Abstract
Cdc6 is essential for eukaryotic DNA replication. We have mutated highly conserved CDK phosphorylation sites in Cdc6. Contrary to their reported phenotypes in human cells, unphosphorylatable ΔCDK mutants fully support DNA replication in Xenopus eggs. WtCdc6 is actively exported from the nucleus, which could explain why nuclear permeabilization is required for reinitiation within one cell cycle. However, ΔCDK mutants are retained in the nucleus, yet surprisingly they still support only one round of replication. As these highly conserved CDK sites are unnecessary for replication once per cell cycle, an alternative checkpoint role for monitoring completion of the S phase is suggested.
Keywords: Cdc6, phosphorylation, nuclear export, Xenopus
Initiation of DNA replication is tightly regulated so that each fragment of genomic DNA is replicated exactly once within each cell cycle (Diffley 1996). One of the mechanisms by which this is achieved relies on the regulated assembly of protein complexes—including the origin recognition complex (ORC), Cdc6, and the minichromosome maintenance (MCM) proteins—onto chromatin during the prereplicative phase of the cell cycle (for review, see Romanowski and Madine 1996, 1997). Studies in yeast (Cocker et al. 1996) and Xenopus (Coleman et al. 1996; Romanowski et al. 1996; Rowles et al. 1996) have shown that prereplicative complexes (pre-RCs) are necessary for the initiation of DNA replication and are disassembled during replication. As neither Cdc6 nor MCMs can rebind to chromatin until mitosis, a second initiation event within the same cell cycle is prevented.
Both Saccharomyces cerevisiae Cdc6 and its Schizosaccharomyces pombe homolog Cdc18 are essential for initiation of DNA replication (Piatti et al. 1995; Nishitani and Nurse 1995) and for loading on chromatin of the MCM protein complex (Tanaka et al. 1997). In fission yeast, Cdc18 is required for initiation of a single round of DNA replication in each cell cycle, yet overexpression leads to multiple rounds of DNA replication without an intervening mitosis (Nishitani and Nurse 1995). However, a similar phenotype was not observed when Cdc6 was overexpressed in S. cerevisiae (Bueno and Russell 1992). Therefore, despite functional similarities, the activity of Cdc6 seems to be differently regulated in the two yeasts.
The binding of Cdc6 protein to chromatin in Xenopus and mammalian in vitro replication systems has been shown to be a critical early step in higher eukaryotic DNA replication (Coleman et al. 1996; Stoeber et al. 1998). Moreover, it has been shown to be required for the subsequent loading of MCM proteins and initiation of DNA replication, because both events are abolished in Xenopus Cdc6-depleted extracts.
Increasing evidence indicates that a cyclin-dependent kinase (CDK)–directed regulation of pre-RCs is required for activation of preexisting complexes throughout the S phase and for inhibition of assembly of new complexes after replication (Schwob et al. 1994; Dahmann et al. 1995). Because of its unique role in promoting the assembly of the pre-RCs, Cdc6 has long been considered an excellent candidate in restricting DNA replication to once per cell cycle by mediating both of the CDK functions. Interestingly, CDK consensus sites are conserved in Cdc6 proteins of different species. Phosphorylation of Cdc6 by CDKs controls protein stability in yeast (Jallepalli et al. 1997; Elsasser et al. 1999), and cell cycle–specific phosphorylation is involved in alteration of HuCdc6 subcellular localization in cultured cells (Jiang et al. 1999; Petersen et al. 1999). However, it is still unclear whether phosphorylation of Cdc6 is a mechanism by which higher eukaryotes inactivate Cdc6 function to prevent overreplication of the genome.
In this study, we constructed two different versions of Xenopus Cdc6 protein, either mutating or deleting CDK consensus sites. We then tested their function in the Xenopus laevis cell-free replication system.
Our work indicates that CDK-dependent phosphorylation of Cdc6 in vertebrates is not required for initiation of DNA replication or for blocking reformation of prereplicative complexes during or after replication, because the unphosphorylatable XCdc6 proteins are functional and able to maintain the once per cell cycle regulation of replication. Export of XCdc6 from nuclei during replication is dependent on CDK consensus sites, but even retention of Cdc6 in the nucleus is insufficient to trigger reinitiation of replication within a single cell cycle.
