Abstract
Cytokinesis ensures the successful completion of the cell cycle and distribution of chromosomes, organelles, and cytoplasm between daughter cells. It is accomplished by formation and constriction of an actomyosin contractile ring that drives the progression of a cleavage furrow. Microinjection experiments and in vitro transfection assays have suggested a requirement for small GTPases of the Rho family in cytokinesis. Yet, the identity of proteins regulating Rho signaling pathways during cytokinesis remains unknown. Here we show that in Drosophila, Pebble (Pbl), a putative exchange factor for Rho GTPases (RhoGEF), is required for the formation of the contractile ring and initiation of cytokinesis. The dynamics of Pbl expression and its distribution during mitosis, as well as structure-function analysis, indicate that it is a key regulatory component of the pathway. pbl interacts genetically with Rho1, but not with Rac1 or Cdc42, and Pbl and Rho1 proteins interact in vivo in yeast. Similar to mutations in pbl, loss of Rho1 or expression of a dominant–negative Rho1 blocks cytokinesis. Our results identify Pbl as a RhoGEF specifically required for cytokinesis and linked through Rho1 activity to the reorganization of the actin cytoskeleton at the cleavage furrow.
Keywords: Cytokinesis, contractile ring, RhoGEF, Rho1 GTPase, pebble
Cytokinesis in animal cells is accomplished by formation and constriction of a cleavage furrow that divides a cell in two daughter cells. The driving force behind this constriction is an actomyosin contractile ring that forms at the cell equator in late anaphase. The assembly of the contractile ring involves local reorganization of actin filaments around the cell equator into a cortical complex consisting, in addition to actin, of myosin, septins, and other cytoskeletal proteins (Goldberg et al. 1998). In Drosophila, genetic analysis and biochemical studies have led to the identification of a number of genes required for the assembly or function of the contractile ring (Field et al. 1999). Structural components of the contractile ring include actin, the regulatory light chain of nonmuscle myosin [encoded by spaghetti-squash (sqh), Karess et al. 1991], septins [Peanut (Pnut) (Neufeld and Rubin 1994), Septin-1 (Fares et al. 1995), and Septin-2 (Field et al. 1996)], and actin-binding proteins cofilin [encoded by twinstar (tsr), Gunsalus et al. 1995], profilin [encoded by chickadee (chic), Giansanti et al. 1998), and anillin (Field and Alberts 1995; Giansanti et al. 1999)]. Mutations in sqh, pnut, tsr, and chic result in a failure of cytokinesis, most likely due to defects in contractile ring organization or function. Yet, little is known about the roles of these proteins in contractile ring assembly, stabilization, constriction, and disassembly or about the molecular machinery regulating actin cytoskeleton reorganization during cytokinesis.
Small guanosine triphosphatases (GTPases) of the Rho family control organization of the cytoskeletal architecture in all eukaryotic cells and have been implicated in many actin-based processes including cell motility, cell adhesion, chemotaxis, axon guidance, and cytokinesis (for review, see Van Aelst and D’Souza-Schorey 1997; Hall 1998). Rho GTPases act as molecular switches, cycling between inactive (GDP-bound) and active (GTP-bound) states. Rho proteins are activated by guanine nucleotide exchange factors (GEFs), which enhance the exchange of bound GDP for GTP, and are inactivated by GTPase-activating proteins (GAPs), which increase the intrinsic GTPase activity of Rho proteins. Activation of Rho GTPases results in a conformational change of the protein revealing structural domains required for the interaction with downstream target proteins. Thus, the intracellular ratio of the GTP/GDP-bound forms of Rho proteins determines the activation of signal transduction pathways regulating the spatial and temporal reorganization of cytoskeletal architecture.
Here, we show that in Drosophila, a putative guanine nucleotide exchange factor (Pbl) and its downstream effector (Rho1 GTPase) are required for the initiation of cytokinesis. We propose a molecular pathway that links a RhoGEF through the activity of small GTPase to reorganization of the actin cytoskeleton at the cell equator leading to the assembly of a contractile ring and initiation of cytokinesis. We provide several lines of evidence suggesting that Pbl is a key regulatory component of this pathway acting upstream of Rho1 to regulate the behavior of cytoskeletal and structural proteins known to be required for cytokinesis.
Results
Mutations in pbl result in the absence of a contractile ring and block of cytokinesis
We and others have demonstrated previously that mutations in pbl lead to a complete block of cytokinesis in mitotic cycle 14 (Hime and Saint 1992; Lehner 1992). Later rounds of nuclear divisions without cytokinesis result in the formation of polyploid multinucleate cells (Fig. 1A,B). Other events of the cell cycle (including nuclear envelope breakdown, chromosome condensation, and assembly and function of mitotic spindle) are not affected (Hime and Saint 1992; Lehner 1992), and during cycle 15 two mitotic figures are formed that independently enter anaphase (Fig. 1C,D). Affected cells fail to initiate a cleavage furrow (Fig. 1D), suggesting a defect in contractile ring function. To further characterize the defects in cytokinesis, we examined the localization of actin, Pnut, and anillin, components of the contractile ring, in pbl mutant cells. In wild-type cells, actin is associated with the cell cortex throughout mitosis and accumulates in the equatorial region of the cell, in which the contractile ring is assembled (Fig. 1E). Anillin, an actin-binding protein required for cytokinesis, is thought to play a role in organizing contractile domains of the actin cytoskeleton (Field and Alberts 1995; Giansanti et al. 1999). In wild-type cells, anillin accumulates at the cleavage furrow at the onset of anaphase and is restricted to the contractile ring during anaphase and telophase (Fig. 1H). Pnut, a member of the septin family of proteins (Neufeld and Rubin 1994), accumulates at the cleavage furrow in late anaphase and is associated with the contractile ring during telophase (Fig. 1J). During cycle 14 in pbl mutant cells, actin (Fig. 1F), anillin (Fig. 1I), and Pnut (Fig. 1K) fail to relocalize from the cell cortex to the cleavage furrow. Subsequently, the cleavage furrow is not initiated (arrows in Fig. 1F,I,K) and cytokinesis fails. Similarly, in later rounds of cell division, actin, anillin, and Pnut fail to accumulate at the equatorial region of the cell, and there are no signs of a cleavage furrow (Fig. 1G; data not shown). These results indicate that in pbl mutant cells the contractile ring does not assemble, leading to a failure of cytokinesis.
Figure 1.
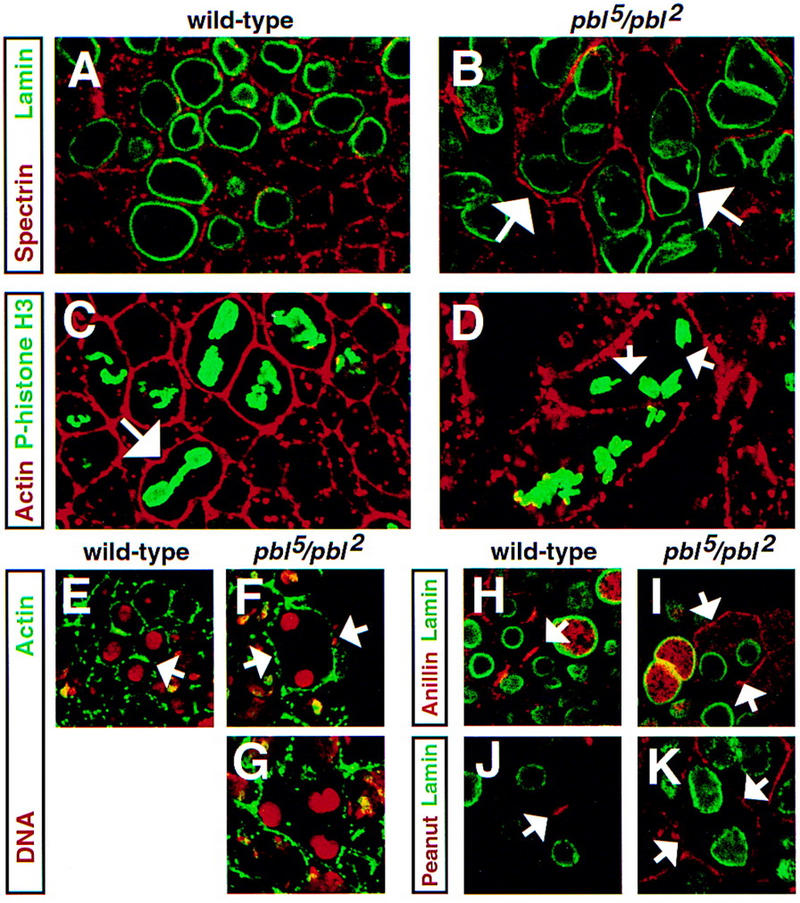
Absence of a contractile ring and failure of cytokinesis in pbl mutants. Wild-type (A) and pbl5/pbl2 (B) embryos stained with anti-α-spectrin (red) to reveal plasma membrane and anti-lamin (green) to mark nuclei. Ectodermal cells of a wild-type embryo (A) contain only one nuclear lamina (nucleus) per cell, whereas several nuclei can be found in pbl mutant cells (B). Absence of cytokinesis in cell divisions following mitotic cycle 14 results in accumulation of multiple nuclei per cell (arrows). Wild-type (C) and pbl5/pbl2 (D) embryos stained with anti-actin (red) to mark plasma membrane and anti-phosphohistone H3 (green) to mark chromosomes during mitosis. Wild-type cells (C) form a single mitotic apparatus per cell and initiate contraction of the cleavage furrow (arrow) in late anaphase. In polyploid pbl mutant cells (D), there are no signs of cleavage, and during mitotic cycle 15, two mitotic apparatuses are formed (arrows) that independently enter anaphase. Wild-type (E,H,J) and pbl5/pbl2 (F,G,I,K) embryos stained with anti-actin (E–G, green), anti-anillin (H,I, red), and anti-Pnut (J,K, red) as markers of a contractile ring. Embryos in E–G were counterstained with propidium iodide (red) to mark DNA, and embryos in H–K were counterstained with anti-lamin (green) to reveal nuclei. In dividing wild-type cells, actin (E), anillin (H), and Pnut (J) accumulate at the equatorial region in which the contractile ring is assembled during late anaphase. They remain enriched at the equator through telophase (arrows in E,H,J). During cycle 14 in pbl mutant cells, actin (F), anillin (I), and Pnut (K) do not accumulate at the equatorial region and remain associated with plasma membrane. Similarly, during later mitotic cycles (G) there are no signs of contractile ring formation.
