Abstract
The tissue polarity mutants in Drosophila include a set of conserved gene products that appear to be involved in the control of cytoskeletal architecture. Here we show that the tissue polarity gene prickle (pk) encodes a protein with a triple LIM domain and a novel domain that is present in human, murine, and Caenorhabditis elegans homologs which we designate PET. Three transcripts have been identified, pk, pkM, and sple, encoding 93-, 100-, and 129-kD conceptual proteins, respectively. The three transcripts span 70 kb and share 6 exons that contain the conserved domains. The pk and sple transcripts are expressed with similar tissue-specific patterns but have qualitatively different activities. The phenotypes of pk mutants, and transgenic flies in which the different isoforms are overexpressed show that the balance between Pk and Sple is critical for the specification of planar polarity. In addition, these phenotypes suggest a tessellation model in which the alignment of wing hairs is dependent on cell shape and need not reflect fine-grained positional information. Lack of both pk and sple transcripts gives a phenotype affecting the whole body surface that is similar to those of dishevelled and frizzled (fz) suggesting a functional relationship between pk and fz signaling.
Keywords: Drosophila, prickle, spiny-legs, polarity, LIM domain
The Drosophila “tissue polarity” genes control the orientation of bristles and hairs in the adult cuticle (Gubb and Garcia-Bellido 1982; Adler et al. 1990; Adler 1992). Mutant flies have the polarity of cells altered in the plane of the epithelium without gross changes in the overall shape of imaginal structures or the distribution of differentiated cell types within them. Other tissue polarity defects include mirror-image duplications of the tarsal joints (Held et al. 1986), rotations of bristle sockets in the leg (Gubb and Garcia-Bellido 1982), and changes in ommatidial polarity (Gubb 1993, 1998; Theisen et al. 1994; Zheng et al. 1995).
There is evidence that these planar polarity phenotypes involve the organisation of the cytoskeleton. For example, localization of the actin fibers that form the prehair is the first observable manifestation of polarity in the pupal wing. Multiple wing hairs are seen in many of the tissue polarity mutants (Gubb and Garcia-Bellido 1982; Wong and Adler 1993) and result from formation of more than one prehair per cell (Wong and Adler 1993). Furthermore, genes identified in other organisms as having major roles in cytoskeletal reorganization, for example, Rac, RhoA, and Cdc42, have been shown to have a role in Drosophila planar polarity (Harden et al. 1995; Eaton et al. 1996; Strutt et al. 1997).
The tissue polarity genes have been divided into three groups (Wong and Adler 1993). Type 1 genes [dishevelled (dsh), frizzled (fz), and prickle (pk)], affect the whole body surface and are therefore believed to directly establish tissue polarity (Shulman et al. 1998). In contrast, the type 2 [inturned (in) and fuzzy (fy) and type 3 [multiple wing hairs (mwh)] genes affect distinct subsets of body areas and are thought to interpret the polarity established by the type 1 genes. A number of tissue polarity genes have been cloned [fz (Vinson et al. 1989); in (Park et al. 1996); fy (Collier and Gubb 1998), and strabismus (stbm; Wolff and Rubin 1998)] and some of these have been shown to have important roles in signal transduction at many points in development. The family of fz homologs are thought to be involved in reception of the Wingless (Wg) signal (Bhanot et al. 1996; Orsulic and Peifer 1996; Cadigan et al. 1997); whereas Dsh acts as an intracellular protein affecting both the Wg and Notch (N) pathways (Klingensmith et al. 1994; Thiesen et al. 1994; Axelrod et al. 1996, 1998)
In this paper we describe the molecular organization of the tissue polarity gene pk. Loss-of-function lesions in alternatively spliced transcripts of this one locus result in two genetically distinct phenotypes, Pk and Spiny-legs (Sple). The pk and sple transcripts encode proteins that contain three LIM motifs and a novel conserved domain that we have called PET (Prickle Espinas Testin). Surprisingly, deletion of the entire gene gives a phenotype that is much weaker than that of either the pk or sple single mutants. This unusual result can be explained by the pk and sple gene products acting in concert. The single-mutant phenotypes result from misactivation, rather than simply blocking, of a pathway of polarity formation. We propose that the correct balance of the Pk and Sple variants is required for normal planar polarity signaling in Drosophila imaginal discs.
Results
Phenotypic analysis of pk and sple
To undertake a thorough analysis of the pk locus we used a variety of genetic strategies to isolate new alleles. These alleles can be divided into three phenotypic classes, Pk, Pk–Sple, and Sple (Table 1; Figs. 1 and 2). None of these classes show any embryonic phenotype (even when homozygous mutant embryos develop from homozygous mutant mothers). Consistent with this lack of either a zygotic or maternal requirement, deletions of the pk gene are fully viable and fertile. The defects of double mutant, pkpk–sple, alleles are the same as those seen with overlapping deletions that remove the entire gene and eliminate all pk functions. Paradoxically, these pkpk–sple alleles do not produce the most severe phenotypes. Instead the single mutant alleles pkpk and pksple give more extreme phenotypes, but in reciprocal regions of the body; pkpk in the wing and notum and pksple in the legs, abdomen, and eyes. Complementation between these classes of allele indicates two subtly different functions at the pk locus.
Table 1.
Molecular and cytological breakpoints of pk mutations and deficiencies
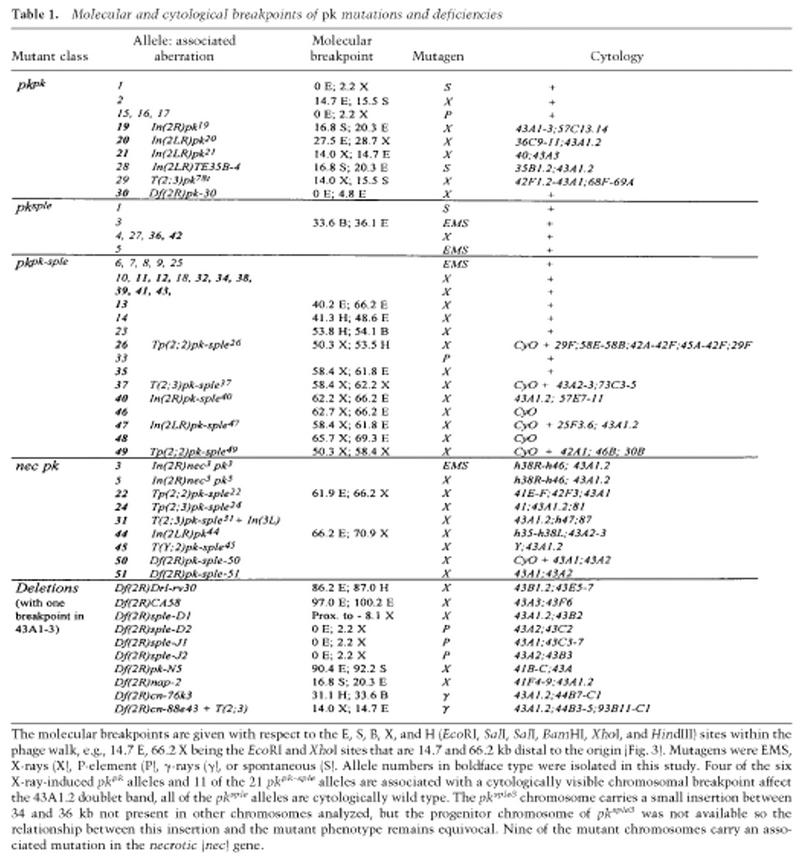 |
The molecular breakpoints are given with respect to the E, S, B, X, and H (EcoRI, SalI, SalI, BamHI, XhoI, and HindIII) sites within the phage walk, e.g., 14.7 E, 66.2 X being the EcoRI and XhoI sites that are 14.7 and 66.2 kb distal to the origin (Fig. 3). Mutagens were EMS, X-rays (X), P-element (P), γ-rays (γ), or spontaneous (S). Allele numbers in boldface type were isolated in this study. Four of the six X-ray-induced pkpk alleles and 11 of the 21 pkpk–sple alleles are associated with a cytologically visible chromosomal breakpoint affect the 43A1.2 doublet band, all of the pksple alleles are cytologically wild type. The pksple3 chromosome carries a small insertion between 34 and 36 kb not present in other chromosomes analyzed, but the progenitor chromosome of pksple3 was not available so the relationship between this insertion and the mutant phenotype remains equivocal. Nine of the mutant chromosomes carry an associated mutation in the necrotic (nec) gene.
Figure 1.
