Abstract
Context:
Whole slide imaging (WSI) for digital pathology involves the rapid automated acquisition of multiple high-power fields from a microscope slide containing a tissue specimen. Capturing each field in the correct focal plane is essential to create high-quality digital images. Others have described a novel focusing method which reduces the number of focal planes required to generate accurate focus. However, this method was not applied dynamically in an automated WSI system under continuous motion.
Aims:
This report measures the accuracy of this method when applied in a rapid continuous scan mode using a dual sensor WSI system with interleaved acquisition of images.
Methods:
We acquired over 400 tiles in a “stop and go” scan mode, surveying the entire z depth in each tile and used this as ground truth. We compared this ground truth focal height to the focal height determined using a rapid 3-point focus algorithm applied dynamically in a continuous scanning mode.
Results:
Our data showed the average focal height error of 0.30 (±0.27) μm compared to ground truth, which is well within the system's depth of field. On a tile by tile assessment, approximately 95% of the tiles were within the system's depth of field. Further, this method was six times faster than acquiring tiles compared to the same method in a non-continuous scan mode.
Conclusions:
The data indicates that the method employed can yield a significant improvement in scan speed while maintaining highly accurate autofocusing.
Keywords: Autofocus, digital pathology, whole slide imaging
INTRODUCTION
Whole slide imaging (WSI) is the digital image acquisition of an entire pathology tissue sample (solid or fluid) from a glass slide.[1,2] Several preliminary studies using first-generation WSI scanners suggest that digital anatomic pathology images are approaching the fidelity of a microscope and may be used in niche clinical applications.[3–10] However, it has been noted that image quality is an important limitation from achieving higher performance.[4,6,9,10] Traditional WSI scanners create digital images by acquiring multiple high-resolution images, or frames, that are subsequently aligned or stitched together to create a complete and seamless representation of the original tissue section. Image quality can be limited by one or any combination of factors including illumination quality, optics, sensors, stage alignment and accuracy, post image processing and focus algorithms.[1] In relation to these aspects, autofocus algorithm development is a nascent discipline, and thus likely a main influencer of image quality for WSI. Accordingly, several studies that implicated image quality as a source of error further implicated poor focus as the main culprit for poor image quality.[4,6,9,10]
Thin tissue sections do not maintain a perfect planar surface when placed on a glass substrate, resulting in variations in the focal height throughout the tissue. A manual microscope allows the user to adjust fine focus to compensate for slight variations in tissue topography. However, automated imaging systems must determine the optimal focal plane for each acquired image. For an automated system to properly calculate focus, as many as 20–50 images (z-stacks) need to be acquired along the optical axis (z-plane dimension) within a single frame dimension.[11–13] Each series of z-stacks is analyzed for a figure of merit, such as edge or contrast, to calculate the ideal z-plane for focus. To repeat z-stack acquisitions and calculations on each tile of a whole slide image would require a prohibitive amount of time for high throughput scanning that is required for routine clinical use. Thus, first-generation WSI scanners do not typically acquire data for the ideal focal height on each frame, but instead rely on a focus map model attained from a series of preselected subsampled focus points. However, ideally, a true focus point would be determined for each frame.
Yazdanfar et al. have previously described a novel method to reduce the amount of time required to perform standard image-based autofocus.[14] A reciprocal Brenner gradient scheme was employed which required as little as three intermediate images along the optical axis to bring a sample into accurate focus. The method was demonstrated to be accurate to within less than 1 μm of the ground truth focal height when compared in a static imaging (stop motion) paradigm. Further, the authors demonstrated a scan time of 36 minutes for a typical hematoxylin and eosin (H and E) stained tissue sample consisting of 441 individual tiles. Although promising for WSI, this method was too slow and not employed in a rapid continuous scanning system. Further, a tile by tile analysis comparing the accuracy of each tile to a ground truth was not determined for a full high throughput continuous scan mode. We extend this method by describing a novel dual camera design that employs the reciprocal Brenner method and significantly decreases scan times from the previous embodiment. We further demonstrate the feasibility and accuracy of this system for high-throughput WSI.
