Abstract
The transcription factor osterix (Osx/Sp7) is required for osteogenic differentiation and bone formation in vivo. While Osx can act at canonical Sp1 DNA-binding sites and/or interact with NFATc1 to cooperatively regulate transcription in some osteoblast promoters, little is known about the molecular details by which Osx regulates osteocalcin (OCN) transcription. We previously identified in the OCN proximal promoter a minimal C/T-rich motif, termed OCN-CxRE (connexin-response element) that binds Sp1 and Sp3 in a gap junction-dependent manner. In the present study, we hypothesized that Osx could act via this non-canonical Sp1/Sp3-binding element to regulate OCN transcription. OCN promoter luciferase reporter assays show that Osx alone is an insufficient activator that requires Sp1, but not Sp3, to synergistically stimulate OCN promoter activity. Moreover, promoter deletion analyses demonstrate that both the Sp1/Sp3-binding OCN-CxRE (−70 to −57) and the −92 to −87 region of the OCN proximal promoter are critical for Osx/Sp1 synergistic activities. Our data show that Sp1 influences Osx activity by enhancing Osx occupancy on the OCN promoter, perhaps via physical interactions between the two transcription factors. Finally, alteration of the expression of the gap junction protein connexin43 modulates the recruitment of both Sp1 and Osx to the OCN promoter. In total, our data are strongly in support of Sp1 as an essential transcription factor required for Osx recruitment and transactivation of the OCN promoter. Further, these data lend insight into a mechanism by which alteration of connexin43 impacts osteogenesis in vitro and in vivo.
Keywords: Osteoblast, Osterix, Sp1, Sp3, Osteocalcin, Gap Junction
INTRODUCTION
Osterix (Osx/Sp7), a member of the Sp family of zinc finger-containing transcription factors, is a master regulator of osteogenesis and bone formation during embryonic development, as it was demonstrated that Osx-null mice form a complete cartilaginous skeleton but fail to form bone [1]. Furthermore, recent findings from Osx postnatal-null mice have concluded that Osx continues to be required for new bone formation in growing and adults bones [2–4]. At the cellular level, Osx is required for the differentiation of bone-forming osteoblasts [1,5], as well as in the maturation and function of osteocytes [3,6]. Osx regulates the expression of several important molecules in osteoblast differentiation including collagen 1(α1), bone sialoprotein, osteopontin and osteocalcin. While Osx shares functional similarities with the Sp/KLF (Specificity protein/Krüppel-Like Factors) superfamily of transcription factors [7,8] and can bind canonical Sp1 cognate elements in vitro [1], the precise DNA binding elements by which Osx regulates osteoblast gene transcription, particularly for the osteocalcin gene, are unclear.
Previously a non-canonical Sp1/Sp3-binding C/T-rich element, located in the −70 to −57 region (relative to transcriptional start) in the rat osteocalcin proximal promoter, was characterized in well-coupled ROS17/2.8 osteosarcoma cells [9]. In that paper, the recruitment of Sp1/Sp3 to the OCN promoter was altered by the overexpression of connexin45 (C×45), a gap junction protein that acts in a dominant negative fashion with respect to C×43 function. The disruption of C×43 function by C×45 overexpression in these cells promoted occupancy of the promoter by the repressor Sp3, instead of the activator Sp1. This unique gap junctions-responsive region of the OCN proximal promoter was termed a connexin-response element or OCN-CxRE. In a follow up study, it was shown that the regulation of Sp1 recruitment to the OCN-CxRE was mediated by ERK signaling and that ERK signaling was affected by modulation of gap junctional communication [10]. Notably, modulation of osteoblast function by C×43-gap junctions has been reported both in vitro and in vivo as genetic ablation of C×43 in mice lead to decreased bone mass due at least in part to a cell autonomous defect in osteoblast differentiation marked by down regulation of markers such as collagen 1(α1), bone sialoprotein and OCN [9,11–16]. Indeed, there is sufficient overlap in the genes affected by disruption of C×43 function and the genes regulated by Osx, to suggest the possibility that C×43 may modulate Osx transcriptional activity.
In the current study, we investigate the role of the non-canonical Sp1 binding OCN-CxRE in transactivation of the osteocalcin promoter by Osx, and Osx function as it relates to the gap junctional regulation of gene transcription. First we examined whether Osx could interact with and regulate transcription from the non-canonical Sp1/Sp3 OCN-CxRE element in the osteocalcin proximal promoter. We further studied if alterations of gap junctional communication in moderately coupled MC3T3 cells could impact Osx regulation of OCN transcription, as it does for Sp1/Sp3. Our data revealed that Osx alone is an insufficient activator and requires Sp1 to cooperatively increase transcription from distinct elements in the OCN proximal promoter. Notably, our results showed that the expression of C×43 dramatically enhanced the recruitment of both Osx and Sp1 to the OCN proximal promoter, and knockdown of C×43 reduced the recruitment of both factors to the promoter. Thus, our study reveals novel information on how Osx regulates OCN gene transcription and expands our understanding of how gap junctional communication influences osteoblast function.
MATERIALS AND METHODS
Chemicals, Reagents and Antibodies
Chemicals and reagents were obtained from Sigma (St Louis, MO) unless stated otherwise. All reagents used for cell cultures were purchased from Cellgro (Herndon, VA). Fetal bovine serum was purchased from HyClone (Logan, UT). The sources of antibodies used were Cell Signaling Technology (Beverly, MA; rabbit anti-Myc-tag; rabbit anti-Lamin A/C), and Millipore (Temecula, CA; rabbit anti-Sp1, rabbit anti-Sp3, rabbit-anti-Osx, mouse anti-GAPDH). Protein G PLUS/Protein A-Agarose beads from Calbiochem (La Jolla, CA) were also utilized in ChIP and CoIP experiments.
Cells and Cell Culture
Experiments for the most part were conducted in Cos-7 cells, an African green monkey kidney fibroblast-like cell line [17]. These naïve fibroblasts are deficient in osteoblastic-related genes such as osterix and osteocalcin, rendering them a simplified model to study OCN promoter transcriptional regulation in the absence of endogenous osteogenic transcription factors. Cos-7 cells were purchased from the American type culture collection, (ATCC; Manassas, VA). The cells were maintained in Dulbecco's modified Eagle medium (DMEM), supplemented with penicillin (50 IU/ml), streptomycin (50 μg/ml) and 10% fetal bovine serum. Cells were seeded initially at a 40,000 cells/cm2 concentration in various vessels relative to each experiment. To validate observations made in Cos-7 cells, MC3T3-E1 (clone 4) osteoblasts, obtained from ATCC were grown in α minimum essential medium (αMEM, 1×) as described previously [18]. Unlike the extremely well coupled ROS-17/2.8 cells used in our initial studies [9,10] MC3T3 cells were used in these studies as they are only moderately coupled by gap junctions [19]. Accordingly, we and others have shown that their communication and resulting signaling can be modulated by both overexpressing and knocking down C×43 function (either by C×45 overexpression [18,20] or siRNA-mediated knockdown [18]. Indeed, Lecanda et al. [20] have shown that the gain or loss of function of C×43 function in ROS17/2.8, MC3T3 and UMR-106 cell lines have a similar overlap in alterations in gene expression patterns. Cultures were kept at 37°C in humidified atmosphere of 95% air and 5% CO2. Culture media were renewed every 3 days and cells were used at passage numbers <15.
