Abstract
The Drosophila gene groucho (gro) encodes a transcriptional corepressor that has critical roles in many development processes. In an effort to illuminate the mechanism of Gro-mediated repression, we have employed Gro as an affinity reagent to purify Gro-binding proteins from embryonic nuclear extracts. One of these proteins was found to be the histone deacetylase Rpd3. Protein–protein interaction assays suggest that Gro and Rpd3 form a complex in vivo and that they interact directly via the glycine/proline rich (GP) domain in Gro. Cell culture assays demonstrate that Rpd3 potentiates repression by the GP domain. Furthermore, experiments employing a histone deacetylase inhibitor, as well as a catalytically inactive form of Rpd3, imply that histone deacetylase activity is required for efficient Gro-mediated repression. Finally, mutations in gro and rpd3 have synergistic effects on embryonic lethality and pattern formation. These findings support the view that Gro mediates repression, at least in part, by the direct recruitment of the histone deacetylase Rpd3 to the template, where it can modulate local chromatin structure. They also provide evidence for a specific role of Rpd3 in early development.
Keywords: Drosophila embryogenesis, pattern formation, groucho, histone deacetylase rpd3, transcriptional repression, corepressors
The Drosophila gene groucho (gro) encodes a transcriptional corepressor with many roles in development (for reviews, see Fisher and Caudy 1998; Parkhurst 1998). This factor contains a conserved WD repeat domain implicated in protein–protein interactions but lacks a DNA-binding domain. (DBD; Hartley et al. 1988; Tata and Hartley 1993; Paroush et al. 1994). Gro is recruited to target promoters via direct interactions with a broad spectrum of sequence-specific DNA-binding transcription factors. For example, Hairy-related basic helix–loop–helix (bHLH) factors, as well as Runt family transcription factors negatively regulate genes controlling neurogenesis, segmentation, and sex-determination via a conserved carboxy-terminal WRPW or WRPY motif through which they recruit Gro to the template (Paroush et al. 1994; Fisher et al. 1996; Alifragis et al. 1997; Aronson et al. 1997; Jimenez et al. 1997; Zhang and Levine 1999).
Other factors lacking WRPW/Y motifs also recruit Gro for transcriptional repression. For example, the Drosophila homeodomain proteins Engrailed and Goosecoid employ a common motif (the Engrailed homology-1 domain) to recruit Gro (de Celis and Ruiz-Gomez 1995; Aronson et al. 1997; Jimenetz et al. 1997; Tolkunova et al. 1998; Jimenez et al. 1999). The Rel-homology domain-containing factor Dorsal appears to recruit Gro with the help of additional DNA-binding transcription factors, which together with Dorsal may form a high affinity template-bound platform for the recruitment of Gro (Dubnicoff et al. 1997; Valentine et al. 1998). In addition, recent studies demonstrate that an interaction between Drosophila Tcf, a transcription factor in the Wingless signaling pathway, and Gro contributes to repression by Tcf, further highlighting the widespread importance of Gro-mediated repression (Cavalo et al. 1998; Levanon et al. 1989; Roose et al. 1998).
Corepressors with homology to Gro have been found in many other eukaryotic organisms. The yeast TUP1 protein may be a homolog of Gro, as it contains a conserved carboxy-terminal WD-repeat domain and also functions as a corepressor in conjunction with a wide array of DNA-binding repressors (Williams and Trumbly 1990; Keleher et al. 1992). In Caenorhabditis elegans, the Gro-like transcription factor UNC-37 seems to function with UNC-4, a homeodomain protein, to repress transcription and govern motor neuron identity (Pflugrad et al. 1997). The human Gro homologs, termed transducin-like Enhancer of split (TLE) proteins, have been shown to interact with the mammalian counterparts of Hairy-related proteins and negatively regulate genes important for neuronal development (Fisher et al. 1996; Grbavec and Stifani 1996; Grbavec et al. 1998). The human Runt-domain repressor encoded by the AML1 gene, one of the most frequent targets for leukemia-associated chromosome translocations, mediates transcriptional repression by interacting with TLE proteins (Imai et al. 1998; Levanon et al. 1998). A recent study also reports that a B-cell-specific repressor, B-lymphocyte induction maturation protein 1 (Blimp1), represses transcription by direct recruitment of TLE-family corepressors to the template (Ren et al. 1999).
Whereas mounting evidence has documented the widespread role of Gro in many development processes, relatively little is known about the mechanisms underlying Gro-mediated repression. The most highly conserved region in Gro other than the WD-repeat motif is the amino-terminal 130 amino acid glutamine-rich region. We have shown previously that this motif mediates Gro homotetramerization and that formation of the tetramer is required for Gro-mediated repression (Chen et al. 1998). Previous studies have also demonstrated that Gro/TLE family corepressors are associated with chromatin in living cells and specifically interact with the amino-terminal tail of histone H3, suggesting that Gro-mediated repression may involve the modulation of local chromatin structure (Palaparti et al. 1997).
Many corepressor complexes, most notably perhaps the Sin3 complex, contain enzymes termed histone deacetylases that remove acetyl groups from lysine residues in the amino-terminal tails of core histones. Conversely, coactivator complexes often contain histone acetyltransferase activity (for recent reviews, see Pazin and Kadonaga 1997; Kuo and Allis 1998; Struhl 1998). By determining the dynamic acetylation state of histones, these enzymes may alter the ability of the general transcriptional machinery to recognize and transcribe genes (Hansen et al. 1998; Luger and Richmond 1998). Hyperacetylated chromatin is usually associated with active transcriptional states, whereas hypoacetylated chromatin is usually associated with repressed transcriptional states (Hebbes et al. 1988; Braunstein et al. 1993).
To further illuminate the mechanism of Gro-mediated repression, we have used Gro as an affinity reagent to purify Gro-binding proteins from Drosophila embryonic nuclear extracts. The histone deacetylase Rpd3 was identified as one protein that specifically interacts with Gro. In vitro protein–protein interaction assays demonstrate that the Gro/Rpd3 interaction is probably direct. Further experiments indicate that Rpd3 and histone deacetylase activity are associated with Gro in vivo and that the deacetylase activity is required for Gro-mediated repression in transient transfection assays. Finally, simultaneous reduction of the maternal gro and rpd3 gene dosage results in synergistic effects on pattern formation and embryonic viability, supporting the idea that the products of these genes function together in development.
Results
Gro binds Rpd3 in vitro and in vivo
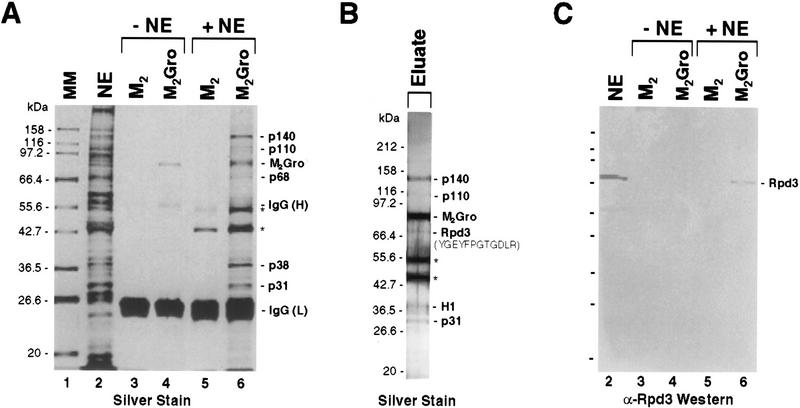
To identify polypeptides that associate with Gro, we used purified Flag-tagged Gro (referred to as M2Gro) as an affinity reagent. In a pilot experiment (Fig. 1A), anti-Flag affinity beads alone (lanes 3,5) or beads containing immobilized M2Gro (lanes 4,6) were incubated with 0- to 12-hr Drosophila embryonic nuclear extracts (lane 2). Following extensive washing of the beads, bound proteins were eluted with SDS, resolved by SDS-PAGE, and visualized by silver staining (Fig. 1A, lanes 5,6). Five polypeptide species with apparent molecular masses of 140, 110, 68, 38, and 31 kD bound to the anti-Flag beads containing M2Gro (lane 6) but not to the beads alone (lane 5).
Figure 1.
Affinity purification of Gro-interacting proteins. (A) Silver-stained SDS–polyacrylamide gel showing results of a small scale affinity purification of Gro-interacting proteins. (Lane 1) Molecular mass markers; (lane 2) unfractionated 0- to 12-hr Drosophila embryonic nuclear extract; (lanes 3–6) SDS elutions of M2 antibody beads lacking (M2, lanes 3,5) or containing (M2Gro, lanes 4,6) epitope-tagged Gro. (Lanes 5,6) The beads were incubated with embryonic nuclear extract and subsequently subjected to extensive washing prior to SDS elution; (lanes 3,4) the nuclear extract was omitted. IgG light (L) and heavy (H) chains are indicated. Polypeptides from the embryonic nuclear extracts that uniquely coprecipitate with the M2Gro beads are indicated (p140, p110, p68, p38, and p31). (*) Two polypeptides that are retained on the beads in both the presence and absence of Gro. (B) Silver-stained SDS–polyacrylamide gel showing an aliquot of the eluate from a large-scale M2Gro affinity column used for affinity purification of Gro-interacting proteins. Elution was achieved by washing the beads with excess Flag peptide. Thus, unlike in A, the immunoglobulin chains were not eluted. However, the same two nonspecific polypeptides (*) that eluted with SDS (A), eluted with the peptide, indicating that they are probably binding directly to the antibody. The amino acid sequence denotes a peptide sequence obtained from an internal lysC fragment of p68. This sequence is a perfect match to a predicted lysC fragment in Drosophila histone deacetylase, Rpd3. P38 has been identified as Drosophila histone H1 (H1). (C) Immunblot with anti-Rpd3 antibody. Lanes are the same as in A.
In a subsequent large-scale purification (Fig. 1B), these five polypeptides were resolved by preparative SDS-PAGE. Then, we attempted to isolate and determine the sequence of lysC peptides from these Gro-associated polypeptides. A lysC peptide recovered from p38 has the sequence ASATPSHPPTQQMVDA, which is identical to a sequence found in Drosophila histone H1 (Croston et al. 1991). A peptide recovered from p68 has the sequence YGEYFPGTGDLR, which perfectly matches a sequence found in Drosophila histone deacetylase Rpd3 (De Rubertis et al. 1996). Immunoblot analysis with affinity purified anti-Rpd3 polyclonal antibody (kindly provided by P. Spierer, University of Geneva, Switzerland) confirms that p68 is Rpd3 (Fig. 1C).
