Abstract
Cytoplasmic polyadenylation element binding (CPEB) proteins bind to and regulate the translation of specific mRNAs. CPEBs from Xenopus, Drosophila, and Spisula participate in oogenesis. In this report, we examine the biological roles of all identifiable CPEB homologs in a single organism, Caenorhabditis elegans. We find four homologs in the C. elegans genome: cbp-1, cpb-2, cpb-3, and fog-1. Surprisingly, two homologs, CPB-1 and FOG-1, have key functions in spermatogenesis and are dispensable for oogenesis. CPB-2 and CPB-3 also appear not to be required for oogenesis. CPB-1 is essential for progression through meiosis: cpb-1(RNAi) spermatocytes fail to undergo the meiotic cell divisions. CPB-1 protein is present in the germ line just prior to overt spermatogenesis; once sperm differentiation begins, CPB-1 disappears. CPB-1 physically interacts with FBF, another RNA-binding protein and 3′ UTR regulator. In addition to its role in controlling the sperm/oocyte switch, we find that FBF also appears to be required for spermatogenesis, consistent with its interaction with CPEB. A second CPEB homolog, FOG-1, is required for specification of the sperm fate. The fog-1 gene produces fog-1(L) and fog-1(S) transcripts. The fog-1(L) RNA is enriched in animals making sperm and is predicted to encode a larger protein; fog-1(S) RNA is enriched in animals making oocytes and is predicted to encode a smaller protein. The relative abundance of the two mRNAs is controlled temporally during germ-line development and by the sex determination pathway in a fashion that suggests that the fog-1(L) species encodes the active form. In sum, our results demonstrate that, in C. elegans, two CPEB proteins have distinct functions in the germ line, both in spermatogenesis: FOG-1 specifies the sperm cell fate and CPB-1 executes that decision.
Keywords: CPEB proteins, C. elegans, spermatogenesis, FOG-1, CPB-1, translational control
The utilization of mRNAs is highly controlled in eukaryotes. Regulation of mRNA localization, stability, and translation determines when, where, and how much protein an mRNA produces. The RNA elements that control these processes often reside in the 3′ untranslated region (3′ UTR) and interact with specific proteins, forming sophisticated regulatory circuits (for review, see Wickens et al. 2000). Many of the regulatory proteins are conserved across species. Moreover, within a single species, multiple members of a single family often exist; whether these family members possess distinct or overlapping functions is an important problem in understanding 3′ UTR-mediated regulation.
We focus here on one multigene family of 3′ UTR regulators, relatives of cytoplasmic polyadenylation element binding (CPEB) protein. CPEB binds to cytoplasmic polyadenylation elements (CPEs) in the 3′ UTRs of specific maternal mRNAs and controls their translational activity (Richter 2000). CPEB family members are characterized by two RRM motifs followed by a diagnostic zinc finger region composed of C4 and C2H2 motifs (called the C/H domain). Both the RRM motifs and C/H domains are required for specific RNA-binding (Hake and Richter 1994). CPEB homologs appear to be specific to the metazoan lineage: They have been identified in humans (Nagase et al. 1999), mice (Gebauer and Richter 1996), zebrafish (Bally-Cuif et al. 1998), clams (Walker et al. 1999), and flies (Lantz et al. 1994), but not in yeast or Arabidopsis. In Xenopus, CPEB has been found to have both positive and negative effects on translation (Richter 2000). It was initially identified as a factor that binds CPEs (Hake and Richter 1994). In vitro experiments argue that CPEB is a positive-acting polyadenylation factor (Richter 2000). Similarly, in vivo, Xenopus oocytes injected with anti-CPEB antibodies fail to polyadenylate and activate c-mos mRNA (Stebbins-Boaz et al. 1996), and Drosophila embryos lacking orb, a CPEB homolog, fail to polyadenylate and activate oskar mRNA (Chang et al. 1999). In contrast, a clam CPEB homolog binds to repressive elements in the 3′ UTRs of specific mRNAs and represses translation in vitro (Minshall et al. 1999; Walker et al. 1999). Xenopus CPEB also appears to repress translation, acting through an eIF-4E binding protein, maskin (Stebbins-Boaz et al. 1996). The dual functions of CPEB mirror those of CPEs, some of which are required for both translational activation (e.g., Fox et al. 1989; McGrew et al. 1989; Varnum and Wormington 1990; Huarte et al. 1992) as well as repression (Sallés et al. 1992; Stutz et al. 1998; de Moor and Richter 1999; Minshall et al. 1999; Ralle et al. 1999; Barkoff et al. 2000).
The biological functions of CPEB are best understood in the female germ line. CPEB mRNA and protein are present in oocytes of many species. Xenopus CPEB, which has been studied in the greatest detail biochemically, binds to multiple maternal mRNAs, including cyclin and CDK mRNAs, and is required for progression of the oocyte through meiosis (Stebbins-Boaz et al. 1996). Orb has multiple roles in oogenesis (Lantz et al. 1994), including activation of oskar mRNA, to which it binds in vivo (Chang et al. 1999). A zebrafish homolog, Zorba, is localized to the animal pole of the oocyte, though its function is unknown (Bally-Cuif et al. 1998). Although most studies of CPEB function have focused on oocytes, transcripts of CPEB homologs can be detected in other tissues of some organisms, including the testes of mice and flies (Lantz et al. 1994; Gebauer and Richter 1996) and somatic tissues of rodents (Wu et al. 1998).
To reveal the roles of all CPEB-related proteins comprehensively in a single organism, we have focused on the CPEB family in the nematode, Caenorhabditis elegans. We selected this species for several reasons. First, its complete genome sequence is available (The C. elegans Sequencing Consortium 1998), ensuring that all CPEB homologs from this single species can be identified. Second, RNA-mediated interference (RNAi) is particularly effective in C. elegans (Fire et al. 1998). Therefore, RNAi can be used to reduce the activity of the CPEB-encoding genes and thereby identify their functions during development. Third, the C. elegans germ line has been studied intensively, making it possible to integrate our analyses of CPEB functions with studies of other germ-line controls (Schedl 1997). In C. elegans, XX animals develop as hermaphrodites. The first germ cells to enter meiosis differentiate as sperm; later in adulthood, germ cells switch fates and become oocytes. Finally, specific cell-fate decisions in the C. elegans germ line rely on 3′ UTR-mediated controls, and are correlated with changes in poly(A) length (for review, see Wickens et al. 2000).
In this report, we demonstrate that two C. elegans CPEB homologs have distinct biological functions, and that, surprisingly, these functions are in spermatogenesis, not oogenesis. One homolog is required for the cell fate decision to become sperm rather than oocyte; this homolog is fog-1, a gene previously identified genetically (Barton and Kimble 1990). Another homolog, cpb-1, is required for progression through spermatogenesis per se. Furthermore, we find that CPB-1 protein binds physically to another 3′ UTR regulatory protein, FBF. We demonstrate that FBF, previously known to mediate the sperm/oocyte switch by binding to the 3′ UTR of fem-3 mRNA (Zhang et al. 1997), appears to be required for spermatogenesis and subsequent embryogenesis. These data emphasize the diversity of functions in a single family of 3′ UTR regulators and demonstrate unexpected roles of CPEB homologs in spermatogenesis, cell fate specification, and differentiation.
Results
Four CPEB-related proteins in C. elegans
The completed sequence of the C. elegans genome predicts four genes with extensive sequence similarity to Xenopus CPEB: C40H1.1, C30B5.3, B0414.5, and a fourth homolog on YAC Y54E10. We refer to the first three homologs as cpb-1, cpb-2, and cpb-3, respectively; the fourth homolog is fog-1, a gene first identified by mutation (Barton and Kimble 1990). The chromosomal locations of these four CPEB homologs are distinct (Fig. 1A). The predicted ORFs of all four genes were confirmed by isolating and sequencing multiple cDNAs. Minor discrepancies in the predicted splicing patterns were found for all four genes [Figs. 5B, 8C, and 10C, below; Wickens laboratory Web site (http://biochem.wisc.edu/wickens/research/publications)].
Figure 1.
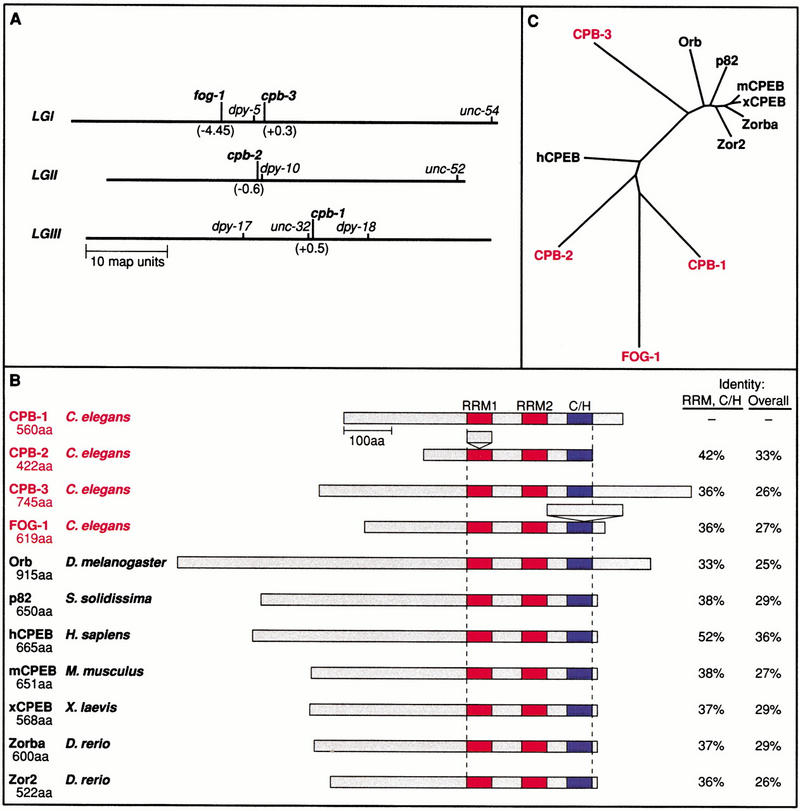
Analysis of the CPEB family of RNA-binding proteins. (A) Genetic map locations of the four C. elegans CPEB homologs. Map positions of cpb-1, cpb-2, and cpb-3 are based on correlation between the physical and genetic maps. Map position of fog-1 was determined previously (Barton and Kimble 1990). (B) Structures of CPEB homologs: conserved motifs. Proteins are drawn to scale; the bar represents 100 amino acids. Red boxes represent an RRM domain; the single blue box represents the two consecutive cysteine/histidine zinc finger motifs, comprising C4 and C2H2 residues. Atypical spacings in RRM1 of CPB-2 and in the zinc finger region of FOG-1 are indicated. Numbers at right indicate the percentage identity at the amino acid level to CPB-1 over either the RNA-binding domain (RRM, C/H) or the entire protein (overall), relative to CPB-1. The number of amino acids in each protein is listed below the protein name. (C) Unrooted tree illustrating the relationship among the conserved RRM and zinc finger regions of the known CPEB homologs. The tree was calculated using the Kimura-corrected distances algorithm with the ClustalX program and drawn using the neighbor-joining method with the Phylip Drawtree application.