Plasmid constructs and Transient Transfection
We used for our investigation various OCN proximal promoter constructs cloned upstream of a luciferase reporter. The −199 osteocalcin promoter-luciferase reporter construct (−199OCN-Luc) contains the −199 to +32 5'-flanking sequence of the rat osteocalcin gene, relative to the transcriptional start site, and spans a single Runx2 binding OSE2 element [21]. The −92OCN-Luc construct covers the −92 to +32 region of the OCN promoter. These promoters regions were provided by Dr. Dwight Towler (Washington University; St Louis, MO) and were subsequently subcloned into the KpnI and MluI restriction sites of pGL3basic luciferase reporter vectors (Promega; Madison, WI). The −92OCNΔCxRELuc construct lacking the OCN-CxRE element, derived from the −92OCN promoter sequence, as previously described [9]. An additional 5'-truncated OCN promoter-luciferase construct (−87OCN-Luc) was generated by amplification of the −87 to +32 region of the rat osteocalcin promoter by PCR with Phusion® high fidelity DNA polymerase (New England Biolabs; Ipswich, MA). Then, PCR products were inserted into pGL3basic vector at KpnI and MluI sites. The −70 to −57 region of the OCN promoter (C/T-rich element) was cloned into an RSV minimal promoter upstream of a luciferase reporter gene to achieve the OCN-CxRE-Luc construct as previously reported [9]. A mutant counterpart (mutOCN-CxRE-Luc) was also made.
Sp1, Sp3 and Osx expression vectors were generated as such: PCR amplifications of Sp1, Sp3 and Osx complete open reading frames derived from their respective mouse cDNA were cloned into the EcoRI and XhoI sites of pcDNA3 vector (pcDNA3-Sp1, pcDNA3-Sp3). A segment of the Osx sequence coding for amino acids 27–428, as described by Nakashima et al. [1], was cloned into EcoRI/XhoI sites of a modified pcDNA3 vector, in frame with a myc-tag cloned into HindIII/BamHI sites, resulting in expression of Myc-tagged Osx proteins (pcDNA3-Osx). All constructs were sequenced to verify fidelity. The pSFFV-C×43 construct [22], which contains a full length cDNA for rat C×43 in the pSFFV-neo backbone plasmid [23], was obtained from Dr. Thomas Steinberg (Washington University, St Louis, MO). This construct has previously been shown to increase electrical [24] and chemical [25] coupling among cells. We have shown that overexpression of this construct in MC3T3-E1 cells can enhance their gap junctional communication and osteocalcin transcription [18]. The empty pSFFV-neo plasmid was a kind gift from Gabriel Nunez (University of Michigan). In addition, a C×43 shRNA plasmid (pRS-gja1; Origene Technologies, Inc.; Rockville, MD) and a negative control shRNA scrambled plasmid (pGFP-V-RS, TR300012) were utilized in gene-silencing experiments. Overexpression and knockdown were routinely confirmed by western blotting.
Upon attachment to the plate surface, either Cos-7 or MC3T3-E1 cells were transiently transfected with the appropriate plasmids (specified in figure legends) using FuGene6 transfection reagent (Roche; Indianapolis, IN), as recommended by the manufacturer's instructions. When luciferase assays were to be performed, cells cultured in 24 well-plates were cotransfected with the indicated amount of expression plasmids and 0.4μg/well of reporter plasmids. Total DNA content was kept constant in all experiments by adding empty vector.
Luciferase Reporter Assays
Gene transcription was monitored 72h post transfection, as previously reported [18]. Briefly, cells were rinsed in Hank's balanced salt solution (HBSS), harvested in reporter lysis buffer and luciferase activities were measured, using a Berthold Centro LB 960 luminometer. Transfection efficiency was determined by cotransfecting the cells with a β-galactosidase reporter (SV-βgal). All experiments were performed in triplicate and repeated a minimum of three times.
Nuclear extraction and Western Blotting
Sp1, Sp3 and Osx protein expression levels in Cos7 cells were assessed by western blotting. Briefly, nuclear extracts were prepared using a NE-PER extraction kit following the manufacturer's directions (Pierce Thermo Fisher Scientific; Rockford, IL). Lysates were resolved in 12% SDS-PAGE gels as described [26]. Following transfer to PVDF membranes (Millipore, Bedford, MA), blots were subsequently probed overnight with the appropriate antibodies. Immunoreactive bands were detected with horseradish-peroxidase-conjugated secondary antibodies and enhanced chemiluminescence detection reagents (Pierce). Blots were acquired and analyzed using an UVP EpiChem gel documentation system (UVP Bioimaging Systems, Upland, CA).
RNA extraction and real time-PCR
Sp1, Sp3 and Osx gene expression levels were assessed in Cos7 cells by real time-PCR. Total RNA was extracted from cells grown on 12-well plates, using TRIzol® Reagent (Invitrogen; Eugene, OR) and subsequently used to prepare first-strand cDNA using RevertAid™ First Strand cDNA Synthesis kit (Fermentas Inc.; Glenn Burnie, MD) according to the manufacturer's recommendations. Newly synthesized cDNA strands were then amplified by real-time PCR with SYBR® Green PCR mix buffer (Applied Biosystems; Foster City, CA), as described previously [27], and the following PCR oligo-primers: mSp1 forward: 5'-CTA CCC CTA CCT CAA AGG AAC AGA-3', mSp1 reverse: 5'-GAG ACT GTG CGG TTC TTG GAA-3'; mSp3 forward: 5'-CAC CTA CAA CTA TAA AAG ATG AAG CT-3', mSp3 reverse: 5'-GGA AGG ACA TAC TGC CCA CTT G-3'; mOsx forward: 5'-TTC TGT CCC CTG CTC CTT CTA G-3', mOsx reverse: 5'-CGT CAA CGA CGT TAT GCT CTT C-3'; 18SrRNA forward: 5'-CAT TAA ATC AGT TAT GGT TCC TTT GG-3', 18SrRNA reverse: 5'-TCG GCA TGT ATT AGC TCT AGA ATT ACC-3'. The conditions applied to allow linear cDNA amplification were as follows: 95.0°C for 10 min., followed by 40 cycles of 15 sec. at 95.0°C and 1 min. at 60.0°C, followed by 15 sec. at 95.0°C, 30 sec. at 60.0°C and 15 sec. at 95.0°C.
Formaldehyde Cross-linking and Chromatin Immunoprecipitation (ChIP)
Formaldehyde cross-linking and ChIP assays were performed on confluent cell culture following modified methods formerly reported [9,18]. Under the present conditions, the quantification of immunoprecipitated genomic DNA was assessed by real-time PCR, using the SYBR® Green methods and primers near the OCN-CxRE element of the osteocalcin promoter (primer forward: 5'-AGC CTG GCA GTC TCC GAT T-3'; primer reverse: 5'-ATG CTG TGG TTG GTG ATT GC-3'). Real time-PCR was performed on an Applied Biosystems 7300 Sequence Detection System (Applied Biosystems; Foster City, CA). Amplification of a single target was confirmed by examination of a dissociation curve. Data are expressed relative to expression levels from the input template for each ChIP reaction. Non-immune IgG was used as a negative control for ChIP reactions.
Co-Immunoprecipitation (CoIP)
Protein-protein interactions between transcription factors were examined by immunoprecipitation assays using similar procedures described previously [26]. Here, nuclear extracts obtained as described above, were used, and immunoprecipitated material was collected using protein A/G plus beads. A fraction of the eluted proteins (beads fraction) was analyzed by western blot with the suitable antibodies.
Statistical Analysis
Data were assessed for statistical significance using a two-tailed t-test (comparison of two data sets) or with analysis of variance (ANOVA, comparison of multiple data sets) followed by a Tukey's or Dunnett's post hoc test. A significant difference was determined as a p-values being 0.05 or less. All experiments were performed independently and repeated 3–5 times with similar results; one representative experiment is then shown or combined experiments when indicated.
