Abstract
Centrosomes organize the mitotic spindle to ensure accurate segregation of the chromosomes in mitosis. The mechanism that ensures accurate duplication and separation of the centrosomes underlies the fidelity of chromosome segregation, but remains unknown. In Saccharomyces cerevisiae, entry into S phase and separation of spindle pole bodies each require CDC4 and CDC34, which encode components of an SCF (Skp1–cullin–F-box) ubiquitin ligase, but a direct (SCF) connection to the spindle pole body is unknown. Using immunofluorescence microscopy, we show that in mammalian cells the Skp1 protein and the cullin Cul1 are localized to interphase and mitotic centrosomes and to the cytoplasm and nucleus. Deconvolution and immunoelectron microscopy suggest that Skp1 forms an extended pericentriolar structure that may function to organize the centrosome. Purified centrosomes also contain Skp1, and Cul1 modified by the ubiquitin-like molecule NEDD8, suggesting a role for NEDD8 in targeting. Using an in vitro assay for centriole separation in Xenopus extracts, antibodies to Skp1 or Cul1 block separation. Proteasome inhibitors block both centriole separation in vitro and centrosome duplication in Xenopus embryos. We identify candidate centrosomal F-box proteins, suggesting that distinct SCF complexes may direct proteolysis of factors mediating multiple steps in the centrosome cycle.
Keywords: Cell cycle, centrosome, DNA replication, SCF ubiquitin ligase, ubiquitin-dependent proteolysis
Animal cell centrosomes or yeast spindle pole bodies (SPBs) are the cell’s microtubule organizing centers and have a central role in organizing the mitotic spindle to separate the chromosomes at mitosis (Winey and Byers 1993; Kellogg et al. 1994; Zimmerman et al. 1999). Like the chromosomes themselves, centrosomes or SPBs must be duplicated once per cell cycle in a semi-conservative manner. Currently, the mechanisms that monitor centrosome number and function are unknown. It has been suggested that if these mechanisms fail, abnormal centrosome number or function can result in the formation of aberrant spindles, and therefore in chromosome missegregation. Tumor cells and p53-negative cells have been found to contain multiple or atypical centrosomes (Fukasawa et al. 1996; Lingle et al. 1998; Pihan et al. 1998), suggesting that centrosome abnormalities contribute to genetic instability in cancer.
Each centrosome has a core of two centrioles, cylindrical structures consisting of nine microtubule triplets. Around the centrioles, the pericentriolar material (PCM) functions to nucleate microtubule growth. During G1 in mammalian cells, the centrioles first separate by a small distance in a process called disorientation (Kuriyama and Borisy 1981). Subsequently, a new daughter centriole or procentriole appears adjacent to each mother centriole. This duplication process continues through S and G2, and into mitosis. At prophase, the newly replicated centriole pairs, and their surrounding PCM, separate and move to opposite sides of the nucleus to form the poles of the mitotic spindle and ultimately the centrosomes of each daughter cell. Therefore, two separation events occur—centriolar separation beginning in G1 and separation of duplicated centrosomes and PCM at prophase. How these processes are controlled is not well understood.
The PCM has been described as a lattice-like structure containing the protein pericentrin and the γ-tubulin ring complex (γ-TuRC), which direct microtubule nucleation and organization (Stearns et al. 1991; Zheng et al. 1991, 1995; Doxsey et al. 1994; Stearns and Kirschner 1994; Moritz et al. 1995b; Dictenberg et al. 1998; Schnackenberg et al. 1998). The γ-TuRC consists of γ-tubulin and associated proteins and is sufficient to nucleate microtubule growth (Moritz et al. 1995b, 1998; Zheng et al. 1995). Components of the PCM, including γ-tubulin, accumulate at the centrosome as cells progress to mitosis. Although centrosomes and yeast SPBs are morphologically different, they share homologous and functionally similar components, including proteins of the γ-TuRC (Geissler et al. 1996; Knop and Schiebel 1997; Martin et al. 1998; Murphy et al. 1998; Tassin et al. 1998).
A number of regulatory proteins are implicated in the stepwise duplication of SPBs and centrosomes (for examples, see Winey et al. 1991; Glover et al. 1995; Lane and Nigg 1996; Fry et al. 1998a,b). Specific connections to the cell cycle machinery and the precise ordering of these steps, however, remain unclear. Recently, studies in Xenopus embryos and using in vitro duplication assays in Xenopus egg extracts, as well as assays based in mammalian cells, have implicated cyclin E/Cdk2 and cyclin A/Cdk2 in centrosome duplication (Hinchcliffe et al. 1999; Lacey et al. 1999; Matsumoto et al. 1999; Meraldi et al. 1999), linking the centrosome cycle to specific cell cycle regulators and therefore to the mitotic cell cycle.
Under some conditions, the centrosome cycle can be dissociated from the mitotic cell cycle. Blocking S phase progression in Chinese Hamster ovary (CHO) cells with hydroxyurea or in fertilized sea urchin eggs with aphidicolin results in the continuation of the centrosome cycle, producing cells with multiple centrosomes (Sluder et al. 1990; Balczon et al. 1995). Treatment of sea urchin or Xenopus laevis embryos with protein synthesis inhibitors also blocks the mitotic cycle but allows the continuation of centrosome duplication (Gard et al. 1990; Sluder et al. 1990). Studies in sea urchins indicate that the capacity for centrosome duplication is present in embryos arrested in S phase but is blocked in M phase (Hinchcliffe et al. 1998). Activated mitotic cyclin B/Cdc2 inhibits centriole separation in vitro (Lacey et al. 1999). Therefore, centrosome duplication is limited in mitosis, although studies in sea urchins indicate that active cyclin B/Cdc2 is not sufficient for this inhibition (Hinchcliffe et al. 1998).
How does the cell normally ensure that the centrosomes are duplicated exactly once each cell cycle? A formally similar mechanism that limits chromosomal replication to once per cell cycle has been described (for reviews, see Dutta and Bell 1997; Stillman 1996). For chromosome replication, cyclin-dependent kinases and ubiquitin-dependent proteolysis are required to maintain the block to rereplication. In budding yeast, the G1–S transition requires a multicomponent ubiquitin ligase complex, called Skp1–cullin–F-box Cdc4p (SCFCdc4p) (for review, see Patton et al. 1998; Peters 1998). SCFCdc4p is so named for its components, the protein Skp1 (Zhang et al. 1995), Cdc53p (Mathias et al. 1996; Willems et al. 1996), which is a member of the small protein family called cullins (Kipreos et al. 1996), and Cdc4p, one of a diverse group of adapter proteins containing a motif called an F-box (Bai et al. 1996). SCFCdc4p associates with the ubiquitin-conjugating enzyme Cdc34p and directs ubiquitination of Sic1p (Feldman et al. 1997; Skowyra et al. 1997), an inhibitor of the cyclin-dependent kinases that drive DNA replication (Schwob et al. 1994). The ubiquitinated Sic1p is then destroyed by the 26S proteasome (for review, see Hochstrasser 1996). Sic1p must be phosphorylated to be recognized by the F-box protein Cdc4, and therefore targeted for destruction (Feldman et al. 1997; Skowyra et al. 1997; Verma et al. 1997). The F-box motif in Cdc4 and other adapter proteins appears to recruit the F-box binding protein Skp1p and the cullin Cdc53p (Bai et al. 1996), whereas conserved domains within cullins may directly bind to E2 enzymes including Cdc34p (Yu et al. 1998; Zachariae et al. 1998). Recent evidence indicates that SCF complexes contain additional components and mediate many cellular events (for reviews, see Patton et al. 1998; Peters 1998; Wolf and Jackson 1998; Koepp et al. 1999; Laney and Hochstrasser 1999).
In Saccharomyces cerevisiae, SPBs duplicate early in G1, but remain together until the onset of mitosis. Mutants in the Cdc4 and Cdc34 genes arrest in G1 with duplicated but unseparated SPBs (for review, see Winey and Byers 1992), suggesting that Cdc4p and Cdc34p may be required for SPB separation. A proteasome cap protein is also required for SPB duplication, again suggesting that proteasome function might be required for duplication and/or separation of SPBs (McDonald and Byers 1997). In these cases, however, connections to specific SPB or centrosome proteins and their functions are not known.
We show here using immunofluorescence and immunoelectron microscopy that mammalian Skp1 is localized to interphase and mitotic centrosomes and to the interphase nucleus in cultured mammalian cells. Skp1 is also present in purified centrosomes. In addition, we find that Cul1 (Kipreos et al. 1996), the human homolog of the yeast cullin Cdc53p (Willems et al. 1996), is present at the centrosome in a form specifically modified by the addition of the ubiquitin-like protein NEDD8 (Kamitani et al. 1997). We find that blocking Skp1 or Cul1 function with specific antibodies inhibits centriole separation in vitro. Further, inhibitors of the 26S proteasome both block centriole separation in vitro and inhibit centrosome duplication when injected into Xenopus embryos. Finally, we identify candidate F-box proteins at the centrosome. These data implicate SCF complexes and ubiquitin-mediated proteolysis in the centrosome duplication process.
Results
Immunofluorescence localization shows that Skp1 is nuclear and centrosomal
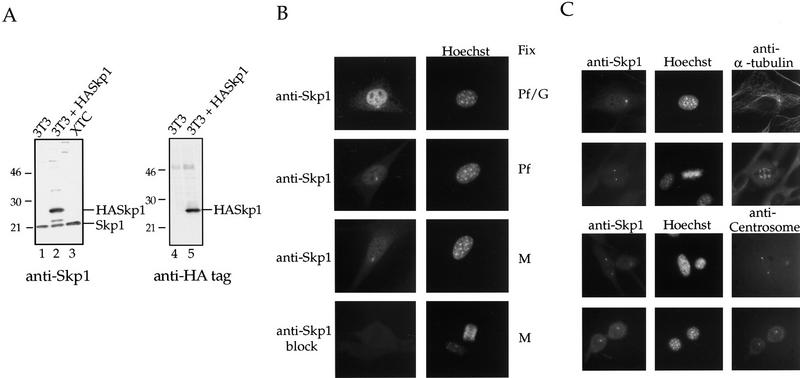
Affinity-purified anti-Skp1 antibodies were produced as described (see Materials and Methods) and tested by Western blotting to verify their specificity (Fig. 1A). Anti-Skp1 antibodies detected a specific 21-kD species corresponding to the endogenous Skp1 in lysates from either NIH-3T3 or Xenopus XTC cells (Fig. 1A, lanes 1,3). Skp1 antibodies also recognized a lower mobility HA-tagged Skp1 protein expressed in transfected NIH-3T3 cells (Fig. 1A, lane 2). A parallel blot probed with anti-HA antibodies demonstrated that the expressed protein present was the HA-tagged Skp1 (Fig. 1A, lane 5). The Skp1 species observed here migrates at 21 kD, slightly larger than the 19-kD species described previously (Zhang et al. 1995). We have confirmed the identity of the Skp1 band by blocking the antibodies with Skp1 protein, and by using sera from different rabbits (data not shown), including the serum originally described (Zhang et al. 1995). The anti-Skp1 reactive species in the Xenopus cells comigrates with that from NIH-3T3 cells, suggesting a highly related protein in Xenopus. Consistent with this observation, we have cloned a Xenopus homolog of Skp1 that is identical to the human protein in amino acid sequence (J. Regan and P. Jackson, unpubl.).
Figure 1.
Characterization of Skp1 antibodies and localization of Skp1 in cells. (A) Western blotting using affinity-purified anti-Skp1 antibodies. (Lanes 1,4) NIH-3T3 cell lysate; (lanes 2,5) lysate of NIH-3T3 cells transiently transfected with HA-tagged Skp1; (lane 3) XTC cell lysate. The blots were incubated with affinity-purified anti-Skp1 antibodies (lanes 1–3) or with rabbit anti-HA antibodies (lanes 4,5), followed by secondary antibodies, and bands visualized using enhanced chemiluminescence (ECL) Western blotting detection reagents. Positions of molecular weight markers are indicated. (B) Immunofluorescence localization of Skp1. Cells were fixed in paraformaldehyde (Pf), a paraformaldehyde/glutaraldehyde (Pf/G) mixture, or methanol (M) as indicated, and labeled with affinity-purified anti-Skp1 antibodies (top three panels) or anti-Skp1 antibodies blocked with excess GST–Skp1 protein (bottom panel) and with Hoechst dye to label DNA. (C) Costaining confirms centrosomal localization. NIH-3T3 cells were fixed in methanol and stained with affinity-purified anti-Skp1 antibodies and either mAb against α-tubulin or human anti-centrosome antiserum as indicated, followed by secondary antibodies and Hoechst dye.
