Abstract
A discrimination method between biologically relevant interfaces and artificial crystal-packing contacts in crystal structures was constructed. The method evaluates protein-protein interfaces in terms of complementarities for hydrophobicity, electrostatic potential and shape on the protein surfaces, and chooses the most probable biological interfaces among all possible contacts in the crystal. The method uses a discriminator named as “COMP”, which is a linear combination of the complementarities for the above three surface features and does not correlate with the contact area. The discrimination of homo-dimer interfaces from symmetry-related crystal-packing contacts based on the COMP value achieved the modest success rate. Subsequent detailed review of the discrimination results raised the success rate to about 88.8%. In addition, our discrimination method yielded some clues for understanding the interaction patterns in several examples in the PDB. Thus, the COMP discriminator can also be used as an indicator of the “biological-ness” of protein-protein interfaces.
Keywords: protein-protein interaction, complementarity analysis, homo-dimer interface, crystal-packing contact, biological interfaces
Introduction
The quaternary structures of proteins are the bases of their physiological functions (Jones and Thornton 1996; Henrick and Thornton 1998; Krissinel and Henrick 2007), and thus it is indispensable to know the biologically relevant complexes of proteins to understand their functions at the molecular level. The structures of proteins are usually determined by X-ray crystallography, and actually 86% of the structures in the Protein Data Bank (PDB) (Berman et al 2000) were obtained by X-ray crystallography, as of May 2008. However, the structures determined by X-ray crystallography could contain nonbiological interactions due to the nature of crystals.
Protein crystals are composed of asymmetric units (ASU), which are the smallest unit of the crystal, and the whole crystal can be generated by rotating and translating the ASU according to the symmetry operators provided for each crystal. The component molecules of each ASU are packed to stabilize the crystal, and they interact with each other both within the ASU and among the adjacent ASUs. The latter interactions are usually designated as crystal-packing, and they are considered to be weaker than the biologically relevant interactions (Janin and Rodier 1995; Carugo and Argos 1997; Dasgupta et al 1997; Janin 1997; Bahadur et al 2004). However, the protein complexes in each ASU are not always the real biological complexes, because the ASU is defined independently of the biological context (Valdar and Thornton 2001; Jefferson et al 2006; Xu et al 2006). For example, a biological molecule can be just a part of an ASU, while on the other hand, a biological complex may be obtained by rotating and translating all or a part of an ASU. In the former case, the part of the interface in the ASU is the biological interface, and in the latter case, the crystal packing can have some biological relevance.
Information about the number and/or kinds of proteins included in a real biological complex, the “biological unit”, is essential to obtain the quaternary structures of the proteins, and therefore, a method to discriminate the biological interfaces from the nonbiological interfaces is needed to use the structure determined by X-ray crystallography (Carugo and Argos 1997; Ponstingl et al 2000; Elcock and McCammon 2001; Valdar and Thornton 2001; Mintseris and Weng 2003; Jefferson et al 2006; Liu et al 2006). When no other information about the quaternary structure is available than that from X-ray crystallography, inferring the biological unit must be done only from the structural data (Carugo and Argos 1997; Ponstingl et al 2000; Ofran and Rost 2003; Ponstingl et al 2003; Levy et al 2006; Krissinel and Henrick 2007). The Protein Quaternary Structure (PQS) server is one of the methods for inferring biological assemblies and is a widely used database, where the data of inferred biological assemblies for all proteins registered in the PDB are stored (Henrick and Thornton 1998). This method composes the biological assemblies by adding the contacts judged as being biological relevant in the crystal. Ponstingl et al improved the PQS method and also constructed the software PITA for inferring the biological interfaces and assemblies (Ponstingl et al 2000, 2003). Generally speaking, the crystal-packing contacts have smaller contact areas as compared to the biological interfaces (Janin and Rodier 1995; Carugo and Argos 1997; Dasgupta et al 1997; Janin 1997; Bahadur et al 2004). Therefore, these discrimination methods that strongly depend on the contact size could achieve modestly high success rates (80%–90%). However, there are some exceptions: crystal-packing contacts can sometimes have larger contact areas than biological interfaces (Janin 1997; Robert and Janin 1998; Elcock and McCammon 2001; Bahadur et al 2004). This indicates that the contact area can be the major factor to discriminate biological interfaces from crystal contacts, as mentioned by Levy and colleagues (2008), but it is not a completely reliable differentiation criterion. Therefore, to improve the discrimination power, a method to determine the biological interfaces that considers other factors than the contact area is needed.
Several studies that use other information than the contact size have already been developed (Elcock and McCammon 2001; Bahadur et al 2004; Krissinel and Henrick 2007; Bernauer et al 2008). Bahadur and colleagues (2004) tried to discriminate between homo-dimers and crystal-packed dimers based on the atomic packing density and the physical-chemical properties of the interfaces (residue propensity, hydrophobic interaction and so on), where the crystal contacts were extracted from the crystals of monomeric proteins. As a result, they obtained the better success rates: 88% for the homo-dimers and 77% for the crystal contacts. Krissinel and Henrik (2007) also tried to predict the biologically relevant macromolecules in crystals by focusing on the binding energy and the entropy of dissociation in the formation of the interface or the assembly, and constructed a PISA database. Their method achieved an 80%–90% success rate using their dataset. Recently, Bernauer and colleagues (2008) have developed the Voronoi tesselation-based SVM for discriminating between homo-dimers and crystal-dimers, with higher accuracy (95%). They prepared 84 parameters (contact area, number of residues, Voronoi volume, frequency of each residue type, frequency of pairs of residues and distance between residues in interfaces) and then reduced them to 27 parameters so that the best performance could be obtained.
In this study, we developed a new method to discriminate biological interfaces from crystal contacts by extending our previous work (Tsuchiya et al 2006). First, we defined the complementarity index of the interface, COMP, so that the set of biological interfaces could be separated from the set of symmetry-related crystal-packing contacts with the highest accuracy, and then a discrimination test between the biological interface and the crystal-packing contact in each crystal was performed. It should be noted that the preparation of the correct set (biological dimer contact set) is not straightforward, because the information about the form of biological assembly is not always provided even in the primary citation of each PDB entry. Therefore, we took a two-step approach. In the first step (discrimination step) we assumed that the interfaces in each ASU are the biological interfaces, and in the following step (evaluation step), we evaluated the discrimination results in detail, to check if the assumption was correct or not. This is because it seems reasonable to assume that there will be a strong tendency that biologically relevant complexes are selected as the ASU. Here we used 282 nonredundant homo-dimer interfaces as correct answers, and 111 crystal contacts as negative ones (see Materials and methods). In the discrimination step, our method displayed modest accuracy (84.8%), and in the subsequent evaluation step, we achieved 88.8% accuracy after literature checks of ambiguous entries. Furthermore, we found some clues to understand the protein-protein interaction patterns occurring in a few confusing cases, through the evaluation step.
Materials and methods
Dataset
We call the biological dimer contact, the correct data, simply as “biological contact”, and the contact generated by symmetry operation, the negative data, as “crystal-packing contact.”
Biological dimer contact set (the correct data set)
We used 393 nonredundant homo-interfaces prepared in our previous work (Tsuchiya et al 2006), in which the PDB entries with two or more chains and with 2.5Å or better resolution were selected and the redundancies were eliminated by selecting one representative from each SCOP family (Murzin et al 1995). These interfaces were included in the homo assemblies within the ASUs of the crystals, which had atomic contacts shorter than 4.0Å between the different protomers. It should be noted that in the case of the homo multimeric assemblies such as a tetramer or octamer, the representative interfaces may be the second or third largest interfaces in the assemblies within the ASU. Moreover, in the case of the homo multimeric assemblies or the case that the biological units of the homologues of the representative are different from that of the representative, such as the two-folded dimer and the dimer of dimers, as discussed by Levy and colleagues (2006, 2008), there may be the different types of interfaces from the representative one in a SCOP family. These interfaces often have small area, and are indistinguishable from the crystal contacts. Therefore, we focused on only one interface in each SCOP family.
