Abstract
Recent studies have demonstrated roles for Spt4, Spt5, and Spt6 in the regulation of transcriptional elongation in both yeast and humans. Here, we show that Drosophila Spt5 and Spt6 colocalize at a large number of transcriptionally active chromosomal sites on polytene chromosomes and are rapidly recruited to endogenous and transgenic heat shock loci upon heat shock. Costaining with antibodies to Spt6 and to either the largest subunit of RNA polymerase II or cyclin T, a subunit of the elongation factor P-TEFb, reveals that all three factors have a similar distribution at sites of active transcription. Crosslinking and immunoprecipitation experiments show that Spt5 is present at uninduced heat shock gene promoters, and that upon heat shock, Spt5 and Spt6 associate with the 5′ and 3′ ends of heat shock genes. Spt6 is recruited within 2 minutes of a heat shock, similar to heat shock factor (HSF); moreover, this recruitment is dependent on HSF. These findings provide support for the roles of Spt5 in promoter-associated pausing and of Spt5 and Spt6 in transcriptional elongation in vivo.
Keywords: Pol II pausing, elongation, heat shock genes, Spt5, Spt6, P-TEFb
The Drosophila melanogaster heat shock gene hsp70 has a promoter architecture that is primed for activation under non–heat shock conditions. The promoter sequences of the uninduced gene are in a nucleosome-free open configuration and are occupied by at least three transcription factors: GAGA factor (GAF), TATA-binding protein (TBP), and RNA polymerase II (Pol II; for review, see Lis 1998). GAF elements have been shown to be important for establishing and maintaining the potentiated chromatin architecture of hsp70 (Lee et al. 1992). It has been proposed that GAF occludes nucleosomes from occupying the hsp70 promoter (Tsukiyama et al. 1994), thereby enabling access to TBP, Pol II, and on heat shock, heat shock factor (HSF) (Shopland et al. 1995). The Pol II molecule at the uninduced hsp70 promoter has been shown to be in a transcriptionally engaged but paused state, having produced short RNA transcripts of ∼21–35 nucleotides (Rougvie and Lis 1988; Rasmussen and Lis 1993). Escape of the paused polymerase is thought to represent the rate-limiting step in transcriptional elongation and is a feature of other Drosophila, human, and viral genes (Laspia et al. 1989; Rougvie and Lis 1990; Krumm et al. 1992; Strobl and Eick 1992; Yankulov et al. 1994).
One hallmark of the paused polymerase molecule is that its C-terminal domain (CTD), which is a substrate for posttranslational modification by protein kinases, is hypophosphorylated (Weeks et al. 1993; O'Brien et al. 1994). Previous studies have demonstrated that the hypophosphorylated form of RNA Pol II (Pol IIa) enters the preinitiation complex, whereas the hyperphosphorylated form (Pol IIo) is actively engaged in transcription elongation (Lu et al. 1991; Dahmus 1994). The CTD of Pol II has a heptapeptide repeat that is conserved from yeast to humans and contains the amino acid sequence YSPTSPS. The second and fifth serines of each repeated unit are major sites of modification by at least two candidate kinases: Cdk7 (Ser 2), a subunit of the general transcription factor TFIIH (Roy et al. 1994), and Cdk9 (Ser 2 or Ser 5), a subunit of positive transcription elongation factor b (P-TEFb; Zhou et al. 2000). P-TEFb is composed of the cyclin-dependent kinase Cdk9 and its cognate cyclin, cyclin T, and has recently been implicated in the regulation of heat shock gene expression (Lis et al. 2000).
Price and colleagues isolated P-TEFb as an elongation factor that stimulated a shift from production of short to long transcripts in vitro (Marshall and Price 1995). These short transcripts are similar in size to those found in vivo at promoters with paused polymerases. The inhibitor of P-TEFb kinase activity, 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), causes the production of short transcripts both in vitro and in vivo (Wada et al. 1998a, and references therein). This DRB effect in vitro requires the presence of a factor termed DRB sensitivity inducing factor (DSIF). DSIF is a heterodimer comprising the mammalian homologues of yeast Spt4 and Spt5 and interacts biochemically and genetically with Pol II through Spt5 (Hartzog et al. 1998; Wada et al. 1998a). This interaction is thought to restrict Pol II to the synthesis of short transcripts in the absence of P-TEFb. Spt5 has several distinct domains, including four regions with similarity to the bacterial transcriptional elongation factor NusG, and two C-terminal elements, CTR1 and CTR2, that have similar composition to the C-terminal domain of the Pol II (Yamaguchi et al. 1999b). P-TEFb can phosphorylate the Spt5 CTR1 domain in vitro, suggesting that P-TEFb-mediated phosphorylation of either the Pol II CTD or the Spt5 CTR1 domain or both leads to removal of DSIF inhibition (Ivanov et al. 2000).
Spt4, Spt5, and a related factor, Spt6, appear to have a positive effect on transcriptional elongation. In vitro transcription using limiting nucleotide concentrations, and analysis of synthetic genetic interactions in yeast, have shown that Spt4, Spt5, and Spt6 promote transcriptional elongation (Hartzog et al. 1998; Wada et al. 1998a). Biochemically, Spt4 and Spt5 have been shown to be in a heterodimeric complex that excludes Spt6; nonetheless, Spt5 and Spt6 are functionally related and have been shown to interact, albeit very weakly, in extracts and pull-down assays (Hartzog et al. 1998). Spt6 could potentially exert its effect on elongation by facilitating transcription through chromatin. It interacts genetically and biochemically with histones H3 and H4 and regulates chromatin structure in vivo and in vitro (Bortvin and Winston 1996). Although these studies have provided insights into the roles of Spt4, Spt5, and Spt6 in transcriptional elongation, their mechanism of action in vivo remains to be clarified.
In this article, we examine the locations of the D. melanogaster homologs of Spt5 and Spt6 proteins on heat shock genes in vivo at high and low resolution. We asked whether their localization patterns were consistent with their proposed roles in establishing a promoter-paused polymerase, facilitating transcriptional elongation, or both. We obtained antibodies to Spt5 (a marker for DSIF) and to Spt6 and determined where these factors were localized on chromosomes. We used polytene immunofluorescence to investigate the low-resolution global and local distribution of Spt5 and Spt6 and crosslinking and immunoprecipitation with specific antibodies to address the high-resolution localization of Spt5 and Spt6 under non–heat shock and heat shock conditions. The immunoprecipitated material was examined using polymerase chain reaction (PCR) or ligation-mediated PCR (LMPCR). From these studies, we propose a role for Spt5 in regulating heat shock gene pausing and activation and roles for both Spt5 and Spt6 in transcriptional elongation. The accompanying study (Kaplan et al. 2000, this issue) also presents an immunofluorescence analysis that corroborates and extends our findings, and strongly supports a role for Spt5 and Spt6 in transcriptional elongation.
Results
Generation and analysis of antibodies to Spt5, Spt6, and HSF
Recent studies have implicated DSIF (Spt4 and Spt5) and Spt6 in the control of transcriptional elongation in yeast and mammals (Hartzog et al. 1998; Wada et al. 1998a,b). We identified the Drosophila homologs of Spt5 and Spt6, which was facilitated by the genome project (Adams et al. 2000), and have generated antibodies to these proteins to evaluate their roles in vivo. To ensure that the signals obtained with the antibodies were specific to these proteins, we performed experiments with antibodies raised in guinea pig, rabbit, and chicken for Spt5 and in both guinea pig and rat for Spt6. Three Spt5 antibodies were generated to non-overlapping fragments of the 1054 amino acid (aa) protein: an N-terminal fragment (aa 112–393) in chicken, a middle fragment (aa 389–733) in rabbit, and a C-terminal fragment (aa 732–1054) in guinea pig. Each of the three Spt5 antibodies predominantly recognizes a ∼150 kD band (Fig. 1A, lanes 2,4,6). Although this is larger than the 117-kD predicted molecular weight, such a reduction in electrophoretic mobility is not surprising for an acidic protein (Wu et al. 1987). The preimmune serum does not detect any bands (Fig. 1A, lanes 1,3,5). Combinations of Spt5 antibodies colocalize mostly on polytene chromosomes (Fig. 2A). This result indicates that although the antibodies to Spt5 were raised in different animals and to different epitopes, they appear to recognize the same protein in vivo. For antibodies against Spt6, the same C-terminal fragment was injected into both animals. Western blotting of whole Drosophila Kc cell culture proteins shows that both Spt6 antibodies recognize a band of ∼215 kD, which is not detected in preimmune serum (Fig. 1A, cf. lanes 7,9 with lanes 8,10). This size is in agreement with the predicted molecular mass of Spt6, which is 209 kD. The few additional bands vary among preparations and appear to be degradation products of this very large protein (data not shown). Coimmunofluorescence staining with both Spt6 antibodies showed a nearly complete colocalization on polytene chromosomes (Fig. 2B), supporting the conclusion that both specifically recognize Spt6 in vivo.
Figure 1.
