the ICM and hypoblast. Oct-4 and OPN mRNA levels were estimated by RT–PCR using primer pairs that span introns, thus eliminating the possibility of bands arising from genomic DNA or unspliced hnRNA. Similar levels of Oct-4 mRNA were detected in morulae and 3.5- or 4.5-day blastocysts (Fig. 3B). In contrast, OPN mRNA was weakly detected in morulae and became clearly present in the two blastocyst stages. Thus, Oct-4 expression precedes the onset of OPN expression in preimplantation mouse embryos but is unlikely to be sufficient to activate OPN expression in morulae.
In situ hybridization experiments were performed to determine whether the expression domains of OPN and Oct-4 overlap in the developing mouse embryo. Embryos of 3.5–6.5 days postcoitum (dpc) were incubated with anti-sense DIG-labeled riboprobes of OPN (Fig. 3C). OPN was expressed weakly but selectively in the ICM of early blastocysts (Fig. 3C, 3.5 dpc). OPN expression was maintained in the ICM and in cells destined to form the hypoblast (Fig. 3C, 4.0 dpc) and was down-regulated in the hypoblast as it formed an epithelial layer of cells (Fig. 3C, 4.5 dpc). Subsequently, expression of OPN is also down-regulated in the ICM/epiblast and becomes undetectable in the embryo by 5.5 dpc (Fig. 3C). By 5.5 and 6.5 dpc OPN expression was confined to the granulated metrial gland cells (GMGC) in the placenta, which do not express Oct-4 (Fig. 3C). Expression of OPN in the GMGCs is in agreement with previous results obtained with 7.5-dpc embryos (Nomura et al. 1988; Waterhouse et al. 1992). OPN and Oct-4 expression only overlap in preimplantation embryos, suggesting that this is the physiological time and space at which OPN may be a target of Oct-4 control.
Coexpression of OPN and Oct-4 in embryonic cell lines and overlapping expression domains in preimplantation embryos, taken in combination with the enrichment of an Oct-4-binding intron of OPN in α4 chromatin immunoprecipitates, clearly suggests a physiological link between the transcription factor Oct-4 and the cell adhesion molecule OPN. However, Oct-4 and OPN transcript levels do not correlate strictly temporally and quantitatively, suggesting that factors in addition to Oct-4 are involved in the regulation of OPN expression during development and in embryonal cell lines.
Oct-4 binds to a 36-bp EC cell-specific enhancer within i-opn
Enrichment of i-opn in α4 chromatin immunoprecipitates strongly suggests that this piece of DNA provides the interface at which the Oct-4 transcription factor controls OPN mRNA levels. The occurrence of a perfect octamer motif (in an inverted dyad configuration with an imperfect octamer motif), a Sox motif, and an engrailed-like motif in a short sequence (+799 to +864 bp) of the i-opn fragment (+758 to +1077 bp) further suggests that these sites and the proteins that interact with them provide the combinatorial regulatory complex that governs the expression of OPN in preimplantation development. Electrophoretic mobility-shift assays (EMSAs) were performed as an initial means to establish which DNA–protein interactions can occur. Bacterially expressed Oct-4 was incubated with an end-labeled fragment of intron 1 (+758 to +1077 bp). Part of the radioactive probe was shifted to a position of lower mobility. Incubation of the mixture with Oct-4-specific antibodies abolished the low mobility band, demonstrating that Oct-4 can bind to i-opn (data not shown; see below).
The i-opn fragment was inserted in front of a minimal herpes simplex virus thymidine kinase (HSV tk) promoter driving the expression of the luciferase reporter gene to test whether it was active in cells expressing Oct-4 (Fig. 4A, i-opn). Point mutations were introduced into the octamer sequence to measure the contribution of the octamer site to the i-opn activity (Fig. 4A, i-opn mut). A short i-opn region (OS), containing only the Sox, engrailed-like, and octamer-binding sites, was also analyzed for activity (Fig. 4A). The i-opn-containing reporter was 4.8 times more active in transiently transfected F9 EC cells than the parental vector without insert (Fig. 4C). Similar activity was obtained with the OS reporter (Fig. 4C). In contrast, the reporter bearing a mutated i-opn (i-opn mut) lacked detectable activity (Fig. 4C). Therefore, i-opn contains cis-acting elements, in particular the octamer site, that confer expression in F9 EC cells.
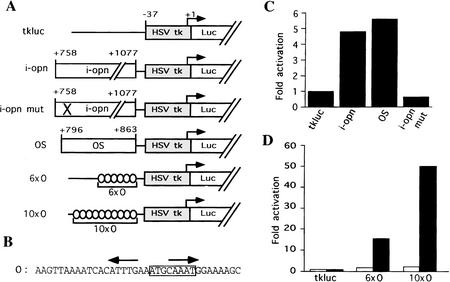
Figure 4.
OPN first intron functions as an EC-specific enhancer. (A) Reporter constructs. HSV tk minimal promoter cloned upstream of the luciferase gene (Luc); i-opn and i-opn mut are PCR fragments of the OPN first intron from +758 to +1077 wild-type and point mutated octamer motif, respectively; OS denotes OPN first intron from +796 to +863. 6 × O and 10 × O denote 6 or 10 copies of the O oligonucleotide, respectively. All fragments were cloned upstream of −37tkluc. (B) Oligonucleotide O represents a short fragment of i-opn that binds to Oct-4. The octamer motif is boxed and the PORE is represented by two inverted arrows. (C,D) F9 (solid bars) and 3T3 (open bars) cells were transfected transiently with 10 μg of respective reporter and 2 μg of human β-actin–LacZ used as an internal standard to normalize luciferase activity. Fold activation refers to the quotient of luciferase activities in the test and control (tkluc) construct.
To assess the activity of the octamer motif in i-opn, short fragments containing the octamer dyad and engrailed-like motif (Fig. 5B, O oligonucleotide; positions +827 to +863) were oligomerized and tested in transactivation assays. Six and 10 direct repeats of the O oligonucleotide were inserted into the tk luciferase reporter plasmid (Fig. 4A). Expression of these reporters was compared to the parental reporter by transient transfection assays. Reporters bearing O repeats showed significantly higher levels of luciferase activity than parental reporters in F9 EC but not in 3T3 cells (Fig. 4D). These results demonstrate that a 36-bp-long region within i-opn can confer EC cell-type specificity when multimerized and suggests that this element provides the interface at which Oct-4 acts to regulate OPN during preimplantation development.
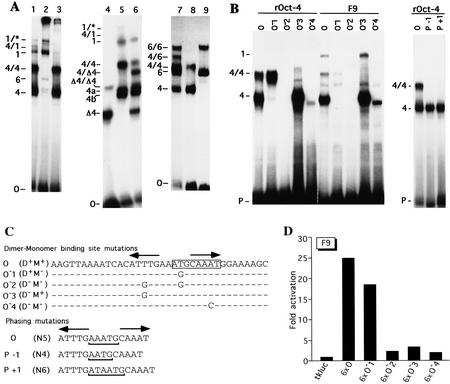
Figure 5.
Oct-4 binding to and transactivation by the PORE (A) EMSA demonstrating Oct-4 homo- and heterodimerization on a PORE-containing oligonucleotide (O). (Lane 1) F9 cell extracts; (lane 2) F9 cell extracts incubated with Oct-4 antibodies; (lane 3) F9 cell extracts incubated with Oct-1 antibodies; (lane 4) COS extract containing an Oct-4 protein lacking 98 amino acids at the amino terminus; (lane 5) F9 EC cell extract; (lane 6) mixture of extracts used in lanes 4 and 5; (lane 7) mixture of purified recombinant Oct-4 and Oct-6; (lane 8) recombinant Oct-4; (lane 9) recombinant Oct-6. (B) EMSA of recombinant Oct-4 (rOct-4) and F9 extracts (F9) with the O oligonucleotide and mutated versions thereof (see O) O−1, O−2, O−3, O−4, P−1, and P+1 in C). (P) Free probe. Complexes in A and B are as follows: (1) Oct-1 monomer; (4) Oct-4 monomer; the complexes labeled 4a and 4b correspond to Oct-4 monomer in mobility and are considered to represent two differentially phosphorylated forms of Oct-4 (V. Botquin, unpubl.). These bands are not clearly resolved in lanes 1 and 3 of A, lane O with F9 extracts of B and therefore, are merely labeled 4. (1/*) Oct-1 homo- or heterodimer; (4/4) Oct-4 homodimer; (Δ4/Δ4) homodimer of truncated Oct-4; (4/Δ4) heterodimer of full-length and truncated Oct-4; (6/6) Oct-6 homodimer; (4/1) heterodimer of Oct-4 and Oct-1; (4/6) heterodimer of Oct-4 and Oct-6. (C) Specific point and phasing mutations in O oligonucleotide to analyze binding and transactivation requirements in B and D. Point mutations were introduced at base pairs predicted to make central contacts with the POUS domain (O−1), the POUHD (O−3, O−4), or both (O−2). Oligonucleotides capable of binding Oct-4 as a dimer or a monomer are denoted by D+ and M+, respectively. (N) Number of nucleotides between the two half sites of the PORE. The octamer motif is boxed and the PORE is labeled by two inverted arrows. Dots represent nucleotides identical to those in oligonucleotide O. (D) Transactivation by mutated versions of oligonucleotide O. 6 × O, 6 × O−1, 6 × O−2, 6 × O−3, and 6 × O−4 are six copies of O, O−1, O−2, O−3, and O−4, respectively, inserted upstream of tkluc. All constructs contain a TATA box at similar distances from the insert. F9 EC cells were transfected transiently with 10 μg of respective reporter and 2 μg of human β-actin LacZ, which served as an internal standard to normalize luciferase activities for transfection efficiency. Fold activation refers to the quotient of luciferase activities in test and control (tkluc) constructs.