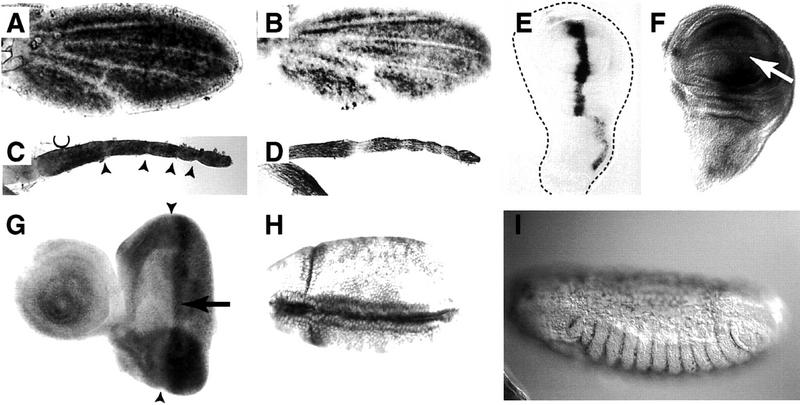
Pk mutant phenotypes. Complete lack of Pk function, in pkpk–sple alleles, gives a weak polarity phenotype in the wing, notum, abdomen, eye, and legs. pkpk alleles cause an extreme polarity phenotype in the wing and notum; pksple alleles affect eye, abdomen, and leg. The pkpk wing phenotype shows a characteristic reversal in the triple-row bristles along the anterior margin, a whorl in the wing hairs near the tip of vein 2, and abrupt discontinuities in hair polarity, e.g., pkpk1 (A). The weak PkPk–sple phenotype shows a slight effect on triple-row bristle orientation and gives gently curved hair polarity vectors, e.g., pkpk–sple13 (B); sple alleles are completely wild type, e.g., pksple1 (C). Arrows indicate the direction along which hairs are aligned; wing veins are designated 1–5 (C). The eye phenotype is wild-type in pkpk1 (D), showing a line of mirror symmetry along the equator (line). On both sides of the equator the R3 photoreceptor cell is aligned towards the pole, in the direction of the arrowhead. In addition to being rotated through 180° ommatidia show reversed chirality around the equator, so that both a rotation and a reflection in the plane of the epithelium is required to superimpose the ommatidial patterns. (E) pksple1 eyes contain a mixture of ommatidia with reversed polarity and chirality in both hemispheres of the eye. These ommatidia remain aligned along the polar axis, but with their R3 photoreceptors directed toward the equator rather than the pole giving rise to D/V mirror-image reversals of the normal rhabdomere pattern. In addition, all the pksple alleles show out ∼1% anteroposterior (A/P) reversed ommatidia (not shown). (F) pkpk–sple13 eyes contain a mixture of chiral forms of ommatidia. Some ommatidia fail to rotate properly, and the resulting imperfections in the hexagonal stacking give a slightly rough eye phenotype. Some ommatidia are aligned at 60° to the equator (black arrows) and some show A/P reversals (white arrows), with the R3 rhabdomere anterior to R4. (G) The tarsi of pkpk1 are wild type; tarsal segments are numbered T1–T5. (H) In pkpk–sple13, the T3 and T4 segments carry medial duplications of the proximal and distal joint structures, with the middle of each segment deleted. This results in alternating reversed-proximal and reversed-distal tarsal joint structures with half the length of a normal segment. (I) In pksple1 the tarsal duplications affect T2, T3, and T4 segments, with an occasional incipient ectopic joint in the distal T1. The distal T5 segment remains unaffected in all mutant alleles.
Figure 2.
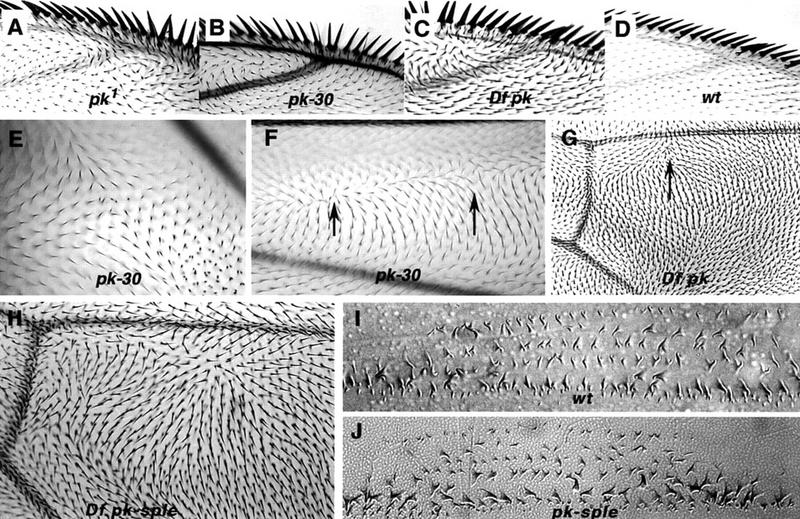
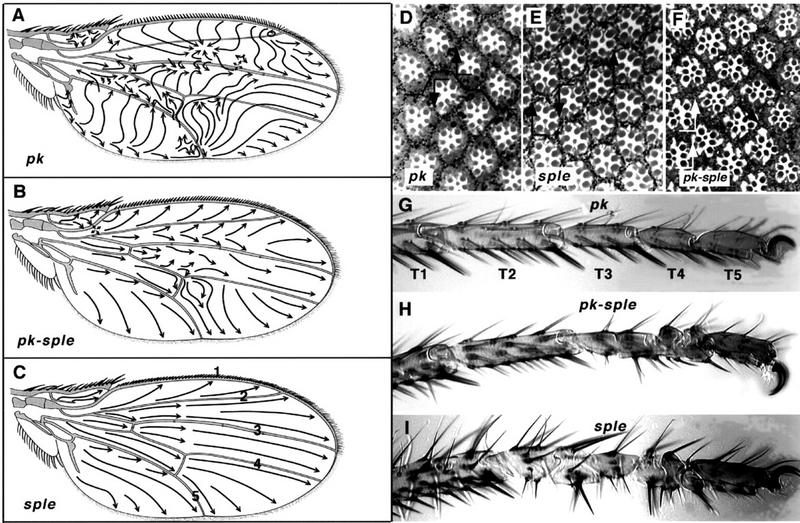
Comparison of wing polarity patterns between different alleles of pkpk. Junction of vein 1 and 2 at anterior margin in pkpk1 (A), Df(2R)pk-30 (B), Df(2R)nap-2/Df(2R)sple-J2 (C), and wild type (D). In pkpk mutant wings, the whorl close to the junction of vein 2 with the wing margin marks a discontinuity in polarity, or stacking flaw. Stacking flaws with different topologies are found in different regions of the wing. (E) Df(2R)pk-30, a cruciform discontinuity posterior to vein 5; note doubled hairs. (F) Df(2R)pk-30, radial and cruciform discontinuities (arrows) between veins 2 and 3. (G) Overlapping pkpk deletions, Df(2R)nap-2/Df(2R)sple-J2. Note triangular dislocation in polarity (arrow) distal to the posterior cross vein as in pkpk1 (Fig. 1A). (H) Overlapping pkpk–sple deletions removing the entire gene Df(2R)pk-N5/Df(2R)sple-J2 (cf. the pkpk–sple13 pattern; Fig. 1B). Embryonic denticle belt in wild type (I) and pkpk–sple13 (J). There is no detectable embryonic phenotype with alleles of pkpk, pksple, or pkpk–sple; in particular, the denticle belt morphology and denticle orientation remains normal.
The duplicated wing hair phenotype typical of most type 1 tissue polarity mutants (Gubb and Garcia-Bellido 1982; Wong and Adler 1993) also affects pkpk alleles, but only 2%–3% of cells show doubled hairs. Where the polarity vectors are changing sharply, however, cells frequently show doubled hairs (Figs. 2E and 3G). After the last cell division in the pupal wing, the cytoskeleton is reorganized. Cells become hexagonal, and prehairs grow out from the distal vertex of each cell (for review, see Eaton 1997). It is not possible, however, to fill an irregular shape such as the wing blade with a perfect hexagonal array of cells, and occasional defects, such as a distorted four-pentagon array, are seen (Fig. 3C). The relationship between hair orientation and the cell shape, implied by the localization of prehair initiation sites, is confirmed by the doubled hairs near stacking flaws (Figs. 2E and 3G) and the lack of regular hexagonal packing in the vicinity of the anterior whorl (Fig. 3B).
Figure 3.