Figure 5.
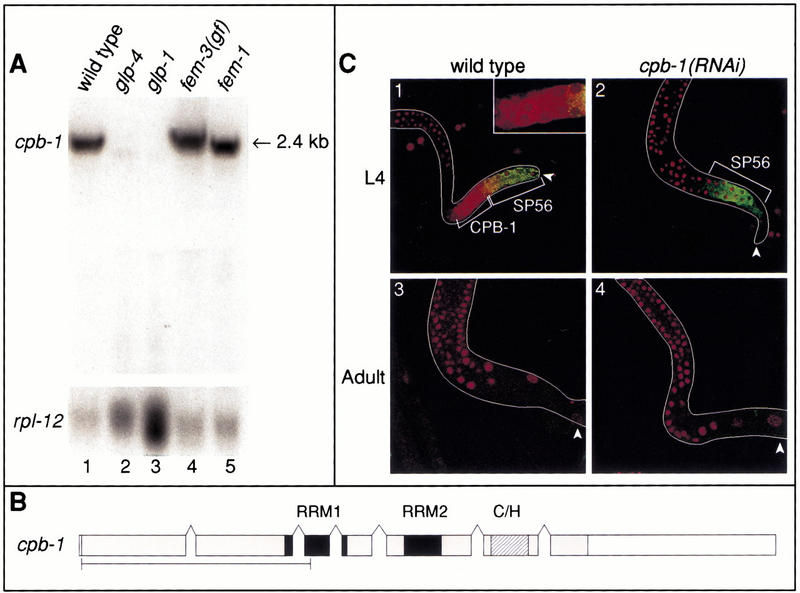
Expression of cpb-1 mRNA and protein. (A) Northern blot showing the tissue distribution of cpb-1 mRNA. Total RNA (15 μg) was isolated from adults of the indicated strains and was probed with a cpb-1-specific probe. The rpl-12 gene encoding ribosomal protein L12 is included as a loading control. The position of cpb-1 mRNA is indicated. (B) Exon diagram of the cpb-1 transcript. The RRMs and zinc fingers are indicated by black and hatched boxes, respectively. Gray boxes represent the coding region; white boxes represent 5′ and 3′ UTR sequences. Bracket indicates the region of CPB-1 used to raise antibodies and also used as bait in the two-hybrid screen. (C) CPB-1 protein expression in the hermaphrodite germ line. Gonads were extruded and stained with affinity-purified CPB-1 antibodies (red) and the sperm-specific SP56 antibody (green). Panels 1 and 2 are L4 gonads. Panels 3 and 4 are adult gonads. (Left) Wild type; (right) cpb-1(RNAi) controls. White arrowheads indicate the proximal end of the gonad. Inset in panel 1 shows an enlargement of the CPB-1-stained region.
Figure 8.
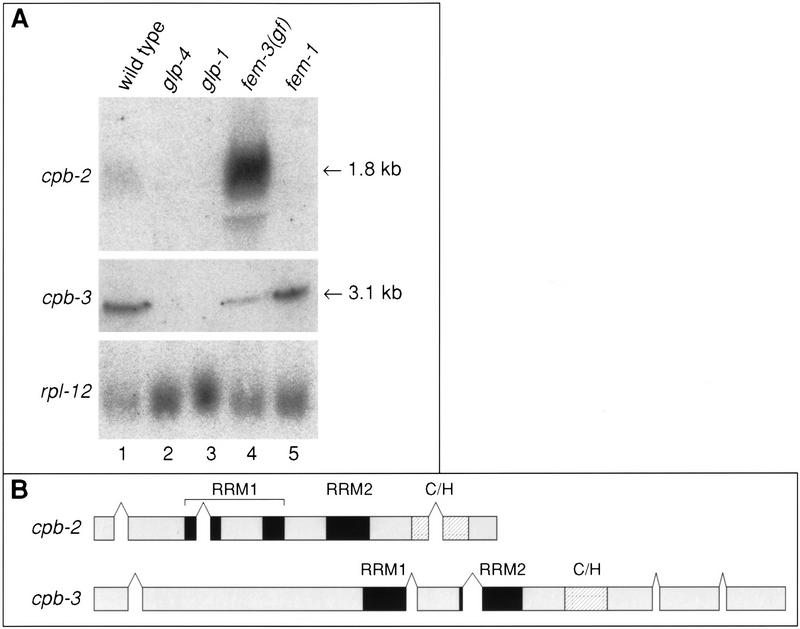
Expression of cpb-2 and cpb-3 mRNAs. (A) Northern blots showing the tissue distribution of the cpb-2 and cpb-3 mRNAs. Total RNA (15 μg) was isolated from adults of the indicated strains and was probed with either cpb-2 or cpb-3-specific probes. The rpl-12 gene encoding ribosomal protein L12 is included as a loading control. The positions of cpb-2 and cpb-3 mRNAs are indicated. (B) Exon diagrams of the cpb-2 and cpb-3 coding regions. The RRMs and zinc fingers are indicated by black and hatched boxes, respectively.
Figure 10.
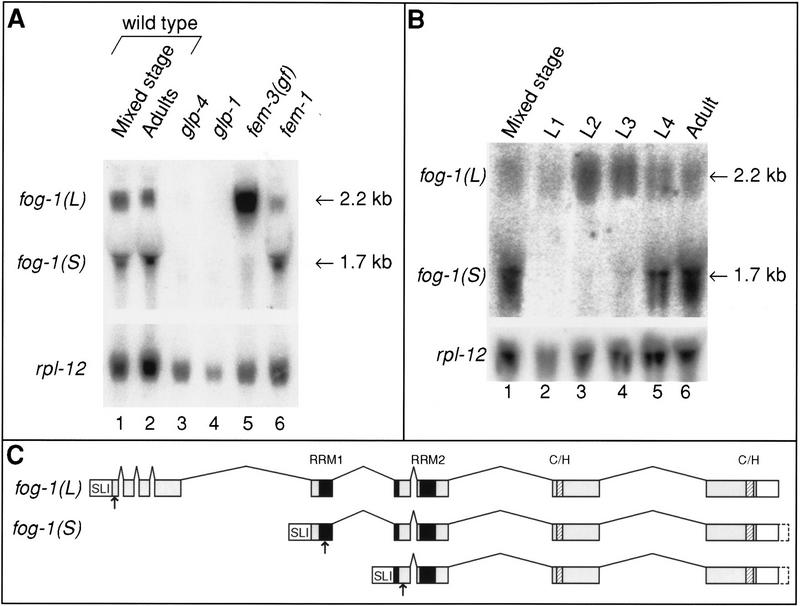
fog-1 transcripts are developmentally regulated. (A) Northern blot showing the tissue distribution of fog-1 mRNA. Total RNA (30 μg) was isolated from adults of the indicated strains and was probed with a fog-1-specific probe. The rpl-12 gene encoding ribosomal protein L12 is included as a loading control. The positions of the fog-1(L) and fog-1(S) mRNAs are shown; as judged by sequencing of RT-PCR products (depicted in C), the fog-1(S) transcripts detected in the Northern blot are heterogeneous. (B) Northern blot showing the temporal distribution of the fog-1(L) and fog-1(S) transcripts during development. Total RNA (15 μg) was isolated from wild-type hermaphrodites at the stages indicated above each lane and was probed with a fog-1-specific probe. The switch from spermatogenesis to oogenesis began in the animals designated L4. Ribosomal protein L12 loading control is shown below. (C) Schematic diagram of the two classes of fog-1 transcripts. SL1 trans-splicing is indicated. Gray boxes represent the coding region.White boxes represent 3′ UTR. The RRM domains and the interrupted zinc fingers are indicated by black and hatched boxes, respectively. Arrows under the fog-1 RNAs indicate the positions of the first in-frame methionines in these transcripts.
The predicted C. elegans proteins are most highly related to the CPEBs of other species in their carboxy-terminal regions (Figs. 1B and 2). This region comprises the diagnostic hallmarks of CPEB family members, including two distinctive RRMs and consecutive C4 and C2H2 zinc fingers (Fig. 1B). However, variations are present in two of the C. elegans homologs: cpb-2 contains a longer than usual spacing between the two RNP motifs of the first RRM, and fog-1 contains an apparent insertion between the second and third cysteines in the first zinc finger domain (Figs. 1B and 2). At present, the significance of these atypical spacings is unknown: In particular, the RNA binding activity of the homologs has not yet been examined.
Figure 2.