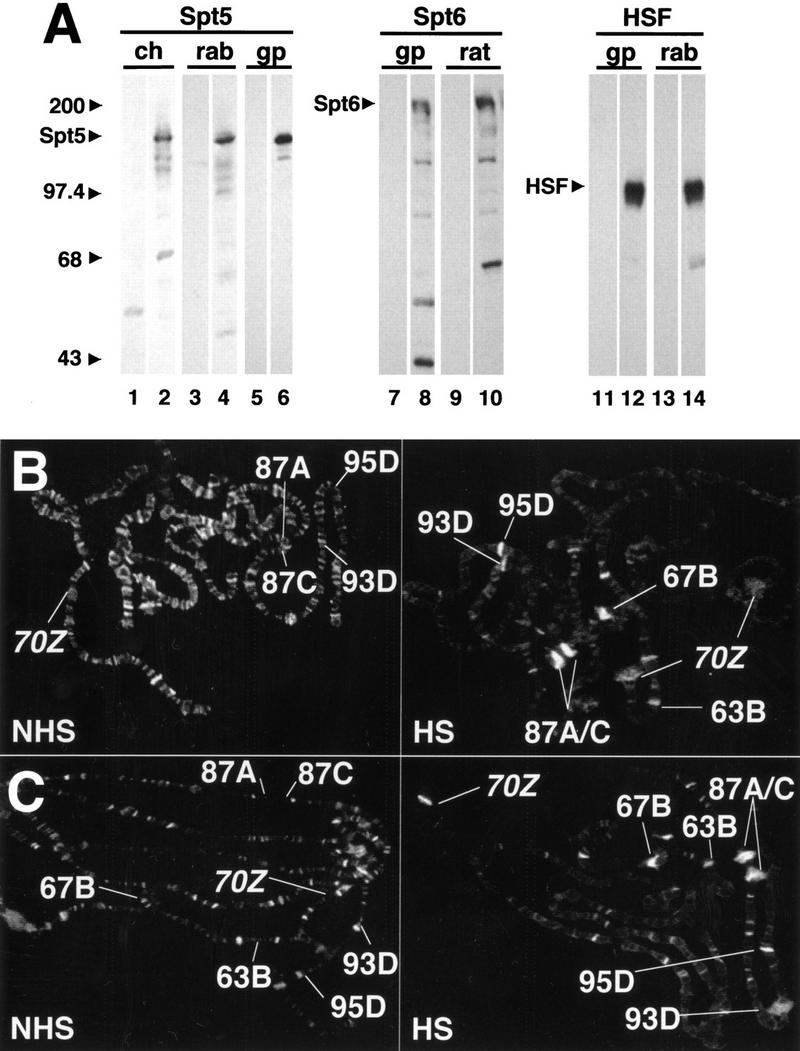
Spt5 and Spt6 are recruited to heat shock puffs on Drosophila polytene chromosomes. (A) Western blot analyses of Spt5, Spt6, and HSF antibodies. Proteins from Drosophila Kc cells were separated on an 8% polyacrylamide gel and transferred to nitrocellulose. Blots were probed with antiserum (even lanes) or with preimmune serum (odd lanes) at a dilution of 1 : 1000. Appropriate secondary antibodies were used and bands visualized using ECL (Amersham). Aligned nitrocellulose strips for each antibody are from the same transferred gels. (Ch) chicken; (rab) rabbit; (gp) guinea pig; (rat) rat. (B,C) Staining of non–heat shock (NHS) and heat shock (HS) polytene chromosomes with rabbit α-Spt5 (B) or guinea pig α-Spt6 (C) antibodies. Major, endogenous heat shock loci (87A, 87C, 63B, 67B, 93D, and 95D) as well as the transgenic sites (hsp70–LacZ [70Z]) are defined. Note the redistribution of Spt5 and Spt6 on heat shock. The staining pattern of both Spt5 and Spt6 is quite similar to that seen previously with cyclin T (Lis et al. 2000).
Figure 2.
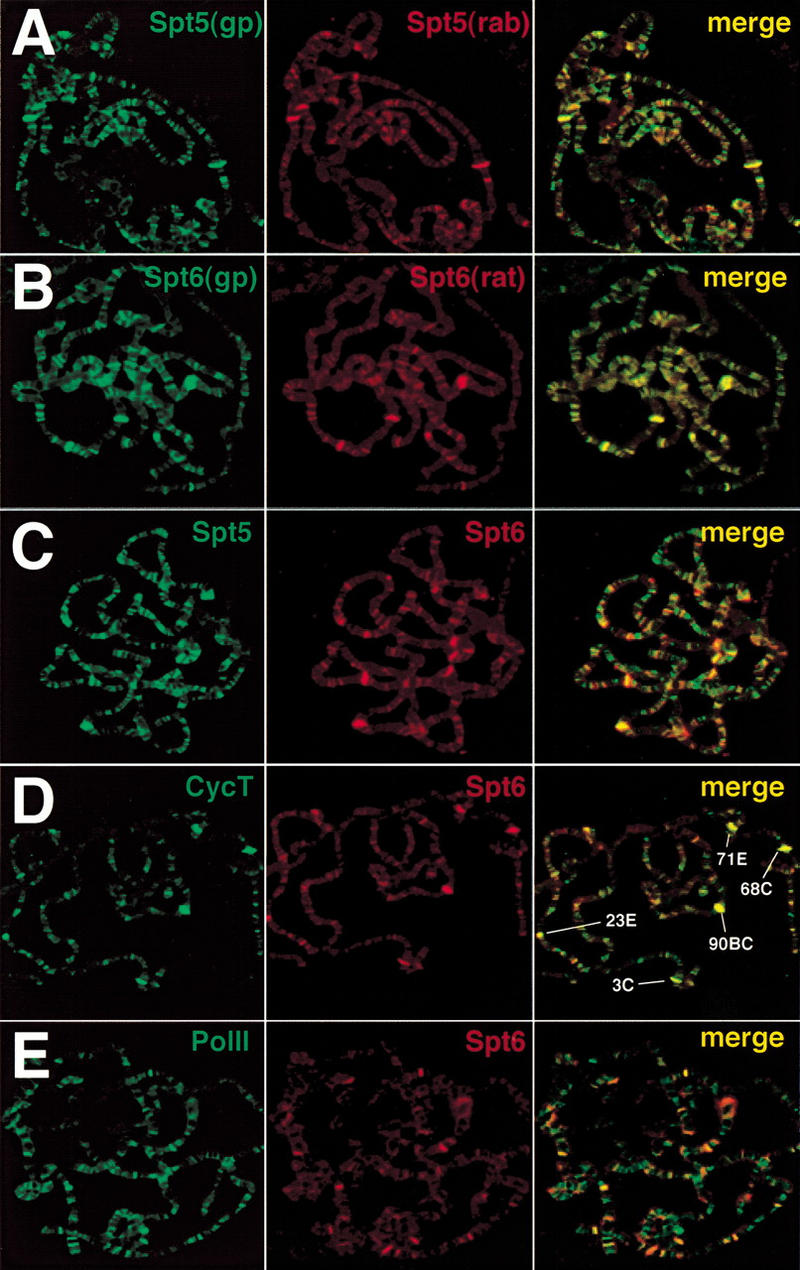
Global immunofluorescence analysis of Spt5 and Spt6 on polytene chromosomes under non–heat shock conditions. (A) Costaining with different Spt5 antibodies. (Rab) Rabbit; (gp) guinea pig. (B) Costaining with different Spt6 antibodies. (C) Costaining with Spt5 (rab) and Spt6 (gp) antibodies. (D) Costaining with Spt6 (gp) and cyclin T (rab) antibodies. (E) Costaining with Spt6 (gp) and Pol II (rab) antibodies. Overlap of the green and red stains appear in the merge as yellow, yellow-green, and orange.
Antibodies were also raised to full-length HSF in both guinea pig and rabbit. These clearly recognize a major band that has the expected mobility of HSF (Wu et al. 1987; Fig. 1A, lanes 12,14). Moreover, the two HSF antibodies gave identical immunofluorescence signals, and preimmune serum produced no signals in Western blots or on polytene chromosomes (Fig. 1A, lanes 11,13; data not shown).
Immunolocalization of Drosophila Spt5 and Spt6 on polytene chromosomes
Clues to the potential function of transcription factors can be discerned by analysis of their distribution on polytene chromosomes. To investigate the possible roles of Spt5 and Spt6, we examined their chromosomal localization by immunofluorescence using polytene chromosomes prepared from either wild-type or transgenic non–heat shocked or heat shocked third instar larvae. Under non–heat shock conditions, Spt5 is distributed throughout the polytene chromosomes and stains the stage-dependent, ecdysone-induced developmental puffs, which are known to be transcriptionally active (Fig. 2A). The Spt5 pattern is dependent on the developmental stage of larvae and is most concentrated at sites of highly active transcription. The pattern closely resembles that observed previously and is mapped in detail for the cyclin T subunit of P-TEFb (Lis et al. 2000; below). Spt6 is also found at developmental puffs and shows an overall pattern that is quite similar to that of Spt5 (Fig. 2, cf. B with A). To determine whether Spt5 and Spt6 were localized to the same chromosomal loci, we performed coimmunofluorescence experiments with various combinations of Spt5 and Spt6 antibodies. Irrespective of the combination of antibodies from different animals that were tested, the results were the same: Spt5 and Spt6 displayed considerable colocalization on polytene chromosomes, especially at the most intensely labeled chromosomal puffs, which are known to be sites of highly active transcription (Fig. 2C). Although the resulting merge of staining patterns produces some bands that are more green or red, close examination shows that most sites are labeled by both to varying degrees. Hence, Spt5 and Spt6 are found together at sites of active transcription.
