Abstract
Transcription initiation at RNA polymerase III promoters requires transcription factor IIIB (TFIIIB), an activity that binds to RNA polymerase III promoters, generally through protein–protein contacts with DNA binding factors, and directly recruits RNA polymerase III. Saccharomyces cerevisiae TFIIIB is a complex of three subunits, TBP, the TFIIB-related factor BRF, and the more loosely associated polypeptide β″. Although human homologs for two of the TFIIIB subunits, the TATA box–binding protein TBP and the TFIIB-related factor BRF, have been characterized, a human homolog of yeast B″ has not been described. Moreover, human BRF, unlike yeast BRF, is not universally required for RNA polymerase III transcription. In particular, it is not involved in transcription from the small nuclear RNA (snRNA)–type, TATA-containing, RNA polymerase III promoters. Here, we characterize two novel activities, a human homolog of yeast B″, which is required for transcription of both TATA-less and snRNA-type RNA polymerase III promoters, and a factor equally related to human BRF and TFIIB, designated BRFU, which is specifically required for transcription of snRNA-type RNA polymerase III promoters. Together, these results contribute to the definition of the basal RNA polymerase III transcription machinery and show that two types of TFIIIB activities, with specificities for different classes of RNA polymerase III promoters, have evolved in human cells.
Keywords: human TFIII, human B″, BRFU, U6 snRNA gene, RNA polymerase III
Correct initiation of transcription by RNA polymerase III requires a number of factors. Of particular interest is the transcription factor IIIB (TFIIIB), because TFIIIB directly contacts RNA polymerase III, and in yeast, once recruited to the promoter, TFIIIB is sufficient to support several rounds of RNA polymerase III transcription (reviewed in Paule and White 2000). Yeast TFIIIB is well defined and consists of three subunits, the TATA box–binding protein TBP (Kassavetis et al. 1992), the TFIIB-related factor BRF (TDS4/PCF4) (Buratowski and Zhou 1992; Colbert and Hahn 1992; López-De-León et al. 1992), and the B′′ protein (TFIIIB90/TFC5/TFC7) (Kassavetis et al. 1995; Roberts et al. 1996; Rüth et al. 1996). In various RNA polymerase III promoters, the mode of TFIIIB recruitment varies. In the gene-internal 5S and transfer RNA (tRNA) promoters, the promoter elements are first recognized by TFIIIA and TFIIIC or directly by TFIIIC, respectively, and this allows the subsequent recruitment of TFIIIB (reviewed in Paule and White 2000). On the yeast U6 promoter, which contains a TATA box ∼25 nucleotides upstream of the transcription start site, TFIIIB is also recruited in a TFIIIC-dependent manner (Brow and Guthrie 1990; Burnol et al. 1993; Eschenlauer et al. 1993), but on naked DNA templates in vitro, it can be recruited by direct binding of the TBP subunit to the TATA box (Moenne et al. 1990; Margottin et al. 1991). In all these cases, however, the same TFIIIB complex, containing the same three subunits, is used (Joazeiro et al. 1994).
In mammalian cells, the situation appears more complicated. Like yeast TFIIIB, mammalian TFIIIB contains TBP (Lobo et al. 1992; Taggart et al. 1992; White and Jackson 1992) and a homolog of yeast BRF, human BRF (hBRF) (Mital et al. 1996), also called TFIIIB90 (Wang and Roeder 1995). These two subunits associate strongly with each other (Wang and Roeder 1995; Mital et al. 1996). A homolog of yeast B′′ has not been identified in mammals or any other organism. In addition, unlike in yeast, there appear to be variants of the TFIIIB complex. In particular, the human U6 promoter, which exemplifies a class of RNA polymerase III promoters whose essential elements are all located within the 5′ flanking sequence of the gene, appears to use another form of TFIIIB than the tRNA-type Ad2 VAI promoter (Lobo et al. 1992; Teichmann and Seifart 1995; Mital et al. 1996; Henry et al. 1998a), which contains a gene-internal promoter. The core U6 promoter, which is sufficient to direct basal levels of transcription in vitro, consists of a proximal sequence element (PSE) centered around position −56 relative to the start site and a TATA box centered around position −27. The U6 promoter is highly similar to the RNA polymerase II small nuclear RNA (snRNA) core promoters, which consist of only the PSE (see Henry et al. 1998a and references therein). The PSE recruits a multisubunit complex called SNAPc (Henry et al. 1995, 1998b) or PTF (Yoon et al. 1995), and the U6 TATA box recruits TBP (Lobo et al. 1991; Simmen et al. 1991). It is not clear which other TFIIIB components aside from TBP are involved in U6 transcription.
Wang and Roeder (1995) showed that depletion of extracts with anti-hBRF antibodies debilitated U6 transcription, but addition of recombinant hBRF did not restore activity. They concluded that hBRF was required for U6 transcription but as part of a complex that somehow differed from the TFIIIB complex required for VAI transcription. On the other hand, we showed that depletion of extracts with antibodies directed against the C-terminal half of hBRF debilitated transcription from the VAI promoter but not from the U6 promoter. Because the antibody treatment removed >95% of the endogenous hBRF present in extracts, the results suggest that hBRF is in fact not required for U6 transcription (Mital et al. 1996). Indeed, we were able also show that upon depletion of extracts with anti-TBP and anti-hBRF antibodies, VAI transcription was only restored by addition of a combination of both TBP and hBRF, whereas U6 transcription could be restored by addition of only recombinant TBP. In fact, U6 transcription was diminished by addition of hBRF (Henry et al. 1998a).
Thus, two main questions remain concerning mammalian TFIIIB. First, it is currently unclear whether a mammalian homolog of yeast B′′ exists, and whether it is required for RNA polymerase III transcription of genes with internal promoters, such as the VAI gene, and genes with external promoters that recruit SNAPc, such as the human U6 gene. Second, the apparent lack of requirement for hBRF in U6 transcription raises the possibility that the mammalian snRNA-type RNA polymerase III promoters use a factor different from, but related to, hBRF. Here, we report the characterization of two new transcription factors, human B′′ (hB′′) and a BRF-related factor we refer to as hBRFU. hB′′ is required for transcription of both the VAI and the U6 genes. hBRFU is a protein with an N-terminal domain related to both hBRF and TFIIB, and a divergent C-terminal domain. The protein is required for U6 but not VAI transcription. These results show that there are two forms of the basal RNA polymerase III transcription factor IIIB in mammalian cells. They also identify the first transcription factors uniquely required for transcription of RNA polymerase III but not RNA polymerase II—snRNA promoters. Together with our previous results indicating that TFIIB is required for transcription of the human RNA polymerase II snRNA genes (Kuhlman et al. 1999), these results suggest that the key event in the determination of RNA polymerase specificity in the human snRNA promoters is the recruitment of hBRFU versus TFIIB.
Results
Characterization of a human homolog of S. cerevisiaeB′′
Both the TBP and BRF subunits of yeast TFIIIB have human homologs, suggesting that a human homolog of yeast B′′ exists. S. cerevisiae B′′ contains a SANT domain. The SANT domain is related to a Myb repeat and was originally identified in a number of proteins, including the SWI3, ADA2, N-Cor, and yeast TFIIIB B′′ proteins (Aasland et al. 1996). In yeast B′′, C-terminal deletions of B′′ that lack most of the SANT domain are inactive for in vitro transcription of a TATA-less tRNA gene, although they are still active for transcription of the TATA-containing U6 snRNA gene (Kumar et al. 1997). We used the S. cerevisiae sequence as a query to search the human and mouse expressed sequence tag (EST) databases and originally identified a short mouse sequence (GenBank accession number AA200560) encoding 21 amino acids with strong similarity to part of the yeast B″ SANT domain. We used this short sequence to design primers, and through successive polymerase chain reactions (PCR) with various libraries and cDNA prepared from HeLa-cell total RNA as well as through database searches, we assembled a sequence encoding a protein with a calculated molecular mass of 156.129 kD, shown in Figure 1A. We also obtained two variations in this sequence. In the first, nucleotides 414–578 (see GenBank accession number AF298151 for the nucleotide numbering), encoding amino acids 109–163, are missing (indicated by vertical brackets in Fig. 1A). Because the nucleotide sequence extending from nucleotides 414 to 578 starts with a GT and ends with an AG, the shorter sequence most probably corresponds to a splicing variant. In the second variation, an additional T residue is inserted at position 4192 (GenBank accession number AF298152). As a result, the last 21 amino acids of the protein in Figure 1A (SEEINNSMIILSLSPTTLKNL) are replaced by the shorter sequence FRGNK. The two sequences may correspond to different alleles. As detailed in Materials and Methods, several recent entries in the database can be aligned with part or most of the sequence shown in Figure 1A.
Figure 1.
