Abstract
DNA methylation patterns were evaluated during preimplantation mouse development by analyzing the binding of monoclonal antibody to 5-methylcytosine (5-MeC) on metaphase chromosomes. Specific chromosome patterns were observed in each cell stage. A banding pattern predominated in chromosomes at the one-cell stage. Banding was replaced at the two-cell stage by an asymmetrical labeling of the sister chromatids. Then, the proportion of asymmetrical chromosomes decreased by one-half at each cell division until the blastocyst stage, and chromosomes became progressively symmetrical and weakly labeled. Our results indicate that chromosome demethylation is associated with each DNA replication and suggest that a passive mechanism predominates during early development.
Keywords: DNA methylation, mammalian chromosomes, preimplantation embryos
The importance of DNA methylation during early stages of mammalian development has been demonstrated by genetic studies. Homozygous mutants for the DNA methyltransferase gene (Li et al. 1992) or for the gene encoding MeCP2 (a protein that binds specifically methylated CpGs) (Tate et al. 1996) are unable to complete embryogenesis.
The pattern of methylation is dynamic. Assays for overall genomic methylation indicate that the genome of the mature oocyte is less methylated than sperm DNA, which is itself less methylated than DNA of somatic tissues (Monk et al. 1987; Razin and Cedar 1993). Nevertheless, individual DNA sequences display specific methylation patterns in gametes (Sanford et al. 1984, 1987; Howlett and Reik 1991; Antequera and Bird 1993; Razin and Cedar 1993; Rubin et al. 1994).
A genome-wide demethylation, which was first demonstrated in mice by analysis of total genomic DNA from eight-cell to blastocyst stages (Monk et al. 1987), characterizes the preimplantation embryo. Single-copy genes, the low-copy gene MUP (murine urinary protein), L1 (long interspersed repeated elements, LINES), and the minor satellite (a component of centromeric heterochromatin) all become demethylated in the embryo prior to implantation (Sanford et al. 1987; Howlett and Reik 1991; Razin and Cedar 1993; Razin and Shemer 1995). However, little is known about the methylation pattern of other highly and mildly repeated sequences that are likely to have a significant effect on the overall genome methylation owing to their GC level (Sanford et al. 1984; Hastie 1996; see also Yoder et al. 1997). These sequences include the major satellite of mouse centromeres and the SINES (short interspersed repeated elements). The Alu sequences of primates and the mouse B1 (Alu equivalent) and B2 sequences belong to this category of elements (Schmid 1996).
The mechanism of genome-wide preimplantation demethylation remains unknown. It is, for example, clear that the promoter region of the APOA1 gene is actively demethylated (Kafri et al. 1993). In contrast, the association between the demethylation of L1 sequences and DNA replication reported by Howlett and Reik (1991) at early stages of preimplantation development suggests a passive demethylation mechanism. More recently, demethylation associated to replication was reported in Xenopus early embryos (Matsuo et al. 1998).
To gain insight into topological aspects of global methylation changes in early mouse embryos, we have employed an immunochemical approach to study metaphase chromosome methylation profiles using monoclonal antibodies recognizing specifically 5-methylcytosines (5-MeCs). The reliability of this technique has been firmly established by a number of independent studies. The antibody has been well characterized and used with success in several laboratories (Reynaud et al. 1991; Miniou et al. 1994; Niveleau et al. 1994; Tweedie et al. 1997). The intensity of antibody labeling correlates with the density of 5-MeCs in particular regions of the genome. The resolution of this approach is not high enough to assess the methylation of individual DNA sequences. However, the antibody-staining profiles of metaphase chromosomes are likely to provide direct information about the methylation status of highly and interspersed repeated sequences (Miniou et al. 1994, 1997). Because only clustered methylated CpGs can be recognized at chromosomal level, CpG islands [which represent only 1% of the total genomic DNA and are unmethylated, except in the X inactive chromosome (Bird 1986)], and single- or low-copy DNA sequences contribute little to the overall staining profile.
In this study anti-5-MeC staining profiles were obtained for metaphase chromosomes at each embryonic mitosis from the pronuclear to the blastocyst stages. The results indicate that there is a programmed progression of methylation events in chromosomes. The methylation pattern of old and new chromatids can be distinguished, suggesting that a replication-dependent mechanism of demethylation occurs during early development.