To verify that Spt5 and Spt6 are recruited to transcriptionally active sites, we examined their localization to heat shock loci. Whereas there is some Spt5 staining at, or near, uninduced heat shock loci, both Spt5 and Spt6 were strongly recruited to the activated major heat shock loci: hsp70 genes at 87A and 87C, small heat shock genes at 67B, hsp83 at 63B, hsp68 at 95D, and the noncoding heat shock transcription units at 93D (Fig. 1B,C). In addition, Spt5 and Spt6 can be observed during heat shock at sites containing transgenic hsp70–LacZ genes located at 61A and 9E or 59B (70Z; Fig. 1B and C, respectively). Thus, Spt5 and Spt6 are recruited to endogenous and transgenic heat shock loci during heat shock gene activation.
Comparative immunofluorescence of Spt5 and Spt6 with HSF, cyclin T, and the largest subunit of RNA polymerase II
We compared the distribution of Spt5 or Spt6 to those of other factors that are critical for transcription elongation: cyclin T and Pol II. The coimmunofluorescence patterns were examined initially under non-heat shock conditions on polytene chromosomes. Because Spt5 co-localizes with Spt6 (Fig. 2C), we present comparisons of only Spt6 with the other factors. We have observed previously that cyclin T (P-TEFb) is concentrated at transcriptionally active loci (Lis et al. 2000). Costaining experiments with Spt6 and cyclin T show that they colocalize most prominently at the highly active developmental loci in noninduced animals (Fig. 2D). At this early third instar stage (puff stage 2), the major sites labeled are 3C, 23E, 68C, 71E, and 90BC. Four out of five of these loci contain the abundantly expressed salivary gland secretory proteins. We observed a similar pattern of colocalization between Spt6 and Pol II, specifically at sites of highly active transcription (Fig. 2E).
HSF rapidly accumulates at major heat shock loci following heat shock. It also concentrates at an additional set of ∼150 loci, most of which are not highly transcribed during heat shock (Westwood et al. 1991). Spt6 does not appear at these additional loci under heat shock conditions (Fig 3A).
Figure 3.
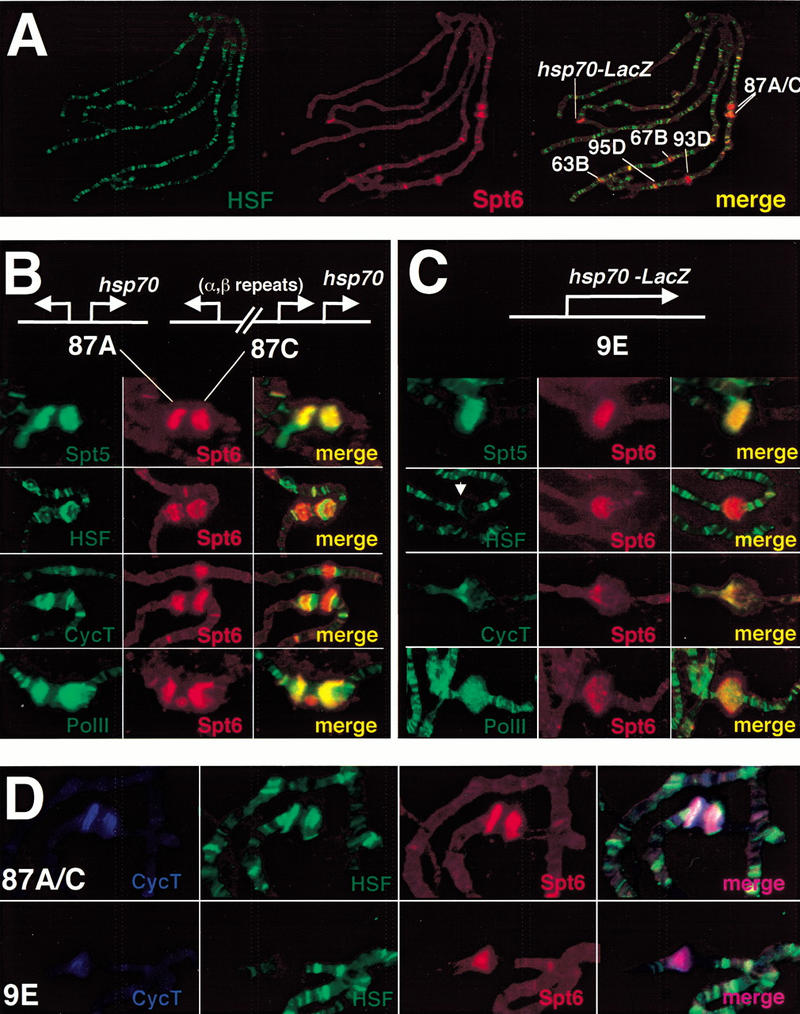
Global and high-resolution analysis of Spt6 at native and transgenic heat shock puffs. (A) Comparative immunofluorescence of Spt6 (rat) and HSF (rabbit) under heat shock conditions. Spt6 is recruited to heat shock loci but not to all sites of HSF localization. (B,C) High-resolution study of Spt6 distribution in comparison to that of HSF, cyclin T, and Pol II. Maps of the native heat shock loci 87A and 87C and the hsp70–LacZ transgene are shown. HSF and cyclin T resolve from Spt6 at both the native and transgenic heat shock loci. At the transgene, these antibodies stain the locus in a region that would be consistent with promoter and 5′ sequences. Note the HSF staining pattern (white arrow). The site above 87A and 87C in the cyclin T and Spt6 costain is the heat shock locus 67B. Spt6 and Pol II show almost complete overlap within the heat shock puffs. (D) Triple label with cyclin T, HSF, and Spt6.
To extend our observation that Spt5 and Spt6 are recruited to heat shock loci and to map the distribution of these factors, we have performed high-resolution examinations at major heat shock puffs (87A and 87C) and at an hsp70–LacZ insert in a transgenic fly line (Simon et al. 1985; Fig. 3B,C). Spt5 and Spt6 stained heavily the endogenous heat shock puffs at 87A and 87C and appear coincident within the puffs. Spt6 likewise colocalizes with Pol II but partially resolves from cyclin T and HSF (Fig. 3B). The transgenic insert allows for an unambiguous assignment of a fluorescent band to a particular DNA segment because it is located at a particular chromosomal site that normally does not contain heat shock genes. Also, the transgenic locus, unlike the endogenous hsp70 loci, contains a single hsp70 gene. Thus, by localizing factors to this transgenic locus, we can make more straightforward interpretations of protein distribution within a puff. Staining this site under heat shock conditions allows us to differentiate the discrete distributions of Spt6 and HSF within the transgenic locus: Spt6 showed a staining pattern within the body of the heat shock puff, whereas HSF was restricted to the edge of the puff (Fig. 3C). Spt6 and cyclin T showed a similar distribution, although cyclin T was more often concentrated in the promoter-proximal half of the puff. The triple immunofluorescence labeling with HSF, cyclin T, and Spt6 antibodies shows that cyclin T and Spt6 significantly overlap within native and transgenic puffs and resolve from HSF (Fig. 3D). In addition, coimmunofluorescence of Spt6 with Pol II always exhibited significant overlap of staining within the body of the transgenic puff. In sum, Spt5 and Spt6 colocalize with Pol II, resolve from HSF, and may partially resolve from cyclin T in endogenous and transgenic heat shock puffs on polytene chromosomes.