Oct-4 binds as a monomer and dimer to the O element
The interaction of Oct-4 and other POU domain transcription factors with the octamer motif, and various synthetic derivatives thereof, has been studied extensively (for review, see Schöler 1991; Verrijzer and van der Vliet 1993; Herr and Cleary 1995; Ryan and Rosenfeld 1997). However, few studies have examined this interaction using octamer motifs embedded in naturally occurring sequence contexts, using physiologically relevant cell extracts. Therefore, the O oligonucleotide was tested in an EMSA for binding with proteins from F9 EC cell extracts (Fig. 5A). Three of the five complexes observed contained Oct-4 because they were removed by addition of Oct-4-specific antibodies (Fig. 5A, cf. lanes 1 and 2). The complex labeled 4/4 corresponds to a dimer of Oct-4 bound to the O oligonucleotide (see below). A third band, labeled 4/1, was composed of Oct-4 and Oct-1 because it was abolished by both Oct-1 and Oct-4 antibodies (lanes 2,3). In addition, two other Oct-1 complexes, labeled 1 and 1/*, were recognized by Oct-1 antibodies (lane 3).
Oct-4-containing complexes were further investigated by using purified recombinant Oct-4 (rOct-4). Bacterially expressed Oct-4 generated two complexes when incubated with the O oligonucleotide (complexes 4 and 4/4; Fig. 5B, lane O, left and right). Both have similar mobilities as Oct-4 complexes obtained with F9 EC cell extracts, suggesting that in EC cells these complexes contained only Oct-4 (Fig. 5B, cf. lanes O in rOct-4 and F9).
To determine the number of Oct-4 molecules within these complexes, two extracts containing Oct-4 proteins of different size were mixed before EMSA (Fig. 5A, lane 6). The shorter version of Oct-4 was expressed in COS cells (Fig. 5A, lane 4, complex labeled Δ4) and lacks 98 amino acids at the amino terminus (16Oct-4; Schöler et al. 1990b). The longer version of Oct-4 was the complete protein from F9 EC cells (Fig. 5A, lane 5, complex labeled 4a/4b). The truncated Oct-4 protein formed two bands (Δ4 and Δ4/Δ4), each with higher mobility than corresponding bands with the complete protein (4a,4b and 4/4; Fig. 5A, cf. lanes 4 and 5). Nontransfected COS cells did not give rise to these complexes, indicating that these complexes contained Oct-4 protein (data not shown). Mixed extracts (Fig. 5A, lane 6) containing both Oct-4 versions gave a set bands that was a composite of the two individual extracts. However, in addition, a complex of intermediate mobility was observed between 4/4 and Δ4/Δ4, which was composed most likely of one short and one long version of Oct-4 (4/Δ4). Thus, Oct-4 can bind to the O fragment of i-opn as a monomer, a homodimer, or, together with Oct-1, as a heterodimer.
To further investigate the ability of Oct-4 to heterodimerize with other POU factors, EMSA was performed using a mixture of purified Oct-4 and rOct-6 (Fig. 5A, lanes 7–9). Similarly to Oct-4, Oct-6 formed monomer and homodimer complexes on the O oligonucleotide (complexes 6 and 6/6; lanes 8,9). In addition to the set of complexes obtained by individual Oct-4 and Oct-6 proteins, an additional band 4/6 of intermediate mobility between 4/4 and 6/6 was observed after mixing both proteins together (Fig. 5A, lane 7). This shows that Oct-4 can heterodimerize with both Oct-6 and Oct-1.
Oct-4 dimer formation requires the palindromic sequence ATTTG N5 CAAAT.
The O fragment contains the canonical octamer motif ATGCAAAT that is bound by several POU proteins. POUS binds to the first and POUHD to the second half of this motif (Klemm et al. 1994). An inverted CAAAT sequence is located 5 nucleotides upstream of the CAAAT in the octamer motif (Fig. 5C). This dyad sequence motif was likely to be required for the formation of Oct-4 dimer complexes. Point mutations were introduced at various positions within the O oligonucleotide to determine which residues were required for Oct-4 monomer and dimer formation. The mutagenesis strategy was designed to reduce specifically binding of either POUS (O−1) or POUHD (O−3, O−4) of Oct-4, or both (O−2) (Fig. 5C, dimer–monomer-binding site mutations). This was performed by altering critical residues of the dyad motif that are predicted, based on the known crystal structure of a POU domain–DNA complex, to make contacts with these two subdomains (Klemm et al. 1994). Mutated oligonucleotides were incubated with either bacterially expressed Oct-4 (rOct-4) or F9 EC cell extracts and analyzed by EMSA (Fig. 5B, left).
rOct-4 binds as a monomer and a dimer to the wild-type oligonucleotide (Fig. 5B, lane O). When a mutation in the first half of the octamer motif was introduced, only rOct-4 monomer formation was affected (lane O−1). In contrast, dimerization of rOct-4 was abolished specifically by a mutation in the ATTTG (inverted CAAAT) sequence located 5′ to the octamer motif (lane O−3). When both mutations were combined neither dimer nor monomer were detected (lane O−2).
A point mutation was introduced into the second half of the octamer motif to test whether the right half site of the palindromic sequence is also required for dimer formation (lane O−4). Indeed, dimer formation on this oligonucleotide was abolished and binding of the monomer was reduced drastically. The mobility of the monomer complex was also slightly increased in comparison to that formed on the wild-type sequence, possibly as a result of a different binding mode. One likely explanation is that the homeodomain now can only bind to the left half site of the palindrome (inverted CAAAT). An altered binding conformation could result in different DNA bending and, as a consequence, in altered mobility of the complex (Verrijzer et al. 1991).
Oligonucleotides with one nucleotide inserted or deleted between the palindromic motifs were also tested with rOct-4 for dimer formation (Fig. 5C, phasing mutations). In neither case was the dimer complex formed, indicating dimerization is highly dependent on proper spacing of the two Oct-4 molecules bound to the palindrome (Fig. 5B, right). The phasing mutations also abolished the formation of Oct-4 homo- and Oct-4/Oct-1 heterodimers in F9 EC cell extracts (data not shown). The specific amino acid side chains required for dimer formation have to be determined.
Oct-4 of F9 EC cell extracts and rOct-4 basically gave the same pattern of complexes with all oligonucleotides (Fig. 5B, cf. left and right). However, dimer formation with rOct-4 was far more efficient than with F9 Oct-4 (cf. both O lanes). In addition, the mutation (O−1) that abolished monomer formation and increased dimer formation with rOct-4 (two- to threefold; compare O and O−1 lanes), reduced monomer but did not increase dimer formation in F9 EC extracts. These results suggest that Oct-4 in F9 EC extracts is modified in a way that interferes with dimerization.
The mutagenesis assays presented above indicate that formation of the monomer complex requires an intact POUS half site and at least one intact POUHD half site. The POUHD half site of the octamer motif results in a more stable monomer complex than the inverted half-site found upstream. In contrast, formation of the Oct-4 dimer complex on the O fragment requires two intact POUHD half sites but does not require an intact POUS half site. Dimerization of Oct-4 depends on the palindromic motif ATTTG–CAAAT, which has to be spaced by five nucleotides. This motif will be subsequently called PORE, for palindromic Oct recognition element.
PORE mediates function of the EC cell enhancer
To determine whether monomer or dimer complexes were mediating the EC cell-specific enhancer function of i-opn, mutagenized oligonucleotides were multimerized and inserted at position −37 in the standard tk luciferase reporter plasmid. The reporter activity of each mutant O-multimer was compared to the activity generated by the wild-type O-multimer (6 × O) in transient transfections in F9 EC cells (Fig. 5D). Hexamers of the O−1 fragment, which only binds Oct-4 as a dimer (Fig. 5B, left), activated the reporter in a manner similar to the hexamer of the wild-type sequence (Fig. 5D, cf. 6 × O with 6 × O−1). In contrast, a hexamer of the O−3 fragment, to which Oct-4 bound only as a monomer, activated transcription only two- to threefold (6 × O−3). This was only slightly above the level of activation by hexamers of O−2, to which Oct-4 could not bind at all (6 × O−2). These results demonstrate that the PORE and not the octamer motif activates transcription of the reporter gene efficiently. They suggest that the PORE is also important for the function of the EC-cell-specific enhancer found in i-opn. The in vitro binding data suggest that PORE activity in vivo depends on Oct-4 homo- or heterodimerization with either Oct-1 or Oct-6.
Sox-2 interferes with Oct-4-mediated transcriptional activation
The variation in OPN expression levels in different ES and EC cell lines (see Fig. 3A) may be attributable to differences in Oct-4 dimerization or binding of other regulatory factors. However, Oct-4 mRNA and protein levels do not vary in the ES and EC cell lines examined so far. In addition, monomer binding is similar in all extracts of these cell types (Yeom et al. 1996; data not shown). As an initial test to determine whether OPN expression is modulated by different dimer levels, in vitro dimer formation was compared in different embryonic cellular extracts (Fig. 6A, left). Oct-4 dimer formation was very similar in all cell extracts and thus cannot explain why OPN levels vary in the ES and EC cells. However, we observed an inverse correlation between OPN expression and the presence of the 4/1 and 1/* complexes, suggesting that an Oct-4/Oct-1 dimer could repress the activity of the i-opn enhancer.
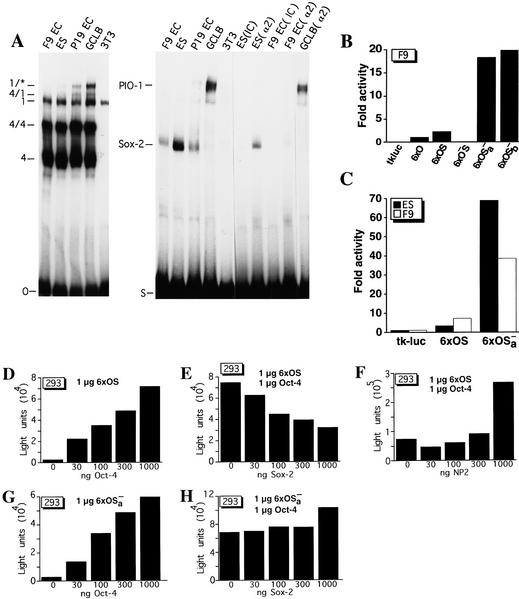
Figure 6.