Results
Chromosome methylation patterns were analyzed after each DNA replication of preimplantation development by indirect immunofluorescence using a monoclonal antibody recognizing 5-MeC.
At the one-cell stage in normal mouse embryos, two distinct sets of chromosomes could be distinguished easily (Fig. 1A,B). One set, which we show below as the maternal set, consisted of 20 elements (in the mouse, 2n = 40 chromosomes) with a heterogeneous fluorescence in euchromatic chromosome arms that corresponded to an R-like banding pattern. This banding pattern indicates that methylated sites recognized by the antibody were clustered in definite chromosome regions along the euchromatic arms. The paternal set of 20 chromosomes was labeled only faintly. The pale staining was not due to the extended length of these chromosomes, as it persisted when chromosomes were condensed by a prolonged colchicine treatment (Fig. 1B).
Figure 1.
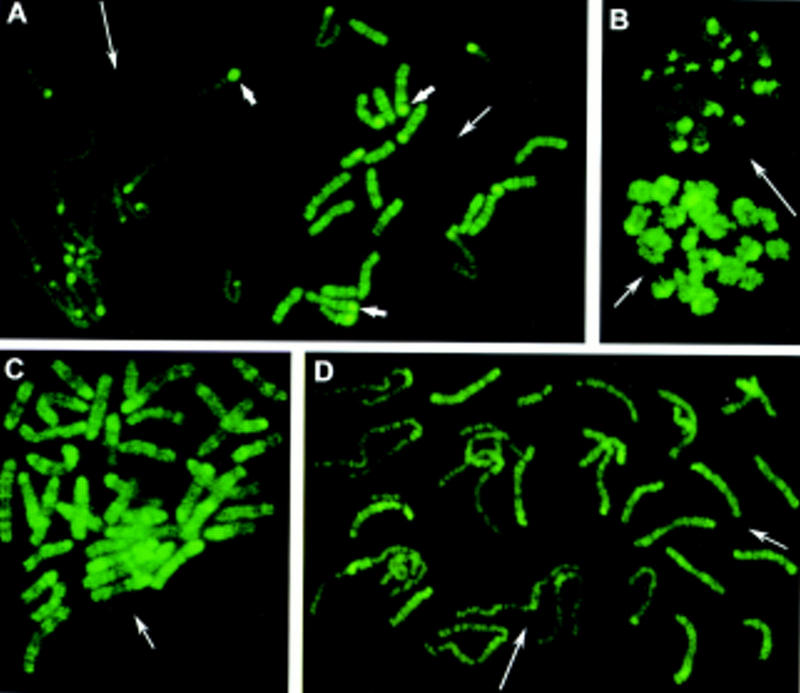
Chromosome methylation patterns of mouse embryos at the one-cell stage. Methylated sites were revealed by indirect immunofluorescence labeling with 5-MeC monoclonal antibody. (A,B) Metaphases from normal mouse embryos obtained without (A) and with (B) colchicine treatment. (A) Two distinct sets of 20 chromosomes each are observed (in mouse 2n = 40). One chromosome set displays an intense R-like banding pattern (right, medium thin arrow). The other set is faintly labeled (left, long thin arrow). Centromeric heterochromatin (small thick arrows) is labeled intensely in only a few chromosomes of the first set, whereas it is labeled in most chromosomes of the second set. (C) Parthenogenetic embryos: all chromosomes display an R-like banding pattern confirming that more labeled chromosomes in normal embryos are of maternal origin. (D) Chromosomes from an embryo carrying Robertsonian translocations of paternal origin: Normal maternal chromosomes (medium thin arrow) are labeled more intensely than paternal translocated chromosomes (long thin arrow).
The parental origin of the two sets of chromosomes was determined in two different ways. First, we examined the chromosomes from parthenogenetic embryos, in which all of the chromosomes are of maternal origin. In these embryos, all 40 chromosomes showed a bright heterogeneous fluorescent pattern similar to R-banding (Fig. 1C). Next, we examined embryos with a number of translocated chromosomes from paternal origin. Here, all of the translocated chromosomes were stained faintly. In favorable examples, it could be seen that paternally inherited chromosomes were also labeled heterogeneously (Fig. 1D). However, the weakness of the labeling made difficult the identification of the banding pattern.