Crosslinking and immunoprecipitation analysis of the heat shock genes hsp70, hsp26, and hsp83
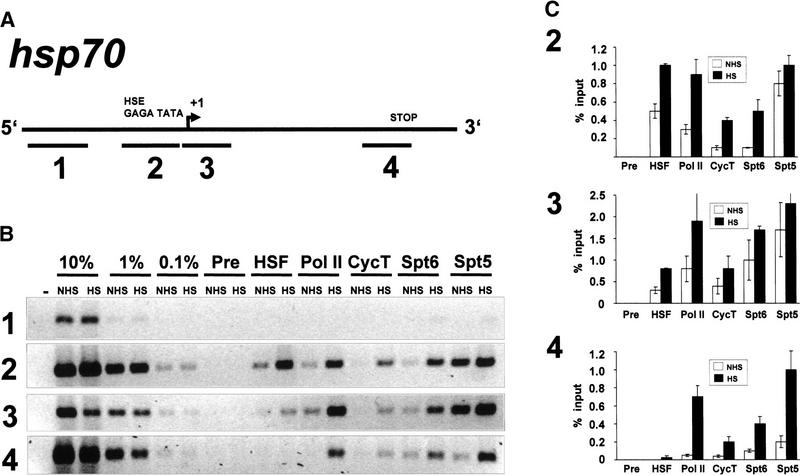
The cytological localization of Spt5 and Spt6 at both native and transgenic heat shock loci indicates that these proteins associate with heat shock genes. To confirm this and to map the location of these proteins more precisely, we performed crosslinking and immunoprecipitation assays. We treated Drosophila tissue culture cells with formaldehyde under non–heat shock or heat shock conditions, isolated crosslinked protein–DNA complexes on CsCl gradients, and immunoprecipitated these complexes using specific antibodies. The coimmunoprecipitated DNA was isolated and analyzed by polymerase chain reaction (PCR) using primers that amplified promoter and 5′- or 3′-transcribed fragments of hsp70 (Fig. 4A, fragments 2,3,4). The amount of coprecipitated DNA was compared to standards consisting of 10-fold serial dilutions of the total DNA used in the crosslinking and immunoprecipitation assays. As a control for immunoprecipitation of specific DNA, we used PCR primers far upstream of the hsp70 promoter (Fig. 4A, fragment 1). This analysis was repeated multiple times, and the ethidium bromide-stained bands in the immunoprecipitates were quantified using imaging software. Results from a typical experiment are shown for each fragment assayed by PCR (Fig. 4B), and the average values of several experiments with standard deviations are also shown (Fig. 4C).
Figure 4.
Crosslinking and immunoprecipitation of Spt5 and Spt6 at hsp70. (A) Schematic representation of hsp70 and the PCR-amplified fragments representing the upstream region (1), the promoter (2), the 5′ end (3), and the 3′ end (4) of hsp70. The promoter elements, transcription start site, and stop are labeled but not drawn to scale. (B) PCR analysis of crosslinked and immunoprecipitated DNA on ethidium bromide (EtBr)-stained 2% agarose gels. PCR reactions using 10%, 1%, and 0.1% of input DNAs are loaded to determine the linear range of signal. NHS, non–heat shock; (HS) heat shock; (Pre) preimmune serum; (−) no DNA. (C) Quantitative analysis of EtBr-stained bands for PCR analysis. Values on the abscissa represent the amount immunoprecipitated as a percentage of total input DNA. Experiments were performed in triplicate, and standard deviations are shown. Note different scale for bar graph of fragment 3 immunoprecipitates.
Previous UV crosslinking studies have demonstrated that in uninduced cells, Pol II is associated with hsp70 promoter-proximal fragments (Gilmour and Lis 1986). Additional studies have demonstrated that this Pol II is in a paused complex that has synthesized 21–35 nucleotides of RNA (Rasmussen and Lis 1993). Here we also observe that Pol II antibody immunoprecipitates the hsp70 promoter and 5′ fragments under non–heat shock conditions (Fig. 4C). This observation is consistent with a paused polymerase occupying the 5′ end of the gene before heat shock. We also observe a threefold increase in the 5′ DNA fragments and a greater than a 10-fold increase in 3′ hsp70 DNA fragments being immunoprecipitated with Pol II antibodies under heat shock conditions. This is consistent with the estimated increase in density of Pol II upon heat shock. The uninduced gene contains a single Pol II associated with the 5′ end of the gene, whereas the induced gene has both a promoter-proximal paused Pol II and, spaced every 100 bp, elongating Pol II complexes (O'Brien and Lis 1993).
Recent in vitro studies have implicated Spt5 and Spt4 in promoter-proximal pausing of RNA polymerase II (Wada et al. 1998a). In order to determine whether this could be the case in vivo, we performed crosslinking and immunoprecipitation assays with Spt5 antibodies and determined whether Spt5 was at the 5′ end of hsp70. With non–heat shocked samples, the Spt5 antibody immunoprecipitated approximately eightfold more hsp70 promoter and 5′ DNA than 3′ fragments. A quantitative comparison of the immunoprecipitated DNA showed that the association of Spt5 with the promoter and 5′ regions in samples prepared from uninduced and induced cells were quite similar but appear to increase on heat shock by ∼30% (Fig. 4C). We observe an approximately fivefold increase in the 3′ fragment being immunoprecipitated by Spt5 antibodies on heat shock. From these results, we conclude that Spt5 is at a relatively high density on the promoter and 5′ region of the hsp70 gene in uninduced cells and that on heat shock, the density of Spt5 on the transcription unit increases dramatically as well.
Heat shock triggers the recruitment of several factors to either the promoter or transcribed regions of heat shock genes. Activated HSF binds tightly to elements in the promoter regions of hsp70; therefore, we examined HSF distribution using the crosslinking and immunoprecipitation assay. HSF antibodies immunoprecipitated DNA fragments representing the promoter and 5′ end of the gene but not the 3′ hsp70 fragment (Fig. 4C). The levels of immunoprecipitated DNA increased approximately twofold in response to heat shock, and the accumulation was restricted to the promoter and 5′ regions (Fig. 4C).
Immunofluorescence studies of polytene chromosomes have shown that both P-TEFb (cyclin T subunit; Lis et al. 2000) and Spt6 (this study) are recruited to heat shock puffs. We examined their recruitment and distribution at higher resolution using crosslinking and immunoprecipitation. Both Spt6 and cyclin T antibodies immunoprecipitate low levels of the promoter fragment in uninduced cells, but four- to fivefold more from induced cells (Fig. 4, C2). Spt6 and cyclin T are concentrated on the 5′ transcribed sequence, as both antibodies precipitate approximately threefold more 5′ fragment than promoter fragment (Fig. 4, cf. C3 with C4). In addition, Spt6 and cyclin T immunoprecipitates show a fourfold increase in the 3′ fragment from non–heat shock to heat shock conditions, suggesting that these factors are also recruited to the 3′ end of the gene (Fig. 4, C4). These data indicate that Spt6 and cyclin T may occupy the uninduced hsp70 gene 5′ region at low levels and accumulate to higher density on the body of the gene following heat shock.
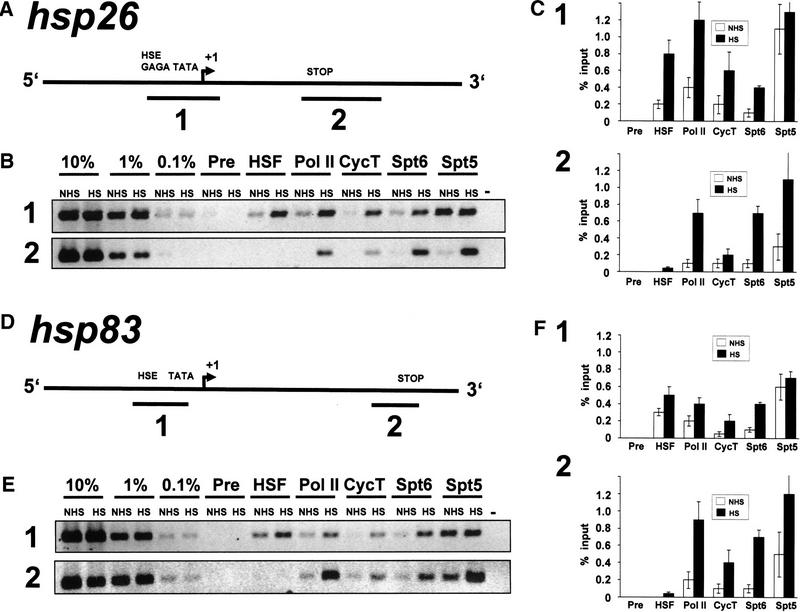
To investigate the distribution of these proteins on other heat shock genes, we performed crosslinking and immunoprecipitation analysis on hsp26 and hsp83 using the same collection of antibodies (Fig. 5). These results with the primers amplifying either the hsp26 promoter or 3′ end are qualitatively similar to those seen with analogous fragments of hsp70. The heat shock gene promoters of hsp26 and hsp83, as at hsp70, are occupied by high levels of Spt5 even before heat shock. In addition, the 5′ and 3′ regions show a low level of association of Spt6, cyclin T, and Pol II in uninduced cells, and their levels increase on heat shock. Analysis of coimmunoprecipitated hsp83 DNA differs slightly from the pattern seen with hsp26 and hsp70 in that more DNA is precipitated with antibody to HSF from uninduced protein–DNA preparations (cf. Fig. 5E with Figs. 4C and 5C). This result is consistent with the immunofluorescence studies showing that hsp83 is the one heat shock locus that is significantly stained with HSF antibody in non–heat shocked flies (Westwood et al. 1991). Thus, these analyses indicate that these different transcription factors show characteristic distributions along the promoter and transcribed regions of heat shock genes: HSF is restricted to the promoter regions, and Spt5, Spt6, cyclin T, and Pol II are more broadly distributed over the induced transcription unit.
Figure 5.
