Abstract
Interferon regulatory factor-3 (IRF-3) was found to specifically interact with HPV16 E6 in a yeast two-hybrid screen. IRF-3 is activated by the presence of double-stranded RNA or by virus infection to form a stable complex with other transcriptional regulators that bind to the regulatory elements of the IFNβ promoter. We show that IRF-3 is a potent transcriptional activator and demonstrate that HPV16 E6 can inhibit its transactivation function. The expression of HPV16 E6 in primary human keratinocytes inhibits the induction of IFNβ mRNA following Sendai virus infection. The binding of HPV16 E6 to IRF-3 does not result in its ubiquitination or degradation. We propose that the interaction of E6 with IRF-3 and the inhibition of IRF-3’s transcriptional activity may provide the virus a means to circumvent the normal antiviral response of an HPV16-infected cell.
Keywords: IRF-3, HPV16, transcription, protein–protein interactions, oncoprotein
The papillomaviruses (PVs) display a specific tropism for squamous epithelial cells and produce benign cutaneous or squamous mucosal proliferative lesions. PVs often establish persistent or latent infections, and the virus productive life cycle is linked to the differentiation program of the infected squamous cell. Several of the PVs, including the human papillomaviruses (HPVs) that have been associated with cervical cancer, encode transforming genes whose functions create a cellular environment that allows replication of the viral DNA. Relatively little is known, however, about other essential viral functions that are necessary to establish a state of persistent infection, to circumvent the cellular anti-viral mechanisms, or to evade the host immune response (Howley 1996).
A close association between cervical cancer and the HPVs has now been established (zur Hausen 1996). Over 70 different types of HPV have been identified and a subset of these has been found in >90% of cervical cancers (Bosch et al. 1995). These high-risk HPV types include HPV16, HPV18, HPV31, and HPV33, and a number of additional related viruses. HPV16 DNA can be found in >50% of cervical cancers. Functional studies of the early region of high-risk HPV virus genomes have demonstrated that two early viral genes, E6 and E7, are both necessary and sufficient for the efficient immortalization of primary human keratinocytes in vitro (Hawley-Nelson et al. 1988; Münger et al. 1989; Hudson et al. 1990). The major mechanism by which E6 and E7 contribute to immortalization is by targeting two distinct cellular tumor suppressor proteins for inactivation or degradation. E7 binds and inactivates the retinoblastoma tumor suppressor protein (pRB) and two closely related proteins, p107 and p130, leading to the activation of E2F responsive genes and the loss of a G1 checkpoint (Jones and Münger 1996). E6 forms a ternary complex with p53 and the E6AP ubiquitin protein ligase resulting in the ubiquitination and degradation of p53 (Scheffner et al. 1990; Werness et al. 1990; Huibregtse et al. 1991, 1993a). Loss of p53 results in deregulated cellular growth and genomic instability, both of which are characteristics of immortalized cells (Hartwell 1992).
We have focused our studies on E6 because several lines of evidence suggest that the E6 protein retains functions in addition to its ability to target the ubiquitination of p53. For instance, whereas p53 deficient mice display normal lens development, transgenic mice expressing HPV16 E6 in the lens are impaired in the normal pattern of differentiation, which include fiber cell denucleation and apoptotic-like DNA degradation (Pan and Griep 1994). In addition, E6 can increase cellular telomerase activity in the absence of p53 degradation (Klingelhutz et al. 1996). Furthermore, it is highly likely that E6 has functions in addition to those that are revealed in transformation or differentiation assays. For instance, E6 can modulate the transcriptional activity of several cellular and viral promoters in both a p53 dependent and independent manner (Desaintes et al. 1992; Etscheid et al. 1994; Shirasawa et al. 1994; Shino et al. 1997). HPV16 E6 like BPV1 E6 can interact with the focal adhesion protein paxillin and, for BPV1 E6, this interaction has been shown to result in the disruption of the actin cytoskeleton (Tong and Howley 1997). Finally, E6 molecules have also been shown to interact with ERC 55, a putative calcium binding protein, although the physiologic consequence of this interaction is unclear (Chen et al. 1995).
Little is known about what regulates the PV life cycle or the antiviral response of squamous epithelial cells to a PV infection. An initial PV infection is associated with little or no inflammation, perhaps because of the lack of induction of cell death and release of viral antigen, or to viral interference with some specific aspect of the host immune response. There is little or no immune recognition of an early PV infection despite the ability of keratinocytes to serve as semiprofessional antigen presenting cells (Frazer 1996). DNA viruses have developed a variety of ways to overcome interferon (IFN) inhibitory effects and to evade host immunity (Vilcek and Sen 1996). To date however, the mechanisms by which the PVs may affect these pathways have not been elucidated.
Type I IFN production is stimulated early in the course of a viral infection, and IFN production is an important determinant of the course of the subsequent disease (De Maeyer and De Maeyer-Guignard 1988; Muller et al. 1994). IFNs act directly on the virally infected cell by interfering with viral replication and by inhibiting cellular proliferation. To carry out these functions, IFNs impinge on many mechanisms ranging from inhibition of viral penetration and uncoating, to reduction of mRNA stability and protein production. Immunomodulatory activities of IFNs also contribute to their antiviral roles. IFNs enhance the expression of cellular proteins such as MHC class I molecules that contribute to immune-mediated lysis of virus-infected cells (De Maeyer and De Maeyer-Guignard 1988). In addition, IFN production, stimulated by viral infection, is responsible for the activation and proliferation of natural killer cells, which act to lyse virus infected cells and to activate the immune system (Brutkiewiewicz and Welsh 1995).
To identify additional cellular targets of HPV16 E6 involved in various aspects of the viral pathogenic mechanism, we carried out a yeast two-hybrid screen. Two independent human cDNAs were identified that interacted with HPV-16 E6. One of these clones was interferon regulatory factor-3 (IRF-3) (Au et al. 1995). A structurally related group of transcription factors, the interferon α-stimulated gene factor (ISGF) or IRF family, has been implicated in the mediation of cellular responses to IFN and to a variety of other cytokines (Vilcek and Sen 1996). IRF-3 was initially identified by its homology to other IRF family members. It was characterized as a transcriptional activator that could bind to the interferon-stimulated response element (ISRE). Its antiviral and interferon signaling activities were initially unclear because IRF-3 mRNA levels were not inducible by IFN or viral infection (Au et al. 1995). Recent studies, however, have shown that IRF-3 is part of a virus activated transcription factor complex and that its transcriptional activity is increased in response to viral infection (Fujita et al. 1989; Schafer et al. 1998; Wathelet et al. 1998; Weaver et al. 1998).
IRF family members are believed to play a critical role in the regulated expression of the IFNα and β genes. IRF-1 in particular can activate transcription from the ISRE, which is present in the promoters of genes activated by IFN and virus infection. In addition, some members of the IRF family are involved in a variety of cellular growth control mechanisms. To date, seven human members of this family have been characterized: IRF-1, IRF-2, IRF-3, IRF-4, IRF-7, IFN consensus sequence binding protein (ICSBP), and ISGF3γ. IRF-1 has been characterized as a transcriptional activator and an antioncogene whose functional loss contributes to aberrant cellular growth (Fujita et al. 1989; Harada et al. 1990, 1993; Reis et al. 1992). The role of IRF-1 as a tumor suppressor is supported by the finding that IRF-1-deficient mouse embryonic fibroblasts readily undergo c-Ha-ras-induced transformation (Tanaka et al. 1994). IRF-2 can repress IRF-1-stimulated transcription and exhibits oncogenic activity (Harada et al. 1990, 1993). Recently, IRF-2 was also characterized as a transcriptional activator that can activate transcription of the human histone H4 gene in a cell-cycle-dependent manner (Vaughan et al. 1995). ICSBP expression is restricted to the immune system and ICSBP can interact with both IRF-1 and IRF-2 at the ISRE to suppress IFN-inducible gene transcription (Driggers et al. 1992; Nelson et al. 1993; Bovolenta et al. 1994). ISGF3γ (p48) is a positive regulator of IFNα-stimulated transcription and forms the ISGF3 complex together with the Stat1 and Stat2 proteins. IRF-4 is a B cell-specific factor that associates with PU.1 and binds to the light-chain gene enhancer (Levy et al. 1988; Eisenbeis et al. 1995). It is essential for B and T cell function and homeostasis (Mittrucker et al. 1997). IRF-7 can repress transcriptional activation by IFN and IRF-1 (Zhang and Pagano 1997). Each member of this family can be stimulated to bind DNA and activate or repress gene transcription upon treatment of cells with cytokines, growth factors, double-stranded RNA, or viral infection.