pbl encodes a novel RhoGEF related to the mouse Ect2 oncoprotein
To clone pbl, we used recently identified P-element alleles of pbl (Salzberg et al. 1997). Genomic fragments flanking the sites of P-element insertions were isolated and used to screen an embryonic cDNA library. Database searches revealed that pbl encodes a novel guanine nucleotide exchange factor for small G proteins of the Rho family (RhoGEF) related to the mouse Ect2 oncoprotein (Miki et al. 1993; Fig. 2A,B). Common functional domains of Pbl and Ect2 include the Dbl homology (DH) and pleckstrin homology (PH) domains (in Pbl, amino acids 390–574 and 600–718, respectively) that are found in tandem in all RhoGEFs (Whitehead et al. 1997). Mammalian members of the RhoGEF family are considered to be potential oncogenes as their truncated forms often induce transformed foci when overexpressed in cultured fibroblasts. The DH domain catalyzes GDP/GTP exchange through direct binding to its effector GTPase (Hart et al. 1994; Zheng et al. 1995; Liu et al. 1998). The PH domain (Shaw 1996), always carboxy-terminal to the DH domain in RhoGEFs, is thought to be required for membrane or cytoskeletal targeting of the protein and in enhancement of the catalytic activity of the adjacent DH domain (Whitehead et al. 1995; Zheng et al. 1996; Liu et al. 1998; Sterpetti et al. 1999). At the amino terminus, Pbl and Ect2 share two BRCT (BRCA1 carboxyl terminus) domains (in Pbl, amino acids 113–198 and 208–291) initially identified in the familial breast and ovarian cancer susceptibility gene BRCA1 (Bork et al. 1997; Callebaut and Mornon 1997). BRCT domains are found in proteins involved in DNA metabolism and cell cycle checkpoint control and are thought to be protein–protein interaction modules. In addition, Pbl contains a putative nuclear localization signal (NLS) (amino acids 306–322) and a PEST sequence (amino acids 371–383) (Fig. 2B), which often serves as a proteolytic signal to target proteins for rapid degradation.
Figure 2.
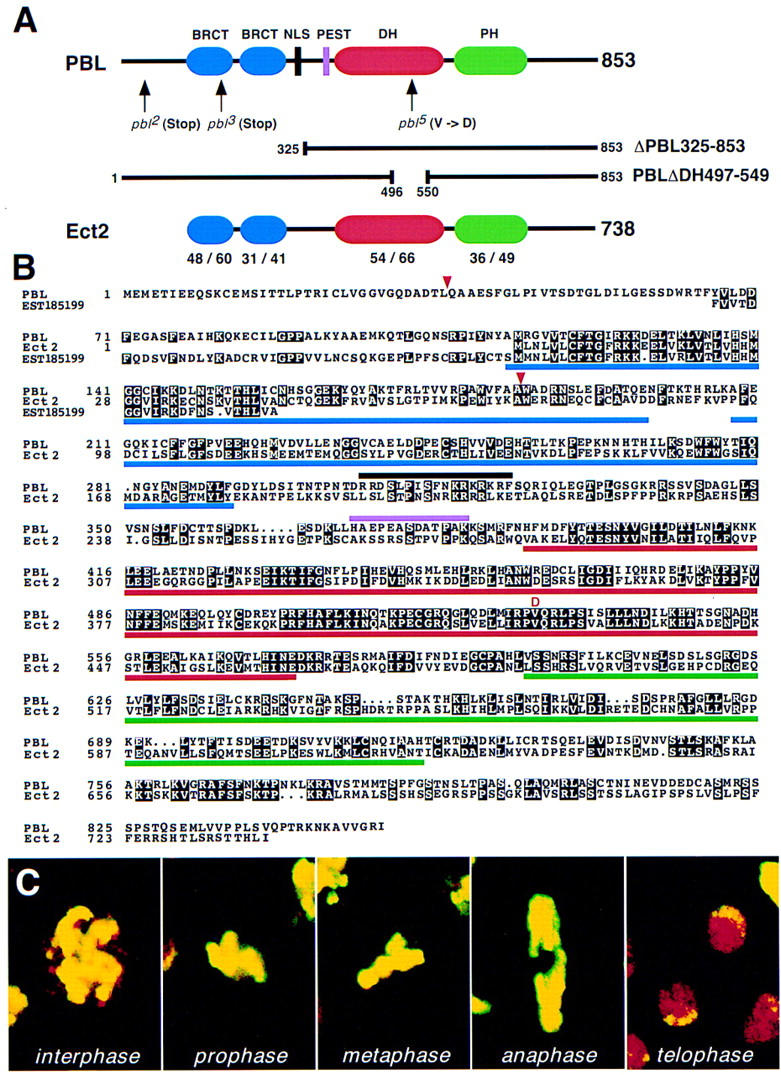
pbl encodes an exchange factor for small G proteins of the Rho family. (A) Domain structure of Pbl and Ect2. The two proteins are represented schematically with functional domains shown in color. The lengths of Pbl and Ect2 are 853 and 738 amino acids, respectively. Overall, the two proteins are 40% identical and 61% similar. Percent identity/similarity between respective domains is shown below the Ect2 protein. Location of mutations and amino acid changes in pbl alleles are indicated by arrows. Lines below the Pbl protein represent constructs used to generate new alleles of pbl in transgenic flies. (BRCT) BRCA1 carboxyl terminus; (DH) Dbl homology; (NLS) nuclear localization signal; (PH) pleckstrin homology. (B) Alignment of amino acid sequences of Pbl, mouse Ect2, and predicted ORF from human EST185199. Black boxes indicate identical amino acids. Common functional domains of Pbl and Ect2 are underlined. The NLS and PEST sequences of Pbl are marked above its sequence. All domains are indicated with the same colors as in A. Amino acids 66–157 of Pbl show 42% identity (56% similarity) to a predicted amino acid sequence of EST185199 (nucleotides 39–314) from human colon carcinoma cell line. In addition, amino acids 44–185 of Pbl show 27% identity (49% similarity) to a predicted amino acid sequence of human EST zs58b08.r1 (nucleotides 248–661; data not shown). Positions of protein truncation caused by nonsense mutations in the pbl2 and pbl3 alleles (Q37Ter and W185Ter, respectively) are marked with red triangles. The red letter D denotes the V531D mutation within the DH domain in the pbl5 allele. The GenBank accession nos. are L11316 (ect2), AA313301 (EST185199), and AA287172 (EST zs58b08.r1). The BRCT domains and DH and PH domains of Ect2 were defined according to Callebaut and Mornon (1997) and Schultz et al. (1998), respectively. (C) Mitotic figures in cycle 14 domains of a wild-type embryo stained with anti-Pbl (red) and anti-phosphohistone H3 (yellow) to mark chromosomes during mitosis. Pbl protein is not detected during late prophase, metaphase, and anaphase. Pbl has the highest levels of expression in telophase and interphase nuclei.
Several lines of evidence suggest that the pbl cDNA we cloned corresponds to the pbl gene. First, four independently generated P-element alleles that fail to complement other pbl alleles are inserted in the 5′-untranslated region (UTR) or open reading frame (ORF) of pbl. Second, each P element is revertible to wild type (Salzberg et al. 1997; this study). Third, the molecular lesions associated with three EMS-induced pbl mutations were identified (Fig. 2A,B). Mutations in pbl2 and pbl3 cause premature termination at Q37 (CAG to TAG) and W185 (TGG to TAG), respectively, whereas a V531D (GTT to GAT) mutation in pbl5 affects a conserved valine within the most highly conserved region (CR3) of the DH domain. A mutation in this residue has been shown to dramatically reduce the nucleotide exchange activity of other RhoGEFs (Liu et al. 1998). We conclude that Pbl is a putative RhoGEF related to the mouse Ect2 protein.
Pbl redistribution during mitosis parallels the onset of cytokinesis
To investigate the subcellular distribution of the protein during the cell cycle, we raised anti-Pbl polyclonal antibodies. Double stainings of wild-type embryos undergoing mitotic cycle 14 with the anti-phospho-histone H3 antibody, which marks chromosomes during mitosis (Hendzel et al. 1997), revealed that Pbl is expressed dynamically during the cell cycle (Fig. 2C). We observed no Pbl protein during late prophase, metaphase, and early anaphase. Pbl protein is first detected in late anaphase, when it colocalizes with separating daughter chromosomes. The highest levels of Pbl staining were found in telophase nuclei, and this staining persisted during interphase of the following mitotic cycle. As we never observed Pbl at the onset of mitosis, it appears that it is rapidly degraded during each cycle of cell division, presumably after being released from the nucleus upon disassembly of the nuclear envelope in prophase. Further analyses with antibodies against lamin to mark nuclei and α-spectrin to stain plasma membranes demonstrated that in late anaphase, there are low levels of Pbl protein at the plasma membrane and in the cytoplasm (Fig. 3A–C). Initiation of cytokinesis, as judged by the appearance of the cleavage furrow (arrowheads in Fig. 3B) and reassembly of nuclear laminae (Fig. 3A), coincides with the accumulation of Pbl at the cleavage furrow. As the cleavage furrow progresses (arrowheads in Fig. 3E), Pbl accumulates at the equator between dividing cells (Fig. 3 cf. arrows in D,F,G with A) and in the nuclei of the daughter cells (Fig. 3G). Pbl translocation to the equatorial region beneath the cleavage furrow (Fig. 3H) parallels the accumulation of anillin (Fig. 3I) and Pnut (Fig. 3J) at the equator in which the contractile ring is assembled. In telophase and early interphase cells, the protein is strongly enriched in the nuclei (Fig. 3K) and is found at low levels at the cortex (data not shown). Hence, during mitosis subcellular distribution of Pbl undergoes dynamic changes and Pbl is associated with the cleavage furrow during cytokinesis. In addition, the levels of protein cycle during each cell cycle with the highest levels of Pbl in late telophase and interphase.
Figure 3.