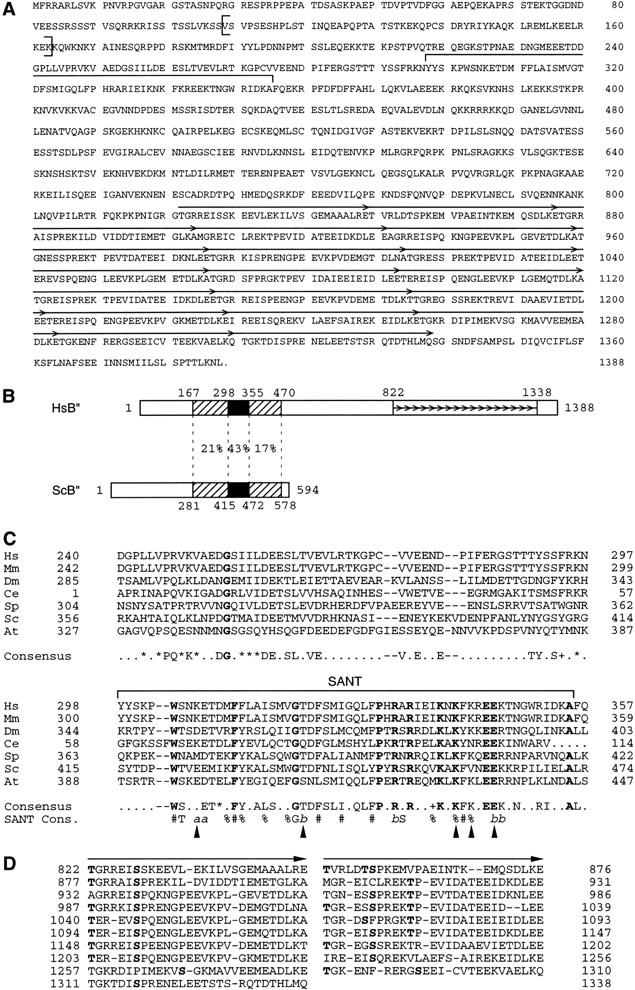
Structure of a hB′′. (A) Predicted hB′′ protein sequence (GenBank accession number AF298151). The SANT domain is indicated by a large horizontal bracket above the sequence. The repeated sequences are indicated by arrows. The sequence between the vertical brackets (amino acids 109–163) is lacking in some cDNAs that probably correspond to splicing variants. (B) Regions of similarity in the Homo sapiens and S. cerevisiae B′′ sequences (HsB′′ and ScB′′, respectively). The black boxes correspond to the SANT domain; the hatched boxes indicate regions of lower but still significant similarity on either side of the SANT domain. The percentages indicate identities between the two proteins in the regions delimited by the dotted lines. The numbers above the boxes indicate aa numbering. The small arrows indicate the repeats in hB′′. (C) Alignment of the SANT domain and region immediately upstream from hB′′ (Hs) and ScB′′ (Sc), as well as putative mouse (Mm), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Schizosaccharomyces pombe (Sp), and Arabidopsis thaliana (At) B′′ homologs. The SANT domain is indicated by the horizontal bracket above the sequence. The region N-terminal of the SANT domain was shown, in ScB′′, to be required for efficient transcription of linear yeast U6 templates in vitro (Kassavetis et al. 1998). The amino acids in boldface are identical in all sequences. The amino acids indicated in the consensus are identical in at least four of the seven sequences. (+) similar amino acids in all sequences; (*) similar amino acids in at least five of the seven sequences. The following amino acids were considered similar: V, I, L, and M; D and E; N and Q; R, K, and H; S and T; and W, F, and Y. The SANT consensus sequence (Aasland et al. 1996) is also shown. In the SANT consensus: (%) semiconserved hydrophobicity; (#) strongly conserved hydrophobicity; (a) conserved acidic residues; and (b) conserved basic residues. The positions marked with arrowheads indicate positions not conserved in the B′′ and SANT consensus sequences. The GenBank accession numbers are as follows: Mm ESTs, AI315861, AI315862, and AI787462; Ce genomic sequences U97016 (B0261) (nucleotides 25059–25185 and 25518–25780); Dm protein, AAF53291.1 (CG9305 gene product); Sp protein, T41239; and At protein T08564 (hypothetical protein T22F8.60). The numberings correspond to the sequence shown in (A) for HsB′′, to the sequences published (Kassavetis et al. 1995; Roberts et al. 1996; Rüth et al. 1996) for ScB′′, and to the sequences corresponding to the protein accession numbers indicated for other sequences. For the Mm sequence, the numbering corresponds to a putative mouse B′′ N-terminal region (amino acids 1–377) assembled from an alignment of various ESTs (GenBank accession numbers AI527964, AI317698, AI787358, AI526613, AI316225, AI639984, AA168265, AW456745, AW049390, AI429656, AI430369, AI450927, W91010, AI787462, AI315861, AI315862, AI892036, and AI315863), and for the Ce sequence, the numbering is arbitrary. (D) Alignment of the repeats within the C-terminal half of hB′′. As indicated by the arrows, each repeat can itself be subdivided into two short imperfect repeats. Residues in boldface indicate potential phosphorylation sites for various kinases, including PKC, PKA, CAMII, and CKII kinases.
When the protein sequence shown in Figure 1A is used as the query to search the Swiss-Prot database, the best match identified is the S. cerevisiae B′′ protein. As detailed below, several other pieces of evidence suggest that this protein is indeed a functional human homolog of yeast B′′, and we therefore refer to it as hB′′.
Figure 1B shows the regions of similarity with yeast B′′ (ScB′′). hB′′ contains a region that is 43% identical with the yeast B′′ SANT domain (black box). In addition, hB′′ shows 21% identity over a 131–amino acid (aa) region upstream, and 17% identity over a 115-aa region downstream, of the SANT domain (hatched boxes).
Figure 1C shows an alignment including a 58-aa region directly upstream of the SANT domain and the SANT domain itself of hB′′, ScB′′, as well as putative B′′ homologs from various organisms (see the legend to Fig. 1C). In all of these sequences, the SANT domain is remarkably conserved, and the mouse and human sequences are identical over this region. The resulting consensus for B′′ SANT domains differs from the SANT domain consensus (Aasland et al. 1996) at several positions, marked by arrowheads in Figure 1C. In addition, the B′′ SANT sequences show absolute conservation of eleven amino acids (indicated in boldface), most of which are not absolutely conserved in all SANT domains. Thus, B′′ SANT domains form a subfamily of SANT domains. The 58-aa region upstream of the SANT domain is also shown, because in ScB′′, small deletions in this region severely debilitate transcription from linear (but not supercoiled) templates in vitro (Kassavetis et al. 1998). Although this region is less conserved, it shows clusters of conserved amino acids, especially around the invariant glycine. Thus, this region may perform a similar function in all B′′ homologs.
Starting at amino acid 822, human B′′ consists of a series of nine long repeats of 53–55 amino acids, each of which can be further divided into two shorter but more degenerate repeats of 26–28 amino acids, followed by one additional short repeat. In a BLAST search of the Swiss-Prot database, regions of hB′′ comprising the repeats and extending as far as upstream of the SANT domain align with proteins belonging to the myosin and intermediate filament families and characterized by long, α-helical domains that can form coiled-coil structures. However, programs that predict coiled-coil regions such as COIL (Lupas et al. 1991) or PAIRCOIL (Berger et al. 1995), and that give very high scores to the proteins that align with hB′′, do not predict similar extended rod-like structures in hB′′, although hB′′ does show short regions with high scores. Thus, the significance of these similarities is not clear. Each of the 19 short repeats contains serine and threonine residues (indicated in boldface in Fig. 1D) that are potential phosphorylation sites for various kinases, including PKC, PKA, CAMII, CDC2, and CKII kinases, suggesting that this part of the molecule may have a regulatory function. This very striking repeated sequence is not present in the yeast protein.
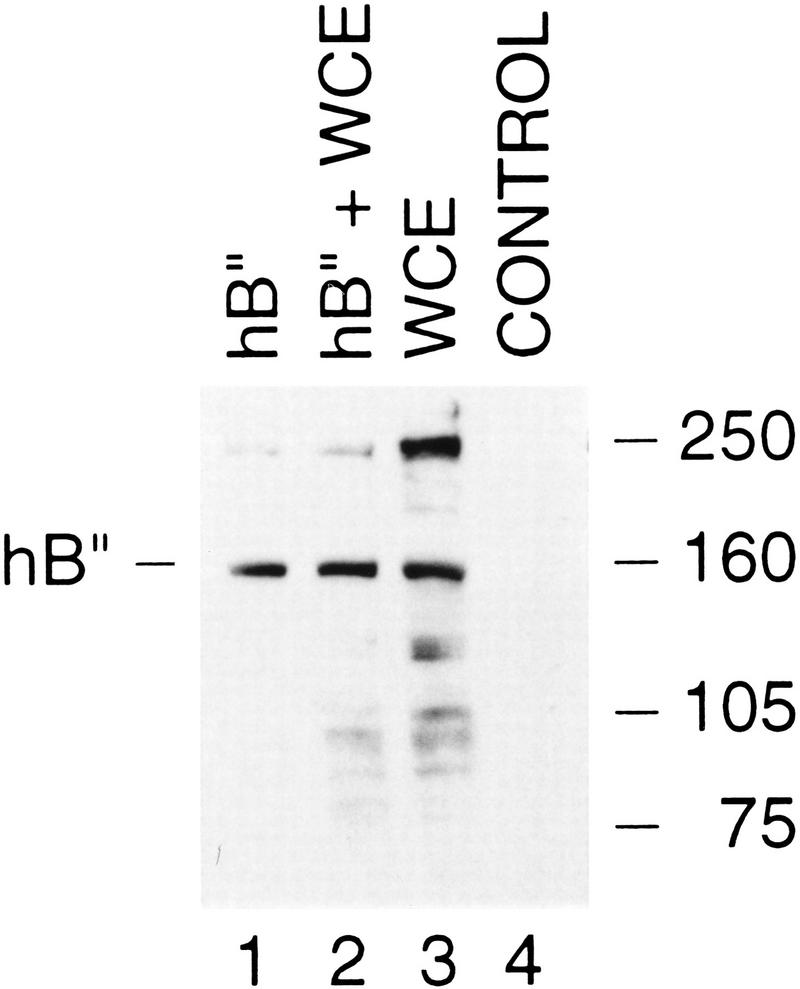
To determine whether the protein shown in Figure 1A corresponds to an endogenous protein, we raised antibodies against several peptides derived from the amino acid sequence shown in Figure 1A (see Materials and Methods). We then performed an immunoblot of the following: hB′′ translated in vitro in a rabbit reticulocyte lysates; whole-cell extract (WCE); a mixture of in vitro–translated hB′′ and whole-cell extract; and unprogrammed reticulocyte lysate (control). As shown in Figure 2, the anti-hB′′ antibody recognized a prominent polypeptide of ∼160 kD in HeLa-cell extracts (lane 3), which comigrated with in vitro–translated hB′′ (lanes 1 and 2). Another polypeptide of ∼250 kD that is prominently detected in the HeLa-cell extract (lane 3) may correspond to a cross-reacting protein or, perhaps, a hyperphosphorylated form of hB′′. No major signal was obtained in the unprogrammed lysate control lane (lane 4). Thus, cells contain a protein that is recognized by anti-hB′′ antibodies and that comigrates with recombinant hB′′ (rhB′′).
