Abstract
The commitment of multipotent cells to particular developmental pathways requires specific changes in their transcription factor complement to generate the patterns of gene expression characteristic of specialized cell types. We have studied the role of the GATA cofactor Friend of GATA (FOG) in the differentiation of avian multipotent hematopoietic progenitors. We found that multipotent cells express high levels of FOG mRNA, which were rapidly down-regulated upon their C/EBPβ-mediated commitment to the eosinophil lineage. Expression of FOG in eosinophils led to a loss of eosinophil markers and the acquisition of a multipotent phenotype, and constitutive expression of FOG in multipotent progenitors blocked activation of eosinophil-specific gene expression by C/EBPβ. Our results show that FOG is a repressor of the eosinophil lineage, and that C/EBP-mediated down-regulation of FOG is a critical step in eosinophil lineage commitment. Furthermore, our results indicate that maintenance of a multipotent state in hematopoiesis is achieved through cooperation between FOG and GATA-1. We present a model in which C/EBPβ induces eosinophil differentiation by the coordinate direct activation of eosinophil-specific promoters and the removal of FOG, a promoter of multipotency as well as a repressor of eosinophil gene expression.
Keywords: C/EBP, FOG, GATA, multipotent progenitor, lineage commitment, eosinophil
In the vertebrate hematopoietic system, the generation of the different blood cell types from hematopoietic stem cells is ongoing throughout life. This process is controlled by an intricate network of transcription factors that both regulate cellular decisions and control the expression of genes characteristic of the different lineages. Each lineage contains a specific complement of transcription factors that generate the gene expression pattern characteristic of the cell type. Elucidating the mechanisms by which these expression patterns are generated and maintained is crucial to understanding how the diverse cell types of the hematopoietic system are formed. Myeloid cell types (neutrophil granulocytes and macrophages) are characterized by the expression of PU.1 and C/EBP transcription factors, which regulate most myeloid-specific promoters (for review, see Tenen et al. 1997), as well as the absence of GATA-1 expression. The latter is important, as GATA-1 is a negative regulator of PU.1 activity (Zhang et al. 1999; Nerlov et al. 2000), and the ectopic expression of GATA-1 in myeloid cells leads to their loss of myeloid gene expression and acquisition of a thrombocytic/multipotent (at high GATA-1 levels) or eosinophil phenotype (at intermediate GATA-1 levels) (Visvader et al. 1992; Kulessa et al. 1995). The eosinophil lineage is characterized by simultaneous expression of GATA-1 and C/EBP factors, and functionally important binding sites for these factors are found in the eosinophil-specific EOS47 and major basic protein promoters (McNagny et al. 1998; Yamaguchi et al. 1999). The requirement of C/EBPα, C/EBPβ, and PU.1 for the correct development and function of neutrophil granulocytes and macrophages has been demonstrated by knockout experiments in mice (Scott et al. 1994; Tanaka et al. 1995; McKerscher et al. 1996; Zhang et al. 1997). In addition, mice lacking the C/EBPα transcription factor have no mature eosinophils (Zhang et al. 1997); so far, the requirement for C/EBPβ or GATA-1 during eosinophil differentiation has not been addressed genetically.
To address the role of PU.1 and C/EBP proteins in hematopoietic lineage commitment, we have taken advantage of an in vitro differentiation system based on the transformation of chicken blood island cells by the E26 leukemia virus, which encodes the Gag–Myb–Ets fusion protein. This system yields clonal multipotent progenitor populations (called MEPs for Myb–Ets progenitors) capable of differentiation along the thrombocytic, erythroid, eosinophil, and granulocyte/macrophage lineages (Graf et al. 1992; Kraut et al. 1994; Frampton et al. 1995; Rossi et al. 1996b). Consistent with the genetic data, in vitro myeloid and eosinophil lineage commitment of MEPs can be achieved through forced expression of PU.1 and C/EBP, respectively (Müller et al. 1995; Nerlov and Graf 1998; Nerlov et al. 1998). In the case of PU.1, myeloid lineage commitment correlates with the ability of PU.1 to down-regulate GATA-1 expression (Nerlov and Graf 1998), whereas the molecular mechanism by which C/EBP induces the differentiation of MEPs into eosinophils is unclear.
MEPs, as well as their counterpart multipotent myeloid/erythroid progenitors isolated from chicken bone marrow, are characterized by their expression of the MEP21 and gpIIb-IIIa antigens and lack of myeloid/eosinophil lineage-specific markers (Graf et al. 1992; Frampton et al. 1995; McNagny et al. 1997; Ody et al. 1999). The MEP21 and gpIIb-IIIa antigens are also highly expressed on cells of the thrombocytic lineage, and MEPs resemble thromboblasts. The recent finding that the GATA-1 cofactor Friend of GATA (FOG) is required for the development of the thrombocytic lineage at a very early stage (Tsang et al. 1998) led us to examine the role of FOG in the generation of MEPs and their differentiation. The FOG protein contains nine zinc fingers, at least two of which are capable of binding to the amino-terminal zinc finger (NF) of GATA-1 (Tsang et al. 1997; Fox et al. 1998, 1999; Crispino et al. 1999). This interaction is necessary for the ability of GATA-1 to promote terminal erythroid differentiation (Crispino et al. 1999), and GATA-1 and FOG cooperate to induce a megakaryocytic phenotype in the 416B myeloid cell line (Tsang et al. 1997).
Here we find that MEPs express high levels of FOG mRNA, whereas their myeloid and eosinophil derivatives express very low levels. Induction of eosinophil differentiation of MEPs through activation of a conditional allele of C/EBPβ leads to loss of FOG expression as well as MEP markers, and up-regulation of EOS47, an eosinophil-specific surface antigen. EOS47 induction was prevented by constitutive expression of FOG in MEPs prior to induction of C/EBPβ, demonstrating that loss of FOG expression is a prerequisite for eosinophil gene expression. FOG blocks the activation of the EOS47 promoter by GATA-1. Finally, ectopic expression of FOG in the 1A1 eosinophil cell line led to down-regulation of EOS47, Mim-1, and C/EBPβ expression and up-regulation of MEP markers. These results show that modulation of GATA-1 activity through removal of FOG provides a developmental switch in hematopoiesis, and indicate that this is part of the mechanism by which C/EBPβ induces eosinophil differentiation.
