Abstract
The myeolomonocytic cell line U937 differentiates into macrophages in response to a variety of agents. Several genes including the cyclin-dependent kinase inhibitor p21waf1/cip1 and the homeobox gene transcription factor HOXA10 are induced at the onset of differentiation. Ectopic expression of either gene results in U937 differentiation. In this paper, we describe a mechanism by which p21 and HOXA10 may act in concert, where HOXA10 can bind directly to the p21 promoter and, together with its trimeric partners PBX1 and MEIS1, activate p21 transcription, resulting in cell cycle arrest and differentiation. These experiments for the first time identify p21 as a selective target for a HOX protein and link the differentiative properties of a transcription factor and a cell cycle inhibitor.
Keywords: myeloid differentiation, homeobox proteins, transcription, cell cycle arrest
Hematopoiesis involves an exquisite coordination of proliferation and differentiation. As multipotent stem cells give rise to developmentally restricted progenitor cells that in turn produce mature, specialized post-mitotic cells, stem and progentior cells are capable of self-renewal and proliferation to replace those cells that have been lost to differentiation. Several endogenous ligands and synthetic compounds can drive this process irreversibly to terminal differentiation. The myeolomonocytic cell line U937 responds to several agents, such as phorbol esters, retinoic acid, and 1,25-dihydroxyvitamin D3 [1,25(OH)2D3], by differentiating along the monocyte/macrophage lineage. We described previously several genes that are induced in response to 1,25(OH)2D3 in this cell line, including HOXA10 and the cyclin-dependent kinase (CDK) inhibitor (CKI) p21waf1,cip1. Ectopic expression of either of these two genes by transient transfection is sufficient to drive a portion of these cells to terminally differentiate into the monocyte phenotype (Liu et al. 1996; Rots et al. 1998). p21 was originally identified as a CKI (Harper et al. 1993) and a target of the p53 gene product (el-Deiry et al. 1993) and subsequently as an inhibitor of proliferating cell nuclear antigen (PCNA) activity (Waga et al. 1994; Luo et al. 1995), all of which reflect antiproliferative properties. p21 has since been shown to possess a number of additional activities, including anti-apoptotic (Asada et al. 1999) and pro-proliferative effects (Mantel et al. 1996; LaBaer et al. 1997; Braun et al. 1998), and these seemingly contradictory characteristics have been demonstrated in myeloid cells at different stages of differentiation (Asada et al. 1998, 1999; Braun et al. 1998; Cheng et al. 2000). A similar paradox exists for HOXA10 expression, which has been associated with cell growth and some types of leukemias (Lawrence et al. 1995), and which we described as a gene with pro-differentiative properties (Rots et al. 1998). These dual effects might reflect the intrinsic balance of proliferation and differentiation taking place during hematopoiesis.
HOXA10 is a member of the homeobox superfamily of transcription factors that contain a 61-amino-acid helix-turn-helix DNA-binding motif. Within this superfamily, the Hox family itself consists of clusters of closely related homeobox genes, which exist in a large number of widely diverse organisms. These genes have been best studied in Drosophila (Antennapedia [ANT] and Bithorax [BX] complexes), where they are known to play key roles in pattern formation. In mammals, Hox genes, among many functions, play important roles in hematopoiesis. A detailed study of the expression of human HOX genes in subpopulations of both CD34− and CD34+ bone marrow cells revealed an expression pattern for HOXA10 that is restricted to all subtypes of CD34+ cells (Sauvageau et al. 1994), which contain the most primitive blood cells, including erythroid and myeloid progenitors and the putative stem cells. Retroviral overexpression of HoxA10 in mouse bone marrow cells dramatically affects myeloid and B-lymphoid differentiation, leading to a cell population devoid of both pre B-lymphoid cells and macrophages (Thorsteinsdottir et al. 1997). A large proportion of mice receiving these cells develop a transplantable acute myeloid leukemia.
Given that HoxA10 expression is elevated during myeloid cell differentiation but also appears to enhance the proliferation of progenitor cells in mice, we reasoned that HOXA10 must also have both anti- and prodifferentiation properties, depending on the cell and temporal context of its expression. In the work reported here, we demonstrate that stable, regulable overexpression of HOXA10 in U937 cells leads to arrest in G1 and subsequent differentiation to the monocytic cell type. This effect closely correlates to elevated levels of p21 that follow HOXA10 induction. Remarkably, we find that HOXA10 protein can bind directly to a region of the p21 promoter and, together with its trimeric partners PBX1 and MEIS1, activate p21 transcription. We believe this link between a transcription factor and a cell cycle regulator is a key component of the complex circuitry that leads to myeloid cell differentiation.