Crosslinking and immunoprecipitation of Spt5 and Spt6 at hsp26 and hsp83. (A,D) Schematic representations of hsp26 and hsp83 and the PCR-amplified fragments representing the promoter (1) and the 3′ end (2). (B,E) PCR analysis of crosslinked and immunoprecipitated DNA on EtBr-stained 2% agarose gels of hsp26 (B) and hsp83 (E). (C,F) Quantitative analysis of EtBr-stained bands for PCR analysis of hsp26 (C) and hsp83 (F). Experiments were performed in triplicate, and standard deviations are shown. (NHS) non–heat shock; (HS) heat shock; (Pre) preimmune serum; (−) no DNA.
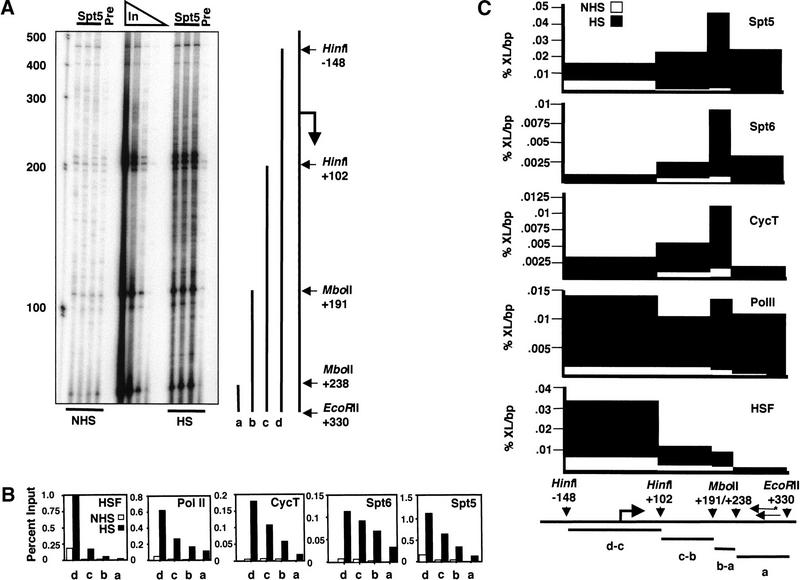
To analyze the distribution of these factors at even higher resolution, we digested the crosslinked protein–DNA complexes with a restriction endonuclease before immunoprecipitation and analyzed the coprecipitated DNA by ligation-mediated PCR (LMPCR; Guzman and Lis 1999). This restriction endonuclease cleavage of protein–DNA complexes allows the location of protein–DNA crosslinks to be defined at the level of a restriction map. The digested DNA fragments, once immunoprecipitated, are processed as described in Materials and Methods. As seen in Figure 6A, partial digestion of the hsp70 DNA, followed by immunoprecipitation and LMPCR, leads to the creation of several specific bands that can be quantified. We calculated and plotted the percentage of immunoprecipitated material relative to totals for each fragment (Fig. 6B) and then plotted the relative protein density on the individual fragments (Fig. 6C). We used this higher-resolution LMPCR technique to map the distribution of the set of transcription factors analyzed by the conventional PCR method described above.
Figure 6.
High-resolution determination of protein occupancy on hsp70. (A) Ligation-mediated PCR analysis on hsp70. hsp70 restriction endonuclease sites and fragments created by partial digestion at HinfI, and MboII sites are shown to right of the gel, and these define one end of the region analyzed: EcoRII was used to cut the DNA to completion and thereby define the other end of the region analyzed. The fragments produced by this digest are shown: a, b, c, and d. The complete analysis, starting with the immunoprecipitation step, was performed in triplicate, and the different samples show very good agreement for all bands that were quantified. “Pre” is an immunoprecipitation performed with a preimmune serum control. (B) Determination of amount of hsp70 DNA immunoprecipitated relative to input DNA. Bands were quantified, and background signals from the preimmune controls were subtracted in each case. The percentage of the total sample that is immunoprecipitated for each of the four fragments is plotted. The triplet shown at HinfI +102 (see A) results from small-length polymorphisms among the hsp70 genes, and the signal from the entire triplet was used to quantify this fragment. Note that HSF and Pol II antibodies coimmunoprecipitate more DNA than do Spt6 and cyclin T antibodies (note the different scales for the abscissae). (C) Determination of protein density on hsp70. The densities were calculated from the measurements in part B. The densities from each interval were derived from subtraction of the next shortest fragment (except for a, which is the shortest), and then dividing by the length of the DNA interval. Note the different scales for the abscissae. (%XL) Percentage crosslinked immunoprecipitated DNA relative to input; (bp) base pair; (*) 32P end-label.
The results with the LMPCR approach are in good agreement with those of the lower-resolution PCR analysis. For example, the promoter/5′ DNA fragment (−148 to +102) coprecipitates with Pol II antibody before heat shock, and its level increases on heat shock (Fig. 6C). These observations are consistent with Pol II being associated with the proximal-promoter region under uninduced conditions and being recruited to high density on heat shock. The −148 to +102 fragment also coprecipitates with Spt5 and HSF antibodies before heat shock, and its level increases on heat shock (Fig. 6C). Neither Spt6 nor cyclin antibodies coprecipitate detectable levels of the −148 to +102 fragment before heat shock, but both do upon heat shock. Whereas our analysis detects a low level of coprecipitated DNA from the +102 to +238 region under non–heat shock conditions for all five antibodies (Fig. 6C), the levels increase dramatically on heat shock. The +238 to +330 segment is the most distal fragment analyzed and is not detectably coprecipitated before heat shock by any of the antibodies analyzed; however, it is efficiently coprecipitated by antibodies to Pol II, Spt5, and Spt6 on heat shock. From these observations, Spt5, Pol II, and HSF appear to occupy the region from −148 to +102 in uninduced cells. Heat shock triggers the strong recruitment of Pol II, Cyclin T, Spt5, and Spt6 into the +102 to +330 region of the hsp70 gene; however, HSF is recruited predominantly to the fragment containing the promoter. Thus, this LMPCR approach confirms and extends the lower-resolution PCR and immunofluorescence analyses.
Rapid recruitment of Spt6 to heat shock loci requires HSF
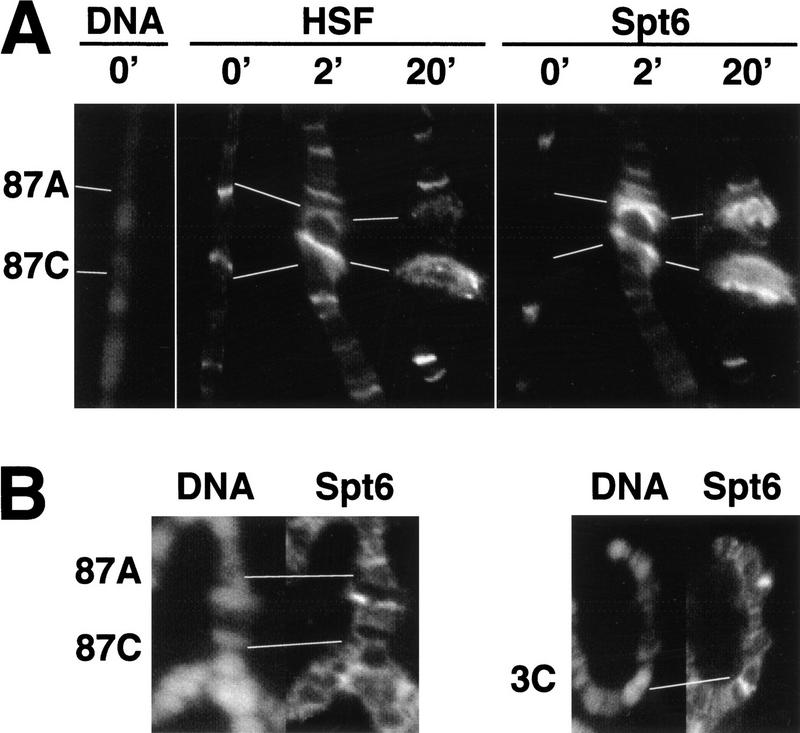
Heat shock induces the binding of HSF to the hsp70 promoter within 2 min of a heat shock treatment (Zimarino and Wu 1987; Weeks et al. 1993). Pol II transcription of hsp70 is stimulated by heat shock with a similar rapidity (O'Brien and Lis 1993). If Spt6 has a critical role in the up-regulation of heat shock genes, then it should be rapidly recruited to these genes on their induction. To test this, we examined the staining of Spt6 at heat shock loci as a function of time following heat shock. As seen in Figure 7A, Spt6 is recruited to endogenous heat shock genes 87A and 87C with a heat shock of 2 min, and its kinetics of recruitment mirrors HSF and what was seen previously for cyclin T (Lis et al. 2000). Moreover, Spt6 and cyclin T persist at these puffs at 20 min, but they are no longer present after a recovery period following heat shock (data not shown). Thus, Spt6 localizes to heat shock loci during their induction with kinetics that suggest a role in the activated transcription of heat shock genes.
Figure 7.