Results
The GATA-1 N-finger is required for reprogramming of myeloid cells into MEPs
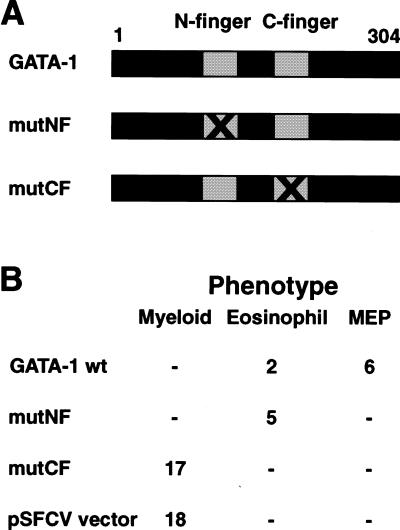
The ability of GATA-1 to reprogram HD50M myeloblasts into eosinophils and MEPs (Kulessa et al. 1995) provides an assay for the function of GATA-1 domains in the establishment of these cell types and was used to test the role of the highly conserved zinc fingers of GATA-1. Expression vectors encoding the neomycin resistance gene and wild-type GATA-1 (pNEO–GATA-1) or derivatives in which the amino-terminal zinc finger (mutNF) or carboxy-terminal zinc finger (mutCF) had been mutated by replacement of two of the zinc-coordinating cysteines with alanine (Fig. 1A), as well as the empty pSFCV vector, were transfected into HD50M myeloblasts. The phenotype of the resulting neomycin-resistant clones was determined by indirect immunofluorescence and flow cytometry (IF/FC) analysis of lineage-specific antigen expression (MEP21 for MEPs, EOS47 for eosinophils, and MYL51/2 for myeloblasts). The results are shown in Figure 1B. Wild-type GATA-1 expression converted the HD50M cells into either MEPs or eosinophils, as reported previously (Kulessa et al. 1995). Mutation of the CF (leading to loss of GATA-1 DNA-binding activity) completely abolished the abilty of GATA-1 to reprogram HD50M cells. Interestingly, although mutation of the NF did not impair the reprogramming into eosinophils, no MEP clones were observed, suggesting that a function harbored in the NF was required for establishment of the MEP phenotype. The NF is dispensable for specific DNA binding, but has been shown to bind the FOG protein (Tsang et al. 1997). Therefore, we tested whether the mutation introduced into this finger abolished interaction between GATA-1 and FOG in GST pull-down assays, and found that mutation of the NF blocked the GATA-1–FOG interaction (data not shown). This was consistent with observations published previously (Tsang et al. 1997) and indicated a role for FOG in the GATA-1-mediated conversion of myeloid cells to a multipotent phenotype. Although the up-regulation of endogenous GATA-1 in reprogrammed myeloblasts (Kulessa et al. 1995) prevents determination of the expression levels of the exogenously supplied GATA-1 proteins, we have found previously that mutations in the NF and CF of GATA-1 do not affect protein stability (Nerlov et al. 2000). Therefore, we do not believe that this is a factor in the effects observed by mutation of GATA-1.
Figure 1.
Effect of zinc finger mutations on myeloblast reprogramming by GATA-1. (A) Schematic drawing of GATA-1 and the two mutants with disrupted Zn-fingers. (B) Phenotypes of HD50M clones transfected with pNEO–GATA-1 or derivatives carrying mutations in either of the GATA-1 zinc fingers. The clones were picked, expanded, and characterized by IIF using monoclonal antibodies against lineage-specific surface antigens. The number of clones with MEP, eosinophil, and myeloblast phenotype is indicated.
MEPs, but not eosinophils or myeloid cells, express high levels of FOG
The expression of FOG in chicken hematopoietic cell types was determined. First, we cloned a partial chicken FOG cDNA by PCR from an MEP cDNA library and used it to probe a Northern blot of erythroid, multipotent, eosinophilic, and myeloid cell lines (Fig. 2). The result showed the highest expression of FOG in the HD50 MEP cell line. Significant FOG expression was seen in HD3 and HD37 erythroid cells, whereas FOG mRNA was very low or absent in 1A1 and HD50/4.8E eosinophils, as well as HD57M and HD50/4.8M myeloblasts. The FOG expression pattern was thus consistent with a role for FOG in establishing the MEP phenotype.
Figure 2.
Expression of FOG in chicken hematopoietic cell lines. A Northern blot was prepared from poly(A)+ RNA of the following chicken cell lines: HD3, HD37 (erythroid); HD50 (MEP); 1A1, HD50/4.8E (eosinophil); HD50/4.8M and HD57M (myeloid). The blot was probed sequentially with 32P-labeled chicken FOG cDNA (top) and chicken GAPDH cDNA (bottom) and signals detected by PhosphorImaging. The positions of the bands corresponding to the two mRNAs are indicated by arrows.
FOG induces eosinophils to acquire a multipotent phenotype
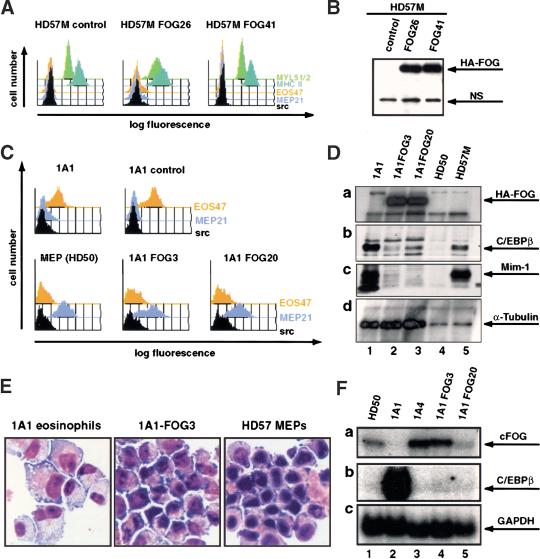
To address the role of cooperation between GATA-1 and FOG in establishment of the multipotent MEP phenotype, we expressed FOG in 1A1 eosinophils (which contain GATA-1, but no FOG) and in HD57M myeloblasts (which express neither GATA-1 nor FOG). The two cell lines were transfected with the pEF–HAFOG–PGKpuro vector, expressing murine FOG tagged with the HA epitope and conferring puromycin resistance, or the corresponding empty vector (pEF–HA–PGKpuro). The phenotype of puromycin-resistant clones was determined by IF/FC as in Figure 1. Whereas no effect of FOG expression was observed in the HD57M myeloblasts (Fig. 3A,B), FOG-expressing 1A1 clones showed a dramatic change in phenotype compared with control clones, as exemplified by the 1A1–FOG3 and 1A1–FOG20 clones (Fig. 3C); they lost expression of the eosinophil-specific EOS47 antigen and expressed the MEP marker MEP21 (Fig. 3C). In addition, they had down-regulated expression of C/EBPβ compared with the parental 1A1 cells, had lost expression of the myeloid/eosinophil-specific Mim-1 protein (Fig. 3D), and acquired an MEP morphology (Fig. 3E). The MEP26 MEP marker was also up-regulated, whereas no expression of the MYL51/2 and MHC II myeloid antigens was observed (data not shown). These data indicated that FOG expression is incompatible with eosinophil gene expression and leads to a conversion of 1A1 eosinophils to an MEP phenotype. Because the 1A1 cell line was derived from myeloblasts through enforced GATA-1 expression (Kulessa et al. 1995) this, along with the failure of GATA-1mutNF to induce MEP formation in myeloblasts, further supported the notion that cooperation between GATA-1 and FOG is important for the generation of MEPs. The ability of wild-type GATA-1 to directly induce the conversion of myeloid cells to an MEP phenotype in the absence of exogenously supplied FOG therefore raised the question of whether this involved up-regulation of endogenous FOG expression. We compared the expression of endogenous FOG in HD50 MEPs with that in 1A1 eosinophils, 1A4 MEPs (obtained by ectopic GATA-1 expression in myeloid cells; Kulessa et al. 1995), and the 1A1–FOG3 and 1A1–FOG20 clones by Northern blot analysis (Fig. 3F). This analysis showed that MEPs obtained both by GATA-1 overexpression in myeloid cells and by FOG expression in eosinophils expressed endogenous FOG at a level similar to that of the HD50 MEP cell line, and had down-regulated expression of C/EBPβ. The up-regulation of endogenous FOG expression was thus a hallmark of the MEP phenotype. Finally, these results also indicated that induction of an MEP phenotype could be obtained through cooperation between GATA-1 and endogenous FOG.