Sox-2 binds i-opn and represses Oct-4-mediated transactivation. (A) EMSA of EC cell (P19 and F9), ES cell, GCLB, and fibroblast (3T3) cell extracts on O and S oligonucleotides. Sox-2 binding was affected with either poly[d(I-C)] (IC) or Sox-2 antibody (α2). Complexes are as follows: (4) Oct-4 monomer; (4/4) Oct-4 homodimer; (4/1) heterodimer of Oct-1 and Oct-4; (1) Oct-1 monomer; (1/*) Oct-1 homo- or heterodimer; (PIO-1) protein binding to intron 1 of osteopontin). (B,C) Transient transfection assays in F9 EC and ES cells with reporters carrying octamer and Sox-binding sites. 6 × O represents a hexamer of oligonucleotide O (PORE only), 6 × OS a hexamer of OS (PORE and Sox-binding sites), 6 × O−S a hexamer of O−S (mutated PORE, intact Sox-2-binding site), 6 × OSa− and 6 × OSb− hexamers of OSa− and OSb−, respectively (intact PORE, mutated Sox-binding site). All hexamers were cloned upstream of tkluc. F9 EC and ES cells were transfected transiently with 10 μg of respective reporter and 2 μg of human β-actin–LacZ as an internal standard. Fold activation refers to the quotient of luciferase activities in test and control (6 × O) constructs. (D) Cotransfection assay in 293 cells of 1 μg of 6 × OS reporter and increasing amount of CMVOct-4 effector. (E) Cotransfection assay in 293 cells of 1 μg of 6 × OS reporter, constant amount of CMVOct-4 and increasing amounts of CMVSox-2 effector. (F) Cotransfection assay in 293 cells of 1 μg of 6 × OS reporter, constant amount of CMVOct-4 and increasing amount of CMVNP2 effector. (G) Cotransfection assay in 293 cells of 1 μg of 6 × OSa− reporter and increasing amount of CMVOct-4 effector. (H) Cotransfection assay in 293 cells of 1 μg of 6 × OSa− reporter, constant amount of CMVOct-4 and increasing amounts of CMVSox-2 effector. All luciferase activities were normalized for β-galactosidase expression. Light units refers to the units of luciferase counts.
Sox-2 is another candidate transcription factor that could modulate Oct-4-mediated activity of i-opn. The Sox-binding site was used in our initial computer analysis to restrict the number of potential Oct-4 target genes (see Fig. 2A). Oct-4 and Sox-2 proteins increase transcription synergistically from the fgf-4 enhancer activity (Yuan et al. 1995). An oligonucleotide spanning the i-opn region with the canonical Sox-binding site (S oligonucleotide) was used to test for differences in Sox-2-binding activity in ES and EC cell lines (Fig. 6A, right). One major complex was formed in F9, P19, and ES cells. A common test for Sox-factor binding is the competition by poly[d(I-C)] (Dailey et al. 1994). Incubation of the reaction mixture with poly[d(I-C)] instead of poly[d(G-C)] resulted in a specific competition of the Sox-2 complex [Fig. 6A, lanes ES(IC) and F9 EC(IC)]. In addition, point mutations within the Sox motif decreased formation of this complex (data not shown; see below). Finally, in both F9 EC and ES extracts, Sox-2 binding was affected specifically by Sox-2 antibodies [Fig. 6A, lanes ES(α2) and F9 EC(α2)]. The Sox-2 complex varies in extracts of the different cell types. The strongest signal was obtained with ES cell extracts, a weaker band with F9 EC and P19 EC (Fig. 6A, right). Finally, no Sox-2 complex could be detected in GCLB and 3T3 (Fig. 6A, right).
A novel low mobility complex [PIO-1 (proteins binding to intron 1 of osteopontin)] formed on the S oligonucleotide when P19 EC and GCLB cell extracts were used (Fig. 6A). The identities of proteins in PIO-1 are unclear. PIO-1 is unlikely to contain Sox-2 because it was not affected by Sox-2 antibodies [Fig. 6A, right, GCLB (α2)]. Sox-2 and PIO-1 binding is found predominantly in ES and EC cells in which OPN expression is low or undetectable, namely in ES, P19 EC, and GCLB. Thus, the presence of the Sox-2 and PIO-1 can be correlated with the expression pattern of OPN in cell lines corresponding to different embryonic cell types.
The activity of the Sox-2-binding site was analyzed in transient transfection assays. The hexamer of the S oligonucleotide in front of the tk luciferase reporter was inactive in F9 EC cells, confirming previous results that a Sox-2-binding site by itself cannot activate transcription (data not shown, Yuan et al. 1996). To study the functional interaction of Sox-2 and Oct-4, longer oligonucleotides spanning both the Oct and the Sox motifs (termed OS) were oligomerized and cloned in front of the standard reporter (position +798 to +863). Tandem hexamers of the native sequence (6 × OS) were inserted upstream of the tk luciferase reporter and compared to tandem hexamers of mutated versions. Mutations were either in the octamer motif (6 × O−S) or in the Sox-binding site (6 × OSa−; complete replacement; 6 × OSb−; point mutated site). In vitro binding analyses indicated that Sox-2 binding to Osa− and Osb− was undetectable or weak, respectively (not shown). Reporter constructs were transfected into F9 EC cells and their activity was analyzed (Fig. 6B). The wild-type hexamer 6 × OS was twice as active as the shorter 6 × O fragment. The activity of 6 × OS depended on intact Oct factor-binding sites, because the mutated counterpart (6 × O−S) was inactive. Strikingly, mutations of the Sox-binding site (6 × OSa− and 6 × OSb) resulted in an additional 9.5- to 10-fold activation (Fig. 6B, cf. 6 × OS with 6 × OSa− and 6 × OSb−). These results indicate that the Sox-binding site is a negative element that interferes with the activity of other elements within the OS fragment, most likely with the activity of the Oct-4-binding sites.
Sox-2-binding intensity differs in ES and in F9 EC cell extracts (Fig. 6A, right). We compared the activity of the wild-type hexamer 6 × OS in these two cell types (Fig. 6C). The 6 × OS reporter was twice more active in F9 EC compared to ES cells and thus the OS activity correlated inversely with Sox-2-binding intensity. In addition, mutation of the Sox-binding site (6 × OSa−) resulted in an additional 20-fold activation in ES compared to a fivefold activation in F9 EC cells (Fig. 6C). This suggests that Sox-2 may decrease the level of OPN expression in ES cells. PIO-1 may act as another negative factor involved in a down-regulation of OPN in GCLB and P19 EC cells (Fig. 6A, right).
To assess directly the effect of Sox-2 on Oct-4, both factors were cloned into CMV vectors and expressed in 293 cells. First, increasing amounts of the Oct-4 expression vector was cotransfected with a constant amount of the 6 × OS reporter (Fig. 6D). Reporter activity was increased in direct proportion with the amount of the expression vector added. The highest amount (1 μg) tested in this experiment stimulated the reporter >20-fold. Subsequently, increasing amounts of the Sox-2 expression vector was cotransfected with a constant amount of both the Oct-4 expression and the 6 × OS reporter vectors. In this case, increased amounts of Sox-2 expression vector steadily decreased the activity mediated by Oct-4 (Fig. 6E). This negative effect depended on an intact Sox element because even the highest amount of Sox-2 did not interfere with Oct-4-stimulated activity of 6 × OSa− reporter (Fig. 6G,H). These results indicate that Sox-2 can reduce levels of Oct-4-mediated transactivation from the 6 × OS reporter and that binding of Sox-2 protein to the canonical Sox motif is required for its repressive effect.
A transactivation domain has been mapped to the carboxy-terminal portion of Sox-2 (Yuan et al. 1995). To determine its contribution to the repressive effect of Sox-2, a truncated version lacking this region was cotransfected with a constant amount of both 6 × OS reporter and Oct-4 expression vectors. Increasing amounts of the truncated Sox-2 protein (NP2) stimulated the reporter up to threefold (Fig. 6F). This activation by NP2 is in striking contrast to the decrease mediated by the complete protein and might indicate an architectural modification of the chromatin-facilitating Oct-4 interaction with the basal transcriptional machinery. The result suggests that transcriptional repression by Sox-2 is not likely attributable to competitive binding of Sox-2 with another transcription factor, rather it is the result of the carboxy-terminal region of Sox-2 that interferes with Oct-4-mediated transcriptional activation.
Oct-4, Sox-2, and OPN expression correlate during hypoblast differentiation
The ICM of the blastocyst, which coexpresses Oct-4, Sox-2, and OPN, differentiates into the epiblast and hypoblast (Gardner 1983; Palmieri et al. 1994; R. Lovell-Badge, pers. comm.; see Fig. 3C). The hypoblast is localized initially to the blastocoelic surface of the ICM and gradually spreads over the inner surface of the blastocoel as it differentiates into parietal and visceral endoderm (Gardner 1983). Oct-4 protein levels increase in cells that form the hypoblast, but decrease drastically as the hypoblast differentiates further (Palmieri et al. 1994). In contrast, Sox-2 expression is down-regulated immediately in cells of the hypoblast lineage (R. Lovell-Badge, pers. comm.).
F9 EC cells are often used as a cellular model to study hypoblast-specific questions (Strickland and Mahdavi 1978; Strickland et al. 1980; Hogan et al. 1981). In a first attempt to compare Oct-4 and OPN expression levels during this specific stage, F9 EC cells were induced to differentiate into parietal and visceral endoderm (Fig. 7A, nonaggregates and aggregates, respectively). In both differentiation modes F9 EC cells are considered to initially pass through a hypoblast-like stage (Adamson 1986). Undifferentiated F9 EC cells expressed moderate levels of Oct-4 and OPN (Fig. 7A, F9 lane 0 in each panel). A transient 2.5-fold increase in both Oct-4 and OPN mRNA levels was observed after 1 day of retinoic acid (RA) treatment regardless whether F9 EC cells were differentiating along visceral or parietal pathways (Fig. 7A, F9 lane 1 in each panel). Oct-4 and OPN mRNA levels decreased to undetectable levels after 2 days in RA (Fig. 7A, F9 lane 2 in each panel). Induction of laminin expression, used as a marker for parietal and visceral endoderm, was observed after 1 day of differentiation of F9 EC cells (data not shown). These results demonstrate a strict correlation between Oct-4 and OPN expression during differentiation of F9 EC. The profile of Oct-4 mRNA up- and down-regulation is closely followed by OPN mRNA levels and recapitulates up- and down-regulation of these genes observed during the hypoblast formation.