In the previous work, we classified all of the homo oligomer interfaces according to the shape and the symmetry of the interfaces. Among them, 297 interfaces with two-fold symmetry and without a tangle were taken as the candidates of the biological contacts, which is based on the assumption that the contact in the ASU is the biological interface as mentioned above. The other interfaces without a symmetrical axis were generally those found in cyclic oligomers, and those with a tangle are very likely to be a biological interface.
Many of the crystal-packing contacts which were generated by symmetry operation as described in the next section, had very small contact areas, and a small number of them had areas as large as those of the biological dimer interfaces. The discrimination will be necessary for the interfaces with contact areas comparable to those of biological interfaces. We thus checked the distribution of the contact areas in the biological contact set and decided to eliminate the entries (contacts) with smaller areas than 5% in the set, which is the first area criterion, 127.4 Å2. In this procedure, 15 biological contacts, which are seven entries that can be monomeric proteins, seven entries with the second or third largest interfaces in the multimeric oligomers, and one entry judged as the dimer protein according to their primary citations, were excluded. The last entry, 3eip (Li et al 1999), contains two subunits of immunity protein Im3 which is a specific inhibitor of colicin E3, in the ASU. The two subunits form the loosely-packed interface, because the zinc and two water molecules mediate the inter-subunit interaction. The colicin binding site exists in the inter-subunit interacting region. The authors of the primary citation mention that it is unclear whether the inter-subunit interaction is biologically important or an artifact caused by the crystallization condition, because the dimer has to dissociate into monomers before binding the colicin. Thus, we consider that the elimination of these 15 entries did not have any problems. Finally, 282 among the 297 biological contacts were used as the correct biological contact set.
Crystal-packing contact set (the negative data set)
All of the contacts in this set were generated from the protomers inside the ASUs by the symmetry operation. Therefore, this set never contains the same contacts as those in the biological contact set. For each contact in the biological contact set, the amino acid sequences of all protein subunits inside the ASU which contains the biological contact, were compared to that of the subunit with the smaller chain ID of the biological contact, by using FASTA (Pearson and Lipman 1988). From the subunits with sequence identity higher than 85% to the subunit of the biological contact, the symmetry-related protomers were generated both in the center unit cell containing the ASU and in the surrounding 26 cells, using the symmetry operators in the header of the PDB entry other than the same operators as those annotated as the “BIOMT” records. Of them, the symmetry-related protomers with atom contacts within distances shorter than 4.0 Å from either of two subunits of the biological contact were picked up: these contacting protomers were considered as the crystal-packed contacting pairs.
The molecular surfaces of both protomers of the pair were generated by Connolly’s algorithm (Connolly 1983). The contacting region of this pair was then defined as a set of pairs of vertices located on different surfaces at a distance shorter than 1.0 Å. Noted that identical interfaces due to crystallographic symmetry were removed and the interfaces lacking two-fold symmetry were also excluded, because we focused on the discrimination of the biological interfaces from crystal contact thus the interface without two fold symmetry are not a problem (Goodsell and Olson 2000). To remove the nonsymmetrical interfaces, we calculated the ratio of the number of the same residues in a protomer of the interface as those in the other protomer to the number of residues in the interfaces (Tsuchiya et al 2006). If the ratio is 1.0, then all of the residues from a protomer of the interface are exactly the same as those in the other protomer. When the ratio is less than 0.6, the interface is considered as nonsymmetrical. Consequently, 308 crystal-packing contacts were obtained.
In order to make a new criteria for discrimination between the biological and crystal-packing contacts, we reduced the above 308 crystal-packing contacts, so that the contact areas were comparable to those of biological interfaces. Thus, 111 crystal-packing contacts, whose interface areas are larger than 127.4 Å2 (same values as the area threshold used in the biological contact set), were finally selected among the above 308 contacts and used in the following analyses.
Complementarity analysis
The basis of the complementarity analyses was originally developed for the classification and analyses of homo-oligomer interfaces in our previous study (Tsuchiya et al 2006). In the analyses, first, the Connolly surface (Connolly 1983) consisting of triangle polygons was constructed for each protomer. Next, the hydrophobicity, calculated by the Ooi-Oobatake method (Ooi et al 1987), and the electrostatic potential, obtained by solving the Poisson-Boltzmann equation numerically with the program SCB (Nakamura and Nishida 1987), were mapped onto each vertex on the Connolly surface. The shape of the surface was also considered using average curvatures at each vertex (Tsuchiya et al 2004). The interacting region on the surfaces was defined as a set of pairs of vertices from different surfaces with a distance shorter than 1.0 Å. Then, complementarity scores, Hcmp, Ecmp, and Scmp for hydrophobicity, electrostatic potential and shape, respectively, were defined as the ratio of the number of complementary vertex-pairs for hydrophobicity (Nhyd, hydrophobic and hydrophobic), electrostatic potential (Nele, opposite sign of the potential) or shape (Nshape, convex and concave), respectively, to the number of all vertex-pairs in the interface, Ntotal (Tsuchiya et al 2006), as follows:
Finally, the complementarity index, COMP, was defined as follows:
| (Eq.1) |
where the weight parameters, Wh, We and Ws, are normalized so that . The weight parameters were optimized by changing them so that the Matthews correlation coefficient (Matthews 1975), MCC, was maximized. The optimization was done by introducing the sub-parameters w1, w2 and w3, so that w1 = Wh × W, w2 = We × W and w3 = Ws × W, where to ensure the constraint of . The sub-parameters were changed from −100 to 100 with intervals of 1, and the MCC was calculated by changing the threshold values of COMP from 0 to 1.0 with intervals of 0.001 in order to judge whether the interface was biological or not.
Discrimination between the biological and crystal-packing contacts
Discrimination step
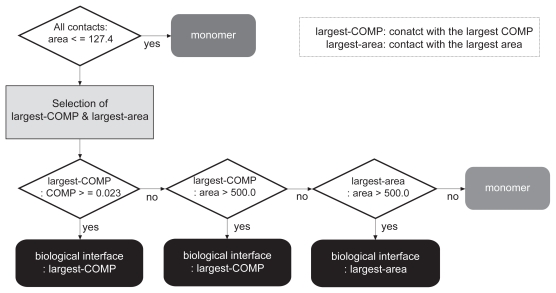
The discrimination between the biological contact and the crystal-packing contact(s) in each entry was carried out according to the selection scheme flowcharted in Figure 1, where the most probable biological interface was selected among the biological and the crystal-packing contacts. As this chart shows, first the contacts with an area larger than the criterion, 127.4 Å2 (described further in the Results and Discussion), were picked among all of the possible contacts in the crystal. If none of the contacts in the crystal meets the area criterion, then the protein is judged to be monomeric. Since all of the contacts in both datasets used in this study had areas larger than this criterion as described above, we skipped this step. Second, the contacts with the largest COMP and with the largest area were searched among the biological contact and the crystal-packing contacts. The most probable biological interface was then chosen from the two contacts, as follows: if the contact with the largest COMP met the threshold of the COMP (0.023) that was determined in the weight optimization of the COMP as described later, then the contact was judged as the most probable biological interface. If the contact with the largest COMP did not meet the threshold, but had an area larger than 500.0 Å2 which is the second area criterion and will be described later, then the contact was judged as the most probable biological interface. When the contact with the largest COMP did not meet the COMP threshold and the second area criterion, but the contact with the largest area had an area larger than 500.0 Å2, then the contact with the largest area was judged as the most probable biological interface. If no contact met the COMP threshold and the second area criterion, then the protein was judged to be monomeric.