RESULTS
Osterix with Sp1 but not Sp3 cooperatively stimulate transcription from the osteocalcin proximal promoter
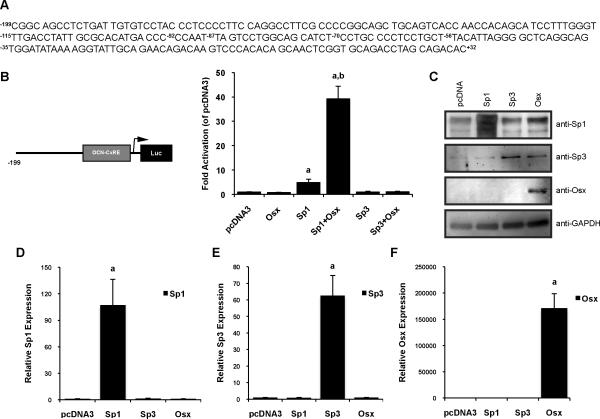
We first investigated Osx, Sp1 and Sp3 transcriptional activities on the osteocalcin (OCN) proximal promoter in Cos-7 cells. We have previously identified a minimal element, located −70 to −57 in the rat OCN proximal promoter, that binds Sp1 and Sp3 [9]. Therefore, Cos-7 cells were transiently transfected with the −199OCN-Luc promoter reporter construct (sequence of the promoter region illustrated in Figure 1A), along with pcDNA3, Osx, Sp1, and Sp3 expression plasmids. As showed in Figure 1B, overexpressing Osx in Cos7 cells had minimal transcriptional effect on the −199OCN-Luc reporter whereas overexpression of Sp1 induced a fivefold stimulation compared to control (pcDNA3) transfected cells. When Osx and Sp1 are expressed simultaneously the transcriptional response is dramatically enhanced, up to 39-fold of promoter basal activity (pcDNA3). In contrast, there was no cooperative induction of transcription by Osx/Sp3-overexpressing cells, compared to control pcDNA3. Our findings corroborate previous reports that identified Sp1 as a transcriptional activator and Sp3 as a transcriptional repressor of the OCN proximal promoter [9]. Moreover we showed that Osx is an insufficient transcriptional activator that requires Sp1 to induce a potent transcriptional response. We confirmed by western blots (Figure 1C) as well as by real time-PCR (Figures 1D–F) that transient transfections of Cos7 cells resulted in efficient protein and mRNA expressions of Sp1, Sp3 and Osx.
Figure 1. Osterix transcriptional activity on the osteocalcin proximal promoter requires Sp1 but not Sp3.
(A) The −199 to +32 rat osteocalcin proximal promoter sequence highlighting the OCN-CxRE element (C/T-rich region) located at −70/−57 bp (numbering is relative to the transcriptional start site). (B) Cos-7 cells were transiently transfected with the -199 to +32 rat osteocalcin promoter, cloned upstream of a luciferase reporter vector (−199OCN-Luc). Additionally cells received pcDNA3 (empty vector), Osx, Sp1 and Sp3 expression plasmids. 72h post-transfection, cells were lysed in reporter lysis buffer and samples were analyzed for luciferase activities. Osx alone has no transcriptional effects on the promoter reporter. When Osx and Sp1 are co-expressed, the transcriptional response is synergistically increased compared to the response induced by Sp1 alone. Such stimulatory effect is not observed with Osx and Sp3 combined. Relative luciferase activities were normalized to pcDNA3 (control empty vector). Data are representative of 3–5 independent experiments and error bars show the standard deviation (SD) of the mean of triplicates. a, significantly different from control pcDNA3 (p< 0.05); b, significantly different from Sp1 alone and indicates more than an additive response (p< 0.05). (C) Western blot analysis from Cos-7 nuclear extracts. Cells were harvested 72h following transient transfection with pcDNA3, Osx, Sp1 and Sp3, as described in the “Experimental Procedures” section. Sp1 and Sp3 protein expressions were analyzed using their respective antibodies. Osx was revealed using an anti-Myc antibody. Sp1, Sp3 and Osx are overexpressed in cells with regard to the expression plasmids they received. GAPDH protein detection ensured equal loading. Levels of gene expression assessed by real time-PCR confirmed overexpression of Sp1 (D), Sp3 (E) and Osx (F) in Cos7 cells. Data are representative of 3 independent experiments and error bars show the standard deviation (SD) of the mean of triplicates. Transcripts expressions were normalized using 18SrRNA. a, significantly different from control pcDNA3 (p< 0.05).
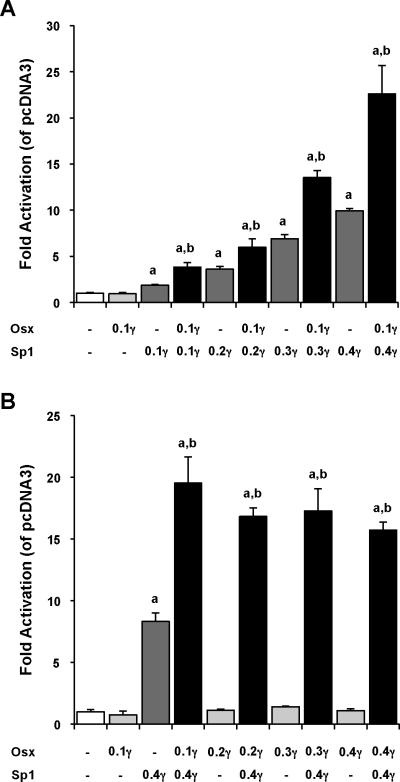
Interestingly, upon increasing the amounts of either Sp1 (Figures 2A) or Osx (Figures 2B), we observed that the amplitude of Osx/Sp1 synergistic responses correlated with the amounts of Sp1 transfected but not with those of Osx received. While Sp1 dose-dependently transactivated the −199OCN-Luc promoter reporter and also enhanced the synergy when paired with Osx (Figures 2A), Osx on the other hand had no significant transcriptional activity regardless of the concentration tested. Noticeably, the Osx/Sp1 cooperative response achieved with the highest dose of Osx (0.4μg) was comparable to those obtained with lower doses (Figures 2B). These results confirm that Osx is an insufficient activator of OCN transcription and that Sp1 is required for the cooperative activation of transcription by Osx.
Figure 2. Effects of various amounts of Sp1 or Osterix on Osx/Sp1 transcriptional synergistic responses.
Cos-7 cells were transiently transfected with the −199OCN-Luc reporter construct along with Osx (0.1μg/well) and increasing amounts of Sp1 (0.1 to 0.4μg/well) expression plasmids (A). Luciferase activities assessed 72h post-transfection as described above, revealed that Sp1 dose-dependently stimulates the −199OCN promoter driven-luciferase activity in addition to Osx/Sp1 synergistic responses. (B) In assays where the −199OCN-Luc was cotransfected with Sp1 (0.4μg/well) and increasing amounts of Osx (0.1 to 0.4μg/well) expression plasmids, the transcriptional effects of Osx expression alone were not significantly different from control pcDNA3 despite the increment of Osx amounts. However, when co-expressed with Sp1 there was a synergistic induction of transcription. Relative luciferase activities were normalized to pcDNA3 (white column). Data are representative of 3 independent experiments and error bars show the standard deviation (SD) of the mean of triplicates. a, significantly different from control pcDNA3 (p< 0.05); b, significantly different from Sp1 alone and indicates more than an additive response (p< 0.05).