We have found that the E6 protein encoded by HPV16 can bind to IRF-3. IRF-3 can stimulate transcription from a luciferase reporter construct containing tandem ISRE sites, and as a fusion protein with the Gal4 DNA binding domain it can activate transcription from a chloramphenicol acetyltransferase (CAT) reporter plasmid containing five repeats of the Gal4 binding site. We show that HPV16 E6 does not target IRF-3 for degradation. The interaction of HPV16 E6 with IRF-3 is specific and results in a marked reduction of the IRF-3 transactivation function in vivo. Finally, HPV16 E6 expression in primary keratinocytes significantly dampens the induction of IFNβ mRNA after viral infection. These results suggest a novel function for E6 that may be relevant to the life cycle of the PV. The interaction of HPV16 E6 with IRF-3 and the inhibition of its transactivation function could contribute to the ability of the virus to disrupt the cellular antiviral response. Furthermore, it is possible that the interaction of E6 with IRF-3 could be related to the oncogenic potential of the virus, affecting either the regulation of cellular proliferation or apoptosis, or through a perturbation of the ability of the immune system to recognize an HPV16-infected cell.
Results
Identification of HPV16 E6-interacting proteins
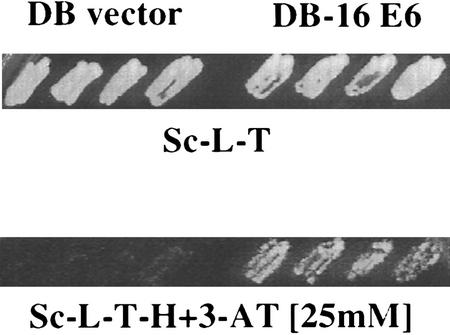
HPV16 E6 protein fused to the Gal4 DNA-binding domain (amino acids 1–147) was employed as the bait in a yeast two-hybrid screen to identify additional cellular proteins whose interactions may be important for E6 functions. A cDNA library from activated human T cells was screened (4 × 106 transformants) for interaction with the Gal4–HPV16 E6 fusion protein. Fifty-six independent colonies were selected for elevated expression of the HIS3 reporter gene on 3-aminotriazole (3AT)-containing plates. The corresponding Gal4-activation domain cDNA encoding plasmids were isolated and retransformed into fresh yeast cells. The interaction of these retransformed clones with HPV16 E6 was tested under a series of selection conditions. In addition to growth on 3AT (Fig. 1), transformants were also tested for β-galactosidase production and growth on uracil-deficient plates (data not shown). One of these cDNAs, found six times in the screen, was identified as IRF-3 by sequence analysis.
Figure 1.
A yeast two-hybrid system was used to identify proteins that interact with HPV16 E6. The four patches of cells on the left of each plate contain empty Gal4 DB vector, pPC97, and prey cDNA–AD vectors (AD–IRF-3). The four patches of cells on the right contain HPV16 E6–DB vector and AD–IRF-3. Patches of cells growing on plates selective for the presence of both plasmids (Sc-L-T-H) were replica plated onto plates lacking histidine and containing 25 mm 3AT (Sc-L-T-H+3AT).
IRF-3 interacts selectively with HPV16 E6
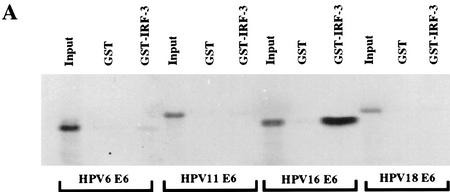
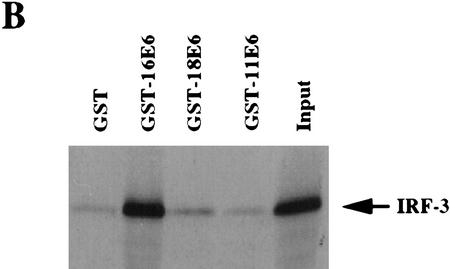
To further characterize the association of IRF-3 with HPV16 E6 and to determine whether the ability to interact with IRF-3 was shared among the E6 proteins encoded by other HPV types, we examined the in vitro interaction of full-length IRF-3 synthesized in Escherichia coli as a GST fusion protein (GST–IRF-3) with E6 proteins from both high and low-risk HPV types. HPV18, HPV16, HPV11, and HPV6 E6 were transcribed and translated in wheat germ extract and tested for interaction with GST–IRF-3. GST–IRF-3 interacted strongly with HPV16 E6, binding up to 41% of the input HPV16 E6 protein in a number of experiments. In these experiments, HPV6, HPV11, and HPV18 E6 proteins interacted poorly with IRF-3, exhibiting only 1%–2% binding in vitro (Fig. 2A). The reciprocal experiment was conducted by use of GST-18 E6, GST-16 E6, and GST-11 E6 proteins and in vitro-translated IRF-3. Strong binding of IRF-3 with GST–16 E6 (53% of input) and lesser binding with GST-18 E6 (5% of input) was detected. No binding was seen between GST-11 E6 and IRF-3 (Fig. 2B). These results indicate that HPV16 E6 has the highest affinity for interaction with IRF-3. The ability of HPV16 E6 to bind IRF-3 cannot be dependent on the cellular factor E6AP because E6AP is not present in wheat germ extract (Huibregtse et al. 1991). Furthermore, in the presence of HPV16 E6, IRF-3 was not brought into a complex with E6AP (data not shown). Finally, HPV16 E6 does not target IRF-3 for ubiquitin-mediated degradation in vitro (data not shown).
Figure 2.
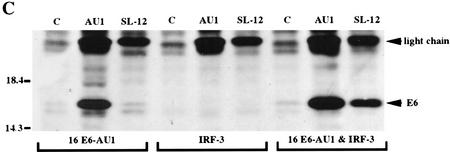
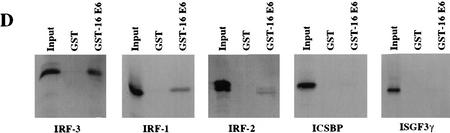
Analysis of the interactions between HPV E6 proteins and interferon regulatory factors (A) The indicated HPV E6 proteins were in vitro-translated in the presence of [35S]cysteine and methionine and mixed with GST–IRF-3 immobilized on glutathione–Sepharose. The complexes were washed to removed noninteracting proteins and resolved by PAGE. Input corresponds to 10% of protein used in the binding experiments. (B) IRF-3 was synthesized in rabbit reticulocyte lysate in the presence of [35S]cysteine and methionine and mixed with indicated GST alone or GST–E6 immobilized on glutathione–Sepharose. Complexes were washed and resolved by PAGE. Input corresponds to 50% of the protein used in the binding reactions. (C) COS-7 cells were electroporated with indicated constructs and after 36 hr, lysates were immunoprecipitated with control C, AU1, or IRF-3 (SL-12) monoclonal antibodies. HPV16 E6 and the light chain are indicated by arrows. (D) Indicated IRF proteins were synthesized in rabbit reticulocyte lysate in the presence of [35S]cysteine and methionine and mixed with either GST or GST–E6 immobilized on glutatione–Sepharose. Complexes were washed and resolved by PAGE. Input corresponds to 50% of protein.
To demonstrate an in vivo interaction of HPV16 E6 and human IRF-3, COS cells were transfected with plasmids expressing each protein individually or together. Figure 2C shows a coimmunoprecipitation of the AU1-tagged HPV16 E6 (Sherman and Schlegel 1996) and IRF-3 with the IRF-3-specific monoclonal antibody SL-12. SL-12 was raised against human IRF-3 and has been used previously to immunoprecipitate endogenous IRF-3 from human cells (Wathelet et al. 1998). SL-12 does not recognize monkey IRF-3 and does not detect IRF-3 in COS cells by Western analysis or by immunoprecipitation (data not shown). In the experiment depicted in Figure 2C, AU1-tagged HPV16 E6 was detected by Western analysis with the AU1 monoclonal antibody. Attempts to coimmunoprecipitate IRF-3 with the AU-1 antibody recognizing the tagged HPV16 E6 were not successful.