Figure 1.
The selection scheme of the most probable biological interfaces. The most probable biological interface in each crystal is selected among the biological contact and the crystal-packing contact(s) according to the scheme shown in this flow chart. The explanation of the scheme is described in the text.
Evaluation step
The discrimination result was then evaluated by referring to the primary citation of the entry regarding whether the contacts judged as the most probable biological interface agreed with the actual biological interfaces that were determined according to the opinions of the authors in the primary citations of the entries.
Comparison of dimer structures determined by the different ways
Comparison of structures determined by X-ray crystallography and NMR techniques
The homo-dimer structures determined by the NMR technique were extracted from the PDB in October 2006. The dimers which consist of the subunits with sequence identity higher than 90% to any protomers in the biological contact set were selected by using FASTA (Pearson and Lipman 1988). Consequently, 14 dimers for five entries in the biological contact set were obtained. In Table 1, the original entries in the biological contact set (X-ray crystal structures) and their counterparts (NMR structures) are listed in the left-hand and right-hand columns, respectively. The comparisons were done by visual inspection of the interface (Kinoshita and Nakamura 2004).
Table 1.
Comparison of the structures determined by X-ray and NMR
| X-ray
|
NMR
|
|||
|---|---|---|---|---|
| PDB Chain ID | Category | PDB Chain ID | Seq IDa | |
| 1 | 1ci4A-B | 1 | 1qckA-B | 97.8 |
| 2ezxA-B | 97.8 | |||
| 2ezyA-B | 97.8 | |||
| 2ezzA-B | 97.8 | |||
| 2 | 1kzkA-B | 1 | 1bveA-B | 91.9 |
| 1bvgA-B | 91.9 | |||
| 3 | 1m1fA-B | 1 | 2c06A-B | 97.3 |
| 4 | 1mkkA-B | 1 | 1katV-W | 91.9 |
| 5 | 1msoB-D | 1 | 1ai0B-D | 100.0 |
| 1aiyB-D | 100.0 | |||
| 2aiyB-D | 100.0 | |||
| 3aiyB-D | 100.0 | |||
| 4aiyB-D | 100.0 | |||
| 5aiyB-D | 100.0 | |||
Note: Sequence identity between the protomer in the X-ray crystal structure and that in the NMR structure.
Comparison of structures determined in the different crystallization conditions
The symmetry-related dimer complexes determined by X-ray crystallography and with 2.5 Å or better resolutions, were extracted from the PDB in October 2006. Among them, we searched for the dimers that have a subunit sharing 100% sequence identity to a protomer in the biological contact set and that are determined in the different crystallization condition from that of the corresponding original entry. Finally, we found 17 dimers for 14 entries in the biological contact set, as listed in Table 2, where the original entries and their counterparts are listed in the left-hand and right-hand columns, respectively. For each dimer, all possible contacts in the crystals of the original entry and the counterparts were generated, and the interfaces with areas smaller than the first area criterion, 127.4 Å2, were removed. Then, the COMP value and area of each contact in the original entry were compared with those of all contacts in the counterparts along with checking the forms of the dimer complexes visually.
Table 2.
Comparison of the structures determined in the different crystallization conditions
| Original entry
|
Different crystal form
|
|||||
|---|---|---|---|---|---|---|
| PDB Chain ID | Category | Evaluationa | Space group | PDB Chain ID | Space group | |
| 1 | 1dj8C-D | 1 | biological | P 1 21 1 | 1bg8A-B | C 1 2 1 |
| 2 | 1f4mA-B | 1 | biological | P 32 | 1f4nA-B | C 1 2 1 |
| 3 | 1j59A-B | 1 | biological | C 2 2 21 | 1i5zA-B | P 21 21 21 |
| 4 | 1jm0E-F | 3 | biological | P 21 21 21 | 1jmbB-C | C 2 2 21 |
| 5 | 1ks2A-B | 1 | biological | P1 | 1lkzA-B | C 2 2 21 |
| 6 | 1m0wA-B | 1 | biological | P1 | 1m0tA-B | C 2 2 21 |
| 7 | 1m1nF-H | 1 | biological | P 1 21 1 | 1m34B-D | C 1 2 1 |
| 8 | 1m7gA-B | 2 | biological | P 21 21 21 | 1d6jA-B | C 2 2 21 |
| 9 | 1msoB-D | 1 | biological | H 3 | 1os4B-D | P 1 |
| 1ev6B-D | P 1 21 1 | |||||
| 1gujB-D | P 21 21 21 | |||||
| 1benB-D | R 3 | |||||
| 10 | 1nmsA-B | 1 | biological | C 1 2 1 | 1nmqA-B | P 21 21 21 |
| 11 | 1o7jB-D | 1 | biological | C 1 2 1 | 1hfkA-C | P 61 2 2 |
| 12 | 1oaoA-B | 1 | biological | C 1 2 1 | 1mjgC-D | P1 |
| 13 | 1oh0A-B | 1 | biological | C 1 2 1 | 1e3vA-B | P 21 21 21 |
| 14 | 1p1jA-B | 1 | biological | C 1 2 1 | 1p1hC-D | P 1 21 1 |
Note: The “biological” means that the contact in the biological contact set was judged as the most probable biological interface in the crystal in the evaluation step.
Results and Discussion
Weight optimization of the complementarity index, COMP
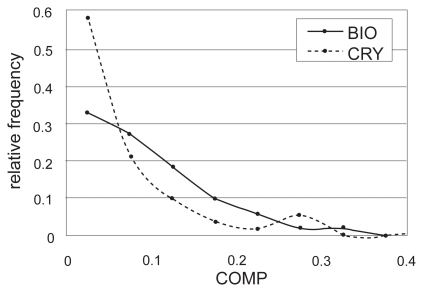
We used the COMP value (Eq.1) to separate biologically relevant interfaces from artificial crystal-packing contacts, based on the idea that the biological interface is more complementary in terms of its physicochemical properties and shape than the crystal-packing contacts. The COMP value is obtained by combining the three complementarities using weights, Wh, We, and Ws. These weights were defined so that the sets of the 282 biological contacts and the 111 crystal-packing contacts could be separated with the highest accuracy measured by the MCC value (Matthews 1975). Consequently, the maximum MCC = 0.33 was obtained with the weight values Wh = 0.99, We = 0.030 and Ws = 0.16 and the COMP threshold = 0.023. The results of the weight optimization are summarized in Table 3. As shown in Figure 2 which indicates the distributions of the COMP values computed using this weight combination for all entries in the biological contact set and the crystal-packing contact set respectively, the distribution in the biological contact set slightly sifted to the larger side.
Table 3.
Results of the weight optimization of the COMP
| w1 | w2 | w3 | Wh | We | Ws | MCC | Threshold | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|---|---|
| 78 | 2 | 13 | 0.99 | 0.030 | 0.16 | 0.33 | 0.023 | 0.75 | 0.89 | 0.40 |
Figure 2.
The relative frequencies of the COMP values in the biological (BIO, thick line) and crystal-packing (CRY, dotted line) contact sets.