A putative Osx/Sp1 DNA-binding region in the OCN proximal promoter supports the transcriptional synergy
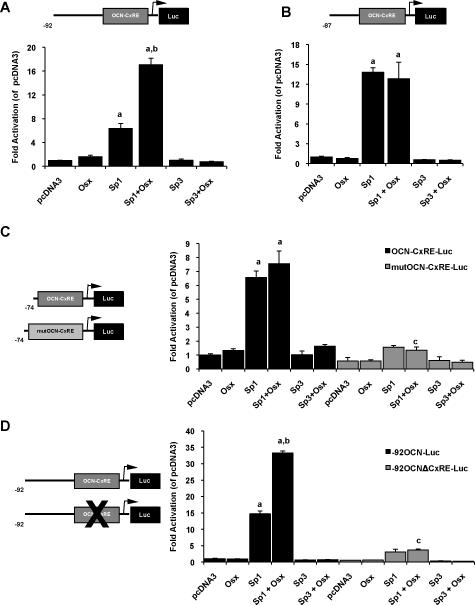
Having established that Osx enhances Sp1-induced OCN promoter activity, we then proceeded to determine an Osx/Sp1 responsive region in the OCN proximal promoter. Similar to the results observed on the −199OCN-Luc reporter, a 6.4-fold Sp1-induction and a 17-fold Osx/Sp1 synergistic response compared to control (pcDNA3) cultures were obtained with the 5'-deletion mutant construct −92OCN-Luc reporter (Figure 3A; referred to Figure 1A for sequence). Conversely, the −87OCN-Luc reporter construct reveals that Osx is no longer able to amplify Sp1 transcriptional stimulatory response on this truncated construct (Figure 3B). However, the −87OCN-Luc reporter remains robustly Sp1 responsive (13.8-fold and 12.8-fold inductions of the OCN promoter activity in the absence or presence of Osx respectively). These results are in favor of a bi-partite regulatory element controlling the Osx/Sp1 transcriptional synergistic response where element(s) necessary for Osx-induced synergy are located upstream (−92 to −87) of the Sp1-responsive region contained within the −87OCN promoter reporter construct. Despite this result, we tested our hypothesis that Osx may act at the non-canonical Sp1/Sp3-binding OCN-CxRE element (highlighted in Figure 1A), which has been previously characterized [9]. Transcriptional activities of Osx, Sp1 and Sp3 from the OCN-CxRE-Luc construct were comparable to those obtained with the −87OCN-Luc reporter (Figure 3C). Sp1 stimulated transcription from this element with no further activation observed when Osx is overexpressed alongside Sp1. Moreover, when using the mutOCN-CxRE-Luc reporter construct (mutated OCN-CxRE), Sp1-induced OCN promoter activity is largely suppressed, confirming the specificity of the OCN-CxRE element for Sp1 (Figure 3C). Furthermore, luciferase activities from the −92OCNΔCxRE-Luc reporter, in which the Sp1/Sp3-binding OCN-CxRE element has been deleted, showed that the Sp1 response is greatly reduced and the cooperative stimulation of transcription by Sp1 and Osx is fully abrogated compared to those obtained with the −92OCN-Luc construct (Figure 3D). Thus, these results further indicate that Osx-mediated transactivation of the OCN proximal promoter requires Sp1, but not Sp3, binding to its cognate OCN-CxRE.
Figure 3. Osterix and Sp1 transcriptional synergistic activities require distinct regulatory elements in the osteocalcin proximal promoter.
Cos-7 cells were transiently transfected with (A) the −92 to +32 rat osteocalcin promoter, upstream of a luciferase reporter (−92OCN-Luc), or (B) the −87 to +32 rat osteocalcin promoter, upstream of a luciferase reporter (−87OCN-Luc) together with pcDNA3, Osx, Sp1 and Sp3 expression plasmids and followed 72h later by lysis and analysis for luciferase activities. The 5'-truncated −92OCN-Luc reporter emulated the results obtained with the −199OCN-Luc reporter construct with Osx synergizing with Sp1 to increase transcription. The −87OCN-Luc remained Sp1 responsive, however Osx is no longer able to amplify the Sp1 transcriptional stimulatory response observed with the −199OCN-Luc or the −92OCN-Luc reporters. (C) Cos-7 cells were transiently transfected with the OCN-CxRE (OCN-CxRE-Luc) or the mutated OCN-CxRE (mutOCN-CxRE-Luc) promoter luciferase reporters, along with pcDNA3, Osx, Sp1 and Sp3 expression plasmids. Similarly, the synergistic transcriptional response induced by Osx and Sp1 is abolished in the OCN-CxRE reporter while this element is still greatly sensitive to Sp1. Additionally the mutOCN-CxRE-Luc reporter abrogates most of the transcriptional activity by Sp1 and fully inhibits the cooperative induction of transcription by Osx in the presence of co-expressed Sp1. (D) Cos-7 cells received either the −92OCN-Luc reporter construct or a mutant reporter in which the OCN-CxRE element has been deleted (−92OCNΔCxRE-Luc), along with pcDNA3, Osx, Sp1 and Sp3 expression plasmids followed by luciferase assays. Unlike with the −92OCN-Luc reporter, when the Sp1-binding element is lacking in this region of the osteocalcin promoter, both Sp1 and Osx/Sp1 responses are greatly diminished, as observed using the −92OCNΔCxRE-Luc construct. Relative luciferase activities were normalized to pcDNA3 (control empty vector). Data are representative of 3–5 independent experiments and error bars show the standard deviation (SD) of the mean of triplicates. a, significantly different from control pcDNA3 (p< 0.05); b, significantly different from Sp1 alone and indicates more than an additive response (p< 0.05) ); c, significantly different from matching groups (p< 0.05)..
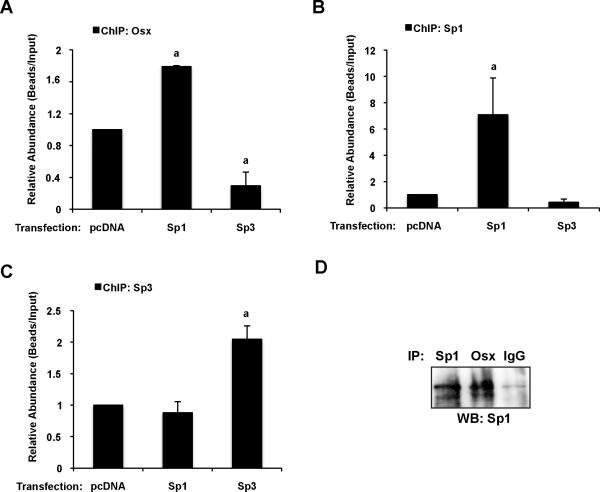
Sp1 enhances the recruitment of Osx to the OCN proximal promoter, thus promoting their functional interactions
Investigation of the mechanisms by which Sp1 might influence Osx transcriptional activity was assessed by ChIP assays using MC3T3-E1 cells overexpressing pcDNA3, Osx (containing a Myc-tag epitope fused to its amino terminus), Sp1 or Sp3 as indicated. In Sp1-overexpressing cells, real time-PCR analysis of the osteocalcin proximal promoter, following immunoprecipitation of DNA-protein complexes with an anti-Myc antibody, revealed that overexpression of Sp1 enhances Osx association with the OCN promoter relative to pcDNA3 transfected control (Figure 4A). Conversely, overexpressing Sp3 in MC3T3-E1 cells reduced Osx occupancy of the OCN promoter. Complementary ChIP assays also showed increased complexes formation between the OCN proximal promoter and Sp1 or Sp3, pulled-down with their respective antibodies, demonstrating binding affinities of Sp1 (Figure 4B) and Sp3 (Figure 4C) to this OCN promoter region. In addition, CoIP experiments were performed to further assess Sp1 and Osx interactions. Nuclear extracts from Cos-7 cells expressing Myc-tagged Osx and Sp1 were immunoprecipitated with anti-Sp1, anti-Myc or non-immune IgG antibodies as indicated and co-precipitated proteins were detected on western blots using anti-Sp1 antibodies. As shown in Figure 4D, Sp1 was co-precipitated with Osx using anti-Myc antibodies, suggesting a physical interaction between Sp1 and Osx.
Figure 4. Sp1 influences Osterix binding activity on the osteocalcin proximal promoter.