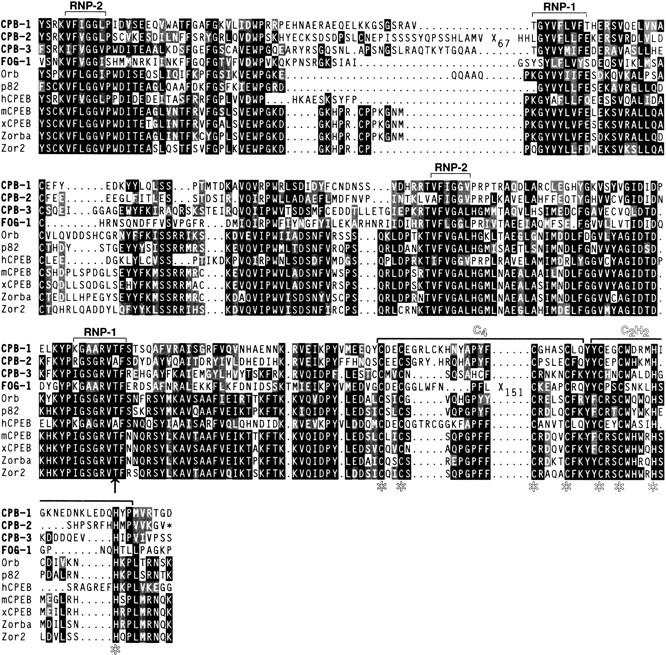
Alignment of the conserved RRM and zinc finger portions of the CPEB homologs. Identical residues are black; similar residues are gray. Brackets indicate the RNP-2 and RNP-1 submotifs of the RRM domains and the conserved C4 and C2H2 zinc fingers. Asterisks mark the diagnostic cysteine and histidine residues. The red arrow indicates the threonine to isoleucine change in the fog-1(q253ts) allele.
To determine the sequence relationship of the C.elegans proteins to CPEB homologs in other species, we used the Kimura-corrected distances algorithm to construct an unrooted phylogenetic tree based on the RRM and zinc finger motifs (Fig. 1C). Of the four C. elegans genes, cpb-3 is most closely related to the vertebrate, clam, and Drosophila homologs, all of which have been implicated in oogenesis (see introductory section).
cpb-1 is required for progression through spermatogenesis
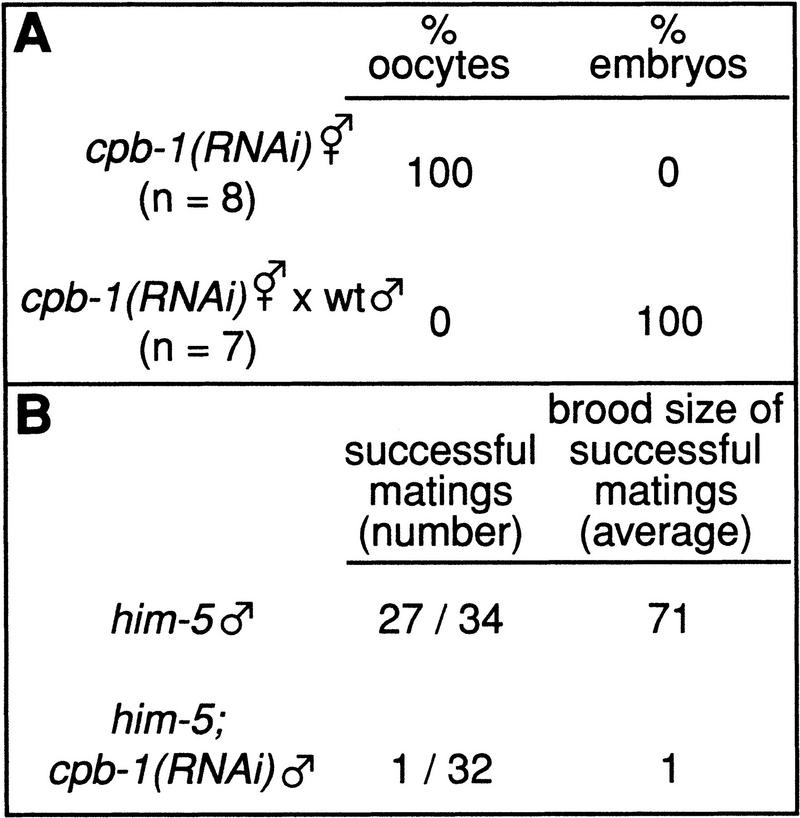
To determine the function of cpb-1, we used RNAi to disrupt cpb-1 gene activity. Adult hermaphrodites were injected with a 1-kb double-stranded (ds) RNA corresponding to cpb-1 cDNA and progeny of injected parents were examined; we refer to such progeny as cpb-1(RNAi) animals. We found that cpb-1(RNAi) hermaphrodites were grossly normal in morphology but did not produce any progeny. Instead, they laid oocytes (Fig. 3A), a phenotype typical of animals that have defective sperm. Consistent with a defect in spermatogenesis, the cpb-1(RNAi) hermaphrodites produced viable embryos when crossed with wild-type males (Fig. 3A). Therefore, cpb-1(RNAi) animals produce fully functional oocytes, but lack functional sperm, suggesting that cpb-1 function is essential for spermatogenesis. To test the effect of reducing cpb-1 function in males, we used a him-5 strain to generate XO males (Hodgkin et al. 1979). The cpb-1(RNAi);him-5 males were almost entirely sterile: Only 1 of 32 animals produced any cross-progeny, and that single animal produced only one embryo (Fig. 3B). By contrast, most control him-5 males produced cross-progeny and yielded much larger brood sizes (Fig. 3B).
Figure 3.
cpb-1 is required for spermatogenesis in hermaphrodites and males. (A) cpb-1(RNAi) hermaphrodites were allowed to self-fertilize or were mated with wild-type males. After fertilization, the percentages of oocytes and embryos laid over a 24-h period were scored. (B) him-5 or him-5; cpb-1(RNAi) adult males were mated to females for 44 h and successful matings were scored. The brood size (average number of progeny) from successful matings is shown.
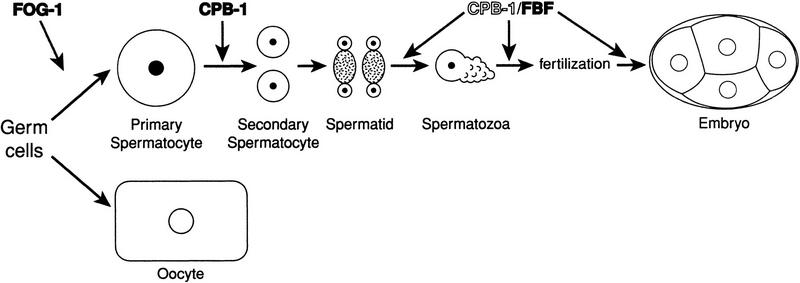
To determine the nature of the sperm defect in cpb-1(RNAi) animals, we examined their sperm by microscopy. In wild-type animals, overt spermatogenesis begins when a primary spermatocyte enlarges and buds away from the syncytial rachis (for review, see L'Hernault 1997). This primary spermatocyte undergoes two meiotic divisions to form secondary spermatocytes, spermatids, and finally, spermatozoa (Fig. 4A). Mature sperm are easily recognized by their small size, characteristically granular cytoplasm, and highly condensed nuclei. Mature sperm were not observed in cpb-1(RNAi) animals; instead, much larger cells occupied the region where spermatogenesis would normally proceed (Fig. 4B). These large cells were of a uniform size that is typical for primary spermatocytes; they stained with the sperm-specific SP56 antibody (see Fig. 5C), indicating that spermatogenesis was initiated in cpb-1(RNAi) animals. Thus, cpb-1 is not required for specifying the sperm cell fate or for the switch from spermatogenesis to oogenesis, but instead is required for progression through the meiotic divisions during spermatogenesis.
Figure 4.