Oct-4 and OPN expression was also compared in RA-treated P19 EC cells, which were induced to differentiate along a neuronal pathway (Robertson 1987). In contrast to F9 EC cells, the level of Oct-4 and OPN transcripts were down-regulated immediately (Fig. 7A, P19). Therefore, the transient increase of OPN mRNA during F9 EC cell differentiation is not likely attributable to a direct effect of RA on OPN gene regulation.
The in vitro binding activities of Oct-4 and Sox-2 were compared with the OPN transcript levels during differentiation of F9 EC and P19 EC cells (Fig. 7B) to determine whether a correlation between factor binding and OPN expression could be established. Oct-4 monomer and dimer binding to the O oligonucleotide encoding the i-opn site was not noticeably altered in F9 extracts prepared after 1 day of differentiation (Fig. 7B, right: F9, cf. lane 0 with lane 1). Extracts prepared after 2 days of differentiation showed similar monomer binding but had a marked reduction in Oct-4 dimer formation (Fig. 7B, left: F9, lane 2). In contrast, the level of Sox-2 decreased immediately after 1 day of differentiation (Fig. 7B, right: F9, lane 1) and only low levels of Sox-2 were detected after 2 days of RA treatment (Fig. 7B, right: F9, lane 2). These results suggest that transiently elevated OPN expression levels observed after 1 day of RA treatment may be attributable to the more rapid decline in Sox-2 binding than in Oct-4 binding. Consequently, the inhibitory action of Sox-2 on Oct-4 in the i-opn fragment (above) is alleviated temporarily, resulting in higher levels of i-opn activity in the brief interval before both factors are absent.
The level of Oct-4 in P19 cells decreased to undetectable levels after 2 days of differentiation (Fig. 7B, left: P19). An additional complex detected at day 1 of RA treatment (Fig. 7B, left: P19, lane 1, mobility between 4 and 4/4 complex) may contain Brn-2 (Fujii and Hamada 1993). In contrast to Oct-4, only a slight decline of Sox-2 levels was observed after 2 days of differentiation (Fig. 7B, left: P19), suggesting that the rapid decrease of Oct-4 protein levels might result in an immediate OPN down-regulation.
Discussion
In this study we show that OPN is likely a direct target gene of Oct-4 in cells of the mouse preimplantation embryo. Oct-4 is thought to play an important role in the establishment of hypoblast-derived cell layers (Palmieri et al. 1994) and OPN is a secreted phosphoprotein containing a RGD motif promoting adhesion and migration in various cell types (for review, see Denhardt and Guo 1993; Denhardt et al. 1995). We have demonstrated that Oct-4 is found in close association to a new EC-cell-specific enhancer of the OPN gene, i-opn, in the chromatin of EC cells. Furthermore, the activity of the i-opn enhancer in these cells depends on at least two transcription factors that are coexpressed in the preimplantation embryo, namely Oct-4 and Sox-2 (Rosner et al. 1990; Schöler et al. 1990b; R. Lovell-Badge, pers. comm.). Oct-4 binds as a dimer to a specific element, termed PORE, in the i-opn enhancer, and transactivates more strongly from this element than from the canonical octamer motif alone. Biochemical analysis of POU protein binding to mutated PORE elements indicates that the Oct-4/PORE complex provides a new paradigm for the interaction of POU proteins with DNA by using only the homeodomain core motifs for binding. Sox-2 also binds to the i-opn enhancer and represses transactivation by Oct-4 and other as yet unidentified factors. Sox-2-mediated repression in i-opn contrasts with Sox-2-mediated activation in the fgf-4 enhancer (Yuan et al. 1995). Sox-2-mediated repression and activation of Oct-4 function in these two enhancers depends on the carboxy-terminal region of Sox-2 (Yuan et al. 1995).
Oct-4 dimer formation on the PORE
The POU transcription factors contain a pair of closely and flexibly linked DNA-binding domains that, in principle, have numerous possible modes of interactions with substrate DNA. The POUS and POUHD domains can be separated and bind DNA independently. However, mixing experiments demonstrate that the separated domains, as well as the linked domains, can bind cooperatively to appropriate DNA substrates (Klemm and Pabo 1996). Analysis of crystals containing POU protein–DNA complexes has revealed that the amino-terminal POUS domain is joined to the POUHD domain by an unstructured linker that would allow each domain to dock onto DNA with few steric constraints imposed by the other domain (Klemm et al. 1994). The linker of Oct-4 is relatively short (17 amino acids) compared to those of other POU proteins (15–56 amino acids) and constrains the distance between and the relative orientation of two half sites for simultaneous binding. The crystal structure of the Oct-1 POU domain bound to the octamer sequence of the histone H2B promoter (Klemm et al. 1994) shows that the POUS domain interacts with the 5′ ATGC portion of the octamer motif and the POUHD with the 3′ A/T-rich portion. The two domains bind to opposite faces of the DNA helix and contact the same base pairs at the center of the octamer motif, but do not interact directly. The fact that POUS and POUHD bind the same base pairs, albeit at opposite sides, has been suggested to be important for mediating cooperative binding of POUS and POUHD through changes in the DNA structure (Klemm and Pabo 1996). According to the length and the conformational flexibility of the linker, it seems possible that binding of two more distantly spaced half sites by the two domains of a POU protein monomer occurs.
A second possibility is that the linker allows the POU protein to bind half sites in different orientations. This is demonstrated by the crystal structure of the Pit-1 POU domain on a palindromic recognition site (ATGTATATACAT; Jacobson et al. 1997). Although the protein folds of each of the two Pit-1 DNA-binding domains, and the docking onto DNA of each domain is very similar to that observed in the Oct-1 crystal, the relative orientation of the two domains in the two crystals is inverted. The Oct-1 POUS domain recognizes the GCAT half-site and the Pit-1 POUS domain binds a corresponding sequence GTAT. However, the latter motif lies on the opposite strand (schema in Fig. 8D). As a consequence, the orientation of POUS relative to POUHD is inverted.
Figure 8.
Computer modeling of Oct-4 POU domain dimer conformation on the PORE binding sequence. (A,B) Lateral and frontal view of Oct-4 POU domain dimer conformation on the PORE binding site. Oct-4 POU domains were modeled into the coordinates of Oct-1. Each Oct-4 molecule has one color. The POUS domain has four α-helices (labeled 1–4) and the POUHD has three α-helices (labeled 1–3). DNA structure is represented by purple for phosphate, red for oxygen, green for carbon, and blue for nitrogen. A and B generated with WHAT IF software. (C) Amino acid sequence alignment of POU specific, linker, and POU homeodomain of Oct-1 and Oct-4. Amino acids numeration are taken from Herr and Cleary (1995). Dots represents identical amino acids found in Oct-4. α-Helices are indicated by boxes. (D) Schematic representation of Oct-4 and Pit-1 homodimers. Each molecule is given one color.
Cooperative binding by domains of two different POU protein molecules provides a second fundamentally different mode of POU protein–DNA interaction. In the Pit-1 crystal, the repeat unit is a homodimer in which a dimerization interface is formed between the amino-terminal part of POUHD helix 3 of one Pit-1 molecule and the amino-terminal end of POUS in the second molecule (helix 1 in conjunction with the loop between helices 3 and 4).
Our data show that in vitro Oct-4 also forms a homodimer on DNA. However, mutagenic analysis of the binding site (Fig. 5A,B) and computer modeling suggest that the Oct-4 dimer is different than the Pit-1 dimer (schema on Fig. 8D) and illustrates yet a third means by which POU proteins can interact with specific DNA sequences. In contrast to Pit-1, which requires the palindromic arrangement of ATGTAT without any spacing, the Oct-4 dimer requires a palindrome of ATTTG with an exact spacing of 5 nucleotides (Figs. 5B and 8D). The second half site of the PORE is contained in the octamer motif, but specific binding of POUS to the ATGC sequence seems not to be required as shown by a mutational analysis (Fig. 5B,C). To determine the arrangement of both Oct-4 POU domains the sequence of Oct-4 POU domain was modeled into the coordinates of Oct-1 POU domain (Fig. 8A,B). Because the POU domains of Oct-1 and Oct-4 are extremely well conserved, it is likely that Oct-4 POU domain protein structure bound to DNA is similar to that of Oct-1 (Herr and Cleary 1995; Fig. 8C). This was done first for the octamer motif that overlaps with the second half site of the PORE and then extended to the other half site (Fig. 8A). The arrangement basically results from a point-symmetrical flipping of the first Oct-4 POU molecule with respect to the second. In case of Oct-4, the amino terminus of POUHD (nonhelical region between linker and helix 1) would interact with the carboxy-terminal part of POUS helix 1 and the loop between helices 1 and 2. Thus, dimerization of Oct-4 on the PORE likely represents a new paradigm for POU protein–DNA interaction.
