Abstract
The Spt4, Spt5, and Spt6 proteins are conserved throughout eukaryotes and are believed to play critical and related roles in transcription. They have a positive role in transcription elongation in Saccharomyces cerevisiae and in the activation of transcription by the HIV Tat protein in human cells. In contrast, a complex of Spt4 and Spt5 is required in vitro for the inhibition of RNA polymerase II (Pol II) elongation by the drug DRB, suggesting also a negative role in vivo. To learn more about the function of the Spt4/Spt5 complex and Spt6 in vivo, we have identified Drosophila homologs of Spt5 and Spt6 and characterized their localization on Drosophila polytene chromosomes. We find that Spt5 and Spt6 localize extensively with the phosphorylated, actively elongating form of Pol II, to transcriptionally active sites during salivary gland development and upon heat shock. Furthermore, Spt5 and Spt6 do not colocalize widely with the unphosphorylated, nonelongating form of Pol II. These results strongly suggest that Spt5 and Spt6 play closely related roles associated with active transcription in vivo.
Keywords: Spt, P-TEFb, CTD, elongation, RNA polymerase, polytene
Several classes of transcription elongation factors have been identified in both prokaryotes and eukaryotes (for review, see Reines et al. 1996; Greenblatt et al. 1998; Roberts et al. 1998; Shilatifard 1998a,b; Parada and Roeder 1999; Reines et al. 1999). The Spt4, Spt5, and Spt6 proteins are conserved factors, originally grouped together because of shared mutant phenotypes (for review, see Hartzog and Winston 1997; Winston and Sudarsanam 1998). Recently, a role in transcription elongation has been demonstrated for these proteins by both yeast and mammalian studies (Hartzog et al. 1998; Wada et al. 1998a,b; Yamaguchi et al. 1998,1999a; Kim et al. 1999; Parada and Roeder 1999; Price 2000). These data suggest that Spt4 and Spt5, functioning as a complex (Spt4/Spt5) work in conjunction with both the positive transcription elongation factor b (P-TEFb) and RNA polymerase II (Pol II) to control transcription elongation. The requirements for, and the functions of Spt4/Spt5 appear to be modulated by phosphorylation of the carboxy-terminal domain (CTD) of the largest subunit of Pol II (Wada et al. 1998b; Yamaguchi et al. 1999b; Price 2000). A possible role for Spt6 in these interactions has not yet been studied.
P-TEFb is a kinase complex that is critical for elongation in many systems (for review, see Price 2000). P-TEFb was identified from Drosophila melanogaster as a factor required for the production of long transcripts by Pol II in vitro (Marshall and Price 1995) and is required for the bypass of an early block to elongation (Kephart et al. 1992; Marshall and Price 1992). P-TEFb consists of two subunits, Cdk9, a cyclin-dependent protein kinase, and cyclin T (for review, see Price 2000). Studies of the general transcription elongation inhibitor, 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB) have shown that it inhibits the P-TEFb protein kinase activity, thereby blocking a step in transcription elongation where P-TEFb is required (Wada et al. 1998b; for review, see Yamaguchi et al. 1998). A substrate for P-TEFb activity is the Pol II CTD (Herrmann and Rice 1995), which participates in an elongation-dependent phosphorylation cycle.
The CTD is a key regulatory domain for the control of transcription and of cotranscriptional mRNA processing. It contains multiple repeats (26 in yeast, 42 in Drosophila, and 52 in mammals) of a heptapeptide sequence (consensus YSPTSPS) and undergoes a cycle of phosphorylation and dephosphorylation that correlates with the activity of Pol II during the process of transcription (for review, see Dahmus 1996). In this cycle, Pol II with a hypophosphorylated CTD (Pol IIa) initiates transcription. The CTD then becomes heavily phosphorylated (Pol IIo) as Pol II enters elongation. The Pol IIo form, although competent for elongation, does not initiate transcription efficiently. Therefore, following elongation, the CTD is dephosphorylated, completing the transcription cycle (Dahmus 1996; Archambault et al. 1998; Cho et al. 1999). Throughout, the state of CTD phosphorylation modulates the recruitment or activity of initiation and elongation factors (Myers et al. 1998; Wada et al. 1998a,b; Cho et al. 1999; Otero et al. 1999; Yamaguchi et al. 1999a) as well as mRNA processing factors (for review, see Bentley 1999). Phosphorylation of the CTD occurs primarily at serines at positions 2 and 5 within the heptad repeat and evidence exists that phosphorylation at each site may play distinct roles (West and Corden 1995; Yuryev and Corden 1996; Patturajan et al. 1998).
Phosphorylation of the Pol II CTD by P-TEFb also relieves the inhibition of Pol II elongation by Spt4/Spt5 in vitro. Spt4/Spt5 from HeLa cells (also called DRB sensitivity inducing factor [DSIF]) and another HeLa negative factor called NELF were identified in cell extracts as being required to confer sensitivity to DRB in vitro (Wada et al. 1998a; Yamaguchi et al. 1999a). The requirement of these factors for DRB sensitivity in vitro suggested that they may be the targets, direct or indirect, of P-TEFb (Wada et al. 1998b; Ivanov et al. 2000). Indeed, P-TEFb antagonizes Spt4/Spt5 and NELF inhibition of Pol II (Wada et al. 1998b) in vitro. P-TEFb may overcome the inhibition conferred by Spt4/Spt5 indirectly by phosphorylation of the Pol II CTD, which renders Pol II insensitive to the negative activity of Spt4/Spt5 (Wada et al. 1998b), or directly by phosphorylation of Spt5 (Ivanov et al. 2000).
Although able to block Pol II elongation in response to DRB, there is biochemical and genetic evidence that Spt4/Spt5 is also a positively acting transcription factor. In Saccharomyces cerevisiae, spt5 mutations cause decreased levels of particular mRNAs (Compagnone-Post and Osley 1996; Hartzog et al. 1998). Under certain in vitro conditions, human Spt4/Spt5 can stimulate the elongation rate of Pol II (Wada et al. 1998a). In addition, Spt4/Spt5, as well as human Spt6 have been implicated in aiding Tat activation of HIV transcription (Wu-Baer et al. 1998; Kim et al. 1999; Parada and Roeder 1999; Ivanov et al. 2000). Finally, much work has shown that P-TEFb is also crucial for Tat activation (for review, see Jones 1997; Garber and Jones 1999). Therefore the in vivo relationship between P-TEFb, Spt4/Spt5, and Spt6 in the regulation of transcription is complex, as Spt4/Spt5 appears both to cooperate with and be antagonized by P-TEFb activity, whereas the role of Spt6 is less well elucidated in these contexts.
Several fundamental aspects of Spt4/Spt5 and Spt6 function are not known. For example, although Spt5 and Spt6 are essential for growth in S. cerevisiae, little is known about the extent of their roles in transcription in vivo. In addition, genetic analyses suggest that Spt5 and Spt6 are closely related functionally, but the two proteins are not stably physically associated (Hartzog et al. 1998); therefore, it is unknown if they actually work together. Finally it is not known whether Spt5 and Spt6 are physically associated with transcription complexes in the context of chromatin. To investigate these issues regarding the Spt4/Spt5 complex and Spt6, we have identified the Spt5 and Spt6 proteins of D. melanogaster and have examined the relationship between Spt5, Spt6, and Pol II transcription on Drosophila polytene chromosomes. Our results show that Spt5 and Spt6 colocalize on polytene chromosomes at a large number of sites. Furthermore, their localization is highly coincident with the localization of elongating, phosphorylated Pol II, suggesting that Spt5 and Spt6 are present at most or all regions of active transcription. Consistent with these findings, Spt5, Spt6, and the P-TEFb subunit cyclin T are recruited to heat shock genes induced by heat shock. We also show that a subset of polytene loci, some of which include the highly transcribed Salivary gland secreted (Sgs) genes, are enriched for unphosphorylated Pol II, cyclin T, Spt5, and Spt6. Finally, we demonstrate that one of these genes, Sgs-4, is a direct target of Spt5, Spt6, and cyclin T association. The relationship between Spt5 and Spt6 with elongating, phosphorylated Pol II and the elongation factor P-TEFb on polytene chromosomes strongly suggests a general role in active transcription for these proteins, most likely at the level of elongation. Results in the accompanying manuscript (Andrulis et al. 2000) also provide strong evidence for an important and general role for Spt5 and Spt6 in transcription elongation.