Figure 2.
rhB′′ comigrates with endogenous B′′ from HeLa cells. The immunoblot was performed with an anti-hB′′ antibody (αhB′′-2, CS913). (Lane 1) 5 μL of hB′′ translated in vitro; (lane 2) 5 μL of hB′′ translated in vitro and 2.5 μL of whole HeLa-cell extract (WCE); (lane 3) 2.5 μL of HeLa-cell extract; and (lane 4) 5 μL of unprogrammed rabbit reticulocyte lysates (control). The position of hB′′ and the positions of molecular weight markers are indicated.
hB′′ is required for transcription from both the VAI and the U6 promoters
To determine whether hB′′ plays any role in transcription by RNA polymerase III, we covalently attached anti-hB′′ antibodies to beads and used these beads to deplete extracts of endogenous hB′′. The extracts were then tested for their ability to support transcription from four types of promoters: the Ad2 VAI gene–internal RNA polymerase III promoter, the RNA polymerase III U6 and RNA polymerase II U1 snRNA promoters, and the Ad2 major late (Ad2 ML) promoter, a typical mRNA-type RNA polymerase II promoter. The results are shown in Figure 3. In all cases, treatment of the extracts with preimmune antibody beads had little effect on transcription (cf. lane 10 with lanes 1 and 5). In contrast, treatment of the extracts with anti-B′′ antibodies directed against two different peptides (antibodies 913 and 843) severely debilitated transcription from both the VAI and U6 RNA polymerase III promoters but had little effect on transcription from the RNA polymerase II U1 and Ad2 ML promoters (cf. lane 2 with lane 1, and lane 6 with lane 5). The effect was specific, because incubation of each antibody with the peptide against which it was raised, but not with a nonspecific peptide, abolished the inhibitory effect (lanes 3, 4, 7, and 8). As a control, we also treated the extracts with an antibody directed against the SNAP43 subunit of SNAPc (Henry et al. 1995). We have shown before that SNAPc is specifically required for transcription from both RNA polymerase II and III snRNA promoters (Sadowski et al. 1993; Henry et al. 1998b). As expected, treatment with the anti-SNAP43 antibody debilitated RNA polymerase III transcription from the U6 snRNA but not the Ad2 VAI promoter, and RNA polymerase II transcription from the U1 snRNA but not the Ad2 ML promoter (lane 9). Note that in the U1 panel, the lower band (U1) corresponds to correctly initiated U1 RNA; the upper band (5' RT) corresponds to a read-through RNA initiating at cryptic promoters within vector sequence (Sadowski et al. 1993).
Figure 3.
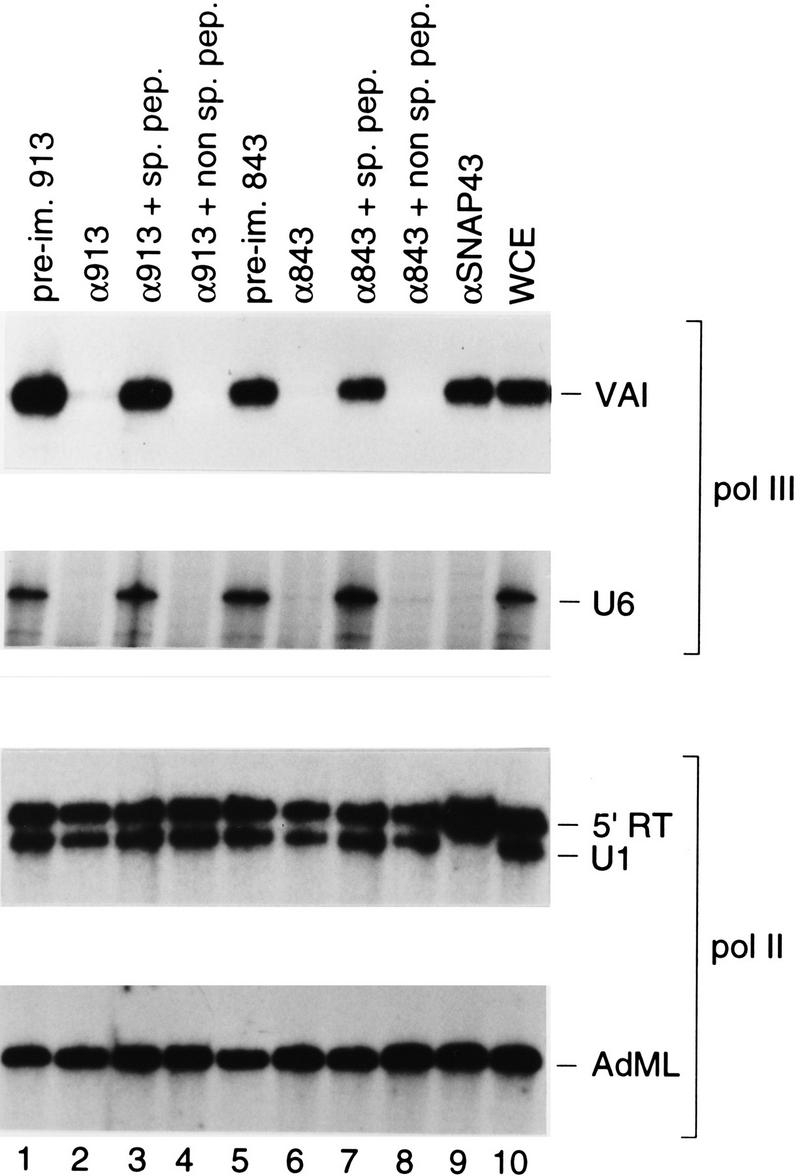
Depletion of hB′′ debilitates RNA polymerase III, but not RNA polymerase II, transcription in vitro. Whole-cell extract was treated with beads coated with either the preimmune antibodies (lanes 1 and 5) or the anti-B′′ antibodies (lanes 2–4 and 6–8) indicated above the lanes (913 is αhB′′-2; 843 is αhB′′-3). In lanes 3 and 7, and in lanes 4 and 8, the antibody beads were preincubated either with the specific peptide against which the antibodies were raised or with nonspecific peptide, as indicated. In lane 9, the extract was treated with beads coated with an antibody directed against the SNAP43 subunit of SNAPc. Aliquots of the variously treated extracts were then tested in parallel for their ability to direct transcription from the Ad2 VAI, human U6, human U1, and Ad2 major late (AdML) promoters. The bands corresponding to correctly initiated RNA are indicated in each panel. In the U1 panel, the band labeled 5′ RT corresponds to RNA initiated within the vector and reading through the U1 snRNA promoter (Sadowski et al. 1993). Lane 10 shows transcription in untreated whole-cell extract.
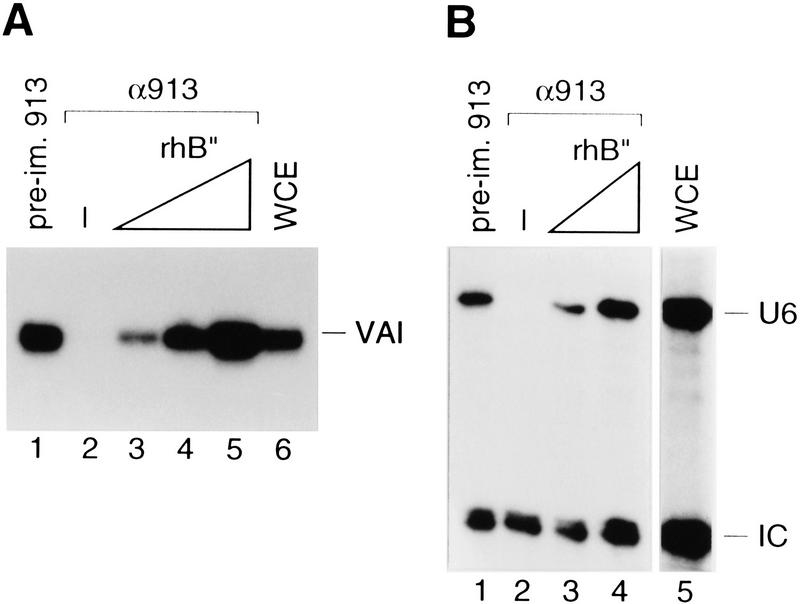
To ensure that the debilitating effect on VAI and U6 transcription observed above upon treatment of transcription extracts with anti-hB′′ antibodies resulted from depletion of hB′′, we tested whether rhB′′ could reconstitute transcription in the depleted extracts. As shown in Figure 4, addition of increasing amounts of recombinant hB′′ (rhB′′) produced in Escherichia coli to depleted extracts restored both VAI (Fig. 4A) and U6 (Fig. 4B) transcription to levels higher than, or comparable to, those observed in the starting extract (cf. lanes 3–5 with lanes 1, 2, and 6 in Fig. 4A, and lanes 3 and 4 with lanes 1, 2, and 5 in Fig. 4B; note that in Fig. 4B, lane 5 was overloaded as evidenced by the intensity of the internal control signal [IC]). These results indicate that hB′′ is an RNA polymerase III transcription factor that, unlike hBRF (Mital et al. 1996), is required for transcription from both TATA-less and TATA-containing promoters.
Figure 4.
hB′′ is required for RNA polymerase III transcription from the Ad2 VAI and human U6 promoters. (A) Whole-cell extract was treated with beads coated with either preimmune (lane 1) or anti-hB′′ (lanes 2–5) antibodies and tested for transcription from the Ad2 VAI promoter. In lanes 3–5, increasing amounts of rhB′′ (rhB′′) expressed in E. coli were added to the depleted extract. Lane 6 shows VAI transcription in untreated whole-cell extract. (B) Whole-cell extract treated with beads coated with the preimmune (lane 1) or anti-hB′′ (lanes 2–4) antibodies and tested for U6 transcription. In lanes 3 and 4, increasing amounts of rhB′′ expressed in E. coli were added to the depleted extract. Lane 5 shows U6 transcription in untreated whole-cell extract. (IC) internal control signal.
hB′′ is found on the U6 promoter region in vivo
These experiments strongly suggest that hB′′ functions in transcription of the human U6 gene. To determine whether hB′′ can be found in the human U6 promoter region in vivo, we used the chromatin immunoprecipitation assay, in which intact cells are first treated with formaldehyde to cross-link proteins to DNA and then chromatin is extracted, extensively sonicated, and used as starting material for immunoprecipitations, with antibodies directed against the protein of interest (see Boyd et al. 1998; Kuo and Allis 1999; and references therein). The presence of specific DNA sequences in the immunoprecipitates can then be ascertained by PCR. We performed immunoprecipitations of cross-linked chromatin with anti-hB′′ and, as a control, preimmune antibodies and anti-TFIIB antibodies (Kuhlman et al. 1999). We then checked the immunoprecipitates for the presence of U6 and, as a control, U1, promoter sequences.