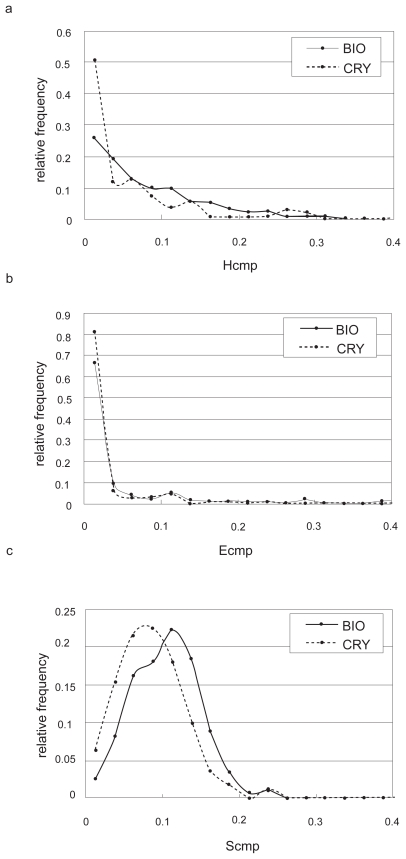
As seen in Table 3, the weight for the electrostatic potential (0.030) is much smaller than those for the hydrophobicity (0.99) and shape (0.16). This may indicate that the complementarity for the electrostatic potential did not contribute as much to the discrimination between the both contact sets. To address this possibility, we checked the distribution of each complementarity (Figure 3). As Figure 3b shows, there was no difference between the distributions of the relative frequencies of Ecmp in the biological contacts and in the crystal-packing contacts, while Hcmp and Scmp had different tendencies (Figures 3a and c). This suggests that the main discrimination factor between these two contact sets would be hydrophobic and shape complementarities, and it seems consistent that a large interface will tend to be a biological interface.
Figure 3.
The relative frequencies of the complementarities for a) hydrophobicity, b) electrostatic potential and c) shape. The thick lines in the three figures indicate the distributions of complementarities in the biological contact set (BIO), and the dotted lines indicate those in the crystal-packing contact set (CRY).
Discrimination between the biological and crystal-packing contacts
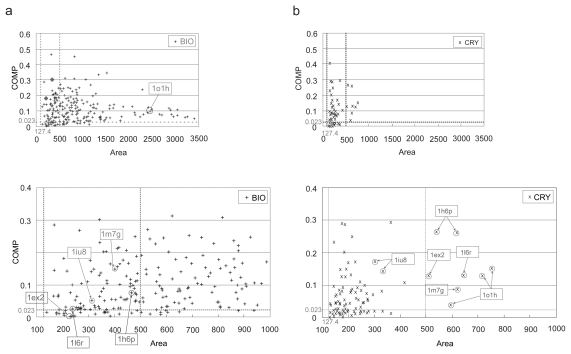
In each entry, the most probable biological interface was chosen among the biological and crystal-packing contacts according to the selection scheme summarized in Figure 1, as described in Materials and Methods. The threshold of the COMP and the two area criteria were used for the judgments in some steps of this scheme. The COMP threshold, 0.023, came from the COMP value with the maximum MCC in the weight optimization. One of the area criteria, 127.4 Å2, was the lower 5% boundary of the biological contact set as described above. The other area criterion, 500.0 Å2, was added to judge a contact with a large area as a biological interface even if its COMP did not meet the threshold. As shown in Figure 4 where the relationship between the COMP and the contact area in each contact is indicated, this is because only a few crystal-packing contacts had areas larger than 500.0 Å2 (Figure 4b), while many biological contacts had larger areas than 500.0 Å2 (Figure 4a), some of them were over 1,000 Å2, as observed previously (Bahadur et al 2003, 2004). It should be noted that the COMP threshold and the weight combination in the calculation of the COMP value were determined in the optimization step with the same data that was used in this discrimination step, due to a small number of entries available. However, the discrimination and the weight optimization are different problems, because the former carried out only within an entry, while the later tried to separate the two sets of interfaces, biological contacts and crystal contacts. Therefore, the use of same data would not affect the results largely.
Figure 4.
The scatter plots between the COMP and the contact area in a) the biological contact set (BIO) and in b) the crystal-packing contact set (CRY). In each figure, each sign indicates each contact, and the horizontal dotted line and the two vertical dotted lines indicate the threshold of the COMP (0.023) and the contact area criteria (127.4 and 500.0 Å2), respectively. The lower figures in both a) and b) show an enlarged display of the region smaller than 1000.0 Å2. Some entries discussed here are marked with their PDBIDs.
To facilitate the understanding of the results, all of the entries were classified into four categories, according to the types of contacts, biological contact or crystal-packing contact, with the largest COMP and with the largest area. In each entry, if the biological contact had both the largest COMP and the largest area, then the entry was classified as category 1. When the contact with the largest COMP was the biological contact and the contact with the largest area was the crystal-packing contact, the entry was classified as category 2. Similarly, the entry with the largest COMP as the crystal-packing contact and the largest area as the biological contact was classified as category 3, and the entry with both the largest COMP and largest area as the crystal-packing contact was classified as category 4.
The results of the discrimination and evaluation are summarized in Table 4, where the numbers of the entries, the contacts judged as the most probable biological interface in the discrimination step, and whether the discrimination agreed with the actual biological state or not, are indicated in each category. As the results shown in Table 4, an 84.8% ( = 239/282) success rate for the discrimination was obtained, where the accuracy was estimated based on the assumption that the biological contact is a biological interface. In the following evaluation step, the discrimination results were reviewed along with the classification of the entries to clarify the results. The details of the evaluation results are summarized in Table 5. Here, we will describe the details of some of the striking examples.
Table 4.
Summary of the classification, the discrimination and the evaluation
| Classification
|
Discrimination
|
Evaluation
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Category | biologicala | crystal-packingb | Numberc | % | biologicald | crystal-packinge | non biof | OKg | NGh | Excludedi |
| 1 | COMP/AREA | – | 255 | 90.4 | 236j | 0 | 19k | 235j/8k | 0j/10k | 1j/1k |
| 2 | COMP | AREA | 3 | 1.1 | 3 | 0 | 0 | 1 | 2 | 0 |
| 3 | AREA | COMP | 16 | 5.7 | 0 | 15l | 1m | 0l/0m | 14l/1m | 1l/0m |
| 4 | – | COMP/AREA | 8 | 2.8 | 0 | 8 | 0 | 3 | 4 | 1 |
| Total | 282 | 100 | 239 (84.8%) | 23 (8.2%) | 20 (7.1%) | 247 (88.8%) | 31 (11.2%) | 4 | ||
Notes: Biological contacts had largest COMP (COMP) and/or largest area (AREA), or did not have both largest COMP and area (−);
Crystal-packing contacts had largest COMP (COMP) and/or largest area (AREA), or did not have both largest COMP and area (−);
Number of the entries;
Number of the entries judged that the biological contact is the most probable biological interface;
Number of the entries judged that the crystal-packing contact is the most probable biological interface;
Number of the entries judged that both the biological and crystal-packing contacts are not biological;
Number of the entries where the discrimination result agreed with the (probable) actual biological state;
Number of the entries where the discrimination result disagreed with the (probable) actual biological state;
Number of the entries which were excluded from the estimation of the success rate in the evaluation step. In category 1, the numbers of entries with “j” or “k” in the Evaluation column come from those with “j” or “k” in the Discrimination column. In category 3, the numbers of entries with “l” or “m” in the Evaluation column come from those with “l” or “m” in the Discrimination column.
Table 5.