Immunoprecipitation assays (Fig. 2A) suggest that endogenous Gro and Rpd3 are associated in Drosophila nuclei, as antibodies against Gro coprecipitated Gro and Rpd3 from both Drosophila embryonic (lane 3) and S2 cell (lane 7) nuclear extracts. Neither protein was precipitated by nonimmune serum (lanes 2,6) or by negative control anti-Flag antibodies (lanes 4,8). In each case, ∼10%–20% of Rpd3 in the nuclear extracts was found to precipitate with Gro. In vitro histone deacetylase assays using 3H-acetyl-labeled histones as the substrate indicates that ∼20% of the histone deacetylase activity in the embryonic extracts coprecipitates with Gro, whereas no significant activity is precipitated by the nonimmune serum or control anti-Flag antibody (Fig. 2B). The histone deacetylase activity in the crude extracts and in the Gro immunoprecipitate is largely inhibited by the histone deacetylase inhibitor trichostatin A (TSA) (Taunton et al. 1996), as is the activity of purified recombinant Rpd3.
Figure 2.
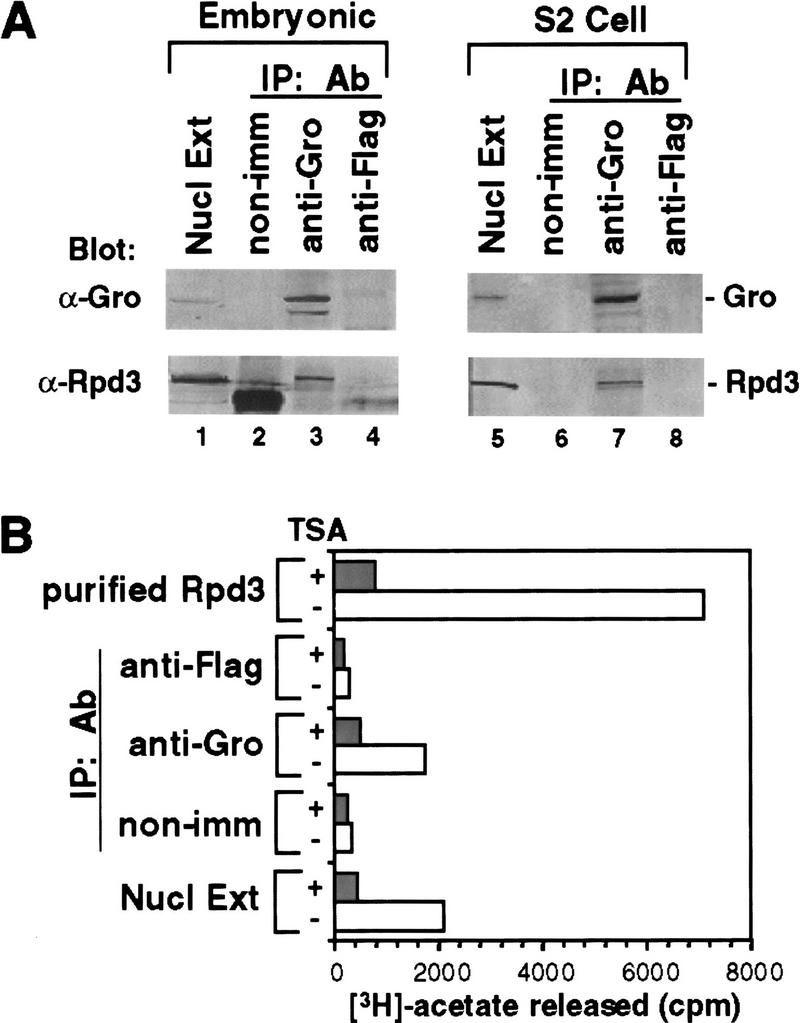
Endogenous Gro and endogenous Rpd3 form a complex in vivo. (A) Immunoprecipitation of embryonic (lanes 1–4) and S2 cell (lanes 5–8) nuclear extracts. (Lanes 1,5) Unfractionated nuclear extracts; (lanes 2,6) immunoprecipitates obtained with nonimmune serum; (lanes 3,7) immunoprecipitates obtained with anti-Gro antibody; (lanes 4,8) immunoprecipitates obtained with anti-Flag antibody. Proteins were analyzed by SDS-PAGE and immunoblotting with anti-Gro (top) or anti-Rpd3 (bottom) antibodies. (B) In vitro histone deacetylase assays of immunoprecipitates from embryonic nuclear extract. Samples assayed are those shown in lanes 1-4 of A. In addition, purified baculovirus-expressed Rpd3 was assayed as a positive control. Each sample was assayed in the presence (+) or absence (−) of the histone deacetylase inhibitor TSA (150 nm).
The glycine/proline-rich domain of Gro is required for the interaction with Rpd3
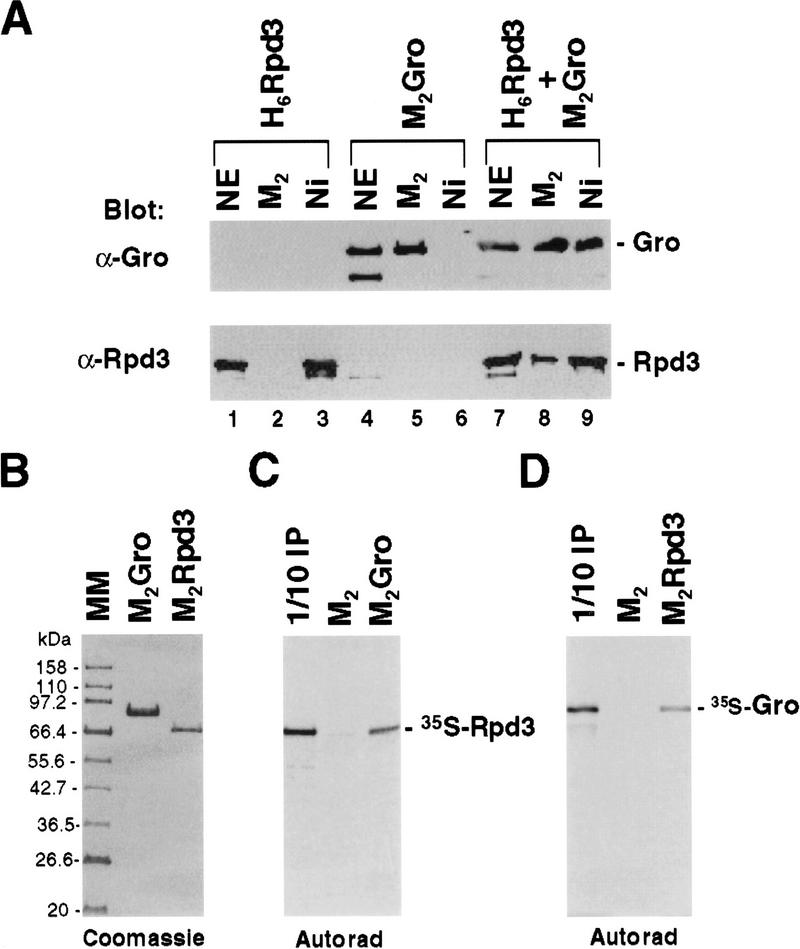
Consistent with the finding that Rpd3 interacts with Gro in vivo, we showed that M2Gro and six histidine-tagged Rpd3 (H6Rpd3) copurified on both Ni2+–NTA-agarose (which binds the histidine tag) and anti-Flag affinity beads, when M2Gro and H6Rpd3 were coexpressed using baculovirus vectors (Fig. 3A). To determine whether the interaction between Gro and Rpd3 is direct, we purified baculovirus-expressed M2Gro and Rpd3 (M2Rpd3) to near homogeneity (Fig. 3B). Equal amounts of these purified proteins were first immobilized on anti-Flag beads and then incubated with in vitro-translated and [35S]methionine-labeled Gro or Rpd3. After extensive washing, bound proteins were separated by SDS-PAGE and visualized by autoradiography (Fig. 3C,D).35S-labeled Rpd3 was retained on the anti-Flag beads containing purified M2Gro but not on anti-Flag beads alone (Fig. 3C). Furthermore, 35S-labeled Gro specifically bound to anti-Flag beads containing purified M2Rpd3 (Fig. 3D). Thus, these findings strongly suggest a direct interaction between Gro and Rpd3, although we cannot completely exclude the possibility that unknown adapter proteins in the rabbit reticulocyte in vitro translation system participate in this interaction.
Figure 3.
A direct interaction between Gro and Rpd3. (A) Copurification of baculovirus-expressed Flag-tagged Gro (M2Gro) and six-histidine-tagged Rpd3 (H6Rpd3). Nuclear extracts (NE) prepared from insect cells expressing either M2Gro, H6Rpd3, or both together were incubated with Ni2+–NTA–agarose (Ni) or anti-Flag affinity beads (M2). After extensive washing of beads, bound proteins were eluted with SDS, resolved by SDS-PAGE, and immunoblotted with anti-Gro (top) or anti-Rpd3 (bottom) antibodies. (B) A Coomassie blue stained SDS–polyacrylamide gel showing baculovirus-expressed and affinity-purified Flag-tagged Gro (M2Gro) and Flag-tagged Rpd3 (M2Rpd3). (C) Equal amounts of anti-Flag M2 beads (M2) or M2 beads containing purified Flag-tagged Gro (M2Gro) were incubated with 35S-labeled Rpd3 produced by in vitro translation. After subjecting the beads to extensive washing, bound 35S-labeled Rpd3 was eluted, and analyzed by SDS-PAGE and autoradiography. (D) In a reciprocal experiment, M2 beads containing purified M2Rpd3 were used to examine the interaction with 35S-labeled Gro. (1/10 IP) Ten percent of total 35S-labeled protein input.