Kinetics of recruitment of Spt6 to heat shock loci. (A) Spt6 recruitment at major heat shock loci 87A and 87C on untreated (0′) polytene chromosomes or on 2- and 20-min heat shock–treated chromosomes. A DNA Hoechst stain is shown for the uninduced sample to allow mapping of the non-puffed heat shock loci. (B) Recruitment of Spt6 to heat shock loci on polytene chromosomes in hsf4 mutant. Shown are the 87A and 87C heat shock loci (left) and the developmentally active locus at 3C (right) after a 20-min heat shock within the same polytene chromosome spread. 87A and 87C display little or no staining with Spt6 antibody, whereas Spt6 staining at the developmentally active locus at 3C persists.
We also addressed whether HSF recruitment was required for recruitment of Spt6 to heat shock genes. To accomplish this, immunofluorescence analysis was performed in a temperature-sensitive (hsf4) strain with Spt6 antibodies. Following a shift to the restrictive temperature in the hsf4 mutant, HSF loses its ability to bind to DNA and is incapable of eliciting a heat shock response (Jedlicka et al. 1997). In the hsf4 strain, puffs are absent following a shift to the nonpermissive temperature, and Spt6 is not recruited to the 87A and 87C loci on heat shock (Fig. 7B). However, Spt6 persists at developmentally expressed loci such as the one at the 3C locus (Fig. 7B). This result indicates that recruitment of Spt6 to heat shock loci is dependent on HSF recruitment and binding.
Discussion
The heat shock inducible gene hsp70 is an extensively characterized model system for studying the regulation of transcriptional elongation, especially in the transition from initiation to a fully competent elongation complex. This is a critical phase in transcription, when Pol II must break its contacts with the promoter and promoter-associated factors and acquire the entourage of proteins needed for efficient elongation and RNA processing. The uninduced hsp70 gene has a promoter-proximal RNA polymerase II molecule that has paused after transcribing a short RNA of 21–35 nucleotides (Lis 1998). The escape of Pol II from this pause to a mature elongating complex appears to be the rate-limiting step in hsp70 transcription. From studies in vitro, Spt5 and Spt4 (DSIF) have emerged as critical factors in generating the pause (Wada et al. 1998a,b), whereas in yeast genetic studies, Spt4, Spt5, and Spt6 have been implicated in the control of transcriptional elongation (Hartzog et al. 1998). Here, we examined the roles of the Drosophila Spt5 and Spt6 in heat shock gene regulation. We analyzed the global distribution of these factors using immunofluorescence on polytene chromosomes and their local occupancy on heat shock genes using crosslinking and immunoprecipitation. We show that these factors localize to heat shock genes in vivo by three independent methods. This study and the accompanying study (Kaplan et al. 2000) reinforce models that Spt5 plays a role in the regulation of polymerase pausing and that Spt5 and Spt6 play roles in transcriptional elongation.
Association of Spt5 and Spt6 with uninduced heat shock genes
Previous studies have demonstrated that Pol II occupies the 5′ end of the uninduced hsp26 and hsp70 heat shock genes (Gilmour and Lis 1986; Rougvie and Lis 1990). Our crosslinking and immunoprecipitation data clearly show that Spt5 also occupies the uninduced heat shock gene promoters and 5′ transcribed regions. One possible explanation for this Spt5 localization is that it may act, perhaps in a complex with Spt4, to retain the paused polymerase. Alternatively, Spt5 may be responsible for either the establishment or maintenance (or both) of the potentiated chromatin architecture associated with promoter-proximal paused polymerase at heat shock genes. Spt6 and cyclin T also occupy the hsp70 5′ end under non–heat shock conditions, but unlike Spt5, these factors reside primarily from +102 to +238 at hsp70 by high-resolution analysis. This is near to, but not overlapping with, the peak of Spt5 and paused Pol II. Spt6 has been shown previously to interact with histones and assemble chromatin in vitro and modulate chromatin structure in vivo (Bortvin and Winston 1996). Therefore, our observation that Spt6 associates with this region of hsp70 is not inconsistent with Spt6 having a role in organizing chromatin architecture at the 5′ end of the uninduced hsp70 gene. Alternatively, the Spt6 and cyclin T present on the uninduced hsp70 gene represents a small portion of these factors that are associated with Pol II molecules that escape the pause.
Surprisingly, some HSF occupies the uninduced hsp70 and hsp26 promoters before heat shock, albeit at a level lower than during heat shock. Although we cannot exclude the possibility that HSF is recruited through a formaldehyde-induced stress response, we observe the same HSF occupancy of the hsp70 promoter using formaldehyde treatments of cells that are as short as 30 sec (data not shown). Perhaps HSF binds the uninduced promoter, although at levels and with an affinity lower than during heat shock.
Spt5 and Spt6 are general transcription elongation factors
The observations that Spt5 is present on the uninduced heat shock promoter suggest that DSIF (Spt5/Spt4) may function early in the process of transcriptional elongation. The DSIF complex causes the formation of short transcripts in an in vitro transcription system (Wada et al. 1998a) that are similar in size to those seen associated with paused Pol II in vivo (Rasmussen and Lis 1993). The addition of P-TEFb to the in vitro transcription system leads to the production of full-length transcripts (Wada et al. 1998a). From these in vitro observations, we predict that Spt5 and cyclin T would colocalize at sites of transcription in vivo—at least at the starts of transcription units where these apparent antagonistic effects of DSIF and P-TEFb are exerted. Spt5, Spt6, cyclin T, and Pol II are associated within the developmentally expressed ecdysone and intermolt puffs and manifest temporal changes consistent with a role in transcription. The immunofluorescence analyses show that these factors are located throughout the puffs, indicating that they have roles within the transcription unit. Thus, these studies support the model that Spt5, Spt6, and P-TEFb may work in concert and have broad roles in transcriptional elongation in vivo.
Spt5 and Spt6 are recruited to heat shock genes
We observed recruitment of Spt5 and Spt6 to both native and transgenic heat shock loci. And, as was observed at developmentally expressed loci, Spt5 and Spt6 localize within the heat shock puffs. The magnitude and kinetics of recruitment of Spt5 and Spt6 are strikingly similar to those of HSF and cyclin T. Recruitment also requires active HSF, as a temperature-sensitive mutation in the DNA binding domain of HSF blocks the association of Spt6 (Fig. 7A) and cyclin T (Lis et al. 2000) with heat shock puffs. These results demonstrate that Spt5 and Spt6 are recruited early in transcriptional activation and may work at any number of steps in transcription. Importantly, recruitment of Spt5 and Spt6 to activated heat shock genes indicates that they are likely to elicit positive effects on these genes. Within native and transgenic heat shock puffs, Spt5 and Spt6 overlap with one another and with Pol II but resolve from cyclin T and HSF. This suggests that Spt5 and Spt6 have functions along the transcription unit that are distinct from the promoter-associated factors.
Higher-resolution crosslinking and immunoprecipitation approaches confirm and extend the immunofluorescence findings. Although the immunofluorescence studies show that Spt5 and Spt6 are recruited to heat shock puffs, crosslinking and immunoprecipitation analysis showed that Spt5 and Spt6 were recruited to DNA segments at both the 5′ and 3′ ends of the hsp70, hsp26, and hsp83 heat shock genes. Pol II is localized throughout the induced heat shock genes, which is also consistent with the immunofluorescence analysis, whereas HSF is restricted to the promoter. Immunofluorescence and crosslinking and immunoprecipitation analysis show that cyclin T predominantly occupies the 5′ half of the puff and the gene, respectively. A higher concentration of P-TEFb at the 5′ end of the gene may reflect the targeting of kinase activity to phosphorylate newly recruited Pol II molecules or the Spt5 polypeptides that appear to associate with the paused polymerase (Wada et al. 1998a,b). Thus, the crosslinking and immunoprecipitation results indicate that in heat shocked cells, Spt5, Spt6, P-TEFb, and Pol II are strongly recruited throughout the transcription unit but are more concentrated on the 5′ region than the 3′ region of hsp70, and HSF is mainly associated with the promoter.
Roles for Spt5 and Spt6 in pausing and transcriptional elongation
What are the roles of Spt5 and Spt6 at uninduced and induced genes? We propose that Spt5, which is present on the 5′ ends of uninduced heat shock of genes, has a role in generating the promoter-proximal paused RNA Pol II. This agrees with previous in vitro observations that show that the DSIF complex causes formation of only short RNAs in vitro that are of a size similar to those associated with the paused Pol II in vivo. Furthermore, the paused Pol II is unphosphorylated (O'Brien et al. 1994), and it is this form that is acted on by DSIF in vitro (Wada et al. 1998b). While the effect of Spt5 on Pol IIa has been considered to be negative, this is in apparent contrast with evidence for the positive roles of Spt5 (and Spt4 and Spt6) in facilitating elongation (Hartzog et al. 1998). We suggest that Spt5 is an elongation factor that provides a surveillance role in the early phases of elongation at the 5′ end of genes: Spt5 surveys the promoter-proximal paused Pol II and ensures that it is modified by and accesorized with factors for productive elongation and RNA processing.