Summary of the evaluation results
| Category (Numbera) | Discriminationb (Numbera) | Evaluation | Bio-dimerc | Resultd | Numbera |
|---|---|---|---|---|---|
| 1 | biological | No crystal-packing contact with the area > the first area criterion. | biological | OK | 177 |
| (255) | (236) | No crystal-packing contact with the COMP > the threshold. | biological | OK | 26 |
| Biological contact has a large area (>500.0 Å2). | biological | OK | 18 | ||
| Only biological contact meets only the second area criterion. | biological | OK | 7 | ||
| Biological contact is an actual biological interface based on the literature. | biological | OK | 7 | ||
| no literature (1pug) (excluded) | – | – | 1 | ||
| nonbio | Biological contact is not a largest interface in multimeric complex. | biological | NG | 7 | |
| (19) | The protein acts as a monomer. | nonbio | OK | 8 | |
| Biological unit is dimeric based on the literature. | biological | NG | 3 | ||
| Biological contact is a part of the biological dimer interface (1jy2). (excluded) | – | – | 1 | ||
| 2 | biological | Biological contact is a biological interface based on the literature (1 m 7 g). | biological | OK | 1 |
| (3) | (3) | The protein acts as a monomer. | nonbio | NG | 2 |
| 3 (16) | crystal-packing | Biological contact is a biological interface based on the literature (1jm0, etc.). | biological | NG | 10 |
| (15) | The protein acts as a monomer | nonbio | NG | 4 | |
| no literature (1o1h) (excluded) | – | – | 1 | ||
| nonbio (1) | Biological unit is dimeric based on the literature | biological | NG | 1 | |
| 4 (8) | crystal-packing (8) | Crystal-packing contact may be biologically relevant (1h6p, 1ex2, 1l6r). | crystal-packing | OK | 3 |
| Biological contact may be biologically relevant (1iu8). | biological | NG | 1 | ||
| no information about the biological assembly (1auv) (excluded) | – | – | 1 | ||
| The protein acts as a monomer. | nonbio | NG | 3 |
Notes: Number of entries;
The entries in the “biological” category were judged that the biological contact is the most probable biological interface in the discrimination step, on the other hand, those in the “crystal-packing” category were judged that the crystal-packing contact is the most probable biological interface. The entries in the “nonbio” category were judged that both biological and crystal-packing contacts are not biological;
The contact concluded as the (probable) actual biological contact in the evaluation step. The “nonbio” means that both biological and crystal-packing contacts are not biological;
OK: the discrimination result agreed with the actual biological state concluded in the evaluation. NG: the discrimination result disagreed with the actual biological state concluded in the evaluation. -: the entry was excluded from the estimation of the success rate.
Category 1 (largest COMP: biological contact, largest area: biological contact)
About 90% of all entries were classified as this category (255 entries, 90.4% = 255/282). In 236 of them (92.5% = 236/255), the contacts in the biological contact set were judged to be biological interfaces, and in the other 19 entries, the proteins were judged to be monomeric.
In the former 236 entries, because 235 ( = 177 + 26 + 18 + 7 + 7) entries contained no crystal-packing contacts that were strongly considered as being biologically relevant, the biological contacts in these entries may be biologically relevant, as listed in Table 5. For the entry, 1pug, we could not find any literatures. We therefore excluded this entry from the estimation of the success rate.
Among the latter 19 entries, seven entries contained biological multimeric oligomers, such as tetramers or octamers, where the biological contacts were not the contacts with the largest area in their multimeric complexes. The contacts without the largest area in the large multimeric complexes may be allowed to have the small COMP and area values. We think that the judgments for these entries, “the contacts in both datasets are not biological”, are reasonable, however, they disagreed with the actual biological states. One other entry (PDBID 1jy2 [Madrazo et al 2001]) contains six subunits in the ASU, which form three homo subunit pairs with two-fold symmetry. We chose one pair of them as a homo-dimer entry. However, the biological oligomer was a symmetry-related homo-dimer. Each monomer of the dimer consists of three subunits, which are three of one-halves of the symmetry-related subunit pairs, according to the primary citation. Thus, the contact in the biological contact set was a part of the biological homo-dimer interface. We therefore decided to exclude this entry from the estimation of the success rate.
In summary, the judgments for 235 entries that the biological contacts were actually biologically relevant and those for 8 entries that the proteins were monomeric, may agree with the actual biological states (96.0% = [235 + 8]/[255–2]), as shown in Table 5.
Category 2 (largest COMP: biological contact, largest area: crystal-packing contact)
Of the three entries classified as category 2 (1.1% = 3/282), only one entry, PDBID 1 m 7 g (adenosine 5′-phosphosulfate kinase with ADP and APS) (Lansdon et al 2002), contains a biological homo-dimer. In this crystal, there were the biological contact (COMP: 0.151, area: 400.4 Å2) and crystal-packing contact (COMP: 0.087, area: 620.1 Å2), and the biological contact may be biologically relevant in spite of the smaller interacting area, according to the primary citation where the authors describe that the active sites exist near the biological contact as shown in Figure 5b. We will describe the biological state of this entry in more detail in the next section.
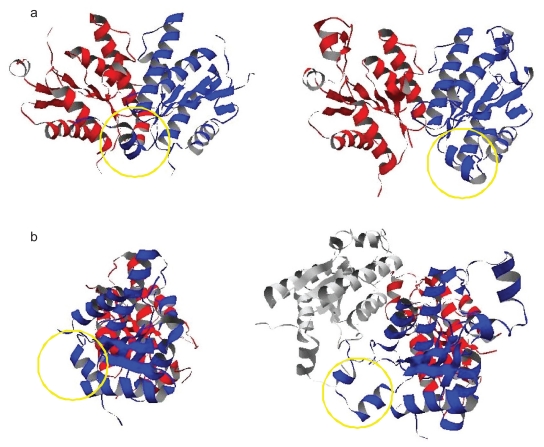
Figure 5.
The dimer structures within the ASUs in 1d6j a) and 1 m 7 g b). The regions circled by the yellow lines indicate the N-terminal regions of one subunits in the both ASU dimers. The lower figures show the rotated dimers in the upper figures by 90 degrees around the two-fold axis. In the lower dimer of 1 m 7 g b), the interaction between the ASU subunit colored in blue and the subunit colored in white which exists in the adjacent cell to the center unit cell corresponds with the crystal-packing contact mentioned in the text.
In summary, only three entries were classified as category 2, where one of them could be judged the biological state correctly by our method. Thus, there may be less number of such PDB entries that the contact in the ASU is not largest in the crystal and is likely to be a biological interface.
Category 3 (largest COMP: crystal-packing contact, largest area: biological contact)
Sixteen entries were classified as category 3 (5.7% = 16/282). In 15 of them, the crystal-packing contacts were judged to be the most probable biological interface, and in the other one entry, the protein was judged to be monomeric.
In 10 of the former 15 entries, including 1jm0 which will be discussed in the next section, the crystal-packing contacts had the small area, most of which were smaller than 200 Å2. Since the complementarity score for each property was normalized by the contact size, the COMP value for a contact with a very small area might have a tendency to be overestimated. Their primary citations show that the contacts in the biological contact set were possibly the biological dimers. Therefore, the crystal-packing contacts in these entries may not be biologically relevant. As shown in Table 5, no entry agrees with the actual biological state.
Category 4 (largest COMP: crystal-packing contact, largest area: crystal-packing contact)
In all of the 8 entries classified as category 4 (2.8% = 8/282), the crystal-packing contacts were judged to be the most probable biological interface.
One example, PDBID 1h6p (human telomeric protein TRF2) (Fairall et al 2001), contained the biological contact (COMP: 0.076, area: 465.1 Å2) and the crystal-packing contact (COMP: 0.261, area: 617.0 Å2). It is known that TRF2 binds to double-stranded telomeric DNA as a homo-dimer, and the authors of the primary citation of this entry also confirmed this experimentally. Furthermore, they mention that the crystal-packing contact which corresponds to the contact included in the crystal-packing contact set is the biological dimer interface and the contact in the ASU corresponding to the biological contact is artificial. This is because the biological dimer interface (the crystal-packing contact) consists of four helix bundles with a crossbrace, which is widely adopted in many other dimer interfaces. This observation agrees with the judgment for this entry.