Figure 3.
Ectopic expression of FOG in the myeloid cell line HD57M and the eosinophilic cell line 1A1. (A) Phenotype of FOG expressing myeloblast cell lines determined by IF/FC. The HD57M parental cell line is compared with the FOGexpressing clones HD57M FOG26 and HD57M FOG41. The graphs show the fluorescence intensity (log scale) obtained after staining with the indicated monoclonal antibodies plotted against the cell number (linear scale). (B) Western blot of lysates from the cell lines analyzed in A. Equal amounts of cellular protein were run on a 7.5% SDS–polyacrylamide gel and Western blotting was performed with the 12CA5 monoclonal antibody, detecting the HA tag on the FOG protein. The band corresponding to FOG is indicated. A nonspecific band (NS) served as a control for equal loading. (C) The surface antigen expression on the 1A1 eosinophil cell line, a 1A1 cell line stably transfected with the pEF–HA–PGKpuro vector (1A1 control), the HD50 MEP cell line, and two FOGexpressing 1A1 clones (1A1–FOG3 and 1A1–FOG20) was analyzed as in A. (D) Western blot of lysates from the cell lines analyzed in C. Proteins were separated by 10% SDS-PAGE and a Western blot sequentially probed with the following antibodies: 12CA5 (anti-HA-tag; a), anti-chicken C/EBPβ (b), anti-chicken Mim-1 (c), and anti-α-tubulin (d). The arrows indicate the positions of the bands corresponding to the antigens. (E) May-Gruenwald-Giemsa staining of 1A1 eosinophils, 1A1–FOG3 cells, and HD57 MEPs, as indicated. (F) Expression of endogenous FOG in MEPs derived by ectopic GATA-1 and FOG expression. A Northern blot of total RNA from HD50 MEPs, 1A1 eosinophils, 1A4 MEPs, and the 1A1–FOG3 and 1A1–FOG20 cell lines was sequentially probed with 32P-labeled cDNA for chicken FOG (a), chicken C/EBPβ (b), and chicken GAPDH (c), and signals detected by PhosphorImaging. The positions of the bands corresponding to the mRNAs are indicated by arrows.
FOG represses the eosinophil-specific EOS47 promoter
The down-regulation of eosinophil-specific genes upon expression of FOG in 1A1 cells led us to examine the molecular mechanism involved. The EOS47 promoter is a well-characterized eosinophil-specific promoter that is cooperatively activated by C/EBP and GATA-1, as well as Myb and Ets-1/Fli-1, (McNagny et al. 1998; Fig. 4A). We tested the wild-type and mutNF alleles of GATA-1 for their ability to activate the EOS47 promoter, as well as their subsequent repression by FOG, in a transient cotransfection experiment in Q2bn fibroblasts (Fig. 4B). This analysis showed that both wild type and GATA-1mutNF could activate the EOS47 promoter, although the latter was somewhat less efficient (see Discussion). Simultaneous expression of FOG repressed activation by wild type, but not mutNF GATA-1, which indicated that FOG expression leads to repression of the EOS47 promoter, most likely through interaction with the GATA-1 NF. To test whether this was also true on the endogenous EOS47 promoter, we used HD57M myeloblasts expressing a fusion between GATA-1 and the hormone-binding domain of the human estrogen receptor (GATA–ER). These cells have a normal myeloblast phenotype in the absence of β-estradiol (βE), but when estrogen is added, myeloid antigens are down-regulated and the eosinophil-specific EOS47 antigen is expressed (Kulessa et al. 1995; Nerlov et al. 2000). HD57M-GATA–ER cells were transfected with the pEF–HAFOG–PGKpuro expression vector and the resulting puromycin-resistant clones were analyzed for FOG expression by Western blotting (Fig. 4C). Four FOG-expressing clones, as well as a resistant control clone expressing no FOG protein, were chosen for further analysis. As expected from the above results, FOG expression induced no change in lineage-specific antigen expression compared with the parental HD57M–GATA–ER cells in the absence of βE (Fig. 4D; top). However, upon βE activation of the GATA-1–ER fusion protein, expression of FOG blocked the induction of EOS47 expression, whereas down-regulation of the myeloid MYL51/2 and MHC II antigen was still observed (Fig. 4D, bottom; data not shown). HD57M–GATA–ER clones with high FOG expression (FOG7, FOG15) showed a lesser degree of EOS47 up-regulation than a clone with lower FOG levels (FOG8) (Fig. 4E; EOS47 up-regulation given as a percent of that observed in the parental HD57M–GATA–ER cells) indicating a correlation between repression of the EOS47 promoter and FOG expression. The nonexpressing FOG2 control clone behaved indistinguishably from the parental HD57–GATA–ER cells. Therefore, these experiments indicate that FOG is able to repress the activity of the eosinophil-specific EOS47 promoter, most likely through direct interaction with GATA-1 bound to the promoter.
Figure 4.