Because the members of the IRF transcription factor family share substantial sequence homology, we next examined whether HPV16 E6 could bind to other members of the IRF/ISGF family of proteins (Fig. 2D). IRF-1, IRF-2, ISGF3γ, and ICSBP were tested for interaction with GST-16 E6 in vitro. HPV16 E6 exhibited weaker interaction with in vitro-translated IRF-1 (binding 7.6% of input IRF-1) compared with IRF-3, and did not strongly interact with any other family members (Fig. 2D). GST-16 E6 bound 3%, 1.5%, and 2.5% of the input IRF-2, ICSBP, and ISGF3γ proteins, respectively. These results suggest that HPV16 E6 specifically interacted with only a subset of the members of the IRF family in vitro, and that the strongest interaction was with IRF-3.
Identification of the E6-binding region within IRF-3
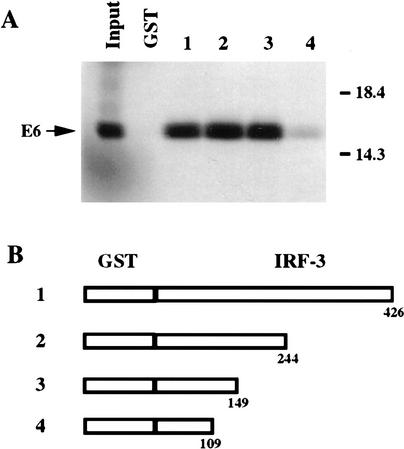
To map the domain of IRF-3 involved in binding to HPV16 E6, a series of carboxy-terminal deletion mutants of IRF-3 fused to GST were synthesized. Equal amounts of the GST fusion proteins (∼0.15 μg) were assayed for their abilities to bind HPV16 E6 by mixing the GST fusion proteins, immobilized on glutathione-Sepharose, with 35S-labeled, in vitro-translated HPV16 E6. Figure 3B shows schematics of the IRF-3 proteins that were tested for interaction with HPV16 E6. Full-length IRF-3 was found to bind 42% of the HPV16 E6 present in the reaction mixture (Fig. 3A, lane 1). The two IRF-3 proteins truncated at amino acids 244 and 149 bound 62% and 55% of the input E6, respectively (Fig. 3A, lanes 2 and 3). The amino-terminal portion of IRF-3 comprising amino acids 2–109 bound only 1% of the input E6 (Fig. 3A, lane 4). On close analysis, the portion of IRF-3 located between amino acids 109 and 149 was found to contain a stretch of amino acids (ELLG) that are present in the E6 binding domain of E6AP (Huibregtse et al. 1993b). This ELLG sequence has also been implicated as an E6 interaction domain by screening of a two-hybrid peptide library in which peptide sequences containing ELLG, or variants of it such as EFLG, ELVG, or DILG, were found to interact with HPV16 E6 (Elston et al. 1998).
Figure 3.
Mapping of the IRF-3 region that directs E6 binding. (A) Binding of HPV16 E6 to GST–IRF-3 proteins 1–5. HPV16 E6 was in vitro-translated in wheat germ extract in the presence of [35S]cysteine and methionine and mixed with the indicated GST–IRF-3 proteins as described previously. Input represents 50% of the E6 protein used in each assay. (B) Schematic representation of the GST-fused carboxy-terminal deletions and site-specific mutation containing indicated portions of IRF-3. (Open boxes) IRF-3 coding sequences.
HPV16 E6 expression does not promote the degradation of IRF-3 in human keratinocytes
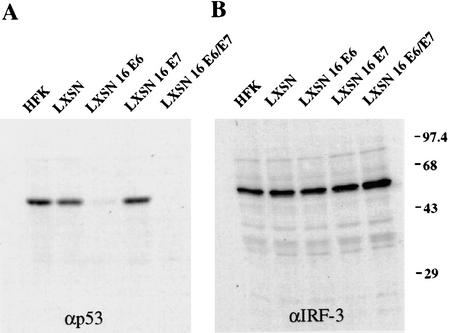
To determine whether interaction of IRF-3 with E6 resulted in its ubiquitination and degradation, primary neonatal human foreskin keratinocytes (HFKs) were isolated and infected with a recombinant retrovirus carrying individual HPV16 genes (Halbert et al. 1991). Western analyses to determine HPV16 E7, p53 and IRF-3 protein levels were conducted on lysates from HFKs infected with viruses expressing vector alone, HPV16 E6, E7, or E6 and E7. E7 expression was demonstrated in these cells by Western analysis (data not shown). Functional E6 production was determined by analysis of p53 protein levels. As expected, E6 expression led to a marked reduction in p53 protein levels (Fig. 4A); however, E6 did not affect the steady-state levels of IRF-3 (Fig. 4B). In addition, in vitro ubiquitination and degradation experiments were carried out and HPV16 E6 did not promote the ubiquitination or degradation of IRF-3 under conditions that led to the proteolysis of p53 (data not shown). Half-life determinations for p53 and IRF-3 were also conducted in HFK cells expressing the HPV viral oncoproteins. E6 expression resulted in a shortened half-life for p53 in agreement with previous reports, but had no effect on the half-life of IRF-3. The IRF-3 protein in HFK was long-lived with a half-life of >3 hr (data not shown).
Figure 4.
HPV16 E6 expression does not promote IRF-3 degradation in vivo. Immunoblot analysis of 100 μg of total protein lysates from HFK cells expressing HPV ORFs as indicated. (A) Western blot probed with p53-specific antibody, Ab6 (Calbiochem). (B) Western blot probed with IRF-3-specific monoclonal antibody, SL-12. High range molecular weight protein standards are indicated (GIBCO-BRL).
IRF-3 is a potent transcriptional activator
The initial published characterization of IRF-3 concluded that the protein did not contain a transcriptional activation domain (Au et al. 1995). In those experiments, the IRF-3 cDNA was fused in-frame to the DNA-binding domain of Gal4. Using a CAT reporter containing five Gal4 binding sites upstream of a minimal thymidine kinase promoter to assay Gal4–IRF-3 transactivation function, Au et al. (1995) found Gal4–IRF-3 to be devoid of intrinsic transactivational activity in the murine fibroblast cell line L929. We also constructed an IRF-3 Gal4 DNA-binding domain fusion protein and assayed it for transcriptional activation capacity. In contrast to the previously published results, our experiments showed Gal4–IRF-3 to be a potent transcriptional activator in each of several different cell types tested, including C33A and U2OS cell lines.
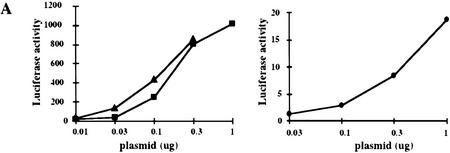
Increasing concentrations of a Gal4–IRF-3 expressing plasmid were cotransfected into L929 cells with either a CAT reporter containing five Gal4 binding sites (5Gal4–TKCAT) upstream of the thymidine kinase promoter, or a luciferase reporter containing five Gal4 binding sites upstream of a TATA box. Figure 5A shows representative luciferase assays comparing the transcriptional activity of Gal4–IRF-3 to that of Gal4–IRF-1 and Gal4–Stat 2. These results indicate that Gal4–IRF-3 is a 10- to 100-fold more potent transcriptional activator than Gal4–IRF-1 and possesses similar transcriptional activity as Gal4–Stat-2. Similar results were observed by use of a 5Gal4–CAT reporter.
Figure 5.