The recruitment of P-TEFb and its phosphorylation of Pol II may be a critical step that increases the rate of escape of the DSIF-paused Pol II into productive elongation. In vitro it is known that the DSIF complex does not prevent Pol IIo from elongating through a transcription unit (Yamaguchi et al. 1999a). The levels of Pol II, Spt5, and Spt6 all increase together in concert over the activated genes, indicating that these factors could be part of the elongation complex. Indeed, Spt6 interacts with Spt5, and both Spt5 and Spt6 have both been implicated in promoting elongation (Hartzog et al. 1998). This elongation complex could potentially act in any of many ways, but the previous demonstration of an interaction between Spt6 and histones (Bortvin and Winston 1996) evokes a model in which it might influence chromatin architecture of transcription unit in a manner that facilitates transcription.
Materials and methods
Plasmid construction
Dong-ki Lee initially identified a Drosophila Spt5 gene by searching the Drosophila genome database (Adams et al. 2000), and Zhuoyu Ni cloned a pGEM–Spt5 plasmid containing the entire Spt5 gene (data not shown), which was generously provided for the subcloning and antibody production below. Three portions of full-length Spt coding region contained in pGEM–SPt5 were amplified by PCR Spt5 aa 112–393 (Spt5N; EAO121a, CGCTCTAGAGGGGGCTTCATCATTGACGAGG CC and EAO122, CGCGTCGACTTAGTGTACTTCACCTC CAATTGCCC), Spt5 aa 389–733 (Spt5M; EAO123a, CGCTC TAGAGGAGGTGAAGTACACTCAGATGG and EAO124, CG CGTCGACTTACACGGCGCCCTTATAAGGTCCTCC), and Spt5 aa 732–1054 (Spt5C; EAO125a, CGCTCTAGAGCCGTGG GAATTGTGAAGGATGCC and EAO126, CGCGTCGACTTA GTCGATGGACTTCATCTTGC) and cloned into pMAL-C2 as XbaI–SalI (New England Biolabs [NEB]) fragments. All oligos were obtained from Operon (Operon Technologies). MBP-Spt6 aa 1349–1831 was constructed by cloning a SalI (NEB) fragment from a GAL4-activation domain hybrid isolated in an unrelated two-hybrid screen (E. Andrulis, unpubl.), into pMAL-C2. MBP-dHSF (aa 1–691) was kindly provided by P. Mason (Cornell University).
Recombinant protein purification and antibody production
Maltose binding-protein fusion constructs were transformed into BL21-DE3 cells, and recombinant proteins were induced using IPTG. Cells were lysed, and recombinant proteins were processed on and eluted from amylose resin according to manufacturer's recommendation (NEB). Protein was quantified either by Bradford assay or direct comparison to BSA standard. 100 μg aliquots of each recombinant factor were sent to Pocono Rabbit Farm and Laboratory for antibody production. Spt5N, Spt5M, and Spt5C were injected into chicken, rabbit, and guinea pig, respectively. Spt6 was injected into rat and guinea pig. HSF was injected into rabbit and guinea pig. Antibodies were tested via Western blot and indirect immunofluorescence to determine titer.
Indirect immunofluorescence
Salivary gland polytene chromosomes were isolated and mounted on slides as previously described (Lis et al. 2000). Polytenes were stained with primary antibodies at the following dilutions: HSF, 1 : 50 or 1 : 100; Spt5, 1 : 100; Spt6, 1 : 50 or 1 : 100; cyclin T, 1 : 20; and Pol II, 1 : 50. Rabbit anti-Pol II (exon 2) was affinity purified and provided by E. Wong (Cornell University). Appropriate secondary antibodies with conjugated labels (fluorescein isothiocyanate or rhodamine) were used, and images were collected. For triple labels, the third secondary antibody was conjugated to Alexa Fluor 350 (Molecular Probes).
Formaldehyde crosslinking of Drosophila cells
Drosophila Kc cells were maintained as previously described Lis et al. 2000). Cells were grown to density of 5 × 106 cells/mL in a spinner flask, and equal amounts of cells for non–heat shock and heat shock samples were removed. Heat shocked cells were grown at 36.5°C for 20 min before processing. Formaldehyde was added to a final concentration of 0.1% and mixed with cells on benchtop or shaker for no more than 3 min. Glycine was added to a final concentration of 125 mM and mixed for 3 min. Cells were centrifuged at 2000g for 5 min to collect cells, and the supernatant was decanted. The cell pellet was resuspended in Kc lysis buffer (100 mM KCl, 50 mM NaCl, 5 mM MgCl2, 10 mM Tris-HCl at pH 7.4, 0.5 mM PMSF). To the slurry, 0.1 volume of 10% nonidet P-40 (NP-40) was added, and the mixture was placed on ice for 5 min with intermittent vortexing. Lysed cells were centrifuged at 2000g for 10 min to collect nuclei. Following removal of the supernatant, nuclei were resuspended completely in suspension buffer (100 mM NaCl, 10 mM Tris at pH 8, 1 mM EDTA, 0.1% NP-40). Nuclei were lysed with the addition of 0.1 volume 20% sarkosyl. The viscous material was transferred to an SS-34 tube and sonicated with a Markson MK40 Ultrasonic Processor, five times at output setting 50, five times at setting 60, and five times at setting 70. All sonication steps were done on ice in a cold room for 20 sec with a 20-sec break between each pulse. Sonicated DNA was checked on an agarose gel to determine approximate size. To remove the insoluble nuclear debris, the material was centrifuged at 2000g for 10 min. This nuclear lysate was overlaid onto cesium chloride step gradient (1.75 g/cm3, 1.5 g/cm3, and 1.3 g/cm3) and centrifuged in SW28 rotor at 26,000 rpm for 30 h at 20°C. Fractions were collected after puncturing the bottom of the tube with an 18-gauge needle. Protein–DNA complexes were identified using a refractometer, and fractions with a refractive index between 1.38 and 1.4 were pooled. Pooled fractions were dialyzed at 4°C against at least three changes of SE buffer (0.1% sarkosyl, 50 mM Tris-HCl at pH 8.0, 2 mM EDTA). These protein–DNA complexes were then aliquoted and stored at −80°C or used immediately for immunoprecipitation.
Immunoprecipitation of protein–DNA complexes
Protein–DNA complexes (1 mL at ∼1 mg/mL) were precleared with 50 μL of a 50% suspension of protein A agarose beads (GIBCO BRL). This was done for 2 h, the beads were spun down, and the supernatant was collected and pooled. Protein–DNA complexes were dispensed in 100-μL aliquots and treated with antibody overnight on a rotating wheel at 4°C. The following day, 50 μL of a 50% suspension of protein A agarose beads was added to the immunoprecipitations and incubated for 4 h. The washing steps were done at room temperature and were subsequently spun down for 1 min at 1000g. Each wash was carried out with 1 mL of wash solution. The beads were washed seven times in Wash Buffer (50 mM HEPES/KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA 1% Triton X-100, 0.1% sodium deoxycholate, 0.5 mM PMSF), once with Wash Buffer with 500 mM NaCl, once with Lithium Wash Buffer (10 mM Tris-Cl at pH 8.0, 0.25 M LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate), and once with TE. After the final wash, the protein–DNA complexes were eluted by the addition of 100 μL of Elution Buffer (50 mM Tris-Cl at pH 8.0, 10 mM EDTA, 1% SDS) and incubation at 65°C for 15 min. The beads were spun down at 6000g, and the supernatant was transferred to a new tube. The beads were resuspended in 150 μL of Elution Buffer and centrifuged at 6000g. The supernatant from this wash was added to the first elution, and the crosslinks were reversed overnight at 65°C. From the starting material, 10% was used as a total DNA control. The total DNA control sample also had its crosslinks reversed by the addition of 230 μL of 1× TE and 10 μL of 10% SDS and incubation at 65°C overnight. The DNA samples were subsequently extracted twice with an equal volume of buffered phenol and once with chloroform. The DNA was then precipitated by the addition of 25 μL of 4 M LiCl, 2 μL of glycogen (10 mg/mL), and 625 μL of 100% ethanol. The coimmunoprecipitated DNA was spun down for 30 min at 15,000g, washed with 70% ethanol, and finally resuspended in 50 μL of TE for use in either PCR or ligation-mediated PCR.