According to this model, several side chains are predicted to interact with each other. The computer model of Oct-4 POU domain binding to the PORE predicts that Ile-21 of the POUS domain of one molecule is within 4 Å of Ser-7 of the POUHD of the other molecule (Fig. 8C, numbering of the POU domain according to Herr and Cleary 1995). Thus, the molecules would be in very close proximity at one position on each side of the dimer. Phosphorylation of Ser-7 in the Oct-1 POUHD inhibits binding to the octamer sequence in the H2B promoter (Segil et al. 1991). DNA binding is also reduced in case of Thr-7 phosphorylation of the Pit-1 POUHD, but the magnitude of the effect is dependent on the DNA sequence (Kapiloff et al. 1991; Caelles et al. 1995). Both Ser and Thr residues can be phosphorylated in vitro by protein kinase A (Kapiloff et al. 1991; Segil et al. 1991) and in vivo by a cell-cycle-dependent kinases (Segil et al. 1991; Caelles et al. 1995). In the case of the Oct-4 dimer, phosphorylation of POUHD Ser-7 should interfere with dimer formation on the PORE. Because Oct-1, Oct-2, and Oct-6 can also form a dimer on PORE (Fig. 5A; unpubl.), a reduction of dimer formation attributable to phosphorylation at position 7 of POUHD could be generally valid for all POU factors. In contrast, phosphorylation at position 7 should not interfere with Pit-1 dimerization on the ATGTATATACAT sequence.
Regulation of OPN by Oct-4 and Sox-2
Our data show that a short fragment of i-opn contains DNA-binding sites for Oct-4 and Sox-2 in close proximity and is sufficient to drive expression of reporters in F9 EC cell lines. The intron fragment of OPN is enriched in α4 immunoprecipitates of cross-linked F9 EC cell chromatin. Because the F9 EC cell line shares many characteristics with cells of the preimplantation embryo, we have proposed that Oct-4 and Sox-2 regulate expression of the OPN gene during preimplantation stages of embryogenesis by binding to specific DNA sequences in i-opn (Strickland and Mahdavi 1978; Strickland et al. 1980; Hogan et al. 1981). Oct-4 mRNA is expressed at comparable levels in cells of the morulae, in the ICM of blastocysts, and in the epiblast of day 5.5 and 6.5 embryo (Schöler et al. 1990a; Yeom et al. 1991). In contrast, OPN expression changes during these early stages of development. Weak OPN expression is detected in the morulae (Fig. 3B). The highest expression level of OPN is observed in the ICM/hypoblast of day 4.0 and 4.5 blastocysts (Fig. 3B,C). After these stages, OPN is down-regulated (Fig. 3C). This could be attributable to additional transcription factors that interact with Oct-4 on the intron enhancer. Either a repressive or an activating cofactor could account for the relatively variable OPN expression during early embryonic development.
Sox-2 is one candidate for a modulator of Oct-4 transactivation. Sox-2 expression varies in different embryonic cell types, which correlates inversely with the expression pattern of OPN in these cells (Fig. 6A). In transient transfection assays, the PORE but not the octamer motif is active (Fig. 5D). The activity of the PORE region is increased two- to threefold when the Sox-binding site is included (Fig. 6B, cf. 6 × O and 6 × OS). However, this increase is not attributable to Sox-2 because a 20-fold activation was achieved when the Sox-2-binding site was mutated (Fig. 6B). Furthermore, cotransfection experiments in differentiated cells indicate that Sox-2 can repress Oct-4-mediated i-opn activity in a dose-dependent manner (Fig. 6E). Repression requires Sox-2 binding to DNA and also depends on the carboxy-terminal region of Sox-2 that has been described previously as a transactivation domain (Fig. 6F,H; Yuan et al. 1995). Repression of the i-opn enhancer by Sox-2 is in contrast to what has been shown for the fgf-4 3′ enhancer, where Sox-2 stimulates Oct-4-mediated activity (Yuan et al. 1995).
Varying the level or activity of Sox-2 and Oct-4 in embryonal cells is likely to result in altered expression of genes such as fgf-4 and OPN. Changes in Oct-4, Sox-2, and OPN expression levels that occur during differentiation of F9 EC cells are consistent with the model of Oct-4 and Sox-2 interaction on i-opn that was established from binding and transactivation data. Sox-2 levels decline rapidly, whereas Oct-4 levels decline more slowly during F9 differentiation (Fig. 7B). Thus, Sox-2-mediated repression of Oct-4 activation of i-opn is relieved briefly, resulting in the transient increase of OPN expression (Fig. 7A). Forty-eight hours after RA treatment, Oct-4 is no longer expressed, leading to the loss of a complex at the PORE, which would likely result in low or undetectable levels of OPN mRNA production (Fig. 7A).
Sox-2 is not likely to be the only transcription factor involved in modulating the Oct-4-mediated transactivation of the intron enhancer. This would explain why the PORE/octamer region is much weaker than a fragment that is longer and contains a mutated Sox element (Fig. 6B, cf. 6 × O and 6 × OS).
Possible role of OPN in the preimplantation embryo
The formation of the hypoblast and its derivatives, parietal and visceral endoderm, depends on interactions between the ECM and integrins (Behrendtsen et al. 1995). OPN binds to cells displaying the αvβ1, αvβ3, and αvβ5 integrins on their surface and contains a GRGDS amino acid motif that is absolutely required for the integrin interaction (for review, see Eble and Kühn 1997). Deletion of the β1 integrin gene in mice results in ICM failure and peri-implantation lethality, indicating that β1 integrin is required for preimplantation development (Fässler and Meyer 1995; Stephens et al. 1995).
OPN may bind to integrins in a way that alters cell–cell adhesion properties selectively. High levels of OPN expression in the 4.0-dpc ICM and forming hypoblast could result in the loosening of cell–cell and cell–ECM contacts. OPN-deficient embryos exhibit no apparent phenotype (Hynes 1996) suggesting that loss of OPN function during embryogenesis can be compensated by other ECM proteins. Bone sialoprotein II (BSP II) is one possible candidate. Further analyses will be done to define other genes that are also up-regulated transiently by Oct-4 in the developing hypoblast.
Materials and methods
Embryo collection, cell culture, and transient transfection
Morulae and blastocysts were flushed from CD1 mice in M2 medium as described in Hogan et al. (1994). COS, 3T3, 293, and F9 EC cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS). P19 EC and GCLB cells were maintained in DMEM supplemented with 10% FCS and 1% nonessential amino acids (Seromed). MBL-1 ES cells were cultured in DMEM [0.45% glucose (wt/vol)], 15% FCS, 1 μm β-ME, and in the presence of 1000 U/ml leukemia inhibitory factor (LIF, GIBCO-BRL). Murine EC and ES cells were grown on 0.1% gelatin-coated tissue culture plates. P19 EC cells were induced to differentiate by treatment with 1 μm all-trans RA (Sigma). Differentiation of F9 cells into parietal endoderm was accomplished by adding 0.1 μm RA to adherent cells. Differentiation of F9 cells into visceral endoderm was performed by growing the cells for 48 hr in suspension using bacterial Petri dishes. Aggregates were transferred to tissue culture dishes allowing cells to attach to a surface for further growth. Medium containing RA was changed every 24 hr.
Cells were transfected using the calcium–phosphate precipitation method at a density of 3 × 105 cells per 6-cm dish. Each dish received a total of 12 μg of DNA including 10 μg of luciferase reporter plasmid and 2 μg of human β-actin–LacZ as internal standard. In cotransfection assays, 1 μg of luciferase reporter plasmid and varied amounts of expression plasmids were used in combination with pBluescript KS (Stratagene) to bring the final DNA concentration to 12 μg. All experiments were carried out four times. After 40 hr of transfection, cells were harvested and 50-μl extracts were prepared in ice-cold 250 mm Tris (pH 7.8), 1 mm DTT. All luciferase activities were normalized by β-galactosidase expression levels.
Plasmid constructions
The i-opn and i-opn mut fragment were generated by PCR amplification of osteopontin intron 1 using 1 μg of F9 genomic DNA, 5′-TATTAGTCCAAATAGAACATC-3′ and 5′-TATTAGTCCAAATAGAACATCTTACTCAAATTCAAAGATATCTTTGTTTCTTTCAGCTTTGTATAATGTAAGTTAAAATCACATTGCACAAGCAAGCGG-3′ as sense primer, respectively, and 5′-CTCTCATCCTTAGCAAGGAA-3′ as antisense primer. The conditions were the same as described for PCR of immunoprecipitated cross-linked chromatin. The i-opn was precloned into the PCR II vector using the TA Cloning kit (Invitrogen). The i-opn was cut out of the PCR II vector by EcoRI and was recloned into the EcoRI site of pBluescript KS. After HindIII–BamHI digestion, the i-opn of pBluescript KS was cloned into HindIII–BamHI site of −37tkluc (kindly provided by A. Hecht, ZMBH, Heidelberg, Germany). The 6 × O, 10 × O, 6 × O−1, 6 × O−2, 6 × O−3, 6 × O−4, 6 × OS, 6 × O−S, and 6 × OS− reporter plasmids were obtained by multimerizing the corresponding oligonucleotides. The 5′ overhang of the multimers were filled in using Klenow polymerase and fragments of 6 or 10 oligonucleotide repeats were precloned into the EcoRV site of pBluescript KS. The HindIII–BamHI fragment containing the oligonucleotide multimer was cut out of the pBluescript KS and was inserted into HindIII–BamHI sites of −37tkluc.
Labeling, cross-linking, and shearing chromatin
Proteins were labeled by adding 175 μCi/ml [35S]methionine (labeling grade; Amersham) 1–2 hr before harvest. In vivo fixation of chromatin was done as described by Orlando and Paro (1993) with several modifications. Cells were removed from the plates by trypsin, resuspended at a density of 5 × 106 cells/ml in DMEM/5% heat-inactivated FCS, and fixed in vivo by adding one-tenth volume of formaldehyde buffer [11% formaldehyde (vol/vol) in 10% methanol, 0.1 m NaCl, 1 mm Na-EDTA, 0.5 mm Na-EGTA (pH 8.0), 50 mm Tris-HCl (pH 8.0)]. The fixation reaction was incubated 10 min at room temperature thoroughly mixed on ice for 40 min, and stopped by adding glycine (125 mm final concentration). Fixed cells were collected by centrifugation (500g for 10 min at 4°C) and resuspended on a roller for 10 min at 4°C in 40 ml Triton-washing buffer [0.25% Triton X-100, 10 mm Na-EDTA, 0.5 mm Na-EGTA, 10 mm Tris-HCl (pH 8.0)] to lyse unfixed cells. Fixed chromatin was collected by centrifugation at 500g for 10 min at 4°C, washed in 40 ml of NaCl washing buffer [200 mm NaCl, 1 mm Na-EDTA, 0.5 mm Na-EGTA, 10 mm Tris-HCl (pH 8.0)] for 10 min at 4°C, centrifuged again, resuspended in 2–3 ml of TE–EGTA buffer [1 mm Na-EDTA, 0.5 mm Na-EGTA, 10 mm Tris-HCl (pH 8.0)], transferred to siliconized Corex-glass tubes, and ∼0.5 ml of glass microbeads (diameter of 0.10–0.11 mm; B. Braun Biotech International) were added to every 3 ml of cell suspension. The mixture was sonified on ice for 15 min with the Branson model B15 at a “duty cycle” of 0.65% with the output control set at 7–7.5 to produce DNA fragments of an average size of 500 bp, with a maximum size of 2.5 kb. Samples were adjusted to 0.5% Sarkosyl and gently swirled for 10 min at room temperature. Cell debris was eliminated by two 15-min centrifugation steps at 15,000g at 4°C.