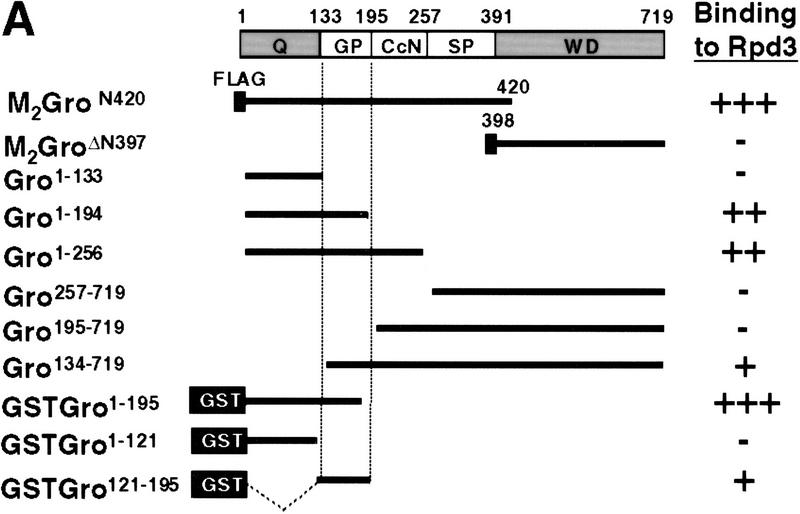
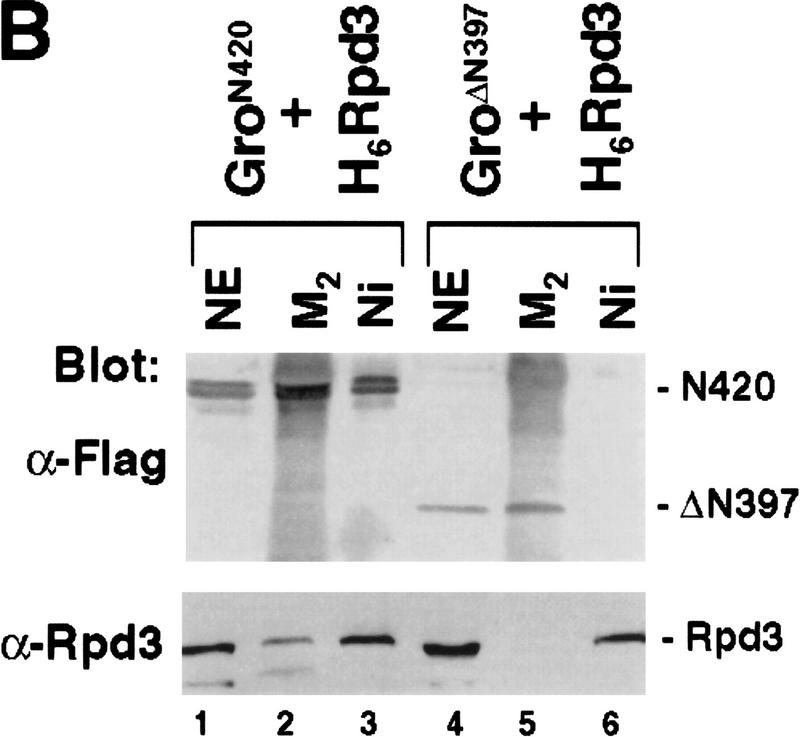
To map the domain(s) of Gro responsible for the interaction with Rpd3, we constructed a series of truncated forms of Gro (Fig. 4A). When M2-Gro lacking the carboxy-terminal WD repeat domain (M2GroN420) was coexpressed with H6Rpd3 in insect cells, we found that M2GroN420 and H6Rpd3 copurified on both Ni2+–NTA–agarose and anti-Flag affinity beads (Fig. 4B, lanes 2,3). In contrast, M22GroΔN397 failed to copurify with H6Rpd3 (Fig. 4B, lanes 5,6). Therefore, the amino-terminal region of Gro is necessary, whereas the WD repeat domain is dispensable for the Rpd3 interaction.
Figure 4.
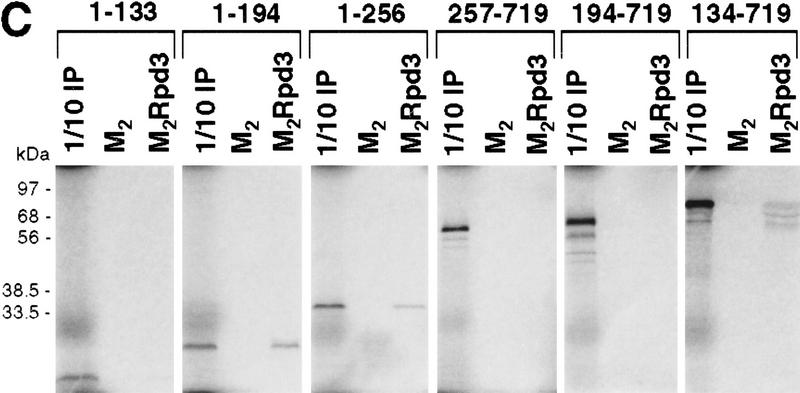
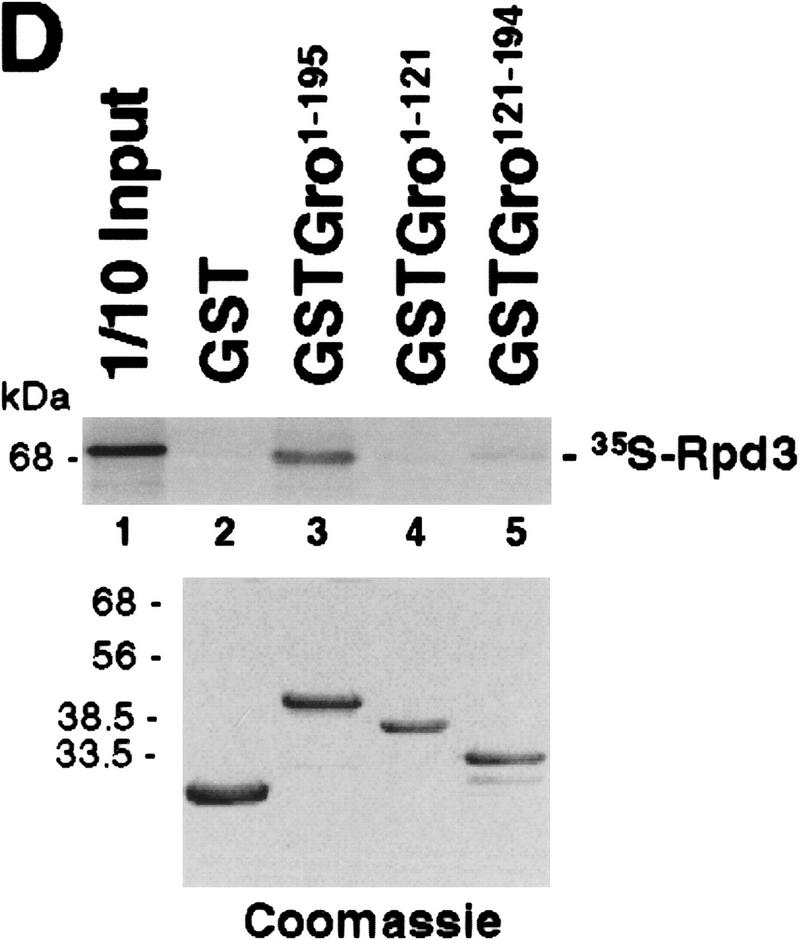
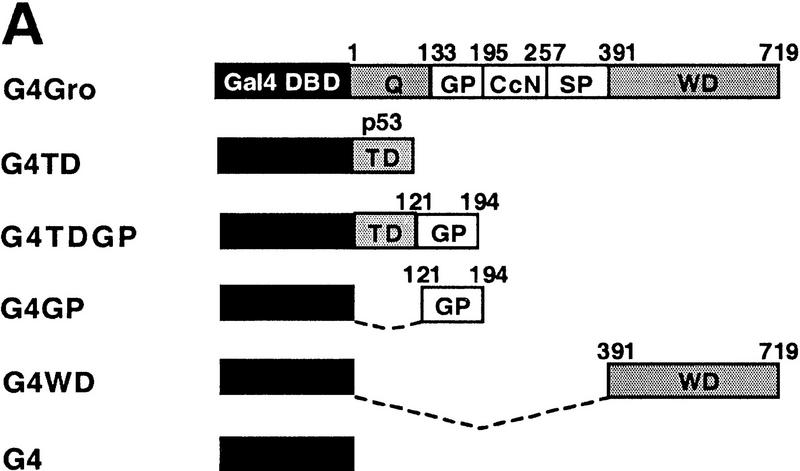
Mapping the Rpd3-interaction domain to the GP region of Gro. (A) Schematic diagram of different Gro deletions used in the protein interaction assays described in B–D. The conserved glutamine-rich (Q) and WD repeat (WD) domains of Gro are shaded. GP and SP denote the glycine/proline and serine/proline rich regions of Gro. CcN represents the motif containing putative cdc2 and casein kinase II phosphorylation sites as well as a nuclear localization signal. (B) The WD-repeat domain of Gro is dispensable for the Rpd3 interaction. Equal amounts of nuclear extracts prepared from insect cells coexpressing a Flag-tagged Gro deletion (M2GroN420 or M2GroΔ397) and H6Rpd3 were affinity purified using Ni2+–NTA–agarose (Ni) or anti-FLAG affinity beads (M2). Purified proteins were immunoblotted with anti-FLAG (top) or anti-Rpd3 (bottom) antibodies. (C) The GP domain of Gro is required for the interaction with Rpd3. Equal amounts of anti-Flag affinity beads alone (M2) or beads containing purified M2Rpd3 were used to examine the interac-tions with the indicated 35S-labeledGro deletions. Bound proteins were resolved by SDS-PAGE and visualized by autoradiography. (1/10 IP) Ten percent of each 35S-labeled protein input. (D) GST pull-down assays confirm the requirement of the GP domain for the Rpd3 interaction. Highly purified GST–Gro fusions were immobilized on glutathione beads and incubated with 35S-labeled Rpd3. After extensive washing of the beads, bound proteins were eluted and separated by SDS-PAGE and visualized by autoradiography (top). (Bottom) A Coomassie blue stained SDS–polyacrylamide gel of the purified GST–Gro fusions.
