Abstract
The condensin complex in frog extracts, containing two SMC (structural maintenance of chromosomes) and three non-SMC subunits, promotes mitotic chromosome condensation, and its supercoiling activity increases during mitosis by Cdc2 phosphorylation. Here, we report that fission yeast has the same five-member condensin complex, each of which is essential for mitotic condensation. The condensin complex was purified and the subunits were identified by microsequencing. Cnd1, Cnd2, and Cnd3, three non-SMC subunits showing a high degree of sequence conservation to frog subunits, are essential for viability, and their gene disruption leads to a phenotype indistinguishable from that observed in cut3-477 and cut14-208, known mutations in SMC4 and SMC2-like subunits. Condensin subunits tagged with GFP were observed to alter dramatically their localization during the cell cycle, enriched in the nucleus during mitosis, but cytoplasmic during other stages. This stage-specific alteration in localization requires mitosis-specific phosphorylation of the T19 Cdc2 site in Cut3. The T19 site is phosphorylated in vitro by Cdc2 kinase and shows the maximal phosphorylation in metaphase in vivo. Its alanine substitution mutant fails to suppress the temperature-sensitive phenotype of cut3-477, and shows deficiency in condensation, probably because Cut3 T19A remains cytoplasmic. Therefore, direct Cdc2 phosphorylation of fission yeast condensin may facilitate its nuclear accumulation during mitosis.
Keywords: Chromosome condensation, SMC, Barren, condensin, nuclear export, S. pombe mitosis
Chromosome condensation is an enigmatic phenomenon in mitosis (Bak et al. 1977). Its occurrence is an essential step before subsequent sister chromatid separation in anaphase (for review, see Murray 1998). Condensed chromosomes at metaphase, containing duplicated sister chromatids and folded many times, are ready for separation when the sister kinetochores are properly associated with the bioriented spindle microtubules (Yanagida 1998). Condensed chromosomes should have structures significantly different from those in interphase, but their detailed features are little understood. DNA topoisomerase II (topo II) and subtypes of SMC (structural maintenance of chromosomes; Strunnikov et al. 1995) proteins are known to be required for condensation (Uemura et al. 1987; Adachi et al. 1991; Hirano and Mitchison 1993,1994; Saka et al. 1994; Strunnikov et al. 1995; for review, see Saitoh et al. 1995). The SMC proteins are highly conserved in evolution (Hirano 1998; Jessberger et al. 1998; Strunnikov 1998), with frog XCAP-E, budding yeast Smc2p, and fission yeast Cut14 constituting one subtype, and XCAP-C, Smc4p, and Cut3 forming another subtype. Condensin complex containing SMC2 and SMC4 was first identified in frog cell extracts as a five-member complex and shown to be essential for an in vitro reconstitution system of mitotic chromosome condensation (Hirano and Mitchison 1994; Hirano et al. 1997). Three other non-SMC subunits of frog condensin were identified by microsequencing individual subunits (Hirano et al. 1997; Kimura et al. 1998; the XCAP-G sequence is available in GenBank, accession no. AF111423). The frog condensin has an ATP-dependent in vitro activity that promotes positive supercoiling of DNA, which is thought to be important for chromosome condensation (Kimura and Hirano 1997).
Fission yeast mutants cut3-477 and cut14-208 are defective in chromosome condensation as well as sister chromatid separation (Saka et al. 1994; Yanagida 1998). Their phenotypes highly resemble that of top2 mutants, deficient in the topo II activity (Uemura and Yanagida 1986). FISH has been applied successfully to visualize condensation in wild-type chromosomes using cosmid probes derived from the arm of chromosome II; severalfold folding of the rod-like arm into the spherical condensed arm was observed in wild-type mitosis, but was entirely deficient in cut14-208 mutant cells (Saka et al. 1994). Nuclear chromatin in these mutant cells was found to be sensitive to single strand-specific S1 nuclease, suggesting that these proteins may have some role in forming a particular chromatin DNA conformation (Sutani and Yanagida 1997). Cut3 and Cut14 formed a stable complex upon overproduction, and the purified heterodimeric complex possessed an extremely efficient in vitro DNA renaturation activity, forming duplex DNA from complementary single-stranded DNAs (Sutani and Yanagida 1997). This activity may have physiological significance as a complex containing mutant protein exhibiting renaturation activity only at permissive temperature. This in vitro activity, however, did not require ATP and was dependent on the presence of the central coiled–coil region. These studies suggested that the unwinding and rewinding of chromosomal DNA is important for chromosome condensation. However, the heterodimeric complex was not sufficient to promote the ATP-dependent supercoiling reaction (T. Sutani and M. Yanagida, unpubl.).
In this study our initial aim was to determine whether fission yeast has the same condensin complex as in frog; prior to this, condensin has not been isolated in other organisms. We show that fission yeast has the same five-member condensin complex. Genes for three non-SMC proteins, Cnd1, Cnd2, and Cnd3, were cloned by microsequencing of isolated proteins. Disruption of these three non-SMC genes revealed that they are essential for viability. Furthermore, gene disruption caused defects in mitotic chromosome condensation, indistinguishable from that of cut3 and cut14 mutants. We propose that the two non-SMC subunits Cnd1 and Cnd3 may function to link Cut3 and Cut14 based on the sequence similarity to AP3-β, which is required for proper clathrin assembly. We show that fission yeast condensin subunits are shuttled between the cytoplasm and the nucleus during the cell cycle. This shift in localization requires the T19 Cdc2 site in the Cut3 protein. We discuss the significance of Cdc2 phosphorylation of condensin for mitotic chromosome condensation in fission yeast.
Results
Immunoprecipitation of HA-tagged Cut3 and Cut14
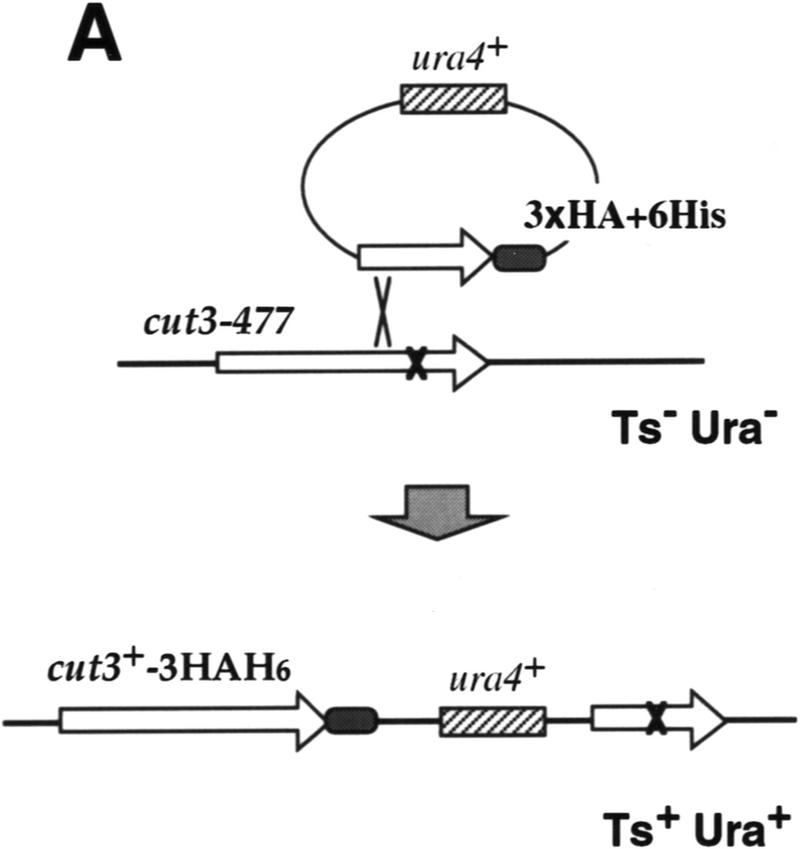
For immunoprecipitation of condensin subunits, plasmid carrying the cut3+ gene tagged with 3× HA and 6× His at the carboxyl-terminus was made and integrated into the chromosome of Schizosaccharomyces pombe mutant cut3-477 using the ura4+ marker (schematized in Fig. 1A. The carboxy-terminal mutation site in cut3-477 is indicated by a cross). Integration of the tagged cut3+ gene was confirmed by the Ts+/Ura+ phenotype of transformed integrated cells (Cut3–3HAH6). Immunoblotting of Cut3–3HAH6 extracts using antibodies against HA revealed a protein band at the expected molecular mass (155 kD; Fig. 1B, lane 1). No band was detected in the nonintegrated wild-type 972 strain (lane 3). Plasmid carrying the cut14+ gene tagged with 3HA–6His was also made, and integrated into the chromosome of the cut14–208 mutant (the mutation site is also in the carboxyl-terminus); the expected Cut14–3HA6His band could be detected in Ts+/Ura+-integrated cells (130 kD; lane 2). These results demonstrate that Cut3–3HAH6 and Cut14–3HAH6 proteins can functionally replace the wild-type proteins.
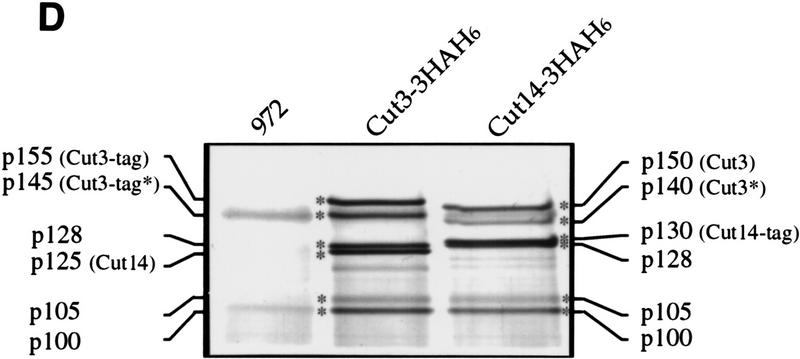
Figure 1.