Results
Identification and sequence analysis of Drosophila homologs of Spt5 and Spt6
To characterize Drosophila Spt5 and Spt6, we obtained Drosophila cDNAs encoding proteins highly homologous to the human and murine Spt5 and Spt6 proteins. For Spt5 we obtained a full-length cDNA and for Spt6 we deduced the full-length cDNA sequence from overlapping partial cDNAs. Drosophila Spt4 (polytene map position 49B) and Spt6 (polytene map position 5E) have also been identified by Chiang et al. (1999) and Spt5 has been identified by the Berkeley Drosophila Genome Project as CG7626 (polytene map position 56D5-6). The predicted Drosophila proteins are conserved throughout with their corresponding homologs (Fig. 1). Among previously noted motifs, the homology between Spt5 proteins from other organisms and the bacterial elongation factor NusG (Hartzog et al. 1998; Wada et al. 1998a; Wu-Baer et al. 1998) is conserved in Drosophila Spt5. For Spt6, all other identified homologs are predicted to have a nonsequence specific DNA binding domain (HhH domain) (Doherty et al. 1996) and all but S. cerevisiae Spt6 have been noted to have an RNA binding domain (S1 domain) (Bycroft et al. 1997); these motifs are conserved in Drosophila Spt6.
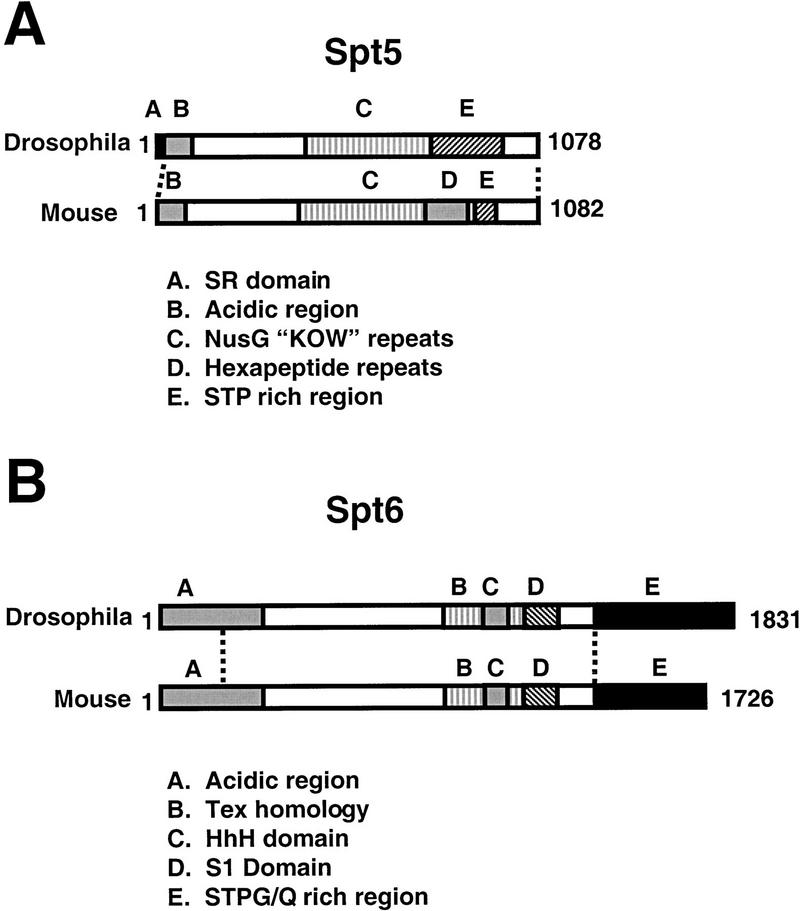
Figure 1.
Conservation and domain structure of the Drosophila melanogaster Spt5 and Spt6 proteins. Overall regions of homology indicated by dashed lines. (A) Drosophila Spt5 domain structure illustrated with murine Spt5 as a comparison. Spt5 proteins have acidic amino termini (region B), sequence homology to Escherichia coli NusG (region C) (Hartzog et al. 1998; Wada et al. 1998a; Wu-Baer et al. 1998), and serine-, threonine-, and proline-rich carboxy-terminal repeat regions noted in Yamaguchi et al. (1999b) and defined as CTR1 and CTR2 in Stachora et al. (1997) (regions D and E). Region E of Drosophila Spt5 has characteristics of CTR1 and CTR2 but the repeats appear degenerate. Homology of D. melanogaster Spt5 determined by BLAST (Altschul et al. 1990) with murine Spt5 is 50% amino-acid identity (E value = 0.0) and with Saccharomyces cerevisiae is 26% amino-acid identity (E value = 1e-58). The amino-terminal RS domain of D. melanogaster (region A) is novel for the Spt5 proteins. (B) D. melanogaster Spt6 domain structure illustrated with murine Spt6 as a comparison. Like Spt5 proteins, Spt6 proteins have acidic amino-termini (region A). Spt6 proteins also have sequence homology with a prokaryotic family of proteins implicated in transcription regulation (region B, named after the Bordetella pertussis Tex protein (Fuchs et al. 1996) and this region contains a conserved helix–hairpin–helix fold (HhH, region C, [Doherty et al. 1996]). Most Spt6 proteins are also predicted to contain RNA-binding S1 domains (region D, [Bycroft et al. 1997]). All Spt6 proteins except S. cerevisiae Spt6 have extended, divergent carboxyl termini rich in certain amino acids such as serine, threonine, and glycine (Drosophila) or glutamine (mouse) (region E and data not shown). Homology (by BLAST) of D. melanogaster Spt6 with murine Spt6 is 48% amino-acid identity (E value = 0.0) and S. cerevisiae Spt6 is 22% amino-acid identity (E value = 3e-79).
In addition to the conserved regions, Drosophila Spt5 and Spt6 have regions not found in their homologs. Drosophila Spt5 is the only known Spt5 homolog that contains a stretch of serine-arginine (RS) repeats at its amino terminus. RS domains are found in several essential accessory mRNA splicing factors and are believed to mediate protein–protein interactions, including phosphorylation-dependent interactions with the Pol II CTD (Yuryev et al. 1996; Kim et al. 1997). This domain may implicate a role for Drosophila Spt5 in splicing. This link of Spt5 to mRNA processing is not the first, as human Spt5 has been shown to modulate the activity of and interact with mRNA capping machinery (Wen and Shatkin 1999). Drosophila Spt6 has a divergent, extended carboxyl terminus that is serine-, threonine-, proline-, and glycine-rich; however, this divergence is also found in Spt6 homologs from Arabidopsis, mouse, and human. S. cerevisiae Spt6 lacks this carboxy-terminal domain (Swanson et al. 1990).
Expression and localization of Spt5 and Spt6 in Drosophila embryos
To characterize expression of Spt5 and Spt6 in embryos, we analyzed their mRNAs and protein products. First, to identify the Spt5 and Spt6 transcripts, we performed Northern analysis of Drosophila embryonic RNA (Fig. 2A). Single RNAs were detected for each gene. Their sizes, ∼3.6 kb for Spt5 and 6.0 kb for Spt6, roughly match those predicted from the cDNA sequences (3436 bp and 6087 bp, respectively), suggesting that the cDNAs are full length. We raised antisera against two nonoverlapping regions in each protein (see Materials and Methods) and used these reagents in Western analysis of Drosophila embryonic extracts. Each of the two antisera raised against Spt5 detect a high molecular weight protein of similar mobility (∼175 kD)(Fig. 2B, cf. lanes 1 and 2) and the two antisera raised against Spt6 detect a high molecular weight band of >200 kD (Fig. 2B, cf. lanes 3 and 4). These results strongly suggest that the two antisera for each protein detect the same protein. The apparent molecular weights for both Spt5 and Spt6 are greater than those predicted from the cDNA sequence. However, as both proteins are highly acidic and therefore expected to migrate abnormally during gel electrophoresis (Takano et al. 1988), this discrepancy is not surprising. Similar differences between the actual and apparent molecular weights have been observed with yeast and human Spt5 proteins (Swanson et al. 1991; Hartzog et al. 1998; Wada et al. 1998a; Wu-Baer et al. 1998). The lower molecular weight bands are likely to be degradation products of the large proteins, although the mobility of some bands may be due to covalent modification. To determine the localization of Spt5 and Spt6 in embryos, the proteins were detected by indirect immunofluorescence (Fig. 2C). These results demonstrate that both Spt5 and Spt6 are nuclear proteins with ubiquitous distribution in early embryos. Later stage embryos show the same ubiquitous distribution (data not shown).
Figure 2.