We further mapped the domain(s) in the amino-terminal region of Gro required for the Rpd3 interaction by incubating in vitro-translated 35S-labeled Gro deletions with anti-Flag affinity beads containing purified M2Rpd3 (Fig. 4C, results summarized in Fig. 4A). Those 35S-labeled Gro variants that contain the glycine/proline-rich (GP) domain bound to beads containing purified M2Rpd3 but not to beads alone. Conversely, those 35S-labeled Gro variants lacking the GP domain failed to associate with M2Rpd3. In addition, a GST pull-down assay using purified GST–Gro fusion proteins (Fig. 4D, bottom) confirmed that the GP domain of Gro was required for the interaction with Rpd3 (Fig. 4D, top; cf. lanes 4 and 5). Furthermore, the deletion of the amino-terminal glutamine-rich (Q) domain of Gro severely reduced the affinity of the interaction (Fig. 4D, cf. lanes 3 and 5). In conclusion, these findings suggest that the GP domain is required for the interaction, whereas the Q domain, which our previous studies have shown is required for Gro tetramerization (Chen et al. 1998), significantly stimulates the interaction.
The interaction between Gro and Rpd3 contributes to Gro-mediated transcriptional repression in cultured cells
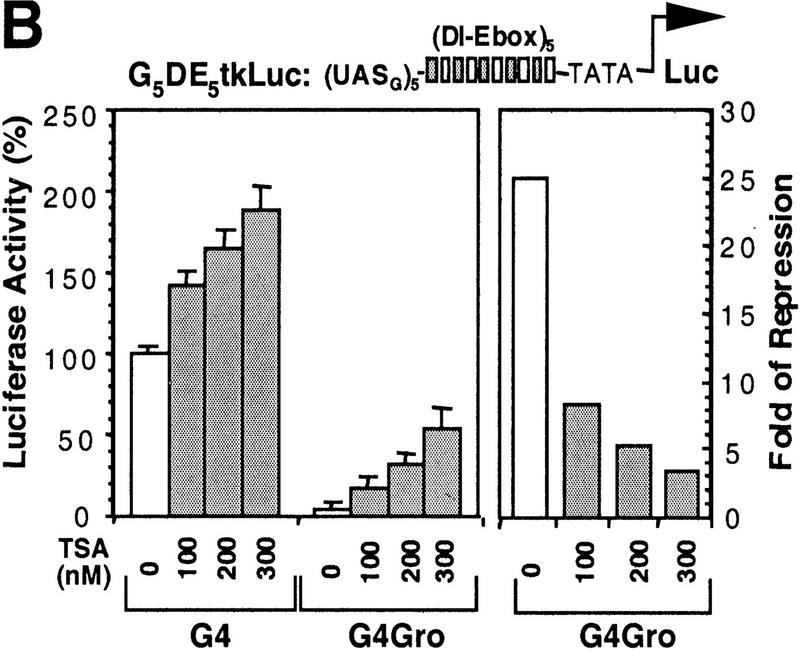
To address whether the interaction between Gro and Rpd3 is functional in Gro-mediated repression, we first determined if histone deacetylase activity was important for transcriptional repression by Gro in cultured cells. We and others have shown that Gro strongly represses activated transcription in S2 cells when directly targeted to a promoter by fusion to the DBD of the yeast transcription factor Gal4 (Fisher et al. 1996; Chen et al. 1998). As shown in Figure 5B (open bars), the Gal4–Gro fusion strongly represses the transcriptional activation promoted by the combination of Dorsal and Twist, when expression vectors encoding these factors were cotransfected with a luciferase reporter (G5DE5tkLuc) driven by a herpes simplex virus thymidine core promoter, an artificial enhancer element containing multimerized Dorsal and Twist binding sites (Dl–Ebox), and multimerized Gal4-binding sites (USAG).
Figure 5.

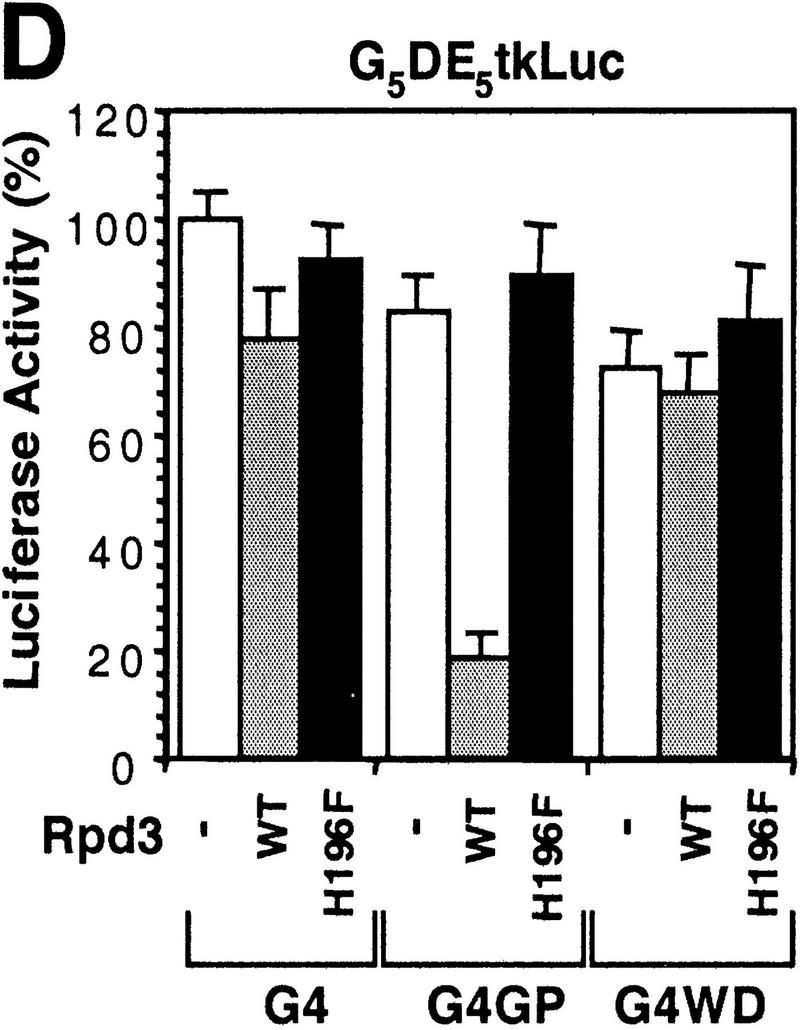
Histone deacetylase activity contributes to Gro-mediated repression in cultured cells. (A) Schematic diagram of various Gal4–Gro fusion constructs used in the cotransfection assays described in B–D. (TD) The tetramerization domain of p53 (residues 309–371). (B) TSA treatment dramatically reduces Gal4–Gro mediated repression. The structure of the firefly luciferase reporter (G5DE5tkLuc) is depicted at top. This reporter and an internal control reporter (p-37tkRLuc) encoding Renilla luciferase were cotransfected into S2 cells with vectors expressing Dorsal, Twist, and the Gal4 DNA-binding domain (G4) or the Gal4–Gro fusion protein (G4Gro). Twenty-four hr post-transfection, cells were treated with the specified amounts of TSA and luciferase activities were measured 10 hr later. All firefly luciferase activities (normalized first to the control Renilla luciferase activities) are normalized to the activity in the presence of Dorsal, Twist, and Gal4 DBD alone, which is set at 100%. Each bar represents the average plus standard deviation of three independent duplicate assays (left). The fold repression (right) reflects the ratio of the activity observed with Gal4 DNA-binding domain alone to that observed with Gal4–Gro at each TSA concentration. (C) The Gro GP domain is able to repress transcription when fused to the Gal4 DBD and the p53 TD (G4TDGP). However, the GP domain alone (G4GP) and the p53 TD alone (G4TD) fail to repress transcription when fused to the Gal4 DNA-binding domain. Cotransfection assays were conducted as described in B. (D) The Gal4–GP fusion protein synergizes with the enzymatically active form of Rpd3 to repress transcription. S2 cells were transfected with the reporters and expression constructs encoding Dorsal, Twist, and the indicated Gal4 fusion protein in the absence (−, open bars) or presence of a vector expressing either wild-type (WT, shaded bars) or single-point mutant (H196 F, solid bars) forms of Rpd3.
Next, we treated transfected S2 cells with the histone deacetylase inhibitor TSA to determine whether deacetylase activity is important for Gal4–Gro-mediated repression. As shown in Figure 5B (shaded bars), TSA treatment dramatically reduced Gal4–Gro-mediated repression, suggesting that histone deacetylation does contribute to this repression. Introduction of TSA resulted in a small (less than 2-fold) increase in the level of reporter activity in the absence of Gal4–Gro, but a much larger (up to 20-fold) increase in the level of reporter activity in the presence of Gal4–Gro (Fig. 5B, left). As a result, the calculated repression by Gal4–Gro decreases from 25-fold in the absence of TSA to ∼3-fold in the presence of 300 nm TSA (Fig. 5B, right). The residual repression observed at high concentrations of TSA suggests that although full repression of transcription by Gro requires histone deacetylase activity, Gro may also utilize histone deacetylase-independent mechanisms for transcriptional repression.
In agreement with the finding that the GP domain of Gro is required for the interaction with Rpd3, this GP domain functions as a repression domain (Fig. 5C, constructs shown in Fig. 5A), when fused to the Gal4 DBD and the tetramerization domain (TD) of p53 (construct G4TDGP). The p53 TD, which itself does not repress transcription when fused to the Gal4 DBD (Fig. 5C, construct G4TD), was utilized in place of the Gro TD to avoid the repression activity that is believed to result from the association of Gal4–Gro TD fusion with endogenous full-length Gro. In the absence of the p53 TD, the Gal4–GP domain fusion (Fig. 5C, construct G4GP) failed to repress transcription. These findings suggest that efficient repression by the GP domain of Gro requires tetramerization. This agrees with our previous finding that the Q domain is required for efficient binding between Gro and Rpd3 (Fig. 4D) and with our previously published results showing that efficient repression by Gro requires a functional tetramerization domain (Chen et al. 1998).
The GP domain synergizes with an enzymatically active form of Rpd3 to repress transcription
To further determine whether histone deacetylase activity is critical for the function of the Gro GP domain, we generated a single point mutation in Rpd3 in which a highly conserved histidine residue (H196) is replaced with a phenylalanine (H196F) (Fig. 6A). Consistent with studies on mammalian histone deacetylases (Hassig et al. 1998), we found that the mutation decreased the specific activity of the enzyme by about sevenfold (Fig. 6B). Additionally, using a GST pull-down assay, we found that both Rpd3WT and Rpd3H196F bound to Gro with comparable affinity (Fig. 6C). Unlike Rpd3WT, which repressed transcription three- to fourfold when fused to the Gal4 DBD, the Gal4–Rpd3H196F fusion failed to repress transcription in a similar assay (Fig. 6D). Therefore, Rpd3H196F represents an enzymatically inactive form of Drosophila histone deacetylase that binds to Gro with the same affinity as wild-type Rpd3.