Centromeric regions of the pale chromosomes were usually strongly labeled, except for a small chromosome, probably the Y. Among the maternally derived chromosomes, only a few were labeled intensely at the centromeric heterochromatin (Fig. 1A).
Important changes in the chromosome staining appeared at the two-cell stage. The chromosomes still fell into two classes, strongly and weakly labeling with the anti-5-MeC antibody, corresponding to their maternal and paternal origin, respectively. But now, the staining was restricted to only one of the sister chromatids of each chromosome (Fig. 2A). This asymmetrical labeling of the chromatid pairs was also evident in the faint stained chromosomes of paternal origin, such as the translocated chromosomes in Figure 2, B and C. The differential staining of the two chromatids is particularly evident after image enhancement. In the parthenogenetic embryos, all 40 (maternally derived) chromosomes showed the strong, asymmetric labeling of the euchromatic arms. As was found for the chromosomes at the one-cell stage, in slightly undercondensed chromosomes, a band-like pattern reminiscent of R-banding could be observed along the brightly labeled chromatids of maternal origin.
Figure 2.
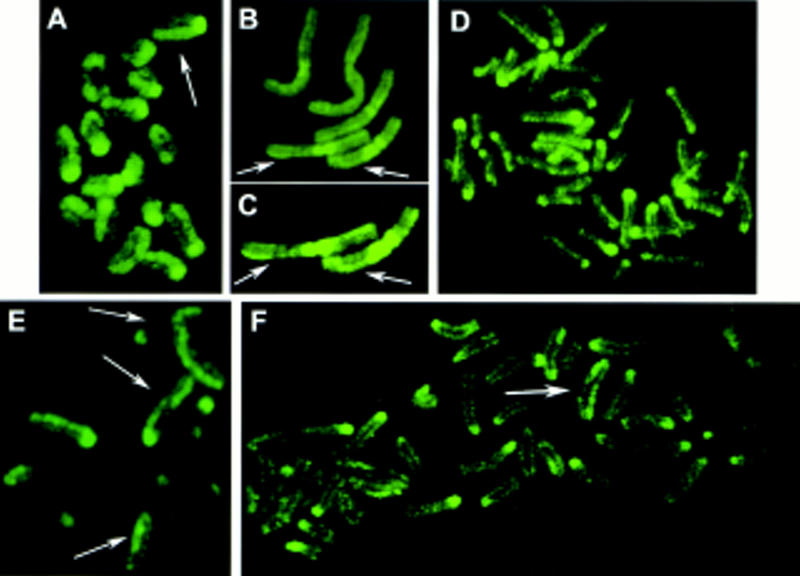
Chromosome methylation patterns of mouse embryos at the two-, four- and eight-cell stages. 5-MeC antibody binding and indirect immunofluorescence detection. (A–D) Chromosomes from two-cell embryos showing an asymmetrical labeling of the two sister chromatids (arrows). In some chromosomes labeling similar to sister chromatid exchanges can be seen. (A) Chromosomes of maternal origin from a normal mouse embryo. (B,C) Chromosomes from embryos carrying Robertsonian translocations of paternal origin. Paternal chromosomes are faintly labeled in relation to maternal chromosomes. In C fluorescence of the same paternal chromosomes showed in B was intensified. (D) Metaphase from a parthenogenetic embryo showing the asymmetrical labeling of maternal chromosomes. (E) Chromosomes from a four-cell embryo: Asymmetrical (arrows) and faintly labeled chromosomes are observed. (F) Metaphase from a eight-cell embryo: Asymmetrical chromosomes are rare (arrow).
In some chromosomes, pale and brightly stained regions alternated in the chromatids, presenting a pattern similar to that resulting from sister chromatid exchanges (SCE) observed after 5-bromodeoxyuridine (BrdU) treatment during two S phases (Zakharov and Egolina 1972; Latt 1973). Similar chromosomes were occasionally observed at later stages (Fig. 2E,F).