Results and Discussion
Stable, regulated expression of HOXA10 induces cell cycle arrest and myeloid differentiation
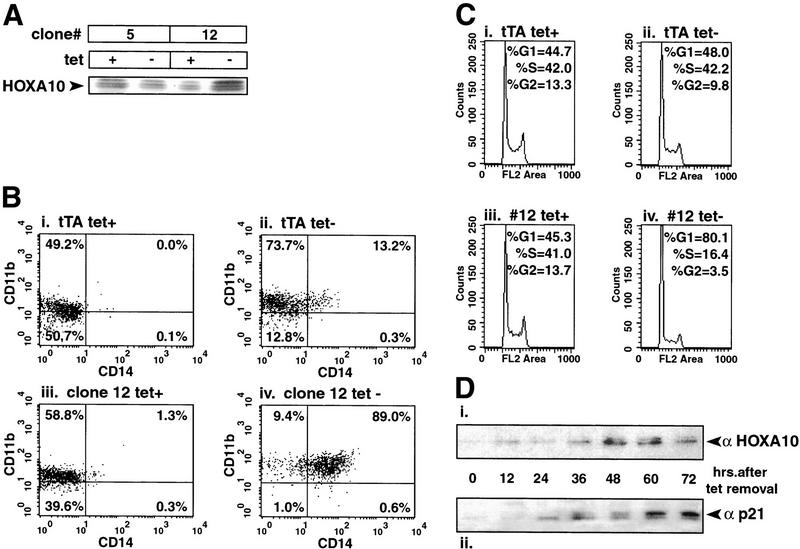
When U937 cells are differentiated with 1,25(OH)2D3, they begin to accumulate detectable HOXA10 protein after only four hours of treatment (Rots et al. 1998). To test the effects of HOXA10 expression on myeloid differentiation independent of inducing agents, we generated a tetracycline (tet)-repressible HOXA10-stable U937 cell line, which makes use of the tet transactivator (tTA)/VP16 fusion protein to activate a gene in the absence of tet (Shockett et al. 1995). We obtained two stable transfectants (clones 12, Fig. 1A; and 30, not shown), which expressed HOXA10 upon removal of tet. These cells were subjected to FACS analysis to detect any changes in expression of the monocyte/macrophage-specific differentiation markers CD11b and CD14. Both the stable clones and the parental tTA cell line expressed CD11b in the absence of any treatments, a phenomenon also observed by others (M. McConnel and J. Licht, pers. comm.). After four days under inducing conditions (tet withdrawal), 13% of parental cells expressed both markers, whereas 89% of the HOXA10-expressing clone 12 cells and 51% of clone 30 cells, were positive for both markers (Fig. 1B, cf. ii and iv, upper right quadrant; data not shown). We used clone 12 to further examine the effects of HOXA10 by characterizing the cell cycle profile of U937 cells following HOXA10 expression. Cell cycle analysis revealed that after four days in the presence of tet, 45% of both the parental tTA cells and the clone 12 cells were in G1 and 42% were in S phase (Fig. 1C, i and iii, respectively). Removing tet had a minimal effect on the cell cycle profile of parental tTA cells (Fig. 1C, ii), but resulted in an increase to 80% of cells in G1 and a concomitant drop to only 16% in S phase for clone 12 cells (Fig. 1C, iv). These results show that HOXA10 expression in U937 cells results in both cell cycle arrest in G1 and myeloid differentiation.
Figure 1.
Stable, regulated expression of HOXA10 induces cell cycle arrest and myeloid differentiation. (A) Candidate clones were subjected to 24 h of incubation in the presence or absence of tet. Equal μg amounts of protein from lysates were subjected to SDS-polyacrylamide gel (10%) electrophoresis and immunoblotted for the presence of HOXA10. Shown are two representative clones, one that overexpresses (clone 12) and one that does not overexpress (clone 5) HOXA10 upon tet removal. (B) HOXA10 overexpression in U937 cells results in differentiation. Parental tTA U937 and U937–HOXA10 (clone 12) cells were subjected to four days of incubation in the presence or absence of tet. FACS analysis was carried out to detect the expression of the monocyte/macrophage terminal differentiation cell surface markers CD11b and CD14. The percentage of cells for each maker is indicated in each quadrant. (C) HOXA10 expression in U937 cells results in G1 cell cycle arrest. Cells from the tet removal experiment in B were subjected to cell cycle analysis by staining nuclei with ethidium bromide and detecting fluoresence using flow cytometry. (D) p21 levels increase following HOXA10 expression in U937 cells. Aliquots of cells were taken at the times indicated (in h) from the same tet removal experiment, and equal μg amounts of whole cell extract from each time point were run on parallel gels, subjected to Western blotting and probed with the indicated antibody.