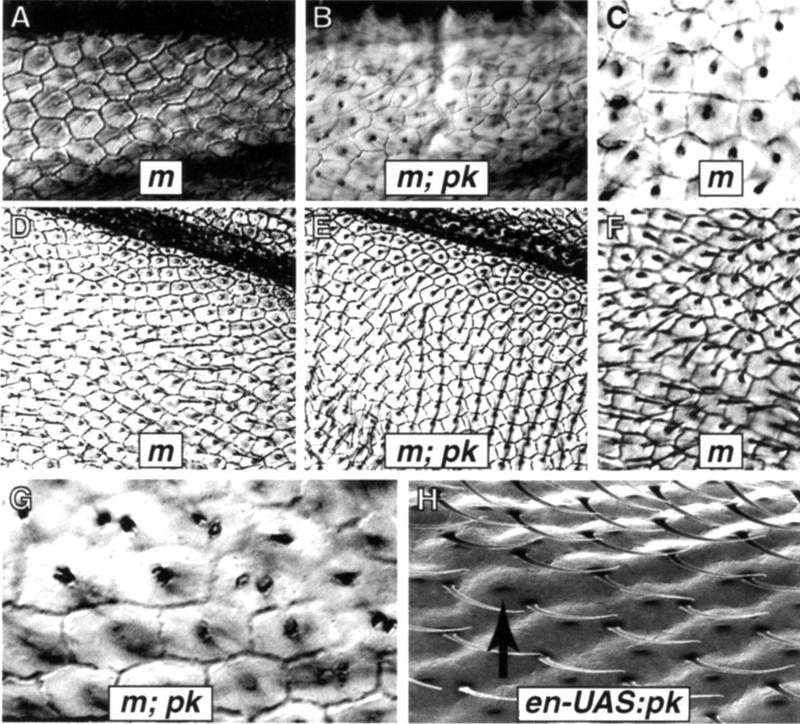
Cell shapes and polarity. The m38c mutation was used to visualize cell boundaries in both wild-type and Df(2R)pk-30 backgrounds. In m38c wings the majority of cells are hexagonal, although occasional pentagonal cells are seen. (A) Distal vein 2 region in m38c wings. (B) The same region in m38c; Df(2R)pk-30 wings, with more irregular cell shapes. (C) A common packing defect consisting of a group of four irregular pentagons with one 90° corner fitted within the surrounding hexagonal array. (D) The region posterior to vein 5 in m38c shows a regular hexagonal array, although cells close to the vein tend to be trapezoidal or square. Wing hairs are aligned along the proximodistal axis. (E) The same region in m38c; Df(2R)pk-30 wings shows parallel rows of hexagons with hairs oriented anteriorly. (F) A small region between veins 3 and 4 in m38c wings shows cuboidal cells, with local polarity disruptions near the transition between cuboidal and hexagonal packing. (G) Cells near the anterior whorl in m38c; Df(2R)pk-30 wings tend to be roughly cuboidal and associated with duplicated hairs. (H) Hairs fail to migrate from the distal vertex of pupal wing cells in en–UAS:pk flies (gal4–en; P[UAS:pk+]). This s.e.m. micrograph of adult wings shows hairs remaining at the distal vertex; the pedicel in the middle of the mature cell (arrow) marks the position that the adult hair would normally occupy.
Molecular organization of the pk locus
The breakpoints of pk-associated mutations map across 70 kb (Fig. 4; Table 1). Single-mutant pkpk alleles carry lesions within the proximal 30 kb, whereas pkpk–sple double-mutant alleles map between 42 and 69 kb and none of the pksple single mutations have been localized. Two putative pk transcripts were detected as weak bands of ∼4.2 and 5.1 kb in embryos and 2-day pupae on developmental Northern blots (Fig. 5). The shorter transcript corresponds to pk and the larger to sple (Fig. 5). The 5′ end of pk hybridizes to the region of the pkpk breakpoints and is separated by a large intron from the remaining 6 exons (Fig. 4). The 5′ end of sple is within the large pk intron. An additional medial transcript, pkM, is only detected in embryonic stages (Fig. 5B). The identity of the pk and sple transcripts was confirmed by rescue experiments (Figs. 7 and 8, below).
Figure 4.
(A) Map showing the location of pk mutant breakpoints with respect to transcripts in the region. The zero coordinate is the EcoRI site 973 bp proximal to the pk transcription start site. Transcripts are indicated below the molecular coordinates. The 5′ start of the pk transcript lies within a cluster of three serpins (serine Proteinase inhibitors) (Spn43Aa, Spn43Ab, and Spn43Ac), one of which corresponds to the necrotic gene, and a transcript with homology to Adenosine kinases maps just distal to pk. Aberrations that break between coordinates 0 and 30 kb cause pk mutations, similar to the homozygous deletion Df(2R)pk-30, which deletes the pk 5′ start and the 3′ end of the medial serpin transcript Spn43Ab. Aberrations between coordinates 40 and 70 kb, which interrupt the common exons, give the weaker PkPk–Sple phenotype typical of pkpk–sple13. Overlapping deletions give an additional necrotic phenotype when the serpin cluster is removed, but deletion of the adenosine kinase transcript [Df(2R)sple–J2/Df(2R)pk-N5] gives no further phenotype. (B) The Pk protein isoforms encode putative 870 (pk), 936 (pkM), and 1206 (sple) amino acid peptides with the conserved PET and LIM domains mapping entirely within the common exons. The first pk exon contains an untranslated leader sequence of 0.8 kb, with the putative translation start site (Cavener and Ray 1991) being 39 bp 5′ to the large intron. The putative translation start sites of the pkM and sple transcripts are within their first and second introns, respectively.
Figure 5.
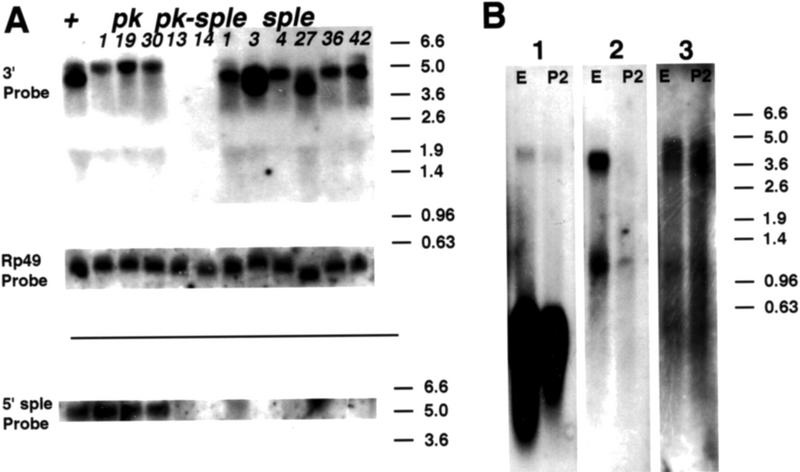
Northern analysis. A probe homologous to sple 5′ sequences detects a 5.1-kb mRNA, but this band is no longer detected, or has an altered length, in the sple mutants. A shorter, 4.2-kb transcript is missing in pkpk mutants. Both 5.1- and 4.2-kb transcripts are detected by a common exon probe in wild-type RNA, and neither are detected in pkpk–sple mutant strains. (A) Total 2-day pupal RNA from homozygous wild-type (+) and mutant flies (pkpk1, pkpk19, Df(2R)pk-30, pkpk–sple13, pkpk–sple14, pksple1, pksple3, pksple4, pksple27, pksple36, pksple42) hybridized with a 3′ common exon probe (3′), an Rp49 loading control (Rp49) and a unique sple exon probe (5′). The 4.2-kb transcript is missing in pkpk and pkpk–splealleles but retained in pksple alleles. The 5.1-kb transcript is present in pkpk1, pkpk19 and Df(2R)pk-30; missing in pkpk–sple13, pkpk–sple14 and lost, or reduced in length, in pksple3, pksple4, pksple27, and pksple36. (B) Three strips cut from a filter carrying wild-type embryonic (E) and 2-day pupal (P2) poly(A)+ RNA at the same sample loading. Individual strips were hybridized with pk 5′ + Rp49 (1), pkM 5′ (2), and sple 5′ (3) probes. The pk and sple transcripts are expressed in embryo and 2-day pupae. The pkM transcript was only detected during the embryonic stage.
Figure 7.