In normal four-cell-stage mouse embryos, most chromosomes were labeled faintly (Fig. 2E). The number of intensely and asymmetrically labeled chromosomes (maternal origin) was lower than at the previous stage, varying from cell to cell with a mean number per cell of 10 (20 metaphases were analyzed). Thus, at this stage, the proportion of asymmetrically stained chromosomes was half that at the two-cell stage. The weak labeling of chromosomes of paternal origin made an accurate counting of the number of asymmetrically labeled chromosomes difficult. Nevertheless, the global fluorescence of the karyotype was lower than at the two-cell stage. Again, chromosomes were often fluorescent in the centromeric region. In parthenogenetic embryos, the number of asymmetrically labeled chromosomes also fell by 50% between the two- and four-cell stages.
At the eight-cell stage, the overall level of methylation of euchromatin was lower than at the four-cell stage. Centromeric heterochromatin was labeled intensely in more than half of the chromosomes. Only five intensely labeled asymmetrical chromosomes were observed per metaphase (Fig. 2F). Thus, the number of asymmetrically labeled maternal chromosomes was again reduced by half. Concordantly, the number of asymmetrically labeled chromosomes observed in parthenogenetic embryos at the eight-cell stage was only 25% of that found at the two-cell stage.
At the morula and blastocyst stages, asymmetrically labeled chromosomes were very rare and the overall anti-5MeC fluorescence was low. Two populations of weakly staining metaphases were observed in normal and parthenogenetic embryos (not shown). In some metaphases, chromosomes had a normal appearance while in others they were thick and apparently less condensed. Micronuclei were frequently (10%–15%) observed (not shown). New experiments are required to clarify the origin of these two populations. Note that at these stages trophectoderm and ICM cells are already differentiated.
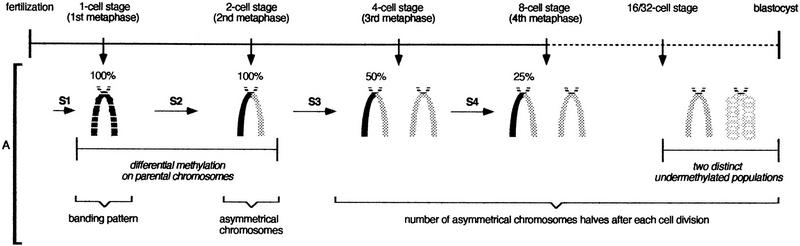
A diagram of the chromosome methylation patterns observed in euchromatin during preimplantation and the DNA modifications compatible with these patterns are shown in Figure 3.
Figure 3.
Diagram of the chromosome methylation changes occurring during preimplantation development of mouse embryos. (A) Chromosome patterns obtained after 5-MeC antibody binding. (B) Summary of DNA methylation alterations that are compatible with the observed chromosome patterns. (A) At the one-cell stage, maternal and paternal chromosomes display heterogeneous but symmetrical labeling of chromatids. Maternal chromosomes are labeled more intensely than paternal chromosomes and display an R-like banding pattern. At the two-cell stage, maternal and paternal chromosomes become asymmetrically labeled, maternal chromosomes always being stained more strongly than paternal chromosomes. From the four-cell stage to blastocyst the number of asymmetrical chromosomes halves after each S phase. At morula (16/32 cells) and blastocyst stages, two cell populations differing by their chromosome condensation can be distinguished: in the first, chromosomes are normally condensed (A, left); in the second, chromosomes are thick and their condensation seems relaxed (A, right). (B) The presence of asymmetrical chromosomes after two S-phases following fertilization (two-cell stage) indicates that after the first S phase (one-cell stage, first metaphase), DNA is hemimethylated in both chromatids, and after the second S phase (two-cell stage, second metaphase), DNA is hemimethylated in one chromatid and demethylated in the other one. As the number of asymmetrical chromosomes halve as cleavage progresses, a failure in maintenance activity on the new DNA strand of each S phase can explain the progression of demethylation. (S) Sphase; (○) unmethylated CpGs; (•) methylated CpGs.
Discussion
The methods currently used for the study of DNA methylation rely on either differential enzymatic or chemical cleavage or on differential sensitivity to chemical conversion of one base to another. Our experimental approach is based on the detection of 5-MeC on mouse metaphase chromosome preparations using a highly specific monoclonal antibody. This method is able to provide a general overview of genomic methylation patterns with a resolution at the level of chromosomal bands. The results obtained help to understand how and where in the genome major methylation changes occur during the first steps of development. However, they give little direct information about the nucleotide sequences involved in these changes. Nevertheless, the R-band-like appearance of the antibody labeling pattern at some stages may suggest that the role of mildly repeated sequences like B1 (the mouse equivalent of the human Alu sequences) or B2 preferentially found in R bands (Boyle et al. 1990) could be important in the establishment of genomic methylation patterns.