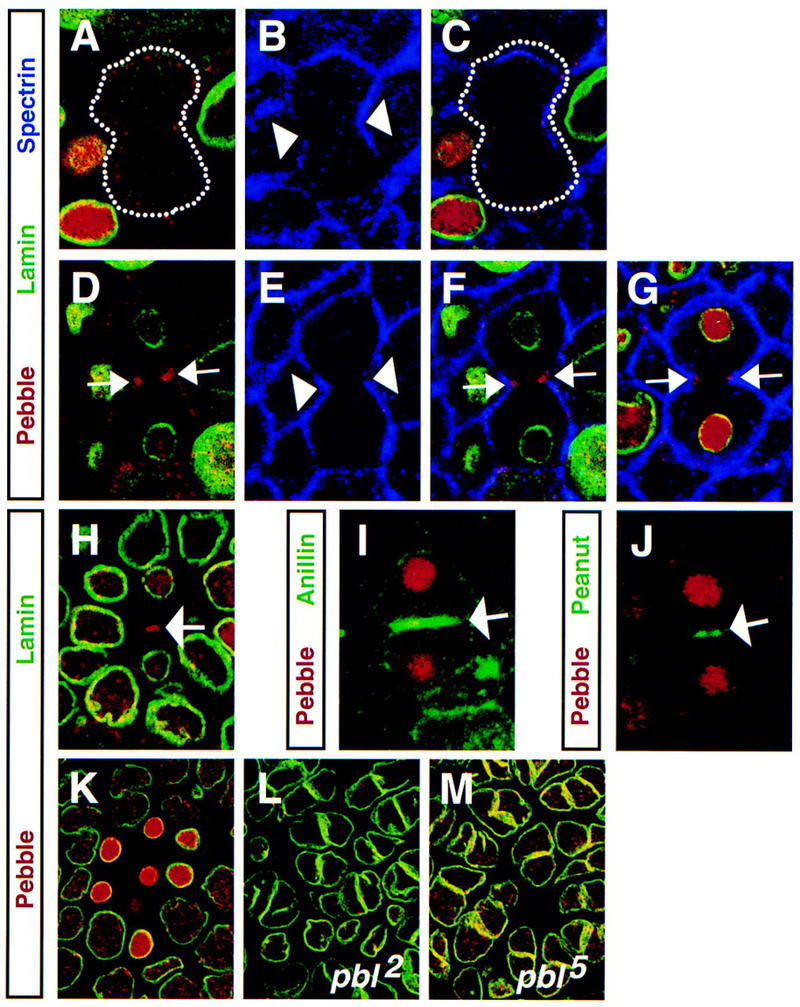
Pbl is localized to the cell cortex and cleavage furrow during cytokinesis. (A–G) Wild-type embryos stained with anti-Pbl (A,D,G, red), anti-lamin (A,D,G, green) to mark nuclei, anti-α-spectrin (B,E,G, blue) to mark plasma membrane, and merged images (C,F). Upon initiation of cytokinesis, Pbl is cortical and cytoplasmic (A–C, the cell is outlined with a dotted line). As the cleavage furrow progresses (arrowheads in B,E), Pbl accumulates as two spots at the equator (arrows in D,F,G) and also in reforming nuclei (D,G). For comparison, images in C and F were not adjusted for brightness, contrast, or gamma. Embryo expressing full-length Pbl (prd–GAL4/pblEP3415) (H) and wild-type embryos (I, J) stained with anti-Pbl (red) and anti-lamin (H, green), anti-anillin (I, green), or anti-Pnut (J, green). Pbl accumulation at the equator between dividing cells (arrow in H) parallels accumulation of anillin (arrow in I) and Pnut (arrow in J) beneath the cleavage furrow in which the contractile ring is assembled. (K) Wild-type, (L) pbl2 homozygous, and (M) pbl5 homozygous embryos stained with anti-Pbl (red) and anti-lamin (green). Unlike in a wild-type embryo (K), cycle 15 cells of the pbl2 embryo have greatly reduced levels of Pbl expression (L) with few remaining speckles of staining scattered in nuclei of affected polyploid cells. Cycle 15 cells of pbl5 embryo (M) have levels of nuclear Pbl staining comparable with wild type.
Embryos homozygous for the pbl2 null allele as well as cytological deficiencies removing pbl [Df(3L)pbl–NR and Df(3L)25] have greatly reduced levels of staining in cycle 15–16 interphase cells (Fig. 3 cf. L with K; data not shown), which is likely to correspond to the remaining maternal contribution. In contrast, pbl5 embryos that carry the V531D missense mutation affecting a conserved valine (Fig. 2A,B) have protein levels comparable with wild type (Fig. 3M). It is likely that the reduction in the levels of cytoplasmic, but not nuclear Pbl, is responsible for failure of cytokinesis in mitotic cycles 14–16, because we have never observed Pbl at the cortex or at the cleavage furrow during anaphase and telophase in cycle 14 pbl mutant cells (pbl2 and pbl5 alleles).
Ectopic expression of amino-terminally truncated Pbl or Ect2 blocks cytokinesis
The dramatic increase in Pbl levels at the onset of cytokinesis and localization of Pbl to the cleavage furrow suggest that Pbl may play a regulatory role in cytokinesis. As a protein, which presumably regulates the activity of Rho GTPases, Pbl may be required for the initiation of cytokinesis or may couple cytokinesis with the mitotic cycle. To investigate this hypothesis, we compared the effects of overexpression of full-length Pbl and of Pbl protein in which the BRCT domains and the NLS sequence were deleted (ΔPbl325–853 allele) (Fig. 2A). The amino-terminal truncation in ΔPbl325–853 resembles a similar mutation in a number of mammalian RhoGEFs (for review, see Whitehead et al. 1997), including Ect2 (Miki et al. 1993), that results in the constitutive activation of their transforming potential. However, the amino-terminal sequences that are lost in different RhoGEFs do not share common domains. Hence, their role in controlling the activity of the full-length protein remains elusive.
Ectopic expression of full-length Pbl (with pblEP3415 or the UAS–Pbl3.2 line) in stage 8–11 embryos with the paired–GAL4 (prd–GAL4) driver did not result in a detectable phenotype (Figs. 4A and 3H). Although all cells in segments overexpressing Pbl had high levels of exogenous protein in their nuclei and cytoplasm, this did not interfere with cytokinesis, completion of mitosis (arrowhead in Fig. 4A), or viability. In contrast, expression of ΔPbl325–853 with the same driver led to 100% embryonic lethality. Most cells within the embryonic segments expressing ΔPbl325–853 became multinucleate (Fig. 4B). This polyploid phenotype was caused by a failure of cytokinesis, because the cleavage furrow was not initiated (Fig. 4C), and, likewise, the contractile ring was not assembled (Fig. 4D; data not shown). We used the same approach to overexpress the amino-terminally truncated form of Ect2, ΔEct2. As for ΔPbl325–853, ectopic expression of ΔEct2 resulted in the absence of the contractile ring and cleavage furrow, leading to formation of multinucleate cells (Fig. 4E,F), and lethality. In contrast, flies expressing full-length Ect2 exhibited no phenotype and were viable (data not shown).
Figure 4.
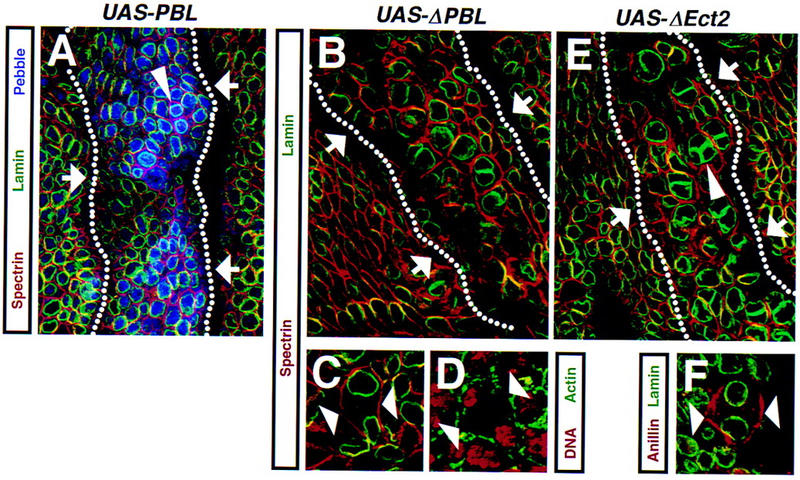
Ectopic expression of amino-terminally truncated Pbl or Ect2 blocks cytokinesis. (A) Stage 9 UAS–Pbl3.2/+, prd–GAL4/+ embryo expressing full-length Pbl in a prd-like pattern stained with anti-α-spectrin (red) to mark plasma membrane, anti-lamin (green) to reveal nuclei, and anti-Pbl (blue). Expression of full-length Pbl does not cause a phenotype during cell division, despite high levels of expression of exogenous Pbl in the affected segment (arrows). Note a pair of young postmitotic cells (arrowhead) with high levels of Pbl in the nuclei. High levels of exogenous Pbl and its apparent association with the nuclear envelope cause the nuclear laminae to appear blue. (B,C) Stage 9 UAS–ΔPbl325–853S4–11/+, prd–GAL4/+ embryo expressing amino-terminally truncated Pbl stained with anti-α-spectrin (red) and anti-lamin (green). (D) A similar embryo stained with propidium iodide (red) to mark DNA and anti-actin to mark contractile ring (green). Expression of amino-terminally truncated Pbl leads to a block of cytokinesis. Most cells within the affected segment (arrows in B) become polyploid. During late anaphase, the cleavage furrow fails to initiate (arrowheads in C) and actin does not redistribute to the equatorial region in which the contractile ring is assembled (arrowheads in D). The ΔPbl325–853 protein is cytoplasmic and cortical (data not shown). (E) Stage 9 prd–GAL4/UAS–ΔEct2S2-1A embryo expressing amino-terminally truncated Ect2 stained with anti-α-spectrin (red) and anti-lamin (green). (F) A similar embryo stained with anti-anillin (red) to mark the contractile ring and anti-lamin (green). Most cells within the segment expressing ΔEct2 (arrows in E) become binucleate. Failure of cytokinesis during later divisions leads to formation of multinucleate cells (arrowhead in E). Similar to ΔPbl325–853, there are no signs of a cleavage furrow and anillin does not accumulate at the equatorial region in late anaphase (arrowheads in F). Dotted lines in A,B, and E denote ectodermal segments expressing Pbl or Ect2.
Thus, it appears that full-length Pbl (as well as Ect2) does not behave in vivo as an active protein, because it does not cause a cytokinetic phenotype when expressed in the embryonic ectoderm. This suggests that activation of the exchange activity of Pbl during cytokinesis may require some initiating event. In contrast, amino-terminally truncated Pbl, when overexpressed in wild-type embryo, has a phenotype similar to lack of pbl. We propose that ΔPbl may behave as an activated protein competing with endogenous Pbl for a rate-limiting target protein (such as Rho GTPase), which results in an inappropriate activation of downstream effectors. Furthermore, the amino-terminal BRCT domains of Pbl, lacking in ΔPbl and known to be protein–protein interaction modules (Yamane et al. 1997; Chai et al. 1999), may be required for its regulation by an upstream signal. Such regulation may be essential for the spatial and temporal control of cytokinesis and for the coupling of signaling pathways initiating cytokinesis with other mitotic events.