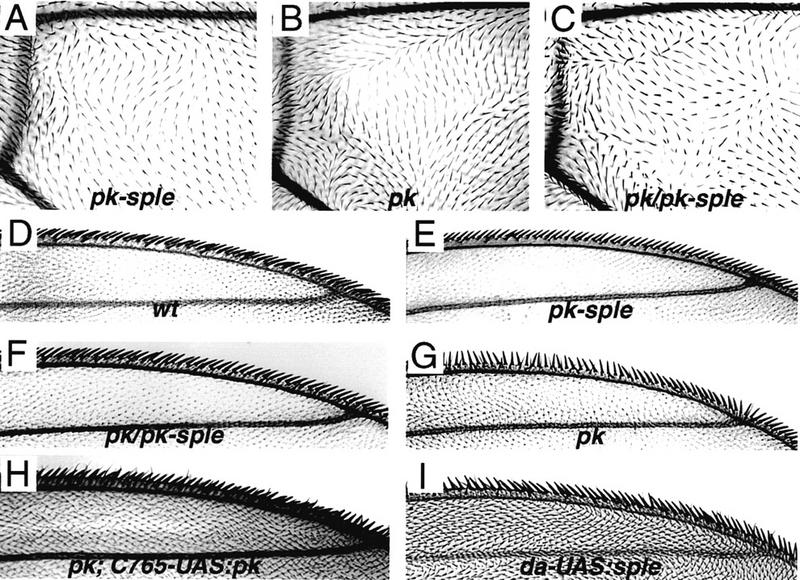
The wing phenotypes associated with titrating the number of intact copies of pk and sple. (A) pk–sple, moderate phenotype resulting from homozygous deletion of the common exons, pkpk–sple13/pkpk–sple13. (B) pk, extreme phenotype resulting from deletion of the pk transcript, Df(2R)pk-30/Df(2R)pk-30. (C) pk/pk–sple, moderate phenotype resulting from deleting pk and halving the number of sple copies in Df(2R)pk-30/pkpk–sple13 flies. The polarity pattern shown here, distal to the posterior cross vein, is stronger than in the remainder of the wing blade, which is closer to pkpk–sple13. (D) polarity of the marginal triple row bristles in wild type. (E) pk–sple triple row (pkpk–sple13/pkpk–sple13). (F) pk/pk–sple triple row [Df(2R)pk-30/pkpk–sple13]; (G) pk triple row [Df(2R)pk-30 /Df(2R)pk-30]. The pk wing phenotype was rescued by driving UAS:pk+ expression with the gal4–C765 driver, which is expressed in the wing and leg disks. (H) The pkpk triple row bristle phenotype is completely rescued and the wing hair polarity is close to wild type in pk; C765–UAS:pk wings [Df(2R)pk-30; gal4–C765/P(UAS:pk+)]. Small regions of abnormal polarity remain within the wing blade (both in pk; C765–UAS:pk and C765–UAS:pk in a wild-type background). sple overexpression also gives a wing phenotype in da–UAS:sple+ wings (I); the wing shown is EP(2)2557/+; gal4–da/+ (EP(2)2557 inserts a UAS driver 80 bp 5′ to the sple first intron), but the same phenotype is shown with a P[UAS:sple+] transformant line, in P[UAS:sple+]/+; gal4–da/+ wings.
Figure 8.
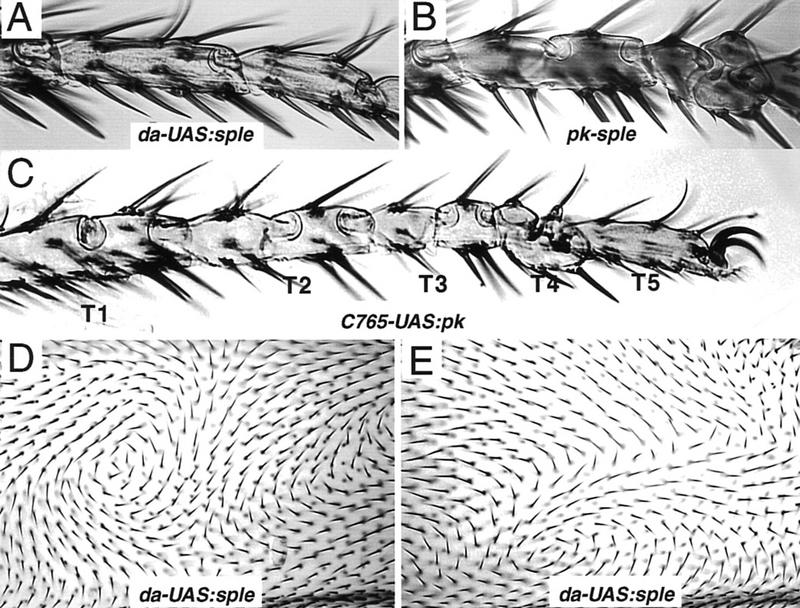
Overexpression phenotypes in leg and wing blade. Overexpression of sple in da–UAS:sple+ [EP(2)2557/+; gal4–da/+] legs gives no mutant phenotype; the wild-type morphology of T3 and T4 tarsal segments is shown in A. For comparison, the duplicated proximal (ball) and distal (socket) joint structures of the T3 and T4 segments resulting from lack of function in pkpk–sple-3 legs is shown in B; the T1, T2, and T5 segments (not shown) remain normal. A more extreme tarsal duplication phenotype affecting T1–T4 segments is given by Pk overexpression in 765–UAS:pk+ (P[UAS:pk]+/gal4–C765) flies (C). (D,E) The same region anterior to vein 4 in two different da–UAS:sple [EP(2)2557/+; gal4–da/+] wings. The polarity in any given region is unpredictable from wing to wing, but hair orientation changes gradually across large fields of cells. Whorls, cruciform, and radial stacking flaws are seen as in pkpk wings, but at variable positions within the wing blade.
The molecular analysis therefore identifies three different transcripts from the pk locus. The relationship between these transcripts and the different classes of lesion associated with mutant alleles explains the three classes of mutant alleles at the pk locus. Breakpoints that interrupt the common exons cause pkpk–sple mutations equivalent to deletions of the entire locus. pkpk mutations map to the proximal region and interrupt the 5′ region of the pk transcript. The pkM transcript appears to have no adult function, as the In(2LR)pkpk20 breakpoint is 3′ to the pkM 5′ exon (Table 1) but gives a similar phenotype to other pkpk alleles.
pk encodes a LIM domain protein
The second to seventh exons of pk are common to the three transcripts (Fig. 4). These common exons encode a peptide sequence containing three LIM domains, but no recognized DNA binding, membrane spanning, or signal peptide motifs (Table 2). LIM domains are cysteine-rich sequences with a double zinc finger motif (for review, see Sanchez-Garcia and Rabbitts 1994; Dawid et al. 1998). Four closely related LIM domain sequences were found by database searches; one murine (Testin, Divecha and Charleston 1995), one human (LMO6, Fisher et al. 1997), and two Caenorhabditis elegans. An additional pk cognate gene in Drosophila, espinas (esn), maps just proximal to the 43A1.2 Serpin cluster (C. Green and D. Gubb, unpubl.). Comparison of these sequences (Table 3) identifies a novel conserved PET domain, 5′ to the LIM domains. Two further incomplete sequences with PET domain homology were identified by database searches, a human EST and a C. elegans ORF. The PET domain is unrelated to other sequence motifs in the protein database and encodes a neutral stretch of amino acids with no obvious distinguishing features. Database searches failed to identify any sequences with significant homology to peptides encoded by the unique 5′ pk, pkM, or sple exons (Table 2).
Table 2.
The translated amino acid sequence of the alternative 5′ and common exons
| pk 5′ |
| MDTPNQMPVELER |
| pkM 5′ |
| MNDSTDNLHADCDGRVSNNNNGNSNTNDGPNNDGD SDEEVIEGMALLEGNYQVLRQWVPPAPNYWDAPPKAII KSAEVR |
| sple 5′ |
| MSSLSTGGGAGGSSGGPGGADAAAAPAAGQATVTATG NMEPAMVPRTANLLACKQWWRVCFLYGDQQKYYRQ LYSKAAAQRLADANQEPDNARDREYDTVDCDLIAGQ LDAVEDADDGIDLGDHSSTPKGGATTAGRPLFPHSSSP RRSKKLLRSLRAHVRGEKLPKNDTTTANVSNEVTQRN ARVTALDDPFLFGIDADHLGDLVVRGKRYSTLDATEN MARFYAEQEATAQVLEIIEQEEESPEQEAPKPALPPKQKQ QRPVPPLPPPPANRVTQDPGTQPAAPQVPLQPLTAGDL QFLNLSLRQRSLPRSMKPFKDAHDISFTFNELDTSAEPEV ATGAAQQESNE |
| Common exons |
| PISRTPLTQISYLQKIPTLPRHFSPSGQGLATPPALGSGGMG LPSSSSASALYAAQAAAGILPTSPLPLQRHQQYLPPHHQ QHPGAGMGPGPGSGAAAGPPLGPQYSPGCSANPKYS NAQLPPPPHHHHQLSPALSTPSPPSLLHHPAGGTSSASA HAPFLGGPHMDMQRQSHSDDDSGCALEEYTWVPPGL RPDQVRLYFSQIPDDKVPYVNSPGEQYRVRQLLHQLPP HDNEVRYCHSLTDEERKELRLFSTQRKRDALGRGNVR QLMS ARPCDGCDDLISTGDIAVFATRLGPNASWHPAC FACSVCRELLVDLIYFHRDGRMYCGRHHAETLKPRCSA CDEIILADECTEAEGRAWHMNHFGCHECDKQLGGQR YIMREGKPYCLHCFDAMFGEYCDYCGEGIGVDQGQM SHDGQHWHATDECFS CNTCRCSLLGRAFLPRRGGIYC SIACSKGEPPTPSDSSGTGMYTTPTPPTQGVRPHPQAPL PARIPSSHASSSPPMSPQQQQQHQATFNQAMYQMQSQ QMEAAGGLVDQSKSYAASDSDAGVVKDLEHGGHMG GGDLTDFSGGRASSTSQNLSPLNSPGDFQPHFLPKPME LQRQLLENPHTASMPELAGKLVAPPAHMQHLSQLHAV SSHQFQQHEYADIVHPPPPPPGEIPELPTPNLSVASTALP PELMGSPTHSAGDRSLNTPMS TQSASHAPPHPVSILSG ASSSSPMSGEPAKKKGVRFEGIPDTLPDHAATPVTERER VVAARGSGIGTRETRREEVAMVTVTPREGEDVASPPPA RRTIAAAAVIAPTRPLERTPMHLRNPILVLPGSTFGSAG CQPGPRVTQPTAEGTGARAGTQGVRGV |
Table 3.