(A–C) Chromatin-immunoprecipitation assays (ChIP) performed with anti-Myc (Osx, A), anti-Sp1 (B) and anti-Sp3 (C) antibodies on Myc-tagged Osx overexpressing MC3T3-E1 cells that had been co-transfected with myc-Osx, pcDNA3, Sp1 or Sp3. (A) Overexpressing Sp1 enhanced Osx recruitment to the OCN promoter relative to pcDNA3 transfected controls. Sp1 (B) and Sp3 (C) antibodies were able to pull down the OCN promoter region in their respective transfected samples. Data represent combination of 3 different experiments and quantification of immunoprecipitated genomic DNA was assessed by real time/PCR, using primers near the OCNCxRE element in the OCN proximal promoter as described in the “Experimental Procedures” section. a, significantly different from control pcDNA3 (p< 0.05). (D) Co-immunoprecipitation (CoIP) assays were performed using nuclear extracts from Cos7 cells overexpressing Myc-tagged Osx and Sp1 as indicated. Immunoprecipitation with an anti-Myc antibody revealed the presence of Sp1 in the immunoprecipitated complexes, detected on western blots with the anti-Sp1 antibody, thus suggesting protein-protein interactions between Osx and Sp1. IP, immunoprecipitation; WB, western blotting.
C×43-gap junctions regulate OCN transcription by altering recruitment of both Osx and Sp1
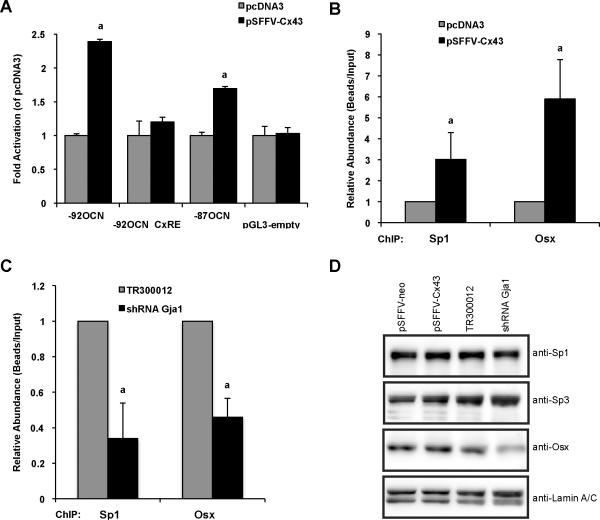
Previously, we have shown that the balance of Sp1/Sp3 on the osteocalcin promoter could be modulated in response to altered gap junctional communication via the OCN-CxRE [9]. Subsequently, we reported that disruption of C×43 function could attenuate ERK signaling, resulting in the preferential recruitment of Sp3 to the osteocalcin promoter [10]. Conversely, under conditions of robust gap junctional communication, ERK activation resulted in the preferential recruitment of the transcriptional activator Sp1 to the osteocalcin promoter. Since Sp1, but not Sp3, is required for Osx transactivation activity, we next examined whether Osx transcriptional activity could be modulated by alteration of C×43-gap junctions. Interestingly, in C×43-overexpressing MC3T3-E1 osteoblasts, luciferase reporter assays revealed that: (i) the −92OCN-Luc promoter reporter, uncovered as the minimal promoter region that supports Osx/Sp1 transcriptional synergy, is up-regulated compared to pcDNA3-transfected cells in response to C×43 overexpression, consistent with the enhanced recruitment of Sp1 and thus Osx to the OCN promoter; (ii) consistent with our previous data [9], when the OCN-CxRE is deleted from the −92OCN-Luc promoter (−92OCNΔCxRE-Luc) the reporter construct becomes non-responsive to C×43 overexpression; (iii) increasing C×43-gap junctions also stimulates transcription from the −87OCN-Luc promoter construct though to a lesser extent compared to the response obtained with the −92OCN-Luc reporter, presumably due to the fact that Sp1 can be recruited to the OCN-CxRE but Osx cannot be co-recruited to its cognate element at −92 to −87 of the promoter (Figure 5A).
Figure 5. Cx43-gap junctions' regulation of the OCN proximal promoter activity affects the recruitment of both Sp1 and Osx to the promoter.
(A) MC3T3 osteoblasts were transiently transfected with pcDNA3 or Cx43 expression plasmids, along with the −92OCN-Luc, −92OCNΔCxRE-Luc and −87OCN-Luc promoter reporter constructs. Subsequent luciferase assays showed that overexpression of Cx43 increases transcription from the −92OCN-Luc and, to a lesser extent, the −87OCN-Luc promoter reporter constructs while remaining ineffective on the −92OCNΔCxRE-Luc reporter (promoterless pGL3basic vector served as luciferase control). a, significantly different from control pcDNA3 (p< 0.05). (B) Activation of DNA-binding activity by Cx43-overpressing MC3T3 cells. Chromatin-immunoprecipitation (ChIP) assays with Sp1 and Myc antibodies followed by real time/PCR analysis for the presence of the OCN proximal promoter region revealed that Cx43 amplifies the recruitment of both Sp1 and Osx on the OCN promoter. a, significantly different from control pcDNA3 (p< 0.05). (C) Conversely, ChIP assays assessed in MC3T3 cells transfected with shRNA Gja1 to knockdown Cx43 expression showed that the abundance of both Sp1 and Osx were significantly reduced on the OCN promoter compared to cells who received the shRNA negative control (TR300012). a, significantly different from control TR300012 (p< 0.05). ChIP data represent combination of 3 different experiments and quantification of immunoprecipitated genomic DNA assessed by real time/PCR, using primers flanking the OCN-CxRE element in the OCN proximal promoter as described in the “Experimental Procedures” section. (D) Western blot analysis from MC3T3 nuclear extracts with the indicated antibodies. Cells were harvested 72h following transient transfection with pSFFV-neo (empty vector), pSFFV-Cx43, pRL-TR300012 (shRNA negative control) and pRL-Gja1 (shRNA Gja1), as described in the “Experimental Procedures” section. Sp1, Sp3 and Osx protein expressions were analyzed using their respective antibodies. Detection of Lamin A/C was used as a loading control.
To validate that alterations of gap junctional communication could impact Osx and Sp1 recruitments to the OCN promoter, we performed ChIP experiments in MC3T3-E1 cells transiently transfected with C×43 or pcDNA3 expression plasmids or shRNA constructs to knockdown C×43 expression in addition to myc-tagged Osx. Our data showed that the expression of C×43 dramatically enhances the recruitment of both Osx and Sp1 to the OCN proximal promoter (Figure 5B). Conversely, when gap junctional communication is disrupted by means of C×43 gene knockdown (shRNA Gja1), ChIP results indicated that Osx and Sp1 occupancies of the OCN promoter significantly diminished up to 65% in this group compared to cells that received the non-effective shRNA scrambled control (TR300012) (Figure 5C).
The protein abundance of endogenous Sp1, Sp3 and Osx was assessed by western blotting of nuclear extracts from MC3T3 cells transfected with empty vector (pSFFV-neo), C×43 overexpression (pSFFV-C×43), scrambled shRNA vector (TR300012), or shRNA directed against C×43 (Figure 5D). Overexpression or knockdown did not significantly alter the expression of Sp1 or Sp3 in these cells. Notably, knockdown of C×43 reduced the expression of Osx, relative to the scrambled shRNA control. In immunoprecipitation experiments, we did not detect phosphorylation of Osx either in the presence of overexpressed C×43 or under conditions of C×43 knockdown (data not shown).
DISCUSSION
During osteogenesis Osx plays a pivotal role in directing osteochondral progenitor cells down the osteoblast lineage. So it is important to understand Osx function, including its transcriptional mechanism and partners. To the best of our knowledge, our study is the first to identify a cognate element in the osteocalcin promoter by which Osx acts and the first to describe a gap junction-sensitive regulation of Osx transcriptional activity involving Sp1. Using luciferase reporter-based experiments, our data showed that Osx alone is an insufficient transcriptional activator and requires Sp1 to synergistically stimulate the OCN proximal promoter. In fact, Osx transcriptional activities remained unchanged despite increasing amounts of Osx assessed. In comparable studies of promoter regulation, strong transcriptional activation by Osx has been largely reported using recombinant Osx proteins consisting of Osx transcription activation domain fused with the DNA-binding domain of GAL4 transcription factor to drive DNA binding [1,28]. Consistent with these studies, our results suggest that it is the DNA affinity of Osx rather than its transcriptional activity that results in Osx limitation to enhance OCN transcription by itself. Indeed, the data suggest that the presence of Sp1 is necessary to direct Osx to its cognate element to drive OCN transcription. Furthermore, Osx cooperative induction with Sp1 but not Sp3 extends our previous findings describing Sp1 and Sp3 as positive and negative transcriptional regulators of the rat OCN proximal promoter, respectively [9].