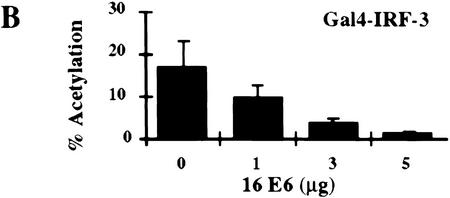
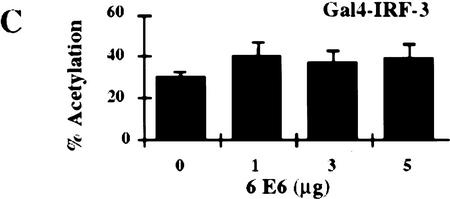
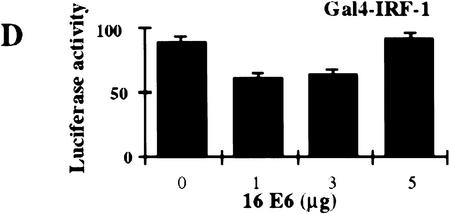
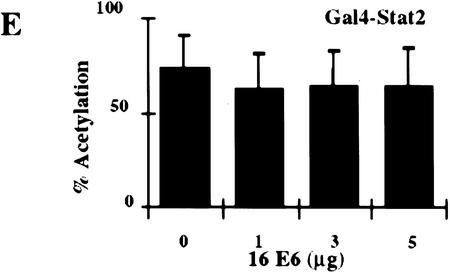
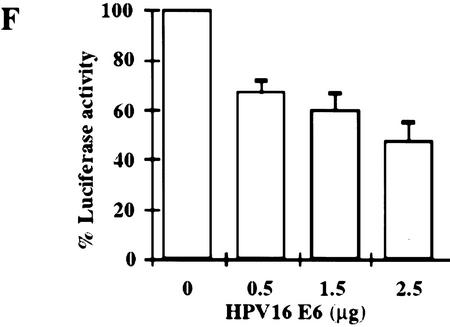
Stimulation of 5Gal4–TK–luciferase expression by Gal4–IRF-3, Gal4–IRF-1, and Gal4–Stat-2 and dose-dependent inhibition of IRF-3 transactivation by HPV16 E6. (A) L929 cells were cotransfected with 2 μg of the 5Gal4–TK–luciferase reporter construct and increasing amounts of Gal4–IRF-3 (▪), Gal4–Stat2 (▴), or Gal4–IRF-1 (•) expression plasmids. (B) L929 cells were cotransfected with 1 μg of SV40-β-gal, 2 μg of the 5Gal4–E1bTATA–CAT reporter (pG4BCAT) and 0.1 μg of Gal4–IRF-3 expression construct and increasing amounts of the HPV16 E6 expression plasmid p1436. (C) L929 cells were cotransfected with 1 μg of SV40–β-gal, 2 μg of the 5Gal4–TKCAT reporter, 0.1 μg of Gal4–IRF-3 expression construct and increasing amounts of p1436. (D) L929 cells were cotransfected with 1 μg of SV40–β-gal, 2 μg of the 5Gal4–TK-luciferase reporter, 1 μg of a Gal4–IRF-1 expression construct and increasing amounts of the HPV6 E6 expression plasmid p1478. (E) L929 cells were cotransfected with 1 μg of SV40–β-gal, 2 μg of the 5Gal4–TKCAT reporter, 0.1 μg of Gal4–Stat2 expression construct and increasing amounts of p1436. (F) C33A cells were cotransfected with 0.5 μg of SV40–β-gal, 0.25 μg of the ISG15–ISRE luciferase reporter, 0.5 μg of pCDNA3–IRF-3 and increasing amounts of p1436. The levels indicated were derived from duplicate samples within each experiment and are means from at least three independent experiments. CAT and luciferase activities were normalized to β-galactosidase expression levels. Error bars, standard error.
HPV16 E6 inhibits IRF-3 transactivation
Several studies have suggested that HPV16 E6 may be able to modulate transcription of certain cellular genes (Dey et al. 1997; Kinoshita et al. 1997; Shino et al. 1997). To examine what effect HPV16 E6 might have on IRF-3 function in vivo, we ascertained whether HPV16 E6 could influence IRF-3 transactivation. In this assay, HPV16 E6 and Gal4–IRF-3 were cotransfected into L929 cells with a Gal4–CAT reporter and a β-galactosidase indicator plasmid. Transfection of increasing concentrations of plasmid DNA expressing HPV16 E6 (p1436) (Münger et al. 1989) resulted in a dose-dependent inhibition of IRF-3 transactivation (Fig. 5B). Similar results were observed by use of a Gal4–luciferase reporter and in the cervical carcimona cell line C33A (data not shown). A reduction of >85% in the levels of IRF-3 transactivation was observed at the highest concentration of HPV16 E6 plasmid. In contrast, transfection with HPV6 E6 at similar plasmid concentrations did not impair IRF-3 transactivation (Fig. 5C). Because of the lack of sensitive antibodies to the E6 proteins, we were unable to measure and compare the HPV16 and HPV6 E6 protein levels in these experiments. However, we have confirmed expression from both constructs by Northern analysis (data not shown) and both constructs have been demonstrated to have activity in human mammary epithelial cell immortalization assays suggesting that both constructs encode a functional E6 protein (Band et al. 1993). The results presented here are consistent with our in vitro binding results that showed binding of HPV16 E6 but not of HPV6 E6 to IRF-3. Because a low level of HPV16 E6 binding to IRF-1 was observed in the GST binding studies (Fig 2C), we next determined what effect HPV16 E6 expression had on IRF-1 transactivation in vivo. Because Gal4–IRF-1 is a relatively weak transactivator, the more sensitive luciferase reporter system was used. Cotransfection of HPV16 E6 with Gal4–IRF-1 had no effect on IRF-1 transactivation (Fig. 5D). To further determine whether HPV16 E6 inhibition of IRF-3 transactivation was specific, experiments were conducted with Gal4–Stat2. HPV16 E6 had no effect on Stat-2 transactivation (Fig. 5E). Taken together, these results suggest that the ability to impair IRF-3 transactivation was specific for a high risk form of E6, and that HPV16 E6 does not have a general inhibitory effect on transactivation. Furthermore, the relatively weak interaction of HPV16 E6 with IRF-1 observed in vitro does not appear to be physiologically significant because HPV16 E6 had no effect on IRF-1 dependent transactivation in vivo.
We have demonstrated that HPV16 E6 can inhibit IRF-3 transactivation when IRF-3 binds DNA via the Gal4 DNA-binding domain. Next, we determined whether HPV16 E6 affected the transactivation capacity of IRF-3 acting on the ISRE. It was demonstrated previously that cotransfection of IRF-3 (not as a Gal4 fusion) with the ISG15 promoter inserted upstream of the CAT reporter gene resulted in a dose-dependent increase in CAT activity (Au et al. 1995). Therefore, we examined the activity of IRF-3 at a promoter containing three copies of the ISG15 ISRE. The HPV negative human cervical carcinoma cell line C33A was used for these experiments to demonstrate that this activity of HPV16 E6 is conserved in a human epithelial cell line. Because the p53 gene is mutated in C33A cells (Scheffner et al. 1991), these experiments also allowed us to determine the effect of HPV16 E6 on IRF-3 transactivation in the absence of wild-type p53. As seen with the Gal4–IRF-3 experiments, increasing concentrations of the HPV16 E6 plasmid resulted in a dose-dependent inhibition of IRF-3 transactivation at the ISRE (Fig. 5F). In these experiments, we consistently observed 40%–70% inhibition of IRF-3 transactivation at the highest transfected concentration of the HPV16 E6 expression plasmid. Comparable results were found in L929 and U2OS cells (data not shown).
E6 inhibits the viral induction of IFNβ mRNA
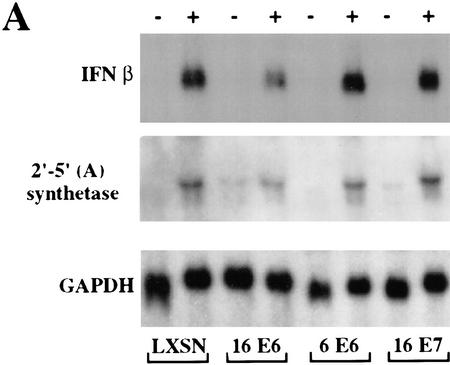
Wathelet et al. (1998) have demonstrated that viral infection leads to the phosphorylation and nuclear translocation of IRF-3 and its incorporation into a complex designated the virus-activated factor (VAF). Once activated, VAF binds to the ISRE-like elements in virus-inducible promoters and promotes transcription (Fujita et al. 1989; Schafer et al. 1998; Wathelet et al. 1998; Weaver et al. 1998). IFNβ then activates a complex antiviral cellular response. Therefore, we asked whether HPV16 E6 had an effect on the induction of IFNβ by virus infection. Primary human keratinocytes stably expressing HPV16 E6, HPV6 E6, or HPV16 E7 were infected with Sendai virus and the induction of IFNβ mRNA was determined by Northern analysis (Fig. 6A). The expression of HPV16 E6 resulted in a 44% inhibition of IFNβ mRNA production. Expression of HPV6 E6 or HPV16 E7 had no effect on the induction of IFNβ mRNA. IRF-3 has been found to be directly activated by viral infection resulting in the production of IFNβ. Type I IFN treatment of cells induces the production of a variety of IFN-inducible transcripts. The 2′–5′ (A) synthetase family of enzymes activates a latent ribonuclease that can cleave single-stranded RNAs (Vilcek and Sen 1996). To determine whether HPV16 E6 could affect the induction of the 2′–5′ (A) synthetase, the Northern blot was stripped and probed with the 2′–5′ (A) synthetase cDNA. In the presence of HPV16 E6, the 2′–5′ (A) synthetase mRNA was induced to 32% of the level found in cells expressing vector alone (LXSN). The induction of 2′–5′ (A) synthase mRNA by viral infection is indirect (Wathelet et al. 1992) and our results suggest that the inhibitory effect of HPV16 E6 on IFNβ production is physiologically significant in that it can impair the induction of secondary antiviral transcripts.