Results and Discussion
XCdc6 binds to sperm chromatin before initiation of DNA replication and disappears from the nucleus during replication
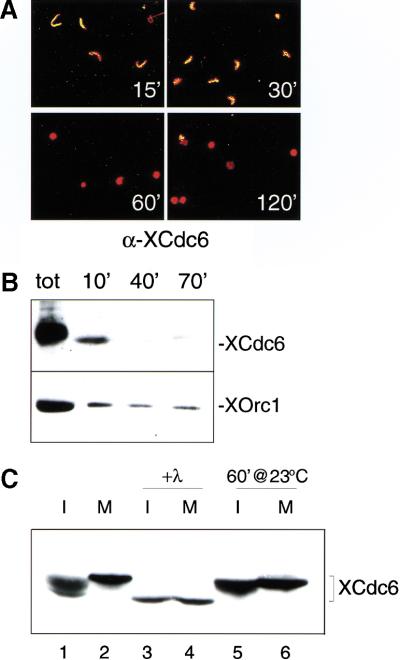
We investigated the behavior of Cdc6 protein in Xenopus in vitro replication both by immunofluorescence localization and by western blotting of the chromatin-bound protein fraction. By immunofluorescence (Fig. 1A), we show that during incubation of sperm chromatin in interphase egg extracts, XCdc6 is accumulated early in the nuclei (15 min) and disappears from the nuclei around the time of initiation of DNA replication (after 30 min). In agreement with this, we also show by blotting the chromatin-bound protein fraction that XCdc6 rapidly binds to chromatin (within 10 min) and is mostly released during replication. Little, if any, Cdc6 remains on chromatin after 40 min of incubation (Fig. 1B, top panel). This is very similar to the behavior of the MCM proteins (Madine et al. 1995b), whereas XOrc1, the largest of the ORC subunits, remains associated to chromatin until the end of replication (70 min, Fig. 1B, bottom panel), in agreement with previously published data (Coleman et al. 1996; Romanowski et al. 1996; Rowles et al. 1996).
Figure 1.
Xenopus Cdc6 is displaced from nuclei during replication in egg extract, and its phosphorylation varies with cell cycle phase. Xenopus sperm nuclei were incubated in egg extract and harvested at various times. (A) Reactions were stopped by fixation after 15, 30, 60, and 120 min, and nuclear XCdc6 was detected by affinity purified anti-XCdc6 antibody. Sperm nuclei were stained with propidium iodide (red), and XCdc6 was revealed by an anti-rabbit fluorescein-linked antibody (green). Panels show merged images (red + green = yellow). (B) Chromatin-bound XCdc6 after 10, 40, and 70 min of incubation detected by immunoblotting (top panel). Chromatin-bound XOrc1 is in the bottom panel. The total amount of XCdc6 and XOrc1 in the extract is shown (tot). (C) XCdc6 is hypophosphorylated during interphase and hyperphosphorylated in mitotic extracts. Immunoblots of XCdc6 show different electrophoretic mobility forms of the protein in interphase (lane 1, I) and mitotic extracts (lane 2, M). The interphase and mitotic extracts were incubated with (lanes 3,4) or without (lanes 5,6) λ-phosphatase.
XCdc6 undergoes phosphorylation during interphase and hyperphosphorylation in mitotic extracts
Next we investigated the phosphorylation states of Cdc6 in egg extracts. By Western blotting, multiple forms of different electrophoretic mobility are detected in interphase extracts and a slow mobility form is present in mitotic extracts, (Fig. 1C, lanes 1,2), potentially corresponding to different phosphorylation states of XCdc6 protein. The mobility of XCdc6 decreases further under in vitro replication conditions (1 hr of incubation at 23°C, Fig. 1C, lanes 5,6), and all the forms become fast migrating after λ-phosphatase treatment (Fig. 1C, lanes 3,4). These conversions in protein mobility on SDS-PAGE are therefore consistent with modifications of XCdc6 caused by phosphorylation, and they correlate with the replication stage of the extract. Moreover, they resemble the reported cell cycle variations of Cdc6 protein caused by phosphorylation in mammalian cells (Fujita et al. 1999; Petersen et al. 1999).
XCdc6 is phosphorylated on CDK consensus sites
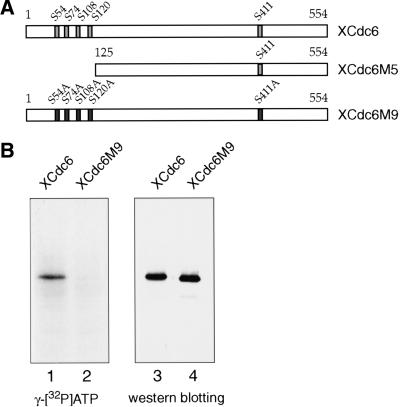
XCdc6 contains five potential full consensus sites for CDK-dependent phosphorylation, four of which are clustered at the amino terminus of the protein (Ser 54, Ser 74, Ser 108, Ser 120, and Ser 411; Fig. 2A). To characterize the functional role of CDK-mediated phosphorylation of XCdc6, we have constructed two different mutated versions of XCdc6 (Fig. 2A). One of them, XCdc6M5 (Δ1–125), is a truncated protein, lacking the four amino-terminally clustered CDK sites; the other, XCdc6M9 (ΔCDKs), was obtained by substituting the serines in each of the CDK consensus sites with an alanine.