Sp1 and Sp3 are ubiquitously expressed in mammalian cells and regulate the expression of a large number of genes that take part in virtually all facets of cellular functions [7] but their roles in bone physiology have not been fully elucidated. Sp1−/− mice display early embryonic lethality before the formation of skeletal elements [29] while in short-lived Sp3−/− mice both endochondral and intramembranous ossification are impaired in embryos at E18.5 [30].
Osx ability to upregulate OCN expression has been previously demonstrated [1,31,32], however the molecular details are not fully elucidated and more importantly an Osx cognate response element has yet to be characterized. As a Sp1-related transcription factor, Osx is predicted to bind to functional GC-rich sequences similar to the consensus binding sites of Sp1 [1]. In that regard, it has been shown that overexpression of Osx in vitro induced expression of markers such as collagen 1(α1) [28], Dkk1 [33], sclerostin [6] and bone sialoprotein [34] reportedly from canonical Sp1 binding sites. However, with the exception of the human OCN promoter, no Sp1 consensus binding motifs have been identified in the mouse or rat OCN proximal promoter [35]. As a result of the characterization of the non-canonical Sp1/Sp3-binding OCN-CxRE element in the rat OCN proximal promoter [9], we speculated that Osx could bind and modulate transcription from this region. From the start, with the −199/+32 region of the OCN proximal promoter (Figures 1 & 2) our results clearly established that Osx-mediated transactivation of the OCN proximal promoter requires Sp1, prompting further investigation of Osx/Sp1 cooperative transcriptional activities using OCN-CxRE-containing osteocalcin promoter constructs. Thus, sequential 5'-deletion analyses established that Osx/Sp1 synergistic activities occur on a minimal region spanning at least −92 to +32 bp region (relative to transcriptional start) in the rat OCN proximal promoter. Notably, Osx and Sp1 likely function through distinct cis-regulatory elements in the OCN promoter as no Osx/Sp1 synergistic response was obtained with the −87/+32 OCN promoter construct while the Sp1-induced OCN promoter activity was preserved. Accordingly, our findings are in support of a bi-partite Osx/Sp1 regulatory element comprising (i) the OCN-CxRE motif (located −70 to −57), which is highly responsive to Sp1 but insufficient to support Osx/Sp1 synergistic activities, and (ii) an Osx-responsive region at −92 to −87, which when truncated prevents the synergy from occurring. In addition, based upon the lack of synergy observed when OCN-CxRE is deleted or mutated in the −92/+32 region of the OCN promoter, our data strongly suggested that Osx transcriptional activity requires binding of Sp1 to its cognate element OCN-CxRE in order to induce OCN transcription. Interestingly, the −92 to −87 region corresponds to a CCAAT-box in the OCN proximal promoter [36] making our study the first to show this consensus element targeted by Osx, distinct from the CCAAT-binding transcription factors or proteins previously described [37,38]. While it is unknown whether Osx directly or indirectly acts at this homeobox binding region of the osteocalcin promoter, ChIP data indicate that Osx is in fact occupying the OCN proximal promoter, suggesting that if the action of Osx is indirect it is likely due to participation in a multiprotein complex, rather than via the indirect regulation of expression of a CCAAT-binding transcription factor. Though essential to confer Osx responsiveness to the OCN proximal promoter through Osx/Sp1 synergistic response, the CCAAT-box is not sufficient for Osx transcriptional activation, as Osx independently failed to stimulate luciferase gene expression as reported earlier. Indeed, our ChIP data suggest that Sp1 overexpression enhances Osx occupancy of the promoter. Conversely, Sp3 overexpression inhibits Osx recruitment to the promoter. These data suggest that the Sp1/Sp3 balance is important for the recruitment of Osx to the CCAAT box-containing −92 to −87 region of the osteocalcin promoter.
Subsequently, we gained further insight into Osx/Sp1 transcriptional regulation with data suggesting that the influence of Sp1 on Osx activity might be dependent on physical interactions as evidenced by co-immunoprecipitation of Osx/Sp1 protein complexes. As a result, Sp1 increases Osx abundance to the OCN proximal promoter leading to a synergistic transcriptional response. Thus, our study demonstrated that Osx, though not directly binding OCN-CxRE as we first hypothesized, binds to an adjacent site and transactivates the rat OCN proximal under Sp1/OCN-CxRE-mediated transcriptional control. Consistent with our study, cooperative transcriptional activities between transcription factor NFATc1 and Osx have also been reported for the regulation of the collagen 1(α1) promoter activity in osteoblasts [28]. In another study, the authors showed that the interactions between Osx and the chromatin protein NO66 resulted in inhibition of transcriptional activation by Osx in promoters of Osx target genes [32].
Notably, our data reveal a gap junction-dependent mechanism affecting the Osx/Sp1 synergistic effects. Gap junctions have been shown to be critical to the full elaboration of the osteoblast phenotype and the acquisition of peak bone mass in vivo [12,14,39–41]. In order to understand how gap junctions affect osteoblast function, we formerly characterized OCN-CxRE as a Cx43-gap junctions-sensitive regulatory region with Sp1 and Sp3 identified as the transcriptional mediators recruited to this element to impact OCN promoter activity, in response to alterations of gap junctional communication by overexpression of Cx45 in well coupled RO17/2.8 cells [9]. Subsequently, we reported that the preferential recruitment of Sp1 to OCN-CxRE in the presence of robust Cx43 function was mediated by the influence of Cx43 on the degree of activation of the ERK signaling cascade [10]. Additionally, we have shown that this OCN-CxRE is sensitive to Cx43 overexpression in moderately coupled MC3T3 osteoblast like cells [18]. Previously, it has been shown that overexpression of Cx43 in MC3T3 cells mimicked the impact of Cx43 overexpression in poorly coupled UMR106 cells on transcription of osteoblastic genes, and conversely, overexpression of Cx45 in MC3T3 cells mimicked the impact of Cx45 overexpression in well-coupled ROS17/2.8 cells or the impact of Cx43 overexpression in poorly coupled UMR106 on transcription of osteoblastic genes [20]. This makes MC3T3 cells an interesting model for observing the effects of both up regulating and down regulating Cx43-mediated gap junctional communication. Importantly, the genes affected by these alterations of gap junctional coupling in osteoblasts are similar to the changes in osteoblast gene expression observed in several mouse models of Cx43 genetic ablation [12,14,39]. Notably, we have also shown similar effects of Cx45 overexpression and Cx43 shRNA knockdown on the ability of MC3T3 cells to activate transcription from a Runx2 cognate DNA-binding element in response to FGF2 [18]. Thus, while the mechanism of action of inhibition of Cx43 function by shRNA-mediated knockdown and Cx45 overexpression are quite different, the net effect on the expression of osteoblastic genes has been quite similar, suggesting an overlap in the effects on signaling and transcription.