FOG represses the eosinophil-specific EOS47 promoter through the NF of GATA-1. (A) Schematic representation of the positions of transcription factor binding sites present in the chicken EOS47 promoter (from McNagny et al. 1998). (B) Activation of the EOS47 promoter by GATA-1 and repression by FOG. The EOS47/-152-LUC reporter (1.0 μg) was cotransfected with pRSV–βGal (0.25 μg) into Q2bn fibroblasts. Expression vectors for Ets-1 (pCRNCM–cEts-1, 0.1 μg), c-Myb (pCRNCM–cMyb, 0.1μg), GATA-1 (pSPCMV–GATA-1, 0.3 μg), mutNF GATA-1 (pSPCMV–GATA-1mutNF, 0.3 μg), FOG (pEF–HAFOG–EFpuro, 0.5 μg), or equivalent amounts of the corresponding empty vectors (pCRNCM; pSPCMV; pEF–HA–PGKpuro, lanes indicated with −) were added as indicated. After 48 hr, luciferase and β-galactosidase activities were measured, and the luciferase activity normalized to the β-galactosidase activity. The basal promoter activity was arbitrarily assigned a value of one. Three independent transfections were carried out for each effector combination and standard deviations are indicated by the error bars. (C) The HD57–GATA–ER cell line was transfected with the pEF–HAFOG–PGKpuro expression vector and stable clones expressing FOG identified (HD57–GATA–ER FOG7, FOG8, FOG15, and FOG16). These, as well as a control nonexpressing clone (FOG2), were subjected to Western blot analysis as in Fig. 3B. Note that the expression level of clone 8 (lane 3) is lower than that of the other HA–FOG-expressing clones (the nonspecific band [NS] served as a control for equal loading). (D) HD57M GATA-1–ER clones not expressing (parental control and FOG2 clone) or expressing FOG (FOG7, FOG8, FOG15, and FOG16 clones) were induced with 0.1 μM βE for 36 hr (bottom) or left uninduced (top). Cells were analyzed for expression of the indicated antigens as in Fig. 3A. (E) The up-regulation of EOS47 antigen expression in D is given as a percent of that observed in the parental HD57M GATA–ER clone.
FOG is down-regulated during C/EBPβ-mediated eosinophil lineage commitment of multipotent progenitors
The differentiation of MEPs along the eosinophil lineage can be efficiently induced by the C/EBPβ transcription factor (Müller et al. 1995; Nerlov et al. 1998). Therefore, we tested the effect of C/EBPβ on FOG expression in MEPs. HD57 MEP cells expressing an estrogen-inducible C/EBPβ allele (C/EBPβ–ER clone 3 (previously designated HD57–NF-M–ER clone 3; Müller et al. 1995), as well as nonexpressing HD57 control cells (clone 12), were subjected to βE treatment and their expression of MEP21 antigen, EOS47 antigen (Fig. 5A), and FOG and GATA-1 mRNA (Fig. 5B) determined by IF/FC and Northern blotting, respectively. The result showed no change in MEP21, EOS47, GATA-1, or FOG expression in the control clone 12 cells, whereas FOG mRNA was rapidly down-regulated in C/EBPβ–ER-expressing clone 3 cells, preceding up-regulation of EOS47 expression. The expression of GATA-1 was not altered by C/EBPβ–ER activation during this time course (Fig. 5B). To correlate the down-regulation of FOG expression to eosinophil lineage commitment, we generated pools of primary MEPs transformed by the E26–βER virus expressing the C/EBPβ–ER fusion (Nerlov et al. 1998), as well as control MEPs transformed by wild-type E26 virus. These pools were subjected to βE treatment. After 0, 1, 2, and 3 d, two aliquots were removed. One was used for preparation of total RNA for Northern analysis, and one was washed extensively in βE-free medium and replated in the absence of βE. After a total of 6 d in culture, the replated cells were analyzed for EOS47 expression to determine the degree of eosinophil lineage commitment at the time of βE removal (Fig. 5C). FOG mRNA down-regulation was observed after 1 d of C/EBPβ–ER induction, and was complete after 3 d (Fig. 5D). Concomitant with FOG down-regulation, we observed the up-regulation of mim-1 expression, which is an eosinophil/myeloid-specific marker (Kulessa et al. 1995). In contrast, GATA-1 expression remained constant over this time course. The slower kinetics of FOG mRNA down-regulation compared with the HD57–C/EBPβ–ER cell line may reflect the somewhat lower levels of C/EBPβ–ER in the primary MEPs. Eosinophil lineage commitment was significant after two days and most cells were committed after three days. FOG mRNA down-regulation thus coincided with eosinophil lineage commitment, consistent with removal of FOG being a prerequisite for eosinophil differentiation to take place.
Figure 5.
Down-regulation of FOG mRNA during eosinophil differentiation. (A) Cells from the C/EBPβ–ER-expressing HD57 clone 3, and the nonexpressing HD57 clone 12 were induced with 1.0 μM β-estradiol, and EOS47 expression determined by IIF after 1, 2, and 3 d of induction. Expression is given as percent of antigen-positive cells. No EOS47 expression was observed in the absence of βE in either clone (data not shown). (B) A Northern blot of total cellular RNA harvested at the timepoints in A was hybridized sequentially to probes for chicken FOG, chicken GATA-1, and chicken GAPDH. Results were analyzed by PhosphorImaging. (C) Primary MEPs were obtained by infection of yolk-sac blood island cells with the E26–WT and E26–βER viruses. After phenotyping to identify pure MEP populations (MEP21+, MEP26+, EOS47−, MYL51/2−, MHC II−), four to six clones for each construct were pooled. The pooled cells were left untreated or exposed to βE (1 μM final concentration). After 1, 2, and 3 d, one aliquot was removed for total RNA preparation (see D), and one aliquot washed free of βE and replated. A total of 6 d after induction, EOS47 antigen expression of the replated cells was determined by IIF, and plotted as a function of the time cells were exposed to βE (days). No change in antigen expression was observed in the absence of βE (data not shown). (D) A Northern blot of total RNA prepared as outlined in C was hybridized sequentially to probes for chicken FOG, chicken GATA-1, mim-1, and chicken GAPDH. Results were analyzed by PhosphorImaging.
FOG down-regulation is required for C/EBPβ-mediated induction of eosinophil gene expression in multipotent progenitors
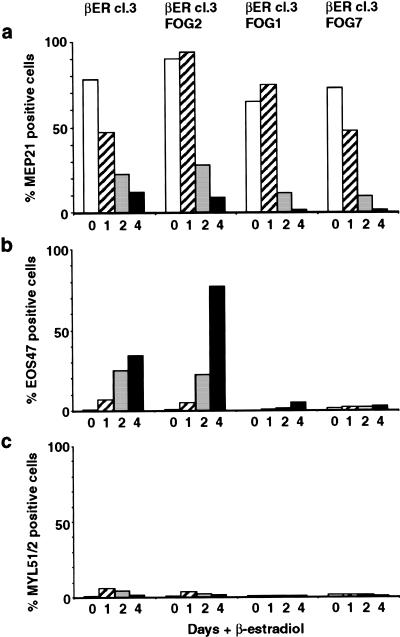
To address this issue directly, we stably expressed FOG in HD57M–C/EBPβ–ER clone 3 cells by transfection with the pEF–HAFOG–PGKpuro expression vector and tested the resulting clones for their ability to up-regulate EOS47 gene expression upon βE induction. Two FOG-expressing clones (βER3–FOG1 and βER3–FOG7) were compared with the parental cell line (βER3) and a nonexpressing control clone (βER3–FOG2), which showed that although both βER3 and βER3–FOG2 cells strongly up-regulated EOS47 expression (35% and 78% EOS47-positive cells after four days of induction, respectively) EOS47 expression for the FOG-expressing clones was only 5% and 2%, respectively (Fig. 6b). However, FOG expression was insufficient to maintain expression of MEP21, which was down-regulated in all C/EBPβ–ER-expressing clones (Fig. 6a), indicating the existence of additional C/EBPβ targets involved in the maintenance of the MEP phenotype. No expression of the myeloid marker MYL51/2 was observed in any of the clones (Fig. 6c), indicating that the inability of the HD57–C/EBPβER cells to express eosinophil-specific genes did not divert them toward a myeloid fate. We conclude that down-regulation of FOG coincides with and is a prerequisite for the C/EBPβ-mediated up-regulation of eosinophil-specific gene expression in MEPs.