Purification, immunoprecipitation, and decross-linking of fixed chromatin fragments
Cross-linked chromatin complexes were separated from free proteins, DNA, and RNA by CsCl isopyknic centrifugation. Samples were adjusted to 1.42 gram/ml CsCl, brought to 5 ml with the TE–EGTA–Sarkosyl buffer, and centrifuged in a Beckman SW55Ti rotor at 40,000 rpm for 72 hr at 20°C. Fractions of 300 μl were collected from the bottom of the gradient using a 0.25-mm capillary needle and 35S content was measured in a Beckman LS 6000 SC scintillation counter. Fractions containing the cross-linked chromatin (∼1.38 gram/ml) were pooled and dialyzed overnight at 4°C against 5% glycerol, 1 mm Na-EDTA, 0.5 mm Na-EGTA, 10 mm Tris-HCl (pH 8.0). Fixed chromatin can be stored in this buffer for at least 3 months at −80°C and small aliquots were used for immunoprecipitations.
Aliquots (200 μg of cross-linked chromatin in 100 μl) were cleared by centrifugation (15 min at 13,000 rpm), mixed with 30 μl H2O and 100 μl 2× TE-EGTA buffer, carefully adjusted to 0.1% SDS (wt/vol) and 0.5 m NaCl, incubated for 5 min at room temperature, adjusted to 1% Triton X-100 (wt/vol), 0.1% Na-Deoxycholate and 0.1% BSA, incubated for 10 min, and cleared again by centrifugation for 15 min at 13,000 rpm. As a preclearing step, supernatants were incubated with 100 μl of Dynabeads coupled to sheep anti-rabbit IgG (6 × 108 to 7 × 108 beads/ml; Dynal) for 1 hr. Supernatants were removed from beads by a magnetic particle concentrator (MPC; Dynal) and 10 μg of specific antibodies against Oct-4 (or IgG of preimmune serum) were added. Samples were rotated for 3 hr at 4°C, 300 μl of sheep anti-rabbit IgG Dynabeads added, incubated with rotation for 2 hr at 4°C, and immunocomplexes were pelleted by magnetic field. Pellets were washed (10 min per wash) five times in 1 ml of washing buffer [1% Triton X-100 (wt/vol), 0.1% Na-deoxycholate (wt/vol), 0.1% SDS (wt/vol), 0.1% BSA (wt/vol), 0.5 m NaCl, 1 mm Na-EDTA, 0.5 mm Na-EGTA, 10 mm Tris-HCl (pH 8.0)], once with 1 ml of LiCl-washing buffer [250 mm LiCl, 0.5% NP-40 (wt/vol), 0.5% Na-deoxycholate (wt/vol), 1 mm Na-EDTA, 0.5 mm Na-EGTA, 10 mm Tris-HCl (pH 8.0)], twice with 1 ml of TE–EGTA buffer, and resuspended in 350 μl of TE–EGTA buffer. One hundred microliters were kept for protein analysis, and 250 μl was treated 30 min at 37°C with DNase-free RNase A (50 μg/ml) and incubated overnight at 37°C in 250 μg/ml proteinase K/0.25% SDS.
PCR on immunoprecipitated cross-linked chromatin
PCR amplification was performed in a final volume of 100 μl, using 1 ng of genomic F9 DNA or 1 ng of immunoprecipitated cross-linked chromatin as template, 10 pmoles of each primer, 2.5 mm dNTPs (Pharmacia), 2.5 units of Taq DNA polymerase (Perkin Elmer-Cetus); and 1× PCR buffer (Perkin Elmer-Cetus). The PCR consisted of 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C. Upstream and downstream primers pairs were as follows: OPN intron 1, 5′-CAAATTCAAAGATATCTTTGTTTC-3′, 5′-CCCCACTATCTGATGTCTCT-3′; OPN exon 7, 5′-ATCCTGATGCCACAGATGAG-3′, 5′-ACTTGTGGCTCTGATGTTCC-3′; G6PD fragment, 5′-AAGCCAAACTAGCAGCTAGG-3′, 5′-GGGCTAGTCTATCATTGCAG-3′.
Preparation of protein extracts and EMSA
Extraction of proteins and EMSA conditions used for Oct-4 and Sox-2-binding analysis were carried out as described in Sylvester and Schöler (1994) and Dailey et al. (1994), respectively.
Oligonucleotides used in this study
Bold indicates introduced mutation, lowercase letters indicates non-OPN sequences. O, 5′-ctgaAAGTTAAAATCACATTTGAAATGCAAATGGAAAAGCaagtcga-3′; O−1, 5′-ctgaAAGTTAAAATCACATTTGAAAGGCAAATGGAAAAGCaagtcga-3′; O−2, 5′-ctgaAAGTTAAAATCACATGTGAAAGGCAAATGGAAAAGCaagtcga-3′; O−3, 5′-ctgaAAGTTAAAATCACATGTGAAATGCAAATGGAAAAGCaagtcga-3′; O−4, 5′-ctgaAAGTTAAAATCACATTTGAAATGCAACTGGAAAAGCaagtcga-3′; OS, 5′-ctgaTCTTTGTTTCTTTCAGCTTTGTATAATGTAAGTTAAAATCACATTTGAAATGCAAATGGAAAAGCaagtcga-3′; O−S, 5′-ctgaTCTTTGTTTCTTTCAGCTTTGTATAATGTAAGTTAAAATCACATTTGAAATGCAACTGGAAAAGCaagtcga-3′; OSa−, 5′-ctgaTGCACTGACCTTTCAGCTTTGTATAATGTAAGTTAAAATCACATTTGAAATGCAAATGGAAAAGCaagtcga-3′; OSb−, 5′-ctgaTCTCTGTGTCTTTCAGCTTTGTATAATGTAAGTTAAAATCACATTTGAAATGCAAATGGAAAAGCaagtcga-3′; P-1, 5′-ctgaAAGTTAAAATCACATTTGAATGCAAATGGAAAAGCaagtcga-3′; P+1, 5′-ctgaAAGTTAAAATCACATTTGATAATGCAAATGGAAAAGCaagtcga-3′.
RNA extraction and Northern blot analyses
Poly(A)+ RNA was isolated from cell lines using the Oligotex mRNA kit (Qiagen). Total cellular RNA was prepared by the guanidinium thiocyanate–phenol extraction procedure described in Chomczynski and Sacchi (1987). Northern blot analysis of poly(A)+ and total RNA was performed as described elsewhere (Kroczek 1993). Hybond-N+ filters (Amersham) were hybridized using a PstI–PstI Oct-4 probe (400-bp fragment corresponding to the 5′ end of the cDNA) (106 cpm/ml), a HindIII–HindIII OPN probe (position +157 to +1144 of the cDNA) (2 × 106 cpm/ml) (2AR plasmid, kindly provided by D. Denhardt, Rutgers University, Piscataway, NJ), or an oligonucleotide 28 S probe: 5′-CAGCGAGCCGGGCTTCTTACCCATTTAAAGTTTGAGAATAGGTGGAGATCG-3′ (0.5 × 106 cpm/ml; kindly provided M. Muckenthaler, EMBL). Northern blots were analyzed for band intensity using the PhosphorImager (Molecular Dynamics).
RNA isolation from preimplantation embryos and RT–PCR
Ten embryos were suspended in 200 μl of RNAzol (Tel-test, Inc.) and 20 μl of chloroform while on ice by shaking. The suspension was left on ice 5 min and cleared by centrifugation (12,000g for 15 min at 4°C), and total RNA was precipitated for 15 min on ice after adding 1 volume of isopropanol and 2 μl of glycogen (Boehringer Mannheim; 20 mg/ml stock), collected by centrifugation (12,000g for 15 min at 4°C), washed with 70% ethanol, and dissolved in 11 μl of DEPC–H2O. Reverse transcription was performed for 1 hr at 37°C in a total volume of 20 μl by adding 9 μl of the following mix to the dissolved RNA: 0.5 μg of oligo(dT) (Promega), 39 units of RNasin (Promega), 1 mm dNTPs (Pharmacia), 1× RT-buffer, and 200 units of reverse transcriptase (M-MLV; Promega). PCR was performed using 5 μl of the RT reaction as template for a 25 μl final reaction volume. Each reaction contained: 10 pmoles of each primer, 2.5 mm dNTPs, 2.5 units of Taq polymerase (Perkin Elmer-Cetus), and 1× PCR buffer (Boehringer Mannheim). The PCR consisted of 35 cycles of 30 sec at 94°C, 1 min at 62°C, and 1 min at 72°C. Upstream and downstream primers were as follows: Oct-4, 5′-GGCGTTCTCTTTGGAAAGGTGTTC-3′ and 5′-CTCGAACCACATCCTTCTCT-3′ (spanning exons 2–3 and 4–5, respectively); OPN, 5′-GCAGACACTTTCACTCCAATCG-3′ and 5′-GCCCTTTCCGTTGTTGTCCTG-3′ (located in exon 6 and exon 7, respectively).