METHODS
Scanner and Scan Parameters
A commercial BX41 Olympus (Olympus Corp, Center Valley, PA, USA) microscope frame was adapted with a Prior stage and controller (Prior Scientific, Rockland, MA, USA). Two CCD based cameras (Imperex, Boca Raton, FL, USA) were co-aligned on a custom mount and imaging was performed with a 20×, 0.75NA UPlanSApo lens (Olympus Corp.). Illumination was accomplished with a Thor Labs (Thor Labs, Newton, NJ, USA) white light emitting diode (LED). All the scanning was performed at 20× magnification, which yielded images of 0.37 μm/pixel.
Image capture was generally performed as previously described with adaptations to allow for interleaving of focus to be performed by one camera and image capture by the second camera. The focus camera was binned and windowed to allow for faster frame rates than the imaging camera. Three tissue types were chosen (liver, brain and prostate) which differed on both structural content and stain uptake. The scan plan was configured to capture approximately 150 tiles per tissue. Each tissue type was scanned with three scan modes: a) ground truth (stop and go with extensive z-plane sampling); b) static 3-point (stop and go with minimal z-plane sampling using only three planes); and c) dynamic predictive focus (stage in continuous motion, minimal z-plane sampling using only three planes). In each case, the optimal focal plane was calculated and the objective moved to that height for acquisition by the tile camera. The z-height for each tile was recorded for analysis. For each scan mode, autofocusing was performed with the Auto-Focus camera, while the image acquisition was performed using the Tile camera. Ground truth scanning was performed by acquiring 50 images along the optical axis at 0.1 μm step sizes and obtaining the position where maximum figure of merit is calculated.[11,14]
Static 3-point scanning was performed by stopping at each tile in the scan plan and allowing the focus camera to acquire three full width frames at 0 μm, +5 μm and –5 μm relative to the nominal focus position of the previous frame. A parabola is fit to the 3 figures of merit to predict the focal z-height.[14] Upon completion, the field of view moves to the next tile in the scan plan, and the autofocus process is repeated until the entire scan plan is covered.
Dynamic predictive focus scanning was performed similarly to stop motion, but the x–y stage was in continuous motion during focus and tile acquisitions. The autofocus camera acquires three consecutive images (spaced along the x-direction as well the z-direction) between adjacent tiles for focusing. In this case, the figure of merit is calculated only on the region of those images which overlap, significantly reducing the region used for calculation.
3-Point Autofocus Simulation
The simulation is performed using a customized tool that uses the mathematical model of the autofocus algorithm. The analysis is performed on a single tile basis. Initially, an image stack along the optical axis is obtained with 0.1 μm spacing (for a total of 20 μm). The ground truth for the tile is obtained by analyzing the stack to calculate at which z-plane the figure of merit is maximal.[14] The z-height of the image with maximal figure of merit value is the ground truth for the tile. To simulate the autofocus during live scanning, the model assumes that the current z-position at each tile will be within 2 μm of the ground truth. After setting the input parameters, such as the binning size, autofocus image size, number of autofocus images, and spacing between images, the tool uses each image in the stack within the 2-μm spacing of the ground truth as the nominal image, and obtains two more images from the stack at ±5 μm from that image. The Brenner figure of merit of each set of three images is used to predict the z-plane for best focus, and the difference of the actual and prediction is calculated to get a prediction error.
To simulate the static 3-point scanning, the input parameters are set to a 2 × 2 binning, full-width autofocus image size, and three autofocus images with 5 μm spacing. The program uses these inputs to obtain the figure of merit values of the three process images and calculates the prediction error. To simulate the dynamic scanning, the same input parameters are used, except that the autofocus image size is reduced to encompass only the region where the three autofocus images would overlap during a dynamic scan, and the prediction error is calculated.