Figure 2.
Mutation of conserved CDK consensus sites abolishes phosphorylation of Cdc6. (A) Schematic representation of XCdc6 and XCdc6 mutants used in this work. XCdc6 contains five full consensus sites for CDKs (Ser 54, Ser 74, Ser 108, Ser 120, and Ser 411). XCdc6M5 (Δ1–125) is an amino terminal deletion mutant lacking four CDK consensus sites. XCdc6M9 (S54A, S74A, S108A, S120A, S411A) was obtained by mutating serines in the CDK sites into alanines. (B) Recombinant wild-type XCdc6 and XCdc6M9 mutant proteins were tested for phosphorylation under replication assay conditions. They were incubated in interphase extracts with [γ-32P]ATP and then immunoprecipitated by using an anti-His antibody. The reactions were split into two aliquots for autoradiography and western blotting.
We asked whether the XCdc6 protein, which is subject to phosphorylation in egg extracts, is a target of CDKs. For this purpose, we incubated XCdc6 proteins, either wild-type or M9XCdc6, in interphase extracts with [γ-32P]ATP. We show that although the recombinant wild-type XCdc6 protein undergoes phosphorylation, as revealed by incorporation of radioactive ATP (Fig. 2B, lane 1), phosphorylation of the CDK mutant is prevented (Fig. 2B, lane 2). Altogether, this shows that the mutant XCdc6M9 functions as an unphosphorylatable XCdc6 protein and confirms the CDK-dependent phosphorylation of wild-type XCdc6.
Phosphorylation of XCdc6 is not required for initiation of DNA replication
The role of CDK-mediated phosphorylation of Cdc6 appears to be different in different organisms. Phosphorylation is involved in degradation of yeast Cdc6/Cdc18, and overexpression of unphosphorylatable and undegradable Cdc18 causes a strong overreplication phenotype (Jallepalli et al. 1997). In striking contrast, overexpression of an unphosphorylatable HuCdc6 protein (analogous to XCdc6M9 in this study) inhibited initiation of DNA replication in human fibroblasts (Jiang et al. 1999).
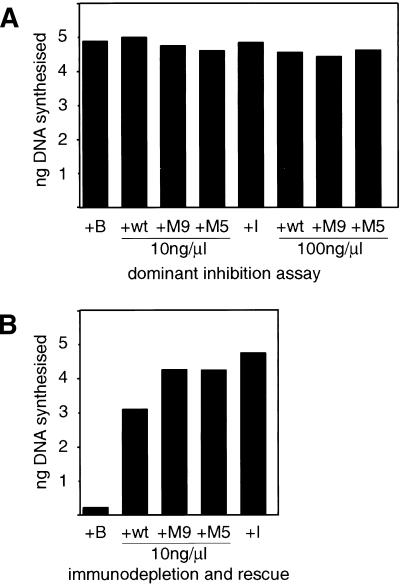
To establish the functional role of phosphorylation of XCdc6 on replication in higher eukaryotes, we have used X. laevis in vitro replication systems. DNA added to Xenopus interphase extracts is assembled into functional nuclei and undergoes a single round of semiconservative DNA replication. First, we looked for a possible dominant inhibitory effect of the XCdc6 unphosphorylatable proteins on replication by adding wild-type XCdc6, XCdc6M9, or XCdc6M5 to a standard replication reaction and checking the amount of DNA synthesized by [α-32P]dATP incorporation. Results from these experiments (Fig. 3A) clearly show that neither of the unphosphorylatable mutant XCdc6 proteins interferes with DNA replication. In fact, we obtained comparable levels of DNA synthesis (corresponding to >95% of genomes replicated) whether the addition was of buffer alone, wild-type XCdc6, either of the unphosphorylatable proteins, or fresh interphase egg extract. We added recombinant proteins at concentrations comparable to the amount of XCdc6 in the egg extract (10 ng/μL) and 10-fold higher concentrations (100 ng/μL).
Figure 3.