The organization of the human U6 genes is not well understood, but U6 genes and pseudogenes appear dispersed in the genome. A BLAST search with the U6 gene we work with (Kunkel et al. 1986) indicated that the promoter region is probably unique; we therefore designed a pair of test primers within the U6 promoter. The control primers hybridized to a region 4 kb upstream of the U6 transcription start site. The U1 snRNA genes number an estimated 30–100 copies per haploid genome, each carried on a tandemly repeated 45-kb fragment (Bernstein et al. 1985). In addition, the human genome contains a large number of U1 pseudogenes. Most have different flanking sequences than the true genes, but the U1 class I pseudogenes have 5′ and 3′ flanking regions very similar to those of the true genes (Denison and Weiner 1982). We designed test primers capable of hybridizing to the true U1 promoters but not to flanking regions of U1 pseudogenes. The control primers hybridized to a region within the 45-kb repeat (but not in repeated sequences such as Alu sequences) at 7 kb upstream of the U1 start site, to ensure that any difference in signal between test and control primers did not result from a difference in copy number of the target DNA sequence. For both the U6 and the U1 control primers, the size of the expected fragment is close to that expected from the test primers, to ensure comparable PCR efficiency.
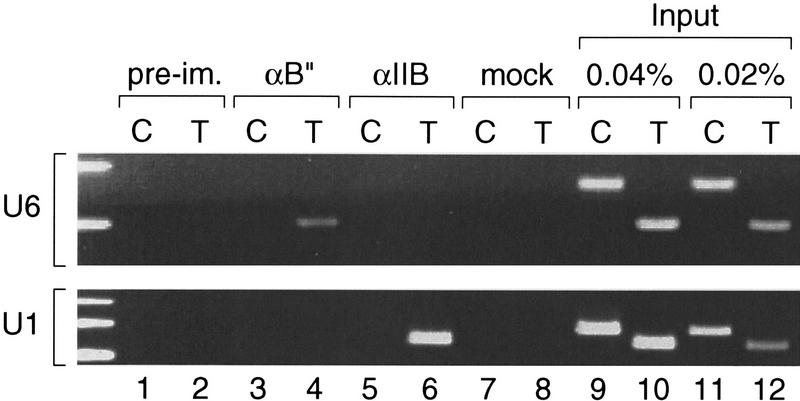
As shown in Figure 5, both the test and control primers gave the expected fragments when used to amplify the starting material for immunoprecipitation (lanes 9–12). As expected, no fragments were obtained with material precipitated by preimmune antibodies or in the absence of antibodies (lanes 1, 2, 7, and 8). However, the material immunoprecipitated by anti-B′′ antibodies contained U6 promoter sequences but not sequences located 4 kb upstream of the U6 transcription start site. U1 promoter sequences were not present above the background obtained with the test primers in this immunoprecipitation. In sharp contrast, U1, but not U6, promoter sequences were present at levels well above the background in material immunoprecipitated with an anti-TFIIB antibody, consistent with our previous observations that TFIIB is required for RNA polymerase II but not RNA polymerase III transcription from snRNA promoters (Kuhlman et al. 1999). Together, these data strongly argue that hB′′ is indeed part of the U6 initiation complex.
Figure 5.
hB′′ is found in the U6 promoter region in vivo. Rapidly growing HeLa cells were treated with formaldehyde; then cross-linked chromatin was extracted, sonicated, and used as starting material for immunoprecipitations with beads coated with either preimmune (lanes 1 and 2), anti-hB′′ (lanes 3 and 4), or anti-TFIIB (lanes 5 and 6) antibodies, or just beads alone (lanes 7 and 8). The immunoprecipitated material was then analyzed by PCR with test (T) primers specific for the U6 (upper panel) or U1 (lower panel) promoters, or control (C) primers hybridizing to a unique region located 4 kb upstream of the human U6 snRNA gene (upper panel) or a unique region within the 45-kb U1 repeat (Bernstein et al. 1985) 7 kb upstream of the U1 gene (see Materials and Methods for details). Lanes 9–12 show PCRs performed with the test (T) or control (C) primers with 0.04% and 0.02% of the input material used for immunoprecipitation, as indicated above the lanes.
Isolation of a cDNA encoding a factor related to both hBRF and TFIIB
We recently generated antibodies directed against various peptides within the TFIIB-related N-terminal domain of hBRF and observed that depletion of extracts with one of these antibodies (1043), unlike depletion with all other anti-hBRF antibodies we have tested, debilitated U6 transcription (see below). This result was similar to the results obtained by Wang and Roeder (1995) and suggested that a protein different from, but related to hBRF was required for U6 transcription. We therefore, searched the database for human proteins with similarity to the N-terminal domain of hBRF. The highest scoring hit after TFIIB was an unnamed protein with a calculated molecular mass of 46.42 kD (GenBank accession number BAA91975). We used the corresponding DNA sequence to design primers and amplify by PCR a sequence from total HeLa-cell cDNA, which can code for the protein shown in Figure 6A (GenBank accession number AF298153). This protein is identical to that encoded by the BAA91975 entry except that a cysteine at position 157 in BAA91975 is replaced by a tyrosine in our sequence.
Figure 6.
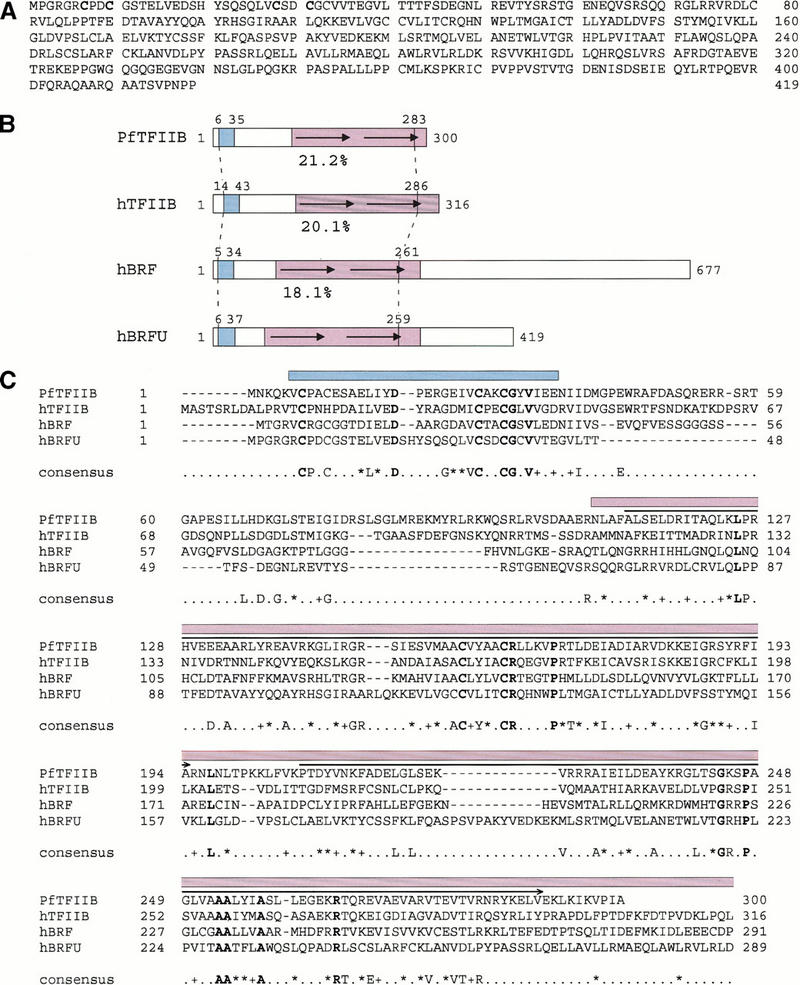
Structure of hBRFU. (A) Predicted hBRFU protein sequence (GenBank accession number AF298153). (B) Regions of similarities in the Pyrococcus furiosus TFIIB homolog (PfTFIIB) and the hTFIIB, hBRF, and hBRFU sequences. The location of the structured zinc ribbon as determined in PfTFIIB by nuclear magnetic resonance (Zhu et al. 1996), and as modeled in BRF and TFIIB (Hahn and Roberts 2000), and the location of the corresponding region in hBRFU are indicated in blue. The location of the structured core domain of TFIIB (Bagby et al. 1995; Nikolov et al. 1995) and that of the corresponding regions in the other proteins are indicated in purple. The region conserved between the proteins is indicated by the vertical stripped lines. The percentages below the sequences indicate percentage of identity between hBRFU and PfTFIIB, hTFIIB, or hBRF within the region bracketed by the vertical stripped lines as calculated from the alignment shown in C. (C) Alignment of PfTFIIB, hTFIIB, and the N-terminal regions of hBRF and hBRFU. The alignment was performed with the ClustalW program and the default parameters. The amino acids in boldface are identical in all sequences. The amino acids indicated in the consensus are identical in three of the four sequences. (+) similar amino acids in all sequences; (*) similar amino acids in three of the four sequences. The following amino acids were considered similar: V, I, L, and M; D and E; N and Q; R, K, and H; S and T; and W, F, and Y. The horizontal arrows above the sequences indicate the location of the structural repeats (Nikolov et al. 1995). The location of the structured zinc ribbon is indicated in blue, and that of the core (hTFIIB amino acids 113–316) in purple.