The other two entries, PDBIDs 1ex2, and 1l6r, are also successful examples. The entry 1ex2 (Bacillus subtilis Maf protein) (Minasov et al 2000) contained the biological contact (COMP: 0.004, area: 233.8 Å2) and the crystal-packing contact (COMP: 0.129, area: 511.1 Å2). The entry 1l6r (phosphoglycolate phosphatase) (Kim et al 2004) had the biological contact (COMP: 0.026, area: 237.6 Å2) and the crystal-packing contact (COMP: 0.130, area: 645.7 Å2). In the primary citations of both entries, the authors describe that the proteins are dimeric under physiological conditions, and nothing about which dimeric assembly is biologically relevant in the crystals. Therefore, we confirmed the number of hydrogen bonded atom pairs for each contact by using the program HBPLUS (McDonald and Thornton 1994). As a result, for both entries, the crystal-packing contacts had larger numbers (1ex2: 19 hydrogen bonded atom pairs, 1l6r: 9 pairs) than those of the biological contacts (1ex2: 10 pairs, 1l6r: 4 pairs). These results support the validity of our discrimination.
PDBID 1iu8 (pyrrolidone-carboxylate peptidase) (Sokabe et al 2002) contained the biological contact (COMP: 0.052, area: 313.7 Å2) and the crystal-packing contact (COMP: 0.143, area: 333.1 Å2). The quaternary state of this protein is dimeric according to the primary citation. This citation also shows that there are the inter-subunit ion cluster with three salt bridges, some hydrogen bonds and the hydrophobic core in the biological contact. The loop structure which is highly conserved and important for the activity of enzyme, also participates in the formation of the dimer, stabilizing the dimer interaction. The crystal-packing contact contains two salt bridges and four hydrogen bonds, and most of the inter-subunit interactions are water mediated hydrogen bonds. The authors imply that the biological contact may be the biological dimer interface for above reason. On the other hand, our complementarity calculation indicated that the crystal-packing contact may be biological because it was more complementary than the biological contact, in spite of having the similar interfaces in size. The other two methods, PQS (Henrick and Thornton 1998) and PISA (Krissinel and Henrick 2007), predicted this entry as biological tetramers. Thus, this entry was not straightforward to predict the biological state.
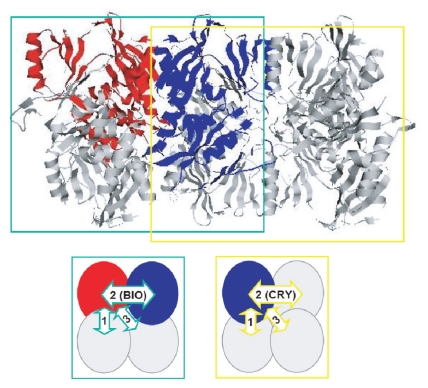
Another entry is 1auv (C domain of Synapsin IA) (Esser et al 1998). The biological state of this protein is a homo-tetramer (a dimer of dimers) which generally has three types of contacts. In this crystal, only two protomers are included in the ASU, and therefore, the other two contacts will be generated by a symmetry operation. In this study, we did not consider any contacts generated by the symmetry operator which is annotated as the “BIOMT” record in the header of the PDB, as biological contacts, because such contacts were often indistinguishable from the artificial crystal-packing contacts due to their small areas. In this entry, the contact inside the ASU was considered as the biological contact (COMP: 0.066, area: 181.6 Å2), which had the second largest area among three contacts in the dimer of dimers and was much smaller than the largest contact (COMP: 0.048, area: 1056.3 Å2). The crystal-packing contact was the contact formed between one protomer inside the ASU in the center unit cell and the symmetry-related protomer belonging to the cell close to the center unit cell, which was identical to the contact formed between two different tetramers. The area of the crystal-packing contact (COMP: 0.250, area: 214.3 Å2) was larger than that of the biological contact. As shown in Figure 6, there are two possible homo-tetrameric assemblies in this crystal. The authors mention in the citation that the left tetramer, surrounded by the green dotted line, is biologically relevant and nothing about the other possibility. The biological contact is the second largest contact in this tetramer. The right tetramer, surrounded by the yellow dotted line, is another possibility; if the right tetramer is considered as the biological assembly, then the crystal-packing contact is the biological second largest contact in the tetramer. We again checked the predicted biological state of this entry by the PQS (Henrick and Thornton 1998) and the PISA (Krissinel and Henrick 2007), however, the different results were obtained. Thus, this entry is not a good example for the discrimination test. We therefore excluded this entry from the estimation of the success rate.
Figure 6.
Two possible tetramers in the crystal of 1auv. In the upper figure, the left complex surrounded by the green line is the biological tetramer according to the primary citation of this entry, and the right one surrounded by the yellow line is another possibility. Both tetramers are tightly packed with each other in the crystal. The lower figures show the biological contacts in these two tetramers by the arrows having the same color as the line surrounding the corresponding tetramer. The green arrow with “2 (BIO)” represents the biological contact which has the second largest area in the left tetramer. The yellow arrow with “2 (CRY)” corresponds to the crystal-packing contact which is the second largest contact in the right tetramer and is also the crystal-packing contact formed between the left tetramer and the neighboring tetramer including the right half of the right tetramer on the left side. The arrows with “1” represent the contacts with the largest area in both tetramers; these two contacts can be similar.
In summary, for category 4, the discrimination results for the three entries, 1h6p, 1ex2 and 1l6r, may agree with the actual biological states. In these entries, the crystal-packing contacts may be the most probable biological interfaces.
Summary of the evaluation
We conclude that the discrimination results in 247 entries may agree with the actual biological states, and those in 31 entries may disagree, as shown in Tables 4 and 5. The success rate rose to 88.8% ( = 247/[282 – 4]) by considering the evaluation result, where the “4” came from the excluded entries. A review of the discrimination results showed that under these circumstances, there is a strong tendency that the contact in the ASU has the largest contact area, along with the largest COMP, and is considered as the biological interface in the crystal structures of dimers stored in the PDB. The discrimination performance based only on the contact size was 93.2% ( = [245 + 11 + 3]/[282 - 4]), where the “245”, “11” and “3” were the numbers of such contacts that had the largest area in the crystal and were judged as being biological, in the categories 1, 3 and 4, respectively (see the 4th and 6th columns in Table 5). It was slightly higher than the success rate based on the COMP. It may indicate that the discrimination using the interface area is an easiest and effective way.
Comparison of dimer structures determined in the different ways
According to our analysis, about 90% of the entries had the biologically relevant interfaces within the ASU, which had the largest area in the crystals. To further confirm this conclusion, we compared the putative biological dimer interfaces of the proteins determined by both X-ray crystallography and NMR (comparison 1), and those in the crystal structures having the different crystal forms (comparison 2), regarding whether the ASU contact in the biological contact set is identical with the putative biological interface in the dimer structure of the same protein which is determined in the different ways. Comparisons of the intra-molecular interactions in the monomeric structures determined by both X-ray crystallography and NMR were made previously (Billeter 1992; Wagner et al 1992; MacArthur et al 1994; Gronenborn and Clore 1995; Andrec et al 2007); however, they never focused on the inter-molecular interactions in the multimeric structures.