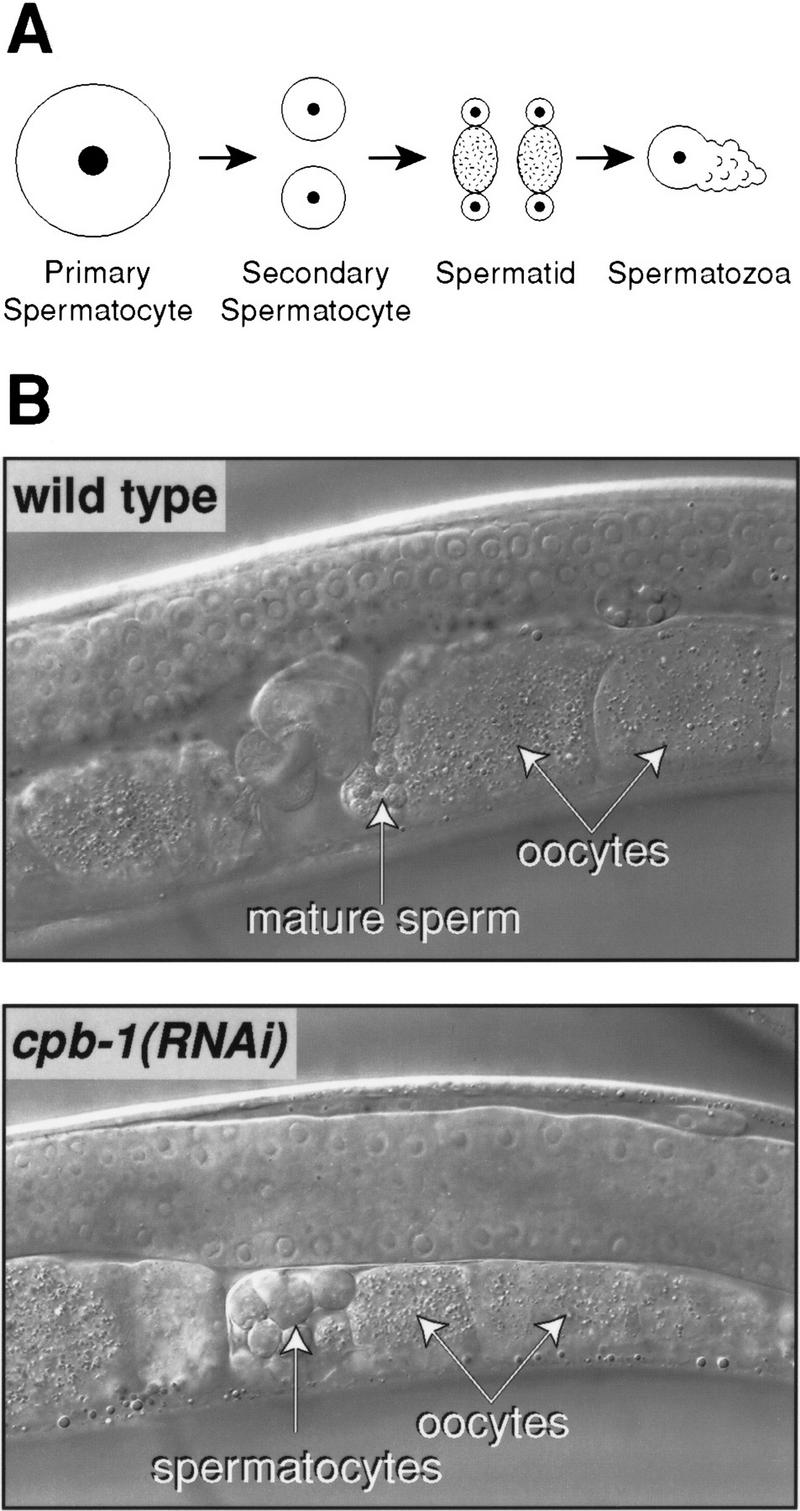
cpb-1-defective sperm arrest during spermatogenesis. (A) Summary of wild-type spermatogenesis in C. elegans. Primary spermatocytes undergo two meiotic divisions to form secondary spermatocytes and finally spermatids. Spermatids bud away from a residual body, grow a pseudopod, and are activated to form mature spermatozoa. (B) Nomarski micrographs of wild-type (top) and cpb-1(RNAi) (bottom) young adult hermaphrodites. Mature sperm are indicated in the wild-type gonad, and the large spermatocytes are indicated in the cpb-1(RNAi) gonad.
cpb-1 mRNA and protein are expressed in the germ line
Because cpb-1 is required for progression through spermatogenesis, we expected that it might be expressed in the spermatogenic germ line. We prepared a Northern blot with RNA isolated from a wild-type strain and from temperature-sensitive mutant strains with various germ-line defects. In wild-type adults, cpb-1 gives rise to a prominent 2.4-kb transcript (Fig. 5A, lane 1). To determine the exon/intron structure of cpb-1 mRNA, we isolated and sequenced multiple cpb-1 cDNAs; from this data, we identified an mRNA of 2302 nucleotides composed of seven exons (Fig. 5B). The 2.4-kb cpb-1 transcript appears to be specific to the germ line, as it was not detected in either glp-4 or glp-1 mutants, which possess virtually no germ line (Fig. 5A, lanes 2 and 3). In contrast, the transcript was abundant in fem-3(gf) worms, which produce only sperm (Fig. 5A, lane 4); this is consistent with the requirement for cpb-1 during spermatogenesis. The 2.4-kb transcript was also present at nearly identical levels in fem-1 worms, which produce only oocytes (Fig. 5A, lane 5). Therefore, cpb-1 mRNA is present in both spermatogenic and oogenic germ lines.
To determine the pattern in which the CPB-1 protein is expressed, we generated affinity-purified polyclonal antibodies against the nonconserved N-terminal 258 amino acids of the protein. We doubly stained extruded gonads of wild-type and cpb-1(RNAi) hermaphrodites with anti-CPB-1 antibodies and the monoclonal SP56 antibody, which recognizes cells undergoing spermatogenesis, beginning at least as early as the primary spermatocyte stage (Ward et al. 1986). CPB-1 protein was detected at high levels in the L4 germ line, in a region just distal to the region that stained by SP56 (Fig. 5C, panel 1). The spatial distributions of CPB-1 and SP56 staining abut but do not overlap significantly. An identical distribution was observed in males (not shown). SP56 staining is still observed in cpb-1(RNAi) animals (Fig. 5C, panel 2), supporting the hypothesis that spermatogenesis is initiated in cpb-1(RNAi) animals.
CPB-1 staining was predominantly cytoplasmic, as indicated by a uniform strong signal that is excluded from nuclei (seen as round “holes” in Fig. 5C, panel 1). This cytoplasmic staining was specific: It was not detectable in cpb-1(RNAi) L4 hermaphrodites (Fig. 5C, panel 2). The same pattern was observed in males (not shown). In contrast, a faint nonspecific staining was detected in nuclei throughout the gonad (Fig. 5C, panels 1–4). In Figure 5C, panels 2–4 are shown at higher contrast to highlight the lack of cytoplasmic staining. The nuclear staining was observed in both wild-type and cpb-1(RNAi) animals (Fig. 5C, panels 1 and 2), suggesting it is not CPB-1; it was also observed in cpb-2(RNAi), cpb-3(RNAi), and fog-1 mutants (data not shown), suggesting that it does not reflect cross-reactivity with other CPEB proteins. We conclude that the nuclear staining is nonspecific background and that CPB-1 is predominantly cytoplasmic.
CPB-1 protein was not detectable in adult germ lines that have switched to oogenesis (Fig. 5C, panels 3 and 4), in accord with cpb-1 having no role in oogenesis. Thus, although cpb-1 mRNA is present in adult hermaphrodites, which are making only oocytes (Fig. 5A), the protein is not. This suggests that the activity of cpb-1 mRNA is regulated post-transcriptionally during oogenesis.
CPB-1 and FBF proteins physically interact
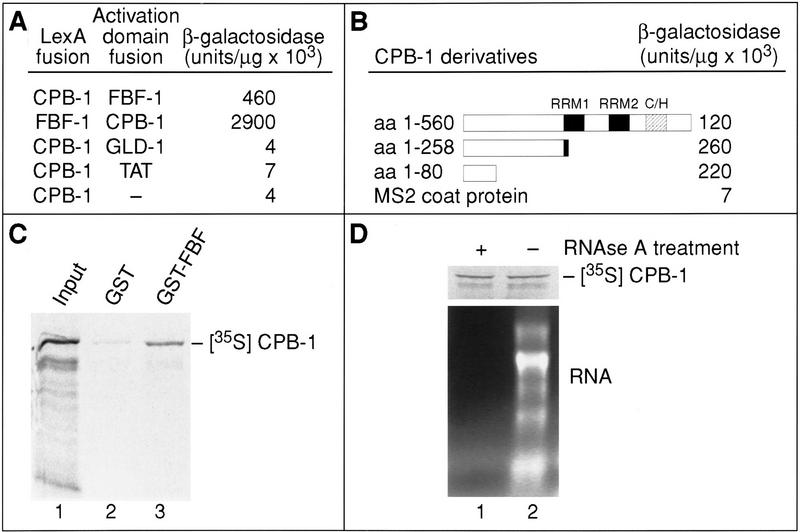
To identify proteins that might interact with CPB-1 and influence its activity during spermatogenesis, we performed a yeast two-hybrid screen. As “bait,” we used a fusion protein between the N-terminal 258 amino acids of CPB-1 (missing most of the RRMs and the zinc fingers; see Fig. 5B) and the LexA DNA-binding domain (LexA/CPB-1). Of 2,000,000 independent cDNA transformants, 10 activated both the HIS3 and lacZ reporters in the presence of LexA/CPB-1 but not in the presence of a fusion protein between the LexA DNA-binding domain and the MS2 coat protein. Two of these positives were identified as FBF-1, a protein that binds to the 3′ UTR of fem-3 mRNA and is required for the sperm–oocyte switch (Zhang et al. 1997). In directed two-hybrid tests, FBF-2 [a protein 91% identical in sequence to FBF-1 (Zhang et al. 1997)] also interacts with CPB-1 (not shown). The interaction between CPB-1 and FBF-1 was specific as replacing FBF-1 with the C. elegans protein, GLD-1, or with the HIV protein, TAT, did not activate the lacZ reporter protein (Fig. 6A); similarly, replacing CPB-1 with FOG-1 eliminated activation (not shown). The specificity of the FBF/CPB-1 interaction was also corroborated by results of a reciprocal two-hybrid screen using LexA/FBF-1 as bait. From 20,000,000 transformants, 19 were recovered that activated both reporter genes, and 5 of these were CPB-1 (data not shown).
Figure 6.
CPB-1 and FBF proteins physically interact. (A) CPB-1, FBF (amino acids 121–614), GLD-1, TAT (the first exon), and MS2 coat protein (amino acids 1–131) were tested in the yeast two-hybrid system using fusions to either the LexA DNA-binding domain or the Gal4 transcriptional activation domain. β-Galactosidase activity per μg of protein is shown (Zhang et al. 1999). (B) The N-terminal 80 amino acids of CPB-1 domain are required for interaction with FBF. C-terminal truncations of the CPB-1 protein were fused to the LexA DNA-binding domain and tested in the yeast two-hybrid system with FBF (amino acids 121–614) fused to the Gal4 transcriptional activation domain. (C) Interaction between a GST–FBF fusion protein and CPB-1. A fusion protein consisting of GST and FBF (GST–FBF) or GST alone was coupled to Sepharose-conjugated glutathione beads and incubated with radiolabeled CPB-1 protein. Bound proteins were eluted from the beads and analyzed by SDS-PAGE. (D) The interaction between FBF and CPB-1 appears to be RNA independent. Radiolabeled CPB-1 protein was made by in vitro translation and the lysate was either treated with RNAse A or mock-incubated without the nuclease. After treatment the lysates were incubated with GST–FBF beads and the bound proteins were eluted and analyzed by SDS-PAGE. RNA was isolated from aliquots of the RNAse or mock-treated lysates and was analyzed by electrophoresis and ethidium bromide staining.