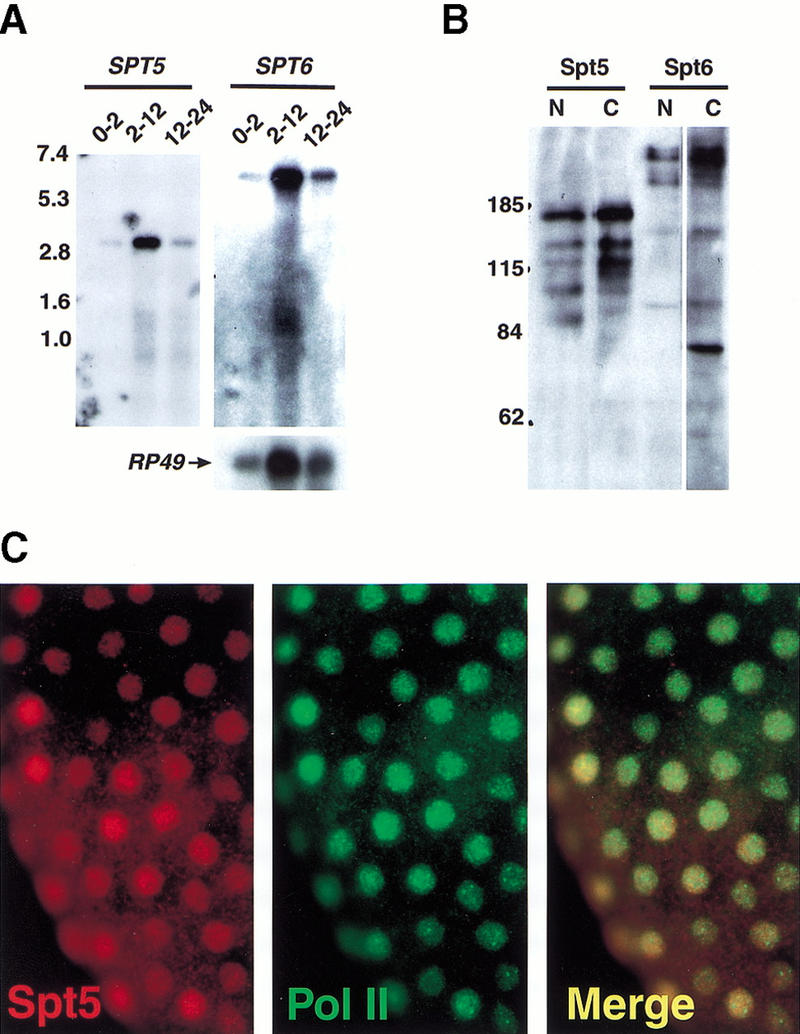
Detection of Spt5 and Spt6 mRNA and protein in Drosophila embryos. (A) Northern analysis of Spt5 and Spt6 transcripts in Drosophila embryos using Poly(A)+ RNA isolated from Drosophila embryos at specific times (indicated in hours) after egg deposition (1.5 μg RNA/lane). The lower panel shows the control probe RP49. Numbers to the left indicate sizes (kb) of molecular weight markers. (B) Western analysis of Spt5 and Spt6. Each lane contains 200 μg of Drosophila 2–12 h embryonic extract. Lanes were then separated and probed with antibodies directed against Spt5, Spt6, or Cyclin T. Antibodies used: lane 1, HMGP11 (directed against amino terminus Spt5); lane 2, HMGP14 (carboxyl terminus Spt5); lane 3, HMGP15 (directed against amino terminus Spt6); lane 4, HMGP17 (carboxyl terminus Spt6). Numbers to the left indicate sizes (kD) of molecular weight markers. (C) Whole mount immunofluorescence detection of Spt5 in Drosophila embryos. A precellularization embryo probed with HMGP11 (left panel); or with an antibody directed against the Pol II CTD as a nuclear marker (middle panel); (right panel) overlay of left and middle panels showing colocalization of red and green signals (appears yellow). All Spt5 and Spt6 antibodies give the same ubiquitous, nuclear staining pattern in both early- and later-staged embryos (data not shown).
Spt5 and Spt6 colocalize with Pol IIo on Drosophila polytene chromosomes
S. cerevisiae Spt5 and Spt6 are essential factors that have been implicated in the control of transcription. Specifically, Spt6 interacts with histones and Spt5 interacts with Pol II (Bortvin and Winston 1996; Hartzog et al. 1998; Wada et al. 1998b) suggesting that there may be a broad requirement for Spt5 and Spt6 in transcription and that they are associated with chromatin, either as general chromatin components or as more direct participants in the transcriptional process. To address these issues and to characterize their possible chromatin association, we examined the localization of both Drosophila Spt5 and Spt6 on salivary gland polytene chromosomes. In control experiments, antisera directed against the two nonoverlapping regions of Spt5 gave the same staining pattern (data not shown); therefore, only antiserum HMGP11 (directed against an amino-terminal region of Spt5 lacking the RS domain, to reduce cross-reactivity to other proteins) was used in subsequent experiments. Similarly, the two antisera raised against nonoverlapping regions of Spt6 gave the same staining patterns and antiserum HMGP15 (directed against an amino-terminal region of Spt6) was used in subsequent experiments. Control experiments show that the signals are dependent on and specific to the primary antibody used, and that there is insignificant or no cross-reactivity of secondary antibodies (data not shown). Our results (Fig. 3) show that Spt5 and Spt6 bind to a large number of sites throughout euchromatin (Fig. 3,A,D). This binding occurs primarily in interband regions of less condensed DNA (data not shown). Furthermore, the two proteins colocalize during larval development as described below.
Figure 3.
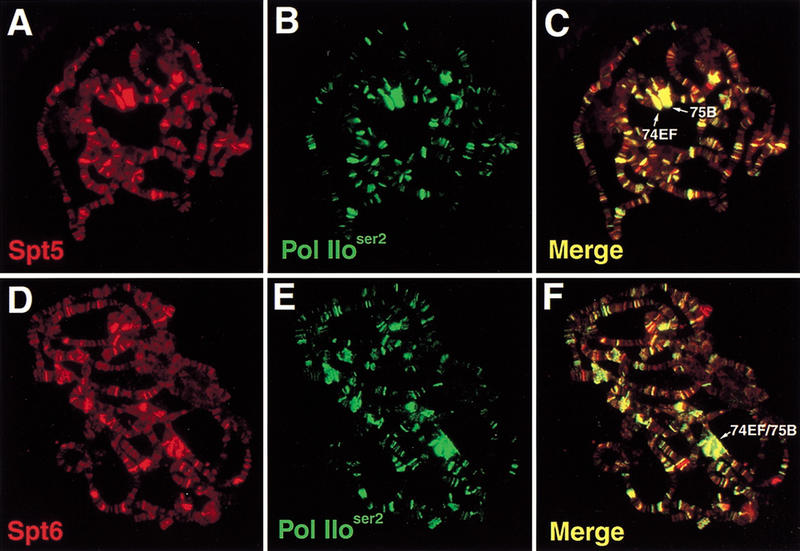
Colocalization of Spt5 and Spt6 with Pol IIoser2 on Drosophila polytene chromosomes: (A) Immunofluorescence detection of Spt5; (B) Pol IIoser2; (C) overlay of A and B; (D) Spt6; (E) Pol IIoser2; (F) Overlay of D and E. Colocalization of red and green signals in C and F appear as yellow.
The broad distribution of Spt5 and Spt6 on polytene chromosomes in interband regions suggested that these two proteins are likely associated with some form(s) of Pol II, which also localize to these areas (Weeks et al. 1993). To address this possibility, we first examined the staining relationship between Spt5, Spt6, and Pol IIo. Serines at positions two and five of the CTD repeat are most commonly phosphorylated, and antisera specific for phosphorylation at either position have been raised (Warren et al. 1992; Bregman et al. 1994). It should be noted that due to heterogeneity of CTD phosphorylation and the repeat sequence, a single Pol II CTD may be recognized by several antibodies specific for different phosphorylation states.
We have performed staining experiments with antibody H5, which recognizes Pol IIo phosphorylated on serine-2 (Pol IIoser2), and H14, which recognizes Pol IIo phosphorylated on serine-5 (Pol IIoser5) (Kim et al. 1997). In our polytene staining experiments, H14 did not produce robust signals, therefore H5 was used primarily. Spt5, Spt6, and Pol IIoser2 are each present at numerous bands (Fig. 3). When staining for Pol IIoser2 is merged with either staining for Spt5 (Fig. 3C) or staining for Spt6 (Fig. 3F), a high degree of colocalization is observed for Spt5, Spt6, and Pol IIoser2. The appearance of yellow in merged images indicates that the red and green signals are roughly equivalent, suggesting a stoichiometric relationship for these factors at their common sites.
We also examined the chromosomal localization of Spt5, Spt6, and Pol IIoser2 with regard to the polytene puffs of the third larval instar stage of development. These polytene puffs are cytologically visible structures that appear and recede in a temporal pattern that reflects increased and decreased transcription, respectively, of loci within the puffs (for review, see Thummel 1990). Previous studies demonstrated that Pol IIo localizes to puffs (Weeks et al. 1993). Puffs at polytene positions 74EF and 75B illustrate this point (Fig. 3). Genes within these puffs are induced to a high level of transcription by a stage-specific increase in the hormone ecdysone (Thummel 1990; Huet et al. 1993). Spt5 and Spt6, as well as Pol IIoser2, localize to these puffs, which are members of a class known as early puffs (for review, see Ashburner et al. 1974). When chromosomes from different developmental stages were examined, Spt5, Spt6, and Pol IIoser2 colocalize at most of the developmentally regulated puffs examined (data not shown).