Figure 6.
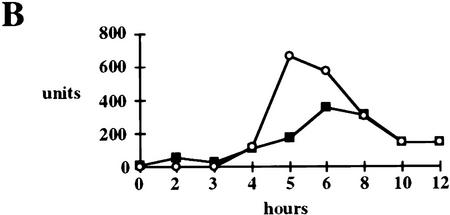
HPV16 E6 impairs virus induction of IFNβ. (A) A Northern blot, containing 7.5 μg of total cell RNA isolated from the indicated cell lines 5 hr after Sendai virus infection, was hybridized with an IFNβ-specific probe (top), with a 2′–5′ (A) specific probe (middle), or with a GAPDH probe (bottom). After hybridization and washing, filters were exposed to X-ray film for 18, 24, and 26 hr, respectively. (B) Northern blots, containing 7.5 μg of total cellular RNA isolated at indicated times after virus infection from HPV16 E6 (▪)- and HPV6 E6 (○)-expressing HKFs, were hybridized with an IFNβ-specific probe or with a GAPDH probe. Specific signals were quantitated by PhosphorImager analysis and the IFNβ levels were normalized to GAPDH levels at each time point.
To determine whether HPV16 E6 had an effect on the kinetics of IFNβ induction as well as the level of expression, we examined IFNβ expression in HFKs expressing HPV16 E6 or HPV6 E6 that had been infected with Sendai virus. Total cellular RNA was isolated at the indicted times and Northern blots were probed for IFNβ and GAPDH mRNAs (Fig. 6B). The levels of IFNβ indicated were normalized by the amount of GAPDH present at each time. Although HPV16 E6 did not have a significant effect on the timing of IFNβ mRNA induction, the extent of induction was impaired by 73% and 38% at 5 and 6 hr after Sendai virus infection.
HPV16 E6 expression does not affect the stability of IRF-3 in human keratinocytes after Sendai virus infection
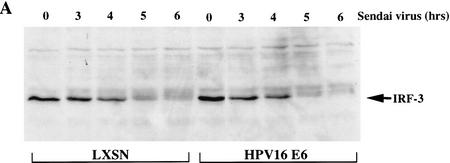
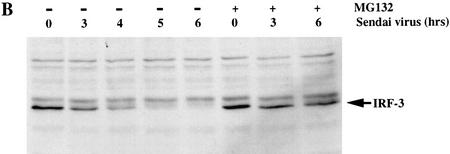
To determine whether the decrease in IRF-3 activity following Sendai virus infection resulted from the HPV16 E6 mediated degradation of IRF-3 protein, we examined the IRF-3 levels in HFK retrovirus vector control (LXSN) or HPV16 E6 expressing cells. Whole cell extracts harvested 0, 3, 4, 5, and 6 hr after Sendai virus infection were analyzed by Western blot with the SL-12 antibody to detect IRF-3 levels. Interestingly, in the parental HFKs as well as the LXSN control HKFs, IRF-3 protein levels dropped dramatically 5 hr after viral infection (Fig. 7A,B). Only a small amount of slower migrating IRF-3 protein could be detected at 5 and 6 hr after infection. IRF-3 is phosphorylated as a consequence of viral infection. It is possible that the slower migrating forms of IRF-3 apparent at 5 and 6 hr corresponded to phosphorylated IRF-3 (Fujita et al. 1989; Wathelet et al. 1998; Weaver et al. 1998). The levels of IRF-3 in HPV16 E6 expressing HFKs were similar to the levels in the LXSN control HFKs at each of the time points (Fig. 7A). To determine whether IRF-3 protein loss resulted from proteosome mediated degradation, parental HFK were treated with the proteosome inhibitor MG132 immediately prior to viral infection (Rock et al. 1994). MG132 stabilized IRF-3 after viral infection, indicating that the degradation of IRF-3 was proteosome mediated. Further experiments will be needed to address whether IRF-3 degradation involves ubiquitination.
Figure 7.
HPV16 E6 expression does not affect the stability of IRF-3 in human keratinocytes after Sendai virus infection. Immunoblot analyses of 100 μg of total protein lysates from HFK cells. (A) Western blot of HFKs expressing vector alone (LXSN) or HPV16 E6 as indicated probed with SL-12. (B) Western blot of the parental HFKs probed with SL-12. Cells were treated with MG132 (40 μm) immediately prior to virus infection. Total cell lysates were harvested at the times indicated after Sendai virus infection.
Discussion
In this study we have found that HPV16 E6 can interact with IRF-3 and inhibit its ability to transactivate. HPV16 E6 is required for the efficient immortalization of human keratinocytes and has been implicated in the initial steps of cellular transformation. IRF-3 was originally identified by its amino acid sequence homology to a family of structurally related transcription factors. The results presented here indicate that IRF-3 is a potent transcriptional activator and that the interaction of HPV16 E6 with IRF-3 inhibits this function in vivo. IRF-3 can activate transcription by binding to ISRE and ISRE-like elements in regulatory regions of genes activated by viral infection. IRF-3 has been shown recently to be an essential component of the VAF complex that transactivates the IFNβ promoter after viral infection (Fujita et al. 1989; Schafer et al. 1998; Wathelet et al. 1998; Weaver et al. 1998). We demonstrate here that E6 inhibition of IRF-3 transcriptional activity impairs the induction of IFNβ in response to viral infection. This represents the first description of a biochemical mechanism by which HPV modulates the antiviral activities of infected cells.
HPV16 E6 inhibited the transactivation of Gal4–IRF-3 by 85% at the highest E6 plasmid concentrations. The inhibitory effect of HPV16 E6 on intact IRF-3 transcriptional activity at the ISRE was more modest, ∼50%. Similar levels of inhibition by HPV16 E6 were seen on the cellular promoter for IFNβ where HPV16 E6 reduced the response to virus infection by 38% to 73% depending on the experiment and time after Sendai virus infection. Multiple signal transduction pathways lead to activation of transcription factors including NFκB, ATF-2, and c-Jun, which converge to affect the expression of the IFNβ gene (Thanos and Maniatis 1995; Kim and Maniatis 1997). In addition, after viral infection, IRF-3 becomes part of a large complex that includes IRF-7, p300, and pCBP. The partial inhibitory effect of HPV16 E6 on the IFNβ promoter may be caused by the complex regulatory pathways that control expression from the IFNβ promoter. As demonstrated by the impaired induction of 2′–5′ (A) synthetase, the inhibitory effect of HPV16 E6 on IRF-3 is sufficient to affect genes regulated by IFNβ, thereby modulating the cellular physiologic response to viral infection.
It has been shown previously that HPV16 E6 can inhibit the transactivation function of p53 (Mietz et al. 1992). That inhibition is presumably the result of the interaction of E6 with the E6AP ubiquitin protein ligase and the formation of a ternary complex with p53, which results in the ubiquitination and degradation of p53. However, the effect of E6 on IRF-3 does not involve its proteolysis. The IRF-3 half-life and steady-state protein levels were unaffected by the expression of HPV16 E6. We have mapped the region of IRF-3 that is involved in HPV16 E6 binding. This region contains a motif similar to the one found within the E6 binding domain of E6AP (Huibregtse et al. 1993; Elston et al. 1998). This suggests that E6 may bind directly to IRF-3, and furthermore suggests that E6AP is not required for the interaction. In support of this, we have found that IRF-3 is not part of a complex with E6AP in the presence or absence of E6 (data not shown). We found that IRF-3 is degraded 5–6 hr after viral infection and that HPV16 E6 expression does not affect this degradation. We have begun to address the mechanism of IRF-3 loss after viral infection. The stabilization of IRF-3 by the proteosome inhibitor MG132 suggests that IRF-3 is targeted for degradation in a proteosome-dependent manner. Interestingly, the timing of the attenuation of IFNβ mRNA production in response to virus infection in HFK mirrors the timing of the degradation of IRF-3 (Figs. 6B and 7A,B). These results suggest that the regulated degradation of IRF-3 may be responsible for shutting off the IFNβ response.