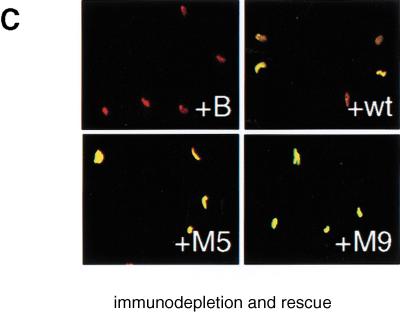
CDK phosphorylation of XCdc6 is not required for initiation of DNA replication. (A) Unphosphorylatable XCdc6 mutants do not inhibit DNA replication when added to interphase extracts at 10 ng/μL or 100ng/μL. Replication was measured by [α-32P]dATP incorporation after addition of buffer alone (+B), XCdc6 (+wt), either of the unphosphorylatable mutants (+M9, +M5), or interphase extract (+I). (B,C) Unphosphorylatable XCdc6 mutants rescue the replication of XCdc6-depleted extracts. Xenopus egg extract was depleted of endogenous XCdc6 and supplemented with buffer (+B), recombinant XCdc6 (+wt), either of the mutants (+M9, +M5), or interphase extract (+I). Proteins were added at a final concentration of 10 ng/μL. Replication was detected by either [α-32P]dATP (B) or biotin-dUTP incorporation (C). For confocal microscopy, nuclei were stained by propidium-iodide (red), and incorporated biotin was detected by fluorescein-linked streptavidin (green). Panels show merged images (red + green = yellow).
As Figure 3A clearly shows that the unphosphorylatable mutants do not inhibit DNA replication, we then asked if they might even be functional. Therefore, we assayed the mutant proteins for their ability to replace the native XCdc6 after immunodepletion of the egg extract. Depletion of the egg extract with antibodies raised against XCdc6 abolishes its ability to support replication of sperm chromatin, and the replication capacity of the extract is restored by addition of recombinant XCdc6 or interphase extract (Fig. 3B,C; see Coleman et al. 1996). When XCdc6-depleted extracts are supplemented with either XCdc6M9 or XCdc6M5 at 10 ng/μL of extract, the mutant proteins fully restore replication ability of the extracts as detected by [α-32P]dATP (Fig. 3B) and biotin-dUTP (Fig. 3C) incorporation. Each experiment was repeated at least three times by using different protein preparations, different egg extracts, and two different anti-XCdc6 antibodies for depletion. Replication of mock-depleted extracts was also included as a control (data not shown). This unequivocally shows that the XCdc6 mutants not only fail to act as dominant negative inhibitors, but also are fully functional. Surprisingly, CDK-mediated phosphorylation of XCdc6 is not required for initiation of DNA replication.
Export of XCdc6 from the nucleus is dependent on CDK-mediated phosphorylation of the protein
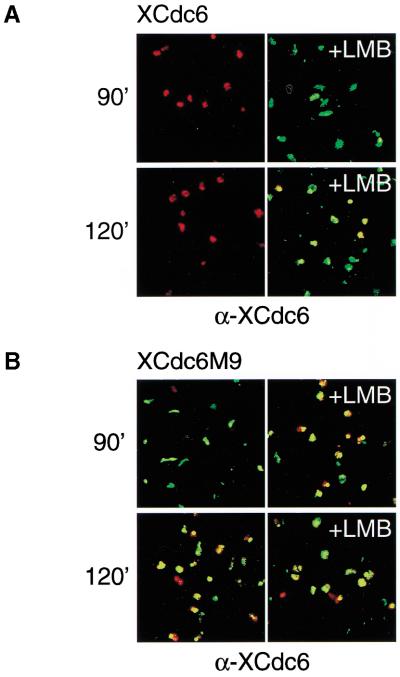
Our observation that XCdc6 mostly disappears from the nuclei during replication and is concurrently phosphorylated might be consistent with either CDK-mediated nuclear degradation or export. Furthermore, Cdc6 is degraded upon phosphorylation in S-phase mammalian cell extracts (Coverley et al. 2000). To discriminate between these two possibilities, we used immunofluorescence to compare the localization of XCdc6 with XCdc6M9 during and after replication in combination with leptomycin B (LMB), a cytotoxin that specifically inhibits nuclear export (Nishi et al. 1994). If export is the reason for Cdc6 disappearance from nuclei, addition of this drug to the replication reaction should result in accumulation of XCdc6 in the nuclei. Alternatively, if nuclear degradation is the mechanism responsible for the disappearance of XCdc6 from the nuclei, LMB should have no effect. To simplify interpretation, endogenous XCdc6 was depleted from the egg extract, and identical concentrations of recombinant XCdc6 or XCdc6M9 were added to the reactions. Importantly, the anti-XCdc6 antibody we used is able to recognize the different phosphorylation forms of the protein (Fig. 1C) and the mutant proteins. Figure 4A shows the behavior of the XCdc6 wild-type protein which, is accumulated in the nuclei and disappears during replication exactly as the endogenous protein (Fig. 4A, left panels). Moreover, the disappearance of XCdc6 from the nuclei is the result of active transport to the cytosol because it is efficiently blocked by 0.46 μM LMB (Fig. 4A, right panels). This effect cannot be attributed to a nonspecific effect of LMB on progression of in vitro DNA replication, because concentrations of LMB ranging from 0.18 to 0.92 μM do not affect replication as measured by [α-32P]dATP incorporation (data not shown). Conversely, XCdc6M9 remains nuclear throughout replication and LMB has no effect on its localization (Fig. 4B). Taken together, these results support a mechanism of active export of XCdc6 to the cytosol during replication, which requires CDK-dependent phosphorylation of the protein. Although we cannot exclude limited nuclear degradation masked by continuous import of XCdc6 from the cytosol and block of export by LMB, we have definitively shown that degradation is not the main mechanism regulating the presence of Cdc6 in nuclei in Xenopus eggs.