The region of CPB-1 that interacts with FBF-1 was further defined by analyzing deletion derivatives of CPB-1 in the two-hybrid system. The smallest portion of CPB-1 that strongly activated the lacZ reporter comprised 80 amino acids at the N terminus of the protein (Fig. 6B). This fragment did not detectably activate the lacZ reporter in the absence of FBF (data not shown). This region contains no identifiable sequence motifs, nor does it share homology with another FBF-interacting protein, NOS-3 (Kraemer et al. 1999). This 80-amino-acid region is highly diverged among CPEB-related proteins, consistent with the failure of FOG-1 to interact detectably with FBF (not shown). The region of FBF used in these binding studies comprises amino acids 121–614 and includes the Puf repeats and flanking Csp regions (Zhang et al. 1997).
To analyze the CPB-1/FBF-1 interaction biochemically, we performed affinity chromatography experiments using glutathione-S-transferase (GST) fusions with FBF-1 and CPB-1 synthesized in vitro. 35S-Labeled, full-length CPB-1 was prepared by in vitro translation in a rabbit reticulocyte lysate and incubated with Sepharose beads bound to either purified recombinant GST–FBF-1 or GST alone. Radiolabeled CPB-1 bound specifically to GST–FBF-1 but not to GST alone (Fig. 6C). Because both CPB-1 and FBF-1 have RNA-binding motifs, we considered the possibility that their interaction might be mediated by an RNA “bridge” in both the two-hybrid and in vitro systems. To address this directly, a lysate containing 35S-labeled CPB-1 protein was treated extensively with RNAse A before incubation with GST–FBF-1. The RNAse A treatment eliminated virtually all of the detectable RNA in the lysate, including rRNA and tRNA, yet did not inhibit the binding of 35S-labeled CPB-1 to GST–FBF-1 (Fig. 6D). This suggests that binding of CPB-1 to FBF occurs in the absence of RNA.
FBF and spermatogenesis
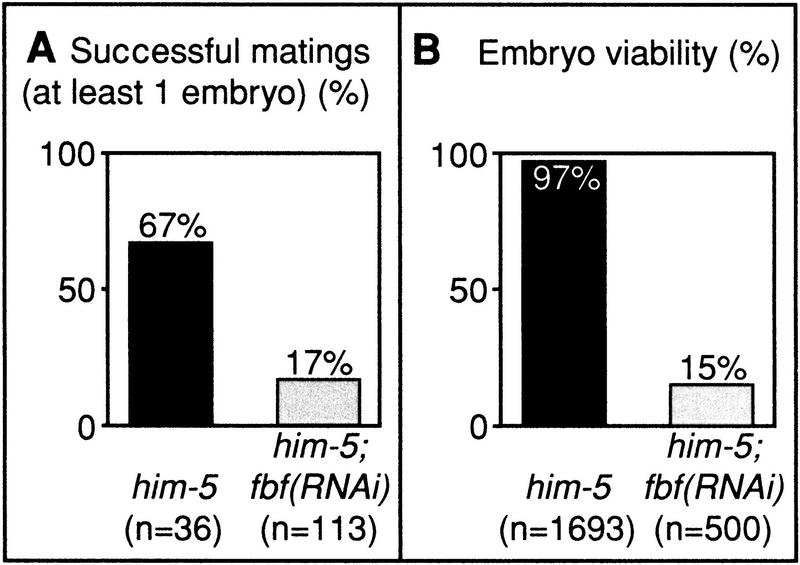
Because CPB-1 and FBF-1 specifically interact and CPB-1 is required during spermatogenesis, we tested whether FBF was essential for spermatogenesis as well. FBF-deficient hermaphrodites have masculinized germ lines and never make the switch to oogenesis (Zhang et al. 1997). The cellular and nuclear morphologies of these sperm are normal as assayed by Nomarski microscopy and DAPI staining (Zhang et al. 1997; not shown). However, because no oocytes are produced in fbf-deficient hermaphrodites, we were unable to test whether or not these sperm were functional. To address this issue, we used RNAi to abolish fbf function in him-5 animals. We then crossed individual fbf(RNAi); him-5 males or buffer-injected control him-5 males into individual fog-1 mutant females and scored the progeny of these matings. We found two major differences. First, fbf(RNAi); him-5 matings were not as successful as controls. Whereas 67% of the control him-5 males produced embryos, only 17% of fbf(RNAi); him-5 males produced any embryos (Fig. 7A). Furthermore, control males produced more embryos than fbf(RNAi); him-5 males (an average of 78 vs 42 embryos per mating). Second, fbf(RNAi); him-5 cross-progeny were much less viable than controls. Whereas 97% of the embryos produced from control males hatched and grew to become viable adults, only 15% of embryos produced from fbf(RNAi); him-5 males hatched (Fig. 7B). The inviable embryos produced by fbf(RNAi); him-5 males often proceeded well into embryogenesis: Embryos elongated and possessed differentiated tissues, such as gut and pharynx (not shown). Embryonic lethality was also observed when fbf(RNAi); him-5 males were mated with fog-2 females (not shown), indicating that embryonic lethality was not a result of an unexpected genetic interaction between fbf and the genotype of the mutant females. The simplest interpretation of these results is that fbf is critical for spermatogenesis. In addition, paternal fbf appears to be important for embryonic viability. This paternal effect may be due to derepression of sperm-specific mRNAs.
Figure 7.
FBF is required for male fertility. (A) him-5 or him-5; fbf(RNAi) adult males were mated to females for 48 h. Successful matings (yielding at least one embryo) were scored. (B) Embryos from successful matings were examined for viability. Embryos were scored as dead if they failed to hatch within 24 h after being laid.
cpb-2 and cpb-3
To identify possible functions of cpb-2 and cpb-3, we used RNAi. Neither cpb-2(RNAi) nor cpb-3(RNAi) animals had any obvious defect; similarly, cpb-2(RNAi); cpb-3(RNAi) double mutants appeared normal. These animals were fertile and had brood sizes comparable to those of control-injected animals (data not shown). To test whether cpb-2 and cpb-3 might have functions redundant with cpb-1 or fog-1, we examined cpb-1(RNAi); cpb-2(RNAi) and cpb-1(RNAi); cpb-3(RNAi) double mutants, as well as cpb-1(RNAi); cpb-2(RNAi); cpb-3(RNAi) and fog-1(RNAi) cpb-3(RNAi); cpb-2(RNAi) triple mutants. These had phenotypes typical of cpb-1 or fog-1, respectively (see below). Furthermore, quadruple mutants lacking all four CPEB homologs had the same phenotype as fog-1 alone. Therefore, we were unable to detect any defect by RNAi for either cpb-2 or cpb-3.
As another approach to gain clues about possible functions for cpb-2 and cpb-3, we examined their mRNAs by Northern analysis. Specifically, we compared wild-type adult RNA with RNA prepared from adults of a variety of germ-line mutants. Comparable data were obtained using RNA from mixed-stage populations of wild-type animals (not shown). Both cpb-2 and cpb-3 mRNAs are germ line-dependent, as neither is expressed at detectable levels in glp-1 or glp-4 mutant animals, which virtually lack germ lines (Fig. 8A, lanes 2 and 3). cpb-2 gave rise to a single 1.8-kb transcript in fem-3(gf) worms, which make only sperm; in contrast, it is undetectable in fem-1 worms, which make only oocytes (Fig. 8A, lanes 4 and 5). The mRNA is present at very low levels in wild-type adults (lane 1); this likely reflects the fact that the mRNA is greatly enriched during spermatogenesis (lane 4), which had ceased in adults (lane 1). Together, these data suggest that cpb-2 mRNA is specific to the spermatogenic germ line.
cpb-3 gives rise to a single 3.1-kb transcript that is present at high levels in fem-1 worms (Fig. 8A, lane 5), and in wild-type adult hermaphrodites (Fig. 8A, lane 1), both of which are producing oocytes. cpb-3 mRNA levels are reduced 3.8-fold in fem-3(gf) vs fem-1 animals (Fig. 8A, lane 4). The likely presence of cpb-3 mRNA in oocytes is consistent with the fact that cpb-3 is the C. elegans CPEB homolog most closely related to the CPEB homologs in other organisms that have been implicated in oogenesis (Fig. 1C). The splicing patterns of the cpb-2 and cpb-3 open reading frames, deduced by the sequencing of multiple cDNAs, are shown in Figure 8B.
Because cpb-2 and cpb-3 mRNAs are readily detected on Northern blots and differentially expressed in masculinized and feminized germ lines, respectively, it seems likely that these genes have some biological function. One possibility is that cpb-2-or cpb-3-deficient animals might have subtle phenotypes that were not detected. Alternatively, cpb-2 and cpb-3 might be relatively resistant to RNAi or act redundantly with other genes that are not CPEB homologs.
fog-1 is a CPEB homolog
To identify the function of the homolog that initially was dubbed cpb-4, we used RNAi and found that cpb-4(RNAi) progeny had a phenotype typical of fog-1 mutants: cpb-4(RNAi) mutant hermaphrodites failed to make any sperm, but instead made only oocytes (Fig. 9A). Similarly, cpb-4(RNAi) males produced only oocytes, not sperm (not shown). Moreover, cpb-4(RNAi) oocytes could be fertilized by wild-type males and produce viable progeny (not shown), as can fog-1 mutant oocytes (Barton and Kimble 1990). No somatic defects were observed.
Figure 9.