Spt5 and Spt6 do not extensively colocalize with Pol IIa
We also determined the extent of colocalization of Spt5 and Spt6 with Pol IIa. Pol IIa is the form of Pol II with a hypophosphorylated CTD, and recent studies have demonstrated that mammalian Spt5 interacts with Pol IIa (Yamaguchi et al. 1999b). Previous studies have shown also that Pol IIa, as determined by an antibody that primarily recognizes unphosphorylated polymerase, is found on polytene chromosomes in a widespread pattern that partially overlaps with that of Pol IIo (Weeks et al. 1993). We assessed the relationship of staining for Spt5 (Fig. 4A, data not shown) and Spt6 (Fig. 4B–D) with Pol IIa in different developmental stages. Our results show that Spt5 and Spt6 staining partially overlaps with, but does not strongly correlate with, Pol IIa staining. In all cases examined (Puff stages [PS] 1–2, 3–4, and 7–8 [Ashburner 1967]), there is only partial overlap between the staining patterns of Spt6 and Pol IIa or the staining patterns of Spt5 and Pol IIa. Moreover, the two patterns of overlap are similar. Note that in PS7–8, highly transcribed puffs that stain strongly for Spt5, Spt6 and Pol IIoser2 do not stain for Pol IIa (cf. bands 74EF and 75B in Fig. 3C,F with Fig. 4D). Furthermore, at most sites where Spt5 and Spt6 do overlap with Pol IIa, the relative levels of Spt5 or Spt6 staining do not correlate well with the level of Pol IIa staining (bands do not appear yellow, data not shown).
Figure 4.
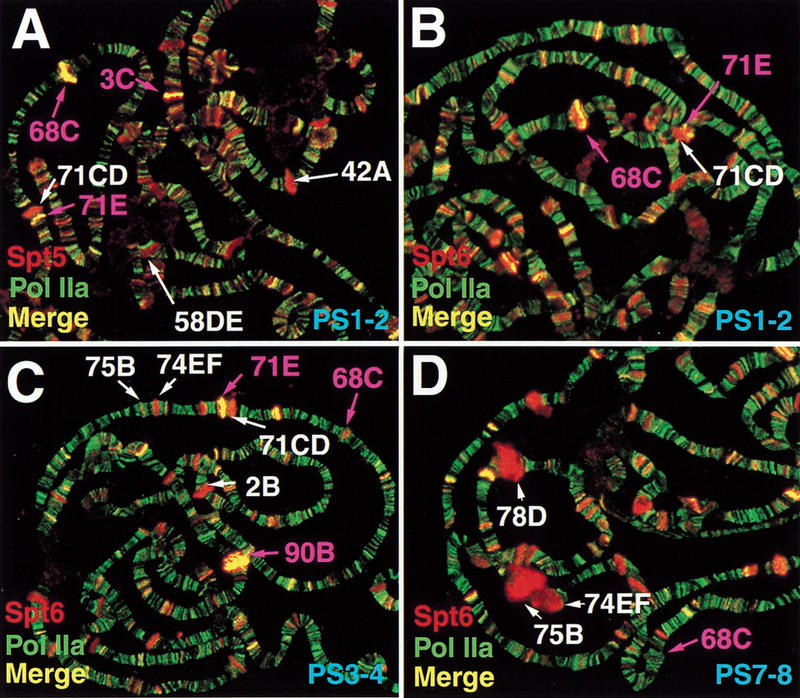
Partial colocalization of Spt5 and Spt6 with Pol IIa. Immunofluorescence detection of Spt5 (red) and Pol IIa (green) (A) or Spt6 (red) and Pol IIa (green) (B) in PS1–2 third instar salivary glands. Detection of Spt6 (red) and Pol IIa (green) in PS3–4 (C) and PS7–8 (D) third instar salivary glands. Loci labeled in magenta correspond to Sgs gene containing intermolt puffs; loci in white correspond to intermolt puffs that do not contain Sgs genes (A) or early or late puffs (B–D). An intermolt puff is located at 71CD (Ashburner 1972). Sgs-6 has been mapped by recombination to 71C-F (Velissariou and Ashburner 1981). Because 71E stains in a similar fashion to other Sgs loci and contains a gene homologous to other Sgs genes (data not shown) we tentatively assign Sgs-6 to 71E and distinguish it from 71CD.
There are, however, a group of bands where a high level of staining for Spt5, Spt6, and Pol IIa does occur. These bands coincide with a subset of intermolt puffs (Ashburner 1972). Intermolt puffs appear in the mid-third instar larval stage and are repressed by high ecdysone levels later in development, prior to the induction of early puffs such as those noted in Figure 3 and Figure 4D. A number of intermolt puffs stain strongly for Spt5, Spt6, and Pol IIa (bands 3C [Fig. 4A], 68C [Fig. 4A,B] and 71E [Fig. 4A–C]; 90B [Fig. 4C]; 25A [data not shown]). This subset of intermolt puffs contains Sgs genes (noted in purple in Fig. 4A–D). Sgs genes are highly transcribed in mid- to late-third instar larvae and encode the glue proteins that adhere pupae to solid substrates (for review, see Lehmann 1996). As shown in Figure 4B–D, Spt6 and Pol IIa localization at Sgs genes decreases (cf. 68C in Fig. 4A,B to 68C in Fig. 4C,D) while Spt6 localization increases at early puffs as development proceeds (e.g., 74EF and 75B in Fig. 4C,D; also noted in Fig. 3C,F). Intermolt puffs at positions 42A and 58DE that do not contain Sgs genes are shown in Figure 4A, and have staining characteristics of early puffs. Further examination of Sgs loci is presented below.
Heat shock induces localization of Spt5, Spt6, and cyclin T to activated heat shock genes
Our data suggest a causal relationship between active transcription and the location of Spt5 and Spt6 on polytene chromosomes. We tested this relationship by asking whether heat-shock mediated alteration of the transcription patterns in polytene chromosomes alters Spt5 and Spt6 localization. Exposure of larvae to high temperatures induces the transcription of heat shock genes while reducing the transcription of many other loci (Lis et al. 1981). As seen previously, we observe that prior to heat shock, Pol IIa but not Pol IIo staining can be detected at uninduced heat shock loci such as those shown at 87A and 87C (Weeks et al. 1993; shown by alternate methods in O'Brien et al. 1994) (Fig. 5A,C,D,F). Whereas a small amount of Spt5 appears to be present at 87A and 87C at normal temperatures, coincident with Pol IIa (Fig. 5B,C), we are unable to detect Spt6 or Pol IIoser2 at these positions (Fig. 5D–F). In contrast, a five-minute heat shock induces Spt5, Spt6, Pol IIo, and the P-TEFb subunit, cyclin T, to localize to the heat shock puffs (Fig. 5G–I). Intense signals for Spt5 (Fig. 5G,H, data not shown), Spt6, and Pol IIoser2 can be seen at heat shock loci located at 63BC, 64F, 87A, 87C, 93D, and 95D (Fig. 5I). The strong recruitment of cyclin T to these loci on heat shock has also been described elsewhere (Lis et al. 2000). These results demonstrate that heat shock causes the recruitment of Spt5 and Spt6, along with Pol IIo and the positive elongation factor cyclin T to heat shock loci.
Figure 5.
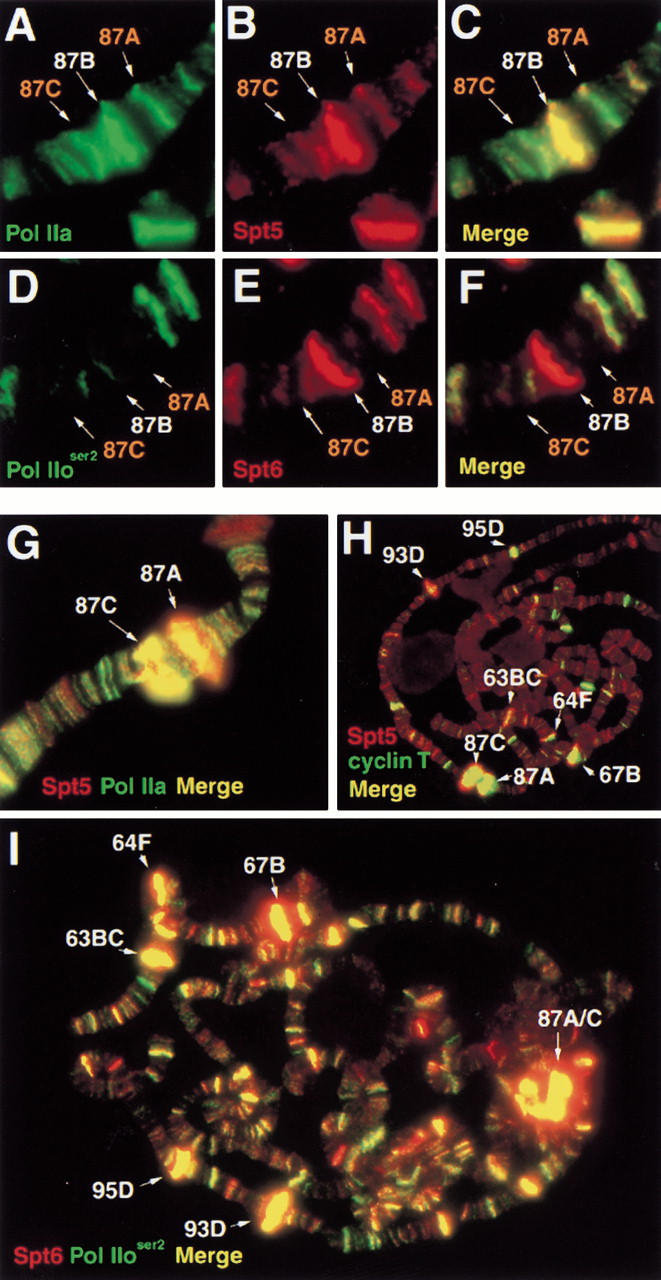
Heat-shock induced localization of Spt5, Spt6, and cyclin T to heat-shock loci. Immunofluorescence detection of Spt proteins at uninduced hsp70 loci: (A) Pol IIa; (B) Spt5; (C) overlay of A and B; (D) Pol IIoser2; (E) Spt6, (F) overlay of D and E. Polytene bands containing hsp70 loci labeled in orange. Panels G–I show recruitment of Spt proteins to heat-shock loci upon 5 min heat-shock: (G) Spt5 (red) and Pol IIa (green); (H) Spt5 (red) and cyclin T (green); (I) Spt6 (red) and Pol IIoser2 (green).