Figure 4.
CDK-dependent phosphorylation of XCdc6 is required for its export from the nucleus. (A) XCdc6 recombinant protein rescues replication of XCdc6-depleted extract and becomes undetectable in the nuclei after replication (90 and 120 min, left panels). Addition of leptomycin B (+LMB, right panels) prevents translocation of XCdc6 to the cytosol. (B) The unphosphorylatable XCdc6M9 mutant is not subject to export but remains in the nuclei throughout and after replication (90 and 120 min, left panels). LMB does not affect the localization of this mutant (+LMB, right panels). Replication was detected by Texas Red-streptavidin (red); antigen, by fluorescein-linked secondary antibody (green). Panels show merged images (red + green =yellow). Nuclei were stained with TOTO-3 iodide (not shown).
Constitutive nuclear localization of XCdc6 and insensitivity to the inhibitory effects of CDK controls do not cause reinitiation within one cell cycle
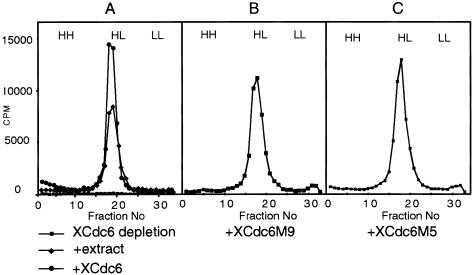
According to the results presented above, the unphosphorylatable XCdc6 proteins used in this study are: (1) Functional, because they are able to rescue DNA replication of sperm chromatin in XCdc6-depleted extracts; (2) present in the nuclei throughout interphase and therefore potentially available for a new round of replication without a previous mitosis; and (3) insensitive to CDK activity and therefore refractory to the inhibitory effects of CDK controls on replication. This, then, poses the question: Does the constitutive presence of mutant XCdc6 in the nuclei allow overreplication without an intervening mitosis as seen in S. pombe (Jallepalli et al. 1997)? To answer this question, we examined the replication products synthesized in the presence of either recombinant XCdc6 protein or unphosphorylatable XCdc6 mutants. For this purpose, we added the thymidine analogue bromodeoxyuridine triphosphate (BrdUTP) and [α-32P]dATP to replication reactions and subsequently separated the replication products on caesium chloride buoyant density gradients. A single round of DNA replication gives rise to DNA substituted on only one strand (hemi-substituted, heavy-light [HL]) whereas DNA substituted on both strands (fully-substituted, heavy-heavy [HH]) would indicate that more than one round of replication must have occurred. Depletion of XCdc6 from the extract abolishes replication, and readdition of interphase extract or physiological concentrations of recombinant XCdc6 (10 ng/μL of extract) results in radioactive nucleotide incorporation into DNA with the buoyant density of HL DNA (Fig. 5A). As expected, this indicates a single round of DNA replication. Readdition to a depleted extract of either of the unphosphorylatable mutants at 10 ng/μL of extract fully rescues replication (as shown above) and, strikingly, produces only HL peaks (Fig. 5B,C). We repeated the rescue of replication and density substitution experiments at least three times with different protein preparations and even using tenfold higher concentrations of recombinant proteins (100 ng/μL of extract). However, HH peaks of overreplicated DNA were never detected. Unphosphorylatable XCdc6M9 does not trigger a second round of DNA replication even after transient kinase inhibition and addition of fresh protein to nuclei that had undergone one round of replication (data not shown). Therefore, although the behavior of Cdc6 suggests that regulated chromatin binding and nuclear export of Cdc6 can contribute to the mechanism that prevents overreplication, this mechanism is sufficiently robust and internally redundant that it withstands multiple simultaneous disruptions. Significantly, the amino terminus domain of XCdc6, which represents more than one quarter of the protein and is considered essential for mediating interactions with CDKs and Orc1 in yeast and human cells (Brown et al. 1997; Saha et al. 1998; Petersen et al. 1999), is completely dispensable for XCdc6 function in replication.