The list of transcription factors that can act as bifunctional regulators of transcription, by activating gene expression from some promoters while repressing others, is quite large. HPV E6 proteins have transcriptional-modulatory activities, some of which are p53 dependent and some are p53 independent (Lamberti et al. 1990; Sedman et al. 1991; Mietz et al. 1992). HPV16 E6 can increase cellular fibronectin gene expression (Shino et al. 1997). HPV16 E6 can also transactivate the prothymosin α, c-myc, and TGF-β1 promoters (Dey et al. 1997; Kinoshita et al. 1997). Furthermore, both high-risk and low-risk HPV E6 proteins can transactivate the adenovirus E2 promoter as well as a number of viral TATA-containing promoters in NIH-3T3 cells (Crook et al. 1991; Sedman et al. 1991; Desaintes et al. 1992). In contrast, HPV16 E6 can inhibit the activity of two viral promoters, the Molony murine leukemia virus LTR and the cytomegalovirus immediate early promoter (Etscheid et al. 1994). HPV16 E6 has been shown to localize to the nucleus as well as the cytoplasm, consistent with the studies implicating it with functions affecting transcription of specific genes (Androphy et al. 1987; Lechner et al. 1992). In this paper we demonstrate that HPV16 E6 interacts very strongly with IRF-3 in vitro whereas HPV18 E6 interacts only modestly with IRF-3. Similarly, in vitro-translated HPV18 E6 interacts less well with E6AP, however, HPV18 E6 does function in vivo to degrade p53. Experiments are ongoing to address whether HPV18 E6 can bind to IRF-3 in vivo and modulate its transcriptional activity.
IRF family members have been shown to be modulators of the cell cycle and of apoptosis, and may have functional similarities to p53. That IRF-1 can function as a tumor suppressor has been most clearly demonstrated in experiments that showed that embryonic fibroblasts from IRF-1 null mice could undergo transformation by the expression of c-Ha-ras alone (Tanaka et al. 1994). In addition, IRF-1 and p53 appear to cooperate in response to DNA damage and in the transcriptional activation of p21 (Tanaka et al. 1996). Both IRF-1 and p53 are essential for DNA damage-induced apoptosis in T lymphocytes and in embryonic fibroblasts (Lowe et al. 1993; Tanaka et al. 1994, 1996; Tamura et al. 1995). Through these studies, the regulators of the IFN pathway have been linked to cellular transformation and apoptosis. HPV16 E6 can functionally impair p53, and from the experiments presented here, E6 can interfere with the function of an IRF family member. It is tempting to speculate that in addition to diminishing the cellular response to viral infection, the ability of E6 to interfere with keratinocyte differentiation and its potential to avert an apoptotic signal in a p53 independent manner may reside in part in its ability to interact with and modulate the activity of IRF-3.
Materials and methods
Plasmids
The Gal4 DNA-binding domain in pSG424 (Sadowski and Ptashne 1989) was fused to full-length IRF-3 residues 2–427. The Gal4–Stat2 plasmid contained residues 670–851 (Bhattacharya et al. 1996). The Gal4–IRF-1 plasmid contained the full-length IRF-1 residues 1–325, IFNβ, and 2′–5′ (A) synthetase cDNA containing plasmids were provided by Marc Wathelet (Wathelet et al. 1992). The β-actin HPV16 E6 (p1436) and β-actin HPV6 E6 (p1478) plasmids were constructed by Karl Munger (Munger et al. 1989). GST–IRF-3 was constructed by cloning an EcoRI–NotI fragment from the yeast prey vector, pPC86 into pGex4T-1. Truncated forms of GST–IRF-3 were constructed by PCR cloning an EcoRI–XhoI fragment containing the indicated amino acids into pGex4T-1. Gal4–IRF-3 was constructed by cloning an EcoI–NotI fragment from the GST–IRF-3 construct into pSG424. The IRF-3 expression plasmid was cloned into pCDNA3 (Invitrogen) by PCR of the first 139 amino acids of IRF-3 containing an EcoRI–XmnI fragment and ligation to the remaining portion of IRF-3 (XmnI–NotI) derived from the yeast prey vector–IRF-3 plasmid. Plasmids used for in vitro translation of other IRF family members, IRF-1, IRF-2, ICSBP, and ISGF3 were kindly provided by Dr. K. Ozato (Bovolenta et al. 1994). Plasmids for in vitro translation of E6 proteins and GST–E6 plasmids have been desccribed previously (Werness et al. 1990; Huibregtse et al. 1993). The ISG15–ISRE luciferase reporter was constructed by insertion of a HindIII–SacI fragment containing three copies of the ISG15 ISRE upstream of the E1b TATA box into the G5luciferase (G5luc) reporter plasmid (Deng and Karin 1993). All constructs generated by PCR were confirmed by DNA sequencing. Plasmids used for transfections were purified by CsCl gradients two times.
Two-hybrid screening
A Gal4-based yeast two-hybrid screen was performed as described previously (Yasugi et al. 1997). The HPV16 E6 bait plasmid was constructed by cloning the full-length HPV16 E6 gene in-frame with the Gal4 DNA-binding domain (amino acids 1–147) in the pPC97 vector. For the library screen, an activated human T cell cDNA library, kindly provided by Dr. Joshua La Baer (MGH), cloned into the Gal4 activating domain (amino acids 768–881; pPC86), was transformed into the yeast host strain MaV103 (Mata ura3-53 leu2-3,112 trp1-901 hisΔ200 ade2-101 gal4gal80Δ GAL1::LacZ GAL1::HIS3 lys2 SPAL10;URA3) carrying the pPC97-16 E6 plasmid. Transformants were replica plated onto plates lacking histidine and with 3AT (synthetic complete medium [Sc]-L-T-H+3AT 30 mm). Potential interactors were picked from the 3AT-selective plates. The pPC86–cDNA plasmids were recovered and reintroduced into yeast MaV103 containing pPC97-16 E6. To confirm the interaction, transformants were plated onto Sc-L-T-H+3AT 30 mm plates, Sc-L-T-U plates, and Sc-L-T plates containing X-gal, a substrate for the lacZ-encoded enzyme.
In vitro binding assays
The HPV E6 in vitro-transcribed and -translated proteins were tested for association with IRF-3 proteins by mixing 7.5 μl of 35S-labeled wheat germ extract-translated HPV16, HPV18, HPV11, or HPV6 E6 and 10 μl of glutathione–Sepharose beads containing GST–IRF-3 protein. The mixture contained 125 μl of 25 mm Tris-HCl (pH 7.4), 50 mm NaCl and 25 μl of lysis buffer containing 0.1 m NaCl, 1% NP-40, and 0.1 m Tris (pH 7.4). The mixtures were rotated at 4°C for 4 hr. The beads were collected by centrifugation, washed three times with lysis buffer, boiled in SDS–gel loading buffer and electrophoresed on SDS–13% polyacrylamide gels. Gels were fixed, dried, and exposed to Kodak XAR film. Binding of in vitro-translated IRF proteins to GST and GST–E6 proteins was carried out in the same manner.
Immunoprecipitations
Electroporated COS-7 cells were washed with PBS, scrapped in PBS, and pelleted by centrifugation. Pellets were frozen in dry ice then resuspended in 4 volumes of lysis buffer [20 mm HEPES (pH 7.9), 0.2 mm EDTA, 0.2 mm EGTA, 10% glycerol, and the protease inhibitor cocktail (Pharmingen)]. KCl (2 m) was added to 400 mm final concentration, and the extracts were rotated at 4°C for 30 min and centrifuged at 12,000 rpm for 10 min. Lysates were precleared with 50 μl of a 50% slurry of protein A–agarose and protein G–agarose (1:1) and 1 μl of normal mouse serum in lysis buffer. The IP was conducted with ∼5 μg of an irrelevant mAb, AU1, or SL-12 mAb. Fifty microliters of a 50% slurry of protein A–agarose and protein G–agarose (1:1) was added to each lysate, and the suspension was rotated for 2 hr at 4°C. The protein A/G–agarose from each sample was washed three times at 4°C with 1 ml of lysis buffer, boiled in SDS-PAGE loading buffer containing DTT and analyzed by PAGE and autoradiography.