Staining of NIH-3T3 cells using affinity-purified anti-Skp1 antibodies showed that the protein was localized to the centrosome in interphase and mitotic cells (Fig. 1B,C). Skp1 was also present in a stippled pattern in the nucleus of interphase cells fixed with a paraformaldehyde/glutaraldehyde mixture (Pf/G: Fig. 1B, top tier), paraformaldehyde (Pf: Fig. 1B, second tier), or methanol (M: Fig. 1B, third tier). Nuclear staining was somewhat more pronounced in the paraformaldehyde/glutaraldehyde fixed cells. Skp1 is a small protein that is abundant in vivo (∼1 μm in 3T3 cells), and very soluble in vitro (see Materials and Methods). We suspect that some soluble Skp1 is lost during fixation, and that the addition of cross-linking fixatives like glutaraldehyde reduces the amount of Skp1 extracted. Interestingly, centrosomal Skp1 was not extracted by any of several fixation methods we tried. Similar Skp1 staining was observed in Xenopus XTC cells, human HeLa, MCF7, and several human tumor cell lines (not shown). Control staining with antibodies blocked by incubation with the Skp1 protein (Fig. 1B, fourth tier), preimmune serum, or no primary antibody (not shown), all gave little staining, demonstrating the specificity of the antibody staining pattern.
We costained cells with anti-Skp1 antibodies and with either anti-α-tubulin (Fig. 1C, top panels) or an anti-human centrosome antiserum (Fig. 1C, bottom panels). Both colocalization experiments confirm that Skp1 staining is centrosomal.
Skp1 is centrosomal during all phases of the cell cycle in NIH-3T3 cells
Costaining using affinity-purified anti-Skp1 antibodies and antibodies directed against α-tubulin (Fig. 2) demonstrated that Skp1 was detectable at the centrosome in interphase (Fig. 2, top tier) and during all phases of mitosis (Fig. 2, tiers 2–6). Skp1 is also present in the nucleus in interphase (Fig. 2, top tier). As described for other centrosomal proteins, Skp1 staining is more pronounced at mitotic than at interphase centrosomes (Stearns et al. 1991; Zheng et al. 1991).
Figure 2.
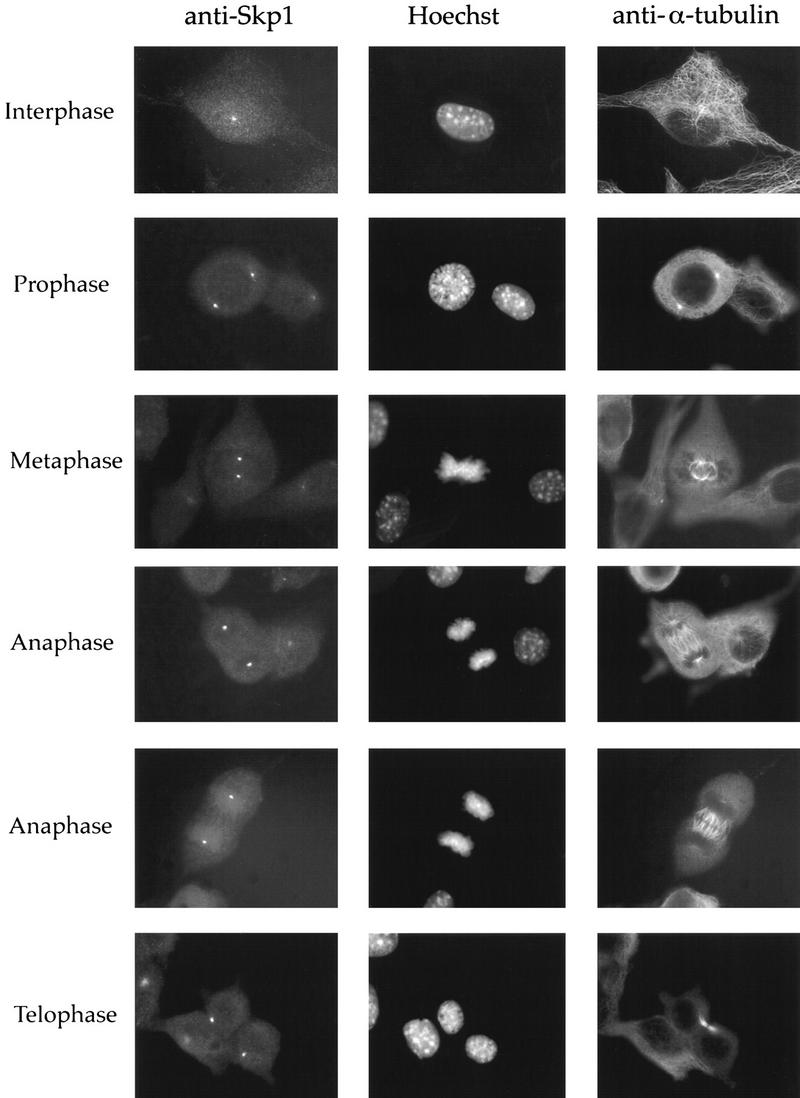
Skp1 is centrosomal during all phases of the cell cycle in NIH-3T3 cells. NIH-3T3 cells were fixed in methanol and labeled with the indicated antibodies or with Hoechst dye to stain the DNA. Cells at specific stages of the cell cycle are indicated.
Localization of Skp1 to centrosomes does not require microtubules and is observed on overduplicated centrosomes in hydroxyurea-treated cells
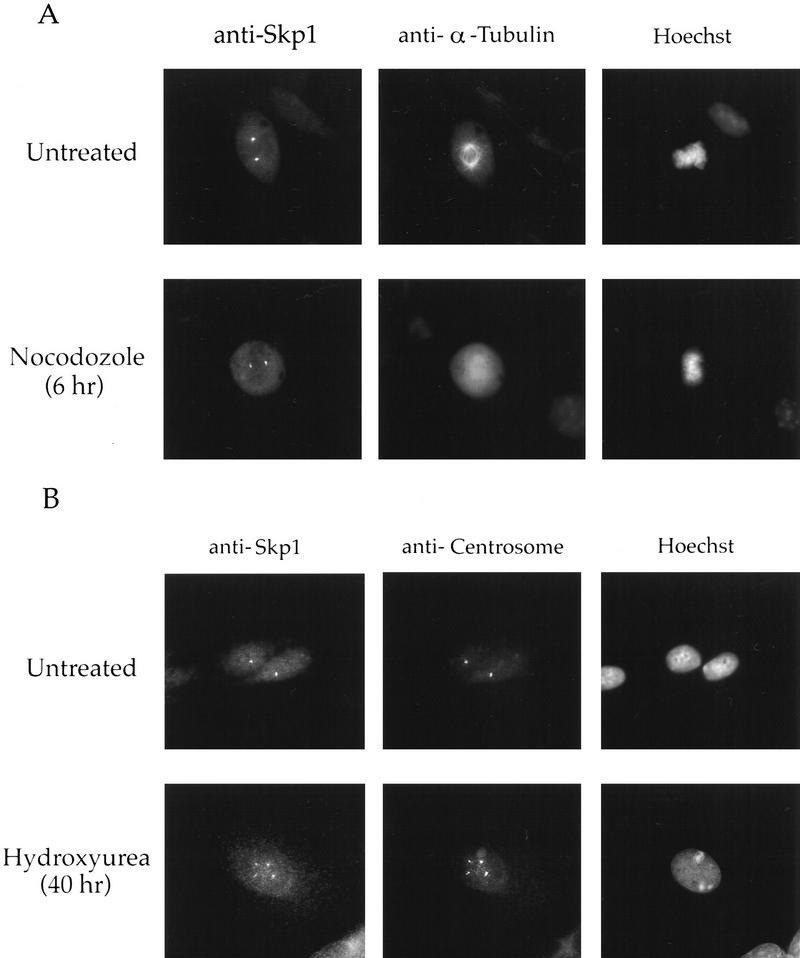
To determine if Skp1 association with the centrosome requires the continued presence of microtubules, we examined the localization of Skp1 in cells treated with nocodazole, a microtubule-depolymerizing drug. After treating cells with nocodazole for 10 min (data not shown) or for 6 hr (Fig. 3A, bottom tier), microtubules were disassembled, but Skp1 remained at the centrosomes, suggesting that Skp1 is an integral component of the centrosome.
Figure 3.
Skp1 is centrosomal in nocodazole-treated and ectopically produced centrosomes. (A) Nocodazole treatment of NIH-3T3 cells. Cells were incubated with nocodazole (0.3 μg/ml) or untreated (control), fixed in methanol, and stained with the indicated antibodies and Hoechst dye. (B) Hydroxyurea treatment of CHO cells. Cells were grown on coverslips and incubated with or without hydroxyurea (2 mm). They were fixed in methanol and stained with the indicated antibodies and with Hoechst dye as above. (Bottom panel) An example of a cell containing multiple centrosomes.
Brinkley and colleagues demonstrated that the centrosome cycle continues in CHO cells treated with hydroxyurea to block DNA synthesis, thereby producing cells with multiple centrosomes (Balczon et al. 1995). We found that Skp1 also localized to these ectopically duplicated centrosomes in CHO cells treated with hydroxyurea (Fig. 3B, bottom tier), as shown by colocalization with human anti-centrosome antibody. These data suggest that Skp1 is assembled onto centrosomes as a part of their intrinsic cycle and does not require specific passage through any cell cycle stage.
Skp1 is pericentriolar in purified centrosomes and in NIH-3T3 cells
We examined the presence of the Skp1 protein in centrosomes purified from CHO cells (see Materials and Methods). Centrosomes were centrifuged onto coverslips, stained with anti-Skp1 antibodies and with anti-α-tubulin antibodies to label the centrioles, and visualized by deconvolution microscopy (Fig. 4A). The majority (97%) of purified centrosomes identified by anti-α-tubulin staining as centriole doublets also reacted with anti-Skp1 antibodies but not with the preimmune serum control (data not shown), supporting the specificity of the anti-Skp1 staining.
Figure 4.
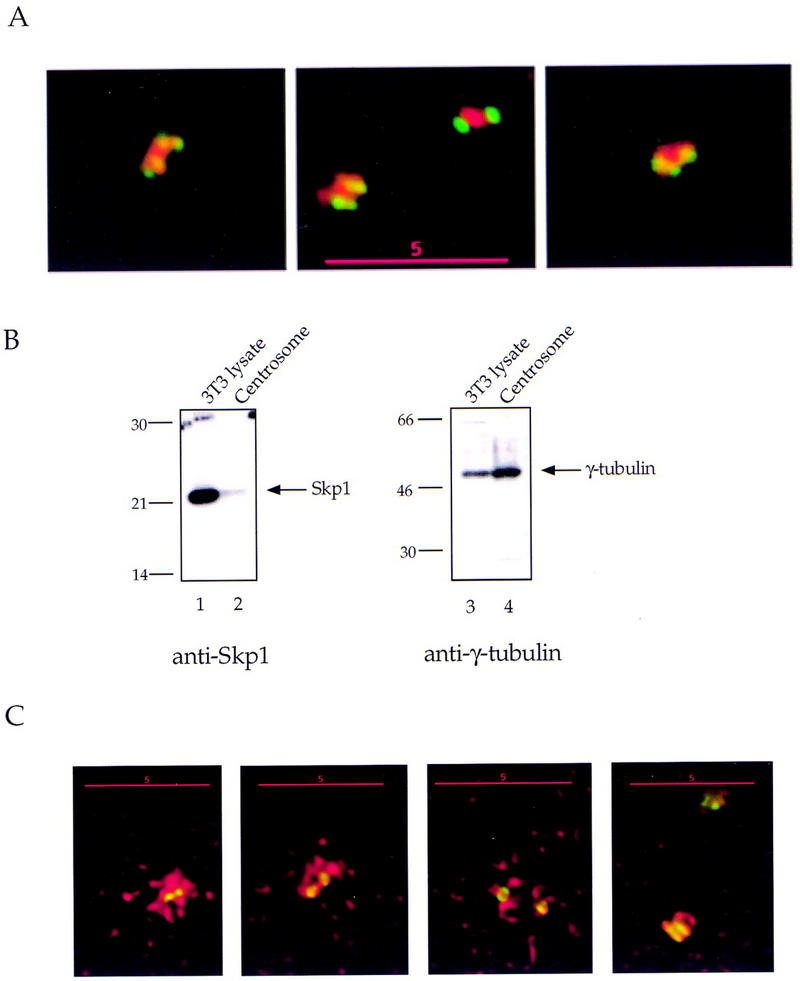
Skp1 is pericentriolar in purified centrosomes and in NIH-3T3 cells. Centrosomes were purified from CHO cells and prepared for immunofluorescence as described (see Materials and Methods). (A) Centrosomes spun onto coverslips were fixed in methanol and labeled with mouse monoclonal antibodies against α-tubulin and affinity-purified rabbit antibodies against Skp1, followed by secondary antibodies. Anti-α-tubulin (green); Anti-Skp1 (red). (B) Immunoblotting detects Skp1 in a purified centrosome sample. An NIH-3T3 cell lysate (lanes 1,3) and a centrosome sample (lanes 2,4) were subjected to Western blotting with either affinity-purified anti-Skp1 antibodies (lanes 1,2) or rabbit antibodies to γ-tubulin (lanes 3,4). Secondary antibody was HRP-conjugated donkey anti-rabbit IgG and bands were detected with ECL reagents as above. Skp1 and γ-tubulin are indicated (arrows). Molecular mass markers are as indicated. (C) NIH-3T3 cells fixed in methanol, and labeled using a mouse mAb against γ-tubulin and with rabbit affinity-purified anti-Skp1 antibodies followed by secondary antibodies. Anti-γ-tubulin (green); anti-Skp1 (red). Deconvolution images of centrosomes in cells at different phases of the cell cycle are pictured (left to right).