RESULTS
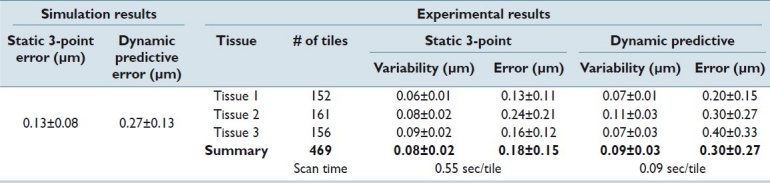
Table 1 shows the simulation and experimental results for focus prediction error of the two scanning methods on the three different tissue types as compared to the ground truth. From the mathematical model, the algorithm is estimated to have a 0.13 μm error for the static scan and 0.27 μm error for the dynamic scan. Each scan mode in the study was repeated 10 times to calculate the system repeatability. There was little difference in the repeatability of each scan mode. The average prediction error is 0.18 μm for the static method and 0.30 μm for the dynamic focusing method, showing close agreement to the simulation data. The total scan time (cumulative for all three tissues) was 257 and 42 seconds for the static and dynamic scan modes, respectively.
Table 1.
Experimental results of focal prediction error
Since there was tile to tile variability in accuracy to ground truth, we plotted the variance from the ground truth (focal prediction error) along the optical axis for each tile and compared to the system's calculated depth of field [Figure 1]. Although the error for the dynamic predictive focus scan is greater when compared to the static 3-point scan, approximately 95% of the tiles are within the system's depth of field (±0.8 μm). This result is expected since the static 3-point method is able to use the full field of view of its binned sensor (i.e. is not restricted due to shifting decreased overlap of autofocus images caused by stage motion during scanning), thus generating better sampling. The static 3-point scan error demonstrates the accuracy a simple 3-point figure of merit fit would give. The dynamic scan error combines the inherent 3-point fit error and the error generated from subsampling, adjacent-tile prediction, and continuous motion.
Figure 1.
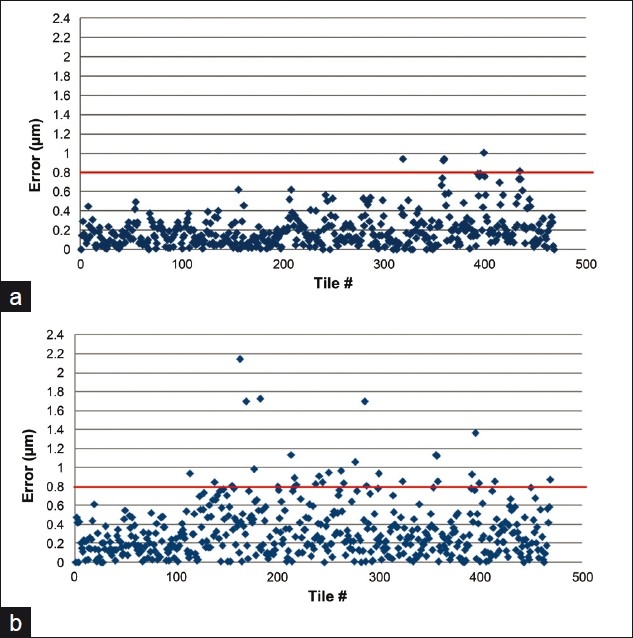
Focal prediction error. The prediction error for each tile with (a) a static 3-point scan and (b) a dynamic predictive focus scan. The line at 0.8 μm indicates the system's theoretical depth of field
To illustrate the number of tiles that fall either inside or outside the system's theoretical depth of field, we plotted each tile against its z-position for both stop and go and continuous scanning [Figure 1]. In the dynamic continuous scan mode, approximately 95% of the tiles are imaged within the system's depth of field, suggesting such errors will not be technically out of focus. The theoretical depth of field is not necessarily where the human observer can notice changes in focus. Figure 2 shows representative images for each tissue set that are at various distances along the optical axis from the ideal focus height. Images that lie beyond 0.8 μm still appear to be in relatively good focus. This confirms other studies performed by our group that the observed depth of field is greater than the theoretical (data not shown) value.
Figure 2.