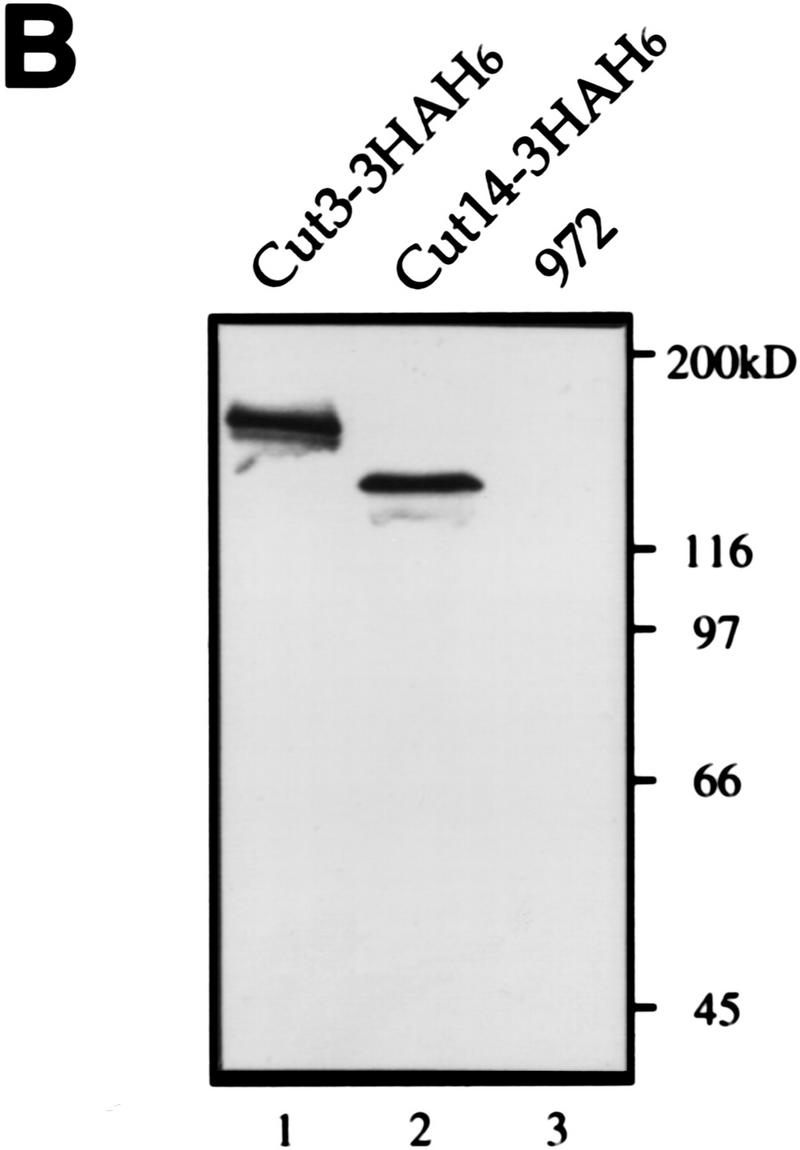
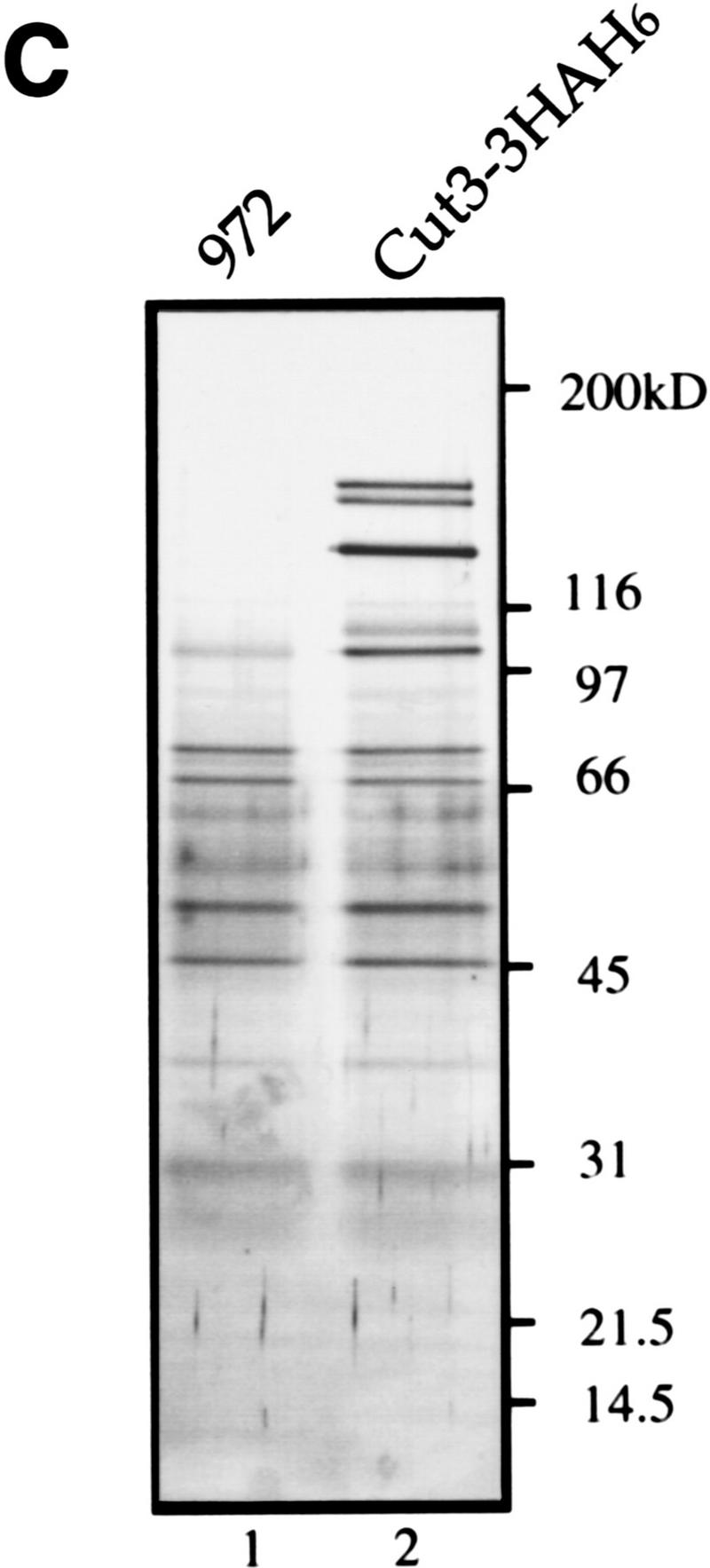
Isolation of condensin subunits. (A) 3× HA+6× His-tagged cut3+ gene was integrated into the chromosome as schematized. Plasmid carrying the carboxy-terminal portion of the tagged cut3+ and the ura4+ marker was introduced into a cut3-477 mutant, which was Ts−/Ura−. The mutation site was located near the carboxyl-terminus (indicated by ×). Ts+/Ura+ integrants were thus obtained. The same strategy was applied for the Cut14–3HAH6 gene. (B) Immunoblotting of Cut3–3HAH6 (lane 1) and Cut14–3HAH6 (lane 2) integrants, and wild-type 972 (lane 3) was done using anti-HA antibodies. Bands at the expected molecular mass positions were obtained. (C) Silver-stained immunoprecipitation patterns of wild type 972 (lane 1) and the integrant Cut3-3HAH6 by anti-HA antibody. (D) Silver-stained immunoprecipitation patterns of wild-type 972, as well as the integrants of Cut3–3HAH6 and Cut14–3HAH6, by anti-HA antibodies. Assignments of individual bands are shown. The bands indicated by asterisks are not present in the 972 wild-type strain (see text). Cut3* is the amino-terminally cleaved form.
Immunoprecipitation of these integrated strains was done using anti-HA antibodies (with wild-type 972 as a negative control). The efficiency of immunoprecipitation was ∼20%–50% (high yields were obtained with increasing amounts of anti-HA antibodies). Individual polypeptides could be distinguished in immunoprecipitates (Fig. 1C,D). Several polypeptides of high molecular masses (Fig. 1D, asterisks) not present in the 972 control could be clearly recognized in extracts of the Cut3–3HAH6 and Cut14–3HAH6 strains. Three polypeptides, designated p128, p105, and p100, were common to the precipitates obtained from the two integrant strains. Three other bands (p155, p145, and p125) were seen only in the precipitates using the Cut3–3HAH6 cell extracts. p155 and p145, respectively, appear to be the full-length and amino-terminally cleaved forms of Cut3–3HAH6, judging from the immunoblot patterns using anti-Cut3 antibodies, whereas p125 represented the untagged Cut14. Three bands, p150, p140, and p130, were specific to the precipitates obtained from the Cut14–3HAH6 strain. p150 and p140 were the untagged Cut3 and its degradation product, respectively, whereas p130 was the tagged Cut14. These predictions were confirmed by immunoblotting (data not shown).
Identification of three unknown protein components, p128, p105, and p100, was done by microsequencing of the polypeptides prepared by large-scale immunoprecipitation (described below). They were likely to be components of the condensin complex as they could be commonly immunoprecipitated with Cut3 and Cut14. One-step affinity chromatography of the tagged Cut14 protein from a Ni column was done using various concentrations of imidazole for elution. The highest purification obtained was ∼100-fold. Because immunoprecipitation gave sufficient purity of the protein bands, the His tag was not used for large-scale preparation.
Identification of three non-SMC components by microsequencing
Immunoprecipitation was done on a large scale using 5 × 1010 cells of Cut3–3HAH6 (described in Materials and Methods). Approximately 1–3 μg each of p128, p105, and p100 proteins was obtained. Polypeptide bands were cut from the gel and subjected to peptide sequence analysis after digestion with an endopeptidase (Achromobacter protease I) specific for the carboxyl-terminus of lysine residues. Two to four peptide sequences were obtained for p100, p105, and p128, as shown in Table 1. The molecular masses of the peptides measured by mass spectrometry were well matched with those calculated for the amino acid sequences determined. The sequences obtained from p100 and p105 showed 100% match with genes already present in the genome database of S. pombe (the Sanger Centre Project). To obtain the identity of the remaining gene corresponding to p128, gene cloning based on the peptide amino acid sequences was necessary (described below).
Table 1.
Peptide sequences of p128, p105, and p100 determined
| Proteins
|
Determined sequencesa
|
Positionsb
|
|---|---|---|
| p128 | 1. kRFYLEALQYIDIVEAGAK | 518–536 |
| 2. klwQVYSYQk | 658–667 | |
| 3. kTQRQwDHVVYAl | 1126–1138 | |
| p105 | 1. kINSTNTWNFALIDYFHDMS | 82–113 |
| LLRDGEDINFQK | ||
| 2. kIyTSRIDSVATETGK | 123–138 | |
| 3. kMSVLPNADGEGqLqLNK | 536–553 | |
| 4. kSFFSTVNQLEETYEK | 674–689 | |
| p100 | 1. kNDYEGEEITVsQK | 453–466 |
| 2. kTQTGDDYIALNxNVxLxLA | 727–760 | |
| NMIFESLPNASEGK |
Uppercase letters indicate residues identified unambiguously; (x) unidentified residues.