Figure 6B shows a schematic comparison of this protein, which we call hBRFU, with the TFIIB homolog from Archaea Pyrococus furiosus (PfTFIIB), human TFIIB (hTFIIB), and hBRF; Figure 6C shows the corresponding alignment. hBRFU is strikingly similar to all three proteins. In the region bracketed by a dotted line in Figure 6B, it is 21.2%, 20.1%, and 18.1% identical with PfTFIIB, hTFIIB, and hBRF, respectively. This region of conservation contains two structured domains, a zinc ribbon (indicated in blue), whose structure was solved by nuclear magnetic resonance (NMR) in PfTFIIB (Zhu et al. 1996) and modeled in yeast TFIIB and BRF (Hahn and Roberts 2000), and a core (indicated in purple) whose structure was solved in human TFIIB by NMR and crystallography (Bagby et al. 1995; Nikolov et al. 1995). Strikingly, the regions in which the hBRFU sequence is most similar to the three other proteins are precisely within these structured regions. Within the zinc ribbon domain, hBRFU is 40.6%, 37.5%, and 31.2% identical with PfTFIIB, hTFIIB, and hBRF, respectively, strongly suggesting that this region of BRFU also adopts a zinc ribbon structure. Within the core domain up to amino acid 259 in hBRFU (the C-terminal end of the core is not conserved), the percentage of identities varies between 18.7% and 19.8%. The hTFIIB core domain consists of two structural repeats (indicated by arrows in Fig. 6B and by arrows above the sequence in Fig. 6C) (Nikolov et al. 1995), which correspond to repeats in the sequence. In hBRF, the sequence repeats are less similar to each other than are those in TFIIB, and in BRFU, the second repeat contains an insertion relative to the first repeat and the repeats are altogether barely recognizable. Nevertheless, the strong similarities in the sequence suggest that the core of BRFU adopts a structure similar to the core of hTFIIB.
To determine whether cells indeed contain a protein corresponding to hBRFU, we fractionated on an SDS–polyacrylamide gel purified recombinant hBRFU (rhBRFU) expressed in E. coli, a P11 B fraction from HeLa-cell extracts, and a mixture of both. The proteins were transferred to nitrocellulose and probed with the antibody capable of debilitating U6 transcription in vitro (antibody 1043). As shown in Figure 7, the antibody recognized rhBRFU as well as a comigrating HeLa-cell protein. Thus, the protein we identified in the database is indeed recognized by the 1043 antibody and is probably expressed in HeLa cells.
Figure 7.
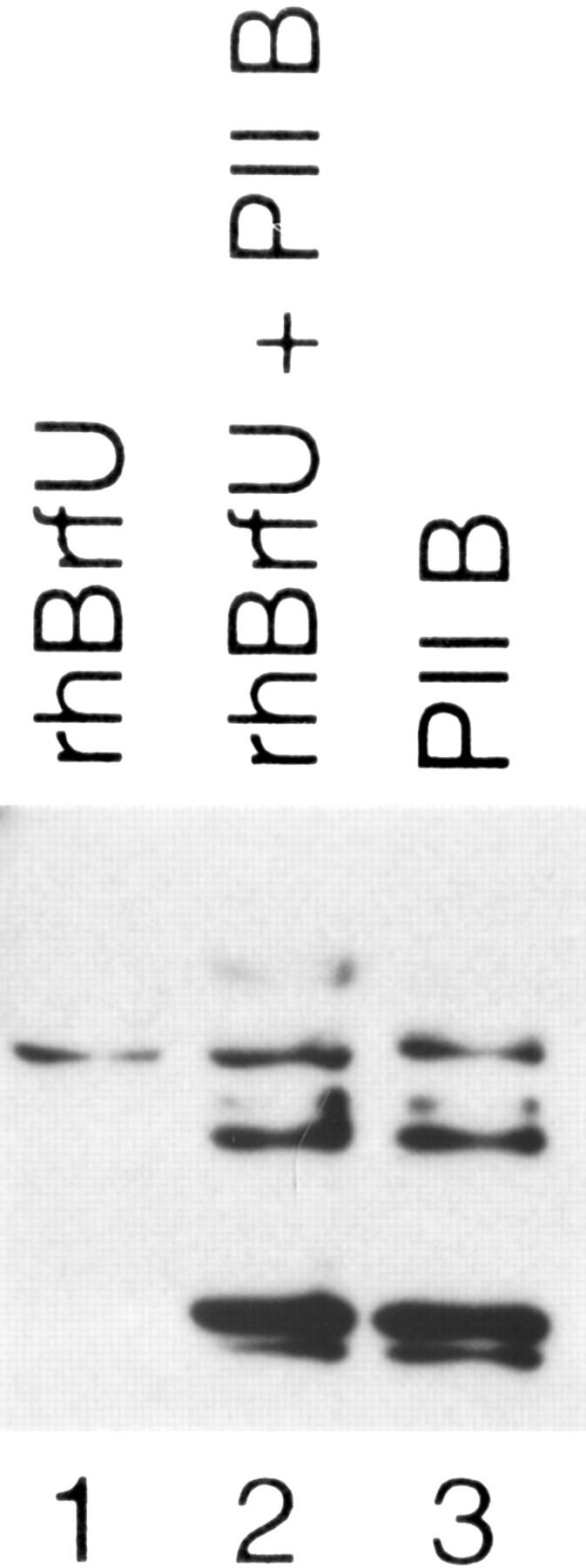
hBRFU is expressed in HeLa cells. The immunoblot was performed with an anti-hBRFU/hBRF antibody (CS1043). (Lane 1) rhBRFU expressed in E. coli; (lane 2) rhBRFU expressed in E. coli and 2 μL of P11 B fraction; and (lane 3) 2 μL of P11 B fraction. rhBRFU migrates as a protein of ∼50 kD. The additional bands seen in the P11 fraction probably correspond to cross-reacting proteins.
hBRFU functions for U6 but not VAI transcription in vitro
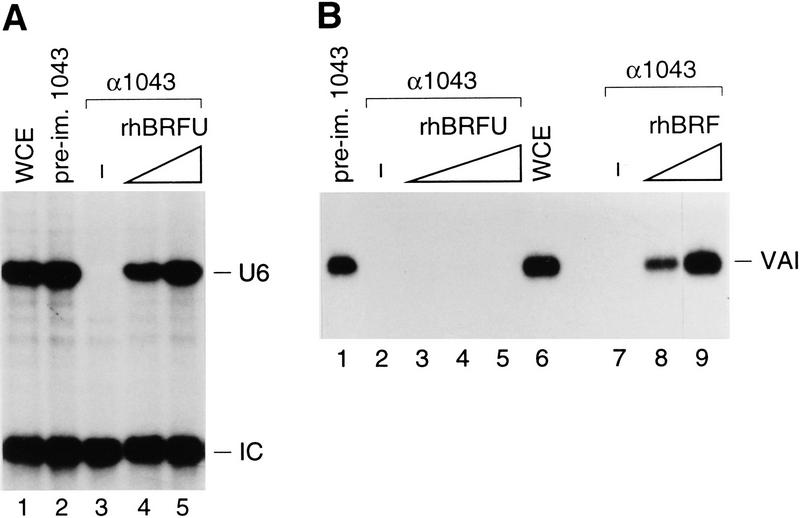
To determine whether hBRFU might be involved in RNA polymerase III transcription from the human U6 promoter, we treated extracts with the 1043 antibodies directed against the N-terminal domain of hBRF. As shown in Figure 8, this treatment resulted in debilitation of transcription from both the U6 (Fig. 8A) and the VAI (Fig. 8B) promoters (cf. lanes 2 and 3 in Fig. 8A, and lanes 1 and 2 in Fig. 8B). We then complemented the extracts with rhBRFU expressed in E. coli. Strikingly, transcription from the U6 promoter, but not the VAI promoter, was restored to levels comparable to those obtained in extracts depleted with preimmune antibody beads or untreated extract (cf. lanes 4 and 5 with lanes 1 and 2 in Fig. 8A and lanes 3–5 with lanes 1 and 6 in Fig. 8B). VAI transcription could be reconstituted by addition of hBRF (lanes 8 and 9 in Fig. 8B) but U6 transcription could not be (not shown), which is consistent with our previous observations (Mital et al. 1996; Henry et al. 1998a). Thus, human cells contain at least two homologs of yeast BRF, each specific for different types of RNA polymerase III promoters.
Figure 8.
hBRFU is specifically required for U6 transcription. (A) A whole-cell extract was treated with beads coated with preimmune (lane 2) or CS1043 (lanes 3–5) antibodies and tested for transcription from the U6 promoter. In lanes 4 and 5, increasing amounts of rhBRFU were added to the depleted extract. Lane 1 shows U6 transcription in untreated whole-cell extract. (B) A whole-cell extract was treated with beads coated with preimmune (lane 1) or CS1043 (lanes 2–5 and 7–9) antibodies and tested for transcription from the VAI promoter. In lanes 3–5, increasing amounts of rhBRFU were added to the depleted extract. In lanes 8 and 9, increasing amounts of rhBRF were added to the depleted extract. Lane 6 shows U6 transcription in untreated whole-cell extract.
Discussion
Unlike the TFIIIB activity in yeast cells, which has been well defined and is apparently unique, the TFIIIB activity in other organisms has not been completely defined. Here, we have characterized a human homolog of yeast B′′, and we have shown that this protein is required for transcription of both TATA-less, tRNA-type promoters and TATA-containing, snRNA-type RNA polymerase III promoters. In addition, we have identified a factor related to both BRF and TFIIB, which is specifically required for transcription from the U6 snRNA-type promoter. These results characterize TFIIIB, the central RNA polymerase III transcription factor, in human cells.