The p21 promoter is transactivated by HOXA10
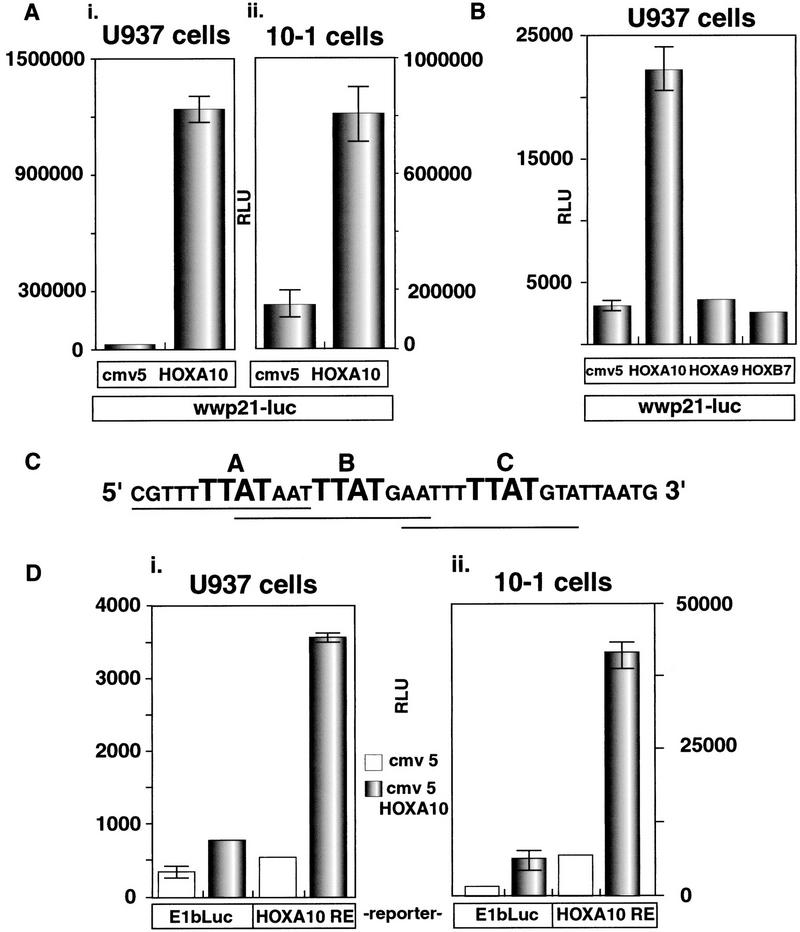
During the course of tet removal, aliquots of cells were collected and subjected to Western blot analysis, revealing a detectable level of HOXA10 protein 12 h after tet withdrawl that continuted to increase before dropping off by 72 h (Fig. 1D, i). Given that HOXA10 mediates G1 arrest and accompanying differentiation, these samples were also analyzed for the presence of the CKI p21. p21 protein could first be detected 24 h after tet withdrawl, closely following the accumulation of HOXA10 (Fig. 1D, ii). Because HOX genes encode transcription factors, this result suggested that p21 could be a direct transcriptional target for HOXA10. To test this, we used a p21 promoter/luciferase reporter construct along with the expression plasmid CMV5–HOXA10 to transiently transfect U937 cells. HOXA10 had a very strong inductive effect on the p21 promoter, exhibiting a 50-fold increase in luciferase activity in response to HOXA10 expression (Fig. 2A, left). We performed the same transfection experiment in the p53-negative fibroblast cell line 10–1 to eliminate any myeloid-specific, indirect effects on the p21 promoter. In this cell background, the p21 promoter was induced sixfold by HOXA10 (Fig. 2A, right). No p21 induction was detected when a truncated version of HOXA10 lacking the DNA-binding homeodomain was overexpressed (data not shown). Moreover, this effect was specific for HOXA10, as two other HOX genes, HOXA9 and HOXB7, which are also important for myeloid differentiation (Lawrence et al. 1997; Care et al. 1999), had no appreciable affect on the p21 promoter (Fig. 2B). This result encouraged us to search for HOXA10-responsive elements in the p21 promoter.
Figure 2.
HOXA10 induces p21 transcription. (A) U937 and 10–1 cells were cotransfected with 1.5 μg of either empty CMV5 expression vector or CMV5–HOXA10 and 5 μg of a p21 promoter/luciferase reporter construct (wwp21-luc, containing 2.4 kb of upstream sequence). All transfections included 1.5 μg of CMV–β-galactosidase (β-gal) expression plasmid, and raw luciferase numbers were normalized to β-gal activity. All DNA amounts indicated are per plate of cells transfected. U937 cells were transfected by electroporation and 10–1 cells were transfected by calcium phosphate precipitation. Each transfection was performed in duplicate and repeated a minimum of five times. Shown are the mean and deviations of one representative experiment. (B) p21 induction is specific to HOXA10. U937 cells were cotransfected with 5 μg of wwp21 luc and 1.5 μg of one of three different plasmids, each expressing one of three indicated HOX genes that are known to be involved in myeloid differentiation. (C) Sequence of a potential p21 responsive region containing three (underlined) partially overlapping close matches to the HOXA10 consensus site. HOXA10 core sequences are TTATs, indicated in large lettering. (D) The p21–A10RE confers HOXA10 induction to a minimal promoter. An oligonucleotide containing the sequence shown in C was inserted into a plasmid containing an E1b minimal TATA box/luciferase reporter and tested for activity by transient transfection of both U937 (i) and 10–1 cells (ii).
A HOXA10 consensus binding site has been defined by random binding site selection (Benson et al. 1995). This site contains a TTAT core sequence shared by a subset of HOX genes and flanked by nucleotides that together make up the 12-bp element 5′-AA(A/T)TTTT ATTAC-3′. We visually scanned the p21 promoter sequence and found three potential sites over ∼3 kb of p21 promoter region, each consisting of three clustered sequences that closely match known HOXA10 sites. We describe here the initial characterization of one of these clustered sites, shown in Figure 2C, which is located at −468 relative to the start of p21 transcription. We cloned this 35-bp fragment into a plasmid containing the E1b TATA box just upstream of a luciferase reporter gene and tested this construct (p21–A10RE/E1b-Luc) for HOXA10 responsiveness by transient transfection (Fig. 2D). This single, nonreiterated fragment was capable of conferring to the E1b/Luc reporter construct a five- to sixfold induction in both U937 cells (Fig. 2D, i) and 10–1 cells (Fig. 2D, ii). Thus, this 35-bp fragment from the p21 promoter is sufficient to confer HOXA10-mediated induction.