Numbers represent the corresponding sequence positions in the predicted proteins deduced from the nucleotide sequences.
p128 is the product of cnd1+ required for condensation
The cloning of the gene for p128 was accomplished with degenerated PCR (Materials and Methods). A 1.8-kb genomic fragment was obtained, and nucleotide sequencing verified the presence of a single reading frame containing all the peptide sequences determined for p128. A 7.3-kb fragment containing the full-length genomic coding region was obtained by subcloning a cosmid c35 from the long arm of chromosome II. Nucleotide sequencing indicated that the gene for p128 contained 1158 amino acids (calculated molecular mass, 131 kD) with two putative introns in the amino-terminus; part of the sequence is shown in Figure 2A. The gene coding for p128 was designated cnd1+ (condensin subunit).
Figure 2.
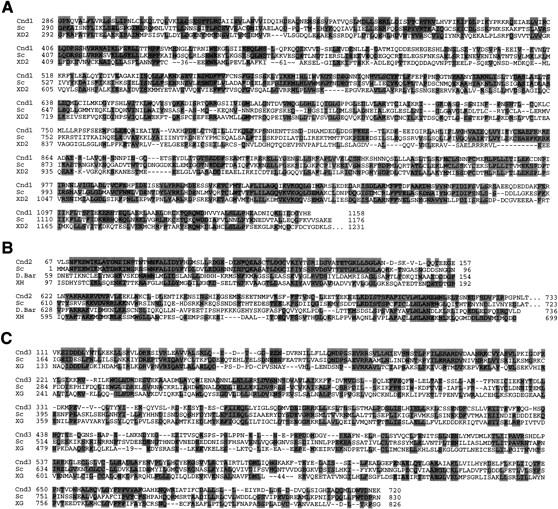
Isolation of the cnd1+, cnd2+, and cnd3+ genes. Partial sequences of the cloned genes are shown. (A) Three amino acid sequences (S. pombe Cnd1, Cnd1; S. cerevisiae YLR272c, Sc; frog XCAP-D2, XD2) are aligned. Identical amino acids are boxed. (B) Amino acid sequence comparison (S. pombe, Cnd2, Cnd2; S. cerevisiae YBL097w, Sc; Drosophila Barren, D. Bar; frog XCAP-H, XH). (C) Amino acid sequence comparison (S. pombe, Cnd3, Cnd3; S. cerevisiae YDR325w, Sc; frog XCAP-G, XG).
Predicted amino acid sequences similar to Cnd1 were found in budding yeast YLR272c and frog XCAP-D2. These sequences are 30%–40% identical, with the highest similarity in the carboxy-terminal regions. They are commonly acidic (pI of Cnd1 is 5.0). Frog XCAP-D2 was reported to be a component of condensin (Kimura et al. 1998), consistent with the present result. No function has been allocated to the budding yeast sequence. The budding yeast and fission yeast sequences are more similar to each other than to the frog XCAP-D2 sequence; the central domains (amino acids 400–550) of the yeast sequences do not resemble that of XCAP-D2.
The cnd1+ gene was disrupted by one-step replacement (Materials and Methods). A linear DNA fragment carrying the disrupted cnd1+ with the S. pombe ura4+ gene was introduced into the chromosome of a Ura− diploid strain by transformation, and a Ura+ heterozygous diploid was obtained. Integration was verified by Southern hybridization. Tetrad dissection of the heterozygous diploid cells indicated that only the two Ura− spores were viable, whereas the two disrupted Ura+ spores did not produce colonies at 26°C and 33°C. These results indicated that cnd1+ is absolutely required for viability.
cnd1-disrupted cells were examined by DAPI staining for DNA, anti-Sad1 (SPB), and anti-tubulin (TUB) antibody staining for SPB and microtubules, respectively, in liquid cultures in the absence of uracil so that only disrupted cells could germinate. Germination took place after 6 hr and cnd1-disrupted cells entered M phase after 10 hr. Most M-phase cells exhibited defects in chromosome condensation (Fig. 3A); the wild-type control is shown in Figure 3D. The spindle was elongated, but extended chromosomes (DAPI) along the spindle neither condensed nor separated. These aberrant mitotic chromosomes were highly similar to those found in cut14-208 or cut3-477 mutants deficient in chromosome condensation.
Figure 3.
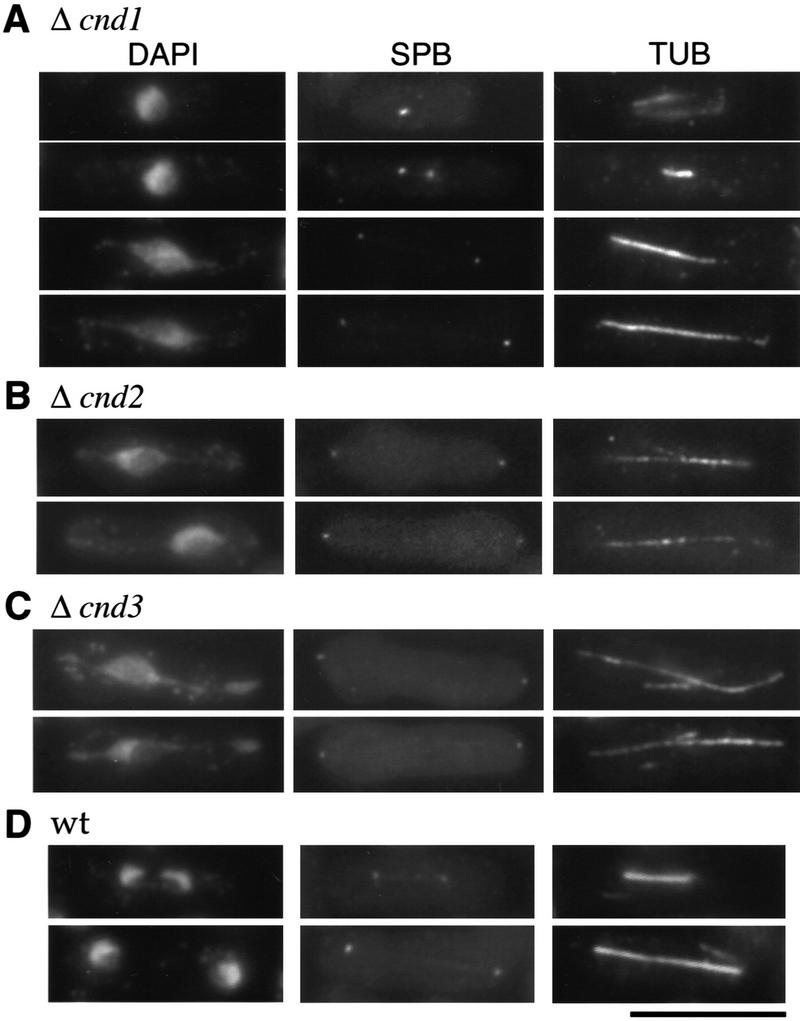
Gene disruption phenotypes of cnd1+, cnd2+, and cnd3+. Gene-disrupted cells were observed after germination by DAPI for DNA, SPB, and TUB antibodies. Mitotic cells were distinguished by the presence of the spindle and the two SPBs. (A) Gene disruption of cnd1+; (B) gene disruption of cnd2+; (C) gene disruption of cnd3+. Chromosome condensation was defective and the spindle was elongated. (D) Wild-type mitotic cells. Bar, 10 μm.
p105, the product of Barren-like cnd2+ is essential for condensation
p105, encoded by cnd2+, consists of 742 amino acids (predicted molecular mass, 83 kD) and resembles Drosophila Barren (Bhat et al. 1996), which has been reported to interact with DNA topo II. Hirano et al. (1997) showed that frog XCAP-H, a component of condensin, was similar to Barren. Related sequences are also present in the databases of budding yeast (YBL097w), Arabidopsis, and human. Portions of the yeast, fly, and frog sequences are compared with Cnd2 in Figure 2B. The amino-terminal domains of the yeast sequences highly resemble each other, whereas the carboxyl termini in all four sequences are similar. These sequences are commonly acidic (pI of Cnd2 is 4.6) as with Cnd1.
Gene disruption of cnd2+ was accomplished by replacement of the 0.9-kb HpaI fragment in the coding region of cnd2+ with the S. pombe ura4+ gene (see Materials and Methods). Sporulation and tetrad dissection of the integrated heterozygous diploid cells (confirmed by Southern blot analysis) indicated that the two Ura− spores were viable, whereas the two Ura+ spores did not produce colonies at 26°C and 33°C, demonstrating that cnd2+ is essential for viability. Disrupted cells were stained with DAPI, SPB, and TUB antibodies. M-phase cells showed defects in both chromosome condensation and separation, indistinguishable from cnd1-null, cut3, and cut14 mutants (Fig. 3B).
Recently, we isolated a temperature-sensitive mutant cnd2–CN385, which was mapped identically at the cnd2 locus and rescued by integration of the cnd2+ gene on the chromosome (Materials and Methods). The temperature-sensitive phenotype of cnd2–CN385 was fully suppressed by plasmid carrying the cnd2+ gene. The mitotic defect of cells cultured at 36°C for 4 hr produced chromosomes deficient in condensation, indistinguishable from that of cnd2 null shown in Figure 3B or cut14-208 at 36°C (Saka et al. 1994).
p100 is the product of cnd3+ required for condensation
The predicted sequence of the cnd3+ gene coding for p100 has 875 amino acids (predicted molecular mass, 101 kD) and is similar to Xenopus XCAP-G and budding yeast YDR325w (see Fig. 2C). A Drosophila partial ORF sequence, resembling Cnd3 is present in the database. They are also acidic proteins (pI of Cnd3 is 4.6). Gene disruption of cnd3+ was done by one-step replacement (Materials and Methods), and heterozygous diploids containing the disrupted gene were verified by Southern hybridization. Analysis revealed that only two Ura− spores were viable, indicating that the cnd3+ gene is also essential for viability. Spores were germinated in liquid culture in the absence of uracil, and germinated cells were observed by DAPI, SPB, and TUB antibody staining (Fig. 3C). Aberrant mitotic cells displaying defects in chromosome condensation were observed at high frequency. Chromosomes were extended along the spindle as seen in disrupted cells of cnd1+, cnd2+, temperature-sensitive cut3, and cut14 mutants. Therefore, the cnd3+ gene is essential for chromosome condensation, like other non-SMC subunits.