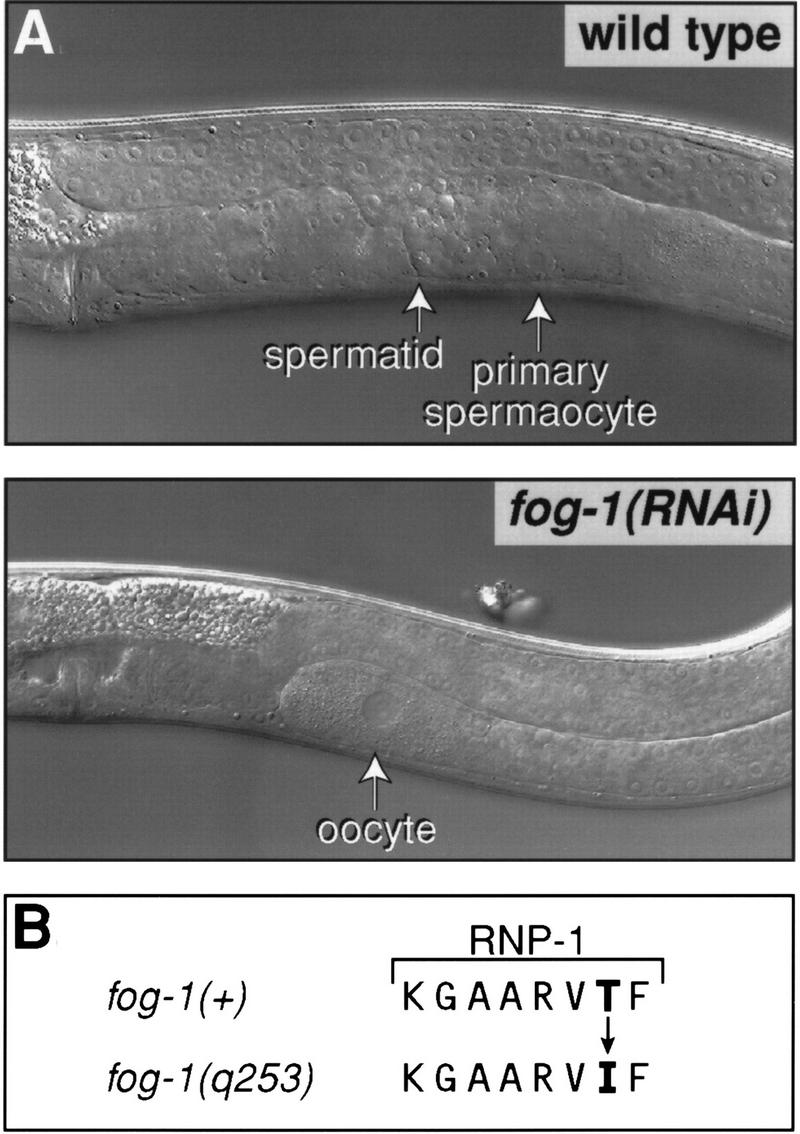
The fog-1 gene encodes a CPEB homolog. (A) Nomarski micrographs showing wild-type (top) and fog-1(RNAi) (bottom) late L4 hermaphrodites. Arrows indicate the relevant germ cells. (B) Sequences of the wild type and q253(ts) allele of the highly conserved RNP-1 submotif of the second RRM of fog-1. The threonine-to-isoleucine change is indicated (also see Fig. 2).
During the course of these experiments, the YAC carrying cpb-4 was found to map to the same chromosomal region as fog-1. To ask directly whether cpb-4 indeed was fog-1, which at the time had not been cloned, we sequenced cpb-4 in DNA prepared from fog-1(q253ts) mutants and detected a single amino acid substitution: A threonine was changed to isoleucine in the highly conserved RNP1 motif of the second RRM (Fig. 9B). Taken together, these data strongly suggest that cpb-4 is indeed fog-1. Experiments performed independently to clone fog-1 corroborate this conclusion (S.-W. Jin and R. Ellis, pers. comm.). We therefore abandoned the name cpb-4 and refer to this CPEB homolog as fog-1.
fog-1 mRNA is present in the germ line and is developmentally regulated
To analyze the tissue-specificity of fog-1 mRNA, we prepared Northern blots using total adult RNA isolated from wild-type and temperature-sensitive mutant strains. In wild-type animals, the fog-1 locus gives rise to two prominent classes of transcripts of 2.2 and 1.7 kb, referred to as fog-1(L) and fog-1(S) mRNAs. Both mRNAs appear to be germ line-specific as they were not detected in either glp-4 or glp-1 mutants, which possess virtually no germ-line tissue (Fig. 10A, lanes 3 and 4). In fem-3(gf) mutants, which produce a vast excess of sperm and never switch to oogenesis, only fog-1(L) was detected (Fig. 10A, lane 5), consistent with a role for fog-1(L) mRNA in spermatogenesis. In fem-1 mutants, which make only oocytes, the level of fog-1(L) mRNA was reduced and fog-1(S) was abundant (Fig. 10A, lane 6).
The results above indicate that fog-1(L) mRNA is enriched during spermatogenesis and fog-1(S) during oogenesis. We therefore examined the relative abundance these two mRNAs during the development of hermaphrodites, in which the germ line switches from spermatogenesis to oogenesis near the L4/adult transition. We prepared RNA from animals at all four larval stages and from adults. The fog-1(L) mRNA first appeared at low levels during the first larval stage (Fig. 10B, lane 2) and was present at high levels through the L3 stage (Fig. 10B, lane 4). At the L4 stage, the level of fog-1(L) mRNA decreased and fog-1(S) appeared (Fig. 10B, lane 5); the same distribution persisted in adults (Fig. 10B, lane 6).
To determine the molecular nature of the two mRNA species, we reverse-transcribed poly(A) + mRNA isolated from either wild-type adult hermaphrodites or fem-3(gf) adults. The majority of mRNAs in C. elegans possess one of two trans-spliced leader sequences at their 5′ ends, designated SL1 or SL2. We used SL1 or SL2 primers, together with fog-1-specific nested primer pairs, to amplify fog-1 cDNAs. Both fog-1(L) and fog-1(S) are SL1 trans-spliced (Fig. 10C). fog-1(L) mRNA consists of nine exons and gives rise to a predicted open reading frame of 619 amino acids. fog-1(S) is composed of at least two mRNAs that differ at both their 5′ and 3′ ends. Relative to fog-1(L) , one fog-1(S) transcript lacks the first four, and another transcript the first five, exons. In addition, fog-1(S) mRNAs also appear to possess heterogeneous 3′ termini. Potential initiator methionines are present in both fog-1(S) mRNA forms (arrows in Fig. 10C) and could potentially be used to initiate translation of N-terminal truncations of the FOG-1 protein. The timing of the appearance of fog-1(S) and the reduction in fog-1(L) correlates well with the timing of the decision to switch from spermatogenesis to oogenesis (see Discussion).
Discussion
In this work, we describe four C. elegans homologs of Xenopus CPEB. Our work leads to five main conclusions. First, two homologs, cpb-1 and fog-1, are required for spermatogenesis. Second, the biological roles of cpb-1 and fog-1 are distinct: cpb-1 controls progression through meiosis during spermatogenesis, whereas fog-1 regulates specification of the sperm fate. Third, CPB-1 physically interacts with FBF, a distinct 3′ UTR regulator of the Puf gene family. Consistent with a possible CBP-1/FBF interaction, we find that spermatogenesis appears to be defective in animals lacking FBF. FBF also is required paternally for embryogenesis. Fourth, two classes of fog-1 mRNA are produced in a fashion that is regulated both during development and by the sex determination pathway. These findings suggest that fog-1(L) mRNA encodes the active form of FOG-1 required for spermatogenesis. Finally, the four CPEB homologs of C. elegans appear to have no essential function in oogenesis.
A conserved CPEB function: progression through meiosis
Xenopus CPEB and C. elegans CPB-1 both control progression through meiosis. In Xenopus, CPEB is required for meiotic maturation during oogenesis. Specifically, CPEB activates translation of c-mos mRNA, which encodes a MAP kinase kinase kinase. Elevation of c-MOS kinase activity triggers a MAPK cascade that leads to activation of MPF and meiotic maturation (Gebauer and Richter 1997; Sagata 1997). Drosophila orb is also required for oogenesis, but its role in the meiotic cell cycle is less clear (Christerson and McKearin 1994; Lantz et al. 1994; Huynh and St Johnston 2000).
Like Xenopus CPEB, the C. elegans cpb-1 gene controls progression through meiosis, but it does so in spermatogenesis rather than oogenesis. Animals deficient for cpb-1 activity possess spermatocytes that have failed to execute either meiosis I or meiosis II. The CPB-1 protein is restricted to a region in which germ cells have entered meiotic prophase I, but have not yet begun to express an early sperm differentiation marker. This arrest of cpb-1(RNAi) animals in early spermatogenesis and the localization of CPB-1 to early meiotic cells together suggest that CPB-1 is critical for initiating meiotic maturation during spermatogenesis. Furthermore, CPB-1 is localized to the cytoplasm, consistent with a role in post-transcriptional regulation. We suggest that CPB-1 protein regulates the translation of mRNAs that are required for progression through spermatogenesis. The target of this regulation is unlikely to be c-mos per se, as no c-mos homolog has been detected in C. elegans, but could be another component of the MAP kinase cascade.