Riboprobe synthesis and whole mount in situ hybridization of 3.5–6.5 dpc embryos
The 1-kb OPN cDNA cloned in pGEM3 (2AR plasmid) was excised with HindIII from the vector and recloned in the same vector so that both orientations were available from the same promoter. Vectors were linearized with EcoRI, both sense and antisense riboprobes were generated by SP6 RNA polymerase and DIG-labeling mix according to the manufacturer (Boehringer Mannheim), reactions were digested with RNase-free DNase I (Promega) for 15 min at 37°C, ethanol precipitated in the presence of 4 m LiCl, and resuspended in DEPC–H2O. The concentration and labeling efficiency of the probes were determined empirically by agarose gel electrophoresis and by dot-blot analyses, respectively.
In situ hybridization of embryos was performed as described by MacPhee et al. (1994) and Rosen and Beddington (1993), with some modifications. Embryos were fixed for 3 hr in 3% paraformaldehyde/0.5% glutaraldehyde in PBS, washed in PBS, permeabilized for 20 min at 4°C in PBT (PBS with 0.1% Triton X-100), digested 15 min with proteinase K (1 μg/ml in PBT), washed twice for 5 min in fresh glycine (2 mg/ml PBT), refixed for 30 min in 4% paraformaldehyde/0.2% glutaraldehyde in PBS, briefly washed in triethanolamine buffer (100 mm, pH 8.0) (Sigma), treated three times for 5 min with 0.25% acetic anhydride (Sigma) in triethanolamine buffer and once for 20 min with 0.1% sodium borohydride (Sigma) in PBT. After prehybridization in 50% formamide, 0.75 m NaCl, 10 mm Pipes (pH 6.8), 1 mm EDTA, 100 mg/ml yeast tRNA, 0.1% BSA, 1 % SDS for 2 hr at 50°C, embryos were hybridized for 34 hr in the same solution containing 1 μg/ml riboprobe. Embryos were rinsed briefly and washed extensively (20 hr at 60°C) in washing buffer 1 [300 mm NaCl, 1% SDS in PE buffer (10 mm PIPES at pH 6.8, 1 mm EDTA], washed two times for 30 min in washing buffer 2 (50 mm NaCl, 0.1% SDS in PE buffer), and incubated once for 30 min at 37°C with 100 μg/ml RNase A diluted in the appropriate buffer [500 mm NaCl, 10 mm PIPES (pH 7.2), 0.1% Triton X-100]. Embryos were then washed for 45 min at 50°C in washing buffer 3 (50% formamide, 300 mm NaCl, 1% SDS in PE buffer), for 30 min at 50°C in washing buffer 4 (50% formamide, 150 mm NaCl, 0.1% Triton X-100 in PE buffer), and finally 30 min followed by 20 min in washing buffer 5 (500 mm NaCl, 0.1% Triton X-100 in PE buffer) at 70°C. Embryos were blocked for 1 hr at room temperature in maleic acid washing buffer [100 mm maleic acid, 150 mm NaCl (pH 7.5)] containing 2% heat-inactivated sheep serum and 2% blocking buffer (Boehringer Mannheim, cat. no. 1175041) and incubated overnight at 4°C with anti-digoxigenin Fab-alkaline phosphatase conjugate antibody (Boehringer Mannheim) diluted 1:2000. Excess antibody was removed by washing the embryos three times for 5 min and then four times for 30 min with 1× TBST at room temperature. Embryos were then equilibrated in alkaline phosphatase buffer [100 mm NaCl, 50 mm MgCl2, 0.1% Tween-20, 100 mm Tris (pH 9.5)] and stained at room temperature in the dark with BM purple (Boehringer Mannheim, cat no. 1442074). Color reaction was stopped by washing the embryos with PBS. Uteri from 5.5 and 6.5 dpc pregnant females were fixed with 4% paraformaldehyde, dehydrated and embedded in paraffin wax. Sections of 6 μm were transferred to silone-coated slides. Treatment of the slides and hybridization were performed using the components and the protocol of Novagen (SureSite II system). After several washing steps, the slides were blocked and incubated overnight at 4°C with 1:500 anti-DIG in blocking buffer. Slides were washed four times for 30 min each with maleic acid washing buffer and stained with BM purple.
Computer searches and modeling
Identification of genes containing combination of octamer and Sox binding motifs was performed using the prepGCG program, findpatterns, and GenBank/EMBL database.
Homology modeling of Oct-4 dimer configuration was based on Oct-1 as template structure (Klemm et al. 1994). Modeling was performed using WHAT IF programme (Vriend 1990) and modeling protocol was used as described in Vriend and Eijsink (1993). There is no structure template available for the amino terminus, carboxyl terminus, and linker. For this reason, these parts of the molecule are not represented on the model.
The DNA was modeled by maintaining the phosphate and the sugar backbone of the DNA in the Oct-1 template structure. The bases were replaced using the corresponding bases found in the PORE DNA and were modeled to fall in the same plane as the corresponding Oct-1 bases with ideal geometry.
The Oct-4 dimer was constructed by optimaly superposing the GCAAAT sequence of the octamer motif on which Oct-4 was modeled, onto the AGTTTA sequence. The RMS deviation of the DNA superposition is ∼1.3 Å, which implies an uncertainty in the overall location of the second Oct-4 with respect to the first Oct-4 of ∼1–12 Å. The average local coordinate error is expected to be ∼1.5 Å (Chinea et al. 1995).
Acknowledgments
We are most grateful to Drs. V. Orlando and R. Paro for a thorough introduction to and sound advice on the chromatin precipitation procedures. We thank Dr. L. Dailey for Sox-2 expression vectors and antibodies, Dr. L. Toldo for computer search, M. Pesce for reading the manuscript, and U. Schibler for a valuable comment. We also thank K. Hübner for Oct-4 protein and antibody, C. Sandberg for RNA purification. H.H. was supported by the Deutsche Studienstiftung and Fond der chemischen Industrie, K.A., V.B., and M.K.G. by the European Community (EU contracts CT92-0073 and BIO4-CT95-0284), G.F. by the CNRS.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL schoeler@embl-heideberg.de; FAX 6221-387 518.
References
- Adamson ED. Cell-lineage-specific gene expression in development. In: Rossant J, Pederson RA, editors. Experimental approaches to mammalian embryonic development. Cambridge, UK: Cambridge University Press; 1986. pp. 321–364. [Google Scholar]
- Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendtsen O, Alexander CM, Werb Z. Cooperative interactions between extracellular matrix, integrins and parathyroid hormone-related peptide regulate parietal endoderm differentiation in mouse embryos. Development. 1995;121:4137–4148. doi: 10.1242/dev.121.12.4137. [DOI] [PubMed] [Google Scholar]
- Caelles C, Hennemann H, Karin M. M-phase-specific phosphorylation of the POU transcription factor GHF-1 by a cell cycle-regulated protein kinase inhibits DNA binding. Mol Cell Biol. 1995;15:6694–6701. doi: 10.1128/mcb.15.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinea G, Padron G, Hooft RW, Sander C, Vriend G. The use of position-specific rotamers in model building by homology. Proteins. 1995;23:415–421. doi: 10.1002/prot.340230315. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, Stevanovic M, Goodfellow PN, Lovell-Badge R. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- Curatola AM, Basilico C. Expression of the K-fgf proto-oncogene is controlled by 3′ regulatory elements which are specific for embryonal carcinoma cells. Mol Cell Biol. 1990;10:2475–2484. doi: 10.1128/mcb.10.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L, Yuan H, Basilico C. Interaction between a novel F9-specific factor and octamer-binding proteins is required for cell-type-restricted activity of the fibroblast growth factor 4 enhancer. Mol Cell Biol. 1994;14:7758–7769. doi: 10.1128/mcb.14.12.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt DT, Guo X. Osteopontin: A protein with diverse functions. FASEB J. 1993;7:1475–1482. [PubMed] [Google Scholar]
- Denhardt DT, Butler WT, Chambers AF, Senger DR. Gene expression and phosphorylation of mouse osteopontin. In: Denhardt DT, Butler WT, Chambers AF, Senger DR, editors. Osteopontin: Role in cell signaling and adhesion. New York, NY: The New York Academy of Sciences; 1995. [Google Scholar]
- Eble JA, Kühn K. In: Integrin-ligand interaction. Eble JA, Kühn K, editors. Heidelberg, Germany: Springer-Verlag; 1997. [Google Scholar]
- Fässler R, Meyer M. Consequences of lack of β1 integrin gene expression in mice. Genes & Dev. 1995;9:1896–1908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- Ferrari S, Harley VR, Pontiggia A, Goodfellow PN, Lovell-Badge R, Bianchi ME. SRY, like HMG 1, recognizes sharp angles in DNA. EMBO J. 1992;11:4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Hamada H. A CNS-specific POU transcription factor, Brn-2, is required for establishing mammalian neural cell lineages. Neuron. 1993;11:1197–1206. doi: 10.1016/0896-6273(93)90231-f. [DOI] [PubMed] [Google Scholar]
- Gardner RL. Origin and differentiation of extraembryonic tissues in the mouse. Int Rev Exp Pathol. 1983;24:63–133. [PubMed] [Google Scholar]
- Giese K, Cox J, Grosscheld R. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Herr W, Cleary MA. The POU domain: Versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes & Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- Hogan BLM, Taylor A, Adamson E. Cell interactions modulate embryonal carcinoma cell differentiation into parietal and visceral endoderm. Nature. 1981;291:235–237. doi: 10.1038/291235a0. [DOI] [PubMed] [Google Scholar]
- Hogan BLM, Beddington R, Constantini F, Lacy E. Section I: In vitro culture of eggs, embryos, primordial sperm cells and teratocarcinoma cells. In: Hogan BLM, Beddington R, Constantini F, Lacy E, editors. Manipulating the mouse embryo. A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. p. 386. [Google Scholar]