Figure 5.
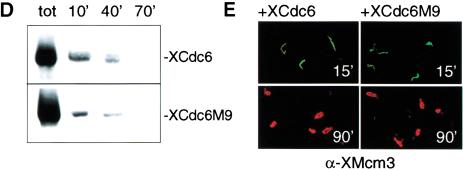
XCdc6 phosphorylation mutants do not cause DNA overreplication. Xenopus sperm chromatin was incubated in (A) XCdc6 depleted extract or depleted extract supplemented with either interphase extract or recombinant XCdc6, (B) depleted extract supplemented with XCdc6M9, and (C) depleted extract supplemented with XCdc6M5. All the recombinant proteins were used at a final concentration of 10 ng/μL. Reactions were incubated for 5 hr in the presence of BrdUTP and [α-32P]dATP. Buoyant density of unreplicated (LL), once-replicated (HL), and overreplicated (HH) DNA are marked at the top of each gradient profile. (D–E) Sperm chromatin was incubated in XCdc6-depleted extract rescued by addition of either XCdc6 or XCdc6M9 (10 ng/μL). (D) After 10, 40, and 70 min, reactions were stopped and sperm chromatin was assayed for the presence of chromatin-bound XCdc6 (top panel) and XCdc6M9 (bottom panel) by immunoblotting. (E) In a parallel experiment, sperm chromatin was assayed for the presence of XMcm3 by immunofluorescence before (15 min) and after (90 min) replication. Replication, nuclei, and antigen were stained as described for Figure 4, with the difference that anti-XMcm3 antibody was used.
Taken together, these results unequivocally show the following: (1) In contrast to mammalian overexpression systems, unphosphorylatable XCdc6 proteins do not inhibit DNA replication in functional assays and (2) phosphorylation of Xenopus Cdc6 by CDKs is not required for regulated DNA replication. The reason for these differences in results between mammalian cells and Xenopus eggs remains to be explained. Species differences in regulation of Cdc6 function or differences in regulation between somatic and embryonic systems may provide explanations.
The lack of phosphorylation does not impair XCdc6 binding to chromatin or MCM protein function
As the unphosphorylatable mutants failed to cause overreplication, we have analyzed the behavior of XCdc6 mutants with respect to chromatin binding and MCM recruitment. To compare results properly, recombinant XCdc6 protein or XCdc6M9 were added to XCdc6-depleted egg extract. The unphosphorylatable protein, which binds to chromatin within 10 min and is released during replication (Fig. 5D) very similarly to the wild-type XCdc6, does not cause any alteration in the dynamic association of MCMs with chromatin (Fig. 5E, right panels). XMcm3 is loaded onto chromatin before replication initiation (15 min) and is then released with DNA synthesis (90 min), exactly as occurs with the recombinant wild-type (Fig. 5E, left panels) or endogenous XCdc6 proteins (Madine et al. 1995b). These data show that CDK-mediated phosphorylation of XCdc6 is not required for XCdc6 binding to chromatin or for its release during replication. In addition, it is not required for regulated loading of MCMs onto chromatin. Taken together, these results strengthen the conclusion that CDK phosphorylation of Cdc6 is not required for Cdc6 function in the initiation of DNA replication.
The observation that free Cdc6 is exported from nuclei during replication could help to explain why it is necessary to permeabilize the nuclear membrane of replicated nuclei to enable them to replicate again without passing through mitosis (Blow and Laskey 1988; Coverley et al. 1993). However, our findings that unphosphorylatable XCdc6 mutants are nuclear throughout replication, without causing further rounds of DNA replication, show that additional factors contribute to the block to reinitiation.
Our observation that CDK phosphorylation of Cdc6 is not required for coupling initiation of DNA replication to the cell cycle in Xenopus focuses attention on alternative functions for these highly conserved phosphorylation sites. We are currently investigating an attractive alternative possibility, namely that these sites play a role in a checkpoint mechanism, for example coupling mitosis to the completion of DNA replication.