Bands that contain Sgs genes have a distinct pattern of Pol IIo, Pol IIa, Spt5, Spt6, and cyclin T staining
Because bands containing Sgs genes stain prominently for Spt5, Spt6, and Pol IIa, we considered that these locations could be sites of negative activity of Spt5 and Spt6 due to their apparent localization with unphosphorylated polymerase. However, Sgs genes are highly expressed at the times observed (for review, see Lehmann 1996). Therefore, we determined the Pol IIoser2 and Pol IIoser5 distribution on polytenes and compared it with that of Spt5 and Spt6 at these stages. Our results show that most of these loci appear to contain less Pol IIoser2 relative to the majority of other loci where Spt5/Spt6/Pol IIoser2 colocalize (Fig. 6A–D). In contrast to the apparent low level of Pol IIoser2 at most of these Sgs containing bands, Pol IIoser5 is enriched (Fig. 6E–G). The difference in the intensity of staining between Pol IIoser2 and Pol IIoser5 relative to Spt6 at bands 23DEF and 25AB (cf. Fig. 6D to 6G) indicates that phosphorylation at serine-2 and serine-5 does not correlate at all sites but that a form of Pol IIo is, indeed, present at these loci.
Figure 6.
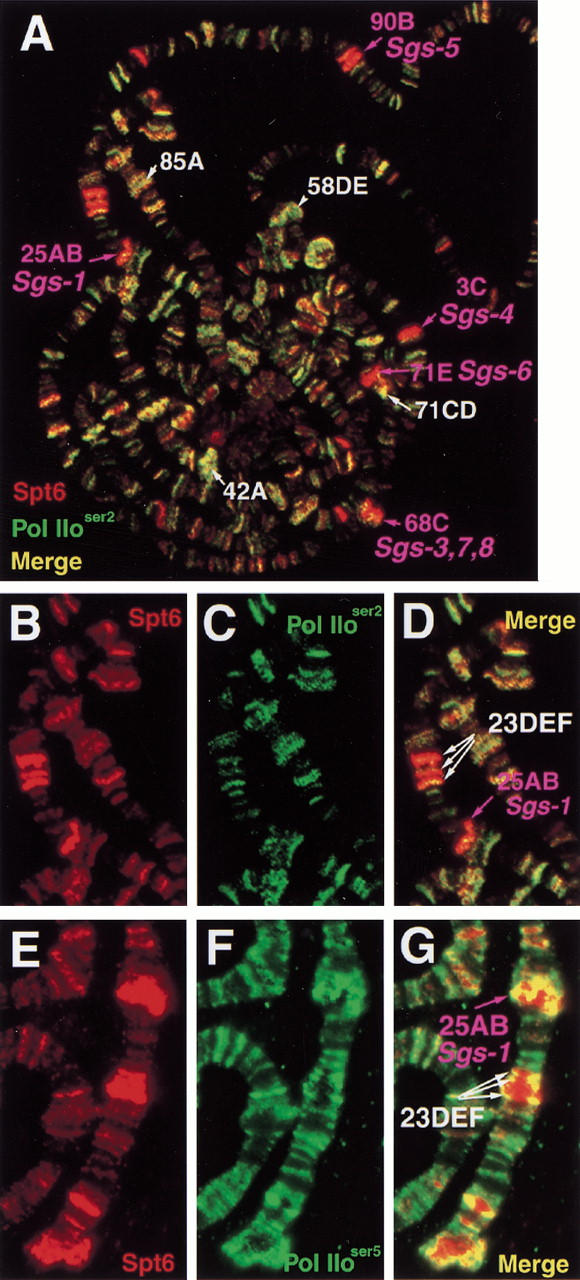
Deficiency of Pol IIoser2 at loci containing Sgs genes and presence of Pol IIoser5. Immunofluorescence detection of Spt proteins and different forms of phosphorylated Pol II on PS1–2 polytenes. (A) Spt6 (red) and Pol IIoser2 (green). Enlargement of chromosome 2L from A showing: (B) Spt6 staining (red); (C) Pol IIoser2 (green); (D) overlay of B and C. Staining of region shown in B–D for Spt6 and Pol IIoser5: (E) Spt6 (red); (F) Pol IIoser5; (G) overlay of E and F. Intermolt puffs containing Sgs genes labeled in magenta. Other intermolt puffs in A labeled in white.
The staining pattern at the Sgs loci is similar to what has been observed at heat shock loci after heat shock, including the presence of Spt5, Spt6, Pol IIa, and Pol IIo. Because the positive elongation factor P-TEFb is also present at heat shock loci upon heat shock, we tested if it is present at Sgs loci during their time of expression. To do this, we assayed binding by the P-TEFb subunit, cyclin T. At the developmental stage shown in Fig. 7 (PS1–2), the sites for the strongest P-TEFb binding correlate with the strongest Spt5 and Spt6 binding (Fig. 7, data not shown). A subset of these bands represents the Sgs loci seen in Figure 4 and Figure 6 that are enriched for Spt proteins, Pol IIa, and Pol IIoser5 but are deficient in Pol IIoser2. The cyclin T staining pattern we observe is similar to that described recently (Lis et al. 2000). Therefore, similar to heat shock puffs, Sgs loci stain heavily for Pol IIa, Pol IIo, Spt5, Spt6, and cyclin T.
Figure 7.
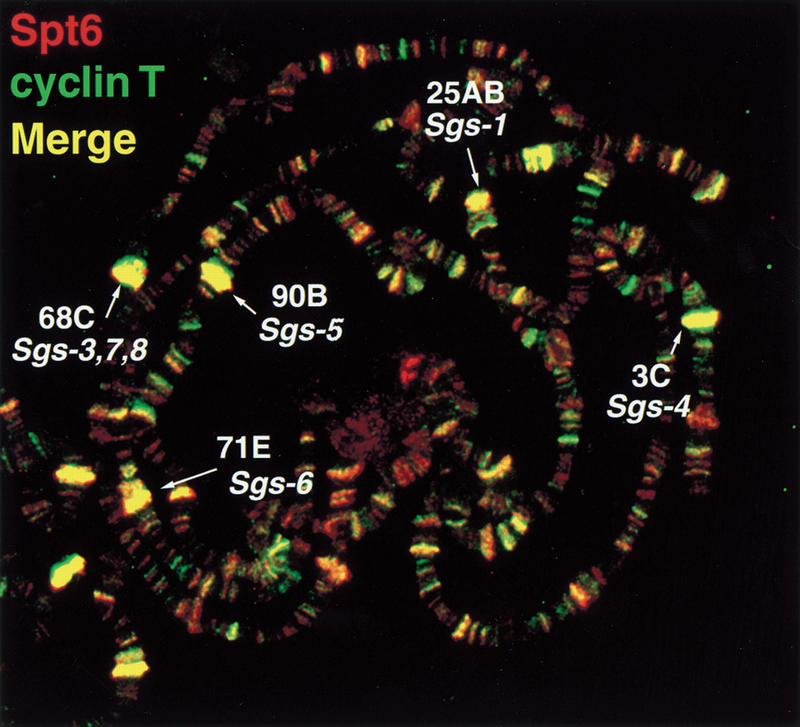
Colocalization of Spt6 and cyclin T and enrichment at loci containing Sgs genes. Immunofluorescence detection of Spt6 (red) and cyclin T (green) on PS1–2 polytenes. Polytene bands containing Sgs genes are labeled.