Like frog CPEB and clam p82, CPB-1 abundance decreases as meiosis proceeds, suggesting a conserved mode of control. In C. elegans spermatocytes, CPB-1 protein is abundant during first meiotic prophase, but drops precipitously once meiosis resumes. In frogs and clams, the level of CPEB similarly decreases upon oocyte maturation, as the translation of several mRNAs increases (Hake and Richter 1994; Walker et al. 1999). We envision three possible models for how CPB-1 controls progression of meiosis in spermatogenesis. First, CPB-1 might repress one or more critical mRNAs (e.g., functional analogs of c-mos) that promote the completion of meiosis; relief of that repression allows meiosis to proceed. Indeed, recent studies suggest that specific phosphorylation of CPEB relieves repression of c-mos in Xenopus (Mendez et al. 2000). Second, CPB-1 may activate one or more mRNAs that initiate meiosis. Finally, CPB-1 might initially repress mRNAs required for meiotic maturation and subsequently activate them. These scenarios all are consistent with destruction of CPB-1 once maturation has begun; having completed its role in launching meiotic maturation, its persistence might interfere with subsequent events in development.
A CPEB homolog is required for cell fate specification
In our analysis of CPEB homologs in C. elegans, we discovered that one CPEB homolog was FOG-1; an independent study designed to clone the fog-1 locus recently reached the same conclusion (S-W. Jin and R. Ellis, pers. comm.). The fog-1 gene is essential for specification of germ cells to the sperm fate; in fog-1 null mutants, germ cells that normally would differentiate as sperm are instead sexually transformed into oocytes (Barton and Kimble 1990). Thus, fog-1 is a germ line-specific sex-determining gene.
The identification of FOG-1 as a CPEB homolog suggests that a translational regulator can specify a specific cell fate. By genetic criteria, fog-1 acts at the end of the sex-determination pathway to specify germ cells as sperm rather than oocytes (Barton and Kimble 1990). Most regulators of terminal cell types act at the transcriptional level. For example, in Saccharomyces cerevisiae, the mating type locus (MAT) encodes transcriptional regulators that specify the a or α fate (Herskowitz 1989), and in vertebrates, myoD-related proteins are transcriptional regulators that specify the muscle fate (Weintraub 1993). In contrast, we suggest that FOG-1 controls gene expression and cell fate at the translational level, by analogy to CPEB functions in other species. Although other translational regulators can influence many cell fates broadly, as in Drosophila sex determination and axial patterning, the cell fate transformation observed in animals lacking FOG-1 is highly specific, suggesting a simple binary switch that determines whether a cell becomes a sperm or an oocyte. Direct tests of this hypothesis will require ectopic expression of FOG-1 to determine whether it is sufficient to drive cells to become sperm.
The fog-1 gene produces at least three transcripts: a longer fog-1(L) transcript and two shorter fog-1(S) transcripts. Both fog-1(L) and fog-1(S) mRNAs encode the conserved RRM and C/H motifs typical of CPEBs, but compared with the fog-1(S) transcripts, fog-1(L) encodes an additional 214 or 269 amino acids in the N-terminal region of the protein. The two fog-1(S) mRNAs contain in-frame methionines and are predicted to encode N-terminally truncated forms of FOG-1. Two experiments suggest that fog-1(L) encodes the active form of FOG-1 that is required for the specification of spermatogenesis. First, in wild-type animals, the fog-1(L) mRNA is abundant during midlarval development when fog-1 activity is required to promote spermatogenesis (Barton and Kimble 1990) and is much reduced in later development once that activity is no longer required; in contrast, fog-1(S) is not detectable during midlarval development and increases later when fog-1 activity is no longer required. Second, in a mutant that produces only sperm in an otherwise female animal, fog-1(L) predominates, whereas in a mutant that produces only oocytes, fog-1(S) is more abundant. One simple interpretation is that the longer mRNA produces active FOG-1 protein and the shorter one produces either nonfunctional FOG-1 or a protein that antagonizes the active form.
Might CPEB specify germ-line fates in other organisms? In C. elegans, this fate decision has been unusually accessible to genetic dissection, because a clean sexual transformation from sperm to oocyte can be obtained when the sex determination pathway is manipulated. The global sex determination pathway specifies male or female fates in all tissues, including all somatic tissues and the germ line (Meyer 1997). In addition, germ line-specific sex-determining genes have been identified (Schedl 1997). By contrast, the controls of germ-line sexual fates in Drosophila and vertebrates are more complex and have been more difficult to analyze. An important challenge for the future is to determine whether the mechanism used for sex determination in the germ line of C. elegans specifies gametes in other metazoans as well.
The C. elegans CPEBs appear to be dispensable for oogenesis
In frogs, flies, and clams, CPEBs are essential during oogenesis. By contrast, in C. elegans, cpb-1 and fog-1 control spermatogenesis and have no essential role in oogenesis: Oocytes that lack fog-1, cpb-1, or both are functional (Barton and Kimble 1990; this paper). Intriguingly, cpb-3 mRNA is enriched during oogenesis and its predicted protein, CPB-3, is most closely related by sequence to Xenopus CPEB and Drosophila Orb. Nonetheless, no effect on oogenesis was observed by cpb-3(RNAi), either alone or in combination with the other homologs. Furthermore, RNAi directed against the remaining CPEB homolog cpb-2, which is actually enriched during spermatogenesis, has no dramatic effect on any tissue, and RNAi directed against cpb-1, cpb-2, cpb-3 into a fog-1 mutant, which is predicted to remove all CPEB activity from the animal, results in only the fog-1 defect. Thus, we have been unable to detect any effect on oogenesis by removing one or more CPEBs singly or in combination. Because these are negative results and rely on RNAi to reduce expression, they are inconclusive. However, RNAi against other CPEB homologs (fog-1 and cpb-1) is effective, as fog-1(RNAi) mimics a fog-1 null mutant, and cpb-1(RNAi) removes CPB-1 protein and yields a completely penetrant and dramatic effect on spermatogenesis.
With this caveat, we suggest that C. elegans CPEBs are not essential for oogenesis, and meiosis of oocytes in particular. In light of CPEB functions in the oocytes of other organisms, what does this mean? We envision two possible explanations. One possibility is that the regulatory machinery controlling meiosis during oogenesis in flies, frogs, and clams has been switched to control meiosis during spermatogenesis in C. elegans. This view gains credibility from the finding that the role of the DAZ RNA-binding protein is reversed in C. elegans compared with flies and humans: In C. elegans, DAZ-1 is required for oogenesis but not spermatogenesis (Karashima et al. 2000), whereasDrosophila DAZ, called boule, is essential for spermatogenesis but not oogenesis (Eberhart et al. 1996), and the human DAZ gene cluster is implicated in spermatogenesis (Reijo et al. 1996). If this idea is right, a sex reversal in the regulatory machinery may have occurred during evolution of C. elegans. An alternative and perhaps more likely explanation is that CPEB and DAZ RNA-binding proteins were used in a common metazoan ancestor to regulate common germ-line processes (such as meiosis) during both oogenesis and spermatogenesis. During evolution, the functions of these genes may have become more specialized. Because much work to date has focused on the role of CPEB in oogenesis, sperm functions may have been missed in other organisms. In particular, Orb protein is present in Drosophila males (Lantz et al. 1994). Although orb mutant males are fertile, a second CPEB homolog has been identified in the fly genome and could play a critical role in spermatogenesis. Similarly, CPEB mRNAs are found in mouse testes (Gebauer and Richter 1996). These findings suggest that spermatogenic functions of CPEB homologs may be common.
CPEBs and FBF during spermatogenesis in C. elegans
A working model depicting the role of CPEBs during C. elegans spermatogenesis is presented in Figure 11. FOG-1 is essential for the initial specification of a germ cell to the sperm fate. Once that fate has been established, CPB-1 is required to initiate maturation of the primary spermatocyte. FOG-1 and CPB-1 therefore act at two nodal decision points during spermatogenesis. Because CPEB can act as both a repressor and activator (for review, see Richter 2000), we suggest that this RNA-binding protein may be particularly well-suited to the role of a molecular switch, consistent with its being used twice in that fashion during C. elegans spermatogenesis.
Figure 11.
A model for the functions of CPEBs during spermatogenesis in C. elegans. Germ cells differentiate as either sperm or oocytes. FOG-1 is required for the decision to become a sperm, whereas CPB-1 is required for implementation of this decision at an early step in spermatogenesis. CPB-1 and FBF proteins interact and FBF is required for a late step in spermatogenesis, suggesting that the CPB-1/FBF interaction promotes this step.
CPB-1 may play a role in spermatogenesis beyond its function in promoting progression through meiosis. This hypothesis is based on the physical interaction of CPB-1 with FBF. The CPB-1/FBF interaction occurs independently of RNA binding, and it is mediated by the first 80 amino acids of CPB-1. FBF is an RNA-binding protein that controls the switch from spermatogenesis to oogenesis and is a homolog of Drosophila Pumilio (Zhang et al. 1997). It binds to a regulatory element in the fem-3 3′ UTR and represses its activity (Zhang et al. 1997). For this process, FBF interacts with NOS-3 protein (Kraemer et al. 1999). We show here that FBF is likely to be required for some step in spermatogenesis in addition to its previously identified role in the sperm/oocyte switch. Because male gametes produced in the absence of CPB-1 arrest early in spermatogenesis, we can not assess whether cpb-1 is also required for subsequent steps. However, as shown in Figure 11, we suggest one simple model—that cpb-1 functions in an fbf-independent fashion for progression through meiosis but then binds to and cooperates with fbf to promote a later step in spermatogenesis. Similarly, orb has multiple roles during oogenesis in Drosophila (Lantz et al. 1994). The interaction of FBF with both CPB-1 and NOS-3 for apparently distinct functions underscores the possibility of a combinatorial network of translational regulators in germ-line development.
In other organisms, CPEB homologs bind to and regulate multiple mRNAs (Richter 2000). The identification of mRNA targets for both FOG-1 and CPB-1 should help illuminate the mechanism by which the CPEB family of regulators control spermatogenesis. Furthermore, analysis of the targets may also reveal how a family of translational regulators diverged during evolution to adopt novel biological roles.