Figure 6.
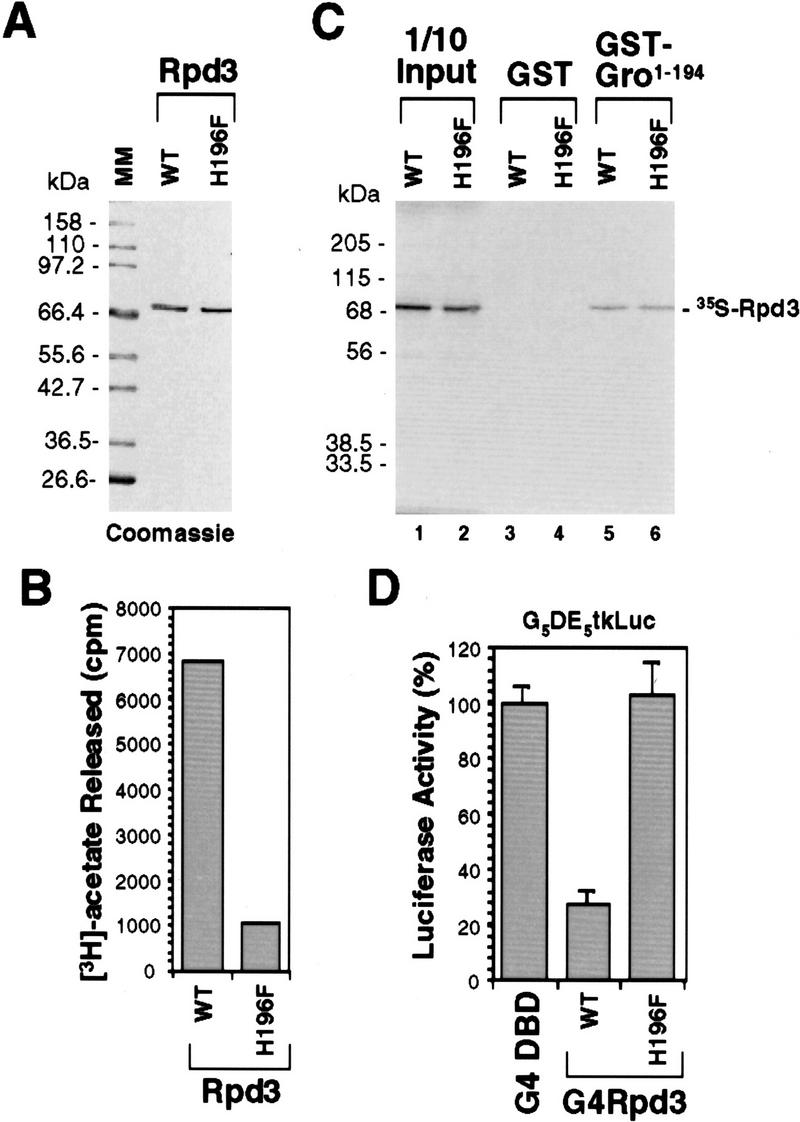
Functional analysis of a single point mutation of Rpd3. (A) A Coomassie blue-stained SDS–polyacrylamide gel showing equal amounts of purified wild-type (WT) and mutant forms of Rpd3. In mutant Rpd3, a conserved histidine residue (H196) has been changed to phenylalanine (H196F). (B) In vitro histone deacetylase assays of purified wild-type (WT) and mutant (H196F) forms of Rpd3. (C) GST pull-down assays show that wild-type (WT) and mutant (H196F) forms of Rpd3 bind to Gro with comparable affinities. GST-pull down assays are conducted as described in Fig. 4D. (D) Gal4 fused to wild-type Rpd3 (WT) is able to repress transcription in S2 cells, whereas Gal4 fused to the single-point mutant form of Rpd3 (H196F) is not able to repress transcription in S2 cells. Cotransfection assays were conducted as described in Fig. 5C.
As shown above, the Gal4–GP domain fusion does not repress activated transcription on its own due to the lack of a tetramerization domain (Fig. 5C). However, we discovered that the Gal4–GP fusion was able to synergize with cotransfected wild-type Rpd3 to repress transcription (Fig. 5D) but not with the mutant catalytically inactive form of Rpd3. As controls, we showed that neither the Gal4 DBD (G4) alone nor the Gal4–WD repeat domain fusion (G4WD) was able to synergize with Rpd3 to repress transcription (Fig. 5D). These results strongly suggest that the GP domain contributes to Gro-mediated repression by interacting directly with the histone deacetylase Rpd3 and that the histone deacetylase activity of Rpd3 is essential for its ability to contribute to Gro-mediated repression.
A genetic interaction between gro and rpd3
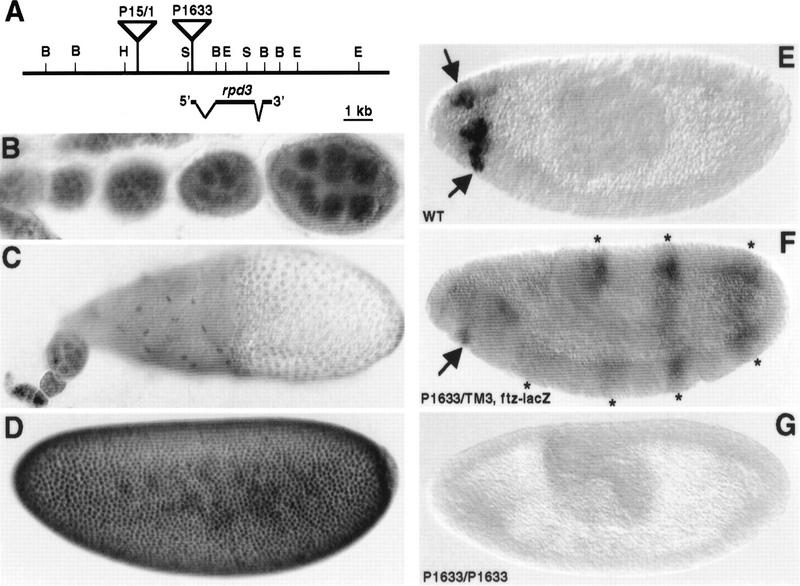
Drosophila gro is a maternally required gene that has critical roles in multiple developmental processes, including anterior/posterior and dorsal/ventral pattern formation, neurogenesis, and sex determination (Fisher and Caudy 1998; Parkhurst 1998). To determine whether or not Rpd3 could possibly interact with Gro to help mediate these processes, we examined the distribution of this protein in ovaries and embryos. Immunostaining of wild-type ovaries indicated that Rpd3 protein was ubiquitously present in the nuclei of both nurse and follicle cells throughout oogenesis (Fig. 7B,C). In addition, Rpd3 protein was detected in all the nuclei of the syncytial blastoderm embryo (Fig. 7D). Rpd3 protein levels dropped significantly and remained low during gastrulation and at later stages of embryogenesis. However, a high level of spatially restricted expression was observed in the head region of stage 9–10 embryos (Fig. 7E, arrows).
Figure 7.
Spatial expression pattern of rpd3 during Drosophila oogenesis and embryogenesis. (A) Genomic organization of the rpd3 gene. The map of the rpd3 transcription unit (at cytological map location 64C1-2) is based on previous reports (DeRubertis et al. 1996; Maixner et al. 1998). BamHI (B), EcoRI (E), HindIII (H), and SalI (S) sites are indicated. The P1633 and P15/1 P-element insertion sites are depicted by inverted triangles. (B–E) Staining of wild-type ovaries and embryos with anti-Rpd3 antibodies. (B) Ubiquitous germ-line nuclear expression of Rpd3 is observed in the wild-type ovary during early oogenesis (before stage 8). (C) Uniform expression of Rpd3 is observed in follicle cell nuclei by stage 10 of oogenesis. (D) Rpd3 is uniformly distributed throughout the nuclei of precellular embryos. (E) Patches of zygotic expression (arrows) are observed in the anterior region of stage 9–10 embryos. (F,G) Characterization of Rpd3 expression in embryos carrying the P1633 P-element insertion. (F) Expression of Rpd3 in the an-terior of stage 9–10 embryos (arrows) heterozygous for the P-element insertion is reduced relative to wild-type embryos (see E). The seven-stripes of lacZ expression due to the ftz–lacZ marker on the TM3 balancer chromosome are indicated with asterisks. (G) Expression of Rpd3 in stage 9–10 embryos homozygous for the P-element insertion is undetectable. Homozygous embryos were recognized by the absence of the ftz–lacZ stripes.
We obtained a strain carrying single recessive lethal P-element insertion (P1633, obtained from the Bloomington Stock Center; Spradling et al. 1995) within the rpd3 transcription unit (47 bp from the transcription start site as determined by the Berkeley Drosophila Genome Project). Embryos heterozygous for this P insertion showed reduced levels of Rpd3 staining in the head (Fig. 7F), whereas homozygous embryos displayed no detectable expression in the head (Fig. 7G). In addition, this P1633 insertion failed to complement a deletion that removes the rpd3 gene entirely, Df(3L)10H. Therefore, P1633 likely represents a null or strong hypomorphic allele of rpd3.
To look for evidence for an interaction between maternally expressed gro and rpd3, we scored the frequency of unhatched embryos produced by females carrying various combinations of gro and rpd3 alleles (Fig. 8A; in this experiment two gro alleles were used: groE48, a strong hypomorphic allele and groBX22, a null allele). Approximately 3%–4% of embryos produced by mothers singly heterozygous for either groBX22, groE48, P1633, or Df(3L)10H failed to hatch. This frequency was not significantly greater than that observed for embryos derived from wild-type mothers. However, embryos laid by females doubly heterozygous for gro and rpd3 alleles [groBX22/P1633, groE48/P1633, groBX22/Df(3L)10H, and groE48/Df(3L)10H] showed a dramatic increase in embryonic lethality—16%–30% of the embryos failed to hatch. These synergistic effects on embryonic lethality suggest that gro and rpd3 function together during Drosophila oogenesis and/or embryogenesis.
Figure 8.