Figure 6.
FOG inhibits C/EBPβ-mediated induction of eosinophil gene expression. HD57-C/EBPβ-ER clone 3 cells were transfected with the pEF–HAFOG–PGKpuro vector and the resulting puromycin-resistant clones analyzed for FOG expression by Western blot analysis (not shown). Two FOG-expressing (FOG1 and FOG7) and a nonexpressing control clone (FOG2) were further analyzed. These cell lines, along with the parental HD57-C/EBPβ-ER clone 3 cells, were induced with 1 μM βE or left untreated, and their expression of surface markers analyzed after 1, 2, and 4 d of exposure to βE. The results for MEP21 (a), EOS47 (b), and MYL51/2 (c) are shown, and are given as the percentage of cells staining positive. MEP26 and MHC II staining gave results similar to those for MEP21 and MYL51/2, respectively.
Discussion
FOG is a C/EBPβ-regulated inhibitor of eosinophil gene expression
In the hematopoietic system, FOG has been shown to play an essential role in the maturation of erythroid cells and in the early development of the thrombocytic lineage (Tsang et al. 1998). Here we find that FOG is highly expressed in chicken multipotent hematopoietic progenitors (MEPs), but not in their derived myeloid and eosinophil cells, similar to the expression pattern observed in mammals (Tsang et al. 1997; Yamaguchi et al. 1999). We demonstrate that down-regulation of FOG in multipotent hematopoietic precursors is an essential step in their C/EBPβ-mediated differentiation along the eosinophil lineage, as constitutive FOG expression in MEPs prevents C/EBPβ from activating eosinophil-specific gene expression. We show that FOG is an inhibitor of eosinophil-specific genes, as introduction of FOG into eosinophils reprograms these to a multipotent phenotype, up-regulating MEP markers and down-regulating EOS47, C/EBPβ, and Mim-1, all markers of the eosinophil lineage. Analysis of the eosinophil-specific EOS47 promoter indicated that this is due to the ability of FOG to repress promoter activity when recruited to a promoter by GATA-1, as repression was not seen upon mutation of the GATA-1 NF, although other effects resulting from the mutation of the NF cannot be ruled out. This result is consistent with the previous findings that FOG can inhibit GATA-1-mediated activation of the eosinophil-specific major basic protein (MBP) promoter (Yamaguchi et al. 1999) and that the GATA-1–FOG interaction is required for FOG to inhibit GATA-1 transactivation (Fox et al. 1999). The EOS47 and MBP are the most well-characterized eosinophil-specific promoters, and both of these are cooperatively activated by C/EBP and GATA-1 (McNagny et al. 1998; Yamaguchi et al. 1999). Therefore, these results suggest a model in which C/EBPβ (and likely C/EBPα as well; Nerlov et al. 1998) coordinately activates and derepresses eosinophil-specific genes by binding to C/EBP sites on their promoters and by down-regulating FOG expression, allowing cooperative activation through C/EBP and GATA sites (Fig. 7A). It is interesting to note that Deconinck et al. (2000) observed that expression of high levels of FOG had an inhibitory effect on erythroid differentiation in Xenopus embryos, possibly by blocking the differentiation of precursor cells, which indicates that modulation of FOG levels may be relevant for several hematopoietic lineages, and suggests that distinct levels of FOG expression are required in different developmental scenarios. We also found that murine FOG and FOG-2 have effects similar to those of Xenopus FOG on blood cell differentiation and embryonic development in Xenopus embryos, indicating a high degree of conservation of FOG function among vertebrates, and supporting the relevance of the effects we observe when expressing murine FOG in chicken cells.
Figure 7.
(A) Model for the induction of eosinophil-specific genes by C/EBP, as exemplified by the EOS47 promoter. In the MEPs, the EOS47 gene is inactive because of the absence of C/EBP, and the presence of FOG, which inhibits the activity of GATA-1 bound to the promoter (indicated by blunt-ended line). Upon induction of C/EBP, FOG is down-regulated, and the C/EBP site occupied (arrow), leading to the synergistic activation of the EOS47 promoter by GATA-1 and C/EBP. (B) Lineage determination by controlled collapse of the MEP phenotype. See text for explanation.
Regulation of FOG by C/EBP: developmental consequences
The six vertebrate GATA factors are all essential for embryonic development (Tsai et al. 1994; Pandolfi et al. 1995; Fujiwara et al. 1996; Kuo et al. 1997; Molkentin et al. 1997; Morissey et al. 1998; Reiter et al. 1999), and modulation of their activity by the FOG and FOG-2 proteins is likely to be of general significance for developmental processes in several tissues, including blood, heart, brain, gut, and the germ line (Tsang et al. 1997, 1998; Svensson et al. 1999; Tevosian et al. 1999). Here we report the first example of a developmental decision that is governed by modulation of GATA factor activity through regulation of FOG expression. The involvement of C/EBP proteins in the differentiation of many different cell types further suggests that the down-regulation of FOG expression by C/EBP may be relevant for developmental decisions in other GATA factor-expressing tissues. One example is the ovary, in which loss of C/EBPβ leads to failure of granulosa cells to mature appropriately with subsequent lack of proper ovulation and female sterility (Sterneck et al. 1997). Studies on the regulation of the steroidogenic acute regulatory protein (StAR) promoter in primary granulosa cells has revealed that it is coordinately activated by GATA-4 and C/EBPβ (Silverman et al. 1999). C/EBPβ protein is absent from naive granulosa cells, but can be induced by treatment with follicle-stimulating hormone (FSH). Importantly, whereas GATA-4 is constitutively present, the activity of the GATA site on the StAR promoter is increased upon FSH stimulation, consistent with the removal of a repressor of GATA activity, such as FOG, when C/EBPβ protein is induced (Silverman et al. 1999). Although no data addressing FOG regulation in granulosa cells are presently available, these results suggest that a mechanism similar to that observed in hematopoietic progenitors may operate in early granulosa cell differentiation. Therefore, other developmental scenarios may exist in which differentiation takes place through the induction of C/EBP on a poised background of GATA and FOG expression, which could be a more general mechanism for committing GATA-expressing cells to a specific developmental fate.