Northern analysis
RNA was harvested from HFK cells with Trizol (GIBCO-BRL). Five micrograms of total RNA was separated on a 1.1% agarose–formaldehyde gel and transferred to Hybond-N+ membrane. The membranes were sequentially hybridized with IFNβ, 2′–5′ (A), GAPDH, 16 E6 or 6 E6 radiolabeled probes. Gene specific signals on each Northern blot were quantified by PhosphorImager analysis (Molecular Dynamics).
Preparation of primary cell culture and infection by retroviral vectors
Primary keratinocytes were isolated from human neonatal foreskins by standard techniques. Briefly, foreskins were cut into several strips and were incubated in dispase (43.7 mg/ml) overnight. Epidermal layers were removed from the dermis and incubated in trypsin two times at 37°C for 15 min. Trypsin containing keratinocytes was removed, pooled, and inactivated by centrifugation through DMEM containing 10% fetal calf serum. Keratinocytes were maintained in serum-free medium supplemented with human-growth hormone and pituitary extract (GIBCO-BRL).
The amphotropic packaging cell line (PA317) was used to produce recombinant retroviruses LXSN, HPV16 E6, HPV6 E6, HPV16 E7, or HPV17 E6/E7 under the transcriptional control of the Moloney leukemia virus promoter–enhancer sequences (kindly provided by Dr. D. Galloway) (Miller and Rosman 1989; Halbert et al. 1991). The LXSN vectors contain the gene conferring neomycin resistance directed from the SV40 promoter. Recombinant virus was generated according to previously described procedures (Halbert et al. 1991). Viruses produced from PA317 cells were used to infect passage 2 human neonatal foreskin keratinocytes. Infected cells were placed under G418 (200 μg/ml) selection for 48 hr and then carried for 8 additional days until selection was complete. To determine expression of viral proteins, cells lysates were prepared in RIPA lysis buffer plus 0.01% PMSF, and 1 μg of aprotinin and leupeptin per milliliter. Lysates were cleared by centrifugation at 15,000g at 4°C for 5 min. One hundred micrograms of protein was separated by SDS-PAGE (12% polyacrylamide gel), and the levels of HPV16 E7, p53, and IRF-3 were determined by Western analysis as described (Dowhanick et al. 1995). The HPV16 E7 antibody was kindly provided by Dr. K. Münger (Harvard Medical School).
Monoclonal antibody preparation and Western blot analysis
Monoclonal antibody SL-12 was prepared by injecting mice four times with GST–IRF-3 (amino acids 56–427) protein. Serum samples from immunized mice were checked for antibody titers by assaying the efficiency with which they immunoprecipitated in vitro-translated IRF-3 protein. Spleen cells from the mouse displaying the best response were fused to NS-1 cells (Harlow and Lane 1988). Positive clones were identified by testing the ability of the hybridoma supernatants to immunoprecipitate in vitro-translated IRF-3, and by their efficiency to detect IRF-3 protein by Western analysis.
Cell culture, transfection, CAT, and luciferase assays
C33A, HeLa, L929, SL-12 hybridoma and SiHa cell lines were maintained in DMEM supplemented with 10% bovine calf serum (GIBCO). Sendai virus (SPAFAS) was used at 200 HAU/ml. MG132 was purchased from Peptides International.
For transient transfections, 60 mm plates of 50% confluent cells were transfected by the calcium phosphate procedure (Dowhanick et al. 1995). The DNA–calcium phospate precipitate was added to the culture and left on cells for 10 to 15 hr. The transfection cocktail contained 2 μg of reporter plasmid, 1 μg of SV40–β-gal (pSVβ; Clontech Laboratories, Inc.) and the indicated amounts of expression vectors. The total amount of DNA added to each plate was kept constant by including the appropriate amount of empty expression vector. Precipitate was removed, cells were washed twice with PBS, refed with complete media, and CAT assays were performed as described after 36–48 hr (Sakai et al. 1996). The percent acetylation was quantified by PhosphorImager (Dynamics) scanning of chromatography plates. Luciferase assays were conducted as described (Bhattacharya et al. 1996). Briefly, six-well plates were seeded with 2.5 E5 cells and the next day were transfected for 15–18 hr by the calcium-phosphate procedure. Luciferase assays were conducted 24 hr after the removal of the DNA/precipitate.
Acknowledgments
We thank Karl Münger, Grace Gill, Steve Finkel, Marc Wathelet, and Tom Maniatis for helpful discussions and critical reviews of the manuscript. We also thank James DeCaprio, Charles Ro, and Jianmin Gan for support in production of the monoclonal antibody and Ed Harlow for his constant support. L.V.R. was supported by grant 5 F32 AI09167-02 from the National Institute of Allergy and Infectious Diseases and by a grant from Aid For Cancer Research. A.Y.K. is a Howard Hughes Medical Institute Predoctoral Fellow. This research was supported by a grants from the National Institutes of Health (PO1-CA-50661-09 and PO1AI42257-01) to P.M.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL phowley@warren.med.harvard.edu: FAX (617) 432-2882.
References
- Androphy EJ, Hubbert NL, Schiller JT, Lowy DR. Identification of the HPV-16 E6 protein from transformed mouse cells and human cervical carcinoma cell lines. EMBO J. 1987;6:989–992. doi: 10.1002/j.1460-2075.1987.tb04849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Moore PA, Lowther W, Jaung YT, Pitha PM. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band V, Dalal S, Delmolino L, Androphy EJ. Enhanced degradation of p53 protein in HPV-6 and BPV-1 E6 immortalized human mammary epithelial cells. EMBO J. 1993;12:1847–1852. doi: 10.1002/j.1460-2075.1993.tb05833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston DM. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-α. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, Schiffman MH, Moreno V, Kurman R, Shah KV, Alihonou E, Bayo S, Mokhtar HC, Chicareon S, Daudt A, Delosrios E, Ghadirian P, Kitinya JN, Koulibaly M, Ngelangel C, Tintore LMP, Riosdalenz JL, Sarjadi-Schneider A, Tafur L, Teyssie AR, Rolon PA, Torroella M, Tapia AV, Wabinga HR, Zatonski W, Sylla B, Vizcaino P, Magnin D, Kaldor J, Greer C, Wheeler C. Prevalence of human papillomavirus in cervical cancer: A world-wide pespective. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- Bovolenta C, Driggers P, Marks M, Medin J, Politis A, Vogel S, Levy D, Sakaguchi K, Appella E, Coligan J, Ozato K. Molecular interactions between interferon consensus sequence-binding protein and members of the interferon regulatory factor family. Proc Natl Acad Sci. 1994;91:5046–5050. doi: 10.1073/pnas.91.11.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutkiewiewicz RR, Welsh RM. Major histocompatibility complex class I antigens and the control of viral infections by natural killer cells. J Virol. 1995;69:3967–3971. doi: 10.1128/jvi.69.7.3967-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Reid CE, Band V, Androphy EJ. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- Crook T, Tidy JA, Vousden KH. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 building and transactivation. Cell. 1991;67:547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- De Maeyer E, De Maeyer-Guignard J. Interferons and other regulatory cytokines. New York, NY: John Wiley and Sons; 1988. [Google Scholar]
- Deng T, Karin M. JunB differs from c-Jun in its DNA-binding and dimerization domains, and represses c-Jun by formation of inactive heterodimers. Genes & Dev. 1993;7:479–490. doi: 10.1101/gad.7.3.479. [DOI] [PubMed] [Google Scholar]
- Desaintes C, Hallez S, Van Alhen P, Burney A. Transcriptional activation of several heterologous promoters by the E6 protein of human papillomavirus type 16. J Virol. 1992;66:325–333. doi: 10.1128/jvi.66.1.325-333.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Atcha IA, Bagchi S. HPV16 E6 oncoprotein stimulates the transforming growth factor-β1 promoter in fibroblasts through a specific GC-rich sequence. Virology. 1997;228:190–199. doi: 10.1006/viro.1996.8363. [DOI] [PubMed] [Google Scholar]
- Dowhanick JJ, McBride AA, Howley PM. Suppression of cellular proliferation by the papillomavirus E2 protein. J Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggers PH, Elenbaas BA, An J-B, Lee IJ, Ozato K. Two upstream elements activate transcription of major histocompatibility complex class I gene in vitro. Nucleic Acids Res. 1992;20:2533–2540. doi: 10.1093/nar/20.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbeis C, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes & Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- Elston RC, Napthine S, Doorbar J. The identification of a conserved binding motif within human papillomavirus type 16 E6 binding peptides, E6AP, E6BP. J Gen Virol. 1998;79:371–374. doi: 10.1099/0022-1317-79-2-371. [DOI] [PubMed] [Google Scholar]