Western blotting also indicated that Skp1 was present in the purified centrosome preparation (Fig. 4B, lane 2), similar to the known centrosomal structural protein, γ-tubulin (Fig. 4B, lanes 3,4). By quantitating the amount of Skp1 in our centrosome preparation, we estimated that each centrosome contains at least ∼500 molecules of Skp1. Because our microscopy suggests that some centrosomal Skp1 is lost during purification (see below), this is a minimum estimate. For comparison, it has been estimated that 400–800 γ-TuRCs (each containing ∼13 γ-tubulin molecules; see Zheng et al. 1995) are present per centrosome in Drosophila (Moritz et al. 1995). Whereas soluble Skp1 may function in multiple complexes and at diverse subcellular locations, Skp1 present at the centrosome may be sufficient for its local role there.
To understand how the SCF is organized at the centrosome, we examined centrosomes in purified preparations and in cells by deconvolution microscopy. In purified centrosomes, Skp1 was localized in a region around and between centrioles (Fig. 4A), distinct from the centriolar location of α-tubulin. When centrosomes costained with antibodies to α- and γ-tubulin were examined, the pattern of γ-tubulin staining was similar to that of Skp1 (data not shown). Therefore Skp1, like γ-tubulin, is a pericentriolar protein.
In NIH-3T3 cells costained with anti-Skp1 antibodies and an anti-γ-tubulin antibody, Skp1 staining is present around the centriole core in an irregular, lattice-like pattern (Fig. 4C). Skp1 distribution extends to a greater radial distance than does that of γ-tubulin, particularly in interphase cells (Fig. 4C, panels 1–3). Similar deconvolution images were obtained using paraformaldehyde-fixed cells (data not shown), indicating that this distribution is not a fixation artifact. We estimate that the centriolar diameter in NIH-3T3 cells is ∼0.25 μm, and that the pericentriolar region occupied by Skp1 is as large as ∼2.0 μm in diameter in some cells. Unlike in purified centrosomes, Skp1 staining in centrosomes in intact cells is more extended than that of γ-tubulin (Fig. 4C), showing that some Skp1 is lost or disorganized during centrosome purification.
Immunoelectron microscopy indicates that Skp1 is present at centriolar appendages and in a large pericentriolar region
To view Skp1 within the ultrastructure of the centrosome, NIH-3T3 cells were labeled with affinity-purified anti-Skp1 antibodies and processed for immunoelectron microscopy (see Materials and Methods). In electron micrographs of anti-Skp1 stained centrosomes, gold particles are concentrated in a region adjacent to the external surface of the centrioles (Fig. 5A,C,D,E), and also extend for some distance radially around the centrioles (Fig. 5E), consistent with the deconvolution images. Two types of centriolar appendages have been described in previous studies of centrosomal ultrastructure (Paintrand et al. 1992). In the centrioles depicted in Figure 5, A and D, such appendages are decorated by gold particles (10 nm gold, see arrows). Fixation with paraformaldehyde/glutaraldehyde (Fig. 5A–C,F) resulted in better preservation of ultrastructure, but also in less dense staining than fixation in methanol (Fig. 5D,E), perhaps because of reduced antigen accessibility. Otherwise, similar images were obtained after either fixation method. Control staining with no primary antibody showed limited staining (Fig. 5B). The distribution of gold particles in the region around centrioles (Fig. 5E, see arrowheads) suggested the presence of two rings of Skp1 staining: one just adjacent to the external side of the centriole (containing ∼6 times background), and a second 1–3 centriolar diameters away (containing ∼3 times background). The extended radial distribution of Skp1 is similar to that seen in deconvolution images (Fig. 4C), and shows some variation between individual centrosomes. We suspect that this variation is a function of cell cycle stage. Finally, a double staining experiment (Fig. 5F) demonstrates anti-γ-tubulin staining (5 nm gold, see arrowheads in Fig. 5F) highly concentrated around the centriole core while anti-Skp1 staining (15 nm gold) decorates the appendages and forms a broader ring around the centriole, in agreement with our deconvolution images (Fig. 4C).
Figure 5.
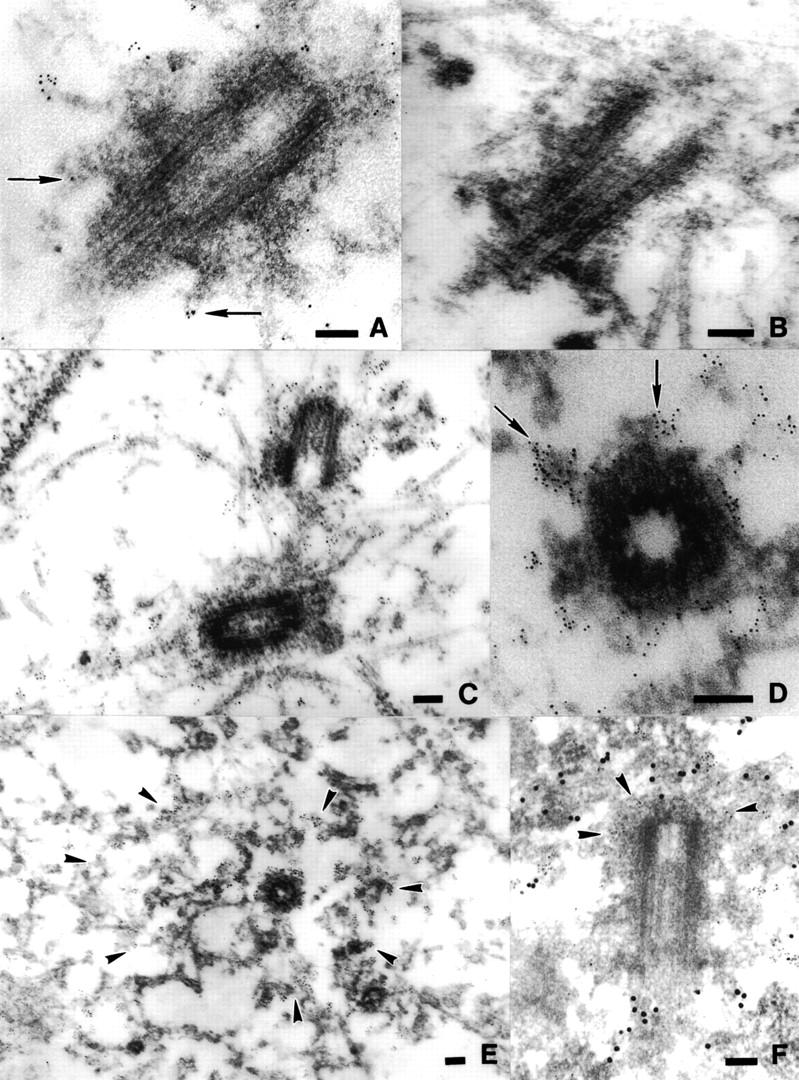
Immunoelectron microscopy of Skp1 stained centrosomes. NIH-3T3 cells were grown on mylar sheets, fixed in glutaraldehyde/paraformaldehyde (A–C, F), or in methanol (D,E) and labeled with no primary antibody (B), with affinity-purified anti-Skp1 antibodies (A,C–E), or with affinity-purified anti-Skp1 antibodies and a mouse monoclonal antibody against γ-tubulin (F), followed by 5 nm gold-conjugated goat anti-rabbit IgG (A–E) or 5 nm gold-conjugated goat anti-mouse IgG and 15 nm gold-conjugated goat anti-rabbit IgG (F). Samples were prepared for electron-microscopy as described (see Materials and Methods). Positions of centriolar appendages in A and D (arrows), concentrations of gold particles in a broad region surrounding the centriole in E (arrowheads), and concentrations of 5 nm gold particles (γ-tubulin staining) around the centriole in F (arrowheads) are indicated. Space bars, 0.1 μm.
A NEDD8-modified form of Cul1 is also present at the centrosome
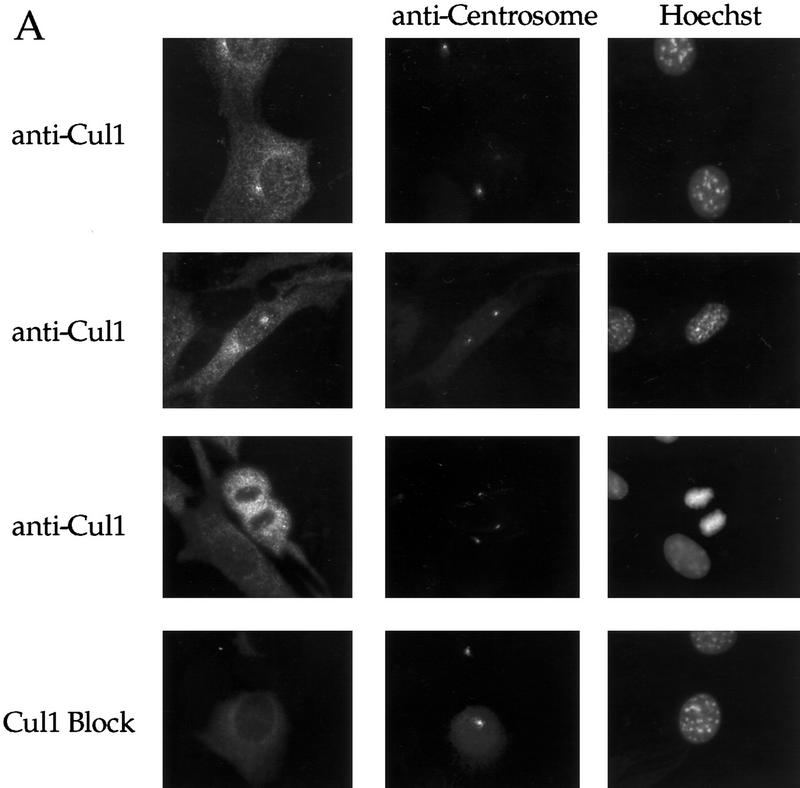
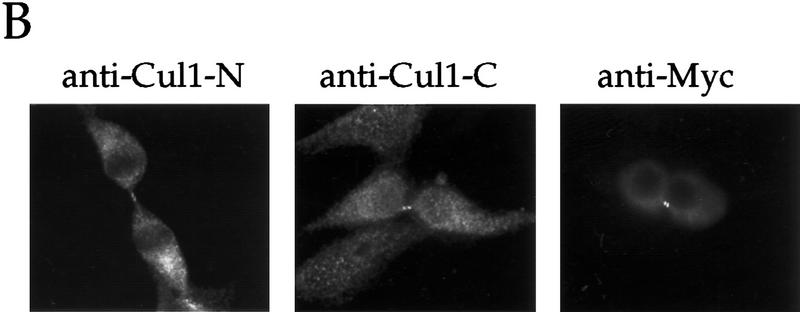
To determine if other SCF components function at the centrosome, we looked first at the localization of Cul1 in murine and human cells. Immunofluorescence microscopy using affinity-purified antibodies to the amino- terminus of human Cul1 indicates that Cul1 is localized to the centrosome in interphase (Fig. 6A, panels 1,2) and mitotic (Fig. 6A, panel 3) NIH-3T3 cells. Control labeling with blocked anti-amino-terminal Cul1 (Fig. 6A, panel 4), preimmune serum, and no primary antibody (data not shown), all resulted in little staining, indicating the specificity of the pattern observed. We also observed nuclear and cytoplasmic staining of Cul1, although Cul1 appeared less prominently nuclear than Skp1. Interestingly, both endogenous and transfected myc-tagged Cul1 localized to the midbody in cells undergoing cytokinesis (Fig. 6B).