Lineage commitment by controlled collapse: PU.1 versus GATA and C/EBP versus FOG
Two lines of evidence suggest that GATA-1 and FOG cooperate in the establishment of a multipotent MEP phenotype. First, expression of FOG leads to MEP formation in GATA-1-expressing eosinophils, but not in GATA-1-negative myeloblasts. Secondly, a GATA-1 molecule deficient in FOG interaction failed to induce MEPs when introduced into myeloblasts. Whereas mutation of the NF may affect GATA functions other than FOG interaction (and the impaired activation potential of GATA-1mutNF on the EOS47 promoter suggests that this is the case), these results nevertheless indicate that collaboration between GATA-1 and FOG is important for maintaining a multipotent phenotype. PU.1 induces myeloid differentiation of MEPs, correlating with its ability to down-regulate GATA-1 expression (Nerlov and Graf 1998). Here we find that FOG is down-regulated during C/EBPβ-mediated eosinophil differentiation of MEPs. In this case, GATA-1 expression is maintained and a C/EBP +GATA no FOG configuration results, as illustrated in Figure 7B. In the case of PU.1-mediated myeloid commitment, loss of GATA-1 is followed by up-regulation of C/EBPβ (Nerlov and Graf 1998), which in turn may down-regulate FOG, leading to the well-known C/EBP +PU.1 no GATA configuration of myeloid cell types. High C/EBP expression is maintained in the presence of GATA-1 (in eosinophils) or FOG (in FOG-expressing myeloblasts), but not when both are present (in MEPs), suggesting that the combined action of FOG and GATA-1 is required for the suppression of C/EBP expression, as was observed in FOG-expressing 1A1 clones and MEPs derived from myeloid cells by GATA-1 overexpression (in which endogenous FOG expression was up-regulated). We have found previously that high levels of PKC activity specifically induces eosinophil lineage commitment of MEPs (Graf et al. 1992; Rossi et al. 1996a), and C/EBPβ is induced under these conditions (C. Nerlov, unpubl.), indicating that PKC signaling can overcome the repression of C/EBPβ expression in MEPs. Low levels of PKC activity, on the other hand, induce mostly myeloid differentiation (Rossi et al. 1996a). The fate of the MEP is thus determined by the manner in which it exits the multipotent state, and this may depend on extracellular signaling, suggesting that specific developmental programs can be initiated in multipotent cells through elimination of one of the factors involved in maintaining multipotency, commitment being, in effect, a controlled collapse of the multipotent state. Knowledge of the factors maintaining a multipotent state may thus help in the elucidation of the regulatory events that commit cells to specific fates. The inability of constitutive FOG expression to maintain the expression of MEP-specific genes upon C/EBPβ induction suggests that factors in addition to FOG and GATA-1 are required for maintenance of the MEP phenotype, and current efforts are directed toward identifying these.
Materials and methods
DNA constructs
The following plasmids have been described previously: pCRNCM–Ets-1, pCRNCM–c-Myb, and pCRNCM expression vectors (Lim et al. 1992); pSPCMV–GATA-1, pSPCMV–GATA-1mutNF, and pSPCMV (McNagny et al. 1998; Nerlov et al. 2000); the EOS47 promoter construct pEOS47/-152-LUC (McNagny et al. 1998); the pRSV–βgal internal control plasmid (Frampton et al. 1996); the E26–WT and E26–βER proviral constructs (Nerlov et al. 1998); the retroviral vector pSFCV (control) and its derivatives pNEO–GATA-1 and pNEO–GATA-ER (Fuerstenberg et al. 1990; Briegel et al. 1993; Kulessa et al. 1995). The FOG expression plasmid pEF–HAFOG–PGKpuro, encoding mouse FOG amino-terminally tagged with the HA epitope, was kindly provided by Dr. Alice Tsang (Harvard Medical School, Boston, MA). Its corresponding control vector (pEF–HA–PGKpuro) was generated by excising the FOG cDNA with BamHI.
PCR cloning
A partial chicken FOG mRNA was cloned from plasmid DNA excised from an HD50 MEP λ-Hybrizap (Stratagene) cDNA library (C. Nerlov, unpubl.) by nested PCR with primers on the basis of homology between FOG and FOG-2 (Tsang et al. 1997; Svensson et al. 1999; Tevosian et al. 1999). The first reaction was performed with Pfu polymerase (Stratagene) and the degenerate primers 5′-GAGATCYTGGCSSAAGAT and 5′-GCGYGSKGCRCAGTARTA. The nested PCR was performed with Taq polymerase (Roche) and the primers 5′-GCYACGTGCTTTGAGTGY and 5′-GATGTTRCARGCYTCRCA. The resulting 390-bp product was cloned into pCR2.1-TOPO using the TOPO TA cloning kit (Invitrogen) and sequenced. Sequence alignment using the ClustalW software showed 48% identity of the predicted amino acid sequence with murine FOG and 30% with murine FOG-2, indicating that the clone represented chicken FOG.
Cell lines and culture conditions
The origin of the cell lines for transfections and as sources of RNA have been described previously as HD3 erythroblasts (Beug et al. 1982), HD37 erythroblasts and HD57 MEPs (Metz and Graf 1991); HD50 multipotent cells and HD57M myeloblasts (Graf et al. 1992), 1A1 eosinophils and 1A4 MEPs (Kulessa et al. 1995). The HD50/4.8E line was derived from the HD50 line after treatment with 20 nM TPA and 0.1 μg/mL of ionomycin and subsequent subcloning, and the myeloblast line HD50/4.8M spontaneously arose from HD50/4.8E. The C/EBPβ–ER-expressing HD57–NF-M–ER clone 3 and nonexpressing control clone 12 have been described (Müller et al. 1995). All cells were grown at 37°C and 5% CO2 in blastoderm medium (McNagny and Graf 1997). Medium for HD57M, HD50/4.8M, HD50/4.8E, and 1A1 cells was supplemented with ∼10 U/mL of recombinant chicken myelomonocytic growth factor (cMGF; Leutz et al. 1989). Q2bn fibroblasts were grown at 37°C and 5% CO2 in DMEM supplemented with 8% FBS and 2% chicken serum (GIBCO BRL).