- Etscheid BG, Foster SA, Galloway DA. The E6 protein of Human Papillomavirus type 16 functions as a transcriptional repressor in a mechanism independent of the tumor suppressor protein, p53. Virology. 1994;205:583–585. doi: 10.1006/viro.1994.1684. [DOI] [PubMed] [Google Scholar]
- Frazer IH. Immunology of papillomavirus infection. Curr Opin Immunol. 1996;8:484–491. doi: 10.1016/s0952-7915(96)80035-5. [DOI] [PubMed] [Google Scholar]
- Fujita T, Kimura Y, Miyamoto M, Yamamoto H, Harada K, Ishihara M, Taniguchi T. Induction of endogenous IFN-α and IFN-β genes by a regulatory transcription factor, IRF-1. Nature. 1989;337:270–272. doi: 10.1038/337270a0. [DOI] [PubMed] [Google Scholar]
- Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type 1 IFN system in EC cells: Transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, Taniguchi T. Anti-oncogenic and oncogenic potentials of interferon regulatory factors -1 and -2. Science. 1993;259:971–974. doi: 10.1126/science.8438157. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor Laboratory, NY: Cold Spring Harbor Press; 1988. [Google Scholar]
- Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- Howley PM. Papillomavirinae: The viruses and their replication. In: Bernard DMK, Fields N, Howley PM, editors. Fundamental Virology. Philadelphia, PA: Lippincott-Raven; 1996. pp. 2045–20076. [Google Scholar]
- Hudson JB, Bedell MA, McCance DJ, Laimins LA. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. J Virol. 1990;64:519–526. doi: 10.1128/jvi.64.2.519-526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Cloning and expression of the cDNA for E6-AP: A protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993a;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Localization of the E6-AP regions that direct HPV E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993b;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DL, Münger K. Interactions of the human papillomavirus E7 protein with cell cycle regulators. Semin Cancer Biol. 1996;7:327–337. doi: 10.1006/scbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- Kim TK, Maniatis T. The mechanism of viral transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Shirasawa H, Shino Y, Moriya H, Desbarats L, Eilers M, Simizu B. Transactivation of prothymosin α and c-myc promoters by Human Papillomavirus type 16 E6 protein. Virology. 1997;232:53–61. doi: 10.1006/viro.1997.8536. [DOI] [PubMed] [Google Scholar]
- Klingelhutz AJ, Foster SA, McDougall JK. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–81. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- Lamberti C, Morrissey LC, Grossman SR, Androphy EJ. Transcriptional activation by the papillomavirus E6 zinc finger oncoprotein. EMBO J. 1990;9:1907–1913. doi: 10.1002/j.1460-2075.1990.tb08317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner MS, Mack DH, Finicle AB, Crook T, Vousden KH, Laimins LA. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J. 1992;11:3045–3052. doi: 10.1002/j.1460-2075.1992.tb05375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Kessler D, Pine R, Darnell J. Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative control. Genes & Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:847–849. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- Mietz JA, Unger T, Huibregtse JM, Howley PM. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 oncoprotein. EMBO J. 1992;11:5013–5020. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AD, Rosman GJ. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–987. [PMC free article] [PubMed] [Google Scholar]
- Mittrucker H-W, Matsuyama T, Grossman A, Kundig TM, Potter J, Shahinian A, Wakeman A, Patterson B, Ohashi PS, Mak TW. Requirment for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- Muller U, Steinhoff U, Reis LFL, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- Münger K, Phelps WC, Bubb V, Howley PM, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N, Marks M, Driggers P, Ozato K. Interferon consensus sequence binding protein, a member of the interferon regulatory factor family, suppresses interferon-induced gene transcription. Mol Cell Biol. 1993;13:588–599. doi: 10.1128/mcb.13.1.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Griep AE. Altered cell cycle regulation in the lens of HPV-16 E6 and E7 transgenic mice: Implications for tumor suppressor gene function in development. Genes & Dev. 1994;8:1285–1299. doi: 10.1101/gad.8.11.1285. [DOI] [PubMed] [Google Scholar]
- Reis L, Harada H, Wolchok J, Tanaguchi T, Vilcek J. Critical role of a common transcription factor, IRF-1, in the regulation if IFN-α and IFN-β inducible genes. EMBO J. 1992;11:185–193. doi: 10.1002/j.1460-2075.1992.tb05041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteosome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539–7540. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Yasugi T, Benson JD, Dowhanick JJ, Howley PM. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J Virol. 1996;70:1602–1611. doi: 10.1128/jvi.70.3.1602-1611.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer SL, Lin R, Moore PA, Hiscott J, Pitha PM. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Munger K, Byrne JC, Howley PM. The state of the p53 and retinoblastoma genes in human cervical carcinoma cell lines. Proc Natl Acad Sci. 1991;88:5523–5527. doi: 10.1073/pnas.88.13.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedman SA, Barbosa MS, Vass WC, Hubbert NL, Hass JA, Lowy DR, Schiller JT. The full-length E6 protein of human papillomavirus type 16 has transforming and trans-activating activities and cooperates with E7 to immortalize keratinocytes in culture. J Virol. 1991;65:4860–4866. doi: 10.1128/jvi.65.9.4860-4866.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman L, Schlegel R. Serum-and calcium-induced differentiation of human keratinocytes is inhibited by the E6 oncoprotein of human papillomavirus type 16. J Virol. 1996;70:3269–3279. doi: 10.1128/jvi.70.5.3269-3279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shino Y, Shirasawa H, Kinoshita T, Simizu B. Human papillomavirus type 16 E6 protein transcriptionally modulates fibronectin gene expression by induction of protein complexes binding to the cylclic AMP response element. J Virol. 1997;71:4310–4318. doi: 10.1128/jvi.71.6.4310-4318.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasawa H, Jin M, Shimizu K, Akutsu N, Shino Y, Simizu B. Transcription-modulatory activity of full-length E6 and E6*I proteins of Human Papillomavirus type 16. Virology. 1994;203:36–42. doi: 10.1006/viro.1994.1452. [DOI] [PubMed] [Google Scholar]
- Tamura T, Ishihara M, Lamphier MS, Tanaka N, Oishi I, Aizawa S, Matsuyama T, Mak TW, Taki S, Taniguchi T. An IRF-1-dependent pathway of DNA damage-induced apoptosis in mitogen-activated T lymphocytes. Nature. 1995;376:596–599. doi: 10.1038/376596a0. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Ishihara M, Kitagawa M, Harada H, Kimura T, Matsuyama T, Lampier M, Aizawa S, Mak T, Taniguchi T. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell. 1994;77:829–839. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Ishihara M, Lampier MS, Nozawa H, Matsuyama T, Mak TW, Aizawa S, Tokino T, Oren M, Taniguchi T. Cooperation of the tumor suppressors IRF-1 and p53 in response to DNA damage. Nature. 1996;382:816–818. doi: 10.1038/382816a0. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Tong X, Howley PM. The papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc Natl Acad Sci. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan PS, Aziz F, van Wijnen AJ, Wu S, Harada H, Taniguchi T, Soprano KJ, Stein JL, Stein GS. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377:362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- Vilcek J, Sen GS. Interferons and other cytokines. In: Fields BN, Howley P M, Knipe DM, editors. Fields virology. 3rd. ed. New York, NY: Raven Press; 1996. pp. 375–400. [Google Scholar]
- Wathelet M, Berr PM, Huez GA. Regulation of gene expression by cytokines and virus in human cells lacking the type-1 interferon locus. Eur J Biochem. 1992;206:901–910. doi: 10.1111/j.1432-1033.1992.tb16999.x. [DOI] [PubMed] [Google Scholar]
- Wathelet M, Lin CH, Parekh B, Ronco LV, Howley PM, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- Weaver BK, Prasanna Kumar K, Reich NC. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1386. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- Yasugi T, Benson JD, Sakai H, Vidal M, Howley PM. Mapping and characterization of the interaction domains of human papillomavirus type 16 E1 and E2 proteins. J Virol. 1997;71:891–899. doi: 10.1128/jvi.71.2.891-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Pagano JS. IRF-7, a new interferon regulatory factor associated with epstein-barr virus latency. Mol Cell Biol. 1997;17:5748–5757. doi: 10.1128/mcb.17.10.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]