Alignment of amino acid sequence of PET and LIM domains within the pk family
 |
Conserved amino acids are shown in uppercase and putative zinc binding ligands in bold. Sequence accession nos., LMO6 (EMBL AJ011654), Testin (SwissProt P47226), C. elegans (bo496.8, EMBL U58748; zk381.7, EMBL U52003; f25h5.1, EMBL z81068) and the human EST (GenBank h90439). The novel PET domain is 101 amino acids in length in pk, esn, LMO6 and f25h5.1 but is 5′ truncated in the other four family members. The length of the individual LIM domains and the unusual spacing of the putative zinc binding ligands in the third LIM motif are conserved features of the pk family. The amino acid sequence homology for both the PET and LIM domains ranges between 40% identity (52% similarity) for bo496.8 compared to pk and 87% identity (92% similarity) for esn compared to pk [percentages calculated using Best Fit (GCG)].
Expression patterns of pk transcripts
The expression patterns of the pk transcripts were investigated on developmental Northern blots and by tissue in situs using probes to the common exons and the unique 5′ exons. Both the temporal and spatial patterns of expression of the three transcripts were indistinguishable, with the exception that the pkM transcript was only detected during the embryonic stages. In 28- to 34-hr pupal wings, pk and sple transcripts are expressed uniformly in intervein cells but leave the presumptive vein regions unstained (Fig. 6A,B). At the same stage in the pupal legs pk and sple transcripts are expressed in a similar pattern, uniform in most cells, but excluded from the segmental boundaries (Fig. 6C,D). In third larval instar imaginal discs, a low level of pk transcripts can be detected in restricted domains that correlate with the places where pk is required. In the eye disc, maximal expression is detected in a stripe of cells behind the morphogenetic furrow (in the region where ommatidial organization and polarity is being specified) (Fig. 6G). In wing discs, pk transcripts are expressed at higher levels along the dorsoventral (D/V) compartment boundary, where the bristles of the wing margin will form (Fig. 6F). In the embryo, pk probes show a dynamic expression pattern in cells engaged in morphogenetic movements, such as invaginating mid-line cells, in the cephalic fold, and at parasegmental boundaries (Fig. 6H,I).
Figure 6.
Tissue in situs. Wild-type pupal leg and wing discs (28–32 hr) probed with pk-specific (A,C) or sple-specific (B,D) 5′ probes. Hybridization is uniform with both probes, with the exception of the wing veins and the tarsal segment boundaries (arrowheads in C). The expression in larval imaginal discs is much weaker; no endogenous signal is detected with a common exon probe at a level at which dpp–UAS:pk (gal4–dpp P[UAS:pk+]) wing discs show strong hybridization within the dpp domain (E) (the dotted line indicates disc boundary). With increasing development times a signal is seen along both sides of the D/V compartment boundary (arrow) in the wing (F) and as a discrete band (arrow) behind the morphogenetic furrow (arrowheads) in the eye disc (G). These preparations show a high background of nonspecific signal (retained in pkpk–sple13 discs and with antisense probes; data not shown). In the embryo, pkM, sple, and common exon probes give similar patterns of expression. Stage 8 embryos (H) show a strong signal in the cephalic furrow and dorsal fold, pkM probe. (I) Stage 14 embryos show signal in the parasegmental folds, common exon probe.
Functional relationships between Pk and Sple proteins
To investigate the functional relationship between the Pk and Sple protein variants, mutant combinations that alter the ratio of the pk and sple transcripts were generated. The strongest phenotypes result from lack of either one of these two transcripts in homozygous mutants. When the single mutant alleles are combined with pkpk–sple alleles or deficiencies, the resulting phenotypes are weaker than the corresponding homozygous single mutant phenotypes. Thus, flies that carry only a single functional copy of the sple transcript [Df(2R)pk-30/pkpk–sple13] or the pk transcript (pksple1/pkpk–sple13), show an intermediate phenotype more similar to pkpk–sple13 than the single mutant (Fig. 7). These results imply that the presence of one transcript without the other (as in pkpk or pksple single mutants) creates an extreme phenotype that is corrected when the dose of the remaining transcript is reduced. The implication is that both the relative and absolute levels of the Pk and Sple proteins are important for the function of putative Pk–Sple homomeric or heteromeric protein complexes.
The importance of the levels of Pk and Sple expression and the balance between them was investigated further with overexpression constructs. When driven by the expression of the uniform drivers gal4–da (daughterless) or gal4–C765, P[UAS:pk+] gives an extreme tarsal duplication phenotype, including a duplicated socket structure in the proximal T1 segment. Overexpression of UAS:sple+ with these drivers gives a Pk phenotype in the wing triple row bristles; the tarsi, however, remain completely wild type (Fig. 8). These results confirm that overexpression of pk gives a phenotype analogous to lack of the sple transcript, whereas overexpression of sple gives a phenotype similar to lack of pk. The da–UAS:pk+ and da–UAS:sple+ wing phenotypes show an unexpected feature that is not seen in mutant alleles. Instead of uniform polarity patterns, swirls are seen in the wing hair orientation with different patterns from wing to wing (Fig. 8).
In addition to phenotypes resembling loss of function, misexpression of pk in engrailed (en)–UAS:pk+ flies blocks the migration of wing hairs from the distal vertex of wing cells to the central position (Fig. 3H).
The relationship of pk to dsh
Deletion of the entire pk gene gives a phenotype over the whole body surface that is similar to dsh and fz. To test whether these mutants affect the same signaling pathway, double mutant combinations were made. Double mutants of pkpk and dsh in the triple row give a Dsh phenotype, suggesting that dsh is epistatic to pkpk (Fig. 9). The situation within the wing blade is less clear, as the double mutants give an intermediate phenotype. The dsh; pksple double mutant retains a Dsh phenotype in the triple row, but the wing hair pattern is altered (Fig. 9), despite the fact that pksple alleles have no wing phenotype. In the leg the Dsh tarsal phenotype is not modified by pkpk. There is a synergistic interaction between dsh and pksple, with the double mutant giving a more extreme mirror-image transformation of the T1 segment than either of the single mutants. This extreme transformation is also seen with UAS:pk+ overexpression and in fz (Figs. 8 and 9).
Figure 9.
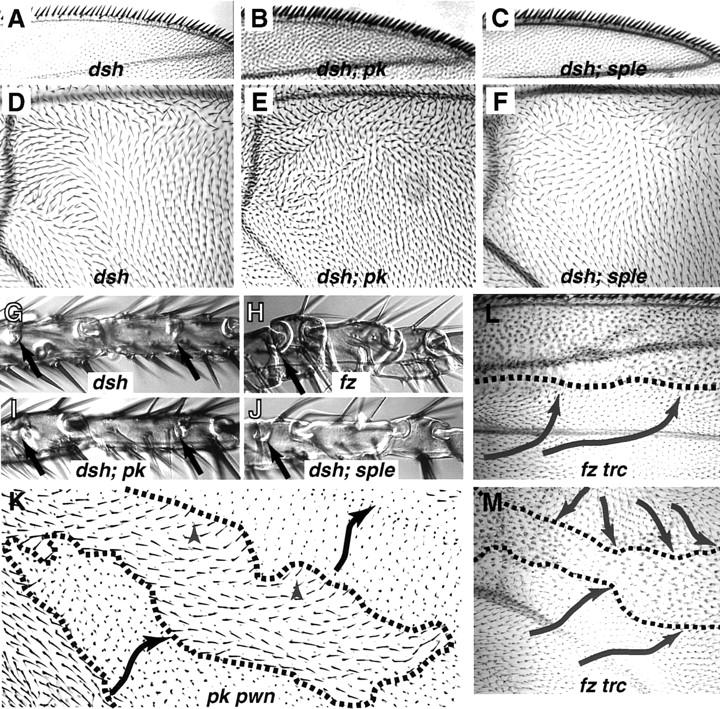
Comparison of different planar polarity phenotypes in wing and tarsi. The triple row bristle phenotype and wing blade polarity patterns of dsh are similar to pkpk–sple13 (A,D). The extreme triple row phenotype of Df(2R)pk-30 is suppressed in dsh; Df(2R)pk-30 flies and becomes phenotypically Dsh in both dsh; Df(2R)pk-30 and dsh; pksple1 double mutants (B,C). In dsh; Df(2R)pk-30 double mutants, the wing blade phenotype is intermediate between the respective single mutants (E). A slight, but consistent modification in the Dsh polarity is also seen in dsh; pksple1 wings, despite the fact that pksple alleles show no wing phenotype (F). The Dsh tarsal phenotype is similar to pkpk–sple13 in giving complete duplications of the T3 and T4 segments; an incipient joint is present in T1 and T2 (arrows), but the external morphology of these segments remains normal (G). This phenotype is unaffected in dsh; Df(2R)pk-30 (I). The most extreme tarsal phenotypes, with complete duplications in T2–T4 and an ectopic joint structure in T1, are seen in fz (H), P[UAS:pk+] overexpression (Fig. 8C) and dsh; pksple (J) mutant strains. In somatic mosaics, the effects of pk and fz are distinct. Within pkpk pwn clones, the polarity pattern is autonomous. A pkpk pwn clone in cell E of the wing is shown in K which expresses pkpk polarity (black arrows) (cf. Fig. 1A); a short range alteration in polarity (gray arrowheads) is seen close to the clone boundary (broken line). This effect is clearest when a small peninsula of pkpk pwn tissue is surrounded by pwn+ tissue. In contrast, frz trc clones cause a long-range nonautonomous alteration in the polarity of surrounding cells, which tend to point toward the mutant clone (L,M).