Transfections
Transient transfections of Q2bn cells and reporter assays were performed as described (Frampton et al. 1996) with 1 μg of pEOS47/-152-LUC reporter and 250 ng of pRSV–βGal plasmid as internal reference. HD57 MEP cells and HD57M myeloblasts were stably transfected by electroporation as described (Kulessa et al. 1995). 1A1 cells were transfected using 2 μl of DMRIE-C (GIBCO BRL) per 5 × 105 cells as described by the manufacturer. Stable clones were selected in 2% Methocel (Fluka) containing blastoderm medium with 1.6 mg/mL G418 (Geneticin, GIBCOBRL; for cells transfected with pNEO–GATA-1 and derivatives) or 0.5 μg/mL puromycin (Sigma; for cells transfected with pEF–HAFOG–PGKpuro and pEF–HA–PGKpuro). Resistant colonies were picked 10–14 d after transfection and expanded in selective medium. Production of and infection with recombinant E26 virus was performed as described (McNagny and Graf 1997). C/EBPβ–ER and GATA–ER fusions were induced by addition of β-estradiol to the medium to a final concentration of 1.0 μM and 0.1 μM, respectively.
Cell staining and flow cytometry
The cells were stained with monoclonal antibodies described earlier (MYL51/2: Kornfeld et al. 1983; c1a anti MHC II: Ewert et al. 1984; EOS47, MEP21, MEP26: McNagny et al. 1997), and phenotyped by indirect IF/FC on a Becton-Dickinson FACSCalibur. Cytospins were fixed in methanol and May-GrünwaldGiemsa stained (Hema Gurr, BDH).
RNA preparation and Northern blotting
Total cellular RNA was prepared according to Chromczynski and Sacchi (1987). Poly(A)-enriched RNA was made using the Oligotex mRNA kit (Qiagen). RNA was separated on 1.2% formaldehyde-agarose gels, transferred to Biodyne B membranes (GIBCO BRL) by capillary blotting, and probed with cDNA labeled by random priming with [α-32P]dCTP (3000 Ci/mmole; Amersham-Pharmacia) and RAD-Prime (GIBCO BRL), and purified on a Nick column (Amersham-Pharmacia). The following cDNAs served as probes: chicken GAPDH cDNA (Dugaiczyk et al. 1983); chicken GATA-1 (Briegel et al. 1993), mim-1 (Ness et al. 1989), chicken C/EBPβ (formerly NF-M; Katz et al. 1993), chicken FOG (this study).
Western blotting
Cells were lysed as described (Kulessa et al. 1995), the debris separated by centrifugation at 4°C, the extracts fractionated by SDS-PAGE, and blotted onto a PVDF membrane (Immobilon-P, Millipore). Blocking was in Tris-buffered saline (20 mM Tris-HCl at pH 7.6, 137 mM NaCl) with 0.1% Tween 20 (TBS-T), and 1% nonfat dry milk. Primary antibodies were: rabbit anti-chicken C/EBPβ (formerly anti-NF-M; Katz et al. 1993; kindly provided by Dr. A. Leutz, Max Delbruck Center for Molecular Medicine, Berlin, Germany) used at a 1:1000 dilution; affinity-purified rabbit anti-Mim-1 antibody (Ness et al. 1989) (1:300 dilution), mouse anti-α-tubulin monoclonal antibody (clone DM 1A, Sigma) (1:300 dilution), and mouse anti-HA epitope monoclonal antibody (clone 12CA5; Roche) (1:5000 dilution). Secondary antibodies were horseradish peroxidase-conjugated anti-rabbit immunoglobulin and anti-mouse immunoglobulin (Amersham-Pharmacia) as appropriate, both 1:5000 in TBS-T. Immunodetection was performed by enhanced chemiluminescence (ECL, Amersham-Pharmacia) as recommended by the manufacturer.
Acknowledgments
We thank Dr. Alice Tsang for providing the pEF–HAFOG–PGKpuro expression vector, Dr. Achim Leutz for anti-chicken C/EBPβ antiserum, and members of the Nerlov and Graf laboratories for discussions. J.D.C. was supported by the Jane Coffin Childs Fund for Medical Research, and S.H.O. is an Investigator of the Howard Hughes Medical Institute. This work was supported by the Deutsche Forschungsgemeinshaft, the Novo Nordisk Foundation, and the Danish Medical Research Council.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL nerlovv@rh.dk; FAX 45-35-45-67-27.
E-MAIL graf@aecom.yu.edu; FAX (718) 430-3305.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.177200.
References
- Beug H, Doederlein G, Freudenstein C, Graf T. Erythroblast cell lines transformed by a temperature sensitive mutant of avian erythroblastosis virus: A model system to study erythroid differentiation in vitro. J Cell Physiol Suppl. 1982;1:195–207. doi: 10.1002/jcp.1041130427. [DOI] [PubMed] [Google Scholar]
- Briegel K, Lim KC, Plank C, Beug H, Engel JD, Zenke M. Ectopic expression of a conditional GATA-2/estrogen receptor chimera arrests erythroid differentiation in a hormone-dependent manner. Genes & Dev. 1993;7:1097–1109. doi: 10.1101/gad.7.6.1097. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single step isolation of RNA by acid guanidinium isothiocyanate method. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: The GATA-1:FOG complex. Mol Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- Deconinck AE, Mead PE, Tevosian SG, Crispino JD, Katz SG, Zon LI, Orkin SH. FOG acts as a repressor of red blood cell development in Xenopus. Development. 2000;127:2031–40. doi: 10.1242/dev.127.10.2031. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A. Cloning and sequencing of a deoxyribonucleic acid copy of glyceraldehyde-3-phosphate dehydrogenase messenger ribonucleic acid isolated from chicken muscle. Biochemistry. 1983;22:1605–1613. doi: 10.1021/bi00276a013. [DOI] [PubMed] [Google Scholar]
- Ewert DL, Munchus MS, Chen CL, Cooper MD. Analysis of structural properties and cellular distribution of avian Ia antigen by using monoclonal antibody to monomorphic determinants. J Immunol. 1984;132:2524–2530. [PubMed] [Google Scholar]
- Fox AH, Kowalski K, King GF, Mackay JP, Crossley M. Key residues characteristic of GATA N-fingers are recognized by FOG. J Biol Chem. 1998;273:33595–33603. doi: 10.1074/jbc.273.50.33595. [DOI] [PubMed] [Google Scholar]
- Fox AH, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 1999;18:2812–2822. doi: 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton J, McNagny K, Sieweke M, Philip A, Smith G, Graf T. v-Myb DNA binding is required to block thrombocytic differentiation of Myb-Ets-transformed multipotent haematopoietic progenitors. EMBO J. 1995;14:2866–2875. doi: 10.1002/j.1460-2075.1995.tb07286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton J, Ramqvist T, Graf T. v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes & Dev. 1996;10:2720–2731. doi: 10.1101/gad.10.21.2720. [DOI] [PubMed] [Google Scholar]
- Fuerstenberg S, Beug H, Introna M, Khazaie K, Munoz A, Ness S, Nordstrom K, Sap J, Stanley I, Zenke M, et al. Ectopic expression of the erythrocyte band 3 anion exchange protein, using a new avian retrovirus vector. J Virol. 1990;64:5891–5902. doi: 10.1128/jvi.64.12.5891-5902.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, McNagny K, Brady G, Frampton J. Chicken “erythroid” cells transformed by the Gag-Myb-Ets-encoding E26 leukemia virus are multipotent. Cell. 1992;70:201–213. doi: 10.1016/0092-8674(92)90096-u. [DOI] [PubMed] [Google Scholar]