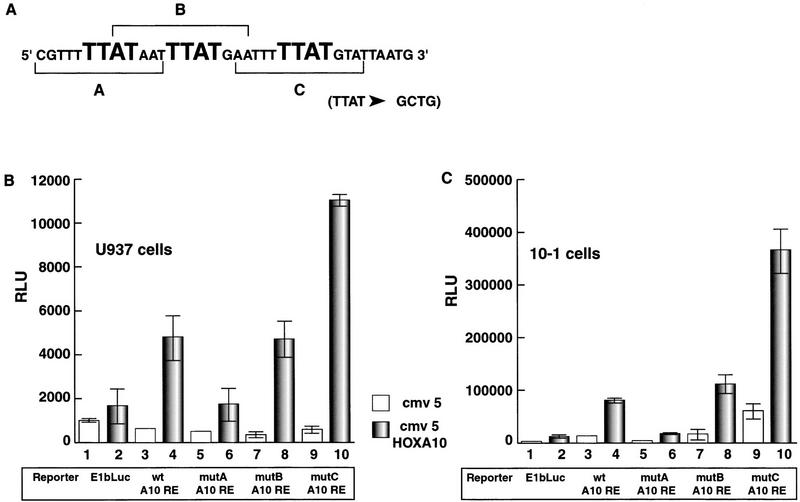
To test which of the three 12-bp sequences within the p21-A10RE are required to mediate HOXA10 regulation, we first subdivided this element into three individual 12-bp subfragments (arbitrarily named A, B, and C, from 5′ to 3′; Fig. 3A) and generated multimers of each smaller fragment as E1b/Luc constructs. None of these elements alone could transactivate in response to HOXA10 in transient transfections (data not shown). We then made a series of mutant E1b/Luc constructs by individually altering each of the three core sites from TTAT to GCTG (mutA, mutB, and mutC), to determine which of these potential 12-bp sequences is involved in binding and activation. When the mutA construct was used to transfect U937 cells, luciferase activity in response to HOXA10 overexpression was severely reduced relative to the wild-type element (wt p21–A10RE) (Fig. 3B). The mutB construct appeared identical to the wt p21–A10RE, whereas mutC resulted in a significant increase in the fold of activation as a consequence of HOXA10 overexpression. Similar results were obtained when we tested these same constructs in the nonmyeloid 10–1 cell line (Fig. 3C). We conclude from these experiments that within the p21 promoter proximal region, the A site is required to confer HOXA10 responsiveness, and the C site may play some sort of negative regulatory role that needs further clarification.
Figure 3.
The p21A10RE contains a cluster of three TTAT core elements. (A) p21 promoter sequence from residues −484 to −450 that contains three TTAT cores. (B) Effect of individual TTAT core mutations on HOXA10 responsiveness. Each core mutation (to GCTG) was tested by transient transfection of U937 cells (B) or 10–1 cells (C) with 5 μg of luciferase construct and 1.5 μg of the expression plasmid CMV5–HOXA10 or CMV-5 vector alone. All transfections included 1.5 μg of CMV–β-gal, and raw luciferase numbers were normalized to β-gal activity.
HOXA10 binds to and transactivates the p21 promoter together with PBX1a and MEIS1b
It has become increasingly evident that HOX proteins may act as part of heterodimeric (van Dijk et al. 1995; Kurant et al. 1998) or heterotrimeric complexes (Berthelsen et al. 1998b; Jacobs et al. 1999; Shen et al. 1999; Schnabel et al. 2000). Two types of such partners have been described, the PBX and MEIS/PREP families. These families both contain homeobox genes that are divergent from HOX genes and contain a three-amino-acid loop extension (TALE) motif important for DNA interaction (Burglin 1997; Piper et al. 1999). The PBX and MEIS/PREP families are currently known to have three (Monica et al. 1991) and four (Nakamura et al. 1996; Berthelsen et al. 1998a) members, respectively. As with all ABD-B-like HOX proteins, HOXA10 has the ability to form dimers with proteins of either of the two TALE families (Shen et al. 1997). In all cases, the TALE-binding portion of the element lies just upstream of the HOX-binding core site, overlapping the 12-bp element described for the binding of HOX proteins. The p21–A10RE, however, does not appear to harbor such an arrangement.
We used expression constructs for the largest splice variants of PBX1 (PBX1a) and MEIS1 (MEIS1b) (Shimamoto et al. 1998; Schnabel et al. 2000) to determine whether they make contributions to HOXA10-mediated p21 transcription. Transient overexpression of either PBX1a (Fig. 4A, lane 2) or MEIS1b alone (lane 3) or both proteins together (lane 4) had no effect on p21–A10RE/E1b-Luc. As observed previously, HOXA10 had an inductive effect (lane 5). Little additional effect on HOXA10 induction was observed in combination with MEIS1b (lane 6). In contrast, when PBX1a was coexpressed with HOXA10, reporter expression nearly doubled relative to HOXA10 alone (cf. lanes 5 and 7). This effect was strongest when MEIS1b was included with both HOXA10 and PBX1a (lane 8). These results suggest a trimeric complex that enhances p21 transcription.