- Hynes RO. Targetes mutations in cell adhesion genes: What have we learned from them? Dev Biol. 1996;180:402–412. doi: 10.1006/dbio.1996.0314. [DOI] [PubMed] [Google Scholar]
- Jacobson EM, Li P, Leon-del-Rio A, Rosenfeld MG, Aggarwal AK. Structure of Pit-1 POU domain bound to DNA as a dimer: Unexpected arrangement and flexibility. Genes & Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- Kapiloff MS, Farkash Y, Wegner M, Rosenfeld MG. Variable effects of phosphorylation of Pit-1 dictated by the DNA response elements. Science. 1991;253:786–789. doi: 10.1126/science.1652153. [DOI] [PubMed] [Google Scholar]
- Klemm JD, Pabo CO. Oct-1 POU domain-DNA interactions: Cooperative binding of isolated subdomains and effects of covalent linkage. Genes & Dev. 1996;10:27–36. doi: 10.1101/gad.10.1.27. [DOI] [PubMed] [Google Scholar]
- Klemm JD, Rould MA, Aurora R, Herr W, Pabo CO. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- Kraft HJ, Mosselman S, Smits HA, Hohenstein P, Piek E, Chen Q, Artzt K, van Zoelen EJ. Oct-4 regulates alternative platelet-derived growth factor alpha receptor gene promoter in human embryonal carcinoma cells. J Biol Chem. 1996;271:12873–12878. doi: 10.1074/jbc.271.22.12873. [DOI] [PubMed] [Google Scholar]
- Kroczek RA. Southern and Northern analysis. J Chromatogr. 1993;618:133–145. doi: 10.1016/0378-4347(93)80031-x. [DOI] [PubMed] [Google Scholar]
- Leger H, Sock E, Renner K, Grummt F, Wegner M. Functional interaction between the POU domain protein Tst-1/Oct-6 and the high-mobility-group protein HMG-I/Y. Mol Cell Biol. 1995;15:3738–3747. doi: 10.1128/mcb.15.7.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Roberts RM. Silencing of the gene for the beta subunit of human gonadotropin by the embryonic transcription factor Oct 3/4. J Biol Chem. 1996;271:16683–16689. doi: 10.1074/jbc.271.28.16683. [DOI] [PubMed] [Google Scholar]
- MacPhee DJ, Barr KJ, De Sousa PA, Todd SD, Kidder GM. Regulation of Na+, K(+)-ATPase alpha subunit gene expression during mouse preimplantation development. Dev Biol. 1994;162:259–266. doi: 10.1006/dbio.1994.1083. [DOI] [PubMed] [Google Scholar]
- Nadijcka M, Hillman N. Ultrastructural studies of the mouse blastocyst substages. J Embryol Exp Morphol. 1974;32:675–695. [PubMed] [Google Scholar]
- Niswander L, Martin GR. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development. 1992;114:755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- Nomura S, Wills AJ, Edwards DR, Heath JK, Hogan BL. Developmental expression of 2ar (osteopontin) and SPARC (osteonectin) RNA as revealed by in situ hybridization. J Cell Biol. 1988;106:441–450. doi: 10.1083/jcb.106.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell. 1990;60:461–472. doi: 10.1016/0092-8674(90)90597-8. [DOI] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Schöler HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Pesce M, Wang X, Wolgemuth DJ, Schöler HR. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev. 1998a;71:89–98. doi: 10.1016/s0925-4773(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Pesce, M., M.K. Gross, and H.R. Schöler. 1998b. In line with our ancestors: Oct-4 and the mammalian germ. BioEssays (in press). [DOI] [PubMed]
- Pruitt SC. Primitive streak mesoderm-like cell lines expressing Pax-3 and Hox gene autoinducing activities. Development. 1994;120:37–47. doi: 10.1242/dev.120.1.37. [DOI] [PubMed] [Google Scholar]
- Rizzino A, Rosfjord E. Transcriptional regulation of the murine k-fgf gene. Mol Reprod Dev. 1994;39:106–111. doi: 10.1002/mrd.1080390116. [DOI] [PubMed] [Google Scholar]
- Robertson E. Teratocarcinomas and embryonic stem cells. A practical approach. Oxford, UK: IRL Press; 1987. Cell culture methods and induction of differentiation of embryonal carcinoma; pp. 19–50. [Google Scholar]
- Rosen B, Beddington RS. Whole-mount in situ hybridization in the mouse embryo: Gene expression in three dimensions. Trends Genet. 1993;9:162–167. doi: 10.1016/0168-9525(93)90162-b. [DOI] [PubMed] [Google Scholar]
- Rosfjord E, Rizzino A. The octamer motif present in the Rex-1 promoter binds Oct-1 and Oct-3 expressed by EC cells and ES cells. Biochem Biophys Res Commun. 1994;203:1795–1802. doi: 10.1006/bbrc.1994.2395. [DOI] [PubMed] [Google Scholar]
- Rosner MH, Vigano MA, Ozato K, Timmons PM, Poirier F, Rigby PW, Staudt LM. A POU-domain transcription factor in early stem cells and germ cells of the mammalian embryo. Nature. 1990;345:686–692. doi: 10.1038/345686a0. [DOI] [PubMed] [Google Scholar]
- Ryan AK, Rosenfeld MG. POU domain family values: Flexibility, partnerships, and developmental codes. Genes & Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Fujii H, Meno C, Sato M, Hirota Y, Nagamatsu S, Ikeda M, Hamada H. Identification of putative downstream genes of Oct-3, a pluripotent cell-specific transcription factor. Genes Cells. 1996;1:239–252. doi: 10.1046/j.1365-2443.1996.d01-237.x. [DOI] [PubMed] [Google Scholar]
- Schöler HR. Octamania: The POU factors in murine development. Trends Genet. 1991;7:323–329. doi: 10.1016/0168-9525(91)90422-m. [DOI] [PubMed] [Google Scholar]
- Schöler HR, Balling R, Hatzopoulos AK, Suzuki N, Gruss P. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 1989a;8:2551–2557. doi: 10.1002/j.1460-2075.1989.tb08393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler HR, Hatzopoulos AK, Balling R, Suzuki N, Gruss P. A family of octamer-specific proteins present during mouse embryogenesis: Evidence for germline-specific expression of an Oct factor. EMBO J. 1989b;8:2543–2550. doi: 10.1002/j.1460-2075.1989.tb08392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: A germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990a;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line-specific protein Oct-4. Nature. 1990b;344:435–439. doi: 10.1038/344435a0. [DOI] [PubMed] [Google Scholar]
- Schöler HR, Ciesiolka T, Gruss P. A nexus between Oct-4 and E1A: Implications for gene regulation in embryonic stem cells. Cell. 1991;66:291–304. doi: 10.1016/0092-8674(91)90619-a. [DOI] [PubMed] [Google Scholar]
- Schoorlemmer J, Kruijer W. Octamer-dependant regulation of the kFGF gene in embryonal carcinoma and embryonic stem cells. Mech Dev. 1991;36:75–86. doi: 10.1016/0925-4773(91)90074-g. [DOI] [PubMed] [Google Scholar]
- Segil N, Roberts SB, Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Larsen PL, Varshavsky A. Mapping protein–DNA interactions in vivo with formaldehyde: Evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Stephens LE, Sutherland AE, Klimanskaya IV, Andrieux A, Meneses J, Pedersen RA, Damsky CH. Deletion of β1 integrins in mice results in inner cell mass failure and peri-implantation lethality. Genes & Dev. 1995;9:1883–1895. doi: 10.1101/gad.9.15.1883. [DOI] [PubMed] [Google Scholar]
- Stickland S, Mahdavi V. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell. 1978;15:393–404. doi: 10.1016/0092-8674(78)90008-9. [DOI] [PubMed] [Google Scholar]
- Strickland S, Smith KK, Marotti KR. Hormonal induction of differentiation in teratocarcinoma stem cells: Generation of parietal endoderm by retinoic acid and dibutyryl cAMP. Cell. 1980;21:347–355. doi: 10.1016/0092-8674(80)90471-7. [DOI] [PubMed] [Google Scholar]
- Sylvester I, Schöler HR. Regulation of the Oct-4 gene by nuclear receptors. Nucleic Acids Res. 1994;22:901–911. doi: 10.1093/nar/22.6.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer CP, van der Vliet PC. POU domain transcription factors. Biochim Biophys Acta. 1993;1173:1–21. doi: 10.1016/0167-4781(93)90237-8. [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, van Oosterhout JA, van Weperen WW, van der Vliet PC. POU proteins bend DNA via the POU-specific domain. EMBO J. 1991;10:3007–3014. doi: 10.1002/j.1460-2075.1991.tb07851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: A molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- Vriend G, Eijsink V. Prediction and analysis of structure, stability and unfolding of thermolysin-like proteases. J Comput Aided Mol Des. 1993;7:367–396. doi: 10.1007/BF02337558. [DOI] [PubMed] [Google Scholar]
- Waterhouse P, Parhar RS, Guo X, Lala PK, Denhardt DT. Regulated temporal and spatial expression of the calcium-binding proteins calcyclin and OPN (osteopontin) in mouse tissues during pregnancy. Mol Reprod Dev. 1992;32:315–323. doi: 10.1002/mrd.1080320403. [DOI] [PubMed] [Google Scholar]
- Wegner M, Drolet DW, Rosenfeld MG. POU-domain proteins: Structure and function of developmental regulators. Curr Opin Cell Biol. 1993;5:488–498. doi: 10.1016/0955-0674(93)90015-i. [DOI] [PubMed] [Google Scholar]
- Werner MH, Huth JR, Gronenborn AM, Clore GM. Molecular basis of human 46X, Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex. Cell. 1995;81:705–714. doi: 10.1016/0092-8674(95)90532-4. [DOI] [PubMed] [Google Scholar]
- Yeom YI, Ha H-S, Balling R, Schöler HR, Artzt K. Structure, expression and chromosomal location of the Oct-4 gene. Mech Dev. 1991;35:171–179. doi: 10.1016/0925-4773(91)90016-y. [DOI] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hübner K, Schöler HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox-2 and Oct-3. Genes & Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Zwilling S, Konig H, Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]