In the present study, Cx43-dependent induction of OCN-CxRE-containing OCN promoters confirmed the involvement of Sp1 in the transcriptional responses. Furthermore, the complete activation obtained with the −92 bp OCN promoter region relative to the partially activated −87 bp promoter region when Cx43 is overexpressed, suggested a contribution of the Osx-responsive region (−92/−87) to the Cx43-dependent transcriptional response. Indeed, ChIP data provided strong evidences indicating that both Sp1 and Osx recruitment to the OCN proximal promoter is affected by modulation of gap junctional communication. Based on our results, a possible mechanism of gap junction regulation of Osx function would involve Sp1, where a gap junction-mediated Sp1 activation would in turn enhance Osx recruitment and subsequent expression of osteoblast genes (Figure 6). Indeed, Cx43-gap junctions involvement in osteogenic gene transcription regulation and by inference bone formation have been reported both in vivo and in vitro [9,11–14,18,42–44]. In addition, cultures of calvarial osteoblasts derived from Cx43-null mice have demonstrated delayed ability to mineralize and reduced expression of osteocalcin, collagen 1(α1) and bone sialoprotein [12,15,16]. We also observed that knockdown of Cx43 expression reduced the expression of Osx. Conversely, Sp1 and Sp3 levels are unaffected. It is likely that this reduction in Osx abundance contributes to the reduced recruitment of Osx to the osteocalcin promoter under conditions of Cx43 knockdown. However, it is likely, based on the COS7 cell data, that the alteration in Sp1 recruitment to the promoter is also a contributor to the ability to co-recruit Osx. The reduction of Osx as a result of Cx43 knockdown is consistent with the recent report from a mouse model of Cx43 deletion in osteoprogenitor cells [39]. Future studies will examine the signal pathways contributing to the modulation of Osx expression by Cx43, as well as the dynamics of the Sp1-Osx interactions. These findings add a new layer of modulation of osteoblast differentiation with the proposed role of gap junctions in the regulation of Osx function and underscore the importance of Cx43 to the elaboration of osteoblast function.
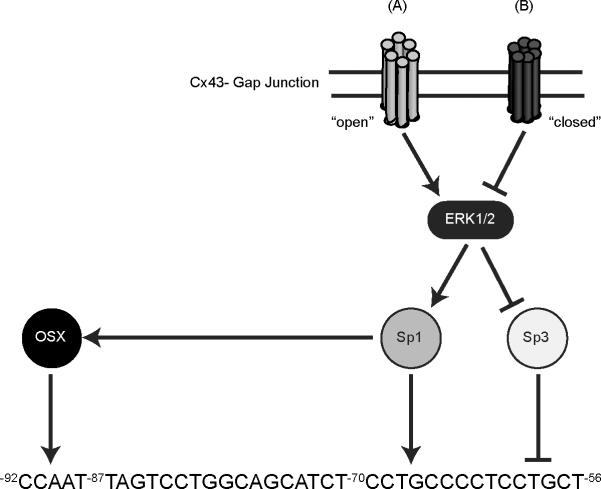
Figure 6. Proposed working model of transcriptional regulation of the −199/+32 region of the rat OCN proximal promoter by Osx, Sp1 and Sp3.
We are proposing a transcriptional regulatory mechanism that involves gap junctions-responsive Sp1/Sp3 transcription factors and Osx. Gap junctions-dependent activation of Sp1 triggers transcription of the OCN proximal promoter once Sp1 binds its cognate element OCN-CxRE (A). When bound to the promoter, Sp1 enhances or stabilizes Osx recruitment (perhaps via direct protein-protein interactions) to an adjacent element (−92 to −87) to further stimulate transcription. Conversely, the binding of Sp3 to the OCN-CxRE box, which is favored when Cx43 function is reduced, prevents the recruitment and subsequent transactivation of the OCN promoter by Osx (B).
In conclusion, our results provide a novel mechanism of Osx transcriptional induction of the OCN proximal promoter involving Sp1-gap junction dependent activation. Ultimately, understanding how Osx and Cx43-mediated gap junctional communication promote osteoblast function will assist in the development of therapeutic tools to improve bone formation.
HIGHLIGHTS
Osterix is an insufficient activator of transcription from the rat osteocalcin proximal promoter
Sp1 (but not Sp3) and Osterix cooperatively stimulate osteocalcin transcription
Sp1 and Osterix act through distinct DNA binding elements located at −70/−57 and −92/−87, respectively, in the osteocalcin proximal promoter
The gap junction protein connexin43, which regulates recruitment of Sp1 to the osteocalcin promoter, also regulates the recruitment of Osterix
ACKNOWLEDGEMENTS
We thank Dwight A. Towler (Washington University, St Louis, MO) for the osteocalcin promoter-luciferase reporter plasmids and Thomas Steinberg (Washington University, St Louis, MO) for the pSFFV-Cx43 construct.
This work was funded by a grant (AR052719) from the National Institutes of Health, National Institute of Arthritis and Musculoskeletal and Skin Diseases.
ABBREVIATIONS
- Osx
osterix
- Sp1/Sp3
specificity protein1/3
- OCN
osteocalcin
- OCN-CxRE
osteocalcin connexin-response element
- C×43
connexin43
- C×45
connexin45
- bp
base pair(s)
- PCR
polymerase chain reaction
- CoIP
co-immunoprecipitation
- ChIP
chromatin immunoprecipitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Nakashima K, Zhou X, Kunkel G, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- [2].Baek WY, Lee MA, Jung JW, et al. Positive regulation of adult bone formation by osteoblast-specific transcription factor osterix. J Bone Miner Res. 2009;24:1055–65. doi: 10.1359/jbmr.081248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhou X, Zhang Z, Feng JQ, et al. Multiple functions of osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A. 2010;107:12919–24. doi: 10.1073/pnas.0912855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Baek WY, de Crombrugghe B, Kim JE. Postnatally induced inactivation of osterix in osteoblasts results in the reduction of bone formation and maintenance. Bone. 2010;46:920–8. doi: 10.1016/j.bone.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nishio Y, Dong Y, Paris M, O'Keefe RJ, Schwarz EM, Drissi H. Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene. 2006;372:62–70. doi: 10.1016/j.gene.2005.12.022. [DOI] [PubMed] [Google Scholar]
- [6].Yang F, Tang W, So S, de Crombrugghe B, Zhang C. Sclerostin is a direct target of osteoblast-specific transcription factor osterix. Biochem Biophys Res Commun. 2010;400:684–8. doi: 10.1016/j.bbrc.2010.08.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Suske G. The sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- [8].Wierstra I. Sp1: Emerging roles--beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- [9].Stains JP, Lecanda F, Screen J, Towler DA, Civitelli R. Gap junctional communication modulates gene transcription by altering the recruitment of Sp1 and Sp3 to connexin-response elements in osteoblast promoters. J Biol Chem. 2003;278:24377–87. doi: 10.1074/jbc.M212554200. [DOI] [PubMed] [Google Scholar]
- [10].Stains JP, Civitelli R. Gap junctions regulate extracellular signal-regulated kinase signaling to affect gene transcription. Mol Biol Cell. 2005;16:64–72. doi: 10.1091/mbc.E04-04-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Vander Molen MA, Rubin CT, McLeod KJ, McCauley LK, Donahue HJ. Gap junctional intercellular communication contributes to hormonal responsiveness in osteoblastic networks. J Biol Chem. 1996;271:12165–71. doi: 10.1074/jbc.271.21.12165. [DOI] [PubMed] [Google Scholar]
- [12].Lecanda F, Warlow PM, Sheikh S, Furlan F, Steinberg TH, Civitelli R. Connexin43 deficiency causes delayed ossification, craniofacial abnormalities, and osteoblast dysfunction. J Cell Biol. 2000;151:931–44. doi: 10.1083/jcb.151.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schiller PC, D'Ippolito G, Balkan W, Roos BA, Howard GA. Gap-junctional communication is required for the maturation process of osteoblastic cells in culture. Bone. 2001;28:362–9. doi: 10.1016/s8756-3282(00)00458-0. [DOI] [PubMed] [Google Scholar]