Identification of Rho1 as a suppressor of pbl
In an effort to identify genes involved in the Pbl signaling pathway, we designed a genetic screen to isolate second-site mutations interacting with pbl. We used the GMR–GAL4 driver which drives expression in all cells posterior to the morphogenetic furrow (Ellis et al. 1993) to express Pbl in the adult fly eye. Overexpression of Pbl (with the UAS–Pbl3.2 line) results in a dominant rough eye phenotype characterized by misshaped ommatidia, disrupted bristle distribution (Fig. 5B,G), misshapen rhabdomeres, and a significant increase in the number of pigment cells (Fig. 5 cf. L with K). In contrast, the external eye morphology of UAS–Pbl3.2 flies was indistinguishable from wild type. Thus, overexpression of Pbl in the eye perturbs normal retinal development. Furthermore, the developing eye seems to be more sensitive than the embryo to overexpression of Pbl, in which it does not cause a phenotype when expressed in ectodermal cells using the prd–GAL4 driver.
Figure 5.
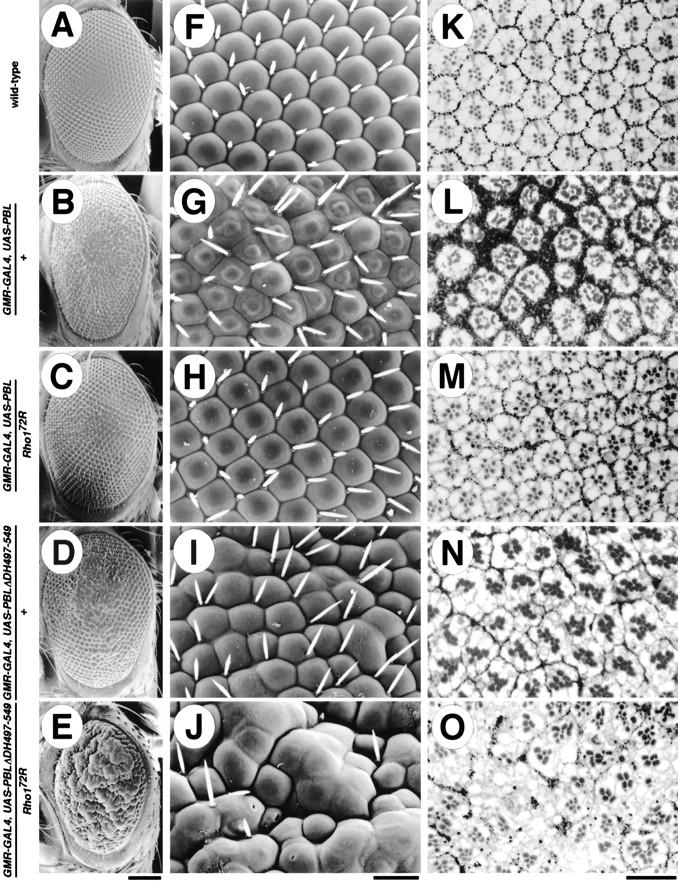
pbl and Rho1 interact in vivo. Scanning electron micrographs (A–J) and methylene blue-stained transverse sections (K–O) of wild-type (A,F,K), GMR–GAL4, UAS–Pbl3.2/+ (B,G,L), GMR–GAL4, UAS–Pbl3.2/Rho172R (C,H,M), GMR–GAL4, UAS–PblΔDH497–5495B, 6A/+ (D,I,N), and GMR–GAL4, UAS–PblΔDH497–5495B,6A/Rho172R (E,J,O) flies. Overexpression of Pbl in the eye induces a disruption of the ordered array of ommatidia (B,G), as well as a disruption of the internal organization of rhabdomeres and a significant increase in the density and number of pigment cells (L). These defects are suppressed dominantly by mutations in Rho1 (C,H,M). Overexpression of PblΔDH497–549 induces disruption of the ordered structure of the eye, with fusions of ommatidia, loss of many bristles (D,I), and a decrease in the number of pigment cells (N). These defects are strongly enhanced in a dominant fashion by mutation in Rho1 (E,J,O). The eye phenotypes of Pbl and PblΔDH497–549 are completely suppressed by the coexpression of the UAS–antisensePbl (data not shown). Flies were raised at 25°C. Calibration bars: (A–E) 100 μm; (F–O) 20 μm.
To identify dominant second-site modifiers of the rough eye phenotype caused by overexpression of Pbl, the GMR–Pbl flies were crossed to males that carry deficiencies covering most of the II chromosome. The resulting F1 progeny were examined for dominant enhancement or suppression of the rough eye phenotype. Df(2R)Jp8 deficiency, identified as the only suppressor in the modifier screen, showed an almost complete suppression of the rough eye phenotype (data not shown). One of the genes uncovered by this deficiency is Rho1. To investigate the possibility that a mutation in Rho1 caused the suppression, we analyzed the phenotype of the progeny from a cross between the GMR–Pbl flies and null alleles of Rho1 (Strutt et al. 1997). Removal of a single copy of Rho1 dominantly suppressed the rough eye phenotype caused by overexpression of Pbl (Fig. 5C,H). Transverse sections showed rhabdomeres of more regular shape, number, and arrangement (Fig. 5 cf. M with L). In addition, the number and density of pigment cells is significantly decreased compared with the GMR–Pbl flies. Hypomorphic alleles of Rho1 cause a similar, but much weaker suppression than Rho1 null mutations (data not shown). In contrast, alleles of Cdc42 or a deficiency removing Rac1 did not suppress or enhance this phenotype (data not shown). These observations suggest that pbl interacts genetically with Rho1 and that the phenotypic suppression is specific for mutations in Rho1, but not for genes encoding other Rho family proteins, Cdc42 and Rac1.
Previously, it was shown that overexpression of Rho1 in the eye results in disruption of both external and internal eye morphology (Hariharan et al. 1995). Mutations in pbl (pbl1, pbl2, pbl3, pbl5, pblP81, pblS23, and pblV58 alleles) caused a strong dominant suppression of the GMR–Rho1-induced rough eye phenotype (data not shown). Because pbl encodes a putative RhoGEF, these results suggest that a reduction in exchange factor activity in pbl alleles is responsible for the suppression of the GMR–Rho1-induced eye phenotype. Similarly, we propose that decreased levels of Rho1, a putative effector of Pbl, account for suppression of the GMR–Pbl-induced eye phenotype by Rho1 mutations.
To further characterize the genetic interaction between pbl and Rho1, we generated a new deletion allele of pbl, PblΔDH497–549, which affects the DH domain only (Fig. 2A). Site-directed mutagenesis in a number of RhoGEFs revealed that point mutations and deletions within the DH domain completely abolish exchange factor activity (Ron et al. 1991; Hart et al. 1994; Whitehead et al. 1995; Steven et al. 1998). Hence, to create an inactive form of Pbl that behaves as a dominant–negative protein, we introduced a short deletion (amino acids 497–549) removing the most highly conserved CR3 region (Soisson et al. 1998) within the DH domain. Mutations within this region dramatically diminish the DH domain function, suggesting that it is responsible for GTPase binding and nucleotide exchange activity. Expression of PblΔDH497–549 in the eye with the GMR–GAL4 driver results in a rough eye phenotype, with pronounced fusion of some ommatidia and a significant reduction in the number of bristles (Fig. 5D,I). Transverse sections show a variation in the number of rhabdomeres per ommatidium and, unlike overexpression of the full-length Pbl, a decrease in the number of pigment cells (Fig. 5 cf. N with L). This phenotype is significantly enhanced in a dominant fashion by mutations in pbl (data not shown). Similarly, mutations in Rho1 act as dominant enhancers of this phenotype, with a dramatic increase in the number of fused ommatidia and loss of many bristles (Fig. 5E,J). Transverse sections show a severe disorganization of the internal architecture of the eye, a complete absence of rhabdomeres in some regions of the eye, and a further decrease in the number of pigment cells (Fig. 5O). Because the PblΔDH497–549 protein is likely to be inactive (see below), we suggest that it may mimic the phenotype of hypomorphic alleles of pbl by sequestering proteins (other than Rho1) that normally bind to or interact with endogenous Pbl. As expected, mutations in Rho1 that interact genetically with pbl enhanced the phenotype of PblΔDH497–549. In summary, pbl shows strong genetic interaction with Rho1, but not with Rac1 or Cdc42, indicating that pbl and Rho1 are in the same genetic pathway in vivo.
Pbl and Rho1 proteins interact in a yeast two-hybrid assay
Exchange factors for Rho proteins are known to activate their downstream targets through direct binding to Rho GTPases (Hart et al. 1994). Hence, the molecular basis for the observed genetic interaction between pbl and Rho1 may be a physical interaction between Pbl and Rho1 proteins. To test this hypothesis, we carried out a yeast two-hybrid assay. We constructed fusions of Pbl (full-length and amino-terminally truncated) as well as Drosophila Rho proteins, Rho1, Rac1, and Cdc42, with both the GAL4 DNA-binding domain (DBD) and GAL4 activation domain (AD). Plasmids were transformed in various combinations, and the resulting colonies were tested for the ability to activate HIS3 and lacZ reporters. Only colonies that carried plasmids expressing both Pbl (full-length or ΔPbl325–853, amino-terminally truncated Pbl) and Rho1 were able to grow on a medium that lacked His, Leu, and Trp (Fig. 6) suggesting that the HIS3 gene was induced. Furthermore, all His-positive colonies expressed β-galactosidase (Fig. 6C, sectors a, d, e, and f). In contrast, double transformants coexpressing Pbl and Rac1, Cdc42, or Pbl instead of Rho1, failed to grow on a medium without His, Leu, and Trp (Fig. 6B, sectors b, c, g, and h). Therefore, we suggest that Pbl and Rho1 proteins interact in vivo and that this interaction is specific for Rho1, but not for Rac1 or Cdc42. Furthermore, the DH domain, but not the amino terminus of Pbl (containing BRCT domains and NLS), is essential for this interaction, because it was abolished by a small deletion within the DH domain (PblΔDH497–549; Fig. 6A), but not by the amino-terminal truncation of Pbl (ΔPbl325–853; Fig. 6B,C, sectors d,f). These results indicate that Pbl and Rho1 form a protein complex in vivo, and that the basis for the genetic interaction between pbl and Rho1 may be a direct interaction between the two proteins.
Figure 6.