Figure 6.
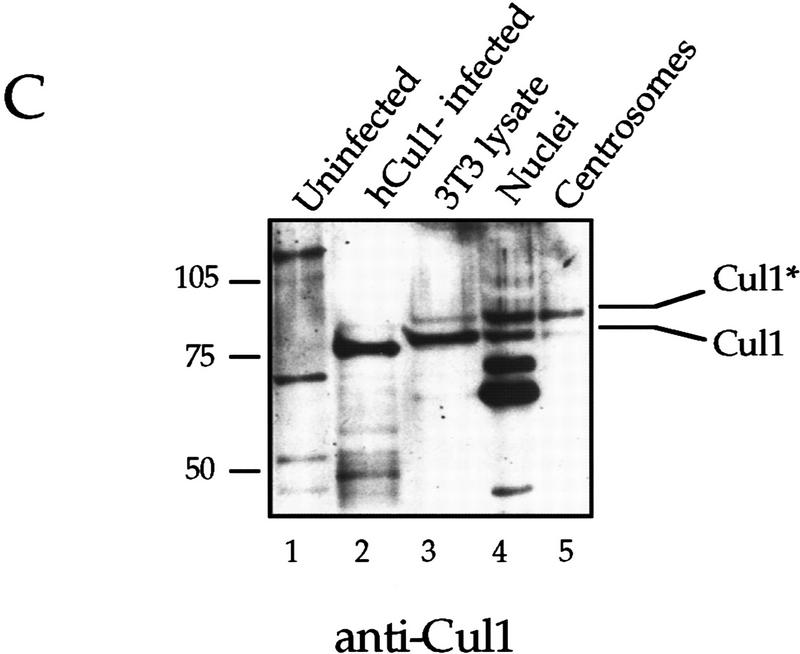
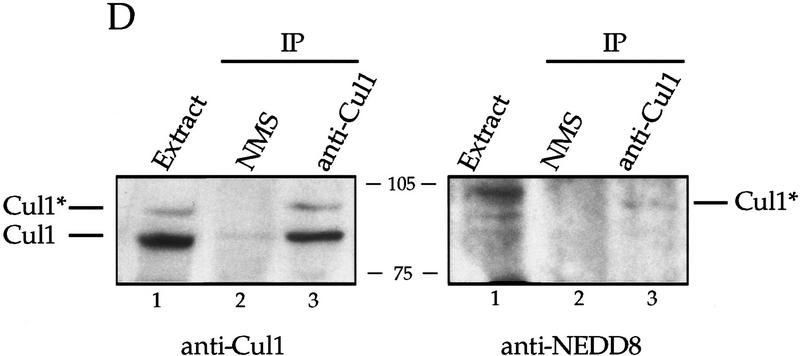
A Nedd8-modified form of Cul1 is present at the centrosome. (A) NIH-3T3 cells were grown on cover slips, fixed in methanol and labeled with affinity-purified antibodies against the amino-terminal portion of human Cul1 and with human anti-centrosome antiserum, followed by secondary antibodies and Hoechst dye. Cul1 staining is shown at the centrosome at different times in the cell cycle (top three panels). Cells were also stained with anti-centrosome antibody in combination with blocked anti-amino-terminal Cul1 antibodies (bottom panel). Cells were visualized and photographed as described (see Materials and Methods). (B) Cul1 localizes to the midbody. (Left and middle) NIH-3T3 cells were grown on coverslips, fixed in methanol, and labeled with affinity-purified rabbit anti-amino-terminal Cul1 antibodies (left) or affinity-purified rabbit anti-carboxy-terminal Cul1 antibodies followed by secondary antibodies(middle). (Right) NIH-3T3 cells expressing Myc-tagged Cul1 were fixed as above and labeled with anti-Myc mAb 9E10 followed by secondary antibodies. (C) Immunoblotting also demonstrates that Cul1 is present at the centrosome. Samples as indicated: (Lane 1) Sf9 cell extract; (lane 2) insect cell extract [Hi5] expressing human Cul1; (lane 3) NIH-3T3 cell lysate; (lane 4) nuclei prepared from CHO cells; and (lane 5) centrosomes purified from CHO cells were subjected to Western blotting with affinity-purified rabbit antibodies to the amino-terminal region of Cul1 followed by HRP-conjugated secondary antibodies. Bands were detected using ECL reagents (Amersham). Two forms of Cul1 detected are indicated (arrows). Positions of migration of molecular mass markers are also indicated. Note also that whole-cell lysate from CHO cells gave a similar result as did 3T3 cell lysate (data not shown). (D) Immunoblotting demonstrates that Cul1 is NEDD8-modified. Rabbit affinity-purified anti-Cul1 (left) and rabbit anti-NEDD8 (right) immunoblots of partially purified Xenopus egg extract (lanes 1), and immunoprecipitates from the same partially purified extract using control normal mouse serum (lanes 2), or mouse anti-Cul1 antibodies (lanes 3). Bands were detected using ECL reagents (Amersham). Two forms of Cul1 and positions of migration of molecular mass markers are indicated.
Western blotting using anti-amino-terminal Cul1 antibodies detected Cul1 in a lysate from insect cells infected with baculovirus expressing human Cul1 (Fig. 6C, lane 2), but not in control insect cells (Fig. 6C, lane 1). A protein of the expected molecular mass was also detected in an NIH-3T3 cell lysate (Fig. 6C, lane 3), in a preparation of nuclei from CHO cells (Fig. 6C, lane 4), and in the purified centrosome preparation (Fig. 6C, lane 5). Lysates from human 293 cells, Xenopus XTC cells and Xenopus eggs all showed similar results, using both affinity-purified rabbit anti-amino-terminal Cul1 antibodies, and mouse anti-Cul1 antibodies (data not shown), supporting that the human, mouse, hamster, and Xenopus forms of Cul1 are highly related proteins.
Two forms of Cul1 are detected in the NIH-3T3 cells. Recognition of both forms is competed by preincubation of the rabbit antibodies with antigen, and both are also detected with mouse anti-Cul1 serum (data not shown). The yeast Cul1 homolog cdc53p becomes modified by the addition of the ubiquitin-like protein Rub1 (Lammer et al. 1998). NEDD8 is a homologous ubiquitin-like protein in human cells. We also detect two forms of Cul1 in Xenopus egg extract (Fig. 6D, left panel, lane 1) and both were immunoprecipitated by mouse anti-Cul1 antibodies (Fig. 6D, left, lane 3). Anti-NEDD8 antibodies detected only the upper form of immunoprecipitated Cul1 (Fig. 6D, right, lane 3), demonstrating that this is NEDD8-modified Cul1. Therefore, NEDD8-modified Cul1 is the major form present at the centrosome, whereas whole cell lysate contains predominantly the faster-migrating species (Fig. 6C, cf. lanes 3 and 5). The selective fractionation of NEDD8-modified Cul1 suggests that this modification may have a role in localizing Cul1 to centrosomes and possibly also to nuclei.
Centriole separation is blocked by anti-Skp1 or anti-Cul1 antibodies and by proteasome inhibitors
To test the function of Skp1 at the centrosome, we used an assay described recently for centriole separation (Lacey et al. 1999). In this assay, purified centrosomes are incubated in extract from fertilized Xenopus eggs (see Materials and Methods). Isolated centrosomes are mostly centriole doublets. When centrosomes are incubated in extract alone, the doublet centrioles separate, resulting in singlet centrioles (Fig. 7A, top panel), which are visualized by staining for both α- and γ-tubulin. Centriole separation in this assay requires ATP, heat- and dilution-sensitive factors, kinase and phosphatase activities, and is time- and temperature-dependent (Lacey et al. 1999). Further, inactivating cyclin E/Cdk2 by immunodepletion or by addition of p21 inhibits centriole separation in this assay (Lacey et al. 1999). Injection of p21 also inhibits centrosome duplication in Xenopus embryos, suggesting that this assay provides a useful in vitro model for processes at the centrosome. The assay is quantitated by counting singlets and doublets after incubation of purified centrosomes in extract, and calculating the fraction converted from doublet to singlet (percent conversion, see Table 1).
Figure 7.
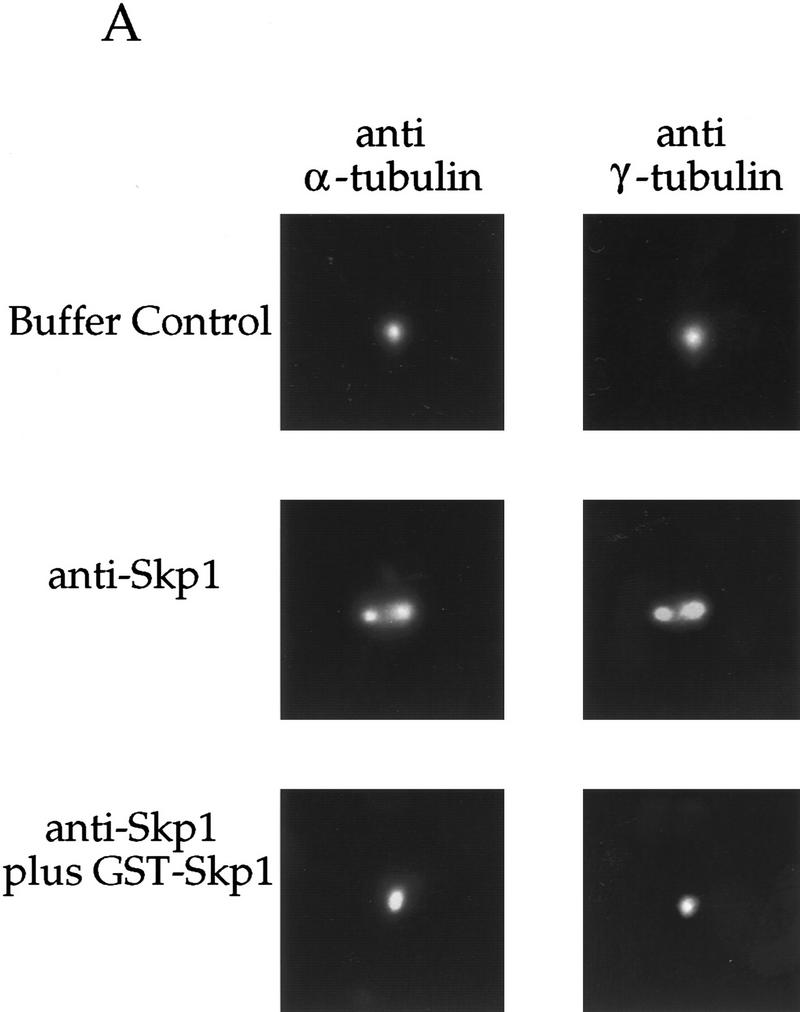
A requirement for Skp1, Cul1, and an SCF complex in centriole separation in vitro. (A) Centrosomes used in the centriole separation assay were pelleted onto coverslips as described (see Materials and Methods), and labeled with antibodies to both α- and γ-tubulin. Rabbit antibodies to Skp1 were used as part of the assay. The staining was done with rat anti-α-tubulin mAb and mouse anti-γ-tubulin mAb (see Materials and Methods). Events were counted and the data tabulated as described in Table 1. (B) Immunodepletion with rabbit anti-amino-terminal Cul1 antiserum was performed (see Materials and Methods), and an aliquot of extract before (lane 1) and after (lane 2) immunodepletion subjected to Western blotting with rabbit anti-amino-terminal Cul1 antibodies. Arrows indicate molecular mass markers and two forms of Cul1. (C) Immunoprecipitation with control normal mouse serum (lanes 2) or mouse anti-Cul1 antiserum (lane 3) was performed using extract prepared as for the centriole separation assay (lane 1) (see Materials and Methods). Starting extract and resultant immunoprecipitates were subjected to immunoblotting with affinity-purified rabbit anti-Cul1 antibodies (top) or affinity-purified anti-Skp1 antibodies (bottom). Bands were detected using ECL reagents. Positions of migration of Cul1, Skp1, and molecular mass markers are indicated.