Sgs-4 is a direct target of Spt5, Spt6, and cyclin T association
The strong localization of Spt5 and Spt6 to polytene bands that contain Sgs genes suggests that Sgs genes might be direct targets of Spt5 and Spt6. To test this possibility for Sgs-4, we studied the localization of Spt5 and Spt6 to two transgenes containing Sgs-4 derivatives (Fig. 8A). The first transgenic line contains wild-type Sgs-4 (P[WT]), (Korge et al. 1990) and the second contains the Sgs-4 enhancer directing transcription of ADH (SAX, A. Hofmann, unpubl.). We found that both types of transgenes recruit high levels of Spt5, Spt6, cyclin T, and Pol IIa (Fig. 8b). In wild-type strains, no staining for these proteins was observed at the sites that contain transgenes in the transformed lines. These results demonstrate that the Sgs-4 enhancer, or the transcription dependent upon it, is sufficient for high level of recruitment of Spt5, Spt6, and cyclin T.
Figure 8.

Localization of Spt5, Spt6, cyclin T, and Pol IIa to Sgs-4 transgenes and recruitment dependent on the Sgs-4 enhancer. (A) Schematic representation of Sgs-4 transgenes. P(WT) contains the wild-type Sgs-4 gene. The Pig-1 gene is divergently transcribed from Sgs-4 under control of a shared set of enhancer elements. Transcription switches from Pig-1 to Sgs-4 during third larval instar development (Mougneau et al. 1993). The SAX transgene contains the Sgs-4/Pig-1 enhancer region fused to ADH (A. Hofmann, pers. comm.). (B) Localization of Spt5, Spt6, Pol IIa, and cyclin T to Sgs-4 transgenes. Top two rows: Spt5 and Pol IIa staining of P(WT) (top) and SAX (bottom). Bottom two rows: Spt6 and cyclin T staining of P(WT) (top) and SAX (bottom). Positions of transgenes indicated by arrows. In the nontransgenic wild-type strain, there is little or no staining for Spt5, Spt6, Pol IIa, or cyclin T at the sites at which transgenes are inserted in the transformed lines (data not shown).
Discussion
Our results suggest that the conserved transcriptional regulators Spt5 and Spt6 play general and related roles in active transcription in D. melanogaster. Spt5 and Spt6 colocalize to polytene chromosomes and their localization strongly correlates with the phosphorylated, actively elongating form of Pol II, Pol IIo. Furthermore, heat shock causes recruitment of Spt5 and Spt6 to heat shock loci with Pol IIo and P-TEFb. These results indicate that a major role for Spt5 and Spt6 in transcription occurs either concurrently with or after phosphorylation of the Pol II CTD and that this role is required during elongation. Results in the accompanying manuscript are consistent with our findings (Andrulis et al. 2000).
Positive and negative roles in transcription for Spt4, Spt5, and Spt6
Several previous studies have suggested a positive role in transcription for Spt4/Spt5 and Spt6. First, genetic studies in S. cerevisiae have demonstrated that all three proteins are required for growth in the absence of the positive elongation factor TFIIS, suggesting that they are required to overcome pausing during elongation (Hartzog et al. 1998). In addition, spt5 mutations in S. cerevisiae cause decreased levels of particular mRNAs (Compagnone-Post and Osley 1996; Hartzog et al. 1998). Second, the human Spt4/Spt5 complex acts as a positive elongation factor in vitro under conditions of limiting nucleotide concentration, a condition that causes a decrease in the rate of polymerase elongation and an increase in pausing (Wada et al. 1998a). Consistent with this activity, Spt5 shares sequence homology with the E. coli elongation factor NusG (Hartzog et al. 1998; Wada et al. 1998a; Wu-Baer et al. 1998) and this region of Spt5 is required for its activity in vitro (Yamaguchi et al. 1999b; Ivanov et al. 2000). NusG has been shown to modulate the elongation rate of E. coli RNA polymerase (Li et al. 1992; Linn and Greenblatt 1992; Burns et al. 1998). Spt4/Spt5 and Spt6 stimulate activation by HIV Tat, again implying a positive role in elongation (Wu-Baer et al. 1998; Kim et al. 1999; Parada and Roeder 1999; Ivanov et al. 2000). Finally, our data extend the possibility of a similar role in most transcription.
Other studies suggest that Spt4, Spt5, and Spt6 also play negative roles in transcription. In vitro studies of the human Spt4/Spt5 complex have shown that it can inhibit elongation in conjunction with another negative factor called NELF (Yamaguchi et al. 1999a) and that this inhibition can be overcome by P-TEFb (Wada et al. 1998b). In addition, in vitro studies have shown an interaction between Spt4/Spt5 and the unphosphorylated form of Pol II, Pol IIa (Wada et al. 1998b; Yamaguchi et al. 1999b). These in vitro studies examine the role of Spt4/Spt5 in the absence of P-TEFb or CTD phosphorylation; however, our results suggest that, in general, P-TEFb and/or CTD phosphorylation are present at sites of Spt4/Spt5 function. Therefore, the negative activity of Spt4/Spt5 may be under constant regulation by factors such as P-TEFb and may have a modulatory role in transcription elongation that is more subtle than that seen in vitro. In S. cerevisiae, spt4, spt5, and spt6 mutations suppress transcriptional defects associated with loss of the Swi/Snf complex and certain promoter mutations, also suggesting a negative role (for review, see Hartzog and Winston 1997, Winston and Sudarsanam 1998). We also observe some limited colocalization of Spt5 and Spt6 with Pol IIa on polytenes, indicating possible negative roles at specific loci in D. melanogaster.
Several variables may determine whether Spt4, Spt5, and Spt6 act positively or negatively. Similar to the requirement for the human factor NELF for the negative activity of Spt4/Spt5 on elongation, different factors may be required for Spt4, Spt5, and Spt6 to function positively in elongation. Two candidate factors are P-TEFb and the elongation complex FACT. P-TEFb may alter the role of Spt proteins either by phosphorylation of Pol II or Spt5 (Ivanov et al. 2000). Mutations that affect Spt4/Spt5/Spt6 and FACT (composed of Spt16/Cdc68 and Pob3) in S. cerevisiae cause some similar mutant phenotypes (Malone et al. 1991) and display genetic interactions (Orphanides et al. 1999). Recent in vitro results suggest a biochemical relationship between Spt4/Spt5 and FACT (Wada et al. 2000), as well.
Many other classes of transcription factors have been shown to act both positively and negatively in transcription, including the E. coli elongation factors NusA and NusG. NusA can increase pausing of polymerase at natural and cryptic pause sites (Linn and Greenblatt 1992; Burns et al. 1998) as well as increase or decrease termination efficiency depending on the type of terminator (Burns et al. 1998) whereas NusG can stimulate elongation as well as termination at rho dependent terminators (Sullivan and Gottesman 1992). Of interest, Spt6 contains two domains found in NusA (Aravind et al. 1999), indicating that along with Spt5, which shows homology to NusG, these factors may play roles in eukaryotic transcription analogous to prokaryotic NusA and NusG. A role for Spt4/Spt5 and Spt6 in termination would represent a negative activity associated with active transcription.
Analysis of Spt4, Spt5, and Spt6 at heat shock and Sgs loci
Study of Spt4, Spt5, and Spt6 function in greater detail at heat shock loci should be informative regarding their mechanism of action. The recruitment of Spt5 and Spt6 to these loci upon heat shock suggests that these factors play an important role in transcriptional activation of these genes. Given that heat shock loci are regulated at the level of elongation (for review, see Lis 1998) and that the appearance of Spt5 and Spt6 correlates closely with the appearance of P-TEFb (our results; Andrulis et al. 2000; Lis et al. 2000), Spt5 and Spt6 likely play a positive role in transcription elongation at these loci. However, because heat shock also causes an increased rate of Pol II initiation we cannot yet discern whether Spt5 and Spt6 function during initiation or elongation at these loci. In addition, the low amount of Spt5 detectable at heat shock loci in the absence of heat shock is consistent with either a positive or negative role. A positive role could involve potentiating transcription, while a negative role could involve either the establishment or maintenance of the paused Pol II found at the 5′ end of heat shock genes. Therefore, Spt4, Spt5, and Spt6 may be playing several important roles at heat shock loci.
Our cytological analysis suggests that Sgs genes are targets for Spt5, Spt6, and P-TEFb activities. Furthermore, the presence of both Pol IIo and Pol IIa at Sgs loci during the time of their expression suggests that these genes are regulated by transcriptional pausing, strikingly similar to heat shock genes. Alternatively, Sgs genes may contain only partially phosphorylated Pol II molecules which are recognized by antibodies against both Pol IIo and Pol IIa. In contrast to heat shock genes, we found that many of the Sgs genes appear to be deficient in Pol IIoser2, although the significance of this difference is unclear. This difference may be caused by the combination of factors that control Sgs-4 transcription, including the Ecdysone Receptor (EcR), Forkhead, and Daughterless/AP-4 (Lehmann 1996; Hofmann and Lehmann 1998). These factors also bind to many other loci on polytene chromosomes; however, these other loci do not exhibit the staining characteristics of Sgs genes with respect to either Spt proteins or Pol II, suggesting a previously unrecognized difference between the activities of these factors at different loci (Mach et al. 1996; King-Jones et al. 1999; Tsai et al. 1999).