- [14].Chung DJ, Castro CH, Watkins M, et al. Low peak bone mass and attenuated anabolic response to parathyroid hormone in mice with an osteoblast-specific deletion of connexin43. J Cell Sci. 2006;119:4187–98. doi: 10.1242/jcs.03162. [DOI] [PubMed] [Google Scholar]
- [15].McLachlan E, Plante I, Shao Q, et al. ODDD-linked Cx43 mutants reduce endogenous Cx43 expression and function in osteoblasts and inhibit late stage differentiation. J Bone Miner Res. 2008;23:928–38. doi: 10.1359/jbmr.080217. [DOI] [PubMed] [Google Scholar]
- [16].Thi MM, Urban-Maldonado M, Spray DC, Suadicani SO. Characterization of hTERT-immortalized osteoblast cell lines generated from wild-type and connexin43-null mouse calvaria. Am J Physiol Cell Physiol. 2010;299:C994–C1006. doi: 10.1152/ajpcell.00544.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–82. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- [18].Lima F, Niger C, Hebert C, Stains JP. Connexin43 potentiates osteoblast responsiveness to fibroblast growth factor 2 via a protein kinase C-delta/Runx2-dependent mechanism. Mol Biol Cell. 2009;20:2697–708. doi: 10.1091/mbc.E08-10-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Donahue HJ, McLeod KJ, Rubin CT, et al. Cell-to-cell communication in osteoblastic networks: Cell line-dependent hormonal regulation of gap junction function. J Bone Miner Res. 1995;10:881–9. doi: 10.1002/jbmr.5650100609. [DOI] [PubMed] [Google Scholar]
- [20].Lecanda F, Towler DA, Ziambaras K, et al. Gap junctional communication modulates gene expression in osteoblastic cells. Mol Biol Cell. 1998;9:2249–58. doi: 10.1091/mbc.9.8.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Towler DA, Bennett CD, Rodan GA. Activity of the rat osteocalcin basal promoter in osteoblastic cells is dependent upon homeodomain and CP1 binding motifs. Mol Endocrinol. 1994;8:614–24. doi: 10.1210/mend.8.5.7914673. [DOI] [PubMed] [Google Scholar]
- [22].Beyer EC, Paul DL, Goodenough DA. Connexin43: A protein from rat heart homologous to a gap junction protein from liver. J Cell Biol. 1987;105:2621–9. doi: 10.1083/jcb.105.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fuhlbrigge RC, Fine SM, Unanue ER, Chaplin DD. Expression of membrane interleukin 1 by fibroblasts transfected with murine pro-interleukin 1 alpha cDNA. Proc Natl Acad Sci USA. 1988;85:5649–53. doi: 10.1073/pnas.85.15.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Veenstra RD, Wang HZ, Westphale EM, Beyer EC. Multiple connexins confer distinct regulatory and conductance properties of gap junctions in developing heart. Circ Res. 1992;71:1277–83. doi: 10.1161/01.res.71.5.1277. [DOI] [PubMed] [Google Scholar]
- [25].Steinberg TH, Civitelli R, Geist ST, et al. Connexin43 and connexin45 form gap junctions with different molecular permeabilities in osteoblastic cells. EMBO J. 1994;13:744–50. doi: 10.1002/j.1460-2075.1994.tb06316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Niger C, Hebert C, Stains JP. Interaction of connexin43 and protein kinase C-delta during FGF2 signaling. BMC Biochem. 2010;11:14. doi: 10.1186/1471-2091-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Niger C, Howell FD, Stains JP. Interleukin-1beta increases gap junctional communication among synovial fibroblasts via the extracellular-signal-regulated kinase pathway. Biol Cell. 2009;102:37–49. doi: 10.1042/BC20090056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Koga T, Matsui Y, Asagiri M, et al. NFAT and osterix cooperatively regulate bone formation. Nat Med. 2005;11:880–5. doi: 10.1038/nm1270. [DOI] [PubMed] [Google Scholar]
- [29].Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–28. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- [30].Gollner H, Dani C, Phillips B, Philipsen S, Suske G. Impaired ossification in mice lacking the transcription factor Sp3. Mech Dev. 2001;106:77–83. doi: 10.1016/s0925-4773(01)00420-8. [DOI] [PubMed] [Google Scholar]
- [31].Matsubara T, Kida K, Yamaguchi A, et al. BMP2 regulates osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem. 2008;283:29119–25. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B. Regulation of the osteoblast-specific transcription factor osterix by NO66, a jumonji family histone demethylase. EMBO J. 2010;29:68–79. doi: 10.1038/emboj.2009.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang C, Cho K, Huang Y, et al. Inhibition of wnt signaling by the osteoblast-specific transcription factor osterix. Proc Natl Acad Sci U S A. 2008;105:6936–41. doi: 10.1073/pnas.0710831105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ortuno MJ, Ruiz-Gaspa S, Rodriguez-Carballo E, et al. P38 regulates expression of osteoblast-specific genes by phosphorylation of osterix. J Biol Chem. 2010;285:31985–94. doi: 10.1074/jbc.M110.123612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yeung F, Law WK, Yeh CH, et al. Regulation of human osteocalcin promoter in hormone-independent human prostate cancer cells. J Biol Chem. 2002;277:2468–76. doi: 10.1074/jbc.M105947200. [DOI] [PubMed] [Google Scholar]
- [36].Heinrichs AA, Banerjee C, Bortell R, et al. Identification and characterization of two proximal elements in the rat osteocalcin gene promoter that may confer species-specific regulation. J Cell Biochem. 1993;53:240–50. doi: 10.1002/jcb.240530309. [DOI] [PubMed] [Google Scholar]
- [37].Heinrichs AA, Bortell R, Bourke M, Lian JB, Stein GS, Stein JL. Proximal promoter binding protein contributes to developmental, tissue-restricted expression of the rat osteocalcin gene. J Cell Biochem. 1995;57:90–100. doi: 10.1002/jcb.240570110. [DOI] [PubMed] [Google Scholar]
- [38].Lian JB, Stein GS, Stein JL, van Wijnen AJ. Osteocalcin gene promoter: Unlocking the secrets for regulation of osteoblast growth and differentiation. J Cell Biochem Suppl. 1998;30–31:62–72. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<62::AID-JCB10>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- [39].Watkins M, Grimston SK, Norris JY, et al. Osteoblast Connexin43 modulates skeletal architecture by regulating both arms of bone remodeling. Mol Biol Cell. 2011 doi: 10.1091/mbc.E10-07-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Flenniken AM, Osborne LR, Anderson N, et al. A Gja1 missense mutation in a mouse model of oculodentodigital dysplasia. Development. 2005;132:4375–86. doi: 10.1242/dev.02011. [DOI] [PubMed] [Google Scholar]
- [41].Dobrowolski R, Sasse P, Schrickel JW, et al. The conditional connexin43G138R mouse mutant represents a new model of hereditary oculodentodigital dysplasia in humans. Hum Mol Genet. 2008;17:539–54. doi: 10.1093/hmg/ddm329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gramsch B, Gabriel HD, Wiemann M, et al. Enhancement of connexin 43 expression increases proliferation and differentiation of an osteoblast-like cell line. Exp Cell Res. 2001;264:397–407. doi: 10.1006/excr.2000.5145. [DOI] [PubMed] [Google Scholar]
- [43].Li Z, Zhou Z, Saunders MM, Donahue HJ. Modulation of connexin43 alters expression of osteoblastic differentiation markers. Am J Physiol Cell Physiol. 2006;290:C1248–55. doi: 10.1152/ajpcell.00428.2005. [DOI] [PubMed] [Google Scholar]
- [44].Taylor AF, Saunders MM, Shingle DL, Cimbala JM, Zhou Z, Donahue HJ. Mechanically stimulated osteocytes regulate osteoblastic activity via gap junctions. Am J Physiol Cell Physiol. 2007;292:C545–52. doi: 10.1152/ajpcell.00611.2005. [DOI] [PubMed] [Google Scholar]