Table 1.
Antibodies to Skp1 and Cul1 block centriole separation
| Condition
|
Percent conversion
|
|---|---|
| Buffer control or extract alone | 100 |
| Anti-GST | 62 |
| Anti-Skp1 | 5 |
| Anti-Skp1 + GST–Skp1 (block) | 49 |
| GST–Skp1 alone | 72 |
| Anti-γ-tubulin mAb | 91 |
| Normal mouse serum | 67 |
| Anti-Cul1 mouse serum | 23 |
| Anti-Cul1 + GST–Cul1 (block) | 100 |
| GST–Cul1 alone | 100 |
| Control (NRS) ID | 93 |
| Cul1 ID | 23 |
| Cul1 ID + GST–Cul1 | 23 |
| Cul1 ID + anti-Cul1 beads | 69 |
Centrioles were counted and classified into three categories: doublets (D), singlets (S), and separated doublets (SD). The percentage conversion is the percentage of doublets that were converted to singlets or separated doublets using the formula [(S + SD) − (S0)]/D0, where S0 is the percentage of singlets and D0 the percentage of doublets in the 0 time point control. Percent conversion is normalized with the buffer control or extract alone for an experiment set at 100%. The data represent individual experiments, and the range of response seen for given treatments is as follows: For anti-Skp1, n = 4, range = <2%–23%; for anti-γ-tubulin, n = 3, range = 81%–97%; for anti-Cul1, n = 3, range = <2%–23%; for Cul1 ID, n = 3, range = <2%–47%; for rescue with anti-Cul1 beads, n = 2, range = 69%–100%. (ID) Immunodepletion; (NRS) normal rabbit serum.
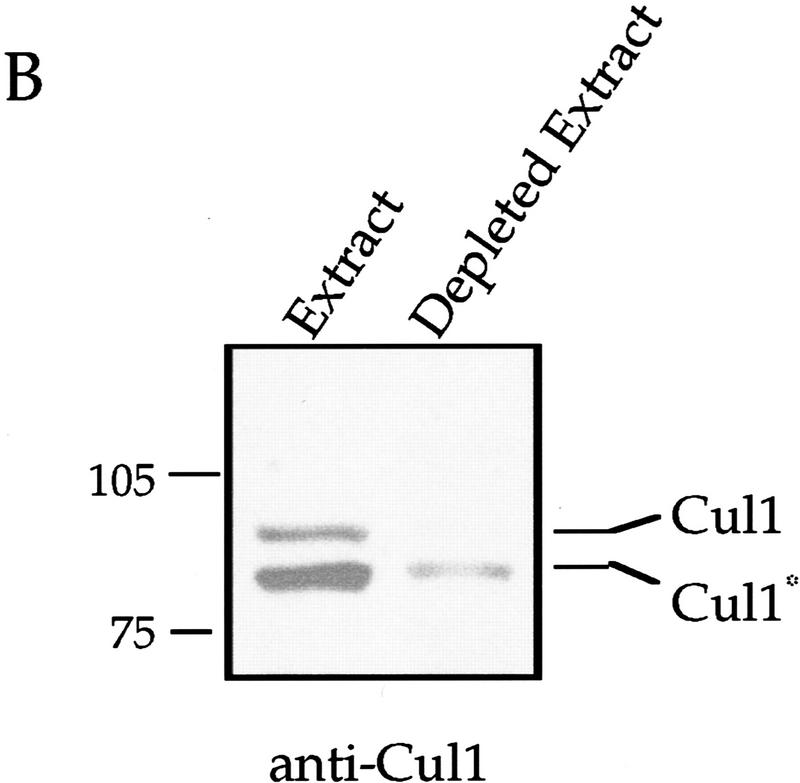
Addition of affinity-purified anti-Skp1 antibodies blocks centriole separation in Xenopus extracts (Table 1; Fig. 7A). Incubation of centrosomes with anti-Skp1 antibodies blocked by pre-incubation with purified GST-Skp1, or with antibodies affinity-purified against GST alone, did not strongly block centriole separation (Table 1; Fig. 7A). Monoclonal antibodies against γ-tubulin, although they bind to the centrosomes, had little effect in this assay (Table 1). These data suggest that Skp1 is required for centriole separation and, therefore, in the centrosome duplication process. Immunodepletion of Skp1 from the extract before incubation with centrosomes had a smaller effect; however, it is likely that the Skp1 already localized to the centrosome is sufficient for separation.
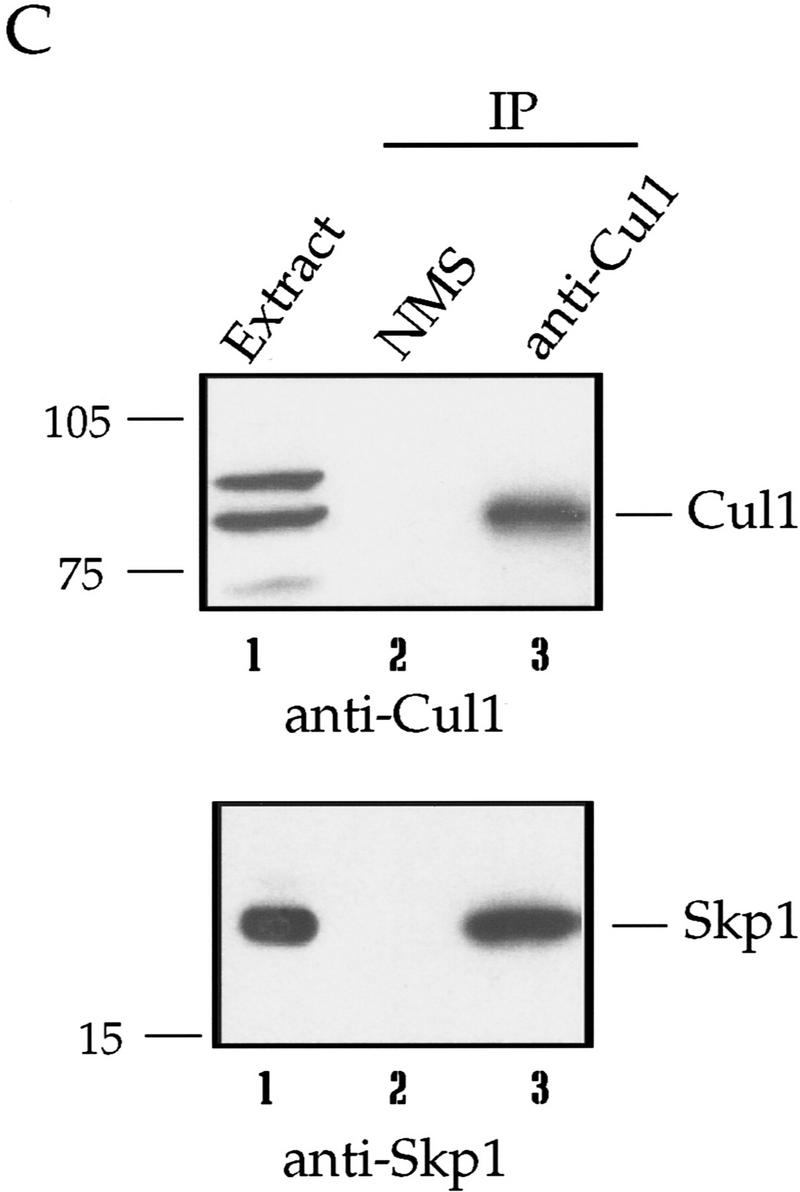
Addition of mouse antiserum raised against the full-length human Cul1 protein also inhibited separation, whereas control mouse serum had little effect (Table 1). In addition, immunodepletion of extract with rabbit antibodies to the amino terminus of Cul1 blocked centriole separation (Table 1). In this experiment, most of the Cul1 protein was immunodepleted (Fig. 7B). Addition of full-length GST–human Cul1 to immunodepleted extract did not rescue activity. Adding back a potential Cul1 complex (see Fig. 7C) on the anti-Cul1 antibody-bound protein A-Sepharose beads used to immunodeplete the extract, however, did result in partial rescue (Table 1). Cul1 immunodepleted from a similar extract contained coprecipitated Skp1 (Fig. 7C), a modified form of Cdc34 (data not shown; see Discussion), and a novel centrosomal F-box protein isolated in our laboratory (data not shown; J. Regan and P. Jackson, unpubl.). These data indicate that depletion of Cul1 removes an SCF complex required in the centrosome duplication process.
If SCF ubiquitin ligases mediate ubiquitination of proteins important for centrosome duplication, we would expect that proteasome-dependent proteolysis is also required. To address this question, we added the proteasome inhibitor clastolactacystin β-lactone (CLBL) to our assay. This compound blocks proteolysis by covalently modifying catalytic components of the proteasome, and is one of the most specific proteasome inhibitors known (Fenteany and Schreiber 1998). Our results indicate that CLBL blocks centriole separation in a dose-dependent manner (Table 2). The related inhibitor lactacystin also blocked centriole separation, but less effectively (data not shown). We have used the fluorogenic proteasome substrate suc-LLVY-7-amido-4-methylcoumarin (suc–LLVY–AMC) to monitor the inhibition of proteasomes by clasto-lactacystin β-lactone in parallel extracts. Significant proteasome inactivation (∼85%) occurred at 300 μm CLBL, but doses of 600 and 800 μm reduced proteasome activity to ∼10% and to background levels, respectively. This suggests that centriole separation is able to proceed with only a small percentage of the normal proteasome activity, or that the proteasomes that subserve this function are not easily reached by the inhibitor. These data are consistent with our observations using proteasome blockers to interfere with DNA replication, another Cdc34- and proteolysis-dependent activity in Xenopus extract (C. Swanson and P. Jackson, unpubl.). Taken together, our data implicate Skp1, Cul1, and SCF-mediated proteolysis in centriole separation.
Table 2.
The proteasome inhibitor clastolactacystin β-lactone blocks centriole separation
| Clastolactacystin β-lactone
|
Percent conversion
|
|---|---|
| Extract alone | 100 |
| DMSO only | 82 |
| 300 μm | 79 |
| 450 μm | 79 |
| 600 μm | 47 |
| 800 μm | 4 |
Results are presented as percent conversion (see Table 1), with numbers normalized to extract alone set at 100%. For clastolactacystin β-lactone, n = 2, range (at 800 μm) = <2%–4%. Lactacystin also inhibited centriole separation but was less effective.
It has been demonstrated recently that cyclin E/Cdk2 is required for centrosome duplication (Hinchcliffe et al. 1999; Lacey et al. 1999; Matsumoto et al. 1999) in this and other assays. Analogous to the requirement for Sic1p proteolysis for initiation of DNA replication, a potential explanation for our results is that the required proteolysis substrate is a cyclin E kinase inhibitor such as the p27Kip1 homolog Xic1 (Su et al. 1995). We addressed this question by determining if excess purified baculovirus-expressed cyclin E/Cdk2 could rescue the effects of anti-Skp1 antibodies in the centriole separation assay. Although cyclin E/Cdk2 was active and could increase the activity of diluted or partially inactive extract (data not shown), it did not rescue the activity of anti-Skp1 inhibited extract (Table 3). Similar results were obtained with anti-Cul1 immunodepleted extract. These data suggest that the proteolysis requirement in centrosome duplication involves substrates other than cyclin E/Cdk2 kinase inhibitors.
Table 3.
Cyclin E/cdk2 does not rescue centriole separation inhibited by anti-Skp1
| Condition
|
Percent conversion
|
|---|---|
| Extract alone | 92 |
| Buffer control | 97 |
| anti-Skp1 | 4 |
| Cyclin E/cdk2 | 92 |
| anti-Skp1 + Cyclin E/cdk2 | 0 |
See Table 1 for details.