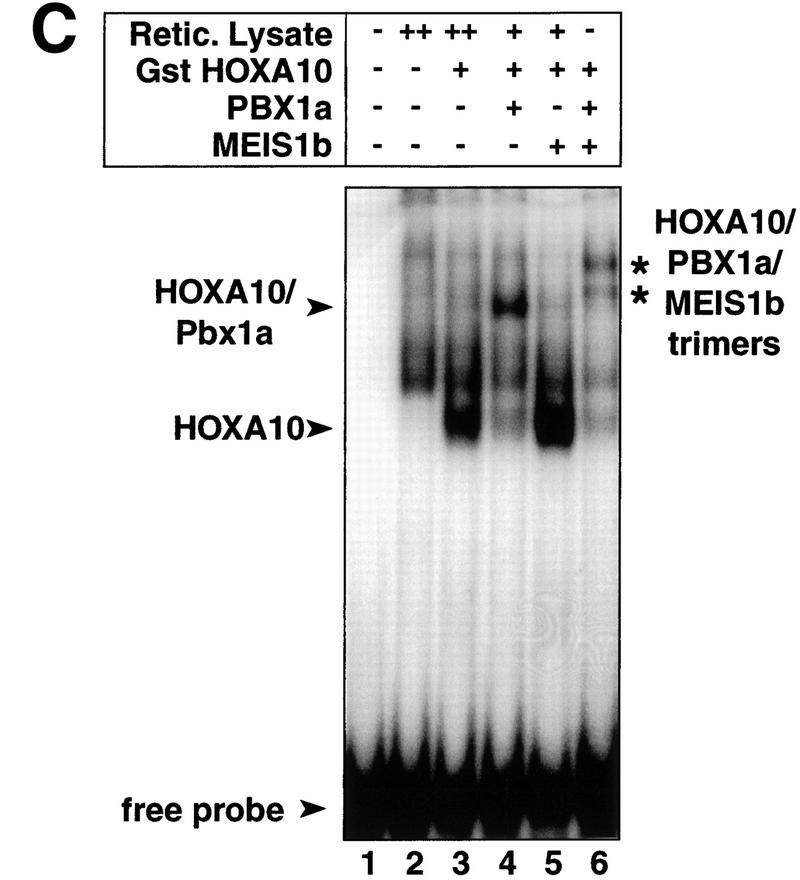
Figure 4.
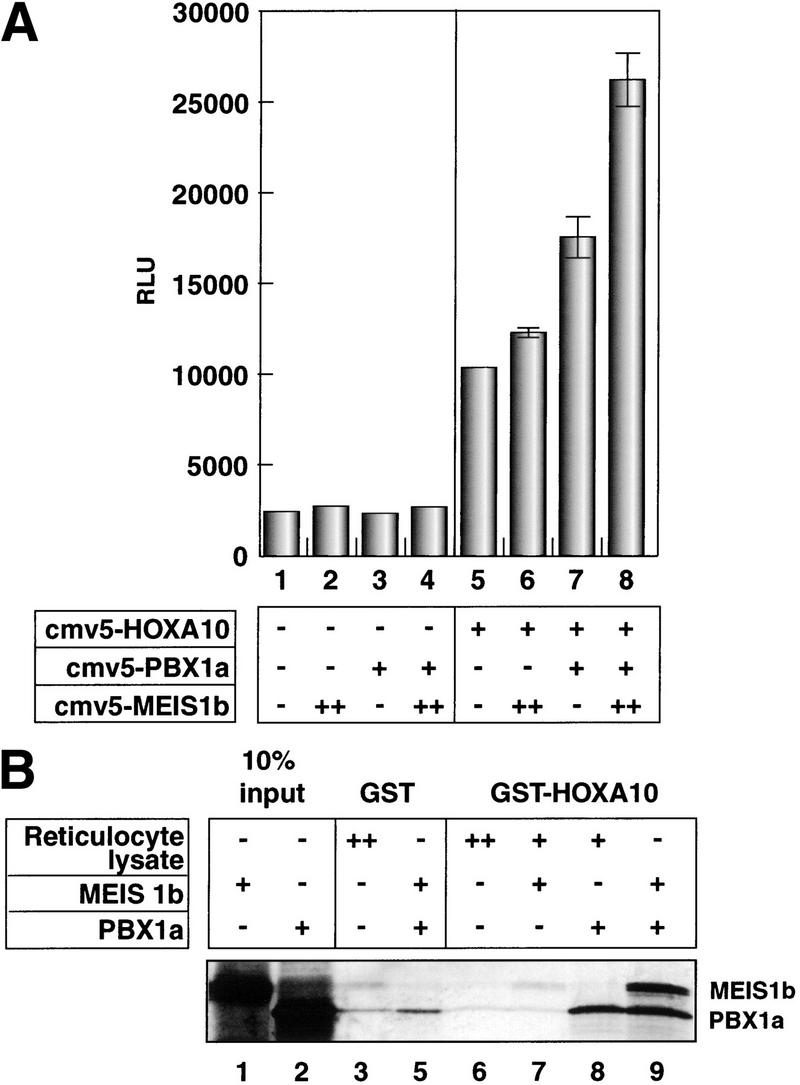
Interactions between HOXA10 and TALE proteins. (A) Maximal induction from the p21–A10RE occurs with HOXA10, PBX1a, and MEIS1b. U937 cells were cotransfected with 5 μg of p21–A10RE/E1b-luc and all combinations of the homeobox constructs as indicated. (+) 1.5 μg of expression plasmid; (++) 3 μg of expression plasmid (all quantities are per plate). Twice the amount of MEIS1b was transfected because this construct expressed approximately half as well as the PBX1a construct (data not shown). All transfections included 1.5 μg of CMV–β-gal, and raw luciferase numbers were normalized to β-gal activity. (B) MEIS1b interaction with HOXA10 is PBX1a-dependent. GST–HOXA10 or GST alone was used as the bait in combination with in vitro-translated proteins or unprogrammed reticulocyte lysate as indicated. Binding reactions were washed extensively and subjected to SDS–PAGE. Gels were dried and subjected to autoradiography. (C) A putative HOXA10/PBX1a/MEIS1b heterotrimer is formed on DNA. The p21–A10RE double-stranded oligonucleotide was used as a probe for a gel mobility shift assay in the presence of the indicated proteins. Unprogrammed reticulocyte lysate was added as indicated to maintain constant amounts of total lysate. Two bands are indicated that represent the putative heterotrimer, because in vitro translation of MEIS1b yields two products of slightly different sizes (not shown).