Clones of fz show a directional nonautonomy that has been interpreted to mean that fz mediates the intercellular communication of a polarity signal (Vinson and Adler 1987). On the other hand, dsh is strictly cell autonomous in clones (Theisen et al. 1994), implying that it is involved in signal reception. To investigate whether pk participates in both reception and transmission of a polarity signal we studied pkpk clones in the wing. Large clones in the wing express the mutant polarity typical of that region of the wing in homozygous flies. There is an occasional nonautonomous disruption of polarity in wild-type cells adjacent to the proximal or lateral margins of a pkpk clone (12 cases in 150 clones induced at 48–72 hr after egg laying). In these cases, a short range perturbation aligns wild-type cells with the mutant polarity pattern (Fig. 9). There is no clear pattern to the position of such clones, but small peninsulas of pwn+ tissue surrounded by pkpk pwn tissue tend to adopt the mutant polarity pattern. Smaller clones, induced later than 72–96 hr, did not alter the polarity of adjacent pawn (pwn+) tissue.
In contrast to the autonomous behavior of pk, clones of fz tricornered (trc) cause long-range domineering nonautonomy both distal (as reported by Vinson and Adler 1987) and lateral to the clone. Wild-type hairs are oriented toward the clone as though it is acting as a polarity ‘sink’ (Fig. 9). Proximal cells are also directed toward the fz trc clone, but as this is the normal orientation for wing cells no polarity changes would be expected.
Discussion
Genetic structure of pk and activity of the different transcripts
The pk gene encodes three transcripts (pk, sple, and pkM) that differ in their 5′ exons (Figs. 4 and 6). That these transcripts have qualitatively different activities in transgenic constructs, together with the intermediate phenotypes of mutant combinations that titrate the number of doses of pk and sple, suggests that the Pk and Sple products are required in stoichiometric ratios. Although pkpk and pksple mutants affect reciprocal regions of the body, their transcripts are expressed with a similar distribution in wing, leg, and eye discs. This implies a function for both transcripts in all tissues that is not obvious from the mutant phenotypes. In overexpression constructs [pk+] gives a Sple phenotype in the tarsi, whereas [sple+] gives a Pk phenotype in the wing (Fig. 8). This antagonism between the pk and sple transcripts implies either that both proteins compete for a target that is present in limiting amounts, or that they form protein complexes with distinct activities. In pkpk and pksple mutants excess homodimers would misactivate polarity signaling. Stoichiometric ratios of the LIM domain proteins Apterous and Chip have been shown to be critical in their developmental function (Fernandez-Funez et al. 1998).
Both the human and mouse homologs, LMO6 and Testin (Divecha and Charleston 1995; Fisher et al. 1997), encode alternative 5′ transcripts that may be a general feature of the pk family.
Sequence analysis and homologies
The LIM domain motif is a zinc binding finger that was originally described in homeodomain proteins but has since been found in combination with kinase or GAP domains (Dawid et al. 1995) and in proteins with no other identified domains. Many nonhomeodomain LIM proteins are associated with the cytoskeleton, several of them being localized primarily in adhesion plaques (Dawid et al. 1998) and having multiple LIM domains. The LIM domain appears to be involved in protein–protein interactions, with the highly charged zinc finger domains binding to target proteins. Homotypic interactions between two LIM domain partner proteins are also thought to occur (Dawid et al. 1998), although it is unclear how two zinc fingers might interdigitate.
There are a number of conserved features within the LIM domains of the pk family proteins. In the first LIM domain, there is a proline residue between the third (H) and fourth (C) zinc binding residues. This internal proline residue might introduce a kink within the zinc binding site. Similar LIMP domains with an internal P at this site are found in the human SLIM 2, SLIM 3, PINCH, Zyxin, and Paxillin proteins and LIM domain kinase 2 (LIK2) from chicken, mouse, rat, and human.
In addition, the length of the consecutive LIM domains is constant in the pk family, with LIM1 containing 57, LIM2 containing 52, and LIM3 containing 56 amino acids. The implication is that the conservation of the triple LIM domain itself is important, rather than the individual LIM domains. In the case of the triple LIM domain protein Zyxin, the individual LIM domains may bind different target proteins and act as a template for the assembly of a number of structural components (Beckerle 1997).
A similar scaffold function for the Pk protein would be consistent with the cell autonomy of pk in clones and the expression of pk transcripts in cells that are changing shape. The blocking of the normal migration of the actin-rich prehair structure to the center of wing cells by overexpression of pk also implies a role in cytoskeletal remodelling. The lack of embryonic phenotype, despite the dynamic expression pattern, implies that pk function is redundant during embryonic development. A putative embryonic pk function could, in principle, be maternally supplied, but pk mutant strains of all three classes are fully fertile and homozygous pk embryos from homozygous mothers remain wild type. Similarly, gal4–da; P[UAS:pk+], and gal4–da; P[UAS:sple+] flies are completely viable and show no embryonic phenotype, despite the embryonic expression of the gal4–da driver.
Tessellation models and fine-grained positional information during planar polarity formation
Perhaps the most surprising feature of the tissue polarity mutants is the precise polarity patterns seen in the wing hairs (Gubb and Garcia-Bellido 1982). Rather than reflecting fine-grained positional information, however, the precision of the final pattern might be dependent on a tessellation mechanism. The orientation of the first cell would determine the alignment of subsequent cells, like sticking tiles on a bathroom wall. In pkpk mutants, the alignment of wing hairs deviates progressively with occasional abrupt changes. In the adult mutant wing, hair polarity alters gradually with sudden topological discontinuities (Fig. 2E–G) that resemble the stacking flaws in liquid crystals (Frank 1958). In Df(2R)pk-30 wings, regions where the wing hairs are uniformly oriented retain a predominantly hexagonal array. Regions surrounding topological discontinuities, such as the anterior whorl, show irregular cell shapes frequently associated with duplicated wing hairs.
Strong support for a tessellation mechanism is given by the overexpression phenotype of Pk and Sple when driven by a ubiquitous promotor (e.g., Fig. 8D,E). Although polarity patterns are variable, hair orientation alters smoothly from cell to cell across the wing surface, indicating that the polarity of cytoskeletal structures is aligned within large fields of cells. It is as if cell packing had nucleated randomly and then spread to neighboring cells until meeting an adjacent domain. The short-range perturbation in polarity that is occasionally seen proximal and lateral to a pkpk clone is consistent with mechanical adjustment of cells to fit against their immediate neighbors, unlike the long-range domineering nonautonomy lateral and distal to fz clones. With both classes of clones, a tessellation mechanism might impose a threshold. Below this threshold, disruptions in polarity signaling would fail to affect the orientation of neighboring cells.
Tarsal duplications and polarity signaling pathways
The tarsal phenotype of dsh is very similar to pkpksple13 causing duplications of the T3 and T4 segments. A more extreme phenotype including complete duplications of T2 to T4 together with a well-developed ectopic T1 joint are seen in dsh; pksple, overexpression UAS:pk+ and fz mutant strains, implying that a similar polarity signaling pathway is affected. It has been suggested that pk and fz are upstream in a signaling pathway leading to dsh (Wong and Adler 1993; Shulman et al 1998). There are several problems with a simple linear pathway, however, and the relationship between pk and fz is unclear. First, neither fz nor pk is clearly epistatic to the other; rather, the double mutant (pkpk; fz) phenotype is intermediate between pkpk and fz (Gubb and Garcia-Bellido 1982; Wong and Adler 1993). Second, whereas fz clones cause a long-range nonautonomous disruption in surrounding tissue (Gubb and Garcia-Bellido 1982; Vinson and Adler 1987) we show here that pkpk clones resemble dsh clones (Theisen et al. 1994) in being almost completely cell autonomous. (Both pksple1 and pkpk–sple13 clones are cell autonomous in the eye (Zheng et al. 1995; Gubb 1998).) On the other hand, the pkpk triple row bristle phenotype (that results from Sple expression in the absence of Pk) is suppressed in dsh; pkpk wings and the dsh polarity pattern is modified in dsh; pksple wings (despite pksple on its own not having a wing phenotype). Finally, complete lack of pk transcripts produces a phenotype very similar to dsh. Taken together, these results indicate that PK is not downstream of Fz but may represent an alternative input into Dsh-mediated planar polarity signaling.