Possible roles for Spt4, Spt5, and Spt6 in RNA Pol II transcription elongation
In summary, several lines of evidence point to roles for Spt5 and Spt6 in transcriptional elongation. Given the previously demonstrated interactions of Spt5 with Pol II (Hartzog et al. 1998; Wada et al. 1998b) and of Spt6 with histones (Bortvin and Winston 1996), a possible positive role for these proteins is promoting Pol II elongation past nucleosomes, which have been shown to inhibit transcription elongation by Pol II in vitro (Chang and Luse 1997; Luse and Samkurashvili 1998). Spt4, Spt5, and Spt6 may also have other activities related to mRNA synthesis, as Spt5 promotes mRNA capping in human cells (Wen and Shatkin 1999) and Drosophila Spt5 contains a domain implicated in the regulation of splicing. Thus, these Spt proteins may also play critical roles in the coordination of mRNA elongation with processing.
Materials and methods
Spt5 and Spt6 cDNAs
cDNAs corresponding to Drosophila Spt5 and Spt6 were identified by TBLASTN homology searches (Altschul et al. 1990) of the Berkeley Drosophila Genome Project EST database using murine Spt5 and Spt6. EST LD10265, obtained from Genome Systems, encodes full-length Spt5. ESTs LD17780, and LD02327 (Genome Systems) and ESTs LP02642 and LD45251 (Research Genetics) are overlapping and together encode full-length Spt6. The Spt5 cDNA sequence has been entered into GenBank under accession no. AF222864.
Antibodies
To raise antisera against Drosophila Spt5 and Spt6, two nonoverlapping regions of each cDNA were fused to glutathione S-transferase. The two Spt5 fusions were created by insertion of a BglII–BglII fragment of LD10265 encoding aa 59–739 of Spt5 into pGEX4T-1 (to generate antisera HMGP11) and by insertion of a BglII–XhoI fragment of LD10265 encoding aa 741–1078 of Spt5 into pGEX5X-1 (to generate antisera HMGP14). The two Spt6 fusions were created by insertion of a BglII–EcoRI fragment of LD17780 encoding aa 53–635 of Spt6 into pGEX1 (to generate antisera HMGP15) and by insertion of an EcoRI–PvuII fragment of LD02327 encoding aa 1046–1723 of Spt6 into pGEX4T-3 (to generate antisera HMGP17). pGEX plasmids are from Promega (pGEX4T-1, 4T-3, and 5X-1 were kind gifts of G. Gill [Harvard Medical School]). These fusions were expressed in E. coli and purified from inclusion bodies (Thompson et al. 1993). The fusion proteins were isolated via SDS-PAGE and injected into Guinea pigs by Cocalico Biologicals (Reamstown, PA). Antisera HMGP11 and HMGP14 were affinity purified over Reactigel beads (Pierce) coupled to the original antigen, following standard protocols (Luo and Wolfe 1995) and the affinity-purified antibodies were used in all experiments shown. Antiserum HMGP15 was also affinity purified; however, because HMGP15 antiserum gave the same results before and after affinity purification, the crude antibodies were used in most experiments. Control experiments with preimmune sera gave no significant signal in Western, embryo staining, or polytene staining assays. Anti-cyclin T antibodies were a kind gift from D. Price (University of Iowa) and were affinity purified using a cyclin T–GST fusion (also a gift of D. Price) (Lis et al. 2000). Polyclonal antiserum recognizing the unphosphorylated Pol II CTD (Pol IIa) was a kind gift from A. Greenleaf (Duke University) (Weeks et al. 1993). Monoclonal antibodies H5 and H14 (BAbCO) directed against phospho-serine-2 and phospho-serine-5, respectively, of the Pol II LS CTD were kind gifts of S. Buratowski (Harvard Medical School). All secondary antibodies were from Jackson Immunoresearch, except HRP-conjugated goat anti-mouse IgG + IgM (BioRad).
Polytene chromosomes and indirect immunofluorescence
Salivary glands were dissected from D. melanogaster third instar larvae and immunostained as described previously (Shopland and Lis 1996). Fixation times were as follows: Spt6, 45 sec; cyclin T, 1 min; Spt5, 80 sec. All other antigens were not sensitive to fixation time. For double staining experiments, longer fixation times were used as appropriate (e.g., any double-staining experiment that included staining for Spt5 was fixed for 80 sec). Antisera dilutions used for polytene staining were 1:250 (purified HMGP11), 1:250 (HMGP15), 1:500 (H5), 1:10 (H14), 1:500 (CTD), and 1:60 (purified anti-cyclin T). For heat shock experiments, larvae were placed in prewarmed 1.7 mL centrifuge tubes in a 37°C water bath for 5 min. Embryo staining was according to Patel (1994) and the antisera dilutions were 1:500 for purified HMGP11 and 1:250 for HMGP15. All polytene spreads were stained with DAPI (15–30 ng/mL for 60 sec) subsequent to staining with secondary antibody, prior to final washes. Degree of colocalization was assessed by the color of the signal in merged images; the appearance of yellow indicates that the red and green signals are roughly equivalent. For all experiments shown, multiple nuclei were examined from at least three independent experiments.
Acknowledgments
We thank David Price for providing the anti-cyclin T antisera, Steve Buratowski for the gift of H5 and H14 antisera, and Arno Greenleaf for anti-Pol IIa antisera. We thank Annemarie Hofmann for Sgs-4 transgenic fly stocks and assistance. We are grateful to Pam Silver and Paul Ferrigno for invaluable aid with microscopy and immunofluorescence, and to Anna Moran for technical assistance. We thank Erik Andrulis and John Lis for communication of results prior to publication and for assistance with polytene staining, and Terry Orr-Weaver for helpful suggestions. We also thank Grant Hartzog for helpful comments on the manuscript and Brad Cairns, Jerry Kaplan, and members of the Winston lab for stimulating discussions. J.M. was supported by the Harvard Society of Fellows. This work was supported by NIH grants GM32967 to F.W. and GMOD5582 to C.-t. Wu.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL winston@rascal.med.harvard.edu; FAX (617) 432-3993.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.831900.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High resolution of Drosophila Spt5 and Spt6 at heat shock genes in vivo: Roles in promoter proximal pausing and transcription elongation. Genes & Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J, Pan G, Dahmus GK, Cartier M, Marshall N, Zhang S, Dahmus ME, Greenblatt J. FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxy-terminal domain of RNA polymerase IIO. J Biol Chem. 1998;273:27593–27601. doi: 10.1074/jbc.273.42.27593. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. I. Autosomal puffing patterns in a laboratory stock of Drosophila melanogaster. Chromosoma. 1967;21:398–428. doi: 10.1007/BF00336950. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Patterns of puffing activity in the salivary gland chromosomes of Drosophila. VI. Induction by ecdysone in salivary glands of D. melanogaster cultured in vitro. Chromosoma. 1972;38:255–281. doi: 10.1007/BF00290925. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Chihara C, Meltzer P, Richards G. Temporal control of puffing activity in polytene chromosomes. Cold Spring Harbor Symp Quant Biol. 1974;38:655–662. doi: 10.1101/sqb.1974.038.01.070. [DOI] [PubMed] [Google Scholar]
- Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, Li Y, Ribisi S, Warren SL. Cytostellin distributes to nuclear regions enriched with splicing factors. J Cell Sci. 1994;107:387–396. doi: 10.1242/jcs.107.3.387. [DOI] [PubMed] [Google Scholar]
- Burns CM, Richardson LV, Richardson JP. Combinatorial effects of NusA and NusG on transcription elongation and Rho-dependent termination in Escherichia coli. J Mol Biol. 1998;278:307–316. doi: 10.1006/jmbi.1998.1691. [DOI] [PubMed] [Google Scholar]
- Bycroft M, Hubbard TJ, Proctor M, Freund SM, Murzin AG. The solution structure of the S1 RNA binding domain: A member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- Chang CH, Luse DS. The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J Biol Chem. 1997;272:23427–23434. doi: 10.1074/jbc.272.37.23427. [DOI] [PubMed] [Google Scholar]
- Chiang PW, Wei WL, Gibson K, Bodmer R, Kurnit DM. A fluorescent quantitative PCR approach to map gene deletions in the Drosophila genome. Genetics. 1999;153:1313–1316. doi: 10.1093/genetics/153.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim TK, Mancebo H, Lane WS, Flores O, Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes & Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagnone-Post PA, Osley MA. Mutations in the SPT4, SPT5, and SPT6 genes alter transcription of a subset of histone genes in Saccharomyces cerevisiae. Genetics. 1996;143:1543–1554. doi: 10.1093/genetics/143.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Doherty AJ, Serpell LC, Ponting CP. The helix-hairpin-helix DNA-binding motif: A structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs TM, Deppisch H, Scarlato V, Gross R. A new gene locus of Bordetella pertussis defines a novel family of prokaryotic transcriptional accessory proteins. J Bacteriol. 1996;178:4445–4452. doi: 10.1128/jb.178.15.4445-4452.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber ME, Jones KA. HIV-1 Tat: Coping with negative elongation factors. Curr Opin Immunol. 1999;11:460–465. doi: 10.1016/S0952-7915(99)80077-6. [DOI] [PubMed] [Google Scholar]