Materials and methods
Plasmids
pHis6-XCdc6 was provided by Dr. T. Coleman and Dr. W. Dunphy. pHis6-XCdc6M5 (XCdc6Δ125–554) and pHis6-XCdc6M9 (XCdc6ΔCDKs: S54A, S74A, S108A, S120A, S411A) were obtained by PCR amplification of the full-length XCdc6 sequence and cloned into the NdeI/EcoRI of the pVL1393N-His6 (Tang et al. 1995). XCdc6 mutant M5 (XCdc6M5) was constructed by using primers 5′-GTGAGACATATGCAAGAGACCCCACCCAGCT-3′ and 5′-CCGGGAATTCTTAAATCCCTGAATTGAG-3′. XCdc6 mutant M9 (XCdc6M9) was generated by annealing and amplifying DNA fragments obtained by PCR, using primers containing the designed mutated sequence to substitute the serine codon in the CDK consensus sites for an alanine. Amplification reactions were performed by using Pwo DNA polymerase (Roche Molecular Biochemicals) according to the manufacturer's recommendations. The constructs were sequenced to confirm the correct introduction of mutations.
Production of recombinant proteins
His6-XCdc6 and His6-XCdc6 mutants were expressed in Sf9 insect cells infected with the corresponding recombinant baculovirus and purified as described in Coverley et al. (2000).
Antibody production and immunodepletions
Antibodies were raised in rabbits using full-length recombinant XCdc6 as an antigen and were affinity-purified (Harlow and Lane 1988). Rabbit anti-XMcm3 and anti-XOrc1 antibodies were the same as described in Madine et al. (1995a) and Romanowski et al. (1996). Monoclonal anti-His antibody (Clontech) was used according to the manufacturer's recommendations. Immunodepletions with anti-XCdc6 antibodies performed as described (Madine and Coverley 1997). Mock-depletions were similarly performed by using preimmune serum or control antibodies (rabbit anti-goat IgG, Sigma).
Using the recombinant protein as a standard, we estimated the concentration of native XCdc6 in the egg extract at approximately 10 ng/μL of extract.
Replication reactions and density substitution experiments
Low-speed Xenopus egg extracts and demembranated sperm nuclei were prepared essentially as reported in Blow and Laskey (1986). Replication reactions were performed exactly as described in Madine et al. (1995b). Replication was detected by incorporation of biotin-16-dUTP (20 μM, Roche) or [α-32P]dATP (100 μCi/mL, Amersham). For density substitution experiments, reactions were incubated in the presence of 0.25 mM BrdUTP and 100 μCi/mL [α-32P]dATP and were processed as in Madine et al. (1995b). Gradient fractions were collected and counted after precipitation with trichloroacetic acid.
Sperm chromatin isolation and detergent washes
For detergent washes before immunoblotting, replication reactions were diluted in 500 μL of buffer A (60 mM KCl, 15 mM Tris-HCl at pH 7.4, 15 mM NaCl, 1mM β-mercaptoethanol, 0.5 mM spermidine, and 0.15 mM spermine) containing 0.2% Triton X-100 and were incubated at room temperature for 5 min. Sperm chromatin was isolated by spinning through a 30% sucrose/buffer A cushion for 5 min at 1500 rpm, and SDS sample buffer was directly added to the protein fraction.
Immunofluorescence microscopy
For immunofluorescence, the replication reactions were diluted in buffer A, fixed for 5 min with 4% freshly depolymerized formaldehyde at room temperature, and spun through a 30% sucrose/buffer A cushion onto poly-lysine coated coverslips (Mills et al. 1989). To analyze chromatin bound proteins, 0.2% Triton X-100 was added to buffer A. Coverslips were blocked for 1 hour in PF buffer (PBS, 0.1% Triton X-100, 0.02 % SDS, and 2% BSA) and then incubated with an appropriate dilution of the primary or secondary antibodies as in Coverley et al. (2000). DNA was counterstained with propidium iodide/RNase A (both at 50 ng/mL, Sigma) or 0.5 μM TOTO-3 iodide (Molecular Probes). Slides were analyzed by confocal fluorescence microscopy.
Miscellaneous methods
λ-Phosphatase treatment was performed for 30 min at 30°C as recommended by the manufacturer (New England Biolabs). For in vitro phosphorylation by interphase extracts, 100 ng of recombinant protein (either XCdc6 or XCdc6M9) was used together 1μCi/μL [γ-32P]ATP and incubated for 30 min at 23°C. Anti-His antibody (Clontech) was used for immunoprecipitation according to the manufacturer's recommendations. Leptomycin B, 0.46 μM, (kindly supplied by Dr. M. Yoshida) was used. Samples were resolved by SDS-PAGE using minigels (Bio-Rad Mini-PROTEAN system), or alternatively, 16-cm gels were used to maximize resolution. Sperm nuclei were permeabilized with 100 μg/mL lysolecithin (Sigma).