Conclusions
We show that the two genetically distinct phenotypes Pk and Sple arise from discrete lesions in a single gene and suggest that the relative and absolute levels of the two gene products are critical to wild-type activity. The developmental expression of a third transcript, pkM, suggests an embryonic function. We have not recovered mutant alleles of pkM, however, and the lack of embryonic phenotype with deletions of the entire pk gene implies that any embryonic function must be redundant.
We have also identified the PET domain, a novel protein motif that is highly conserved in metazoan evolution. We speculate that the PET domain is involved in interactions with as yet unidentified proteins to form a planar polarity signaling complex. Potential partners could include the currently known tissue polarity genes and actin cytoskeletal components. Overexpression of Pk transcripts produces mutant phenotypes similar to recessive lack of function. This unusual property is also shown with overexpression of Fz (Krasnow and Adler 1994) and Dsh (Axelrod et al. 1998). If Dsh is acting as a scaffolding or adapter protein (Axelrod et al. 1998) then it would be required in stoichiometric ratios to its target proteins, and overexpression would prevent assembly of functional complexes. It may be that the Pk protein isoforms are components of this protein complex that are expressed in cells remodeling their cytoskeletal architecture.
Materials and methods
Drosophila stocks
The original alleles of pkpk and pksple are spontaneous. Subsequent alleles were induced in this study (see below and http://www.gen.cam.ac.uk/dept/gubb.html) or described in FlyBase (http://flybase.bio.indiana.edu). The EP line (Rorth 1996), EP(2)2557 maps in the pk region and gives a recessive polarity phenotype in notum and tarsus (B. Reed, pers. comm.). Gal4 driver strains were gal4–pannier (/) pnrMD237; gal4–C765; gal4–dpp (decapentaplegic), gal4–en, and gal4–daG32 (FlyBase 1999). The boundaries of adult wing cells were visualized using the miniature (m) mutant strain In(1)m38c (Newby et al. 1991).
To extend the analysis of pk, new alleles were recovered in a variety of screens. Four pkpk (19, 20, 21, 30) and four pkpk–sple (11, 12, 13, 31) alleles were isolated among 4207 X-irradiated chromosomes, heterozygous with either Df(2R)pk78k or pr pk cn sp. Twenty further alleles of pkpk–sple (22, 24, 26, 28, 32, 34, 35, 37, 38, 39, 40, 41, 43–51) and two of pksple (36, 42) were recovered among 34, 491 X-irradiated chromosomes heterozygous with pksple1. Together with preexisting alleles, this gave a collection of eleven pkpk alleles, 28 pkpk–sple alleles, and seven pksple alleles (Table 1).
Mutagenesis
New X-ray-induced alleles of pkpk and pkpk–sple were recovered heterozygous with Df(2R)pk78k, or pr pk cn sp following the methods of Heitzler et al. (1993). Additional X-ray-induced pkpk–sple and pksple alleles were screened for the pksple phenotype heterozygous with pksple1.
Molecular techniques
A chromosome walk of 150 kb was isolated using the phage genomic insert library of John Tamkun (University fo California, Santa Cruz). The walk was extended from a 3-kb genomic fragment with weak homology to raf (Mark et al. 1987) proximally to FP11/3 and distally to FP0/4, until the Df(2R)sple-J2 and Df(2R)Drl-rv30 breakpoints defining the region had been uncovered. The complete restriction maps of these phage for EcoRI, SalI, BamHI, XhoI, and HindIII and other details are available at the http://www.gen.cam.ac.uk/dept/gubb.html web site.
Chromosome breakpoints were mapped relative to the genomic phage inserts by in situ hybridization with biotin-labeled probes and genomic Southern blots (as described in Gubb et al. 1997). Tissue in situs used riboprobes to the unique exons and the 3′ common exons, made with the Boehringer DIG RNA labeling kit (SP6/T7).
Identification of transcripts and recovery of cDNAs
As the walk progressed, gel-purified genomic fragments were used to probe developmental Northern blots and cDNA library filters. Although three Serpin transcripts and a putative adenosine kinase were identified on Northern blots using random primed genomic insert probes, the much less abundant pk transcripts were not detected. A single putative full-length pk cDNA, Pk4a3, was finally recovered from 5 × 105 clones of an embryonic library (Brown and Kafatos 1988). Additional pk transcripts were identified from embryonic cDNA using nested oligonucleotides from the second exon of Pk4a3 with the Marathon (Clontech)-modified RACE protocol. Products were cloned into the pCR2.1 vector with a TA cloning kit (Invitrogen). This generated three fragments of 1.8, 0.88, and 0.65-kb, which were hybridized to the genomic walk. The 0.88-kb product is equivalent to the 5′ end of Pk4a3; whereas the other two products represent alternative 5′ exons. The 1.8-kb product corresponds to the 5′ sple transcript, whereas the 0.65-kb fragment identifies a third, medial transcript, which we have designated pkM.
Northern analysis
Total RNA was isolated from staged wild-type (Canton-S) flies (Ashburner 1989). Poly (A)+ RNA was prepared using the PolyAT tract mRNA isolation kit (Promega). Developmental Northern blots used ∼0.5 μg of Poly(A)+ RNA from each stage. For pk mutants, 25 μg per lane of total RNA from 2-day pupae was used. Gels were blotted on nylon membrane and hybridized with random-primed 32P-radiolabeled pk common exon probe (the Pk4a3 PstI–XhoI 617-bp plus XhoI–NotI 737-bp fragments) and a Rp49 probe following standard procedures. Single-stranded antisense RNA probes for the alternative 5′ exons were synthesized from the pCR2.1 vector.
Nucleotide sequence analysis of cDNA, genomic clones, and exonic boundaries
Genomic fragments and cDNAs were subcloned into pBluescript SK+ (Stratagene Ltd.). SauIIIA and TaqI restriction fragments of the Pk4a3 cDNA and six-cutter fragments of the sple 5′ cDNA were sequenced from vector primers using the Prism Ready Reaction DyeDeoxy Terminator Cycle Sequencing Kit (Perkin-Elmer Corporation). Sequence gaps were filled using specific oligonucleotide primers.
The exon structure of a full-length 4.2-kb cDNA, Pk4a3, was mapped to the genomic walk by Southern blot and sequence analysis. Similarly, the 1.8-, 0.88-, and 0.65-kb 5′ RACE products were mapped against the genomic walk before being subcloned and sequenced. The precise boundaries of the pk 5′ exon were deduced by comparison to sequence from subcloned genomic fragments from our walk, whereas the 5′ exonic structure of pkM and sple was obtained by comparison of the cDNA sequence of the 1.8- and 0.65-kb RACE products to genomic sequence from the Berkeley Drosophila Genome project (P1 clone DS08462; AC005198).
Transformation constructs
Transformation constructs were cDNA sequences subcloned in the pUAST vector to utilize the gal4 expression system (Brand and Perrimon 1993). The UAS [pk+] transgenic construct was made from the Pk4a3 cDNA, whereas UAS [sple+] was synthesised from Pk4a3 by replacing the 5′ HindIII–SmaI fragment with the sple 5′ HindIII–SmaI fragment from a pBluescript subclone of the 1.8-kb RACE product.
Analysis of somatic clones
Somatic mosaics of pkpk1 marked with the cell autonomous mutation pwn were generated using a P[FRT]42D pkpk1 pwn chromosome following the method of Xu and Rubin (1994) and fz trc as in Gubb and Garcia-Bellido (1982).
Acknowledgments
We thank Jose-Felix de Celis for advice and help with tissue in situs, Michael Ashburner for the cytological analysis of In(2LR)CyO Tp(2;2)pk–sple26, and Carol McKimmie for injection of the P-element transformation constructs. Sarah Bray, Adelaide Carpenter, Jose-Felix de Celis, Miriam Hirshberg, Jean-Marc Reichhart, Steven Russell, and Josh Shulman are acknowledged for discussion and critical reading of the manuscript; Bruce Reed for sharing unpublished results; and John Tamkun and Nick Brown for phage and plasmid libraries. We thank the FlyBase consortium, particularly Rachel Drysdale, for rulings on the pk–sple nomenclature. This work was funded by Medical Research Council programme grants to Michael Ashburner, David Gubb, and Steven Russell.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Note
The EMBL accession numbers for the cDNA and genomic clone sequences in this paper are adenosine kinase (AJ236864), pk isoform (AJ243708), pkM isoform (AJ243709), and sple isoform (AJ243710).
Footnotes
E-MAIL dg27@mole.bio.cam.ac.uk; FAX 44-1223-333992.