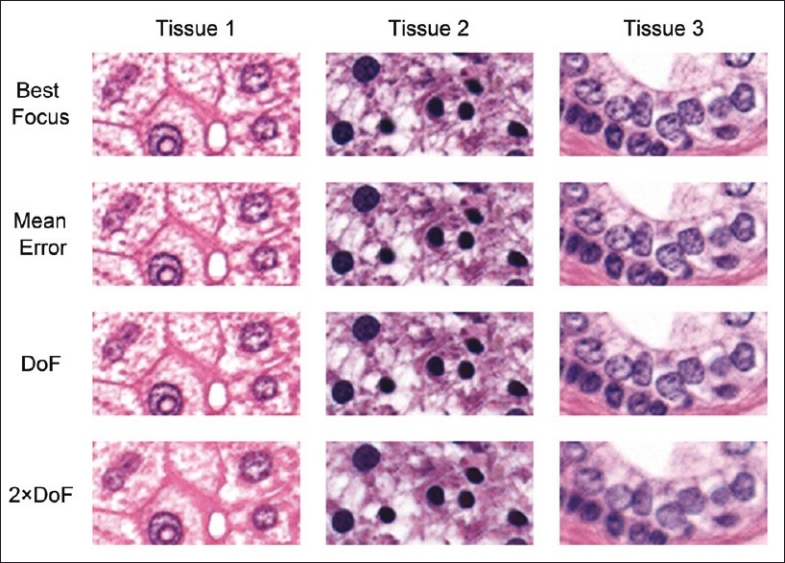
Focus quality. Representative images from the three tissues used in this study at various focal height errors. Best = the ideal focal plane from the ground truth data set. Mean error = the focal plane at the mean focal error of the dynamic scanning system (0.3 μm from best focus). DoF = at the system's theoretical depth of field (±0.8 μm). 2 × DoF = twice the system's theoretical depth of field (±1.6 μm)
DISCUSSION
Using a novel dual camera imaging system coupled with a novel autofocus method, we have demonstrated the feasibility of a highly accurate and rapid scanning system with potential application to digital pathology. In this study, the system displayed an average autofocus accuracy of 0.30 (± 0.27) μm compared to ground truth, which is well within the system's depth of field. On a tile by tile assessment, approximately 95% of the tiles were within the system's depth of field, across three different tissue types, indicating a highly accurate autofocus capability. Further, of the 5% of tiles not within the depth of field, more than half were no more than 0.2 μm outside of this limit, suggesting that most tiles outside the depth of field would not be considerably out of focus, thus the practical capability may be greater than reported here.
In dynamic scan mode, the system is six times faster than using the same focus method in a static “stop and go” paradigm, indicating its application to high-throughput laboratory environments. To put this into a practical context, an average tissue sample consisting of 400 tiles would require an acquisition time (focusing and acquiring tiles only) of 220 seconds using the static method described in this study. Conversely, in continuous dynamic scan mode, the same amount of tiles would require 36 seconds to focus and acquire tiles. This is a significant improvement in scan speed compared to the current generation of WSI scanners.[1]
The tissue scan plan used in this study was set manually to deliberately avoid the edge of tissue or “white space”, and encompassed an area of roughly 9 mm by 9 mm . It is possible that the method may be less accurate when such areas are taken into consideration. However, it is possible that special focus algorithms could be designed to handle tiles with significant white space. We did observe a variance in focus accuracy among the three different tissue types, with almost a twofold difference between tissue 1 and tissue 3. Although it does not appear to be statistically significant, this trend could suggest different tissue types affect focus algorithm accuracy. One explanation for this discrepancy is the apparent lack of nucleated cells in some regions of tissue 3 compared to tissue 1. Such tissues would provide less contrast, which is an important aspect of image-based autofocusing. Further investigation is needed to understand the relationship between focus algorithm accuracy and tissue type. Additionally, due to this tissue dependence, it would not be surprising to find stain dependence as well. While this study concentrates only on hematoxylin and eosin stained tissue sections, evaluation of conventional immunohistochemistry or special stains is warranted and will be the subject of additional studies.