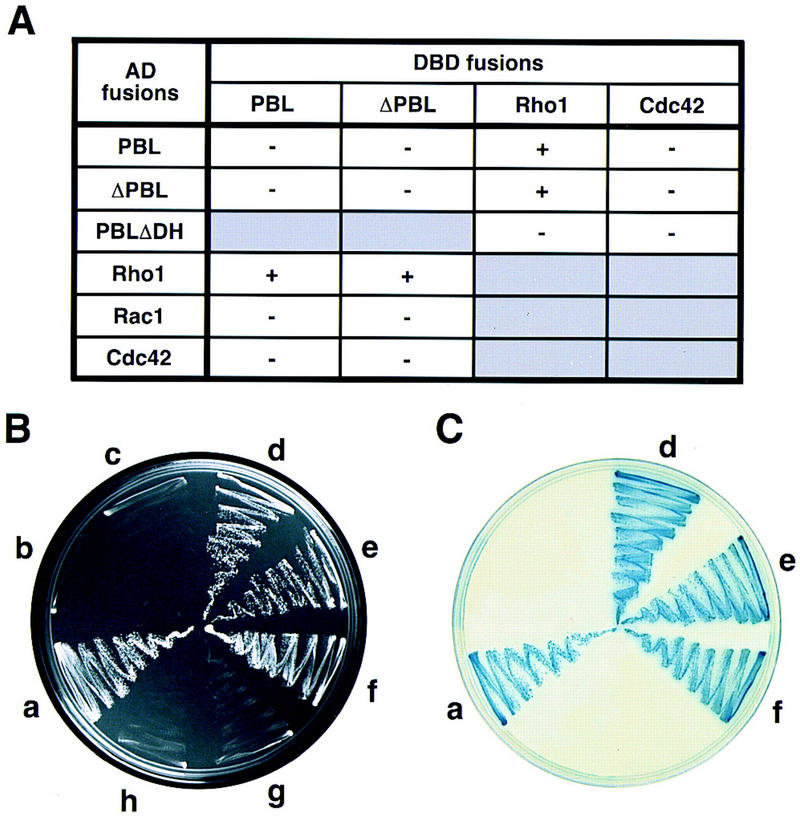
Pbl and Rho1 proteins interact in a yeast two-hybrid assay. (A) Summary of pairwise interactions between Pbl and Drosophila Rho1, Rac1, and Cdc42. Minus and plus signs indicate the absence or presence of the activity of the GAL–HIS and GAL–lacZ reporters. (B,C) Representative results of assays for histidine (B, on SC−His−Leu−Trp) and β-galactosidase (C, on SC−Leu−Trp) activity. (a) pAS2/Pbl and pACT2/Rho1; (b) pAS2/Pbl and pACT2/Rac1; (c) pAS2/Pbl and pACT2/Cdc42; (d) pAS2/ΔPbl and pACT2/Rho1; (e) pAS2/Rho1 and pACT2/Pbl; (f) pAS2/Rho1 and pACT2/ΔPbl; (g) pAS2/Cdc42 and pACT2/Pbl; (h) pAS2/Pbl and pACT2/Pbl. (AD) Activation domain; (DBD) DNA-binding domain; (Pbl) Pbl1-853; (PblΔDH), PblΔDH497–549; (ΔPbl) ΔPbl325–853.
Rho1 is required for cytokinesis
To investigate the possibility that mutations in Rho1 cause defects during cell division, we stained Rho1 embryos with antibodies against lamin and α-spectrin (Fig. 7A,B). Embryos homozygous for null alleles of Rho1 (Rho172O and Rho172R) showed many binucleate cells in the head region (Fig. 7A). Occasionally, we also observed a few polyploid cells in thoracic or abdominal segments (Fig. 7B). Although the latter phenotype was observed with low penetrance, we could never find binucleate ectodermal cells in wild-type embryos. The relatively weak phenotype caused by Rho1 mutations (unlike that of pbl) is most likely due to maternally provided Rho1 protein. Thus, it appears that Rho1 is required for cytokinesis, and that decreased levels of Rho1 may account for the observed phenotype in cytokinesis.
Figure 7.
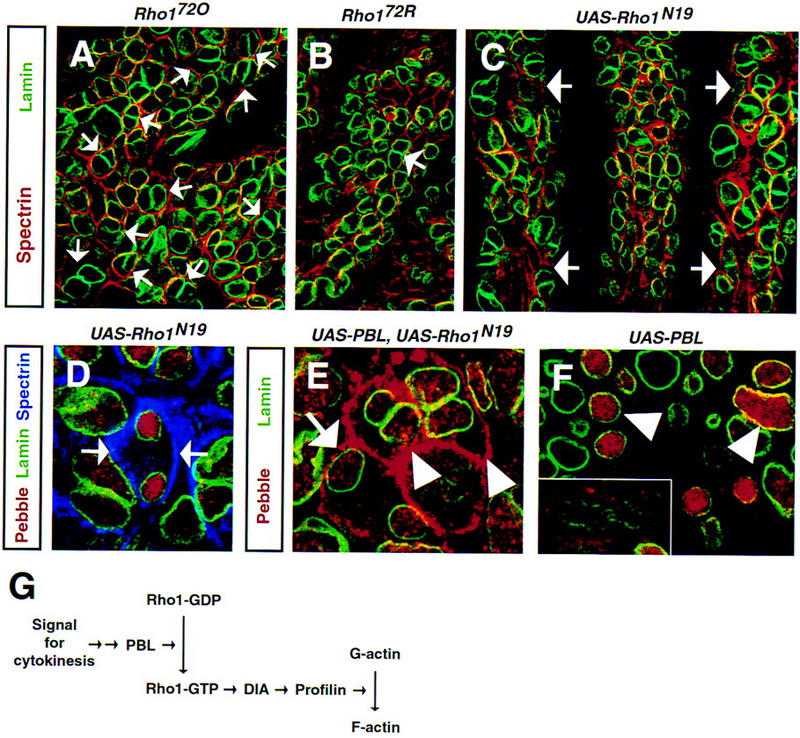
Mutations in Rho1 or expression of a dominant–negative Rho1 abolish cytokinesis. Rho172O homozygous (A, stage 16), Rho172R homozygous (B, stage 13) embryos, and an embryo expressing Rho1N19 in a paired-like pattern (C,D, UAS–Rho1N19/+, prd–GAL4/+, stage 9) stained with anti-α-spectrin (red in A–C, blue in D) to mark plasma membrane, anti-lamin (green) to reveal nuclei, and anti-Pbl (red in D). (A,B) Null mutations in Rho1 block cytokinesis in affected cells. Many cells in the head region of the embryo (A) become polyploid and contain two nuclei per cell (arrows). In addition, polyploid cells are found occasionally in thoracic or abdominal segments of Rho1 embryos (arrow in B, T2 segment). Homozygous Rho1 embryos were identified on the basis of their anterior open phenotype. (C,D) Ectopic expression of a dominant–negative Rho1 leads to formation of polyploid cells. Almost every cell within the affected segments (arrows in C) becomes binucleate. Despite high levels of nuclear Pbl in telophase, cleavage furrow fails (arrows in D) and cytokinesis is not initiated. (E,F) Embryo expressing Pbl and Rho1N19 (E, UAS–Pbl3.2/+, prd–GAL4/UAS–Rho1N19) and embryo ex pressing Pbl alone (F, UAS–Pbl3.2/+, prd–GAL4/+) stained with anti-Pbl (red) and anti-lamin (green). (E) Coexpression of Pbl and Rho1N19 results in accumulation of Pbl at the cortex in telophase (arrow) and especially in interphase (arrowheads) cells. (F) In contrast, distribution of Pbl in cells ectopically expressing Pbl alone is similar to wild-type cells with protein localized to the nuclei during interphase (arrowheads) and to the cortex in early anaphase (inset). (G) Proposed signaling pathway initiating cytokinesis. (Dia) Diaphanous; (F-actin) fibrous actin; (G-actin) globular actin; (Rho1–GDP) inactive GDP-bound Rho1; (Rho1–GTP) active GTP-bound Rho1. For details see Discussion.
To further demonstrate the role of Rho1 in cytokinesis, we analyzed the effects of expression of a dominant–negative form of Rho1, Rho1N19 (Strutt et al. 1997). It was shown that a dominant–negative form of H-Ras, H-RasN17, cannot interact with downstream target proteins (Farnsworth and Feig 1991; Feig 1999). In addition, H-RasN17 competes with normal H-Ras for binding to rasGEF, which results in formation of inactive H-RasN17–rasGEF complexes and depletion of the pool of the endogenous rasGEF leading to a dominant–negative effect (Schweighoffer et al. 1993; Feig 1999). Therefore, a dominant–negative Rho1 may produce a phenotype similar to or stronger than loss-of-function alleles of Rho1. Ectopic expression of Rho1N19 creates a phenotype that is much worse than loss of zygotic Rho1 (Fig. 7 cf. C with A and B). Expression of Rho1N19 in ectodermal stripes leads to a complete block of cytokinesis (Fig. 7D) leading to polyploidy of almost every cell within the affected segment. In contrast, expression of wild-type Rho1 or dominant–negative Rac1 (Rac1N17 or Rac1L89) did not affect cytokinesis or completion of mitosis (data not shown). Interestingly, coexpression of Rho1N19 and Pbl (unlike Pbl alone) results in the mislocalization of Pbl to the cell cortex in interphase and late mitotic cells (Fig. 7E,F) further indicating that the two proteins interact during cytokinesis. These results, together with other data, suggest that activation of the Rho1 GTPase by Pbl, its putative exchange factor, is required for the initiation of cytokinesis, because mutations in either protein or overexpression of an inactive protein result in accumulation of multinucleate cells.
Discussion
Pbl is a putative exchange factor required for cytokinesis
Rho GTPases have been implicated in a variety of cellular processes and developmental events. Often the same Rho protein is involved in several interwoven signaling pathways activated within the same cell. The dynamic coordination of these molecular pathways during morphogenesis implies a complex level of regulation of Rho proteins, both spatially and temporally, by guanine nucleotide exchange factors and other molecules (Van Aelst and D’Souza-Schorey 1997). However, our knowledge about the biological functions of RhoGEFs is limited mainly to the studies of mammalian Dbl proteins and their roles in cell growth and oncogenic transformation (Whitehead et al. 1997). Genetic analyses in Drosophila and Caenorhabditis elegans have led to the identification of exchange factors required for axon guidance and cell migration (UNC-73; Steven et al. 1998), organization of actin cytoskeleton in synaptic terminals (SIF; Sone et al. 1997), and for coordination of cell shape changes during gastrulation (DRhoGEF2; Barrett et al. 1997; Häcker and Perrimon 1998).
The data presented here indicate that Pbl is the first identified putative RhoGEF specifically required for cytokinesis. First, mutations in all pbl alleles result in the same phenotype—absence of a contractile ring and failure of cytokinesis. Second, during embryogenesis the highest levels of Pbl are found in dividing cells. Third, at the onset of cytokinesis, Pbl is associated with the cleavage furrow. As mitosis progresses, Pbl, localized initially at the cortex and in the cytoplasm, accumulates at the cleavage furrow, where assembly of the contractile ring takes place. We propose that Pbl at the cleavage furrow is required for the initiation of cytokinesis by interacting with and regulating proteins involved in the assembly of the contractile ring. Although the highest levels of Pbl are found in late telophase and young postmitotic nuclei, nuclear Pbl does not seem to play a role in the initiation of cytokinesis. The accumulation of Pbl in telophase nuclei follows the initiation of the cleavage furrow and the beginning of the reassembly of the nuclear envelope and, hence, occurs too late in mitosis to be an instructive signal for cytokinesis. This conclusion is further supported by our observation that amino-terminally truncated Pbl, ΔPbl, lacking the NLS and localized to the cell cortex and cytoplasm is able to block cytokinesis. ΔPbl may compete with the endogenous Pbl at the cleavage furrow for Rho1 or, possibly, other proteins that bind to Pbl.