Identification of a functional human homolog of yeast B′′
hB″ is a functional homolog of yeast B″ by several criteria. First, a BLAST search of the protein database with hB″ as the query returns yeast B″ as the highest scoring match; second, hB′′ is functional for RNA polymerase III transcription from both the VAI and the U6 promoters in vitro; and third, it can be localized to the U6 promoter in vivo. We have shown in Figure 1C putative B″ homologs from a number of species, including Mus musculus, D. melanogaster, C. elegans, S. pombe, and A. thaliana. Although there are no functional data concerning these proteins, the observations that (1) they show sequence conservation upstream of the SANT domain, in a region known to be functionally important in yeast B′′ (Kumar et al. 1997), and that (2) their SANT domains are more similar to each other than to SANT domains from other proteins, suggest that they indeed correspond to functional B″ proteins.
The C-terminal region of hB′′ consists of 19 repeats of 26–28 amino acids, each of which contains a number of potential serine and threonine phosphorylation sites. This is reminiscent of the C-terminal domain of the largest subunit of RNA polymerase II, which for human RNA polymerase II consists of 52 heptad repeats with serine, threonine, and tyrosine phosphorylation sites. In the case of the largest RNA polymerase II subunit, the C-terminal domain is involved in a number of functions, including release of the transcription complex from the promoter (promoter clearance), recruitment of mRNA processing factors, and perhaps inactivation of transcription at mitosis (see Orphanides et al. 1996; Parsons and Spencer 1997; McCracken et al. 1998; and references therein). RNA polymerases III and I do not contain domains similar to the RNA polymerase II CTD, and the RNA polymerase III initiation complex, unlike the RNA polymerase II initiation complex, is not known to contain any kinase activity. The hB′′ C terminus may, however, be involved in regulatory functions. For example, inactivation of RNA polymerase III transcription at mitosis is known to occur through inactivation of TFIIIB, but the TFIIIB components affected have not been defined (Gottesfeld et al. 1994; White et al. 1995; Leresche et al. 1996). Perhaps this inactivation involves phosphorylation of the repetitive C-terminal domain of hB′′. Moreover, TFIIIB activity is regulated during interphase and has been shown to associate with the retinoblastoma protein (Rb) and the p107 and p130 pocket proteins, as well as with p53, all of which can downregulate RNA polymerase III transcription (Larminie et al. 1997; Cairns and White 1998; Sutcliffe et al. 1999). Perhaps some of these proteins associate with hB′′, and the association may be regulated in part by phosphorylation of the hB′′ C-terminal domain.
Identification of hBRFU
The generation of the anti-peptide antibody 1043, which is capable of debilitating U6 transcription when used to deplete transcription extracts, allowed us to identify hBRFU. The N-terminal domain of hBRFU is related to the N-terminal domains of both TFIIB and BRF, and thus hBRFU is a new member of the TFIIB family of proteins, which includes TFIIB and BRF of various species. Very strikingly, unlike TFIIB and hBRF, which do not function for U6 transcription (Mital et al. 1996; Henry et al. 1998a; Kuhlman et al. 1999), hBRFU efficiently restores transcription from the U6 promoter in a depleted extract. Thus, TFIIIB activities in human cells contain TBP, a factor required very generally for initiation by all three RNA polymerases, hB′′, a more specialized factor required for initiation by RNA polymerase III at all or many types of RNA polymerase III promoters, and hBRF or hBRFU, very specialized factors required for initiation by RNA polymerase III at subclasses of RNA polymerase III promoters. Figure 9 illustrates the composition TFIIIB on TATA-less and TATA-containing RNA polymerase III promoters.
Figure 9.
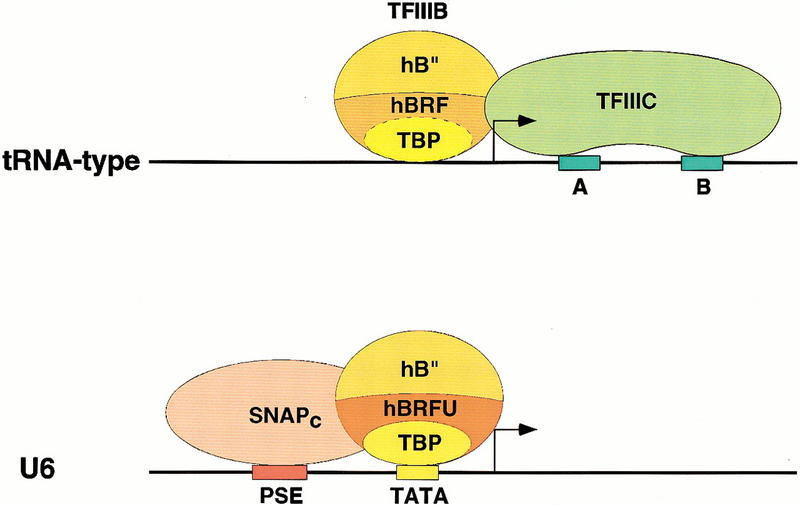
Model of the initiation complexes formed on tRNA-type and U6-type promoters. The upper panel shows a tRNA-type promoter with the gene internal A and B box promoter elements recruiting TFIIIC, which in turn can recruit through protein–protein contacts the TFIIIB components TBP, hBRF, and hB′′. The dotted line separating TBP and hBRF indicates that these two factors are tightly associated with each other. The solid line separating hB′′ from the TBP–hBRF complex indicates that hB′′ is only weakly associated with the rest of the complex. The lower panel shows that for the human U6 gene, the external PSE and TATA box core promoter elements recruit SNAPc and the TFIIIB component TBP, respectively. The binding of these factors probably allows the recruitment of hB′′ and a TFIIIB component specific for PSE-containing RNA polymerase III promoters, hBRFU. The solid lines separating TBP, hBRFU, and hB′′ indicate that in this case, the TFIIIB components are either not associated or only weakly associated with each other.
At TATA-less promoters of the tRNA type, the internal A and B boxes recruit the transcription factor IIIC (upper panel). This allows the subsequent recruitment of TFIIIB, which in this case consists of TBP, hBRF, and hB′′. TBP and hBRF are tightly associated to each other in HeLa-cell extracts (Wang and Roeder 1995; Mital et al. 1996), as indicated by the dotted line separating the two factors. In contrast, hB′′ appears only weakly associated with the TBP/hBRF complex, if at all, as indicated by the solid line. Indeed, depletion of extract with anti-hB′′ antibodies did not remove TBP or hBRF (Fig. 4), and in immunoprecipitations of endogenous hB″, we could detect only low levels of TBP and no hBRF (data not shown). This is similar to the yeast TFIIIB complex, which consists of strongly associated TBP and BRF, and much more loosely associated B′′ (Kassavetis et al. 1991; Huet et al. 1994).
At a TATA-containing promoter such as the human U6 promoter, the gene external core promoter PSE and TATA elements recruit SNAPc and TBP, respectively (lower panel). This presumably allows the subsequent binding of hBRFU and hB′′. In this case, the three TFIIIB components TBP, hBRFU, and hB′′ do not appear to associate strongly with each other in solution, as indicated by the solid lines separating the three factors in Figure 9. Indeed, depletion of extracts with anti-TBP, anti-hBRFU, or anti-hB′′ antibodies does not, in each case, remove significant amounts of the two other factors (Figs. 4 and 8; see also Mital et al. 1996; Henry et al. 1998a). Recently, Takada et al. (2000) showed that in Drosophila cells, the TFIIIB activity contains a hBRF homolog and a TBP-related factor called TRF1. Together with our results, these results suggest that there is a wide range of TFIIIB activities in different species, which contain various TBP-like and BRF-like subunits.
Determination of RNA polymerase specificity at the human snRNA promoters
In mRNA promoters, TFIIB is the factor that directly recruits RNA polymerase II (Orphanides et al. 1996). TFIIB also functions on RNA polymerase II snRNA promoters, as evidenced by the observations that depletion of extracts with anti-TFIIB antibodies debilitates RNA polymerase II transcription from the U1 promoter but not RNA polymerase III transcription from the U6 promoter, and that U1 transcription can be restored by addition of recombinant TFIIB (Kuhlman et al. 1999). Moreover, TFIIB can be localized to U1 promoter regions in vivo by chromatin immunoprecipitation (Fig. 5). Thus, although we have not excluded the possibility that hBRFU might also be functional at RNA polymerase II snRNA promoters, it seems likely that on the human snRNA promoters, RNA polymerase specificity is determined by the recruitment of TFIIB or hBRFU.
Although the N-terminal half of BRF is very similar to TFIIB, the functional domain arrangement of the two factors is different. In TFIIB, the zinc ribbon domain (indicated in blue in Fig. 6) is absolutely required for recruitment of RNA polymerase II and/or TFIIF, and the core region (indicated in purple in Fig. 6) is required for association with the TBP/DNA complex (Orphanides et al. 1996). BRF, like TFIIB, contacts directly the RNA polymerase, in this case RNA polymerase III (Khoo et al. 1994; Wang and Roeder 1995; Brun et al. 1997; Ferri et al. 2000), but the zinc ribbon domain of BRF is not required for RNA polymerase III recruitment (Kassavetis et al. 1997, 1998; Hahn and Roberts 2000). Instead, the BRF zinc ribbon domain, which is essential for both viability in yeast and efficient RNA polymerase III transcription in vitro, plays a role at a later step in open complex formation (Hahn and Roberts 2000). It will be of great interest to determine how hBRFU interacts with TBP, and whether the zinc ribbon domain of hBRFU is required for recruitment of RNA polymerase III and thus functionally resembles that of TFIIB, or whether it is required for open complex formation and thus functionally resembles that of BRF.