References
- Adler PN. The genetic control of tissue polarity in Drosophila. BioEssays. 1992;14:735–741. doi: 10.1002/bies.950141103. [DOI] [PubMed] [Google Scholar]
- Adler PN, Vinson C, Park WJ, Conover S, Klein L. Molecular structure of frizzled, a Drosophila tissue polarity gene. Genetics. 1990;126:401–416. doi: 10.1093/genetics/126.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. Drosophila, a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Axelrod JD, Matsuno K, Artavanis-Tsakonas S. Interaction between Wingless and Notch signaling pathways mediated by Dishevelled. Science. 1996;271:1826–1832. doi: 10.1126/science.271.5257.1826. [DOI] [PubMed] [Google Scholar]
- Axelrod JD, Miller JR, Shulman JM, Moon RT, Perrimon N. Differential recruitment of Dishevelled provides signaling specificity in the planar cell polarity and Wingless signaling pathways. Genes & Dev. 1998;12:2610–2622. doi: 10.1101/gad.12.16.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerle MC. Zyxin: Zinc fingers at sites of cell adhesion. BioEssays. 1997;19:949–956. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse RA. A new member of the frizzled family from Drosophila functions as a wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Brown NH, Kafatos FC. Functional cDNA libraries from Drosophila embryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Fish MP, Rulifson EJ, Nusse R. Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell. 1997;93:767–777. doi: 10.1016/s0092-8674(00)81438-5. [DOI] [PubMed] [Google Scholar]
- Cavener DR, Ray SC. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S, Gubb D. Drosophila tissue polarity requires the cell-autonomous activity of the fuzzy gene, which encodes a novel transmembrane protein. Development. 1998;124:4029–4037. doi: 10.1242/dev.124.20.4029. [DOI] [PubMed] [Google Scholar]
- Dawid IB, Toyama R, Taira M. LIM domain proteins. CR Acad Sci Paris Ser III. 1995;318:295–306. [PubMed] [Google Scholar]
- Dawid IB, Breen JJ, Toyama R. LIM domains: Multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- Divecha N, Charleston B. Cloning and characterisation of two new cDNAs encoding murine triple LIM domains. Gene. 1995;156:283–286. doi: 10.1016/0378-1119(95)00088-n. [DOI] [PubMed] [Google Scholar]
- Eaton S. Planar polarization of Drosophila and vertebrate epithelia. Curr Opin Cell Biol. 1997;6:860–866. doi: 10.1016/s0955-0674(97)80089-0. [DOI] [PubMed] [Google Scholar]
- Eaton S, Wepf R, Simons K. Roles for Rac1 and Cdc42 in planar polarization and hair outgrowth in the wing of Drosophila. J Cell Biol. 1996;135:1277–1289. doi: 10.1083/jcb.135.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Funez P, Lu CH, Rincon-Limas DE, Garcia-Bellido A, Botas J. The relative expression amounts of apterous and its co-factor dLdb/Chip are critical for dorso-ventral compartmentalization in the Drosophila wing. EMBO J. 1998;17:6846–6853. doi: 10.1093/emboj/17.23.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase. The FlyBase database of the Drosophila Genome projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. http://flybase.bio.indiana.edu http://flybase.bio.indiana.edu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Ciccodicola A, Tanaka K, Curci A, Desicato S, D’Urso M, Craig IW. Sequence-based exon prediction around the Synaptophysin locus reveals a gene-rich area containing novel genes in human proximal Xp. Genomics. 1997;45:340–347. doi: 10.1006/geno.1997.4941. [DOI] [PubMed] [Google Scholar]
- Frank FC. On the theory of liquid crystals. Discussions Faraday Soc. 1958;25:19–28. [Google Scholar]
- Gubb, D. 1993. Genes controlling cellular polarity in Drosophila. Dev. (Suppl.) 269–277. [PubMed]
- Gubb D. Cellular polarity, mitotic synchrony and axes of symmetry during growth. Where does the information come from? Int J Dev Biol. 1998;42:369–377. [PubMed] [Google Scholar]
- Gubb D, Garcia-Bellido A. A genetic analysis of the determination of cuticular polarity during development in Drosophila melanogaster. J Embryol Exp Morphol. 1982;68:37–57. [PubMed] [Google Scholar]
- Gubb D, Roote J, Trenear J, Coulson D, Ashburner M. Topological constraints on transvection between white genes within the transposing element TE35B in Drosophila melanogaster. Genetics. 1997;146:919–937. doi: 10.1093/genetics/146.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden N, Loh HY, Chia W, Lim L. A dominant inhibitory version of the small GTP-binding protein Rac disrupts cytoskeletal structures and inhibits developmental cell shape changes in Drosophila. Development. 1995;121:903–914. doi: 10.1242/dev.121.3.903. [DOI] [PubMed] [Google Scholar]
- Held LI, Duarte CM, Derakhshanian K. Extra tarsal joints and abnormal cuticular polarities in various mutants of Drosophila melanogaster. Wilhelm Roux Arch Dev Biol. 1986;195:145–157. doi: 10.1007/BF02439432. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Coulson D, Saenz-Robles M-T, Ashburner M, Simpson P, Roote J, Gubb D. Genetic and cytogenetic analysis of the 43A-E region containing the segment polarity gene costa and the cellular polarity genes prickle and spiny-legs in Drosophila melanogaster. Genetics. 1993;135:105–115. doi: 10.1093/genetics/135.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnow RE, Adler P. A single Frizzled protein has a dual function in tissue polarity. Development. 1994;120:1883–1893. doi: 10.1242/dev.120.7.1883. [DOI] [PubMed] [Google Scholar]
- Klingensmith J, Nusse R, Perrimon N. The Drosophila segment polarity gene dishevelled encodes a novel protein required for response to the wingless signal. Genes & Dev. 1994;8:118–130. doi: 10.1101/gad.8.1.118. [DOI] [PubMed] [Google Scholar]
- Mark GE, MacIntyre R J, Digan ME, Ambrosio L, Perrimon N. Drosophila melanogaster homologs of the raf oncogene. Mol Cell Biol. 1987;7:2134–2140. doi: 10.1128/mcb.7.6.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby LM, White L, DiBartolomeis SM, Walker BJ, Dowse HB, Ringo JM, Khuda N, Jackson FR. Mutational analysis of the Drosophila miniature-dusky (m-dy) locus: Effects on cell size and circadian rhythms. Genetics. 1991;128:571–582. doi: 10.1093/genetics/128.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsulic S, Peifer M. An in vivo structure-function study of armadillo, the beta-catenin homologue, reveals both separate and overlapping regions of the protein required for cell adhesion and for wingless signaling. J Cell Biol. 1996;134:1283–1300. doi: 10.1083/jcb.134.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WJ, Liu J, Sharp EJ, Adler PN. The Drosophila tissue polarity gene inturned acts cell autonomously and encodes a novel protein. Development. 1996;122:961–969. doi: 10.1242/dev.122.3.961. [DOI] [PubMed] [Google Scholar]
- Rorth P. A modular misexpression screen in detecting tissue-specific phenotypes. Proc Natl Acad Sci. 1996;93:12418–12422. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia I, Rabbitts TH. The LIM domain: A new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Shulman JM, Perrimon N, Axelrod JD. Frizzled signaling and the developmental control of cell polarity. Trends Genet. 1998;14:452–458. doi: 10.1016/s0168-9525(98)01584-4. [DOI] [PubMed] [Google Scholar]
- Strutt DI, Weber U, Mlodzik M. The role of RhoA in tissue polarity and Frizzled signalling. Nature. 1997;387:292–295. doi: 10.1038/387292a0. [DOI] [PubMed] [Google Scholar]
- Theisen H, Purcell J, Bennett M, Kansagara D, Syed A, Marsh JL. dishevelled is required during wingless signaling to establish both cell polarity and cell identity. Development. 1994;120:347–360. doi: 10.1242/dev.120.2.347. [DOI] [PubMed] [Google Scholar]
- Vinson CR, Adler PN. Directional non-cell autonomy and the transmission of polarity information by the frizzled gene of Drosophila. Nature. 1987;329:549–551. doi: 10.1038/329549a0. [DOI] [PubMed] [Google Scholar]
- Vinson CR, Conover S, Adler PN. A Drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains. Nature. 1989;338:263–264. doi: 10.1038/338263a0. [DOI] [PubMed] [Google Scholar]
- Wolff T, Rubin GM. strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development. 1998;125:1149–1159. doi: 10.1242/dev.125.6.1149. [DOI] [PubMed] [Google Scholar]
- Wong LL, Adler PN. Tissue polarity genes of Drosophila regulate the subcellular location for prehair initiation in pupal wing cells. J Cell Biol. 1993;123:209–221. doi: 10.1083/jcb.123.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1994;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Zheng L, Zhang J, Carthew RW. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development. 1995;121:3045–3055. doi: 10.1242/dev.121.9.3045. [DOI] [PubMed] [Google Scholar]