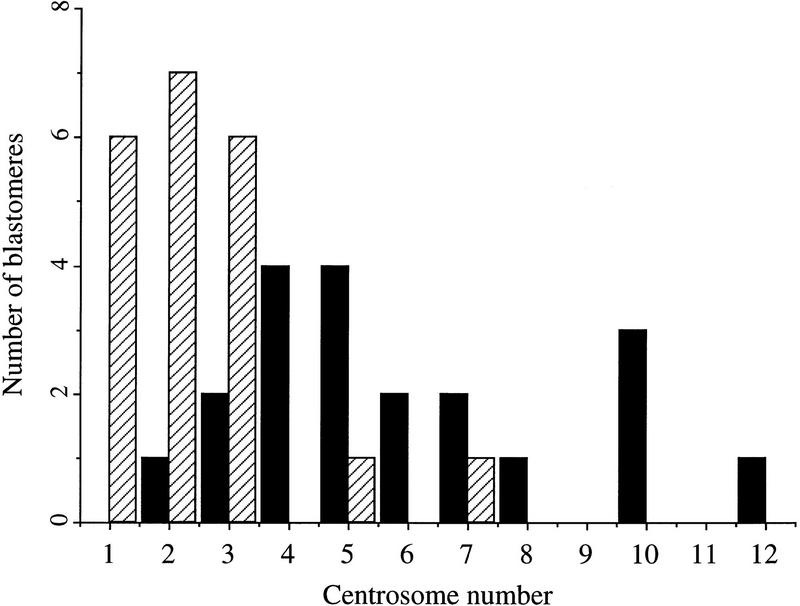
Finally, we looked at the ability of the proteasome inhibitor CLBL to block centrosome separation when injected into cycloheximide-treated Xenopus embryos. In this assay, Xenopus embryos at the 16- to 64-cell stage are incubated with the protein synthesis inhibitor cycloheximide to block translation of cyclin B and the mitotic cell cycle, but allowing centrosome duplication to continue (Gard et al. 1990; Lacey et al. 1999). Single cells from embryos were injected with CLBL, the embryos incubated in cycloheximide, and then assayed by confocal microscopy for the extent of centrosome duplication. FITC-conjugated dextran was coinjected as a visible marker for injected cells. As shown in Figure 8, control- injected blastomeres (DMSO alone) contained an average of 6.0 (s.e.= 0.62) centrosomes, whereas CLBL-injected blastomeres contained an average of 2.4 (s.e.= 0.34) centrosomes. This extent of inhibition is typical for other agents that strongly inhibit centrosome duplication, including Cdk2 inhibitors (Lacey et al. 1999). Therefore, proteasome activity is also required for centrosome duplication in the early Xenopus embryo.
Figure 8.
Injection of proteasome inhibitors into Xenopus blastomeres blocks centrosome duplication. Animal pole cells from frog embryos were injected with DMSO alone (solid bars) or DMSO containing CLBL (hatched bars), and with FITC-conjugated dextran and subsequently incubated in cycloheximide, fixed and stained with antibodies to α-and γ-tubulin. Centrosomes in individual injected cells were counted and the data represented in histogram form.
Known centrosomal proteins contain F-box motifs
To discover candidates for F-box-containing proteins that might be relevant to centrosome duplication and to address the complexity of SCF-mediated proteolysis at the centrosome, we conducted a search of known centrosomal proteins for the presence of the F-box motif (see Materials and Methods). Minimally, two proteins localized previously to the centrosome contain putative F-box sequences (see Materials and Methods). These proteins are C-Nap1 (Fry et al. 1998a) and ninein (Bouckson-Castaing et al. 1996). We have also shown that a novel F-box protein isolated in our laboratory is present at the centrosome (J. Regan, E. Freed, and P. Jackson, unpubl.).
Discussion
The centrosome cycle requires the replication of centrioles and accumulation of PCM, separation of centrioles and PCM at prophase to form the poles of the mitotic spindle, and segregation of centrosomes to daughter cells at mitosis. Studies in tissue culture cells and embryos suggest that there is an intrinsic centrosome duplicating machinery and a mechanism to couple that machinery to the mitotic cycle. Our data support a model where an SCF ubiquitin–ligase complex functions locally at the centrosome to direct at least one step of the centrosome duplicating cycle. Both the Skp1 and Cul1 SCF components can be visualized at the centrosome by conventional and electron microscopy and both Skp1 and a NEDD8-modified form of Cul1 are present in purified centrosomes. Further, an in vitro assay for centriole/centrosome separation demonstrates that Skp1, Cul1, and proteasome activity are required for centriolar/centrosome separation. Finally, blocking proteasome activity in vivo also inhibits centrosome duplication in cycloheximide-treated Xenopus embryos.
The centrosome duplicating machinery is active during S phase and inhibited during mitosis (Hinchcliffe et al. 1998). The requirement for cyclin/Cdk2 to promote centrosome duplication (Hinchcliffe et al. 1999; Lacey et al. 1999; Matsumoto et al. 1999; Meraldi et al. 1999) and the ability of the mitotic state to block duplication suggest how the intrinsic duplicating machine is linked to the cell cycle. These findings help to explain why cells blocked in S phase, when Cdk2 is active but mitotic cyclin/Cdk kinases are not, continue to duplicate their centrosomes. A formally similar mechanism works to ensure that each segment of the DNA in a chromosome is replicated once and only once (for review, see Stillman 1996; Dutta and Bell 1997; see also Fang and Newport 1991; Strausfeld et al. 1994; Jackson et al. 1995; Yew and Kirschner 1997). In budding yeast, S-phase cyclin-dependent kinases and an SCF complex trigger the initiation of DNA replication by firing pre-initiation complexes at replication origins (Schwob et al. 1994; Drury et al. 1997), but the mitotic cyclin-dependent kinases block the formation of these pre-initiation complexes (Dahmann et al. 1995). Similarly, in fission yeast, an apparent SCF complex containing the adapter proteins Pop1 and Pop2 functions to limit replication of each local segment, whereas the mitotic cyclin/Cdc2 kinase is required to block the establishment of replication competence (Broek et al. 1991; Correa-Bordes and Nurse 1995; Kominami and Toda 1997; Jallepalli et al. 1998; Wolf et al. 1999). Our findings suggest a similarity between DNA replication and centrosome duplication, such that an SCF ubiquitin–ligase and the Cdk2 kinase complexes may regulate the intrinsic centrosome cycle, and that the mitotic state, although not necessarily Cdc2 itself, may block the establishment of conditions necessary to reinitiate centrosome duplication.
Skp1 and Cul1 localize to the centrosome and nucleus
We have demonstrated that both Skp1 and Cul1 are centrosomal proteins throughout the cell cycle. Like many centrosomal proteins, the abundance of Skp1 at the centrosome increases at mitosis. Further, Skp1 localization to the centrosome is stable to various fixation conditions and independent of microtubules, suggesting that Skp1, together with γ-tubulin, pericentrin, Nek2, and Drosophila CP190 and CP60, is a core component of the centrosome (Stearns et al. 1991; Doxsey et al. 1994; Oegema et al. 1995, 1997; Fry et al. 1998b).
Biochemical purification of the centrosome demonstrated the presence of Skp1 and a NEDD8-modified form of Cul1. Evidence from budding yeast suggests that the covalent attachment of Rub1p, the S. cerevisiae homolog of NEDD8, to Cdc53p may promote SCF function (Lammer et al. 1998). Recently, human cullins have been shown to be NEDD8-modified (Osaka et al. 1998; Liakopoulos et al. 1999; Wada et al. 1999). Our data do not distinguish whether modification targets Cul1 to the centrosome or nucleus, or is a consequence of Cul1 being localized to these sites. Cells with a mutation in the NEDD8-activating enzyme SMC1, however, demonstrate both DNA rereplication and a failure to separate centrosomes (Handeli and Weintraub 1992). Therefore, it is possible that the addition of NEDD8 to Cul1 is important for limiting DNA replication and centrosome/centriole duplication.
Skp1 was also present in a punctate pattern in the nucleus, as was particularly apparent in aldehyde-fixed material (Fig. 1B). Previous studies on the human SCF complex have indicated a role for Cdc34, Cul1, Skp1, and the F-box/adapter protein Skp2 in DNA replication (Zhang et al. 1995; Lisztwan et al. 1998; Marti et al. 1999; Montagnoli et al. 1999; Tsvetkov et al. 1999); therefore, we might expect Skp1 to be nuclear. Cul1 was also localized in part to the nucleus, consistent with its suggested role in DNA replication. We have observed the F-box protein Skp2 in the nucleus (E. Freed and P.K. Jackson, unpubl.) but did not observe Skp2 at the centrosome.
Studies of Skp1 in S. cerevisiae support a role in assembling the kinetochore (Stemmann and Lechner 1996; Kaplan et al. 1997). We did not observe any Skp1 staining at the mammalian kinetochore, although Skp1 epitopes at the kinetochore may be masked. We also observed endogenous and overexpressed Cul1 staining at the midbody in cells completing cytokinesis, suggesting a potential SCF role there.
Deconvolution microscopy and immunoelectron microscopy indicated that Skp1 is present both near the centriole and in an annular region 1–3 centriolar diameters away from the centriole core, possibly further than the pericentriolar region defined by pericentrin (Doxsey et al. 1994; Dictenberg et al. 1998). These images also demonstrate Skp1 labeling in a pattern of fibers around the centrioles, possibly in the recently described pericentriolar lattice containing γ-tubulin and pericentrin (Dictenberg et al. 1998; Schnackenberg et al. 1998). Immunoelectron micrographs show Skp1 on the distal appendages that emanate from the centriole core (Fig. 5). The function of these structures is not known.
Skp1 and Cul1 function in the centrosome duplication cycle
Centriole separation occurs around the G1–S transition and requires cyclin E/Cdk2 and cyclin A/Cdk2 kinases (Lacey et al. 1999), which apparently have complimentary roles in this process. Therefore, centriole separation may be an early regulated step in the centrosome cycle. In our experiments, blocking Skp1, Cul1, or proteasome function each blocked centrosome duplication at this specific step. Injection of proteasome inhibitor into cycloheximide-treated Xenopus embryos also blocked centrosome duplication at an early step. This is consistent with a proteolytic requirement for centriolar separation, but does not exclude the possibility of additional proteolytic steps in the centrosome cycle.
The activity of extracts immunodepleted of Cul1 could be partially rescued by the addition of the anti-Cul1 beads used in the immunodepletion. These Cul1 immunoprecipitates contain Skp1 (Fig. 7C), Cdc34, and a novel centrosomal F-box protein isolated in our laboratory (J. Regan and P. Jackson, unpubl.), indicating that Cul1 depletion removes an SCF complex. The idea that the Cul1 immunoprecipitates contain an active ubiquitin ligase is suggested by the presence of a slower mobility form of Cdc34 (∼48 kD) than the major 34-kD Cdc34 species present in extract, consistent with the previously described Cdc34 autoubiquitination seen in active SCF complexes (R. Deshaies, unpubl.; Goebl et al. 1994; Skowyra et al. 1999). Nonetheless, Cul1 has also been shown to interact with the proteasomal subunit HsN3, (Lyapina et al. 1998), suggesting that Cul1 immunodepletion may remove other components important for proteolysis, which are also rescued by addition of the immunoprecipitates.
Excess active cyclin E/Cdk2 did not rescue the effects of anti-Skp1 antibodies or Cul1 immunodepletion, suggesting that a proteolysis target other than a cyclin E/Cdk2 inhibitor is required for centrosome duplication. The presence of specific F-box proteins at the centrosome might provide a means to identify their targets. We suggest that at least two known centrosomal proteins contain putative F-box sequences. One of these proteins, C-Nap1, has been shown to largely disappear from centrosomes at prophase, perhaps by a proteolytic mechanism. Our novel centrosomal F-box protein, called SBP5, has been used as bait in an additional two-hybrid screen and has identified three potential centrosomal targets (J. Regan and P.K. Jackson, unpubl.). The presence of F-box proteins at the centrosome suggests that distinct SCF complexes may mediate different steps in the centrosome duplication process, or direct mitotically controlled processes such as microtubule nucleation and spindle assembly.