Determination of protein–DNA interactions by PCR
For PCR analysis, 1 μL of coimmunoprecipitated DNA was incubated in the presence of 5 pM primers, 5 mM dNTPs, 10× Taq buffer (0.2 M KCl, 0.1 M Tris at pH 8.0), 25 mM MgCl2, 2U Taq DNA polymerase, and water to a final volume of 20 μL. A typical PCR reaction would be: 94°C, 1 min; 60°C, 30 sec; and 72°C, 30 sec for 25 cycles followed by one cycle at 72°C for 10 min. PCR primer are listed here in pairs: HSP70-998, GCCCCTTT GAGTTCTAACCATCC and HSP70-572, GGTTTCTTTGCTT AATTAAACGC; HSP70-365, GGCCTTTCTGGCGGACAAC ATCC and HSP70-3, CGCTCCGTCGACGAAGCGCC; HSP70-5, GCGTCAATTCAATTCAAACAAGC and HSP70+412, CCT GGTTCTTGGCCGGTTCGCCG; HSP70+1758, CGACAAGG GACGCCTCTCGCAGG and HSP70+2182, GGCCTTAGTCG ACCTCCTCGACC; HSP26-382, CCGTTAGCCGGCTGTTT CTTTTGCG and HSP26+113, AACTTGTTTGACTTGTAAG CAAAGG; HSP26+702, CGCATCATTCAAATTCAGCAAGT GG and HSP26+1195, GGTGAACTATTTTCGGACACCAA GC; HSP83-458, CCACCAAGTCTCTGAAACTCTGG and HSP83-18, GCGTCTGCTTTTATAGAAAAACC; HSP83+1620, GGTGCCGACATTTCCATGATCGG and HSP83+2030, CCT CATCCTCGCCAACATCCTCG. PCR reactions (20 μL) were loaded onto a 2% agarose gel, and bands were detected by staining with ethidium bromide (EtBr). EtBr-stained bands were quantified using QuantityOne imaging software with the Bio-Rad Gel Doc 2000.
Determination of protein–DNA interactions by LMPCR
Crosslinked material was cut to completion with EcoRII that cut upstream of the HSP70aDP1 primer (see below for sequence) in order to minimize the contribution of upstream signals. Per immunoprecipitation reaction, 100 μL of crosslinked material was used. To cut the DNA, 900 μL of TE, 120 μL of Restriction Buffer 2 (NEB), 12 μL of 10 mg/mL bovine serum albumin, and 10 μL of NP-40 was added to each sample. The tubes were preincubated at 37°C for 15 min before restriction enzymes were added. After the preincubation, 40 U of restriction enzyme EcoRII was added to cut the DNA to completion. The reactions were incubated at 37°C for 30 min. At this point, 20 U of MboII and HinfI were added and the reactions incubated for another 30 min at 37°C for the partial digests. After the second incubation, 50 μL of 0.5 M EDTA (final concentration of 20 mM EDTA) was added to each reaction in order to stop the digestion. Each reaction was precleared with 50 μL of a 50% suspension of protein A agarose beads. This was done for 1–2 h and then the beads were spun down and the supernatant was collected and pooled. The pooled material was dispensed in 1-mL aliquots and treated with antibody. At this point, the protein–DNA complexes were processed as described above for immonoprecipitation of protein–DNA complexes.
For LMPCR analysis of the 5′ end of the hsp70 gene, primers were constructed within the body of hsp70 (DP1 +299 to +275 and DP2 +282 to +256); HSP70aDP1 (TGGTAGACACCCACG CAGGAGTAGG) and HSP70a DP2 (GGAGTAGGTGGTGC CCAGATCGATTCC). Linker primers were L1, GCGGTGAC CCGGGAGATCTGAATTC and L2, GAATTCAGATC. For ligation-mediated PCR, 9 μL of immunoprecipitated DNA was mixed with 3 μL of Buffer 2 (200 mM Tris-Cl at pH 7.7, 250 mM NaCl), and 3 μL of HSP70aDP1 at a concentration of 0.5 pmole/μL. The sample was incubated at 95°C for 2 min and then transferred to a 60°C heat block and incubated for 30 min. After annealing, the sample was then spun down to collect the condensation on the top of the tube and 7.5 μL of Buffer 3 (20 mM MgCl2, 20 mM DTT, 0.2 mM dATP, 0.2 mM dCTP, 0.2 mM dGTP, 0.2 mM dTTP) was added. The primers were extended by the addition of 1.5 μL of Sequenase that had been diluted 1 : 5 in TE and by incubation at 45°C for 5 min. The reaction was then stopped by transferring the reaction to 60°C and incubating for 5 min. At this point, 6 μL of Buffer 4 (310 mM Tris-Cl at pH 7.7) was added and the reaction was incubated for an additional 5 min. Finally, the reaction was cooled on ice and 20 μL of Buffer 5 (17.5 mM MgCl2, 42.3 mM DTT, 125 μg/mL BSA) and 25 μL of Buffer 6 (10 mM MgCl2, 20 mM DTT, 3 mM ATP, 50 μg/mL BSA, 50 mM Tris-Cl at pH 7.7, 100 pmoles annealed linkers, 3 U T4 DNA ligase) were added. This was incubated overnight at 17°C. The next morning, the ligation was precipitated by the addition of 8.5 μL of 3 M sodium acetate, 1 μL of 10 mg/mL yeast tRNA, and 200 μL of 100% ethanol. The precipitation was carried out for 30 min on ice and subsequently spun down for 30 min at 4°C. The pellet was resuspended in PCR Master Mix (20 nmoles dATP, 20 nmoles dCTP, 20 nmoles dGTP, 20 nmoles dTTP, 10 pmoles HSP70aDP1, 10 pmoles linker primer L1, 40 mM NaCl, 10 mM Tris-Cl at pH 8.9, 5 mM MgCl2, 0.01% gelatin, 5 U Taq polymerase) and amplified in a thermal cycler. The DNA went through 20 rounds of amplification followed by five rounds of labeling. This was accomplished by the addition of 10 μL of PCR Master Mix 2 (2 mM dATP, 2 mM dCTP, 2 mM dGTP, 2 mM dTTP, 1–10 pmoles 32P end-labeled HSP70aDP2, 40 mM NaCl, 10 mM Tris-Cl at pH 8.9, 5 mM MgCl2, 0.01% gelatin, 2.5 U Taq polymerase) after the first PCR was complete and an additional five rounds of amplification. Once both PCR steps were complete, the sample was chloroform extracted, ethanol precipitated, and resuspended in 16 μL of formamide loading dye. Typically, 4–6 μL were run out on a 5.1% denaturing sequencing gel.
Acknowledgments
We thank Dong-ki Lee for initiating studies on the role of Spt5 in promoter-paused complexes and Zhuoyu Ni for the full-length Spt5 plasmid; Paul Mason for HSF and Brian Schwartz for preliminary work with Spt5; Michael Seidman for primer design; and David Price for the cyclin T antibody. We would also like to thank Fred Winston and Craig Kaplan for discussing their results before publication. We are grateful to Mariana Wolfner, W. Lee Kraus, Jeffrey Roberts, and Lis laboratory members for comments on the manuscript and scientific discussions. This work was supported by National Institutes of Health grant GM25232 to J.T.L. and National Research Service Award grant GM19999 to E.D.A.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mbgdepartment@cornell.edu; FAX (607) 255-2428.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.844200.
References
- Adams MD, Celniker SE, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Dahmus ME. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog Nucleic Acid Res and Mol Biol. 1994;48:143–179. doi: 10.1016/s0079-6603(08)60855-7. [DOI] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp-70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman E, Lis JT. Transcription factor TFIIH is required for promoter melting in vivo. Mol Cell Biol. 1999;19:5652–5658. doi: 10.1128/mcb.19.8.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Mortin M, Wu C. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J. 1997;16:2452–2462. doi: 10.1093/emboj/16.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan CD, Morris KR, Wu C-t, Winston F. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in Drosophila melanogaster. Genes & Dev. 2000;14:2623–2634. doi: 10.1101/gad.831900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes & Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- Laspia MF, Rice AP, Mathews MB. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- Lee HS, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of Hsp70. Genes & Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes & Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- O'Brien T, Lis JT. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol Cell Biol. 1993;13:3456–3463. doi: 10.1128/mcb.13.6.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced Hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- ————— Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990;10:6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Adamczewski JP, Seroz T, Vermeulen W, Tassan JP, Schaeffer L, Nigg EA, Hoeijmakers JH, Egly JM. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes & Dev. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- Simon JA, Sutton CA, Lis JT. Localization and expression of transformed DNA sequences within heat shock puffs of Drosophila melanogaster. Chromosoma. 1985;93:26–30. doi: 10.1007/BF01259442. [DOI] [PubMed] [Google Scholar]
- Strobl LJ, Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 1992;11:3307–3314. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Becker PB, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 1998a;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998b;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Locus-specific variation in phosphorylation state of RNA polymerase II in-vivo: Correlations with gene activity and transcript processing. Genes & Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- Westwood JT, Clos J, Wu C. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature. 1991;353:822–827. doi: 10.1038/353822a0. [DOI] [PubMed] [Google Scholar]
- Wu C, Wilson S, Walker B, Dawid I, Paisley T, Zimarino V, Ueda H. Purification and properties of Drosophila heat shock activator protein. Science. 1987;238:1247–1253. doi: 10.1126/science.3685975. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999a;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999b;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- Yankulov K, Blau J, Purton T, Roberts S, Bentley DL. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II C-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimarino V, Wu C. Induction of sequence-specific binding of Drosophila heat shock activator protein without protein synthesis. Nature. 1987;327:727–730. doi: 10.1038/327727a0. [DOI] [PubMed] [Google Scholar]