- Katz S, Kowenz-Leutz E, Muller C, Meese K, Ness SA, Leutz A. The NF-M transcription factor is related to C/EBPβ and plays a role in signal transduction, differentiation and leukemogenesis of avian myelomonocytic cells. EMBO J. 1993;12:1321–1332. doi: 10.1002/j.1460-2075.1993.tb05777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld S, Beug H, Doederlein G, Graf T. Detection of avian hematopoietic cell surface antigens with monoclonal antibodies to myeloid cells. Exp Cell Res. 1983;143:383–394. doi: 10.1016/0014-4827(83)90065-4. [DOI] [PubMed] [Google Scholar]
- Kraut N, Frampton J, McNagny KM, Graf T. A functional Ets DNA-binding domain is required to maintain multipotency of hematopoietic progenitors transformed by Myb–Ets. Genes & Dev. 1994;8:33–44. doi: 10.1101/gad.8.1.33. [DOI] [PubMed] [Google Scholar]
- Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes & Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anadappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA-4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes & Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Leutz A, Damm K, Sterneck E, Kowenz E, Ness S, Frank R, Gausepohl H, Pan Y-C, Smart J, Hayman M, et al. Molecular cloning of the chicken myelomonocytic growth factor (cMGF) reveals relationship to interleukin 6 and granulocyte colony stimulating factor. EMBO J. 1989;8:175–181. doi: 10.1002/j.1460-2075.1989.tb03362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim F, Kraut N, Framptom J, Graf T. DNA binding by c-Ets-1, but not v-Ets, is repressed by an intramolecular mechanism. EMBO J. 1992;11:643–652. doi: 10.1002/j.1460-2075.1992.tb05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- McNagny KM, Graf T. Production and analysis of retrovirus-transformed multipotent hematopoietic progenitors. In: Lefkovits I, editor. Immunology methods manual. New York: Academic Press; 1997. pp. 2183–2198. [Google Scholar]
- McNagny KM, Pettersson I, Rossi F, Flamme I, Shevchenko A, Mann M, Graf T. Thrombomucin, a novel cell surface protein that defines thrombocytes and multipotent hematopoietic progenitors. J Cell Biol. 1997;138:1395–1407. doi: 10.1083/jcb.138.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNagny KM, Sieweke MH, Doderlein G, Graf T, Nerlov C. Regulation of eosinophil-specific gene expression by a C/EBP–Ets complex and GATA-1. EMBO J. 1998;17:3669–3680. doi: 10.1093/emboj/17.13.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz T, Graf T. v-myb and v-ets transform chicken erythroid cells and cooperate both in trans and in cis to induce distinct differentiation phenotypes. Genes & Dev. 1991;5:369–380. doi: 10.1101/gad.5.3.369. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA-4 for heart tube formation and ventral morphogenesis. Genes & Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Tang Z, Sigrist K, Lu MM, Jiang F, Ip HS, Parmacek MS. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes & Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C, Kowenz-Leutz E, Grieser-Ade S, Graf T, Leutz A. NF-M (chicken C/EBPβ) induces eosinophilic differentiation and apoptosis in a hematopoietic progenitor cell line. EMBO J. 1995;14:6127–6135. doi: 10.1002/j.1460-2075.1995.tb00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes & Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, McNagny KM, Doderlein G, Kowenz-Leutz E, Graf T. Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes & Dev. 1998;12:2413–2423. doi: 10.1101/gad.12.15.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1 dependent transcription. Blood. 2000;95:2543–2551. [PubMed] [Google Scholar]
- Ness SA, Marknell Å, Graf T. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- Ody C, Vaigot P, Quere P, Imhof BA, Corbel C. Glycoprotein IIb-IIIa is expressed on avian multilineage hematopoietic progenitor cells. Blood. 1999;93:2898–2906. [PubMed] [Google Scholar]
- Pandolfi PP, Roth ME, Karis A, Leonard MW, Dzierzak E, Grosveld FG, Lindenbaum MH. Targeted disruption of the GATA-3 gene causes severe abnormalities in the nervous system and in fetal liver hematopoiesis. Nat Genet. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DYR. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes & Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F, McNagny M, Smith G, Frampton J, Graf T. Lineage commitment of transformed haematopoietic progenitors is determined by the level of PKC activity. EMBO J. 1996a;15:1894–901. [PMC free article] [PubMed] [Google Scholar]
- Rossi F, McNagny KM, Logie C, Stewart AF, Graf T. Excision of Ets by an inducible site-specific recombinase causes differentiation of Myb–Ets-transformed hematopoietic progenitors. Curr Biol. 1996b;6:866–872. doi: 10.1016/s0960-9822(02)00610-3. [DOI] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Silverman E, Eimerl S, Orly J. CCAAT enhancer-binding protein β and GATA-4 binding regions within the promoter of the steroidogenic acute regulatory protein (StAR) gene are required for transcription in rat ovarian cells. J Biol Chem. 1999;274:17987–17996. doi: 10.1074/jbc.274.25.17987. [DOI] [PubMed] [Google Scholar]
- Sterneck E, Tessarollo L, Johnson PF. An essential role for C/EBPβ in female reproduction. Genes & Dev. 1997;11:2153–2162. doi: 10.1101/gad.11.17.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson EC, Tufts RL, Polk CE, Leiden JM. Molecular cloning of FOG-2: A modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Cantor AB, Rieff HI, Fujiwara Y, Corfas G, Orkin SH. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc Natl Acad Sci. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early hematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Fujiwara Y, Hom DB, Orkin SH. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes & Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Elefanty AG, Strasser A, Adams JM. GATA-1 but not SCL induces megakaryocytic differentiation in an early myeloid line. EMBO J. 1992;11:4557–4564. doi: 10.1002/j.1460-2075.1992.tb05557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Nishio H, Kishi K, Ackerman SJ, Suda T. C/EBPβ and GATA-1 synergistically regulate activity of the eosinophil granule major basic protein promoter: Implication for C/EBPβ activity in eosinophil gene expression. Blood. 1999;94:1429–1439. [PubMed] [Google Scholar]
- Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc Natl Acad Sci. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radomska HS, Auron PE, Tenen DG, Sun Z. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]