Comparison of the structures determined by X-ray crystallography and NMR
Only 5 cases could be found for comparison 1 as listed in Table 1. In all cases, the entries of the crystal structures were classified as Category 1. Among them, only one entry (PDBID: 1kzk) had a crystal-packing contact with the area larger than the first area criterion. However, because the area of the crystal-packing contact was much smaller (166.8 Å2) than that of the biological contact (1014.3 Å2), the biological contact may be biologically relevant. Thus, in all 5 entries the contacts in the biological contact set are considered as the most probable biological interfaces. The comparison (see the Materials and Methods) indicated that in all cases, the original dimer structures including the biological contacts were almost the same as those determined by the NMR. This suggests that the biological contacts in these crystal structures have a high possibility of being biological interfaces.
Comparison of the structures determined in the different crystallization conditions
For comparison 2, 14 cases were found. In 12 of them, the biological contacts of the original entries had the largest COMPs and areas (Category 1) and were judged to be biologically relevant as listed in Table 2. The dimer interfaces inside the ASU of the counterparts whose dimer forms were similar to those of the original dimers including the contacts in the biological contact set, also had the largest COMPs and areas in the crystals.
In the case of 1jm0 and 1jmb (Di Costanzo et al 2001), the original entry, 1jm0, was classified as Category 3. The form of the ASU dimer in the same molecule but with the different crystal group, 1jmb, is almost the same as that of the dimer having the biological contact in 1jm0. Moreover, the COMP value and area of the ASU contact of 1jmb were similar to those of the biological contact of 1jm0. The contacts in the ASU dimers of both the original and the counterpart may be the biological interfaces according to the primary citation of their crystal structures, contrary to our judgments that the crystal contacts are biologically relevant as described in the section of “Category 3”.
Another case is the pair of 1 m 7 g (Lansdon et al 2002) and 1d6j (MacRae et al 2000), containing the structures of adenosine 5′-phosphosulfate kinases, as shown in Figure 5. The original entry (1 m 7 g) was classified as Category 2 as mentioned in the above section. The entry (1 m 7 g) is the ligand-bonded (holo) form, and 1d6j is the ligand-free (apo) form. This kinase is supposed to be a homo-dimer under physiological conditions, because the active site is formed in between two protomers. The dimer structure in the ASU of the apo form is similar to that including the biological contact in the holo form, and the active sites exist near the interfaces in the ASU in the both forms. In addition, the ASU contacts of the holo form (COMP: 0.151, area: 400.4 Å2) that consists of the blue and red subunits in Figure 5b, and the apo form (COMP: 0.133, area: 870.9 Å2) that consists of those in Figure 5a, had the largest COMP values in their crystals. Our method judged that in the both forms the ASU contacts are biologically relevant.
However, although the ASU contact in the apo form had the largest area in the crystal, that in the holo dimer was not largest. This is because the N-terminal region of one subunit, which is located close to the dimer interface, is shifted away from the other subunit. This resulted in the formation of a new intra-subunit contact mediated by a sulfate ion, which was derived from the ammonium sulfate used in the sample preparation. The corresponding region in another subunit is disordered. The shift in the former subunit and the disorder in the latter resulted in the loss of the interacting area in the holo dimer. The shift of the N-terminal region also generated the additional symmetry-related crystal-packing contact with the subunit existing in the adjacent cell to the center unit cell, which consists of the blue and white subunits in Figure 5b. This additional contact is the contact in the crystal-packing contact set in the holo form which had the largest area in the crystal (COMP: 0.087, area: 620.1Å2). Thus, although the biological contact of the 1 m 7 g does not have the largest area, the contacts in the ASUs in both 1 m 7 g and 1d6j could be the biological dimer interfaces of this kinase.
In conclusion, the comparisons 1 and 2 indicate that the contacts inside the ASUs, which have the largest area except for 1 m7 g, could be the actual biological interfaces, at least in the cases of five entries for comparison 1 and 14 entries for comparison 2.
Conclusion
We developed a method for discriminating biologically relevant interfaces from artificial crystal-packing contacts, based on the complementarities of the physicochemical properties and the shapes of the protein surfaces. We obtained a success rate of approximately 89% by reviewing the discrimination results in detail. A web server that selects the most probable biological interface among all possible contacts in the crystal of the query protein has also been constructed (Tsuchiya et al 2006).
Our discrimination and subsequent evaluation found several confusing cases; the additional crystal-packing contact made the discrimination difficult as the case of 1 m 7 g. There was no clear difference particularly in size between the biological contacts and crystal-packing contacts in some entries. In the other entries, the contacts formed between the monomeric proteins had a large area and a larger COMP value than the threshold. These contacts seem to be biological homo-dimer interfaces, and as expected, they were judged as the probable biological interfaces in 9 entries. Thus, the discrimination between biological interfaces and crystal-packing contacts in crystals is a difficult task (Carugo and Argos 1997; Henrick and Thornton 1998; Ponstingl et al 2000; Elcock and McCammon 2001; Valdar and Thornton 2001; Mintseris and Weng 2003; Ponstingl et al 2003; Bahadur et al 2004; Krissinel and Henrick 2007). As shown in this study, however, the evaluation of the protein-protein interfaces from several aspects is essential to understand the biological interactions, particularly in the cases where the contact area does not contribute to the discrimination of biological interfaces from crystal contacts. Our method could discriminate the biological interfaces with the almost same performance as that by the method based on the contact area. We think that the complementarity values can be used as the scoring function to select the native-like complexes in the prediction of the proteinprotein complex structures, such as the CAPRI experiments (Janin et al 2003).
Acknowledgments
This work was partially supported by a Research Fellowship from the Japan Society for the Promotion of Science for Young Scientists to YT. KK was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 17081003). HN was supported by a Grant-in-Aid for Scientific Research on Priority Areas (No. 17017024) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was also supported by Japan Science and Technology Corporation for Strategic Japan-UK Cooperative Program to HN, KK and YT.