What is the function of nuclear Pbl? The translocation of Pbl to the nuclei may be a regulatory mechanism to turn off the downstream signaling cascade by removing a RhoGEF from the cellular compartment in which the cascade was initiated. Alternatively, there may be two pools of Pbl protein—a cytoplasmic/cortical pool required for the initiation of cytokinesis and another pool targeted to nuclei in which Pbl plays some other role unrelated to cytokinesis.
Pbl interacts specifically with Rho1
Recent experimental data suggest that Rho proteins are required, in addition to their well-established roles in cell adhesion, cell motility, chemotaxis, and axon guidance, for the regulation of cytoskeletal events at the cleavage furrow during cytokinesis. Microinjection or expression of a Rho-specific inhibitor or activated forms of RhoA or Cdc42 as well as regulators (rho GDI; Kishi et al. 1993) or downstream effectors (Rho kinase; Yasui et al. 1998) of Rho proteins in sand dollar eggs (Mabuchi et al. 1993), Xenopus embryos (Drechsel et al. 1996), and mammalian cells (Dutarte et al. 1996; O’Connell et al. 1999) result in the uncoupling of mitosis from cytokinesis, regression of the cleavage furrow, and formation of multinucleate cells. However, genetic evidence supporting the role of Rho family proteins in cytokinesis is limited to the Dictyostelium racE gene that is required for cleavage furrow constriction and completion of cytokinesis (Gerald et al. 1998). Here, we provide in vivo evidence that in Drosophila, both Rho1 and its putative exchange factor are required for the assembly of the contractile ring and, hence, initiation of cytokinesis. Mutations in both Rho1 and pbl result in a similar phenotype—absence of the contractile ring and formation of polyploid cells. We observed similar defects when we ectopically expressed a dominant–negative Rho1, Rho1N19. Drosophila Rho1 has been shown to be required for cellularization (Crawford et al. 1998), gastrulation (Barrett et al. 1997; Häcker and Perrimon 1998), dorsal closure (Harden et al. 1999), and generation of tissue polarity (Strutt et al. 1997), but not for cytokinesis. Our data suggest that the molecular basis of the genetic interaction between pbl and Rho1 is a physical interaction between Pbl and Rho1 proteins, because the two proteins interact in a yeast two-hybrid assay. Both genetic data and two-hybrid assay results indicate that this interaction is specific for Rho1, but not Rac1 or Cdc42. This interaction presumably results in the activation of the Rho1 GTPase by Pbl and induction of the signaling cascade that initiates the assembly of the contractile ring.
Our results illustrate the complexity of the intracellular signaling pathways involving Rho GTPases and their GEFs. In Drosophila, Rho1 has also been identified as a putative effector of DRhoGEF2 (Barrett et al. 1997; Häcker and Perrimon 1998), indicating that Rho1 is regulated by at least two RhoGEFs. Given the diversity of developmental roles of Rho1, one would expect that there is a complex hierarchy of molecules regulating its activity, both spatially and temporally, within a cell. Such differential regulation of Rho1 by upstream signals may be achieved through tissue-specific expression of regulatory molecules (such as GEFs and GAPs) restricted to a particular developmental or cell cycle stage. An additional level of regulation within a cell can be achieved through subcellular compartmentalization of the molecular machinery (such as Rho effectors) required for the activation of distinct pathways mediated by Rho GTPases.
Molecular pathway initiating cytokinesis: new insights
Our observations suggest that Pbl is a key regulatory component of the signaling pathway initiating cytokinesis and therefore plays a unique role, different from other cytoskeletal and structural proteins known to be required for cytokinesis (Goldberg et al. 1998). First, unlike other proteins required for cytokinesis, the levels of Pbl protein cycle during each round of cell division. Second, Pbl accumulates at the cleavage furrow at the time when cytokinesis is initiated. Third, Pbl is a putative RhoGEF and our knowledge about mammalian RhoGEFs (Whitehead et al. 1997) clearly implicates them in the regulation of signaling pathways involving Rho GTPases. Fourth, mutations within the catalytic DH domain inactivate Pbl function (pbl5 allele; Figs. 2B and 3M) and abolish Pbl interaction with Rho1 (PblΔDH497–549 allele; Fig. 6A). In addition, ectopic expression of PblΔDH497–549 in the embryo or in the eye imaginal disk blocks cytokinesis and results in the formation of polyploid cells (data not shown).
The data presented here together with earlier observations allow us to propose a signaling pathway required for cytokinesis (Fig. 7G). We suggest that activation of Rho1 GTPase by Pbl is required for initiation of a molecular pathway leading to assembly of a contractile ring. Such activation is likely to be achieved through direct binding of Pbl and Rho1 at the cleavage furrow. This is consistent with data from other species, in which Rho proteins have been shown to localize to the plasma membrane or cytosol in resting cells, but to translocate to the cleavage furrow and midbody during cytokinesis (Adamson et al. 1992; Takaishi et al. 1995; Nishimura et al. 1998). Pbl has a similar distribution during mitosis, being initially cortical but accumulating at the cleavage furrow at the onset of cytokinesis. Significantly, coexpression of Pbl and a dominant–negative form of Rho1 results in the accumulation of Pbl at the cell cortex in late mitotic cells, suggesting that the two proteins interact during cytokinesis.
Interaction of Rho1 with its effectors is likely to occur at the mouse Diaphanous (Dia) furrow as suggested previously for mouse RhoA and p140mDia, a formin-related protein required for cytokinesis (Watanabe et al. 1997). Both p140mDia and its Drosophila homolog localize to the cleavage furrow during cytokinesis (Watanabe et al. 1997; Wasserman 1998; Field et al. 1999). Furthermore, mutations in dia result in the absence of a contractile ring (Giansanti et al. 1998) and failure of cytokinesis (Castrillon and Wasserman 1994). Interestingly, null alleles of dia dominantly suppress the rough eye phenotype of the GMR–Rho1 and GMR–Pbl flies, but dominantly enhance the eye phenotype of the GMR–PblΔDH flies (data not shown), suggesting that the three genes are in the same pathway. In addition, ectopic expression of activated Rho1, Rho1V14, but not wild-type Rho1, abolishes cytokinesis (data not shown). Because mouse p140mDia interacts preferentially with activated RhoA, RhoAV14, but not with wild-type RhoA (Watanabe et al. 1997), we suggest that Rho1V14 blocks cytokinesis by titrating out one of its limiting downstream effectors, such as Dia, required for the initiation of cytokinesis. Mouse and Drosophila Dia (Watanabe et al. 1997; Wasserman 1998), as well as other formin-related proteins implicated in cytokinesis [Bni1p in Saccharomyces cerevisiae (Evangelista et al. 1997) and cdc12p in Schizosaccharomyces pombe (Chang et al. 1997)] bind profilin, an actin-binding protein that regulates F-actin polymerization, in vitro and in vivo. In Drosophila, mutations in profilin [encoded by the chic gene] block the assembly of a contractile ring during meiotic cytokinesis (Giansanti et al. 1998), and in fission yeast, mutations in both genes—cdc12 (formin) and cdc3 (profilin) show a synthetic lethal genetic interaction (Chang et al. 1997).
In conclusion, we propose a linear signaling pathway linking a RhoGEF with the actin cytoskeleton and, hence, the actomyosin contractile ring (Fig. 7G). All of the components of this pathway have been implicated in cytokinesis and were shown to localize to the cleavage furrow during cytokinesis in Drosophila or other organisms. Furthermore, all of the proteins in the pathway interact in vivo or in vitro with their neighboring partners. We conclude that the activation of Rho1 by Pbl is likely to be an initiating event for the assembly of the contractile ring at the onset of cytokinesis. Identification of upstream components of this pathway will shed light on how cytokinesis is coordinated with the mitotic cycle and on the nature of the signal required for cytokinesis.
Materials and methods
Fly stocks
Alleles of pbl used in this study are pbl1, pbl2, pbl3, pbl5 (Jürgens et al. 1984), pblP81, pblS23, pblV58 (J.B. Skeath and C.Q. Doe, pers. comm.), pblS008320, pblS054203 (Salzberg et al. 1997), P{PZ}l(3)0964509645, and EP(3)3415 (Berkeley Drosophila Genome Project, unpubl.) that we renamed pbl09645 and pblEP3415, respectively. pbl09645 and pblEP3415 are revertible P-element insertions that fail to complement other alleles of pbl and map to 66B1-5 and 66A20-22, respectively. The Cdc423 and Cdc424 alleles (Fehon et al. 1997) and Rho172O and Rho172R alleles (Strutt et al. 1997) were obtained from R. Fehon and M. Mlodzik, respectively. The Df(3L)Ar14-8 deficiency was used as an allele of Rac1. The UAS–Rho1N19 transgenic lines (Strutt et al. 1997) and UAS–Rho1–Sph lines carrying a wild-type Rho1 transgene were kindly provided by M. Mlodzik. The UAS–Rac1N17 and UAS–Rac1L89 lines (Luo et al. 1994) were received from L. Luo. The GMR–Rho11Rho13 flies (Hariharan et al. 1995) were obtained from J. Settleman. The dia2 allele (Castrillon and Wassermant 1994) was provided by S. Wasserman. The published breakpoints of the Df(2R)Jp8 deficiency (52F5-9; 53A1) (FlyBase: http://flybase.bio.indiana.edu/) are incorrect, because it fails to complement Rho1 mutations that map at 52E3-6. Canton-S was used as the wild-type strain.
Histology, scanning electron microscopy, immunohistochemistry, and confocal microscopy
For histological analyses, adult eyes were incubated in periodate/lysine/paraformaldehyde for 30 min, fixed in 2.5% glutaraldehyde/0.1m sodium phosphate (pH 7.2) overnight, postfixed in 2% osmium tetroxide, washed in water, and dehydrated in acetone. Specimens were mounted in epoxy resin, sectioned at 2 μm, and stained with methylene blue. For scanning electron microscopy, adult eyes were dehydrated through an acetone series, dried in a critical point drier, coated with gold/palladium in a vacuum evaporator, and viewed with a field emission scanning electron microscope (Phillips).