To examine potential interactions between these three proteins, we performed a GST pulldown experiment using bacterially expressed GST fused to the C-terminal half of HOXA10 (Fig. 4B). This construct contains the homeobox and the heterodimerization motif ENAANWLTAKS (Benson et al 1995). Full-length MEIS1b and PBX1a proteins were generated by in vitro translation. PBX1a interacted with HOXA10 directly (lane 8), but MEIS1b could only interact with HOXA10 when PBX1 was also present (cf. lanes 7 and 9). These results are consistent with the transfection experiment depicted in Figure 4A, which showed that MEIS1b affects HOXA10-mediated p21 induction only when PBX1a is also present. We cannot rule out, however, that MEIS1b may interact directly with the portion of HOXA10 that is not present in the truncated GST–HOXA10 construct used for our in vitro studies.
To investigate whether a trimeric complex can bind to the p21–A10RE, we performed a gel mobility shift assay using the three proteins (Fig. 4C). HOXA10 alone bound the probe with a relatively high affinity (lane 3), and a strong higher order complex formed upon addition of PBX1a (lane 4) but not MEIS1b (lane 5). When all three proteins were present however, a putative heterotrimer was formed (lane 6). Two bands appear representing the heterotrimer, because in vitro-translated MEIS1b yielded two slightly variant sizes of the protein. The nature of this binding appears similar to work described on artificial composite Meis/Hox and Pbx/Hox elements, which can support trimer formation by dimer binding to DNA, with the third protein tethered to the dimer but not bound directly to DNA (Shanmugam et al. 1999; Shen et al. 1999).
We have described here a potential mechanism by which HOXA10 contributes to the growth arrest and differentiation of myeloid cells. Interestingly, various studies indicate that both p21 and HOXA10 have opposing effects on myeloid cells at different stages. In the earliest progenitors, p21 slows growth in order to maintain the stem cell pool (Cheng et al. 2000), whereas a proliferative effect has been described for more mature myeloid progenitors (Mantel et al. 1996; Braun et al. 1998). In U937 cells, p21 expression leads to G1 arrest and differentiation (Liu et al. 1996; Asada et al. 1998). HOXA10 expression is also associated with cell growth and some leukemias and has an antidifferentiative effect in early progenitors (Thorsteinsdottir et al. 1997), whereas we have shown here a differentiative and anticycling effect in the intermediate-staged U937 cells. It is tempting to conclude that the various roles of p21 at different stages of myeloid differentiation account for the apparent paradoxical effects of HOXA10. It is becoming clear, however, that HOX proteins have their own mechanism for generating specificity, by making use of heterodimers and trimers and varying composite cis-acting elements for function. We will only understand how HOXA10 exerts its various cellular effects when its other stage- and tissue-specific target genes are identified.
Materials and methods
Cell culture, transient transfection assays
U937 cells were grown in RPMI with 10% FBS, 2 mM glutamine, 1 mM Na+ pyruvate, and Penn/Strep at 37°C in 5% CO2 and maintained at a density of 2 × 105 to 4 × 105 cells/mL. Cells were transfected by electroporation. Cells [400 μL (1 × 107 cells)] in RPMI 1640 were mixed with 20–30 μg of DNA (typically 10 μg reporter, 3 μg CMV–β-gal to normalize, 3μg additional producers, and purified carrier DNA to 20 μg total) and electroporated in a 4-mM gap cuvette (BTX), using the following settings: capacitance/resistance T = 500 V, capacitance C = 2800 μFarads, resistance R = 72 Ohms (R4), and charging voltage S = 250 V (or 225 V in the presence of 30 μg DNA), using the BTX ECM600 electroporator. After 10 min, cells were transferred to 20 mL of complete media, divided into two 10-cm dishes, and harvested after 24 h at 37°C, in 5% CO2. 10–1 cells were grown in DME with 10% FBS and Penn/Strep at 37°C in 5% CO2, and transfected by calcium phosphate precipitation as described previously (Liu et al. 1996). All reporter assays were carried out as described previously (Liu et al. 1996).