T19 Cdc2 kinase site in Cut3 is functionally essential
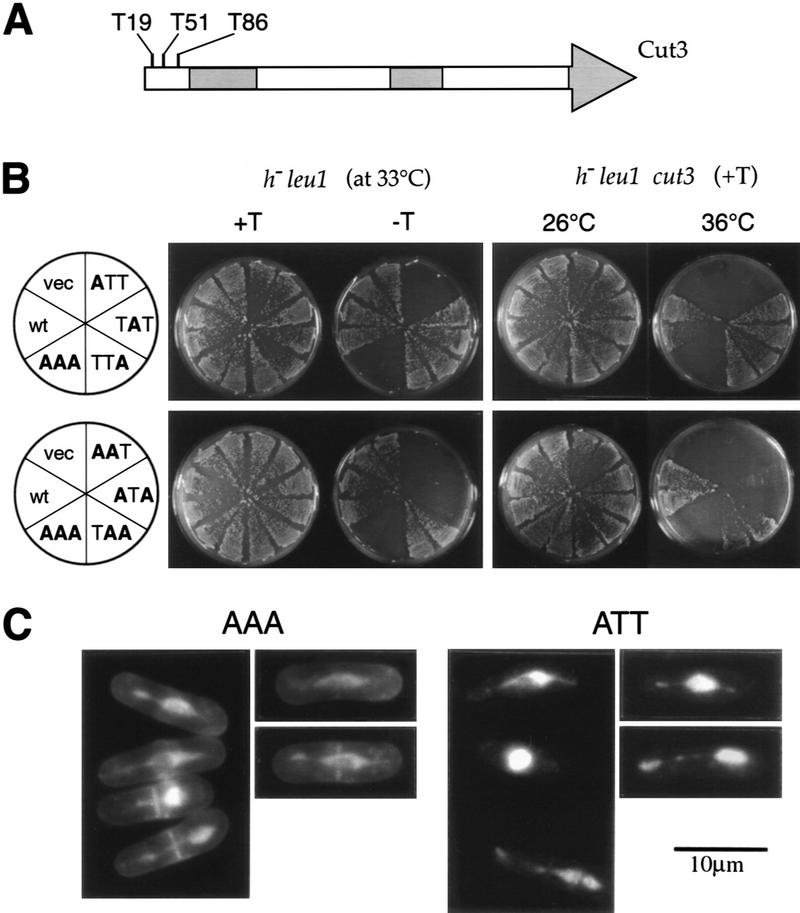
To examine whether fission yeast condensin might be regulated by Cdc2 kinase, we first searched for Cdc2 kinase consensus sites within the sequences of all five components. Three sites exist (19TPDR, 51TPSR, and 86TPSK) in Cut3 and one (47TPRR) in Cnd2, but none in the other three subunits. We then made systematic alanine substitutions for the Cdc2 sites in Cut3 (Fig. 4A), namely, single T19A (ATT), T51A (TAT), and T86A (TTA) and double (AAT, ATA, TAA) or triple (AAA) substitutions. These mutant cut3 genes were placed under the inducible REP81 promoter and expressed in the wild type (h− leu1) at 33°C (Fig. 4B, left). In the presence of thiamine (+T) the promoter is inactive, whereas in the absence of thiamine (−T) the promoter switches on, resulting in the production of ∼10-fold the endogenous level of Cut3; REP81 has the weakest activity among the three promoters (REP81, REP41, and REP1) used (Basi et al. 1993). ATT, AAT, ATA, and AAA mutants were toxic in the absence of thiamine (−T), inhibiting colony formation of the wild-type h− leu1 strain at 33°C, whereas TAT, TTA, and TAA were not inhibitory, indicating that the substitution of T19A was the cause of the inhibition when overproduced. Substitutions of two other Cdc2 sites did not produce any recognizable defect. Observation of cells inhibited by overexpressed T19A mutations AAA or ATT revealed many aberrant mitotic chromosomal DNAs highly similar to those in cut14 or cut3 mutants deficient in chromosome condensation (Fig. 4C). Overproduced T19A mutant protein thus led to the loss of the ability for condensation.
Figure 4.
Dissection of putative Cdc2 kinase sites in Cut3. (A) Cut3 has three Cdc2 kinase sites in the amino terminus. The shaded areas represent homologous regions among SMC proteins: the amino-terminal ATP binding, the central hinge, and the carboxy-terminal DA box. (B) Plasmid carrying the wild-type cut3+ (wt) or one of the substitution mutant genes (see text) placed under the inducible promoter REP81 were introduced into the wild type (h− leu1): resulting transformants were plated at 33°C in the presence of thiamine (+T, the promoter repressed) or the absence of thiamine (−T, the promoter induced). The same plasmids were introduced into cut3–477 mutant and plated at 26°C and 36°C under the repressed promoter condition (+T). (C) Wild-type cells overexpressing triple AAA or single ATT substitution mutant protein under the REP81 promoter were stained by DAPI. Phenotypes were indistinguishable from those of cut3 or cut14 mutant. Bar, 10 μm.
Mutations in the Cut3 T51 or T86 positions (TAT, TTA, or TAA) could still rescue the temperature-sensitive phenotype of the cut3 mutant at 36°C in the presence of thiamine (Fig. 4B; right), but the T19A substitutions (ATT, AAT, ATA, and AAA) failed to suppress the temperature-sensitive phenotype in the presence of thiamine. These results indicated that the T51A and T86A mutants are functional but T19A is not.
T19, the site of Cdc2 kinase in Cut3, is phosphorylated in mitosis
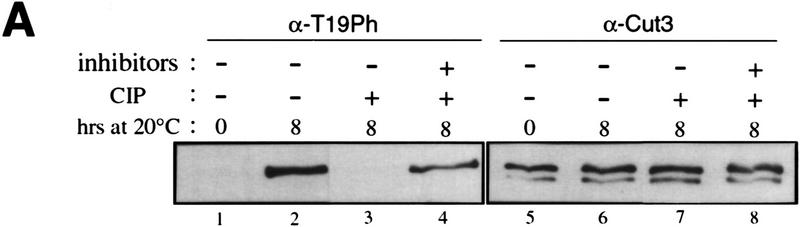
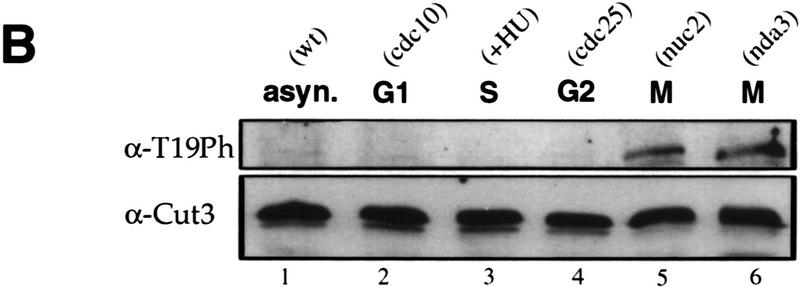
To determine whether T19 is actually phosphorylated, polyclonal antibodies (α-T19PhCut3) were prepared against a chemically synthesized phosphopeptide PSIVDVphosphoT19PDRGER. Mitotically arrested (8 hr at 20°C) cell extracts of nda3–KM311 that contained the integrated cut3+–HA gene were immunoprecipitated by anti-HA antibodies. Immunoprecipitates were immunoblotted by the affinity-purified phosphopeptide antibody α-T19PhCut3 or polyclonal antibody α-Cut3 against a Cut3 recombinant protein (Fig. 5A). An intense band (lane 2) was observed in mitotically arrested extracts (8 hr) by α-T19PhCut3 at the identical position to the major Cut3 band recognized by α-Cut3 (lane 6) but not in interphase cells (0 hr; lane 1). This band disappeared if immunoprecipitates were treated with protein phosphatase [calf intestinal phosphatase (CIP); lane 3], but persisted in the presence of phosphatase inhibitors β-glycerophosphate and p-nitrophenylphosphate (lane 4). The band detected by α-Cut3 showed the same intensity after phosphatase treatment. These results established that α-T19PhCut3 can detect specifically phosphorylated Cut3.
Figure 5.
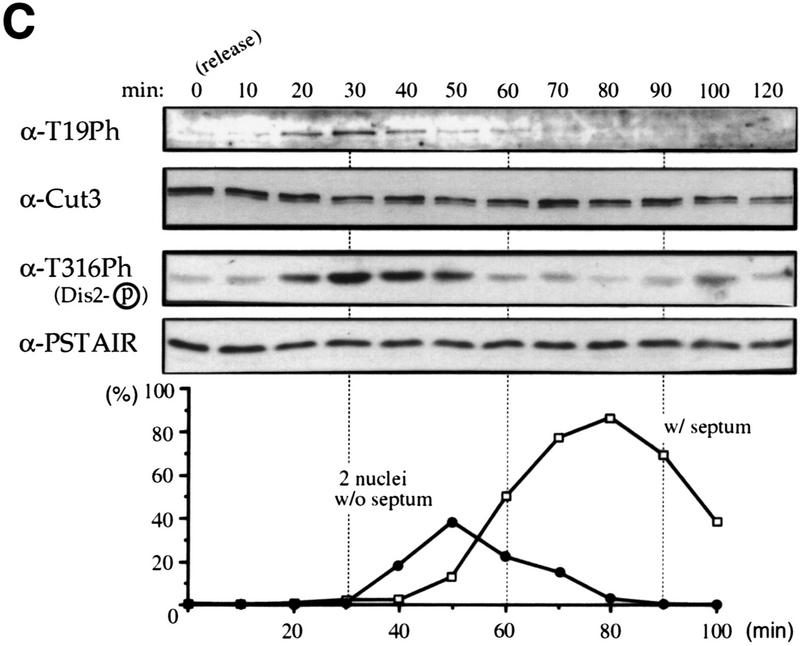
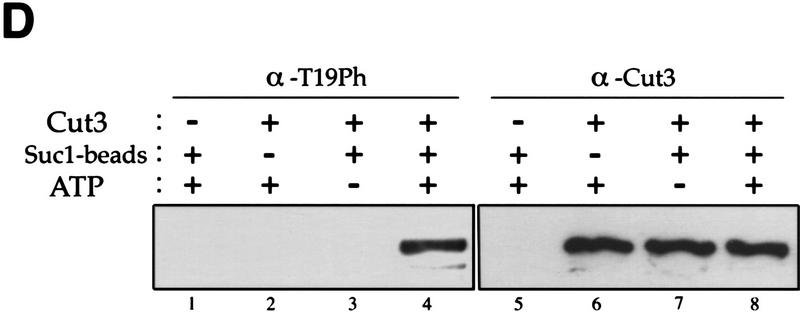
Phosphorylation at T19 identified by phosphopeptide antibodies. (A) Cells of cold-sensitive nda3–KM311 containing the integrated Cut3–3HAH6 gene were grown at 33°C (0 hr) and then arrested in mitosis by shifting to 20°C for 8 hr. Immunoprecipitation was done for extracts by anti-HA antibodies. Precipitates were immunoblotted using phosphopeptide antibodies (α-T19Ph) raised against chemically synthesized phosphopeptide PSIVDVT(PO3H2) PDRGER or polyclonal anti-Cut3 antibodies (α-Cut3). A band corresponding to the full-length Cut3 position recognized by α-T19Ph was obtained only in mitotically arrested cells. α-Cut3 detected the full-length and amino-terminally cleaved forms. Immunoprecipitates prepared above were phosphatase treated (CIP, +) in the presence or the absence of phosphatase inhibitors. The band detected by α-T19Ph disappeared after phosphatase treatment; the band recognized by α-Cut3 remained at the same intensity. (B) Immunoblotting of extracts prepared from exponentially growing wild-type (asyn. wt), G1-arrested cdc10 mutant, HU-treated wild type blocked in S phase, G2-arrested cdc25 mutant, and M-phase-arrested nuc2 and nda3 mutant cells using α-T19Ph and α-Cut3 antibodies. Only M-phase-arrested cell extracts showed the T19-phosphorylated Cut3 band. (C) Synchronous culture by block and release of cdc25 mutant. G2-arrested cdc25–22 cells cultured at 36°C for 4.25 hr were released to 26°C, and cells were collected every 10 min. Extracts were immunoblotted using α-T19Ph, α-Cut3, α-T316Ph (against phosphorylated Dis2 at T316, Ishii et al. 1996) and α-PSTAIR (against Cdc2). The frequencies of cells with the septum and containing two nuclei without the septum were also measured. T19 phosphorylation peaked at metaphase. (D) Purified Cut3 protein (Sutani and Yanagida 1997) was incubated with Cdc2 kinase bound to Suc1 beads in the presence or absence of ATP, and detected by α-T19Ph or α-Cut3 antibodies. Cut3 was in vitro phosphorylated by Cdc2 kinase as detected by α-T19Ph.