Materials and methods
Cloning of cDNAs encoding hB′′ and hBRFU
At the time we started isolating a human homolog of yeast B″, the only entry in the database that showed some similarity to the yeast factor was a mouse EST (GenBank accession number AA200560) encoding 21 amino acids with strong similarity to part of the yeast B″ SANT domain. We used this sequence information to design primers and then used these primers in combination with primers hybridizing to the arms of λ to screen various human cDNA libraries. Several rounds of screening, each time with new primers located near the end of the largest fragment obtained, resulted in the isolation of a partial cDNA encoding amino acids 1–1020 of hB″. The sequence contained a stop codon upstream of the first ATG (see GenBank accession number AF298151), indicating that it encoded the very N terminus of the protein. At this point, a new entry was deposited in the database (GenBank accession number AB033067). Nucleotides 822–2243 of the AB033067 sequence were, with a few exceptions, identical to the last 1422 nucleotides (encoding amino acids 547–1020) of our sequence, but then the AB033067 sequence extended a further 3.5 kb downstream, the first 1104 nucleotides of which extended the open reading frame another 368 amino acids in the C-terminal direction. The 5′ end of the AB033067 sequence was divergent. We used the AB033067 sequence to design primers hybridizing to a region downstream of the stop codon and to a region within the sequence we had obtained, and performed PCR reactions from cDNA prepared from total HeLa-cell RNA. The resulting fragment was cloned and sequenced, and was nearly identical (except for a few amino acid changes) to entry AB033067. The sequence of some portions of hB′ was then further confirmed by the generation, with the high fidelity Pfu polymerase (Stratagene), of PCR fragments from cDNA made from total HeLa-cell RNA, followed by sequencing of the resulting fragments. A very recent entry in the database (GenBank accession number AJ238520) matches our sequence over its entire length almost perfectly. An examination of genomic sequences in the database indicates that the entire human B″ DNA sequence is found on chromosome 5 (GenBank accession number AC008851) and that other entries in the database (accession numbers AJ238520 and AB033067) most likely correspond to alternatively spliced products from the same region. A large region corresponding to the C-terminal half of the human B″ sequence can also be found in genomic DNA from chromosome 2 (GenBank accession number AC025985).
hBRFU was identified in the database as the human protein most related to hBRF after TFIIB (GenBank accession number BAA91975). The sequence information was used to design primers for PCRs with cDNA made from total HeLa-cell RNA and the high fidelity Pfu polymerase (Stratagene). The amplified sequence (GenBank accession number AF298153) was identical to the entry in the database except for one silent nucleotide change and one nucleotide change that changed the codon encoding cysteine 157 in BAA91975 to a codon encoding tyrosine.
Expression of recombinant proteins in E. coli
hBRF was expressed as an N-terminal glutathione S-transferase fusion protein from a modified pSBET vector (Schenk et al. 1995). Induction with IPTG was for 1 h at 37°C, and the glutathione S-transferase moiety was cleaved with thrombin. Both hB″ and hBRFU were expressed in E. coli as N-terminal His-tagged and C-terminal FLAG-tagged proteins with the T7 RNA polymerase expression system (Studier et al. 1990), from modified pSBET (Schenk et al. 1995) vectors. Briefly, the constructs were transformed into BL21 cells and single colonies grown in M9ZB (Studier et al. 1990) at 37°C to an OD600 of 0.8. Induction was with 0.4 mM IPTG for 4 h at room temperature. The cells were pelleted and resuspended in 1:10 of the culture volume in lysis buffer: 10 mM Tris-HCl (pH 8), 250 mM NaCl, 1%Triton X-100, 50 mM NaH2PO4, 20 mM imidazole, and protease inhibitors (1 mM PMSF, 0.5 mM sodium bisulfate, and 0.5 mM benzamidine). The cells were frozen in liquid nitrogen and thawed three times, and homogenized with a dounce homogenizer on ice. The cell debris was pelleted by centrifugation at 15,000 rpm for 15 min at 4°C in a SS34 rotor (Sorvall). The supernatant was then incubated with nickel agarose beads (Qiagen) overnight at 4°C. The beads were washed three times with five-bead volume of wash buffer 1 (20 mM Tris-HCl [pH 8], 500 mM NaCl, 20% glycerol, 0.1% Tween 20, 20 mM imidazole, and protease inhibitors), twice with five-bead volume of wash buffer 2 (10 mM Tris-HCl [pH 8], 100 mM NaCl, 20% glycerol, 0.01% Nonidet P-40, and protease inhibitors), and the bound proteins were eluted with wash buffer 2 containing 300 mM imidazole. The eluate was dialyzed against TBS (20 mM Tris-HCl [pH 8], 150 mM NaCl, and protease inhibitors) and then incubated in the same buffer with flag agarose beads (Sigma). The beads were then washed with TBS containing protease inhibitors and eluted with the same buffer containing 200 μg/mL flag peptide. The eluate was dialyzed against buffer D100: 20% glycerol, 20 mM HEPES-KOH (pH 7.9), 0.5 mM EDTA, and 100 mM KCl.
Antibodies and immunoblots
The rabbit polyclonal anti-hB′′ antibodies were directed against peptides corresponding to amino acids 139–153 (peptide hB′′-2, CS913) and 392–410 (peptide hB′′-3, CS843) of hB′′. The rabbit polyclonal anti-hBRF antibody that cross-reacts with hBRFU (rabbit CS1043) was raised against a peptide (R2A) corresponding to amino acids 229–248 of hBRF (Mital et al. 1996). The anti-TFIIB antibody was from rabbit CS396 (anti-TFIIB-4) (Kuhlman et al. 1999).
The protein samples were fractionated on 7.5% SDS–polyacrylamide gels and transferred to nitrocellulose membranes. The membranes were blocked for 1 h at room temperature with TBS-T (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.1% Tween 20) containing 5% BSA and 5% nonfat milk, washed twice for 10 min in TBS-T, and probed with anti-hB′′ (CS913) or anti-hBRF/hBRFU (CS1043) antibody at a 1:1000 dilution in TBS-T containing 5% BSA overnight at 4°C. The membranes were washed twice for 10 min in TBS-T and probed with the secondary antibody (sheep anti-rabbit) at a 1:4000 dilution in TBS-T containing 5% BSA and 2.5% nonfat milk for 1 h at room temperature. The blots were developed with ECL (Amersham Life Science) and exposed to film.
In vitro transcriptions
In vitro transcriptions were performed as described by Kuhlman et al. (1999) with the following modifications. The U6 transcription reactions were performed in a total volume of 40 μL containing 75 ng of the pU6/Hae/RA.2 template, 500 ng of poly (dG-dC) · poly (dG-dC), and 8 μL of whole-cell extract diluted 1:1, and were complemented with 4 μL of a 10-fold ATP mix (0.5 M ATP, 10 μg/mL phosphocreatine kinase, and 0.5 M creatine phosphate) and 5–10 ng of recombinant TBP. The VAI transcription reactions were performed in a total volume of 20 μL, contained 250 ng of the pSBM13 +VAI template and 8 μL of whole-cell extract diluted 1:1, and were incubated for 1 h at 37°C. The Ad2 major late (p119MLP[C2A]) transcription reactions were performed in a total volume of 30 μL and contained 10 μL of whole-cell extract diluted 1:1. The U1 (pU1*G-) transcription reactions were performed in a total volume of 40 μL containing 15 μL of whole-cell extract diluted 1:1. The whole-cell extract contained 28 mg of protein per mL.
For depletions, 200 μL of extract was mixed with 200 μL of protein A agarose beads covalently linked to the antibody of interest overnight at 4°C. The beads were then pelleted by centrifugation, and the depleted extract (diluted 1:1 relative to the starting extract) was used directly for transcriptions.
Chromatin immunoprecipitations
The formaldehyde cross-linking and immunoprecipitations were performed as described by P. Farnham and colleagues (Boyd et al. 1998), except that the NaCl concentration during immunoprecipitation was raised to 150 mM and the immunoprecipitations were performed overnight at 4°C. To design the control primers for the U6 promoter, we used the sequence in a 22-kb contig (GenBank accession number AC009292, contig extending from 140423 to 162578) that contains the U6 gene we work with. The control primers (U6C1UP and U6C1L) hybridized to a sequence located 4 kb upstream of the U6 transcription start site, and the test U6 primers (U64UP and U62L) bracketed the core U6 promoter. The PCR reaction consisted of 29 cycles.
Acknowledgments
We thank Ryuji Kobayashi, Winship Herr, Ping Hu, Beicong Ma, Xinyang Zhao, and Xuemei Zhao for discussion and reagents, and Julie Wells and Peggy Farnham for providing us with a protocol to perform chromatin immunoprecipitations. We also thank Michael Ockler, James Duffy, and Philip Renna for artwork and photography. This work was funded in part by National Institutes of Health grant GM38810. P.S.P. is supported by a Leukemia Society of America Special Fellowship Award (GN 629100). N.H. and Y.S. are supported by the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hernande@cshl.org; FAX (516) 367-6801.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.836400.