The following primary antibodies were used: anti-actin (C4, 1:20000, ICN Biomedicals), anti-anillin (1:1000; Field and Alberts 1995), anti-lamin A (T47, 1:20; Frasch et al. 1986), anti-lamin Dm0 (611A3A6, 1:20–1:50; Harel et al. 1989), anti-lamin Dm0 (Rb294, 1:50; Fisher and Smith 1988), anti-Peanut (4C9H4, partially purified immunoglobulin, 1:20, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City; Neufeld and Rubin 1994), anti-phospho-histone H3 (1:1000, Upstate Biotechnology; Hendzel et al. 1997), and anti-α-spectrin (Ab#354, 1:400; Byers et al. 1987). Secondary antibodies used were Cy3 (1:500), Cy5 (1:250, Jackson ImmunoResearch Laboratories), Alexa488 (1:400, Molecular Probes) IgG (H+L) conjugates, and biotinylated IgG (H+L) (1:200, Vector Laboratories) with streptavidin–Cy5 conjugate (1:1000, Jackson ImmunoResearch Laboratories). DNA was stained with 100 μg/ml propidium iodide (Sigma). Specimens were mounted in Vectashield mounting medium (Vector Laboratories) and analyzed with the MRC-1024 confocal imaging system (Bio-Rad). Images were processed with LaserSharp 3.0 software (Bio-Rad).
Cloning of pbl and sequencing of point mutations
Genomic sequences flanking pblS008320 and pblS054203 P-element insertions were isolated by plasmid rescue. Genomic fragments from the 0.8-kb A6 EcoRI (from pblS008320) and 2.8-kb B1 BamHI (from pblS054203) rescues were used to isolate the 1A (3140 bp) and 1C (1642 bp) cDNAs from an embryonic (9–12 hr) cDNA library (Zinn et al. 1988). Both cDNAs map to 66B, detect two transcripts of 4.0 and 5.5-kb on Northern blots, which are expressed throughout Drosophila development, have the same expression pattern during embryonic development, and correspond to the same gene. Alignment of the sequences of plasmid rescues from pbl09645 (GenBank accession no. AQ073344), pblEP3415 (GenBank accession no. AQ254711), pblS054203, and pblS008320 demonstrates that the P elements are inserted in the 5′ UTR, 492, 457, and 149 nucleotides upstream of the AUG start codon, and in the ORF of the 1A cDNA, respectively. The 1A pbl cDNA encodes a predicted ORF of 853 amino acids.
To sequence pbl EMS alleles, total RNA isolated from pbl/TM3 flies was used as a template for first strand cDNA synthesis (Superscript II, GIBCO BRL) with a primer complementary to the 3′ end of the pbl coding sequence (5′-CGCGGATCCTCAGCTCTAAATGCGGCCCACAAC-3′). The entire coding sequence was amplified (Elongase, GIBCO BRL) with the same 3′ primer and the 5′ primer (5′-CGCGGATCCTGATGGAAATGGAGACCATTGAAGAG-3′) and cloned subsequently in BamHI sites of pBluescript KS+ (Stratagene). Two independent mutant clones were isolated and sequenced for each pbl allele. Natural polymorphisms made it possible to distinguish mutant clones from those derived from the balancer chromosome. The presence of the CAG to TAG mutation in the pbl2 allele disrupting the PstI site was confirmed by Southern analysis using the 2-kb EcoRI–BglII fragment of the 1A pbl cDNA as a probe.
Generation of antibodies to Pbl
A 2.4-kb EagI fragment of the 1A pbl cDNA corresponding to amino acids 96–853 was cloned into pET-28b(+) (Novagen) to express amino-terminally tagged His fusion protein. The induced protein was resolved on an SDS–polyacrylamide gel, and protein-containing acrylamide fragments were homogenized and injected into rats and guinea pigs. The following observations suggest that the four immune sera produced (rat R8 and R9 and guinea pig GP14 and GP15) are specific. First, all antibodies give the same pattern of staining in wild-type embryos and on Western recognize a 120-kD protein and several bands of lower molecular mass that are likely to be degradation products. Second, the immune sera recognize an ectopically expressed protein. Third, the antigen detected by immune sera is mislocalized, exhibits decreased levels, or is lacking in pbl mutant embryos. The R8 rat anti-Pbl immune serum was used at 1:800–1:1000 for indirect immunofluorescence and at 1:5000 for Western.
Generation of pbl and ect2 transgenic lines
The 1A pbl cDNA was cloned in forward or reverse orientation into pUAST to generate UAS–Pbl and UAS–antisensePbl transgenic lines, respectively. A fragment of the pbl cDNA corresponding to amino acids 325–853 was amplified (Elongase, GIBCO BRL) and cloned into pUAST to generate UAS–ΔPbl325–853 transgenic lines. To generate UAS–PblΔDH497–549 transgenic lines, 1.94-kb EcoRI–ScaI and 1.04-kb SpeI(filled-in)–EcoRI fragments of the 1A pbl cDNA were coligated with pUAST digested (Miki et al. 1993) with EcoRI. Full-length ect2 cDNA (from pCEV27–ect2-F; and amino-terminally truncated ect2 cDNA (from p3N–11/ATG; Miki et al. 1993) were cloned into pUAST to generate UAS–Ect2 and UAS–ΔEct2 transgenic lines, respectively. To assess the relative strength of different lines with independent insertions of a transgene, they were crossed to GMR–GAL4, dpp–GAL4, and 32B–GAL4 drivers, and adult flies were analyzed for abnormalities in eye and/or wing development. The phenotypes ranged from lack of visible defects to 100% pupal lethality. For overexpression studies, the following transgenic lines were selected: UAS–Pbl3.2, UAS–ΔPbl325–853S4-11, UAS–ΔPbl325–853S4-48D, UAS–PblΔDH497–5495B, UAS–PblΔDH497–5496A, UAS–Ect2S3-19, UAS-ΔEct2S2-1A, and UAS–ΔEct2S2-3. The RG1 prd–GAL4 driver was used for ectopic expression of proteins in stage 8–11 embryos. GMR–Pbl (GMR–GAL4, UAS–Pbl3.2) and GMR–PblΔDH (GMR–GAL4, UAS–PblΔDH497–5495B, 6A) are recombinant II chromosomes carrying the GMR–GAL4 driver and one UAS–Pbl transgene (from line 3.2) and two copies of the UAS–PblΔDH497–549 transgene (from lines 5B and 6A), respectively. Expression of pbl transgenes in eye imaginal disks and in embryos was confirmed immunohistochemically.
Yeast two-hybrid interaction assay
ORFs of Rho1, Rac1, Cdc42, ΔPbl325–853, PblΔDH497–549, and 1A pbl cDNA were cloned into pAS2 and pACT2 to create GAL4 DBD and AD fusions, respectively. Plasmids were cotransformed in various combinations in yeast strain PJ69-4A (James et al. 1996), and transformants were selected on SC−His−Leu−Trp medium. His+ colonies were tested for β-galactosidase activity on SC−Leu−Trp medium.
Acknowledgments
We thank D. Branton, C. Doe, R. Fehon, C. Field, P. Fisher, D. Glover, Y. Gruenbaum, Y. Hotta, L. Luo, T. Miki, M. Mlodzik, C. Nüsslein-Volhard, J. Settleman, S. Wasserman, and K. Zinn for providing reagents. We thank Manzoor A. Bhat, David M. Glover, Scott Goode, Anthony L. Lau, and Shelley Sazer for critically reviewing the manuscript and Michelle Coombe for excellent technical assistance. This work was supported in part by the Australian Research Council. L.O. and L.P. are supported by the University of Adelaide Postgraduate Award and an Australian Postgraduate Award, respectively. H.J.B. is Associate Investigator of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Note
The GenBank accession number for the sequence of pbl described in this paper is AF136492.
Footnotes
E-MAIL hbellen@bcm.tmc.edu; FAX (713) 798-8515.