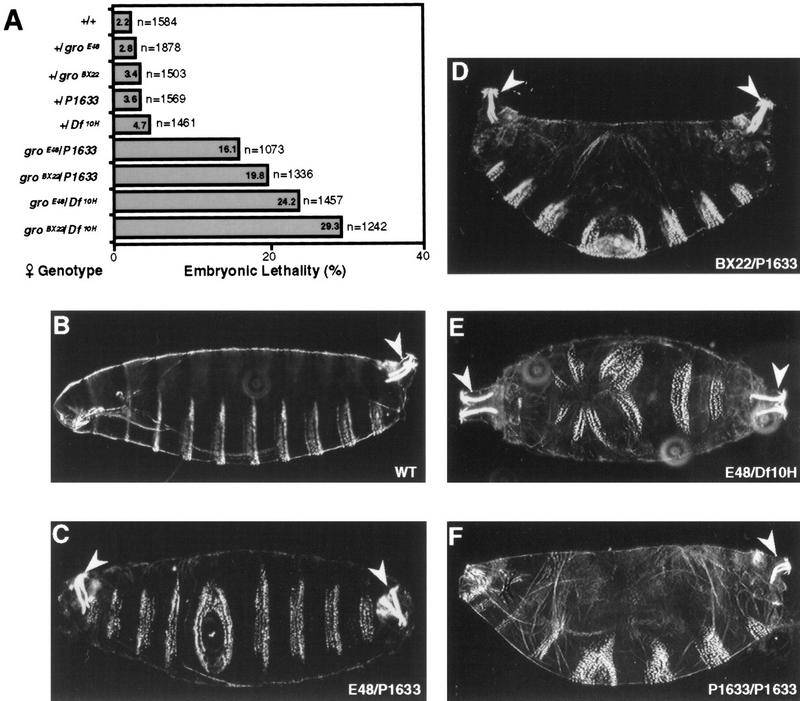
Genetic interaction between gro and rpd3. (A) Embryos were collected from females of the indicated genotypes that had been mated with wild-type males. After 48 hr at 28°C, unhatched embryos were scored to calculate embryonic lethality. The number of embryos scored and percent lethality (%) are indicated to the right and the inside of each bar, respectively. (B) Cuticle of an embryo derived from a wild-type mother (WT). (C,D) Cuticles of unhatched embryos derived from mothers trans-heterozygous for either of two gro alleles (E48 or BX22) and the P-insertional allele of rpd3 (P1633) showing the bicaudal phenotype including a duplicated posterior spiracle (arrowheads) and a mirror-image duplication of the posterior abdominal segments in place of normal anterior segments. (E) Cuticle of unhatched embryo derived from mother trans-heterozygous for groE48 and a deficiency that removes rpd3 (Df10H). Anterior duplication of the posterior spiracle (arrowheads) is accompanied by disorganized denticle belts. (F) Cuticle of an embryo derived from a female containing germ-line clones homozygous for P1633, the P-insertional allele of rpd3. The embryo exhibits a pair-rule phenotype.
Cuticles prepared from the unhatched progeny derived from females singly heterozygous for either gro or rpd3 were indistinguishable from wild type (Fig. 8B; data not shown). However, those unhatched embryos produced by mothers doubly heterozygous for gro and rpd3 often displayed cuticles with striking abnormalities in anterior/posterior pattern formation (Fig. 8C–E). We consistently observed that the majority of the unhatched embryos (>70%) generated by the groE48/P1633 and groBX22/P1633 trans-heterozygous mothers displayed replacement of anterior embryonic segments by a mirror-image duplication of the three to five posterior-most segments (Fig. 8C,D). Mirror-image duplicated structures included denticle belts and posterior spiracles. The remaining unhatched embryos were normal or only had minor defects in head structures (data not shown).
The unhatched embryos collected from the groBX22/Df(3L)10H, and groE48/Df(3L)10H trans-heterozygous mothers displayed more severe cuticle phenotypes. Most (50%–60%) had no cuticle, whereas 5%–10% showed a mirror-image duplication of the posterior spiracle and disordered denticle belts (Fig. 8E; data not shown). The remaining unhatched embryos displayed cuticles that were nearly wild type or had minor defects in head structures (data not shown).
We have also analyzed the cuticle phenotype of embryos derived from mosaic females containing P1633 homozygous germ-line clones. Consistent with previously described results (Perrimon et al. 1996), we observed that the majority of the embryos (>65%) lacking maternally expressed rpd3 failed to hatch, and most of those unhatched embryos (>80%) exhibited variable pair–rule segmentation defects (Fig. 8F). In particular, we observed partial or complete fusion of adjacent denticle belts resulting in embryos with five to eight thoracic and abdominal segments. In addition, variable, often severe, defects were observed in the head skeleton. Gro has been shown to interact physically with the pair–rule gene products Hairy and Runt and to be required for their function as transcriptional repressors. Therefore, the pair–rule phenotype observed in embryos lacking maternal Rpd3 suggests that like Gro, Rpd3 may also be involved in the repression mediated by certain pair–rule gene products.
Discussion
In this report we describe the use of Gro as an affinity reagent to purify proteins from Drosophila embryo extracts that bind Gro in a specific manner. One of these proteins was found to be the histone deacetylase Rpd3. An analysis involving protein–protein interaction, cotransfection, and genetic assays suggests that Gro and Rpd3 form a complex in vivo, that they interact via the GP-rich domain in Gro, and that this interaction contributes to transcriptional repression. Thus, one mechanism by which Gro mediates transcriptional repression appears to involve the direct recruitment of Rpd3 to the template, where it can modulate local chromatin structure.
A physical interaction with the histone deacetylase Rpd3 contributes to Gro-mediated transcriptional repression
A breakthrough in our understanding of the role of chromatin remodeling in repression came with the finding that a mammalian nuclear histone deacetylase exhibits extensive homology with yeast Rpd3 (Taunton et al. 1996), a transcriptional corepressor. Nuclear histone deacetylases were found to exist in large multiprotein complexes that include the well-known Sin3 corepressor and a tightly associated WD-repeat-containing cofactor, RbAp48. Histone deacetaylase complexes are capable of repressing transcription when they are recruited to the template by a variety of DNA-binding repressors (e.g., Ume6, Mad, and YY1). These complexes are also found in association with other corepressors, for example, NCoR, which mediates transcriptional repression by the nuclear hormone receptors (for review, see Pazin and Kadonaga 1997; Struhl 1998).
In this study we have found that a GP-rich region in Gro appears to mediate the recruitment of Drosophila Rpd3 to the template. The recruitment of mammalian histone deacetylases by YY1 also requires a glycine-rich motif, which, like the Gro GP domain, is rich in positively charged residues (Yang et al. 1996). It will be interesting to determine if these domains are representative of a widespread structural motif involved in repression. Because a glycine/proline rich domain is found in most Gro/TLE family proteins, it will also be interesting to determine whether the GP domains in mammalian Gro/TLE proteins are capable of repressing transcription via interactions with histone deacetylases.
Previously, we have identified a highly conserved putative paired amphipathic α-helix (PAH) motif in the amino-terminal Q domain of Gro. The PAH motif mediates Gro tetramerization, which appears to be indispensable for Gro-dependent repression in vivo (Chen et al. 1998). In this study we demonstrate that the Q domain is also required for efficient interaction with Rpd3, although the Gro GP domain itself seems to be sufficient for the interaction. Whereas the Q domain may form a part of the interface with Rpd3, the requirement for the tetramerization domain may simply reflect a need for multiple copies of the GP-rich domain to form a high-affinity Rpd3 interaction surface. Support for this idea comes from our findings that the p53 tetramerization domain can substitute for the Q domain in transcriptional repression and that overexpression of Rpd3 eliminates the need for the Q domain.
Enzymatic activity of histone deacetylase is required for Gro-mediated repression
We have generated and characterized a point mutation of Rpd3 in which a highly conserved histidine is converted to phenylalanine. It has been suggested previously that this histidine has a direct role in catalysis, perhaps by coordinating a catalytically active metal ion (Hassig et al. 1989; Kadosh and Struhl 1998). Mutation of the histidine not only dramatically reduces histone deacetylase activity but also abolishes the ability of a Gal4–Rpd3 fusion to mediate transcriptional repression in S2 cells. Furthermore, although overexpressed wild-type Rpd3 synergistically represses transcription in combination with the Gal4–GP domain fusion protein, the mutant form of Rpd3 does not synergize with the Gal4–GP fusion even though the mutation does not interfere with the binding of Gro to Rpd3. These findings are consistent with previous studies showing that the corresponding histidine in mammalian histone deacetylases is critical for catalytic activity and transcriptional repression (Hassig et al. 1998; Kadosh and Struhl 1989; Zeng et al. 1998).
Implications of the association between histone H1 and Gro
In addition to Rpd3, Drosophila histone H1 was also found to associate with Gro. H1 is a linker histone that stabilizes higher-order chromatin structure (Ramakrishnan 1997; Belikov and Karpov 1998). Several previous findings suggest that the interaction between Gro and H1 may be functionally relevant to Gro-mediated repression. Drosophila H1 was purified and identified as a general inhibitor of transcription by RNA polymerase II in vitro (Croston et al. 1991; Laybourn and Kadonaga 1991). Genetic analysis has indicated that H1 does not appear to affect global transcription but, instead, functions as a gene-specific transcriptional repressor (Shen and Gorovskiy 1996). In addition, the expression of genes encoding various H1 subtypes is developmentally regulated. Thus, it is possible that the interaction between Gro and H1 functions in a developmentally regulated manner to facilitate chromatin condensation and/or establish a repressive chromatin enviroment for transcription.
The functional role of Rpd3 in Drosophila development
Our results indicate that simultaneous reduction of the maternal gro and rpd3 gene dosage results in synergistic effects on embryonic viability. In particular, we found that embryos produced by females doubly heterozygous for either of two alleles of gro and for a P-insertional allele of rpd3 (P1633) or a deficiency that removes rpd3 exhibited greatly increased embryonic lethality relative to embryos produced by wild-type or single mutant females. The unhatched embryos often exhibited a bicaudal defect. This defect is similar to that observed for maternal effect mutations in a number of genes including bicaudal and Bicaudal D. Interestingly, the distribution of phenotypes among the embryos produced by gro/P1633 females appears to be bimodal in nature. Sixteen percent to 20% of these embryos failed to hatch. Of these unhatched embryos, ∼70% exhibit the bicaudal defect, whereas most of the remaining embryos have wild-type or near wild-type cuticles. Very few cuticles with intermediate phenotypes are observed. This bimodal distribution is extremely similar to what has been reported for hypomorphic alleles of bicaudal (Nüsslein-Volhard 1979).