Previous work suggested a requirement for proteolysis for the duplication and subsequent separation of yeast spindle pole bodies (Winey and Byers 1992; McDonald and Byers 1997). Interestingly, recent observations in mammalian cells have localized components of the proteasomal machinery to the centrosome (Wigley et al. 1999). Our work localizes SCF components to the centrosome in animal cells, and demonstrates a requirement for one or more SCF complexes in separation of centrioles in G1, and possibly in other yet unidentified steps in the centrosome cycle. As suggested for the separation of S. cerevisiae SPBs, it seems likely that centrosome separation at prophase may also require proteolytic steps. The identification of specific centrosomal targets and the use of additional assays will be required to elucidate the role of SCF-dependent proteolytic events in the centrosome cycle. By controlling the replication of both centrosomes, which organize the spindle to segregate the chromosomes, and the replication of the chromosomes themselves, SCF-dependent proteolysis appears to be broadly required for maintaining ploidy and genomic stability.
Materials and methods
Cell lines and culture
NIH-3T3 cells were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% heat-inactivated donor bovine calf serum, penicillin/streptomycin, and l-glutamine (2 mm). CHO cells were obtained from ATCC and maintained in McCoy’s 5A medium supplemented with 10% fetal bovine serum, penicillin/streptomycin, l-glutamine, and sodium pyruvate (1 mm).
Immunofluorescence microscopy
Cells were grown on cover slips, and except where otherwise noted, fixed by 5-min incubation in methanol (−20°C), allowed to dry briefly, washed twice with phosphate-buffered saline minus Ca2+ and Mg2+ (PBS−) and then one to two times with immunofluorescence (IF) wash buffer (PBS− containing 0.1 % Triton X-100 and 3% bovine serum albumin). Cells were blocked in 5% normal donkey serum (30 min) in IF wash buffer. Primary antibodies were diluted in IF wash buffer and used at the following concentrations: anti-Skp1 (0.5–1 μg/ml); anti-α-tubulin (DM1α: 1:100); human anti-centrosome serum (5947), kindly provided by Stephen Doxsey (University of Massachusetts, Worchester) (1:900); anti-γ-tubulin mAb (Sigma, Clone GTU-88: 1:500); affinity-purified anti-amino-terminal Cul1 (0.5 μg/ml); affinity-purified anti-carboxy-terminal Cul1 (0.5 μg/ml); and anti-myc mAb 9E10 (1:1000). Texas Red or fluoresceinconjugated secondary antibodies (Jackson Immunoresearch) were used at 1:100 dilution. DNA was visualized by counterstaining with Hoechst dye (5 μg/ml).
Cells pictured in Fig. 1B (top two panels) were fixed at room temperature in paraformaldehyde (4%) in PBS−× 10 min or in paraformaldehyde (2%)/glutaraldehyde (0.1%) in PBS− (30 min). After fixation, aldehyde-fixed cells were permeabilized by 10 min incubation in PBS− containing 0.2% Triton X-100, and subsequently washed and labeled as described above.
Fluorescent cells were visualized with a Zeiss Axioskop microscope using a 63× Neofluor (N.A.=1.3) lens, and photographed using a Princeton Instruments PentaMAX digital camera controlled by computer using Metamorph software (Universal Imaging Corporation). Purified centrosomes were visualized with a 100× Neofluor (N.A.=1.3) lens.
Deconvolution microscopy
Cells or centrosomes were fixed and labeled as above. Deconvolution microscopy was performed using an Applied Precision DeltaVision v. 2.0 system using an Olympus IX-70 inverted microscope equipped with a 100× lens. Optical sections of 0.15 or 0.20 μm were used. The data were deconvolved using the Agard and Sedat inverse matrix algorithm (Agard et al. 1989).
Electron microscopy
Cells were grown on mylar sheets and fixed either in methanol as above, or permeabilized, fixed in paraformaldehyde/glutaraldehyde and treated with sodium borohydride as described (Stearns et al. 1991), with the following changes. Permeabilization before fixation was in 0.2% Triton X-100 and the concentration of fixative was 2% paraformaldehyde/0.1% glutaraldehyde. Fixed cells were washed with PBS− and IF wash buffer with 0.3% normal goat serum (NGS). Cells were blocked with normal goat serum (5% in IF wash buffer for 30 min, Fig. 5A–E) and incubated in affinity-purified anti-Skp1 (10 μg/ml, Fig. 5A,C; 30 μg/ml, Fig. 5D,E for 3.5 hr) in IF wash buffer/0.3% NGS. For double labeling (Fig. 5F), fixation was performed at room temperature for 15 min, the blocking conditions were 1% BSA/0.1% NGS, anti-γ-tubulin mAb (Sigma, Clone GTU-88) was used at 1:6 dilution and anti-Skp1 at 60 μg/ml. AuroProbe gold-labeled goat anti-rabbit IgG (Amersham, 10 nm, Fig. 5A–E; 15 nm, Fig. 5F) and goat anti-mouse IgG (Amersham, 5 nm, Fig. 5F) secondary antibodies were used at 1:3 (Fig. 5A–C,F) or 1:10 (Fig. 5D,E) dilution.
After labeling, cells were fixed in 2.5% glutaraldehyde, 2% paraformaldehyde in phosphate buffer for 12 hr at 4°C, washed several times with 0.1 m sodium cacodylate buffer (pH 7.4), and postosmicated with 1% OsO4 for 3 hr. The mylar sheets were dehydrated in a series of ethanols, followed by several washes of propylene oxide, and infiltrated with LX-112 (Ladd Research Industries, Burlington, VT). The sheets were placed, cell side down, onto a pre-polymerized plate of LX-112 and cured at 60°C. Thin sections were cut en face, on a Reichert OMU3, and lightly stained with uranyl acetate and lead citrate. Electron micrographs were taken with a Seimens Elmiskop 101.
Western blotting
Western blotting was performed by standard methods using HRP-conjugated secondary antibodies (Jackson Immunoresearch) and enhanced chemiluminescence (ECL) detection reagents (Amersham). Antibodies used for blotting were affinity-purified anti-Skp1 antibodies (0.5 μg/ml), rabbit anti-HA-tag (Medical and Biological Laboratories, Nagoya, Japan, 1:100), rabbit anti-γ-tubulin (XGC-1-4, 1:500; Stearns et al. 1991), affinity-purified rabbit anti-amino-terminal Cul1 (∼0.5 μg/ml; Lyapina et al. 1998), and rabbit anti-NEDD8 (Alexis Biochemicals, 1:400).
The number of Skp1 molecules per centrosome was calculated as follows. A known volume of centrosomes was spun onto a coverslip and counted to determine the concentration of centrosomes in that preparation. A sample from the same preparation was compared with known amounts of purified Skp1 protein on Western blots using anti-Skp1 antibodies and densitometric scanning.
Production of proteins and affinity purification of antibodies
For production of anti-Skp1 antibodies, rabbits were injected with bacterially expressed glutathione-S-transferase (GST)–Skp1 fusion protein. GST–Skp1 was soluble at concentrations of > 60 mg/ml. Antisera were affinity-purified by standard procedures (Harlow and Lane 1988), using maltose-binding protein (MBP)–Skp1 coupled to cyanogen bromide-activated Sepharose (Pharmacia). Rabbit anti-Cul1 antibodies were generated as described (Lyapina et al. 1998).
Centrosome purification and labeling
CHO cell centrosomes were purified by the method of Mitchison with modifications as described by Bornens et al. (Mitchison and Kirschner 1986; Bornens et al. 1987). For immunofluorescence labeling, centrosomes were thawed and aliquots diluted (1:50) into PE buffer (10 mm PIPES at pH 7.2, 1 mm EDTA, 8 mm 2-mercaptoethanol), layered over 30% glycerol in PB buffer (80 mm PIPES at pH 6.8, 1 mm EGTA, 1 mm MgCl2) and centrifuged onto glass coverslips at 25,000g for 15 min. Antibodies used for staining are indicated above. To prepare centrosomes for resolution on SDS gels, aliquots were diluted, pelleted at 100,000g, and resuspended in SDS-PAGE sample buffer.
In some experiments (see centrosome in Fig. 4 right), centrosomes were diluted 1:10 into a high speed supernatant from Xenopus egg extract, incubated for 1 hr at room temperature, and treated with nocodazole (30 min, 4°C) before spinning onto coverslips.
Centriole separation assay
Frog embryo extract was prepared from fertilized eggs or from ionophore-activated eggs (Murray 1991). Immunodepletion was performed by incubating extract for 1 hr with rabbit anti-amino-terminal Cul1 antibodies (Fig. 8B) prebound to protein A–Sepharose beads, as described (Jackson et al. 1995). For antibody blocking experiments, centrosomes prepared from Xenopus XTC cells (Mitchison and Kirschner 1986) were preincubated at room temperature with the indicated antibodies or with buffer alone for 15 min (10 μl centrosomes plus 7–10 μl of antibody or buffer). For the experiment summarized in Table 3, cyclin E/Cdk2 was purified from baculovirus-infected Sf9 cells as described (Lacey et al. 1999), and incubated with centrosomes for 10 min, anti-Skp1 antibodies added and the mixture incubated for an additional 10 min prior to incubation with extract. Extract (100 μl) was added to the centrosome mixtures for 1 hr at 25°C. For proteasome inhibitor treatments and for rescue experiments, the extracts were preincubated with protein, protein A–Sepharose beads, or inhibitor (15 min) before addition of centrosomes. Reactions were stopped by the addition of 300 ml 10 mm Tris at pH 8.0, and nocodazole to 75 μg/ml, and placed on ice for 30 min. Samples were then layered over 30% glycerol in PB, centrifuged onto coverslips and processed for immunofluorescence as described (Stearns and Kirschner 1994).
Centriole separation was scored by counting singlets and doublets after staining for both α- and γ-tubulin. In experiments where no antibody treatment was involved, staining was performed with a mouse mAb to α-tubulin (DM1α, 1:100) and rabbit antiserum to γ-tubulin (XGC-1-4, 1:500), and secondary antibodies FITC-conjugated donkey anti-mouse and Texas Red-conjugated goat anti-rabbit (Jackson Immunoresearch, 1:100). When rabbit antibodies were part of the experimental condition, staining was with a rat mAb to α-tubulin (Sera-lab, Clone YL1/2, 1:50) and a mouse mAb to γ-tubulin (Sigma, Clone GTU-88, 1:500). When mouse antibodies were used in the experiment, rat anti-γ-tubulin mAb (see above) was used with rabbit anti-γ-tubulin (XGC-1-4). Centriole singlets or doublets were scored if both antibodies coincidentally stained.
Immunoprecipitation
Immunoprecipitation (Figs. 7D and 8C) was from Xenopus embryo extract prepared as for centriole separation assays (see above) and diluted in immunoprecipitation (IP) buffer (100 mm NaCl, 50 mm β-glycerophosphate, 5 mm EDTA, 0.1% Triton X-100 at pH 7.2) prior to the addition of mouse anti-Cul1 antiserum. Immune complexes were collected using protein G–Sepharose 4B (Sigma) preblocked by incubation in IP buffer containing 3% BSA. For Figure 6D, the extract was first fractionated on a Hytrap Q column, and fractions containing Cul1 pooled and subjected to immunoprecipitation as above.
Proteasome assays
Proteasome activity was measured by an assay using suc-LLVY-MCA (Bachem) as a fluorogenic peptide substrate for the proteasome as described (Glas et al. 1998). After treatment with clasto-lactacystin β-lactone, extracts were diluted 1:20 into 100 μl of reaction buffer (50 mm Tris at pH 7.4, 5 mm DTT, 5 mm MgCl2, 10 μm suc–LLVY–MCA), and incubated at 37°C for 1 hr. Fluorescence was measured using an Aminco–Bowman Series II spectrofluorimeter.
Frog embryo injection and imaging
Embryo injection experiments were carried out as described (Lacey et al. 1999). Briefly, animal pole cells from frog embryos at the 16- to 64-cell stage were injected with either DMSO alone or DMSO containing 50 mm CLBL together with FITC-conjugated dextran, then incubated in cycloheximide-containing medium for 4 hr before fixation and labeling with antibodies to both α- and γ-tubulin. Centrosomes were counted and data represented in graphic form.
Centrosomal F-box protein search
The sequences of known centrosomal proteins were downloaded and used in a preliminary search for F-box-like sequences. Candidates were then compared with Hidden Markov Models created from known F-box sequences (for alignments and scores, see http://csb.stanford.edu/brenner/tmp/misc/f-box/ho.pfam-f-box.f-box-put and http://csb.stanford.edu/brenner/tmp/misc/f-box/ho.f-box-conf.f-box-put).