References
- Adamson P, Paterson HF, Hall A. Intracellular localization of the P21rho proteins. J Cell Biol. 1992;119:617–627. doi: 10.1083/jcb.119.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett K, Leptin M, Settleman J. The Rho GTPase and a putative RhoGEF mediate a signaling pathway for the cell shape changes in Drosophila gastrulation. Cell. 1997;91:905–915. doi: 10.1016/s0092-8674(00)80482-1. [DOI] [PubMed] [Google Scholar]
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- Byers TJ, Dubreuil R, Branton D, Kiehart DP, Goldstein LS. Drosophila spectrin. II. Conserved features of the alpha-subunit are revealed by analysis of cDNA clones and fusion proteins. J Cell Biol. 1987;105:2103–2110. doi: 10.1083/jcb.105.5.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP. From BRCA1 to RAP1: A widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Chai Y, Cui J, Shao N, Reddy ESP, Rao VN. The second BRCT domain of BRCA1 proteins interacts with p53 and stimulates transcription from the p21WAF1/CIP1 promoter. Oncogene. 1999;18:263–268. doi: 10.1038/sj.onc.1202323. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JM, Harden N, Leung T, Lim L, Kiehart DP. Cellularization in Drosophila melanogaster is disrupted by the inhibition of rho activity and the activation of Cdc42 function. Dev Biol. 1998;204:151–164. doi: 10.1006/dbio.1998.9061. [DOI] [PubMed] [Google Scholar]
- Drechsel DN, Hyman AA, Hall A, Glotzer M. A requirement for Rho and Cdc42 during cytokinesis in Xenopus embryos. Curr Biol. 1996;7:12–23. doi: 10.1016/s0960-9822(06)00023-6. [DOI] [PubMed] [Google Scholar]
- Dutartre H, Davoust J, Gorvel JP, Chavrier P. Cytokinesis arrest and redistribution of actin-cytoskeleton regulatory components in cells expressing the Rho GTPase CDC42Hs. J Cell Sci. 1996;109:367–377. doi: 10.1242/jcs.109.2.367. [DOI] [PubMed] [Google Scholar]
- Ellis MC, O’Neill EM, Rubin GM. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fares H, Peifer M, Pringle JR. Localization and possible functions of Drosophila septins. Mol Biol Cell. 1995;6:1843–1859. doi: 10.1091/mbc.6.12.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth CL, Feig LA. Dominant inhibitory mutations in the Mg(2+)-binding site of RasH prevent its activation by GTP. Mol Cell Biol. 1991;11:4822–4829. doi: 10.1128/mcb.11.10.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon RG, Oren T, LaJeunesse DR, Melby TE, McCartney BM. Isolation of mutations in the Drosophila homologues of the human Neurofibromatosis 2 and yeast CDC42 genes using a simple and efficient reverse-genetic method. Genetics. 1997;146:245–252. doi: 10.1093/genetics/146.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA. Tools of the trade: Use of dominant-inhibitory mutants of Ras-family GTPases. NatCell Biol. 1999;1:E25–E27. doi: 10.1038/10018. [DOI] [PubMed] [Google Scholar]
- Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–178. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field CM, Al-Awar O, Rosenblatt J, Wong ML, Alberts B, Mitchison TJ. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J Cell Biol. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field C, Li R, Oegema K. Cytokinesis in eukaryotes: A mechanistic comparison. Curr Opin Cell Biol. 1999;11:68–80. doi: 10.1016/s0955-0674(99)80009-x. [DOI] [PubMed] [Google Scholar]
- Fisher PA, Smith DE. Affinity purification of antibodies using antigens immobilized on solid supports. Biochem Soc Trans. 1988;16:134–138. doi: 10.1042/bst0160134. [DOI] [PubMed] [Google Scholar]
- Frasch M, Glover DM, Saumweber H. Nuclear antigens follow different pathways into daughter nuclei during mitosis in early Drosophila embryos. J Cell Sci. 1986;82:155–172. doi: 10.1242/jcs.82.1.155. [DOI] [PubMed] [Google Scholar]
- Gerald N, Dai J, Ting-Beall HP, De Lozanne A. A role for Dictyostelium racE in cortical tension and cleavage furrow progression. J Cell Biol. 1998;141:483–492. doi: 10.1083/jcb.141.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Bonaccorsi S, Williams B, Williams EV, Santolamazza C, Goldberg ML, Gatti M. Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes & Dev. 1998;12:396–410. doi: 10.1101/gad.12.3.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Bonaccorsi S, Gatti M. The role of anillin in meiotic cytokinesis of Drosophila males. J Cell Sci. 1999;112:2323–2334. doi: 10.1242/jcs.112.14.2323. [DOI] [PubMed] [Google Scholar]
- Goldberg ML, Gunsalus KC, Karess RE, Chang F. Cytokinesis. In: Endow SA, Glover DM, editors. Dynamics of cell division. London, UK: Oxford University Press; 1998. pp. 270–316. [Google Scholar]
- Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML. Mutations in twinstar, a Drosophila gene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häcker U, Perrimon N. DRhoGEF2 encodes a member of the Dbl family of oncogenes and controls cell shape changes during gastrulation in Drosophila. Genes & Dev. 1998;12:274–284. doi: 10.1101/gad.12.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Harden N, Ricos M, Ong YM, Chia W, Lim L. Participation of small GTPases in dorsal closure of the Drosophila embryo: Distinct roles for Rho subfamily proteins in epithelial morphogenesis. J Cell Sci. 1999;112:273–284. doi: 10.1242/jcs.112.3.273. [DOI] [PubMed] [Google Scholar]
- Harel A, Zlotkin E, Nainudel-Epszteyn S, Feinstein N, Fisher PA, Gruenbaum Y. Persistence of major nuclear envelope antigens in an envelope-like structure during mitosis in Drosophila melanogaster embryos. J Cell Sci. 1989;94:463–470. doi: 10.1242/jcs.94.3.463. [DOI] [PubMed] [Google Scholar]
- Hariharan IK, Hu KQ, Asha H, Quintanilla A, Ezzell RM, Settleman J. Characterization of rho GTPase family homologues in Drosophila melanogaster: Overexpressing Rho1 in retinal cells causes a late developmental defect. EMBO J. 1995;14:292–302. doi: 10.1002/j.1460-2075.1995.tb07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MJ, Eva A, Zangrilli D, Aaronson SA, Evans T, Cerione RA, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Hime G, Saint R. Zygotic expression of the pebble locus is required for cytokinesis during the postblastoderm mitoses of Drosophila. Development. 1992;114:165–171. doi: 10.1242/dev.114.1.165. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, Wieschaus E, Nüsslein-Volhard C, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome. Wilhelm Roux’s Arch Dev Biol. 1984;193:283–295. doi: 10.1007/BF00848157. [DOI] [PubMed] [Google Scholar]
- Karess RE, Chang X, Edwards KA, Kulkarni S, Aguilera I, Kiehart DP. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell. 1991;65:1177–1189. doi: 10.1016/0092-8674(91)90013-o. [DOI] [PubMed] [Google Scholar]
- Kishi K, Sasaki T, Kuroda S, Itoh T, Takai Y. Regulation of cytoplasmic division of Xenopus embryo by rho p21 and its inhibitory GDP/GTP exchange protein (rho GDI) J Cell Biol. 1993;120:1187–1195. doi: 10.1083/jcb.120.5.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF. The pebble gene is required for cytokinesis in Drosophila. J Cell Sci. 1992;103:1021–1030. doi: 10.1242/jcs.103.4.1021. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang H, Eberstadt M, Schnuchel A, Olejniczak ET, Meadows RP, Schkeryantz JM, Janowick DA, Harlan JE, Harris EA, et al. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell. 1998;95:269–277. doi: 10.1016/s0092-8674(00)81757-2. [DOI] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes & Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Mabuchi I, Hamaguchi Y, Fujimoto H, Morii N, Mishima M, Narumiya S. A rho-like protein is involved in the organisation of the contractile ring in dividing sand dollar eggs. Zygote. 1993;1:325–331. doi: 10.1017/s0967199400001659. [DOI] [PubMed] [Google Scholar]
- Miki T, Smith CL, Long JE, Eva A, Fleming TP. Oncogene ect2 is related to regulators of small GTP-binding proteins. Nature. 1993;362:462–465. doi: 10.1038/362462a0. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, Rubin GM. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Nakano K, Mabuchi I. Localization of Rho GTPase in sea urchin eggs. FEBS Lett. 1998;441:121–126. doi: 10.1016/s0014-5793(98)01531-2. [DOI] [PubMed] [Google Scholar]
- O’Connell CB, Wheatley SP, Ahmed S, Wang Y. The small GTP-binding protein Rho regulates cortical activities in cultured cells during division. J Cell Biol. 1999;144:305–313. doi: 10.1083/jcb.144.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Zannini M, Lewis M, Wickner RB, Hunt LT, Graziani G, Tronick SR, Aaronson SA, Eva A. A region of proto-dbl essential for its transforming activity shows sequence similarity to a yeast cell cycle gene, CDC24, and the human breakpoint cluster gene, bcr. New Biol. 1991;3:372–379. [PubMed] [Google Scholar]
- Salzberg A, Prokopenko SN, He Y, Tsai P, Pal M, Maroy P, Glover DM, Deak P, Bellen HJ. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: Mutations affecting embryonic PNS development. Genetics. 1997;147:1723–1741. doi: 10.1093/genetics/147.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighoffer F, Cai H, Chevallier-Multon MC, Fath I, Cooper G, Tocque B. The Saccharomyces cerevisiae SDC25 C-domain gene product overcomes the dominant inhibitory activity of Ha-Ras Asn-17. Mol Cell Biol. 1993;13:39–43. doi: 10.1128/mcb.13.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G. The pleckstrin homology domain: An intriguing multifunctional protein module. BioEssays. 1996;18:35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- Soisson SM, Nimnual AS, Uy M, Bar-Sagi D, Kuriyan J. Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein. Cell. 1998;95:259–268. doi: 10.1016/s0092-8674(00)81756-0. [DOI] [PubMed] [Google Scholar]
- Sone M, Hoshino M, Suzuki E, Kuroda S, Kaibuchi K, Nakagoshi H, Saigo K, Nabeshima Y, Hama C. Still life, a protein in synaptic terminals of Drosophila homologous to GDP-GTP exchangers. Science. 1997;275:543–547. doi: 10.1126/science.275.5299.543. [DOI] [PubMed] [Google Scholar]
- Sterpetti P, Hack AA, Bashar MP, Park B, Cheng S, Knoll JHM, Urano T, Feig LA, Toksoz D. Activation of the Lbc Rho exchange factor proto-oncogene by truncation of an extended C terminus that regulates transformation and targeting. Mol Cell Biol. 1999;19:1334–1345. doi: 10.1128/mcb.19.2.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven R, Kubiseski TJ, Zheng H, Kulkarni S, Mancillas J, Ruiz Morales A, Hogue CW, Pawson T, Culotti J. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- Strutt DI, Weber U, Mlodzik M. The role of RhoA in tissue polarity and Frizzled signalling. Nature. 1997;387:292–295. doi: 10.1038/387292a0. [DOI] [PubMed] [Google Scholar]
- Takaishi K, Sasaki T, Kameyama T, Tsukita S, Takai Y. Translocation of activated Rho from the cytoplasm to membrane ruffling area, cell-cell adhesion sites and cleavage furrows. Oncogene. 1995;11:39–48. [PubMed] [Google Scholar]
- Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes & Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- Wasserman S. FH proteins as cytoskeletal organizers. Trends Cell Biol. 1998;8:111–115. doi: 10.1016/s0962-8924(97)01217-8. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch BM, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead I, Kirk H, Tognon C, Trigo-Gonzalez G, Kay R. Expression cloning of lfc, a novel oncogene with structural similarities to guanine nucleotide exchange factors and to the regulatory region of protein kinase C. J Biol Chem. 1995;270:18388–18395. doi: 10.1074/jbc.270.31.18388. [DOI] [PubMed] [Google Scholar]
- Whitehead IP, Campbell S, Rossman KL, Der CJ. Dbl family proteins. Biochim Biophys Acta. 1997;1332:F1–F23. doi: 10.1016/s0304-419x(96)00040-6. [DOI] [PubMed] [Google Scholar]
- Yamane K, Kawabata M, Tsuruo T. A DNA-topoisomerase-II-binding protein with eight repeating regions similar to DNA-repair enzymes and to a cell-cycle regulator. Eur J Biochem. 1997;250:794–799. doi: 10.1111/j.1432-1033.1997.00794.x. [DOI] [PubMed] [Google Scholar]
- Yasui Y, Amano M, Nagata K, Inagaki N, Nakamura H, Saya H, Kaibuchi K, Inagaki M. Roles of Rho-associated kinase in cytokinesis; mutations in Rho-associated kinase phosphorylation sites impair cytokinetic segregation of glial filaments. J Cell Biol. 1998;143:1249–1258. doi: 10.1083/jcb.143.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Hart MJ, Cerione RA. Guanine nucleotide exchange catalyzed by dbl oncogene product. Methods Enzymol. 1995;256:77–84. doi: 10.1016/0076-6879(95)56011-4. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Zangrilli D, Cerione RA, Eva A. The pleckstrin homology domain mediates transformation by oncogenic dbl through specific intracellular targeting. J Biol Chem. 1996;271:19017–19020. doi: 10.1074/jbc.271.32.19017. [DOI] [PubMed] [Google Scholar]
- Zinn K, McAllister L, Goodman CS. Sequence analysis and neuronal expression of fasciclin I in grasshopper and Drosophila. Cell. 1988;58:577–587. doi: 10.1016/0092-8674(88)90574-0. [DOI] [PubMed] [Google Scholar]