Materials and methods
Supplementary information is available at the journal web site (http://www.genesdev.org/supplemental/) and at the Wickens laboratory web site (http://biochem.wisc.edu/wickens/research/publications). The supplementary information includes methods of cDNA isolation and analysis, sequencing mutant alleles, RNAi, antibody preparation and staining, two-hybrid screens, and affinity chromatography.
Strains
C. elegans N2 variety Bristol was used as wild type. Strains were cultured as described by Brenner (1974). The following genes and mutations were also used: glp-4(bn2) I, glp-1(q224) III, fem-3(q20gf) IV, fem-1(hc17) IV, fog-1(q253) I, him-5(e1490) V, and unc-13(e1091) fog-3(q469) I. All strains were maintained at 20°C except the temperature-sensitive strains, which were maintained at 15°C and shifted to 25°C for analysis.
Northern analysis
Wild-type and temperature-sensitive strains of worms were grown at the permissive temperature (15°C) and a synchronous population of embryos was isolated by bleaching the adults. The embryos were then shifted to the restrictive temperature (25°C) and grown to adulthood. RNA was isolated using the Tri-Reagent (Molecular Research) as recommended by the manufacturer. The RNA was glyoxalated, run on a 1.2% agarose gel, and transferred to Hybond-N membrane (Amersham) by capillary action as described (Sambrook et al. 1989). For cpb-1, cpb-2, cpb-3 and fog-1, the membrane was probed with antisense 32P-labeled ssRNA probes prepared by run-off transcription. The probes for these genes correspond to the same regions used for RNAi (see Supplemental information). For rpl-12, the membrane was probed with a PCR-generated 32P-labeled dsDNA probe corresponding to nucleotides 161–284 of the open reading frame of cosmid JC8.3.
Analysis of the roles of cpb-1 and fbf in males
him-5(e1490) hermaphrodites were subjected to RNAi directed against either cpb-1 or fbf. Male cpb-1(RNAi) progeny were mated to unc-13(e1091) fog-3(q469) females at 20°C for 44 h and successful matings were scored. A successful mating is defined as production of at least one embryo over this period. Male fbf(RNAi) progeny were mated to fog-1(q253ts) females at 25°C for 48 h and successful matings were scored. Embryos from these matings that did not hatch within 24 h after being laid were considered dead.
Acknowledgments
We are grateful to Suk-Won Jin and Ron Ellis for communication of results prior to publication. We also thank Yuji Kohara, Sam Ward, and Quinn Mitrovich for providing reagents. Discussions with the Wickens and Kimble laboratories were extremely beneficial. The work was greatly facilitated by information from the C. elegans Genome Consortium. We appreciate the assistance of Anne Helsley-Marchbanks and the UW-Biochemistry Media Lab in preparing the manuscript and figures. The work was supported by an NIH research grant (GM50942 to M.W.). J.K. is an investigator of the Howard Hughes Medical Institute (HHMI). Predoctoral fellowships from HHMI, the UW Molecular Biosciences Training Grant (NIH), and the UW Biotechnology Training Grant (NIH) supported C.L., M.G., and B.K., respectively.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL wickens@biochem.wisc.edu; FAX (608) 262-9108.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.831700.
References
- Bally-Cuif L, Schatz WJ, Ho RK. Characterization of the zebrafish Orb/CPEB-related RNA binding protein and localization of maternal components in the zebrafish oocyte. Mech Dev. 1998;77:31–47. doi: 10.1016/s0925-4773(98)00109-9. [DOI] [PubMed] [Google Scholar]
- Barkoff AF, Dickson KS, Gray NK, Wickens M. Translational control of cyclin B1 mRNA during meiotic maturation: Coordinated repression and cytoplasmic polyadenylation. Dev Biol. 2000;220:97–109. doi: 10.1006/dbio.2000.9613. [DOI] [PubMed] [Google Scholar]
- Barton MK, Kimble J. fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics. 1990;125:29–39. doi: 10.1093/genetics/125.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JS, Tan L, Schedl P. The Drosophila CPEB homolog, orb, is required for oskar protein expression in oocytes. Dev Biol. 1999;215:91–106. doi: 10.1006/dbio.1999.9444. [DOI] [PubMed] [Google Scholar]
- Christerson LB, McKearin DM. orb is required for anteroposterior and dorsoventral patterning during Drosophila oogenesis. Genes & Dev. 1994;8:614–628. doi: 10.1101/gad.8.5.614. [DOI] [PubMed] [Google Scholar]
- de Moor CH, Richter JD. Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 1999;18:2294–2303. doi: 10.1093/emboj/18.8.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhart CG, Maines JZ, Wasserman SA. Meiotic cell cycle requirement for a fly homologue of human Deleted in Azoospermia. Nature. 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fox CA, Sheets MD, Wickens MP. Poly(A) addition during maturation of frog oocytes: Distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes & Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Richter JD. Mouse cytoplasmic polyadenylylation element binding protein: An evolutionarily conserved protein that interacts with the cytoplasmic polyadenylylation elements of c-mos mRNA. Proc Natl Acad Sci. 1996;93:14602–14607. doi: 10.1073/pnas.93.25.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Synthesis and function of Mos: The control switch of vertebrate oocyte meiosis. BioEssays. 1997;19:23–28. doi: 10.1002/bies.950190106. [DOI] [PubMed] [Google Scholar]
- Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989;342:749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- Hodgkin JA, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte J, Stutz A, O'Connell ML, Gubler P, Belin D, Darrow AL, Strickland S, Vassalli JD. Transient translational silencing by reversible mRNA deadenylation. Cell. 1992;69:1021–1030. doi: 10.1016/0092-8674(92)90620-r. [DOI] [PubMed] [Google Scholar]
- Huynh J, St Johnston D. The role of BicD, Egl, Orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development. 2000;127:2785–2794. doi: 10.1242/dev.127.13.2785. [DOI] [PubMed] [Google Scholar]
- Karashima T, Sugimoto A, Yamamoto M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development. 2000;127:1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr Biol. 1999;9:1009–18. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- L'Hernault SW. Spermatogenesis. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 271–294. [PubMed] [Google Scholar]
- Lantz V, Chang JS, Horabin JI, Bopp D, Schedl P. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes & Dev. 1994;8:598–613. doi: 10.1101/gad.8.5.598. [DOI] [PubMed] [Google Scholar]
- McGrew LL, Dworkin-Rastl E, Dworkin MB, Richter JD. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes & Dev. 1989;3:803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Meyer BJ. Sex determination and X chromosome dosage compensation. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 209–240. [PubMed] [Google Scholar]
- Minshall N, Walker J, Dale M, Standart N. Dual roles of p82, the clam CPEB homolog, in cytoplasmic polyadenylation and translational masking. RNA. 1999;5:27–38. doi: 10.1017/s1355838299981220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. XIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6:63–70. doi: 10.1093/dnares/6.1.63. [DOI] [PubMed] [Google Scholar]
- Ralle T, Gremmels D, Stick R. Translational control of nuclear lamin B1 mRNA during oogenesis and early development of Xenopus. Mech Dev. 1999;84:89–101. doi: 10.1016/s0925-4773(99)00078-7. [DOI] [PubMed] [Google Scholar]
- Reijo R, Seligman J, Dinulos MB, Jaffe T, Brown LG, Disteche CM, Page DC. Mouse autosomal homolog of DAZ, a candidate male sterility gene in humans, is expressed in male germ cells before and after puberty. Genomics. 1996;35:346–352. doi: 10.1006/geno.1996.0366. [DOI] [PubMed] [Google Scholar]
- Richter JD. Influence of polyadenylation-induced translation on metazoan development and neuronal synaptic function. In: Hershey JWB, Mathews M, Sonenberg N, editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 785–806. [Google Scholar]
- Sagata N. What does Mos do in oocytes and somatic cells? BioEssays. 1997;19:13–21. doi: 10.1002/bies.950190105. [DOI] [PubMed] [Google Scholar]
- Sallés FJ, Darrow AL, Oconnell ML, Strickland S. Isolation of novel murine maternal messenger RNAs regulated by cytoplasmic polyadenylation. Genes & Dev. 1992;6:1202–1212. doi: 10.1101/gad.6.7.1202. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schedl T. Developmental genetics of the germ line. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 241–269. [PubMed] [Google Scholar]
- Stebbins-Boaz B, Hake LE, Richter JD. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- Stutz A, Conne B, Huarte J, Gubler P, Volkel V, Flandin P, Vassalli JD. Masking, unmasking, and regulated polyadenylation cooperate in the translational control of a dormant mRNA in mouse oocytes. Genes & Dev. 1998;12:2535–2548. doi: 10.1101/gad.12.16.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Varnum SM, Wormington WM. Deadenylation of maternal mRNAs during Xenopus oocyte maturation does not require specific cis-sequences: A default mechanism for translational control. Genes & Dev. 1990;4:2278–2286. doi: 10.1101/gad.4.12b.2278. [DOI] [PubMed] [Google Scholar]
- Walker J, Minshall N, Hake L, Richter J, Standart N. The clam 3′ UTR masking element-binding protein p82 is a member of the CPEB family. RNA. 1999;5:14–26. doi: 10.1017/s1355838299981219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Roberts TM, Strome S, Pavalko FM, Hogan E. Monoclonal antibodies that recognize a polypeptide antigenic determinant shared by multiple Caenorhabditis elegans sperm-specific proteins. J Cell Biol. 1986;102:1778–1786. doi: 10.1083/jcb.102.5.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. The MyoD family and myogenesis: Redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Wickens M, Goodwin EB, Kimble J, Strickland S, Hentze M. Translational control in developmental decisions. In: Hershey JWB, Mathews M, Sonenberg N, editors. Translational control of gene expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 295–370. [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kraemer B, SenGupta D, Fields S, Wickens M. Yeast three-hybrid system to detect and analyze interactions between RNA and protein. Methods Enzymol. 1999;306:93–113. doi: 10.1016/s0076-6879(99)06007-3. [DOI] [PubMed] [Google Scholar]