Acknowledgments
We thank T. Coleman and W. Dunphy for plasmid pHis6-XCdc6 and M. Yoshida for supplying leptomycin B. We are grateful to A. Mills for help with Xenopus extracts and confocal microscopy and to K. Marheineke for help with insect cells. We also thank A. Mills, M. Swietlik, and D. Santamaria for reading the manuscript. This work was supported by the Cancer Research Campaign and the Louis Jeantet Foundation. C.P. was supported by a postdoctoral Human Frontiers Science Program fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cp230@mole.bio.cam.ac.uk; FAX 44-1223-334089.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.176300.
References
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- ————— A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Brown GW, Jallepalli PV, Huneycutt BJ, Kelly TJ. Interaction of S phase regulator cdc18 with cyclin-dependent kinase in fission yeast. Proc Natl Acad Sci. 1997;94:6142–6147. doi: 10.1073/pnas.94.12.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno A, Russell P. Dual functions of CDC6: A yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992;11:2167–2176. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JFX. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Coverley D, Downes CS, Romanowski P, Laskey RA. Reversible effects of nuclear membrane permeabilization on DNA replication: Evidence for a positive licensing factor. J Cell Biol. 1993;122:985–992. doi: 10.1083/jcb.122.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Pelizon C, Trewich S, Laskey RA. Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J Cell Sci. 2000;113:1929–1938. doi: 10.1242/jcs.113.11.1929. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Diffley JF, Nasmyth KA. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Diffley JF. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes & Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Yamada C, Goto H, Yokoyama N, Kuzushima K, Inagaki M, Tsurumi T. Cell cycle regulation of human CDC6 protein intracellular localization, interaction with the human MCM complex, and CDC2 kinase mediated hyperphosphorylation. J Biol Chem. 1999;274:25927–25932. doi: 10.1074/jbc.274.36.25927. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Jallepalli PV, Brown GW, Muzi-Falconi M, Tien D, Kelly TJ. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes & Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wells NJ, Hunter T. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc Natl Acad Sci. 1999;96:6193–6198. doi: 10.1073/pnas.96.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine MA, Coverley D. Xenopus replication assays. In: Dunphy WG, editor. Cell Cycle Control: Methods in Enzymology. New York: Academic Press; 1997. pp. 535–549. [DOI] [PubMed] [Google Scholar]
- Madine MA, Khoo CY, Mills AD, Laskey RA. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature. 1995a;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- Madine MA, Khoo CY, Mills AD, Musahl C, Laskey RA. The nuclear envelope prevents reinitiation of replication by regulating the binding of MCM3 to chromatin in Xenopus egg extracts. Curr Biol. 1995b;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- Mills AD, Blow JJ, White JG, Amos WB, Wilcock D, Laskey RA. Replication occurs at discrete foci spaced throughout nuclei replicating in vitro. J Cell Sci. 1989;94:471–477. doi: 10.1242/jcs.94.3.471. [DOI] [PubMed] [Google Scholar]
- Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Lukas J, Sorensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P, Madine MA. Mechanisms restricting DNA replication to once per cell cycle: MCMs, pre-replicative complexes and kinases. Tr Cell Biol. 1996;6:184–188. doi: 10.1016/0962-8924(96)10015-5. [DOI] [PubMed] [Google Scholar]
- ————— Mechanisms restricting DNA replication to once per cell cycle: The role of Cdc6p and ORC. Tr Cell Biol. 1997;7:9–10. doi: 10.1016/S0962-8924(97)30077-4. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JPJ, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Saha P, Chen JJ, Thome KC, Lawlis SJ, Hou ZH, Hendricks M, Parvin JD, Dutta A. Human CDC6/cdc18 associates with Orc1 and cyclin–cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol Cell Biol. 1998;18:2758–2767. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Stoeber K, Mills AD, Kubota Y, Krude T, Romanowski P, Marheineke K, Laskey RA, Williams GH. Cdc6 protein causes premature entry into S phase in a mammalian cell- free system. EMBO J. 1998;17:7219–7229. doi: 10.1093/emboj/17.24.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Tang Z, Coleman TR, Dunphy WG. Two distinct mechanisms for negative regulation of the wee1 protein kinase. EMBO J. 1995;12:3427–3436. doi: 10.1002/j.1460-2075.1993.tb06017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]