Traditional WSI scanners that are commercially available today employ a “focus map” method of autofocus. Such methods rely on a predetermined amount of focus points selected by the system automatically or manually by the user. From these focus points, a theoretical map of the tissue topography is established and a scan plan determined. Thus, the majority of the scan relies on modeled data, which is only as good as the location and amount of pre-focus points. The method described in this report establishes a focus point calculation for each tile; thus, a tissue section with 1000 tiles will have 1000 focus points. Although the number of pre-focus points varies for scan map methods, the number is rarely greater than 30 for a typical tissue section. Thus, the method described in this report generates far more data points while maintaining a reasonably short scan time. Due to the inability to manipulate commercially available scanners for establishing and recording ground truth z-plane data, the accuracy of the method described in this report compared to traditional WSI scanner could not be determined. Thus, it is not clear if predictive focusing is more or less accurate than traditional methods. However, we have demonstrated that it is possible to acquire an order of magnitude greater focus sampling in approximately the same or less period of time than traditional scanning systems.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2011/2/1/38/84231
REFERENCES
- 1.Rojo MG, Garcia GB, Mateos CP, Garcia JG, Vicente MC. Critical comparison of 31 commercially available digital slide systems in pathology. Int J Surg Pathol. 2006;14:285–305. doi: 10.1177/1066896906292274. [DOI] [PubMed] [Google Scholar]
- 2.Weinstein RS, Graham AR, Richter LC, Barker GP, Krupinski EA, Lopez AM, et al. Overview of telepathology, virtual microscopy, and whole slide imaging: prospects for the future. Hum Pathol. 2009;40:1057–69. doi: 10.1016/j.humpath.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Dangott B, Parwani A. Whole slide imaging for teleconsultation and clinical use. J Pathol Inform. 2010;1:pii–7. doi: 10.4103/2153-3539.65342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans AJ, Chetty R, Clarke BA, Croul S, Ghazarian DM, Kiehl TR, et al. Primary frozen section diagnosis by robotic microscopy and virtual slide telepathology: The University Health Network experience. Hum Pathol. 2009;40:1070–81. doi: 10.1016/j.humpath.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Fine JL, Grzybicki DM, Silowash R, Ho J, Gilbertson JR, Anthony L, et al. Evaluation of whole slide image immunohistochemistry interpretation in challenging prostate needle biopsies. Hum Pathol. 2008;39:564–72. doi: 10.1016/j.humpath.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Gilbertson JR, Ho J, Anthony L, Jukic DM, Yagi Y, Parwani AV. Primary histologic diagnosis using automated whole slide imaging: A validation study. BMC Clin Pathol. 2006;6:4. doi: 10.1186/1472-6890-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho J, Parwani AV, Jukic DM, Yagi Y, Anthony L, Gilbertson JR. Use of whole slide imaging in surgical pathology quality assurance: Design and pilot validation studies. Hum Pathol. 2006;37:322–31. doi: 10.1016/j.humpath.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Jara-Lazaro AR, Thamboo TP, Teh M, Tan PH. Digital pathology: Exploring its applications in diagnostic surgical pathology practice. Pathology. 2010;42:512–8. doi: 10.3109/00313025.2010.508787. [DOI] [PubMed] [Google Scholar]
- 9.Wilbur DC, Madi K, Colvin RB, Duncan LM, Faquin WC, Ferry JA, et al. Whole-slide imaging digital pathology as a platform for teleconsultation: A pilot study using paired subspecialist correlations. Arch Pathol Lab Med. 2009;133:1949–53. doi: 10.1043/1543-2165-133.12.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Massone C, Peter Soyer H, Lozzi GP, Di Stefani A, Leinweber B, Gabler G, et al. Feasibility and diagnostic agreement in teledermatopathology using a virtual slide system. Hum Pathol. 2007;38:546–54. doi: 10.1016/j.humpath.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Brenner JF, Dew BS, Horton JB, King T, Neurath PW, Selles WD. An automated microscope for cytologic research a preliminary evaluation. J Histochem Cytochem. 1976;24:100–11. doi: 10.1177/24.1.1254907. [DOI] [PubMed] [Google Scholar]
- 12.Firestone L, Cook K, Culp K, Talsania N, Preston K., Jr Comparison of autofocus methods for automated microscopy. Cytometry. 1991;12:195–206. doi: 10.1002/cyto.990120302. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, Duthaler S, Nelson BJ. Autofocusing in computer microscopy: Selecting the optimal focus algorithm. Microsc Res Tech. 2004;65:139–49. doi: 10.1002/jemt.20118. [DOI] [PubMed] [Google Scholar]
- 14.Yazdanfar S, Kenny KB, Tasimi K, Corwin AD, Dixon EL, Filkins RJ. Simple and robust image-based autofocusing for digital microscopy. Opt Express. 2008;16:8670–7. doi: 10.1364/oe.16.008670. [DOI] [PubMed] [Google Scholar]