Generation of HOXA10 stable line
tTA U937 cells were grown under the same conditions as described for our nontransfected U937 cells, with the exception that 0.5 μg/mL of puromycin was added to select for the presence of the tTA/VP16 construct and 0.5 μg/mL tet was added to repress tTA/VP16 transcription. Cells were transfected by electroporation as described above. In brief, 2 × 107 cells were transfected with 40 μg of HOXA10–pUHD/hygro–r DNA (a construct containing a tTA-dependent promoter upstream of the HOXA10 cDNA and harboring a hygromycin-resistance gene) and resuspended in 40 mL of complete media. After 24 h, hygromycin at a final concentration of 0.3 mg/mL, as well as tetracycline (tet) and puromycin, was added to the media. After two weeks, selection was done by plating cells at a density of 0.1 cells per well (one 96-well plate), in 10% conditioned complete media. The resulting cultures were expanded and screened by running 10 μg of whole-cell extracts from cells grown in the absence and presence of tet for two days in culture on an SDS-polyacrylamide gel, Western blotting with anti HOXA10 antibody (BAbCO), and comparing HOXA10 expression levels.
Protein expression
HOXA10 was expressed in Escherichia coli as an N-terminal GST fusion to a truncated HOX protein (residues 355–496) containing 55 amino acids N-terminal to the heterodimerization domain, the heterodimerization domain itself (an ENAANWLTAKS motif; Benson et al. 1995), and all amino acids C-terminal to it, including the homeodomain. PBX1a and MEIS1b were in vitro-translated in reticulocyte lysates. 35S-Labeled and unlabeled reactions were performed side by side and 5 μL-labeled reactions were run on an SDS-polyacrylamide gel to verify the presence of the desired protein.
EMSA
Binding reactions were done in 10-μL volumes in a buffer consisting of 10 mM Hepes (pH 7.5), 50 mM KCl, 2 mM BME, 2.5 mg/mL BSA, 5 μg/mL polyd(I-C), and 20% glycerol. Reactions were done by incubation of the indicated proteins for 10 min on ice, followed by addition of probe and an additional incubation of 30 min on ice, run on a 6% polyacrylamide gel at 4°C, 300 V for ∼1.5 h, dried, and subjected to autoradiography.
GST pull-downs
35S-Labeled proteins were generated using the TNT SP6 Reticulocyte Lysate System (Promega). GST–HOXA10 was used as bait, and GST alone was used as a negative control. Each was bound to glutathione Sepharose beads (Amersham/Pharmacia Biotech) and blocked 2 h at 4°C with gentle rocking in a solution containing 20 mM Tris (pH 7.9), 180 mM KCL, 20% glycerol, 0.2 mM EDTA, 0.05% NP-40, 0.5 mM PMSF, 1 mM DTT, 0.05 μg/mL leupeptin, and 1mg/mL BSA. Either PBX1a, MEIS1b, or both proteins were added to a 100-μL volume (35 μL packed) beads. Binding reactions were incubated at 4°C with gentle rocking for 1 h, washed extensively in the binding buffer described above (without BSA and with 0.1% NP40), and run on a 10% SDS-polyacrylamide gel. The gel was soaked twice for 10 min in 1 M Na+ citrate, dried, and subjected to autoradiography.
FACS and cell cycle analyses
Cells (1 × 106 to 2 × 106) were washed twice and resuspended in 75 μL of PBS containing 1% BSA. FITC-conjugated CD11b and PE-conjugated CD14 antibodies (Caltag) (2 μL each) were added and the cells were incubated 1 h on ice in absence of light. These cells were washed twice and then resuspended in 1 mL of PBS with 1% BSA and subjected to FACS analysis (FACScan, Becton Dickinson). Cell cycle analysis was performed as described (Giaretti and Nusse 1994).
Acknowledgments
We thank M. McConnel, J. Licht, M. Cleary, and C. Largman for materials. We also thank D. Domingo and T. Delohery for technical assistance with FACS analysis and flow cytometry. This work was supported by grants from the NIH to L.P.F. V.C.B. was supported in part by the Bruce Forbes Fellowship and is a Jack and Susan Rudin Scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL l-freedman@ski.mskcc.org; FAX (212) 717-3298.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.817100.