Immunoblotting of wild-type and various mutant extracts showed that phosphorylated Cut3 detected by α-T19PhCut3 was also present in nuc2-663 cells arrested at metaphase (Nuc2 codes for an APC/cyclosome subunit), but not in G1-arrested cdc10, S-phase-blocked cells by hydroxyurea (HU), or in G2-arrested cdc25 mutant cells (Fig. 5B).
To further confirm that the T19 site is specifically phosphorylated during mitosis, the block and release experiment of the cdc25 mutant was done (Fig. 5C). The intensity of the band detected by α-T19PhCut3 antibodies became maximal 30 min after the release to 26°C, the same timing for T316 Dis2 phosphorylation (Dis2 is a type 1 phosphatase catalytic subunit and highly phosphorylated at T316 during metaphase; Ishii et al. 1996), whereas the overall intensity of Cut3 and Cdc2 detected by α-Cut3 and PSTAIR antibodies, respectively, remained constant. The frequencies of cells containing two daughter nuclei without septation and cells with the septum, respectively, peaked at 50 and 80 min. These results established that the T19 site is maximally phosphorylated during metaphase.
Although the T19 residue is in the Cdc2 consensus, phosphorylation of T19 by Cdc2 kinase was tested experimentally in vitro. Purified Cut3 (Sutani and Yanagida 1997) was mixed and incubated with Cdc2 kinase bound to Suc1 beads in the presence of ATP (Fig. 5D). α-T19PhCut3 antibodies could recognize Cut3 only after incubation with Cdc2 kinase in the presence of ATP, whereas control α-Cut3 antibodies detected Cut3 under any conditions. These results showed that T19 can be phosphorylated by Cdc2 kinase in vitro.
Non-SMC subunits shuttle between the cytoplasm and the nucleus
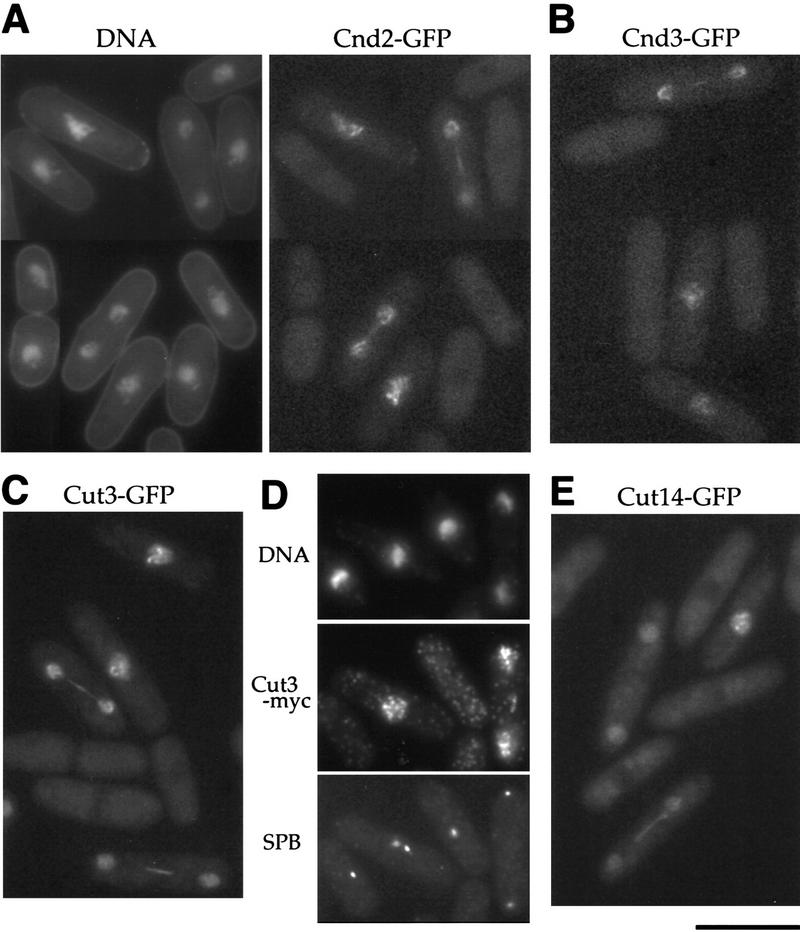
To determine intracellular localization, the cnd2+ gene was tagged with green fluorescent protein (GFP) at the carboxyl terminus and integrated into the chromosome under its native promoter, and its fluorescence was observed in living wild-type cells (Fig. 6A). Cnd2–GFP expressed from the integrated gene appeared to alter its localization dramatically during the cell cycle. The GFP signal was cytoplasmic during interphase but became intensely nuclear during mitosis, judging from comparison with the stained nuclear DNA. Enhanced GFP fluorescence was found throughout the nucleus after mitotic entry and before septation. The rod-like structure seen at the end of nuclear division might represent a narrow midbody-like channel. Then, we examined localization of Cnd3–GFP using plasmid that expresses Cnd3–GFP under the repressed promoter of REP81 (in the presence of thiamine) in the background of the cnd3 null (the plasmid suppresses the cnd3-null phenotype). Cnd3–GFP was highly enriched in the mitotic nucleus like Cnd2–GFP, whereas it was located in the whole cell during interphase (Fig. 6B). Localization of Cnd1, which was tagged with GFP at the carboxyl terminus, integrated on the chromosome, and expressed by the native promoter, was also examined. Fluorescent signals of Cnd1–GFP were located in the cytoplasm during interphase but enriched in the nucleus during mitosis (data not shown). All three non-SMC subunits were thus shuttled between the cytoplasm and the nucleus.
Figure 6.
Cell cycle stage-specific nuclear location of condensin subunits. (A) Living cells expressing integrated Cnd2-GFP (right). The nuclear signal was found in mitotic cells from early mitosis to late anaphase. DNA was stained by Hoechst 33342 (left). (B) cnd3-null cells containing plasmid with the cnd3+–GFP gene under the repressed REP81 promoter in the presence of thiamine. (C) Living wild-type cells expressing Cut3–GFP under the native promoter. (D) Fixed wild-type cells expressing integrated Cut3–Myc. DNA and SPB were stained by DAPI and SPB antibodies, respectively. Anti-myc antibodies were used to detect Cut3–Myc. (E) Living wild type cells expressing integrated Cut14–GFP. Bar, 10 μm.
Cut3 and Cut14 also accumulate into the nucleus during mitosis
The above results suggested that the condensin complex alters localization during the cell cycle. To examine such a notion, localization of two SMC subunits, Cut3 and Cut14, was determined. For this purpose, the cut3+–GFP gene under its native promoter was integrated into the chromosome, and Cut3–GFP was observed in living cells. Cut3–GFP was found to shuttle between the nucleus in mitosis and the cytoplasm in interphase (Fig. 6C). This cytoplasmic–nuclear mobilization was confirmed by immunofluorescence microscopy of fixed cells integrated with the cut3+ gene tagged with Myc using anti-myc and SPB antibodies (Fig. 6D). Cut3–Myc was found in the nucleus of mitotic cells (containing two SPBs). Similar results were obtained for Cut14-GFP, the gene of which was integrated and expressed under the native promoter (Fig. 6E). These results showed that the two SMC subunits also accumulate into the nucleus during mitosis and behave similar to non-SMC subunits. These results are not in agreement with our previous observations obtained by polyclonal anti-cut3 antibody staining, which revealed nuclear localization throughout the cell cycle (Saka et al. 1994). Although affinity purification of the antibodies was done in the previous study, reexamination of those antibodies indicated that the signal detected previously was due to a contaminating nuclear antigen. Localization of the present GFP- and Myc-tagged Cut3 is consistent with that of the tagged Cut14, Cnd1, Cnd2, and Cnd3.
Mitotic nuclear entry of condensin subunits depends on T19 Cdc2 site
To determine whether mitotic phosphorylation of Cut3 is involved in cell cycle-dependent localization, the alanine-substituted T19A Cut3 tagged with GFP was expressed in wild-type cells under the REP81 promoter in the absence of thiamine. Interestingly, Cut3 T19A–GFP signal did not accumulate in the nucleus during mitosis (Fig. 7A). Mitotic chromosomes were aberrantly stretched instead, apparently defective for condensation (indicated by arrow). Consistent with this dominant-negative effect of overproduced T19A Cut3, integrated Cut14–GFP in cells, which overexpressed T19A Cut3, was also not able to accumulate in the nucleus (Fig. 7B), suggesting that Cut14–GFP colocalizes with Cut3–T19A by direct association. Therefore, the T19 phosphorylation site might play a role in correctly localizing condensin components during mitosis.