References
- Aasland R, Stewart AF, Gibson T. The SANT domain: A putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. [PubMed] [Google Scholar]
- Bagby S, Kim S, Maldonado E, Tong KI, Reinberg D, Ikura M. Solution structure of the C-terminal core domain of human TFIIB: Similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein LB, Manser T, Weiner AM. Human U1 small nuclear RNA genes: Extensive conservation of flanking sequences suggests cycles of gene amplification and transposition. Mol Cell Biol. 1985;5:2159–2171. doi: 10.1128/mcb.5.9.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. c-Myc target gene specificity is determined by a post–DNA binding mechanism. Proc Natl Acad Sci. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes & Dev. 1990;4:1345–1356. doi: 10.1101/gad.4.8.1345. [DOI] [PubMed] [Google Scholar]
- Brun I, Sentenac A, Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997;16:5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology with TFIIB. Cell. 1992;71:221–230. doi: 10.1016/0092-8674(92)90351-c. [DOI] [PubMed] [Google Scholar]
- Burnol A-F, Margottin F, Huet J, Almouzni G, Prioleau M-N, Mechali M, Sentenac A. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature. 1993;362:475–477. doi: 10.1038/362475a0. [DOI] [PubMed] [Google Scholar]
- Cairns CA, White RJ. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 1998;17:3112–3123. doi: 10.1093/emboj/17.11.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert T, Hahn S. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes & Dev. 1992;6:1940–1949. doi: 10.1101/gad.6.10.1940. [DOI] [PubMed] [Google Scholar]
- Denison RA, Weiner AM. Human U1 RNA pseudogenes may be generated by both DNA- and RNA-mediated mechanisms. Mol Cell Biol. 1982;2:815–828. doi: 10.1128/mcb.2.7.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschenlauer JB, Kaiser MW, Gerlach VL, Brow DA. Architecture of a yeast U6 RNA gene promoter. Mol Cell Biol. 1993;13:3015–3026. doi: 10.1128/mcb.13.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri ML, Peyroche G, Siaut M, Lefebvre O, Carles C, Conesa C, Sentenac A. A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol Cell Biol. 2000;20:488–495. doi: 10.1128/mcb.20.2.488-495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld JM, Wolf VJ, Dang T, Forbes DJ, Hartl P. Mitotic repression of RNA polymerase III transcription in vitro mediated by phosphorylation of a TFIIIB component. Science. 1994;263:81–84. doi: 10.1126/science.8272869. [DOI] [PubMed] [Google Scholar]
- Hahn S, Roberts S. The zinc ribbon domains of the general transcription factors TFIIB and Brf: Conserved functional surfaces but different roles in transcription initiation. Genes & Dev. 2000;14:719–730. [PMC free article] [PubMed] [Google Scholar]
- Henry RW, Sadowski CL, Kobayashi R, Hernandez N. A TBP-TAF complex required for transcription of human snRNA genes by RNA polymerases II and III. Nature. 1995;374:653–657. doi: 10.1038/374653a0. [DOI] [PubMed] [Google Scholar]
- Henry RW, Ford E, Mital R, Mittal V, Hernandez N. Crossing the line between RNA polymerases: Transcription of human snRNA genes by RNA polymerases II and III. Cold Spring Harbor Symp Quant Biol. 1998a;63:111–120. doi: 10.1101/sqb.1998.63.111. [DOI] [PubMed] [Google Scholar]
- Henry RW, Mittal V, Ma B, Kobayashi R, Hernandez N. SNAP19 mediates the assembly of a functional core promoter complex (SNAPc) shared by RNA polymerases II and III. Genes & Dev. 1998b;12:2664–2672. doi: 10.1101/gad.12.17.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J, Conesa C, Manaud N, Chaussivert N, Sentenac A. Interactions between yeast TFIIIB components. Nucleic Acids Res. 1994;22:3433–3439. doi: 10.1093/nar/22.16.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Kassavetis GA, Geiduschek EP. Identical components of yeast transcription factor IIIB are required for transcription of TATA box–containing and TATA-less genes. Mol Cell Biol. 1994;14:2798–2808. doi: 10.1128/mcb.14.4.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Bartholomew B, Blanco JA, Johnson TE, Geiduschek PE. Two essential components of the Saccharomyces cerevisiaetranscription factor TFIIIB: Transcription and DNA-binding properties. Proc Natl Acad Sci. 1991;88:7308–7312. doi: 10.1073/pnas.88.16.7308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Joazeiro CAP, Pisano M, Geiduschek PE, Colbert T, Hahn S, Blanco JA. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- Kassavetis GA, Nguyen ST, Kobayashi R, Kumar A, Geiduschek EP, Pisano M. Cloning, expression, and function of TFC5, the gene encoding the B′′ component of the Saccharomyces cerevisiaeRNA polymerase III transcription factor TFIIIB. Proc Natl Acad Sci. 1995;92:9786–9790. doi: 10.1073/pnas.92.21.9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Bardeleben C, Kumar A, Ramirez E, Geiduschek EP. Domains of the Brf component of RNA polymerase III transcription factor IIIB (TFIIIB): Functions in assembly of TFIIIB-DNA complexes and recruitment of RNA polymerase to the promoter. Mol Cell Biol. 1997;17:5299–5306. doi: 10.1128/mcb.17.9.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Kumar A, Letts G, Geiduschek EP. A post-recruitment function for the RNA polymerase III transcription-initiation factor IIIB. Proc Natl Acad Sci. 1998;95:9196–9201. doi: 10.1073/pnas.95.16.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo B, Brophy B, Jackson SP. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes & Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- Kuhlman TC, Cho H, Reinberg D, Hernandez N. The general transcription factors IIA, IIB, IIF, and IIE are required for RNA polymerase II transcription from the human U1 snRNA promoter. Mol Cell Biol. 1999;19:2130–2141. doi: 10.1128/mcb.19.3.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Kassavetis GA, Geiduschek EP, Hambalko M, Brent CJ. Functional dissection of the B′′ component of RNA polymerase III transcription factor IIIB: A scaffolding protein with multiple roles in assembly and initiation of transcription. Mol Cell Biol. 1997;17:1868–1880. doi: 10.1128/mcb.17.4.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel GR, Maser RL, Calvet JP, Pederson T. U6 small nuclear RNA is transcribed by RNA polymerase III. Proc Natl Acad Sci. 1986;83:8575–8579. doi: 10.1073/pnas.83.22.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein: DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- Larminie CG, Cairns CA, Mital R, Martin K, Kouzarides T, Jackson SP, White RJ. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leresche A, Wolf VJ, Gottesfeld JM. Repression of RNA polymerase II and III transcription during M phase of the cell cycle. Exp Cell Res. 1996;229:282–288. doi: 10.1006/excr.1996.0373. [DOI] [PubMed] [Google Scholar]
- Lobo SM, Lister J, Sullivan ML, Hernandez N. The cloned RNA polymerase II transcription factor IID selects RNA polymerase III to transcribe the human U6 gene in vitro. Genes & Dev. 1991;5:1477–1489. doi: 10.1101/gad.5.8.1477. [DOI] [PubMed] [Google Scholar]
- Lobo SM, Tanaka M, Sullivan ML, Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell. 1992;71:1029–1040. doi: 10.1016/0092-8674(92)90397-u. [DOI] [PubMed] [Google Scholar]
- López-De-León A, Librizzi M, Puglia K, Willis IM. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:211–220. doi: 10.1016/0092-8674(92)90350-l. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Margottin F, Dujardin G, Gerard M, Egly J-M, Huet J, Sentenac A. Participation of the TATA factor in transcription of the yeast U6 gene by RNA polymerase C. Science. 1991;251:424–426. doi: 10.1126/science.1989075. [DOI] [PubMed] [Google Scholar]
- McCracken S, Rosonina E, Fong N, Sikes M, Beyer A, O'Hare K, Shuman S, Bentley D. Role of RNA polymerase II carboxy-terminal domain in coordinating transcription with RNA processing. Cold Spring Harb Symp Quant Biol. 1998;63:301–309. doi: 10.1101/sqb.1998.63.301. [DOI] [PubMed] [Google Scholar]
- Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and Adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–7042. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moenne A, Camier S, Anderson G, Margottin F, Beggs J, Sentenac A. The U6 gene of Saccharomyces cerevisiae is transcribed by RNA polymerase C (III) in vivo and in vitro. EMBO J. 1990;9:271–277. doi: 10.1002/j.1460-2075.1990.tb08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Parsons GG, Spencer CA. Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol Cell Biol. 1997;17:5791–5802. doi: 10.1128/mcb.17.10.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule M, White R. Survey and summary: Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S, Miller SJ, Lane WS, Lee S, Hahn S. Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J Biol Chem. 1996;271:14903–14909. doi: 10.1074/jbc.271.25.14903. [DOI] [PubMed] [Google Scholar]
- Rüth J, Conesa C, Dieci G, Lefebvre O, Dusterhoft A, Ottonello S, Sentenac A. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- Sadowski CL, Henry RW, Lobo SM, Hernandez N. Targeting TBP to a non-TATA box cis-regulatory element: A TBP-containing complex activates transcription from snRNA promoters through the PSE. Genes & Dev. 1993;7:1535–1548. doi: 10.1101/gad.7.8.1535. [DOI] [PubMed] [Google Scholar]
- Schenk PM, Baumann S, Mattes R, Steinbiss H-H. Improved high level expression system for eucaryotic genes in E. coliusing T7 RNA polymerase and rare Arg tRNAs. Biotechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- Simmen KA, Bernues J, Parry HD, Stunnenberg HG, Berkenstam A, Cavallini B, Egly J-M, Mattaj IW. TFIID is required for in vitro transcription of the human U6 gene by RNA polymerase III. EMBO J. 1991;10:1853–1862. doi: 10.1002/j.1460-2075.1991.tb07711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JE, Cairns CA, McLees A, Allison SJ, Tosh K, White RJ. RNA polymerase III transcription factor IIIB is a target for repression by pocket proteins p107 and p130. Mol Cell Biol. 1999;19:4255–4261. doi: 10.1128/mcb.19.6.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AKP, Fisher TS, Pugh BF. The TATA-binding protein and associated factors are components of pol III transcription factor TFIIIB. Cell. 1992;71:1015–1028. doi: 10.1016/0092-8674(92)90396-t. [DOI] [PubMed] [Google Scholar]
- Takada S, Lis JT, Zhou S, Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- Teichmann M, Seifart KH. Physical separation of two different forms of human TFIIIB active in the transcription of the U6 or the VAI gene in vitro. EMBO J. 1995;14:5974–5983. doi: 10.1002/j.1460-2075.1995.tb00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Roeder RG. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2–related domains. Proc Natl Acad Sci. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Jackson SP. Mechanism of TATA-binding protein recruitment to a TATA-less class III promoter. Cell. 1992;71:1041–1053. doi: 10.1016/0092-8674(92)90398-v. [DOI] [PubMed] [Google Scholar]
- White RJ, Gottlieb TM, Downes CS, Jackson SP. Cell cycle regulation of RNA polymerase III transcription. Mol Cell Biol. 1995;15:6653–6662. doi: 10.1128/mcb.15.12.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-B, Murphy S, Bai L, Wang Z, Roeder RG. Proximal sequence element–binding transcription factor (PTF) is a multisubunit complex required for transcription of both RNA polymerase II- and RNA polymerase III-dependent small nuclear RNA genes. Mol Cell Biol. 1995;15:2019–2027. doi: 10.1128/mcb.15.4.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Zeng Q, Colangelo CM, Lewis M, Summers MF, Scott RA. The N-terminal domain of TFIIB from Pyrococcus furiosusforms a zinc ribbon. Nat Struct Biol. 1996;3:122–124. doi: 10.1038/nsb0296-122. [DOI] [PubMed] [Google Scholar]