The most likely explanation for the bicaudal phenotype is the misexpression and/or mislocalization of components of the maternal posterior pattern forming system, although this may be difficult to assess directly due to the low overall penetrance of the phenotype. Thus, these findings suggest a heretofore unknown role for Gro in regulating the expression of genes involved in the localization of maternal determinants during oogenesis. Consistent with this possibility, both gro and rpd3 are expressed at the protein level in the maternal germ line.
Embryos derived from female germ-line clones homozygous for the P1633 allele of rpd3 exhibit a pair–rule segmentation defect (Fig. 8F; Perrimon et al. 1996). gro is also required for segmentation, and, in particular for the function of the pair–rule genes hairy and runt (Paroush et al. 1994), suggesting that gro and rpd3 may function together in segmentation. Although the maternal effect phenotypes of gro and rpd3 indicate roles for both factors in segmentation, the elimination of maternal Gro results in additional pleiotropic effects. In particular, the complete elimination of maternal Gro activity results in embryos that fail to gastrulate normally and that fail to deposit cuticle. These more extreme phenotypes probably result, in large part, from the roles of Gro in the dorsal/ventral and terminal pattern forming systems (Dubnicoff et al. 1997; Paroush et al. 1997).
There are several possible explanations for the observation that the rpd3 germ-line clone phenotype is weaker than the gro germ-line clone phenotype. First, the P1633 allele, which represents a P-element insertion into the 5′-untranslated leader of rpd3, may be leaky. Although we do not detect Rpd3 expression in a homozygous P1633 genetic background, it is possible that some expression below the level of detection persists. Second, there may be redundant histone deacetylases that can partially compensate for the absence of Rpd3. Both vertebrates and yeast contain multiple histone deacetylases (Grozinger et al. 1999). In Drosophila, three such enzymes, Rpd3, DmHDAC2, and DmHDAC3 (Johnson et al. 1998 and unpubl.), have been reported, and given the large number of histone deacetylases in other eukaryotes, the fruit fly genome may encode additional members of this family. Finally, perhaps Rpd3 is only required for some, but not all of Gro’s roles in development. For example, perhaps the form of Gro recruited to the template by Hairy, a transcription factor involved in segmentation, utilizes Rpd3 for repression, whereas the form of Gro recruited to the template by Dorsal, a transcription factor involved in dorsal/ventral pattern formation, functions in an Rpd3-independent manner. In other words, Dorsal and Hairy may recruit different Gro-containing multiprotein complexes to the template, and these different complexes might repress transcription in different ways. In support of this idea, we note that Hairy recruits Gro via a simple WRPW motif, whereas Dorsal lacks such a motif. Furthermore Dorsal-mediated repression requires additional DNA bound repressor proteins, whereas Hairy-mediated repression does not.
Drosophila rpd3 was originally identified through the characterization of a P-element insertion (P15/1) that enhances position-effect variegation (PEV) (De Rubertis et al. 1996). This suggests that Rpd3 functions to counteract the silencing that results from the insertion of genes in or near heterochromatin. This contrasts with the general idea that histone deacetylases promote the repressed state. However, the P1633 P-element insertion does not have any effect on PEV (G. Chen, unpubl.), suggesting that the P15/1 allele may not be a simple loss-of-function rpd3 allele.
A model for Gro-mediated transcriptional repression
The findings reported here, as well as those from previous studies showing that Gro family proteins make specific contacts with the amino-terminal tails of core histones (Palaparti et al. 1997 and unpubl.), suggest that Gro represses transcription by inducing a silenced chromatin conformation. We thus propose that after a direct interaction with DNA-binding transcription factors brings Gro to the template, the known ability of Gro to oligomerize together with the known favorable interactions between Gro and core histones and/or histone H1 result in the nucleation of a Gro polymer that spreads along the template. Template-bound Gro may then provide an interface for the recruitment of key chromatin-modifying enzymes including histone deacetylases to the template. These enzymes may then serve to modulate local higher order chromatin structure to establish a transcriptionally silenced domain. It is known that the interaction between Tup1 (a possible yeast homolog of Gro) and core histones is actually enhanced by histone deacetylation (Edmondson et al. 1996), so it is possible that histone deacetylation also serves to facilitate the further recruitment of the corepressor to the template, thereby reenforcing the transcriptionally repressed state.
Although histone deacetylation contributes to Gro-mediated repression, there are likely to be additional mechanisms by which Gro represses transcription. For example, our cotransfection experiments show that Gro possesses some repression activity that is resistant to the deacetylase inhibitor TSA. In addition, the WD repeat domain of Gro functions as a weak repression domain (Fisher et al. 1996 and unpubl.), and this repression activity appears to be independent of Rpd3. Consistent with the idea that corepressors can modulate transcription by multiple mechanisms, histone deacetylase-independent repression has also been observed for the Sin3 corepressor (Laherty et al. 1997). It will thus be interesting to determine the identities of additional Gro-interacting proteins to see if they provide any clues to these possible alternative mechanisms of repression.
Materials and methods
Plasmids
Gro constructs used for producing in vitro-translated Gro and Gro deletions have been described before (Chen et al. 1998). Plasmids pGEX4T1–Gro(1–194), –Gro(1–21), and –Gro(121–194) for Escherichia coli expression of GST–Gro fusion proteins were constructed by inserting PCR-generated DNA fragments encoding regions of Gro into the BamHI–NotI sites of pGEX4T1 (Pharmacia). Plasmids pVL1392–M2Gro(1–420) and pVL1392–M2Gro(398–719) for baculovirus expression of Flag-tagged Gro truncations were generated by digesting pVL1392–M2Gro (kindly provided by J. Zwicker and R. Tjian, UC, Berkeley) with NdeI or EcoRI, respectively, and then religating the larger DNA fragments. Gal4–Gro fusion constructs, the pG5DE5tkLuc reporter and the p-37tkRLuc internal control reporter have been described previously (Chen et al. 1998).
Plasmid pGEM3Zf(+)–Rpd3 for producing in vitro-translated Rpd3 protein was generated by inserting SacI–KpnI DNA fragment encoding full-length Rpd3 into pGEM3Zf(+) vector (Promega). Plasmids pAcHLT–B–Rpd3 and pVL1392–M2Rpd3 for baculovirus expression of six-histidine-tagged and Flag-tagged Rpd3 were constructed by cloning either a SacI–KpnI or a NdeI–XbaI DNA fragment encoding Rpd3 into pAcHLT–B (Pharmingen) and pVL1392–M2Gro, respectively. For expression of the Gal4–Rpd3 fusion and Flag-tagged Rpd3 in S2 cells, a PCRproduced DNA fragment encoding full-length Rpd3 was cloned into the KpnI site of pActGal4(1–147)–SK and pPac–Flag. pActGal4(1–147)–SK was constructed by inserting a double-standed oligonucleotide polylinker containing SacI and KpnI sites into the BamHI–BglII site of pActGal4–Gro (Fisher et al. 1996). Similarly, pPacFlag was generated by inserting a polylinker containing the Flag epitope coding sequence and multiple cloning sites (KpnI–NotI–SacI–XhoI–BamHI) into the BamHI site of the pPac vector.
The single-point mutation in Rpd3 (pGEM3Zf(+)–Rpd3H196F) was generated with the QuickChange site-directed mutagenesis kit (Stratagene) using pGEM3Zf(+)–Rpd3 as template. The sequence of the coding strand-mutagenic oligonucleotide primer was 5′-ATGACTGTCAGCTTCTTCAAGTACGGAGAGTAT-3′ (mutated base pairs are underlined). The point mutation was confirmed by DNA sequencing.
Fly stocks
The fly stocks used in this study are as follows: P[ry+ = pZ, l(3)04556], ry+506/TM3, ryRK (referred to as P1633; obtained from Bloomington Stock Center); Df(3L)10H[src−]/TM6B [referred to as Df(3L)10H, provided by C.S. Goodman, UC, Berkeley); groE48/TM3, a hypomorphic gro point-mutant line; groBX22/TM3, a deficiency line that lacks gro and several neighboring genes in the Enhancer of split complex; TM6 Tb/TM3 ftz–lacZ Sb used for generating P1633/TM3 ftz–lacZ Sb line (a gift from J. Lengyel, UCLA). For making female P1633 homozygous germ line clones, P1633 FRT2A/TM3 Sb was kindly provided by N. Perrimon (Harvard Medical School, Boston) and other ovoDFRT2A and hspFLP22 lines were ordered from the Bloomington Stock Center.
Antibodies and coimmobilization assays
The anti-Gro monoclonal antibody (mouse) was a gift from S. Stifani (McGill University, Montreal, Quebec, Canada), and affinity-purified anti-Rpd3 polyclonal antibody (rabbit) was kindly provided by P. Spierer. The anti-Flag M2 monoclonal antibody (mouse) and anti-Flag M2 affinity resin were purchased from Sigma. The anti-β-galactosidase monclonal antibody was purchased from Promega.
Baculovirus expression and purification of Flag-tagged Gro or Rpd3 and six-histidine tagged Rpd3 using anti-Flag affinity beads or Ni2+–NTA–agarose beads were performed as described previously (Valentine et al. 1998; Chen and Courey 1999). Immunoprecipitations with anti-Gro antibody were conducted using the IMMUNOcatcher system (CytoSignal). In vitro immunoprecipitations using M2 affinity beads immobilized with purified Flag-tagged proteins to analyze protein–protein interactions were conducted as described previously (Chen et al. 1998). Bacterial expression and purification of GST–Gro fusions, as well as GST pull-down assays, were conducted as described before . In vitro-translated and [35S]methionine-labeled proteins were produced using the TNT T7-coupled reticulocyte lysate system according to the procedure provided by the manufacturer (Promega).