Acknowledgments
We thank Klaus Bensch and Irma and Herbert Daehne for their generous help with electron microscopy. We also thank Steve Murphy and Lenore Urbani for providing reagents and for helpful discussions; David Beach for Skp1 and Skp2 reagents; Steve Doxsey for providing anti-centrosome serum, Julie Regan for unpublished information about Skp1 and its partners; Margaret Fuller for critical reading of the manuscript; and members of the Stearns and Jackson laboratories for helpful discussions. This work was supported by National Institutes of Health R01 GM54811, a Howard Hughes Medical Institute junior investigator award (P.J.), and a grant from the Donald E. and Delia Baxter Foundation. E.F. was supported in part by U.S. Public Health Service training grant CA 09151 awarded by the National Cancer Institute, Department of Health and Human Services.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pjackson@cmgm.stanford.edu; FAX 650-725-6902.
References
- Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. In: Taylor DL, Wang Y-l, editors. Fluorescence microscopy of living cells in culture. San Diego, CA: Academic Press; 1989. pp. 353–377. [DOI] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M, Paintrand M, Berges J, Marty M-C, Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Mobility Cytoskeleton. 1987;8:238–249. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- Bouckson-Castaing V, Moudjou M, Ferguson DJ, Mucklow S, Belkaid Y, Milon G, Crocker PR. Molecular characterization of ninein, a new coiled-coil protein of the centrosome. J Cell Sci. 1996;109:179–190. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]
- Broek D, Bartlett R, Crawford K, Nurse P. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis [see Comments] Nature. 1991;349:388–393. doi: 10.1038/349388a0. [DOI] [PubMed] [Google Scholar]
- Correa-Bordes J, Nurse P. p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell. 1995;83:1001–1009. doi: 10.1016/0092-8674(95)90215-5. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Diffley JF, Nasmyth K. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JF. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Bell SP. Initiation of DNA replication in eukaryotic cells. Annu Rev Cell Dev Biol. 1997;13:293–332. doi: 10.1146/annurev.cellbio.13.1.293. [DOI] [PubMed] [Google Scholar]
- Fang F, Newport JW. Evidence that the G1-S and G2-M transitions are controlled by different cdc2 proteins in higher eukaryotes. Cell. 1991;66:731–742. doi: 10.1016/0092-8674(91)90117-h. [DOI] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Fenteany G, Schreiber SL. Lactacystin, proteosome function, and cell fate. J Biol Chem. 1998;273:8545–8548. doi: 10.1074/jbc.273.15.8545. [DOI] [PubMed] [Google Scholar]
- Fry AM, Mayor T, Meraldi P, Stierhof Y-D, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol. 1998a;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, Meraldi P, Nigg EA. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 1998b;17:470–481. doi: 10.1093/emboj/17.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- Gard DL, Hafezi S, Zhang T, Doxsey SJ. Centrosome duplication continues in cycloheximide-treated Xenopus blastulae in the absence of a detectable cell cycle. The J. Biol Chem. 1990;110:2033–2042. doi: 10.1083/jcb.110.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment[published erratum appears in15:15124[ EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Glas R, Bogyo R, McMaster JS, Gaczynska M, Ploegh H. A proteolytic system that compensates for loss of proteasome function. Nature. 1998;392:618–622. doi: 10.1038/33443. [DOI] [PubMed] [Google Scholar]
- Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- Goebl MG, Goetsch L, Byers B. The Ubc3 (Cdc34) ubiquitin-conjugating enzyme is ubiquitinated and phosphorylated in vivo. Mol Cell Biol. 1994;14:3022–3029. doi: 10.1128/mcb.14.5.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handeli S, Weintraub H. The ts41 mutation in Chinese hamster cells leads to successive S phases in the absence of intervening G2, M, and G1. Cell. 1992;71:599–611. doi: 10.1016/0092-8674(92)90594-3. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hinchcliffe EH, Cassels GO, Rieder CL, Sluder G. The coordination of centrosome reproduction with nuclear events of the cell cycle in the sea urchin zygote. J Cell Biol. 1998;140:1417–1426. doi: 10.1083/jcb.140.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-Cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Jackson PK, Chevalier S, Philippe M, Kirschner MW. Early events in DNA replication require cyclin E and are blocked by p21CIP1. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Tien D, Kelly TJ. sud1(+) targets cyclin-dependent kinase-phosphorylated Cdc18 and Rum1 proteins for degradation and stops unwanted diploidization in fission yeast. Proc Natl Acad Sci. 1998;95:8159–8164. doi: 10.1073/pnas.95.14.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Yeh ETH. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- Kaplan KB, Hyman AA, Sorger PK. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell. 1997;91:491–500. doi: 10.1016/s0092-8674(00)80435-3. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu Rev Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Harper JW, Elledge SJ. How the cyclin became a cyclin: Regulated proteolysis in the cell cycle. Cell. 1999;97:431–434. doi: 10.1016/s0092-8674(00)80753-9. [DOI] [PubMed] [Google Scholar]
- Kominami K, Toda T. Fission yeast WD-repeat protein pop1 regulates genome ploidy through ubiquitin-proteasome-mediated degradation of the CDK inhibitor Rum1 and the S-phase initiator Cdc18. Genes & Dev. 1997;11:1548–1560. doi: 10.1101/gad.11.12.1548. [DOI] [PubMed] [Google Scholar]
- Kuriyama R, Borisy GG. Centriole cycle in chinese hamster ovary cells as determined by whole-mount electron microscopy. J Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes & Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;97:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A. Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von hippel-lindau tumor suppressor function. Proc Natl Acad Sci. 1999;96:5510–5515. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle W, Lutz WH, Ingle J, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: Implications for genomic stability and cell polarity. Proc Natl Acad Sci. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisztwan J, Marti A, Sutterlutly H, Gstaiger M, Wirbelauer C, Krek W. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45(SKP2): Evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina SA, Correll CC, Kipreos ET, Deshaies RJ. Human Cul1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with Skp1 and a F-box protein. Proc Natl Acad Sci. 1998;95:7451–7456. doi: 10.1073/pnas.95.13.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti A, Wirbelauer C, Scheffner M, Krek W. Interaction between ubiquitin-protein ligase SCFSkp2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol. 1999;1:14–19. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]
- Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: A gamma tubulin-associated protein with an essential role in gamma tubulin ring complex (gammaTuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias N, Johnson SL, Winey M, Adams AE, Goetsch L, Pringle JB, Byers B, Goebl MG. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S- phase transition and identifies a conserved family of proteins. Mol Cell Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- McDonald HB, Byers B. A proteasome cap subunit required for spindle pole body duplication in yeast. J Cell Biol. 1997;137:539–553. doi: 10.1083/jcb.137.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatics cells requires E2F and Cdk2-Cyclin A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Isolation of mammalian centrosomes. Methods Enzymol. 1986;134:261–268. doi: 10.1016/0076-6879(86)34094-1. [DOI] [PubMed] [Google Scholar]
- Montagnoli A, Fiore F, Eytan E, Carrano AC, Draetta GF, Hershko A, Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes & Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Fung JC, Sedat JW, Alberts BM, Agard DA. Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J Cell Biol. 1995a;130:1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995b;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Moritz M, Zheng Y, Alberts BM, Oegema K. Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian γ-tubulin complex contains homologues of the yeast spindle pole body components Spc97p and Spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Cell Cycle Extracts. In: Kay BK, Peng HB, editors. Xenopus laevis: Practical uses in cell and molecular biology. San Diego, CA: Academic Press; 1991. pp. 581–605. [Google Scholar]
- Oegema K, Whitfield WG, Alberts B. The cell cycle-dependent localization of the CP190 centrosomal protein is determined by the coordinate action of two separable domains. J Cell Biol. 1995;131:1261–1273. doi: 10.1083/jcb.131.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Marshall WF, Sedat JW, Alberts BM. Two proteins that cycle asynchronously between centrosomes and nuclear structures: Drosophila CP60 and CP190. J Cell Sci. 1997;110:1573–1583. doi: 10.1242/jcs.110.14.1573. [DOI] [PubMed] [Google Scholar]
- Osaka F, Kawasaki H, Aida N, Saeki M, Chiba T, Kawashima S, Tanaka K, Kato S. A new NEDD8-ligating system for cullin-4A. Genes & Dev. 1998;12:2263–2268. doi: 10.1101/gad.12.15.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: Their sensitivity to divalent cations. J Struct Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: Don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- Peters JM. SCF and APC: The Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- Pihan GA, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey SJ. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- Schnackenberg BJ, Khodjakov A, Rieder CL, Palazzo RE. The disassembly and reassembly of functional centrosomes in vitro. Proc Natl Acad Sci. 1998;95:9295–9300. doi: 10.1073/pnas.95.16.9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Koepp DM, Kamura T, Conrad RC, Conaway RC, Conaway JW, Elledge SJ, Harper JW. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–665. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- Sluder G, Miller FJ, Cole R, Rieder CL. Protein synthesis and the cell cycle: Centrosome reproduction in sea urchin eggs is not under translational control. J Biol Chem. 1990;110:2025–2032. doi: 10.1083/jcb.110.6.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. γ-tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: The central role of gamma-tubulin [see Comments] Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Stemmann O, Lechner J. The Saccharomyces cerevisiae kinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO J. 1996;15:3611–3620. [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Strausfeld UP, Howell M, Rempel R, Maller JL, Hunt T, Blow JJ. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr Biol. 1994;4:876–883. doi: 10.1016/s0960-9822(00)00196-2. [DOI] [PubMed] [Google Scholar]
- Su JY, Rempel RE, Erickson E, Maller JL. Cloning and characterization of the Xenopus cyclin-dependent kinase inhibitor p27XIC1. Proc Natl Acad Sci. 1995;92:10187–10191. doi: 10.1073/pnas.92.22.10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Celati C, Moudjou M, Bornens M. Characterization of the human homologue of the yeast spc98p and its association with gamma-tubulin. J Cell Biol. 1998;141:689–701. doi: 10.1083/jcb.141.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov LM, Yeh K-H, Lee S-J, Sun H, Zhang H. p27Kip1 ubiquitination and degradation is regulated by the SCFSkp2 complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Wada H, Yeh ET, Kamitani T. Identification of NEDD8-conjugation site in human cullin-2. Biochem Biophys Res Commun. 1999;257:100–105. doi: 10.1006/bbrc.1999.0339. [DOI] [PubMed] [Google Scholar]
- Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, Thomas PJ. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Lanker S, Patton EE, Craig KL, Nason TF, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- Winey M, Byers B. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- Winey M, Byers B. Spindle Pole Body of Saccharomyces cerevisiae: A model for genetic analysis of the centrosome cycle. In: Kalnins VI, editor. The centrosome. San Diego, CA: Academic Press; 1992. pp. 197–218. [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: Novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DA, Jackson PK. Cell cycle: Oiling the gears of anaphase. Curr Biol. 1998;8:R636–R639. doi: 10.1016/s0960-9822(07)00410-1. [DOI] [PubMed] [Google Scholar]
- Wolf DA, McKeon F, Jackson PK. F-box/WD-repeat proteins pop1p and Sud1p/Pop2p form complexes that bind and direct the proteolysis of cdc18p. Curr Biol. 1999;9:373–376. doi: 10.1016/s0960-9822(99)80165-1. [DOI] [PubMed] [Google Scholar]
- Yew PR, Kirschner MW. Proteolysis and DNA replication: The CDC34 requirement in the Xenopus egg cell cycle. Science. 1997;277:1672–1676. doi: 10.1126/science.277.5332.1672. [DOI] [PubMed] [Google Scholar]
- Yu H, Peters JM, King RW, Page AM, Hieter P, Kirschner MW. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, Mann M, Nasmyth K. Mass spectrometric analysis of the anaphase-promoting complex from yeast: Identification of a subunit related to cullins. Science. 1998;279:1216–1219. doi: 10.1126/science.279.5354.1216. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Jung MK, Oakley BR. Gamma-tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- Zimmerman W, Sparks CA, Doxsey SJ. Amorphous not longer: The centrosome comes into focus. Curr Opin Cell Biol. 1999;11:122–128. doi: 10.1016/s0955-0674(99)80015-5. [DOI] [PubMed] [Google Scholar]