- Greenblatt J, Mah TF, Legault P, Mogridge J, Li J, Kay LE. Structure and mechanism in transcriptional antitermination by the bacteriophage lambda N protein. Cold Spring Harbor Symp Quant Biol. 1998;63:327–336. doi: 10.1101/sqb.1998.63.327. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Winston F. Nucleosomes and transcription: Recent lessons from genetics. Curr Opin Genet Dev. 1997;7:192–198. doi: 10.1016/s0959-437x(97)80128-1. [DOI] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann CH, Rice AP. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxy-terminal domain of the large subunit of RNA polymerase II: Candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Lehmann M. The transcriptional switch between the Drosophila genes Pig-1 and Sgs-4 depends on a SEBP1 binding site within a shared enhancer region (published erratum appears in Mol. Gen. Genet. 1999, 261: 428) Mol Gen Genet. 1998;259:656–663. doi: 10.1007/s004380050860. [DOI] [PubMed] [Google Scholar]
- Huet F, Ruiz C, Richards G. Puffs and PCR: The in vivo dynamics of early gene expression during ecdysone responses in Drosophila. Development. 1993;118:613–627. doi: 10.1242/dev.118.2.613. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA. Taking a new TAK on tat transactivation. Genes & Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- Kephart DD, Marshall NF, Price DH. Stability of Drosophila RNA polymerase II elongation complexes in vitro. Mol Cell Biol. 1992;12:2067–2077. doi: 10.1128/mcb.12.5.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Yamaguchi Y, Wada T, Handa H, Sharp PA. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol Cell Biol. 1999;19:5960–5968. doi: 10.1128/mcb.19.9.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K, Korge G, Lehmann M. The helix-loop-helix proteins dAP-4 and daughterless bind both in vitro and in vivo to SEBP3 sites required for transcriptional activation of the Drosophila gene Sgs-4. J Mol Biol. 1999;291:71–82. doi: 10.1006/jmbi.1999.2963. [DOI] [PubMed] [Google Scholar]
- Korge G, Heide I, Sehnert M, Hofmann A. Promoter is an important determinant of developmentally regulated puffing at the Sgs-4 locus of Drosophila melanogaster. Dev Biol. 1990;138:324–337. doi: 10.1016/0012-1606(90)90200-3. [DOI] [PubMed] [Google Scholar]
- Lehmann M. Drosophila Sgs genes: Stage and tissue specificity of hormone responsiveness. Bioessays. 1996;18:47–54. doi: 10.1002/bies.950180110. [DOI] [PubMed] [Google Scholar]
- Li J, Horwitz R, McCracken S, Greenblatt J. NusG, a new Escherichia coli elongation factor involved in transcriptional antitermination by the N protein of phage lambda. J Biol Chem. 1992;267:6012–6019. [PubMed] [Google Scholar]
- Linn T, Greenblatt J. The NusA and NusG proteins of Escherichia coli increase the in vitro readthrough frequency of a transcriptional attenuator preceding the gene for the beta subunit of RNA polymerase. J Biol Chem. 1992;267:1449–1454. [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Lis JT, Neckameyer W, Dubensky R, Costlow N. Cloning and characterization of nine heat-shock-induced mRNAs of Drosophila melanogaster. Gene. 1981;15:67–80. doi: 10.1016/0378-1119(81)90105-0. [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes & Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Luo J, Wolfe BB. Use of insoluble fusion proteins to purify antibodies. Biotechniques. 1995;19:544–546. , 548, 550. [PubMed] [Google Scholar]
- Luse DS, Samkurashvili I. The transition from initiation to elongation by RNA polymerase II. Cold Spring Harbor Symp Quant Biol. 1998;63:289–300. doi: 10.1101/sqb.1998.63.289. [DOI] [PubMed] [Google Scholar]
- Mach V, Ohno K, Kokubo H, Suzuki Y. The Drosophila fork head factor directly controls larval salivary gland-specific expression of the glue protein gene Sgs3. Nucleic Acids Res. 1996;24:2387–2394. doi: 10.1093/nar/24.12.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone EA, Clark CD, Chiang A, Winston F. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5710–5717. doi: 10.1128/mcb.11.11.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- Mougneau E, von Seggern D, Fowler T, Rosenblatt J, Jongens T, Rogers B, Gietzen D, Beckendorf SK. A transcriptional switch between the Pig-1 and Sgs-4 genes of Drosophila melanogaster. Mol Cell Biol. 1993;13:184–195. doi: 10.1128/mcb.13.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, Kornberg RD. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Parada CA, Roeder RG. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 1999;18:3688–3701. doi: 10.1093/emboj/18.13.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Meth Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reines D, Conaway JW, Conaway RC. The RNA polymerase II general elongation factors. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- ————— Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr Opin Cell Biol. 1999;11:342–346. doi: 10.1016/S0955-0674(99)80047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, Roberts CW. Antitermination by bacteriophage lambda Q protein. Cold Spring Harbor Symp Quant Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 1998a;12:1437–1446. doi: 10.1096/fasebj.12.14.1437. [DOI] [PubMed] [Google Scholar]
- ————— The RNA polymerase II general elongation complex. Biol Chem. 1998b;379:27–31. doi: 10.1515/bchm.1998.379.1.27. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Lis JT. HSF recruitment and loss at most Drosophila heat shock loci is coordinated and depends on proximal promoter sequences. Chromosoma. 1996;105:158–171. doi: 10.1007/BF02509497. [DOI] [PubMed] [Google Scholar]
- Stachora AA, Schafer RE, Pohlmeier M, Maier G, Ponstingl H. Human Supt5h protein, a putative modulator of chromatin structure, is reversibly phosphorylated in mitosis. FEBS Lett. 1997;409:74–78. doi: 10.1016/s0014-5793(97)00486-9. [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Gottesman ME. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell. 1992;68:989–994. doi: 10.1016/0092-8674(92)90041-a. [DOI] [PubMed] [Google Scholar]
- Swanson MS, Carlson M, Winston F. SPT6, an essential gene that affects transcription in Saccharomyces cerevisiae, encodes a nuclear protein with an extremely acidic amino terminus. Mol Cell Biol. 1990;10:4935–4941. doi: 10.1128/mcb.10.9.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat (published erratum appears in Mol. Cell. Biol. 1991, 11: 4286) Mol Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano E, Maki M, Mori H, Hatanaka M, Marti T, Titani K, Kannagi R, Ooi T, Murachi T. Pig heart calpastatin: Identification of repetitive domain structures and anomalous behavior in polyacrylamide gel electrophoresis. Biochemistry. 1988;27:1964–1972. doi: 10.1021/bi00406a024. [DOI] [PubMed] [Google Scholar]
- Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- Thummel CS. Puffs and gene regulation–molecular insights into the Drosophila ecdysone regulatory hierarchy. BioEssays. 1990;12:561–568. doi: 10.1002/bies.950121202. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Kao HY, Yao TP, McKeown M, Evans RM. SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol Cell. 1999;4:175–186. doi: 10.1016/s1097-2765(00)80365-2. [DOI] [PubMed] [Google Scholar]
- Velissariou V, Ashburner M. Cytogenetic and genetic mapping of a salivary gland secretion protein in Drosophila melanogaster. Chromosoma. 1981;84:173–185. doi: 10.1007/BF00399129. [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 1998a;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998b;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Orphanides G, Hasegawa J, Kim D-K, Shima D, Yamaguchi Y, Fukuda A, Hisatake K, Oh S, Reinberg D, et al. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol Cell. 2000;5:1067–1072. doi: 10.1016/s1097-2765(00)80272-5. [DOI] [PubMed] [Google Scholar]
- Warren SL, Landolfi AS, Curtis C, Morrow JS. Cytostellin: A novel, highly conserved protein that undergoes continuous redistribution during the cell cycle. J Cell Sci. 1992;103:381–388. doi: 10.1242/jcs.103.2.381. [DOI] [PubMed] [Google Scholar]
- Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: Correlations with gene activity and transcript processing. Genes & Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- Wen Y, Shatkin AJ. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes & Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Sudarsanam P. The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harbor Symp Quant Biol. 1998;63:553–561. doi: 10.1101/sqb.1998.63.553. [DOI] [PubMed] [Google Scholar]
- Wu-Baer F, Lane WS, Gaynor RB. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J Mol Biol. 1998;277:179–197. doi: 10.1006/jmbi.1997.1601. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Handa H. Interplay between positive and negative elongation factors: Drawing a new view of DRB. Genes Cells. 1998;3:9–15. doi: 10.1046/j.1365-2443.1998.00162.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999a;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999b;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- Yuryev A, Corden JL. Suppression analysis reveals a functional difference between the serines in positions two and five in the consensus sequence of the C-terminal domain of yeast RNA polymerase II. Genetics. 1996;143:661–671. doi: 10.1093/genetics/143.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]