Affinity purification and peptide sequencing
For large-scale affinity purification of Gro-interacting proteins, anti-Flag M2 affinity resin containing ∼0.6 mg of purified M2Gro was incubated at 4°C overnight with 60 ml of nuclear extracts (Soeller et al. 1988) from ∼260 grams of 0- to 12-hr Drosophila embryos. Beads were then washed extensively with the HEMG buffer [25 mm HEPES at pH 7.6, 0.1 mm EDTA, 2.5 mm MgCl2, 10% glycerol, 1 mm dithiothreitol (DTT), 1 mm sodium metabisulfite, 0.2 mm phenylmethylsulfonyl fluoride (PMSF)] containing 100 mm KCl. After brief washing with HEMG buffer containing 0.4 m KCl, bound proteins were eluted with HEMG buffer containing 0.4 m KCl, 0.02% NP-40, and 2 mg/ml Flag peptide. The eluted fractions were pooled, concentrated, and resolved on a Coomassie blue-stained preparative SDS–polyacrylamide gel. The excised polypeptide bands were digested with lysC endoprotease, further fractionated by reverse-phase high-pressure liquid chromatography, and sequenced by automated Edman degradation.
In vitro histone deacetylase assays and cotransfection assays
In vitro histone deacetylase assays were performed using 3H-labeled acetyl-labeled histones purified from HeLa cells (kindly provided by A.A. Carmen and M. Grunstein) and a modified form of a previously described procedure (Carmen et al. 1996). Fifty microliters of immunoprecipitated protein or ∼0.5 μg of purified Rpd3 was incubated with 20 μg of purified histones (∼20,000 cpm) in an assay buffer containing 75 mm Tris-HCl (pH 7.0), 200 mm NaCl, and 0.1 mm EDTA (total volume 200 μl) at 37°C for 30 min. The reactions were stopped by the addition of 0.12 m acetic acid and 0.72 m HCl, and extracted with 2 volumes of ethylacetate. Released 3H-labeled acetate in 200 μl of ethylacetate was measured in a liquid scintillation counter. Assays were performed in duplicate and the data represent the average of two independent duplicate assays.
Calcium phosphate cotransfections into Drosophila S2 cells and dual-luciferase reporter assays (Promega) were performed as described previously (Chen et al. 1998).
Immunohistochemistry
Whole-mount immunohistochemical staining of ovaries was performed as described previously (Suter and Steward 1991), and egg chambers were staged according to Spradling (1993). Immunohistochemical detection of Rpd3 in embryos was performed according to standard procedures using a 1:400 dilution of anti-Rpd3 antibody and 1:1000 dilution of anti-β-galactosidase antibody in combination with appropriate alkaline phosphatase-coupled secondary antibodies. The embryos were staged as described by Campos-Ortega and Hartenstein (1985).
Acknowledgments
We thank P. Spierer for providing the anti-Rpd3 antibody and Rpd3 cDNA, S. Stifani for anti-Gro and pan-TLE antibodies, A.A. Carmen and M. Grunstein for purified and 3H-labeled histones, J. Zwicker and R. Tjian for M2-baculovirus transfer vectors, N. Perrimon for the P1633 FRT2A strain, and C.S. Goodman for the Df(3L)10H strain. We are grateful to J. Lengyel for her invaluable assistance in analyzing the cuticle preparations and for her very helpful comments on the manuscript. This work was supported by a grant to A.J.C. from the National Institutes of Health (GM44522).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
Since submission of this paper, Mannervik and Levine (Proc. Natl. Acad. Sci. 96: 6797–6801) have published an analysis of the rpd3 germ-line clone phenotype that suggests an essential role for Rpd3 in Even-skipped–directed repression.
Footnotes
E-MAIL courey@chem.ucla.edu; FAX (310) 206-4038.
References
- Alifragis P, Poortinga G, Parkhurst SM, Delidakis C. A network of interacting transcriptional regulators involved in Drosophila neural fate specification revealed by the yeast two-hybrid system. Proc Natl Acad Sci. 1997;94:13099–13104. doi: 10.1073/pnas.94.24.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen JP. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikov S, Karpov V. Linker histones: Paradigm lost but questions remain. FEBS Lett. 1998;441:161–164. doi: 10.1016/s0014-5793(98)01512-9. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embyronic development of Drosophila melanogaster. Berlin, Germany: Springer-Verlag; 1985. [Google Scholar]
- Carmen AA, Rundlett SE, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, Peifer M, Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chen G, Courey AJ. A baculovirus transfer vector for eukaryotic expression and affinity purification of Gal4-fusion proteins. Biotechniques. 1999;26:808–814. doi: 10.2144/99265bm02. [DOI] [PubMed] [Google Scholar]
- Chen G, Nguyen PH, Courey AJ. A role for Groucho tetramerization in transcriptional repression. Mol Cell Biol. 1998;18:7259–7268. doi: 10.1128/mcb.18.12.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croston GE, Kerrigan LA, Lira LM, Marshak DR, Kadonaga JT. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Ruiz-Gomez M. groucho and hedgehog regulate engrailed expression in the anterior compartment of the Drosophila wing. Development. 1995;121:3467–3476. doi: 10.1242/dev.121.10.3467. [DOI] [PubMed] [Google Scholar]
- De Rubertis F, Kadosh D, Henchoz S, Pauli D, Reuter G, Struhl K, Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature. 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- Dubnicoff T, Valentine SA, Chen G, Shi T, Lengyel JA, Paroush Z, Courey AJ. Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes & Dev. 1997;11:2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Caudy M. Groucho proteins: Transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes & Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbavec D, Stifani S. Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem Biophys Res Commun. 1996;223:701–705. doi: 10.1006/bbrc.1996.0959. [DOI] [PubMed] [Google Scholar]
- Grbavec D, Lo R, Liu Y, Stifani S. Transducin-like Enhancer of split 2, a mammalian homologue of Drosophila Groucho, acts as a transcriptional repressor, interacts with Hairy/Enhancer of split proteins, and is expressed during neuronal development. Eur J Biochem. 1998;258:339–349. doi: 10.1046/j.1432-1327.1998.2580339.x. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JC, Tse C, Wolffe AP. Structure and function of the core histone N-termini: More than meets the eye. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- Hartley DA, Preiss A, Artavanis-Tsakonas S. A deduced gene product from the Drosophila neurogenic locus, enhancer of split, shows homology to mammalian G-protein beta subunit. Cell. 1988;55:785–795. doi: 10.1016/0092-8674(88)90134-1. [DOI] [PubMed] [Google Scholar]
- Hassig CA, Tong JK, Fleischer TC, Owa T, Grable PG, Ayer DE, Schreiber SL. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc Natl Acad Sci. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Kurokawa M, Tanaka K, Friedman AD, Ogawa S, Mitani K, Yazaki Y, Hirai H. TLE, the human homolog of groucho, interacts with AML1 and acts as a repressor of AML1-induced transactivation. Biochem Biophys Res Commun. 1998;252:582–589. doi: 10.1006/bbrc.1998.9705. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes & Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G, Verrijzer CP, Ish-Horowicz D. A conserved motif in Goosecoid mediates Groucho-dependent repression in Drosophila embryos. Mol Cell Biol. 1999;19:2080–2087. doi: 10.1128/mcb.19.3.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CA, Barlow AL, Turner BM. Molecular cloning of Drosophila melanogaster cDNAs that encode a novel histone deacetylase dHDAC3. Gene. 1998;221:127–134. doi: 10.1016/s0378-1119(98)00435-1. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes & Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Laybourn PJ, Kadonaga JT. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- Maixner A, Hecker TP, Phan QN, Wassarman DA. A screen for mutations that prevent lethality caused by expression of activated sevenless and Ras1 in the Drosophila embryo. Dev Genet. 1998;23:347–361. doi: 10.1002/(SICI)1520-6408(1998)23:4<347::AID-DVG9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C. Maternal effect mutations that alter the spatial coordinates of the embryo. Symp Soc Dev Biol. 1979;37:185–211. [Google Scholar]
- Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- Parkhurst SM. Groucho: Making its Marx as a transcriptional co-repressor. Trends Genet. 1998;14:130–132. doi: 10.1016/s0168-9525(98)01407-3. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley R, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Wainwright SM, Ish-Horowicz D. Torso signalling regulates terminal patterning in Drosophila by antagonising Groucho-mediated repression. Development. 1997;124:3827–3834. doi: 10.1242/dev.124.19.3827. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- Perrimon N, Lanjuin A, Arnold C, Noll E. Zygotic lethal mutations with maternal effect phenotypes in Drosophila melanogaster. II. Loci on the second and third chromosomes identified by P-element-induced mutations. Genetics. 1996;144:1681–1692. doi: 10.1093/genetics/144.4.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrad A, Meir JY, Barnes TM, Miller DR. The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development. 1997;124:1699–1709. doi: 10.1242/dev.124.9.1699. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. Histone structure and the organization of the nucleosome. Annu Rev Biophys Biomol Struct. 1997;26:83–112. doi: 10.1146/annurev.biophys.26.1.83. [DOI] [PubMed] [Google Scholar]
- Ren B, Chee KJ, Kim TH, Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the groucho family of proteins. Genes & Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Shen X, Gorovsky MA. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- Soeller WC, Poole SJ, Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes & Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Spradling AC. Developmental genetics of oogenesis. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1–70. [Google Scholar]
- Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: An integral component of the Drosophila genome project. Proc Natl Acad Sci. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Suter B, Steward R. Requirement for phosphorylation and localization of the Bicaudal-D protein in Drosophila oocyte differentiation. Cell. 1991;67:917–926. doi: 10.1016/0092-8674(91)90365-6. [DOI] [PubMed] [Google Scholar]
- Tata, F. and D.A. Hartley. 1993. The role of the enhancer of split complex during cell fate determination in Drosophila. Dev. Suppl. 139–148. [PubMed]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Tolkunova EN, Fujioka M, Kobayashi M, Deka D, Jaynes JB. Two distinct types of repression domain in engrailed: One interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol Cell Biol. 1998;18:2804–2814. doi: 10.1128/mcb.18.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine SA, Chen G, Shandala T, Fernandez J, Mische S, Saint R, Courey AJ. Dorsal-mediated repression requires the formation of a multiprotein repression complex at the ventral silencer. Mol Cell Biol. 1998;18:6584–6594. doi: 10.1128/mcb.18.11.6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams FE, Trumbly RJ. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6500–6511. doi: 10.1128/mcb.10.12.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Tang CM, Yao YL, Yang WM, Seto E. Cloning and characterization of the mouse histone deacetylase-2 gene. J Biol Chem. 1998;273:28921–28930. doi: 10.1074/jbc.273.44.28921. [DOI] [PubMed] [Google Scholar]
- Zhang H, Levine M. Groucho and dCtBP mediate separate pathways of transcriptional repression in the drosophila embryo. Proc Natl Acad Sci. 1999;96:535–540. doi: 10.1073/pnas.96.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]