References
- Asada M, Yamada T, Fukumuro K, Mizutani S. p21Cip1/WAF1 is important for differentiation and survival of U937 cells. Leukemia. 1998;12:1944–1950. doi: 10.1038/sj.leu.2401228. [DOI] [PubMed] [Google Scholar]
- Asada M, Yamada T, Ichijo H, Delia D, Miyazono K, Fukumuro K, Mizutani S. Apoptosis inhibitory activity of cytoplasmic p21(Cip1/WAF1) in monocytic differentiation. EMBO J. 1999;18:1223–1234. doi: 10.1093/emboj/18.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson GV, Nguyen TH, Maas RL. The expression pattern of the murine Hoxa-10 gene and the sequence recognition of its homeodomain reveal specific properties of Abdominal B-like genes. Mol Cell Biol. 1995;15:1591–1601. doi: 10.1128/mcb.15.3.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelsen J, Viggiano L, Schulz H, Ferretti E, Consalez GG, Rocchi M, Blasi F. PKNOX1, a gene encoding PREP1, a new regulator of Pbx activity, maps on human chromosome 21q22.3 and murine chromosome 17B/C. Genomics. 1998a;47:323–324. doi: 10.1006/geno.1997.5086. [DOI] [PubMed] [Google Scholar]
- Berthelsen J, Zappavigna V, Ferretti E, Mavilio F, Blasi F. The novel homeoprotein Prep1 modulates Pbx-Hox protein cooperativity. EMBO J. 1998b;17:1434–1445. doi: 10.1093/emboj/17.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun SE, Mantel C, Rosenthal M, Cooper S, Liu L, Robertson KA, Hromas R, Broxmeyer HE. A positive effect of p21cip1/waf1 in the colony formation from murine myeloid progenitor cells as assessed by retroviral-mediated gene transfer. Blood Cells Mol Dis. 1998;24:138–148. doi: 10.1006/bcmd.1998.0181. [DOI] [PubMed] [Google Scholar]
- Burglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care A, Valtieri M, Mattia G, Meccia E, Masella B, Luchetti L, Felicetti F, Colombo MP, Peschle C. Enforced expression of HOXB7 promotes hematopoietic stem cell proliferation and myeloid-restricted progenitor differentiation. Oncogene. 1999;18:1993–2001. doi: 10.1038/sj.onc.1202498. [DOI] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Giaretti W, Nusse M. Light scatter of isolated cell nuclei as a parameter discriminating the cell-cycle subcompartments. Methods Cell Biol. 1994;41:389–400. doi: 10.1016/s0091-679x(08)61730-6. [DOI] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurant E, Pai CY, Sharf R, Halachmi N, Sun YH, Salzberg A. Dorsotonals/homothorax, the Drosophila homologue of MEIS1, interacts with extradenticle in patterning of the embryonic PNS. Development. 1998;125:1037–1048. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. New functional activities for the p21 family of CDK inhibitors. Genes & Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- Lawrence HJ, Sauvageau G, Ahmadi N, Lopez AR, LeBeau MM, Link M, Humphries K, Largman C. Stage- and lineage-specific expression of the HOXA10 homeobox gene in normal and leukemic hematopoietic cells. Exp Hematol. 1995;23:1160–1166. [PubMed] [Google Scholar]
- Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89:1922–1930. [PubMed] [Google Scholar]
- Liu M, Lee MH, Cohen M, Bommakanti M, Freedman LP. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes & Dev. 1996;10:142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- Luo Y, Hurwitz J, Massague J. Cell-cycle inhibition by independent CDK and PCNA binding domains in p21Cip1. Nature. 1995;375:159–161. doi: 10.1038/375159a0. [DOI] [PubMed] [Google Scholar]
- Mantel C, Luo Z, Canfield J, Braun S, Deng C, Broxmeyer HE. Involvement of p21cip-1 and p27kip-1 in the molecular mechanisms of steel factor-induced proliferative synergy in vitro and of p21cip-1 in the maintenance of stem/progenitor cells in vivo. Blood. 1996;88:3710–3719. [PubMed] [Google Scholar]
- Monica K, Galili N, Nourse J, Saltman D, Cleary ML. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol. 1991;11:6149–6157. doi: 10.1128/mcb.11.12.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Jenkins NA, Copeland NG. Identification of a new family of Pbx-related homeobox genes. Oncogene. 1996;13:2235–2242. [PubMed] [Google Scholar]
- Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. Structure of a HoxB1-PBX1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–597. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- Rots NY, Liu M, Anderson EC, Freedman LP. A differential screen for ligand-regulated genes: identification of HOXA10 as a target of vitamin D3 induction in myeloid leukemic cells. Mol Cell Biol. 1998;18:1911–1918. doi: 10.1128/mcb.18.4.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, Largman C, Lawrence HJ, Humphries RK. Differential expression of homeobox genes in functionally distinct CD34 + subpopulations of human bone marrow cells. Proc Natl Acad Sci. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel CA, Jacobs Y, Cleary ML. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene. 2000;19:608–616. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- Shanmugam K, Green NC, Rambaldi I, Saragovi HU, Featherstone MS. PBX and MEIS as non-DNA-binding partners in trimeric complexes with HOX proteins. Mol Cell Biol. 1999;19:7577–7588. doi: 10.1128/mcb.19.11.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WF, Rozenfeld S, Lawrence HJ, Largman C. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with PBX1a on a novel DNA target. J Biol Chem. 1997;272:8198–8206. doi: 10.1074/jbc.272.13.8198. [DOI] [PubMed] [Google Scholar]
- Shen WF, Rozenfeld S, Kwong A, Kom ves LG, Lawrence HJ, Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol Cell Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto T, Ohyashiki K, Toyama K, Takeshita K. Homeobox genes in hematopoiesis and leukemogenesis. Int J Hematol. 1998;67:339–350. doi: 10.1016/s0925-5710(98)00024-3. [DOI] [PubMed] [Google Scholar]
- Shockett P, Difilippantonio M, Hellman N, Schatz DG. A modified tetracycline-regulated system provides autoregulatory, inducible gene expression in cultured cells and transgenic mice. Proc Natl Acad Sci. 1995;92:6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, Largman C, Humphries RK. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1997;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk MA, Peltenburg LT, Murre C. Hox gene products modulate the DNA binding activity of PBX1 and Pbx2. Mech Dev. 1995;52:99–108. doi: 10.1016/0925-4773(95)00394-g. [DOI] [PubMed] [Google Scholar]
- Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]