Figure 7.
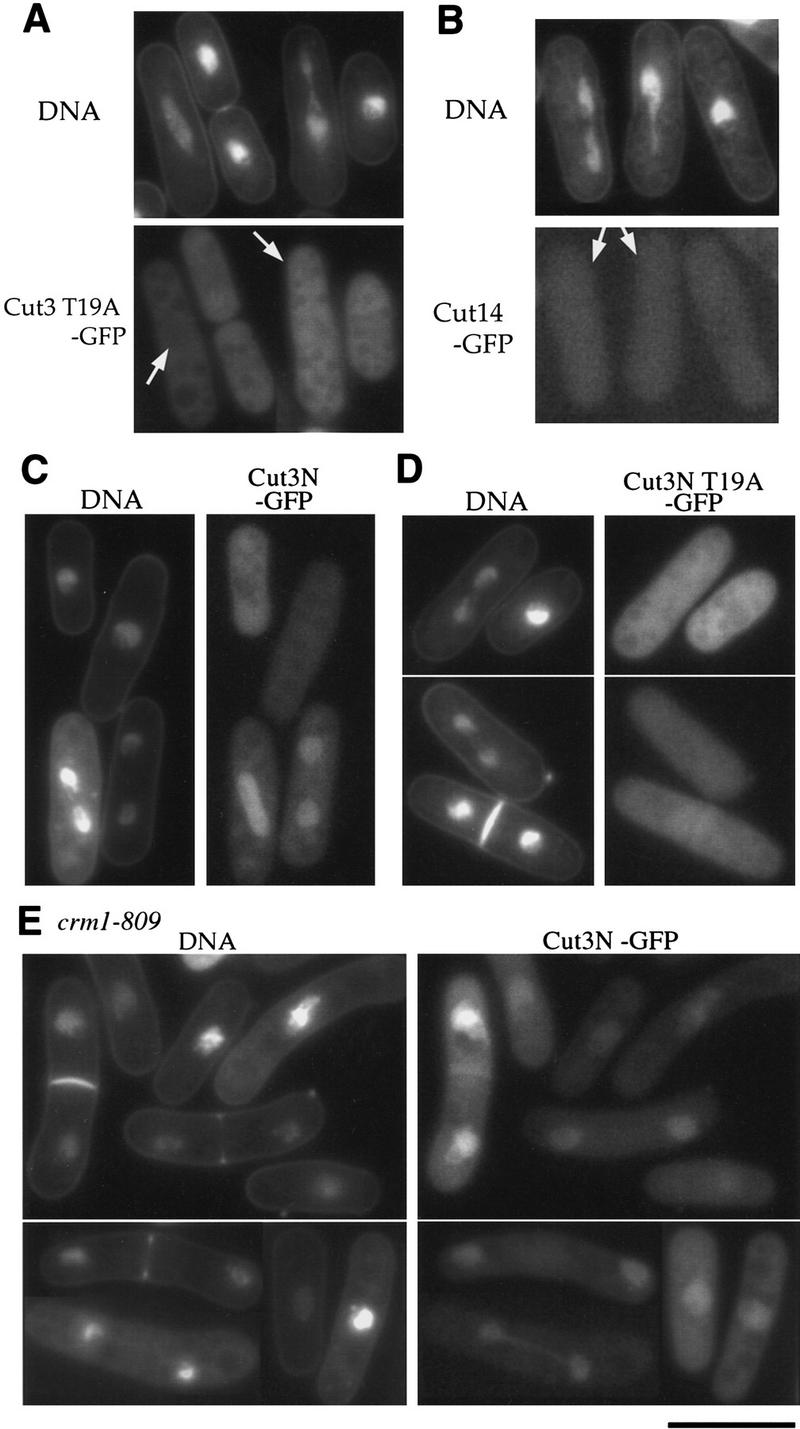
Cell cycle stage-specific nuclear accumulation depends on T19 of Cut3 and Crm1. (A) Living wild-type cells expressing T19A Cut3 under the REP81 promoter in the absence of thiamine. Cut3 T19A was cytoplasmic. DNA was stained by Hoechst 33342 in A–E. (B) Localization of Cut14–GFP in cells overproducing T19A Cut3 mutant protein under the REP81 promoter. Cut14–GFP was cytoplasmic. (C) The amino-terminal fragment of Cut3 tagged with GFP was expressed in wild-type cells. The nuclear signal was found in mitotic cells. (D) The amino-terminal fragment of Cut3 substituted with alanine at T19 and tagged with GFP was expressed in wild-type cells. GFP signal was cytoplasmic. (E) The amino-terminal fragment of Cut3 tagged with GFP was expressed in crm1-809 cultured at the restrictive temperature (20°C) for 6 hr. Crm1 is required for nuclear export. GFP signal accumulated in both interphase and mitotic nuclei. Bar, 10 μm.
Cut3 protein required for cell cycle-regulated localization
To examine whether Cut3 is required for localization of other condensin subunits in mitotic nuclei, mutant cells that deleted the cut3+ gene but expressed Cnd2–GFP were observed. To this end, a heterozygous diploid strain containing a cut3 null allele (Saka et al. 1994) was integrated chromosomally with the cnd2+–myc gene (integration confirmed by PCR) and sporulated. Cells germinated in the culture medium lacking both leucine and uracil produced Cnd2–Myc in the genetic background of cut3 null. In these cells stained with anti-Myc antibodies, Cnd2–Myc was found to be cytoplasmic throughout the cell cycle. Nuclei in germinated cells, which showed aberrant, condensation-defective mitotic chromosomes, lacked the signal for Cnd2–Myc (data not shown). Control cells germinated in the culture medium containing uracil, which produced Cut3 protein, showed the nuclear Cnd2-Myc signal in mitotic cells. Cut3 is thus required for nuclear accumulation of Cnd2 in mitosis.
We found that the amino-terminal region of Cut3 has an important role for cell cycle-regulated localization. A small truncated amino-terminal 1- to 528-amino acid fragment expressed in wild-type cells was still able to shuttle between the nucleus and the cytoplasm (Fig. 7C). Only mitotic cells contained the nuclear GFP signal. The amino-terminal fragment was thus sufficient for cell cycle-dependent nuclear accumulation. However, the same 1- to 528-amino acid fragment, but containing the T19A substitution, expressed in the wild type could not accumulate in the mitotic nuclei (Fig. 7D), resulting in GFP signal throughout the cell. The ability of the fragment for nuclear localization may thus require mitotic phosphorylation of T19.
An intriguing result was observed when the 1- to 528-amino acid fragment was expressed in a crm1–809 mutant at the restrictive temperature for 6 hr. It became localized in the nucleus at different cell cycle stages, including interphase (Fig. 7E). The crm1+ gene is essential for nuclear export (e.g., Fukuda et al. 1997). Addition of leptomycin B (an inhibitor of Crm1) yielded a similar result. The amino-terminal-region of Cut3 contains one putative nuclear export signal (NES), 480LNSEIADLSL489. Whether full-length Cut3 and other condensin subunits are also controlled by the crm1+ gene is under investigation.
Discussion
This study shows that the five-member condensin complex, first identified in frog (Hirano et al. 1997), is present in S. pombe, a eukaryotic microbe in which mitotic chromosome condensation takes place, albeit to a lesser extent than in higher eukaryotes (Umesono et al. 1983; Saka et al. 1994). The S. pombe condensin complex was isolated by immunoprecipitation and analyzed by microsequencing. It contains, in addition to the two known SMC subunits Cut3 and Cut14, three more subunits—Cnd1, Cnd2, and Cnd3. The calculated total molecular mass of the fission yeast condensin subunits (without the tag) was 600 kD. Sucrose gradient centrifugation indicated that the majority of the condensin subunits detected by anti-Cut3 and anti-Cut14 antibodies sedimented at ∼13S (Sutani and Yanagida 1997), identical to the value reported for the frog condensin (Hirano et al. 1997); this demonstrated that a greater part of SMC subunits are in the form of the complex. A spherical protein complex of 600 kD should sediment around 17S–18S, suggesting that the fission yeast condensin has an asymmetrical shape, perhaps reflecting the coiled–coil structure of SMC proteins (Melby et al. 1998). The Cut3–Cut14 complex (calculated molecular mass, 290 kD) formed by overproduction of the two proteins peaked around the 8S position (Sutani and Yanagida 1997).
Cnd1, Cnd2, and Cnd3 are similar to frog XCAP-D2, XCAP-H, and XCAP-G, respectively (Hirano et al. 1997; Kimura et al. 1998; GenBank accession no. AF111423). In this study we showed for the first time that these non-SMC subunits of condensin are essential for cell viability and also for mitotic chromosome condensation in vivo. These results are important in understanding condensation mechanisms, because the role of condensin in condensation has been explained often in relation to the shape and motifs of the SMC subunits. The Barren-like Cnd2 attracts attention because Drosophila Barren is reported to interact with topo II (Bhat et al. 1996). The relaxing activity of topo II is enhanced by the addition of Barren. The frog condensin isolated from mitotic cells demonstrates ATP-dependent activity for positive supercoiling of DNA (Kimura and Hirano 1997). It remains to be determined whether S. pombe condensin shows the same in vitro activity; currently we are trying to purify condensin by overproducing the five gene products within the same cells. Therefore the in vitro DNA renaturation activity reported for the Cut3–Cut14 complex should represent only a part of the whole activity of condensin, and the positive supercoiling activity, which was not detected in the Cut3–Cut14 complex, might require non-SMC subunits.
One common property of the non-SMC subunits of fission yeast and other organisms is that they are acidic (pI < 5.5). They may interact with basic components in chromatin such as histones or high mobility group proteins. Interestingly, only Cut14 is basic (pI = 8.1) among the five subunits, and Cut14 appears to be directly bound to DNA in vitro (T. Sutani and M. Yanagida, unpubl.). The essential role of non-SMC Cnd1 and Cnd3 is still not well understood. PSI–BLAST search (Altschul et al. 1997) revealed a weak homology between Cnd1 or Cnd3 and the β subunit of the AP3 complex (Fig. 8A). The adapter protein (AP) complex is required for vesicle transport and is involved in the assembly of clathrin (Schmid 1997; Odorizzi et al. 1998). Homology exists between the AP3–β subunit and Cnd1 and Cnd3 in the different regions spanning ∼200 amino acids. Note that clathrin consists of two rod-like regions linked by a hinge-like region called knee (Schmid 1997), and the role of AP3–β is presumed to assist in the assembly of rod-like clathrin molecules. Similarly, Cnd1 and Cnd3 might serve for the correct assembly of the rod-like Cut3 and Cut14 subunits (and possibly also Cnd2) into a higher-order structure (a cartoon is shown in Fig. 8B). When purified Cut3 was mixed with Cut14 in vitro, they formed a stable complex but at a rather low efficiency (Sutani and Yanagida 1997). Cnd1 and Cnd3 might have a role in linking the subunits together into the functional condensin architecture. The Drosophila Barren-like Cnd2 might have a specific role in interacting with chromosomal DNA.