References
- Andrec M, Snyder DA, Zhou Z, et al. A large data set comparison of protein structures determined by crystallography and NMR: statistical test for structural differences and the effect of crystal packing. Proteins. 2007;69:449–65. doi: 10.1002/prot.21507. [DOI] [PubMed] [Google Scholar]
- Bahadur RP, Chakrabarti P, Rodier F, et al. Dissecting subunit interfaces in homodimeric proteins. Proteins. 2003;53:708–19. doi: 10.1002/prot.10461. [DOI] [PubMed] [Google Scholar]
- Bahadur RP, Chakrabarti P, Rodier F, et al. A dissection of specific and non-specific protein-protein interfaces. J Mol Biol. 2004;336:943–55. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernauer J, Bahadur RP, Rodier F, et al. DiMoVo: a Voronoi tessellation-based method for discriminating crystallographic and biological protein-protein interactions. Bioinformatics. 2008;24:652–8. doi: 10.1093/bioinformatics/btn022. [DOI] [PubMed] [Google Scholar]
- Billeter M. Comparison of protein structures determined by NMR in solution and by X-ray diffraction in single crystals. Q Rev Biophys. 1992;25:325–77. doi: 10.1017/s0033583500004261. [DOI] [PubMed] [Google Scholar]
- Carugo O, Argos P. Protein-protein crystal-packing contacts. Protein Sci. 1997;6:2261–3. doi: 10.1002/pro.5560061021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–13. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Iyer GH, Bryant SH, et al. Extent and nature of contacts between protein molecules in crystal lattices and between subunits of protein oligomers. Proteins. 1997;28:494–514. doi: 10.1002/(sici)1097-0134(199708)28:4<494::aid-prot4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Di Costanzo L, Wade H, Geremia S, et al. Toward the de novo design of a catalytically active helix bundle: a substrate-accessible carboxylatebridged dinuclear metal center. J Am Chem Soc. 2001;123:12749–57. doi: 10.1021/ja010506x. [DOI] [PubMed] [Google Scholar]
- Elcock AH, McCammon JA. Identification of protein oligomerization states by analysis of interface conservation. Proc Natl Acad Sci U S A. 2001;98:2990–4. doi: 10.1073/pnas.061411798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser L, Wang CR, Hosaka M, et al. Synapsin I is structurally similar to ATP-utilizing enzymes. Embo J. 1998;17:977–84. doi: 10.1093/emboj/17.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L, Chapman L, Moss H, et al. Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol Cell. 2001;8:351–61. doi: 10.1016/s1097-2765(01)00321-5. [DOI] [PubMed] [Google Scholar]
- Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–53. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- Gronenborn AM, Clore GM. Structures of protein complexes by multidimensional heteronuclear magnetic resonance spectroscopy. Crit Rev Biochem Mol Biol. 1995;30:351–85. doi: 10.3109/10409239509083489. [DOI] [PubMed] [Google Scholar]
- Henrick K, Thornton JM. PQS: a protein quaternary structure file server. Trends Biochem Sci. 1998;23:358–61. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- Janin J, Rodier F. Protein-protein interaction at crystal contacts. Proteins. 1995;23:580–7. doi: 10.1002/prot.340230413. [DOI] [PubMed] [Google Scholar]
- Janin J. Specific versus non-specific contacts in protein crystals. Nat Struct Biol. 1997;4:973–4. doi: 10.1038/nsb1297-973. [DOI] [PubMed] [Google Scholar]
- Janin J, Henrick K, Moult J, et al. CAPRI: a Critical Assessment of PRedicted Interactions. Proteins. 2003;52:2–9. doi: 10.1002/prot.10381. [DOI] [PubMed] [Google Scholar]
- Jefferson ER, Walsh TP, Barton GJ. Biological units and their effect upon the properties and prediction of protein-protein interactions. J Mol Biol. 2006;364:1118–29. doi: 10.1016/j.jmb.2006.09.042. [DOI] [PubMed] [Google Scholar]
- Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Yakunin AF, Kuznetsova E, et al. Structure- and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum. J Biol Chem. 2004;279:517–26. doi: 10.1074/jbc.M306054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K, Nakamura H. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 2004;20:1329–30. doi: 10.1093/bioinformatics/bth073. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lansdon EB, Segel IH, Fisher AJ. Ligand-induced structural changes in adenosine 5′-phosphosulfate kinase from Penicillium chrysogenum. Biochemistry. 2002;41:13672–80. doi: 10.1021/bi026556b. [DOI] [PubMed] [Google Scholar]
- Levy ED, Pereira-Leal JB, Chothia C, et al. 3D complex: a structural classification of protein complexes. PLoS Comput Biol. 2006;2:e155. doi: 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy ED, Boeri Erba E, Robinson CV, et al. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–5. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhao D, Djebli A, et al. Crystal structure of colicin E3 immunity protein: an inhibitor of a ribosome-inactivating RNase. Structure. 1999;7:1365–72. doi: 10.1016/s0969-2126(00)80026-x. [DOI] [PubMed] [Google Scholar]
- Liu S, Li Q, Lai L. A combinatorial score to distinguish biological and nonbiological protein-protein interfaces. Proteins. 2006;64:68–78. doi: 10.1002/prot.20954. [DOI] [PubMed] [Google Scholar]
- MacArthur MW, Laskowski RA, Thornton JM. Knowledge-based validation of protein structure coordinates derived by X-ray crystallography and NMR spectroscopy. Curr Opinion Struct Biol. 1994;4:731–7. [Google Scholar]
- MacRae IJ, Segel IH, Fisher AJ. Crystal structure of adenosine 5′-phosphosulfate kinase from Penicillium chrysogenum. Biochemistry. 2000;39:1613–21. doi: 10.1021/bi9924157. [DOI] [PubMed] [Google Scholar]
- Madrazo J, Brown JH, Litvinovich S, et al. Crystal structure of the central region of bovine fibrinogen (E5 fragment) at 1.4-A resolution. Proc Natl Acad Sci U S A. 2001;98:11967–72. doi: 10.1073/pnas.211439798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975;405:442–51. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–93. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Minasov G, Teplova M, Stewart GC, et al. Functional implications from crystal structures of the conserved Bacillus subtilis protein Maf with and without dUTP. Proc Natl Acad Sci U S A. 2000;97:6328–33. doi: 10.1073/pnas.97.12.6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintseris J, Weng Z. Atomic contact vectors in protein-protein recognition. Proteins. 2003;53:629–39. doi: 10.1002/prot.10432. [DOI] [PubMed] [Google Scholar]
- Murzin AG, Brenner SE, Hubbard T, et al. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–40. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Nishida S. Numerical calculations of electrostatic potentials of protein-solvent systems by the self consistent boundary method. J Phys Soc Jpn. 1987;56:1609–22. [Google Scholar]
- Ofran Y, Rost B. Analysing six types of protein-protein interfaces. J Mol Biol. 2003;325:377–87. doi: 10.1016/s0022-2836(02)01223-8. [DOI] [PubMed] [Google Scholar]
- Ooi T, Oobatake M, Nemethy G, et al. Accessible surface areas as a measure of the thermodynamic parameters of hydration of peptides. Proc Natl Acad Sci U S A. 1987;84:3086–90. doi: 10.1073/pnas.84.10.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85:2444–8. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponstingl H, Henrick K, Thornton JM. Discriminating between homodimeric and monomeric proteins in the crystalline state. Proteins. 2000;41:47–57. doi: 10.1002/1097-0134(20001001)41:1<47::aid-prot80>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- Ponstingl H, Kabir T, Thornton JM. Automatic inference of protein quaternary structure from crystals. J Appl Cryst. 2003;36:1116–22. [Google Scholar]
- Robert CH, Janin J. A soft, mean-field potential derived from crystal contacts for predicting protein-protein interactions. J Mol Biol. 1998;283:1037–47. doi: 10.1006/jmbi.1998.2152. [DOI] [PubMed] [Google Scholar]
- Sokabe M, Kawamura T, Sakai N, et al. The X-ray crystal structure of pyrrolidone-carboxylate peptidase from hyperthermophilic archaea Pyrococcus horikoshii. J Struct Funct Genomics. 2002;2:145–54. doi: 10.1023/a:1021257701676. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Kinoshita K, Nakamura H. Structure-based prediction of DNA-binding sites on proteins using the empirical preference of electrostatic potential and the shape of molecular surfaces. Proteins. 2004;55:885–94. doi: 10.1002/prot.20111. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Kinoshita K, Ito N, et al. PreBI: prediction of biological interfaces of proteins in crystals. Nucleic Acids Res. 2006;34:W320–4. doi: 10.1093/nar/gkl267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Kinoshita K, Nakamura H. Analyses of homo-oligomer interfaces of proteins from the complementarity of molecular surface, electrostatic potential and hydrophobicity. Protein Eng Des Sel. 2006;19:421–9. doi: 10.1093/protein/gzl026. [DOI] [PubMed] [Google Scholar]
- Valdar WS, Thornton JM. Conservation helps to identify biologically relevant crystal contacts. J Mol Biol. 2001;313:399–416. doi: 10.1006/jmbi.2001.5034. [DOI] [PubMed] [Google Scholar]
- Wagner G, Hyberts SG, Havel TF. NMR structure determination in solution: a critique and comparison with X-ray crystallography. Annu Rev Biophys Biomol Struct. 1992;21:167–98. doi: 10.1146/annurev.bb.21.060192.001123. [DOI] [PubMed] [Google Scholar]
- Xu Q, Canutescu A, Obradovic Z, et al. ProtBuD: a database of biological unit structures of protein families and superfamilies. Bioinformatics. 2006;22:2876–82. doi: 10.1093/bioinformatics/btl490. [DOI] [PubMed] [Google Scholar]