Figure 8.
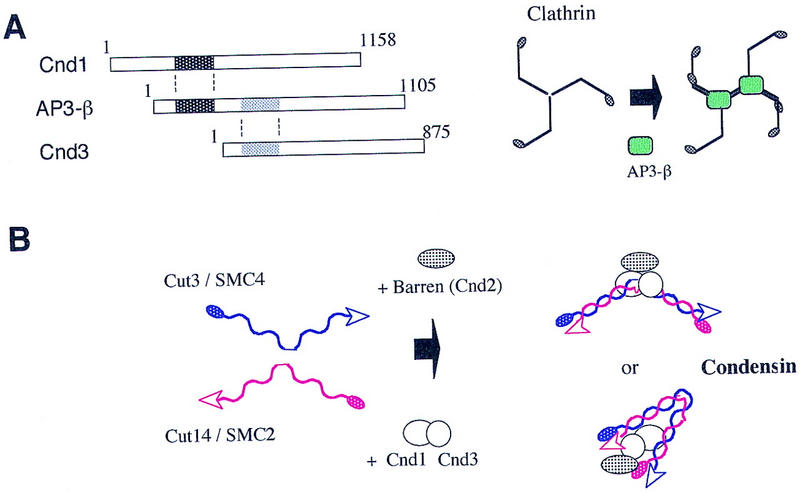
A hypothesis for condensin assembly. (A) Weak homology exists between AP3-β and Cnd1 or Cnd3 (hatched). AP3-β assists in the assembly of clathrin (Schmid 1997), which contains rod and hinge regions. (B) Cnd1 and Cnd3 might assist in the assembly of Cut3, Cut14, and Cnd2 proteins into the five-member condensin complex.
All five subunits tagged with GFP were found to dramatically alter their localization during the cell cycle, the patterns of which were very similar to each other, probably reflecting the behavior of a complex rather than free subunits. Because these genes, except for cnd3+, were integrated into the chromosome and could thus substitute for the wild-type genes, their behavior should reflect the actual localization changes of the functional condensin complex in vivo. The S. pombe condensin accumulates in the nucleus at an early stage of mitosis and reduces its nuclear level at the end of mitosis. The role for condensation is consistent with nuclear localization in early to mid-mitosis of S. pombe, as fungal mitosis occurs within the closed nucleus. Condensin might be needed for chromosome decondensation, which occurs in late anaphase and telophase, although such a concept has no experimental support yet. It would be difficult to discern such a role in mutant cells, as condensation defects would appear first.
What is the possible physiological significance for shuttling of condensin subunits between the cytoplasm and the nucleus? In frog- and chicken-cultured cells, the majority of condensin SMC subunits are localized in nuclei even in interphase (Hirano and Mitchison 1994; Saitoh et al. 1994) so that the S. pombe localization behavior is not universal. A possible explanation is that the S. pombe condensin complex might be active throughout the cell cycle so that its nuclear localization has to be regulated and occurs only in mitosis. In frog, however, it has been established that XCAP-D2 and XCAP-H subunits have to be phosphorylated to activate the in vitro condensin activity (Kimura et al. 1998). Consistent with this, XCAP-D2 has multiple Cdc2 sites that Cnd1 does not have. Alternatively, the S. pombe condensin may change its localization if its association in the interphase chromosome interferes with other chromatin proteins such that it would inhibit certain DNA metabolisms during the S and G2 phases. Another possibility is that the S. pombe condensin activity is regulated, as in the frog, and its shuttling between the nucleus and the cytoplasm may represent dispensable behavior. Regulation of Cnd2 may occur by phosphorylation because the amino terminal Cdc2 site is present as in the frog homolog.
We showed that the T19 site of Cut3 is actually phosphorylated in mitosis, perhaps by Cdc2 kinase, peaking at metaphase, and that the T19 phosphorylation is essential for mitotic accumulation of condensin components in the nucleus. The T19A substitution mutant protein remains in the cytoplasm throughout the cell cycle and fails to rescue the temperature-sensitive phenotype of the cut3 mutant. However, the mechanism by which phosphorylation of T19 leads to nuclear accumulation of Cut3 is still unclear. T19-phosphorylated Cut3 might acquire the ability to interact with other proteins, which ensures nuclear accumulation. Our results suggest that the short amino-terminal sequence of Cut3 contains the nuclear export signal (e.g., Fukuda et al., 1997), but it remains to be determined whether the signal controls localization of the condensin complex. After Cdc2 kinase is inactivated in anaphase, the S. pombe condensin complex is still briefly retained until septation. An additional unknown mechanism might be needed to retain condensin in the postanaphase nucleus.
In summary, this study demonstrates that all of the non-SMC subunits of condensin are absolutely required for mitotic condensation and that Cdc2 kinase promotes chromosome condensation through phosphorylation of the Cut3/SMC4 subunit, which ensures proper cellular positioning during the cell cycle in fission yeast. Whether the regulated activation of condensin by phosphorylation occurs in this organism during mitosis remains to be determined.
Materials and methods
Strains and media
An S. pombe haploid wild-type strain 972 h− and its derivative strains were used: cut3-477, cut14-208 (Saka et al. 1994), cdc10-129, cdc25-22 (Nurse et al. 1976; Fantes 1979), nda3-KM311, and nuc2-663 (Hiraoka et al. 1984; Hirano et al. 1988). For gene disruption, a diploid strain was made by crossing between haploids ST485 (h+ leu1 ura4 ade6–210) and ST486 (h− leu1 ura4 ade6–216). The complete YPD medium (1% yeast extract, 2% BactoPeptone, and 2% glucose) and the minimal EMM2 (Mitchison 1970) were used. The CN385 strain was isolated by its characteristic nuclear phenotype in cells cultured at 36°C for 4 hr and stained by DAPI (Yamashita et al. 1999, and unpubl.). The temperature-sensitive phenotype was suppressed by plasmid carrying the cnd2+ gene but not by those carrying the cnd1+ or the cnd3+ gene. Integration rescue of the temperature-sensitive phenotype showed that CN385 was an allele of the cnd2 locus. To produce Cnd2–Myc in cut3 null, a heterozygous diploid containing a cut3 null allele (the ura4+ marker; Saka et al. 1994) was transformed with the integration plasmid containing the cnd2+–myc gene (the Saccharomyces cerevisiae LEU2 marker). Integration was confirmed by the PCR method. Resulting integrant diploid cells were sporulated, and spores were germinated in the culture medium lacking both uracil and leucine or lacking only leucine. Spores containing Cnd2–Myc in cut3 null could germinate in the medium lacking both uracil and leucine. Cells were fixed and stained by anti-Myc and SBP antibodies 10 hr after germination.
Plasmids
Plasmid pREP1 carrying the nmt1 promoter and its derivative pREP81 (Maundrell 1990; Basi et al. 1993) were used for inducible overproduction. Site-directed mutagenesis was performed by the Sculptor in vitro mutagenesis system (Amersham). All constructs were verified by nucleotide sequencing.
Epitope tagging of cut3+ and cut14+
NotI restriction sites were introduced at the carboxyl termini of cut3+ and cut14+ genes by the PCR method, and the resulting carboxyl termini were ligated with a triple tandem HA plus hexahistidine (3HA+His6) epitope. Integration plasmid pST337, a derivative of pBluescript KS(+), containing a part of the cut3+ coding region (1.4 kb PstI–NotI), 3HA+His6 tag, and the S. pombe ura4+ gene, was used for transformation of the strain h− ura4 cut3–477. Stable Ura+/Ts+ transformants were obtained, and one of them was designated Cut3–3HAH6. In this cell lysate, the epitope-tagged Cut3 protein with the expected molecular mass was detected by immunoblotting. Another integration plasmid pST1421, a derivative of pBluescript KS(+), containing a part of the cut14+ coding region (2.3-kb XhoI–NotI), 3HA+His6 tag, and the S. pombe ura4+ gene, was used for chromosomal integration in the strain of cut14–208. The resulting Ura+/Ts+ integrant strain was designated Cut14–3HAH6. GFP (S65T) and 8× Myc tag, instead of 3HA+His6 epitope, were used for GFP and Myc tagging, respectively.
Immunoprecipitation and peptide sequencing
Exponentially growing cells (3 × 108) of Cut3–3HAH6 or Cut14–3HAH6 strains were lysed in 200 μl of HB buffer [25 mm Tris-HCl (pH 7.5), 15 mm MgCl2, 15 mm EGTA, 60 mm β-glycerophosphate, 15 mm p-nitrophenylphosphate, 0.1 mm NaF, 0.5 mm Na3VO4, 1 mm DTT, 0.1% NP-40, 1 mm PMSF] containing protease inhibitors (1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin A). Extracts were centrifuged twice (10 and 20 min, 17,000g), and BSA (Boehringer) was added to a final concentration of 5 mg/ml. Extracts were incubated with protein A–Sepharose (50 μl) for 1 hr and then with mAb 12CA5 cross-linked to protein A–Sepharose (10 μl of 12CA5 beads) for 3 hr. Beads were washed five times with 1 ml of HB buffer, and proteins eluted with 20 mm NaOH were separated on an SDS–polyacrylamide gel and detected by immunoblotting using anti-cut3 and anti-HA antibodies or silver staining. To isolate p128, p105, and p100 polypeptides, Cut3–3HAH6 cells (5 × 1010) were broken in 17 ml of HB buffer containing protease inhibitors. Extract was centrifuged twice (20 min at 20,000g) and incubated with protein A–Sepharose (3 ml) for 1 hr with 0.4 ml of 12CA5 beads for 4 hr. Beads were washed seven times with 10 ml of HB. Proteins bound to the beads were eluted with 20 mm NaOH and separated on an SDS–polyacrylamide gel. Protein bands visualized by Coomassie brilliant blue R250 were excised and treated with 0.2 μg of a lysyl bond-specific protease, Achromobacter protease I [a gift from Dr. Masaki (Masaki et al. 1981)] in 0.1 m Tris-HCl (pH 9.0) containing 0.1% SDS and 1 mm EDTA at 37°C overnight. The peptides generated were extracted from the gel and separated on tandem columns of DEAE-5PW (2 × 20 mm; Tosoh) and Mightysil RP-18 (2 × 50 mm; Kanto Chemical) with a model 1100 liquid chromatograph (Hewlett Packard). Selected peptides were subjected to Edman degradation using a model 477A automated protein sequencer (Perkin Elmer) and to matrix-assisted laser desorption ionization time of flight mass spectrometry on a Reflex (Bruker-Franzen Analytik).
Isolation of cnd1+, cnd2+, and cnd3+
To isolate the genomic fragment of the cnd1+ gene, mixed oligonucleotides were made as primers for degenerated PCR. They were 5′-CA(AG)TA(CT)AT(CAT)GA(CT)AT(CAT)GTIGA-3′ (encoding QYIDIVE) and 5′- AC(AG)TG(AG)TCCCA(CT)TGIC(TG)(CT)TGIGT-3′(encoding TQRQWDHV on the complementary strand). An 1.8-kb genomic DNA fragment was cloned and used as the hybridization probe for an S. pombe cosmid library. Four cosmids (35, 776, 1509, 1699; Mizukami et al. 1993) were obtained, which were located on the long arm of chromosome II. Subcloning showed that the 4.7-kb SacI–BamHI + 2.6-kb BamHI fragment contained the coding sequence for cnd1+. The nucleotide sequence of the isolated cnd1+ gene was determined with the DyeDeoxy Terminator Cycle Sequencing Kit, 373A sequencer and 310 Genetic Analyser (Perkin Elmer). Peptide sequences derived from p105 matched 100% with an S. pombe hypothetical ORF located on cosmid 306 and 1681 (the short arm of chromosome III). The subcloned 3.8-kb BamHI–BlnI fragment contained the coding sequence for cnd2+. Peptide sequences derived from p100 matched 100% with an S. pombe hypothetical ORF, SPAC1B9.04. It is present in cosmids 1B9, 188, 1434, and 1762 (the long arm of chromosome III). The 4.8-kb BlnI–HpaI fragment contained the coding region for cnd3+. The nucleotide sequence data of cnd1+, cnd2+, and cnd3+ will appear in the DDBJ/EMBL/GenBank nucleotide sequence databases with the accession numbers AB030212, AB030213, and AB030214, respectively.
Gene disruption
One-step gene replacement (Rothstein 1983) was used. For cnd2 gene disruption, a 0.9-kb HpaI fragment in the coding region of cnd2+ was replaced with the 1.8-kb S. pombe ura4+ gene, and the resulting disrupted cnd2 gene was introduced into the chromosome of diploid cells by homologous recombination. Gene disrupted cells yielded 4.8 kb of ClaI and 8.0 kb of NcoI fragments by genomic Southern hybridization. For cnd3 gene disruption, a DNA fragment was used, in which the 1.3-kb XbaI–SpeI region in cnd3+ was replaced with the ura4+ gene. Gene-disrupted cells yielded the expected 5.1-kb BglII fragment by genomic Southern hybridization. For cnd1 gene disruption, the ura4+ gene was inserted at the first NspV site within the cnd1 gene, and resulting fragment was used. Gene-disrupted heterozygous diploids showed the expected 8.2-kb PstI fragment by Southern hybridization. They were sporulated and the tetrads dissected. To examine the phenotype of gene-disrupted cells, heterozygous diploid cells were sporulated at 26°C. Ura+ (gene-disrupted) spores were germinated in EMM2 liquid medium lacking uracil at 33°C. Germinated cells were collected after 6, 10, and 14 hr, fixed, and stained with DAPI and SPB and TUB antibodies as described (Hagan and Hyams 1988; Funabiki et al. 1993). TUB antibody (TAT1) was the gift of Dr. K. Gull (University of Manchester, UK).
Immunochemical methods
Anti-T19Ph Cut3 antibodies were obtained by injecting phosphopeptide PSIVDV T(PO3H2) PDRGER (Peptide Institute, Inc., Osaka, Japan) conjugated with keyhole limpet cyanin (KLH) to a rabbit. Antibodies were affinity purified by using phosphopeptide coupled to FMP-activated cellulofine (Seikagaku) and used for immunoblotting. CIP treatment of immunoprecipitants was performed as described (Pereia et al. 1998). As phosphatase inhibitors, 60 mm β-glycerophosphate and 15 mm p-nitrophenylphosphate were used. Antibodies against the carboxy-terminal phosphorylated sequence T316Ph Dis2 and Anti-Cut3 antibodies were described previously (Saka et al. 1994; Ishii et al. 1996). PSTAIR was the gift of Drs. M. Yamashita (Hokkaido University, Sapporo, Japan) and Y. Nagahama (National Institute for Basic Biology, Okazaki, Japan).
Light microscopy
Strains expressing GFP-fused proteins were cultured at 26°C in EMM2 medium to log phase, mounted directly on a glass slide without fixation, covered with a coverslip, and observed by a Zeiss Axiophot microscope, using a Zeiss blue filter set and a Hamamatsu C5985 Chilled CCD camera. For visualizing DNA, Hoechst 33342 was added to a final concentration of 50 μg/ml immediately before observation. Immunolocalization was performed using anti-Myc mAb (9E10; Calbiochem) based on procedures described previously (Hagan and Hyams 1988). The fixation method using glutaraldehyde was employed.
Acknowledgments
This work was supported by the CREST Research Project of the Japan Science Technology (JST) Corporation and a grant from the Human Frontier Science Program Organization (HFSPO). T.S. is the recipient of a special fellowship of the Japan Science Promotion Society. We thank Dr. Ruth T. Yu for editing the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
This paper is dedicated to the memory of Dr. Kazuhiko Umesono, deceased on April 12, 1999.
E-MAIL yanagida@kozo.biophys.kyoto-u.ac.jp; FAX 81 75 753 4208.
References
- Adachi Y, Luke M, Laemmli UK. Chromosome assembly in vitro: Topoisomerase II is required for condensation. Cell. 1991;64:137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak AL, Zeuthen J, Crick FH. Higher-order structure of human mitotic chromosomes. Proc Natl Acad Sci. 1977;74:1595–1599. doi: 10.1073/pnas.74.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Bhat MA, Philp AV, Glover DM, Bellen HJ. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell. 1996;87:1103–1114. doi: 10.1016/s0092-8674(00)81804-8. [DOI] [PubMed] [Google Scholar]
- Fantes P. Epistatic gene interactions in the control of division in fission yeast. Nature. 1979;279:428–430. doi: 10.1038/279428a0. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hirano T. SMC protein complexes and higher-order chromosome dynamics. Curr Opin Cell Biol. 1998;10:317–322. doi: 10.1016/s0955-0674(98)80006-9. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosome assembled in Xenopus egg extracts. J Cell Biol. 1993;120:601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A heteromeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- Hirano T, Hiraoka Y, Yanagida M. A temperature-sensitive mutation of the Schizosaccharomyces pombe gene nuc2+ that encodes a nuclear scaffold-like protein blocks spindle elongation in mitotic anaphase. J Cell Biol. 1988;106:1171–1183. doi: 10.1083/jcb.106.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: A cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Ishii K, Kumada K, Toda T, Yanagida M. Requirement for PP1 phosphatase and 20S cyclosome/APC for the onset of anaphase is lessened by the dosage increase of a novel gene sds23+ EMBO J. 1996;15:6629–6640. [PMC free article] [PubMed] [Google Scholar]
- Jessberger R, Frei C, Gasser SM. Chromosome dynamics: The SMC protein family. Curr Opin Genet Dev. 1998;8:254–259. doi: 10.1016/s0959-437x(98)80149-4. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: A biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Masaki T, Tanabe M, Nakamura K, Soejima M. Studies on a new proteolytic enzyme from Achromobacter lyticus M497-1. I. Purification and some enzymatic properties. Biochim Biophys Acta. 1981;660:44–50. doi: 10.1016/0005-2744(81)90106-6. [DOI] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: Long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison JM. Physiological and cytological methods for Schizosaccharomyces pombe. Methods Cell Physiol. 1970;4:131–165. [Google Scholar]
- Mizukami T, Chang WI, Garkavtsev I, Kaplan N, Lombardi D, Matsumoto T, Niwa O, Kounosu A, Yanagida M, Marr TG, Beach D. A 13kb resolution cosmid map of the 14Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell. 1993;73:121–132. doi: 10.1016/0092-8674(93)90165-m. [DOI] [PubMed] [Google Scholar]
- Murray AW. How to compact DNA. Science. 1998;282:425–427. doi: 10.1126/science.282.5388.425. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen & Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Cowles CR, Emr SD. The AP-3 complex: A coat of many colours. Trends Cell Biol. 1998;8:282–288. doi: 10.1016/s0962-8924(98)01295-1. [DOI] [PubMed] [Google Scholar]
- Pereira G, Knop M, Schiebel E. Spc98p directs the yeast γ-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of spindle pole body. Mol Biol Cell. 1998;9:775–793. doi: 10.1091/mbc.9.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein RJ. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Goldberg IG, Wood ER, Earnshaw WC. ScII: An abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Goldberg I, Earnshaw WC. The SMC proteins and the coming of age of the chromosome scaffold hypothesis. BioEssays. 1995;17:759–766. doi: 10.1002/bies.950170905. [DOI] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL. Clathrin-coated vesicle formation and protein sorting: An integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV. SMC proteins and chromosome structure. Trends Cell Biol. 1998;8:454–459. doi: 10.1016/s0962-8924(98)01370-1. [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes & Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yanagida M. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature. 1997;388:798–801. doi: 10.1038/42062. [DOI] [PubMed] [Google Scholar]
- Uemura T, Yanagida M. Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: Uncoordinated mitosis. EMBO J. 1986;5:1003–1010. doi: 10.1002/j.1460-2075.1986.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura Y, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Umesono K, Hiraoka Y, Toda T, Yanagida M. Visualization of chromosomes in mitotically arrested cells of the fission yeast Schizosaccharomyces pombe. Curr Genet. 1983;7:123–128. doi: 10.1007/BF00365637. [DOI] [PubMed] [Google Scholar]
- Yamashita, Y.M., Y. Nakaseko, K. Kumada, T. Nakagawa, and M. Yanagida. 1999. Fission yeast APC/cyclosome subunits, Cut20/Apc4 and Cut23/Apc8, in regulating metaphase-anaphase progression and cellular stress responses. Genes Cells (in press). [DOI] [PubMed]
- Yanagida M. Fission yeast cut mutations revisited: Control of anaphase. Trends Cell Biol. 1998;8:144–149. doi: 10.1016/s0962-8924(98)01236-7. [DOI] [PubMed] [Google Scholar]