In accordance with previous studies (Monk et al. 1987), we observe a gradual demethylation of the genome. However, each developmental stage has its own characteristic chromosome pattern. At the one-cell stage, half of the chromosomes in normal embryos are labeled more intensely and display an R-like banding pattern. Chromosome analysis of parthenogenetic embryos and embryos carrying Robertsonian translocations of paternal origin shows that the more strongly labeled chromosomes are those of maternal origin. Earlier studies concluded that the genome is undermethylated in mature oocyte and methylated in sperm (Monk et al. 1987). Thus, our observation is unexpected, raising the possibility of a differential accessibility of the antibody to DNA. Independent confirmation is clearly needed.
Nevertheless it should be noted that major sperm chromatin remodeling seems to occur before the replication of male and female pronuclei (McLay and Clarke 1997), and chromosomes were analyzed after this first S phase. In addition, we still observed parental differences at the two-cell stage. A lower overall methylation level of the paternal genome would correlate well with the reported higher transcriptional activity and higher hyperacetylated H4 histone content of the paternal pronucleus in fertilized oocyte (Ram and Schultz 1993; Wiekowski et al. 1993; Adenot et al. 1997). Finally, the study of Monk et al. (1987) deals with unmethylated HpaII cutting sites, whereas our approach emphasizes the detection of clustered abundant methylated sites.
At the two-cell stage, chromosome labeling of parthenogenetic embryos and embryos carrying paternally inherited translocated chromosomes indicates that maternal chromosomes are still labeled more intensely, but the signal is found asymmetrically on only one of the two sister chromatids of each chromosome. In less condensed maternal chromosome preparations, the visible banding on asymmetrically labeled chromatids indicates that methylated sites are still clustered in definite regions. Paternal chromosomes are also asymmetrically labeled, although the intensity of the label is very weak.
In embryos at the four-cell stage the number of asymmetrically labeled chromosomes decreases by half, and by the blastocyst stage, asymmetrically labeled chromosomes are very rare and most have become pale and symmetrically stained. Therefore, the distinction between parental chromosomes as detected by the monoclonal antibody is progressively lost between fertilization and implantation.
Asymmetrically stained chromosomes and sister chromatid exchanges have been described in somatic tissues following treatment of normal cells for two successive S phases with the thymidine analog BrdU (Zakharov and Egolina 1972; Latt 1973) and the demethylating agent 5-azadeoxycytidine (5-aza-dC) (Haaf et al. 1986). The incorporation of 5-aza-dC into DNA disturbs DNA methyltransferase activity at replication, resulting in chromosomes with hemimethylated DNA in both chromatids (Jüttermann et al. 1994). After a second replication cycle, chromosomes have one chromatid with unmethylated DNA and the other one with hemimethylated DNA. These chromosomes are stained asymmetrically. If the incorporation of the base analog is continued, the average number of asymmetrical chromosomes halves at each additional cell cycle.
By analogy, if there is a failure of methylation activity during the first DNA replication in the zygote, both new chromosomes will have two hemimethylated chromatids and 5-MeC antibody labeling will then be symmetrical. This is observed for the first metaphase after fertilization (pronuclear or one-cell embryos). After the second cycle, if there is again no methylation activity, the chromosomes will be labeled asymmetrically due to the presence of one hemimethylated and one unmethylated chromatid. Asymmetrically labeled chromosomes are observed at the two-cell stage. The number of asymmetrical chromosomes thereafter decreases by one-half after each cell cycle, indicating that the failure of maintenance methylation persists during DNA replications until blastocyst. The preferential cytoplasmic localization of DNA methyltransferase in embryos (Carlson et al. 1992) is compatible with a failure in maintenance activity. However, methylation activity in chromosomes is not inhibited completely during preimplantation development, as centromeric heterochromatin remains labeled until blastocyst stage.
Loss of DNA methylation may also result from active mechanisms involving either an enzymatic activity, which removes methylcytosine and replaces it by cytosine, or a deamination of methylated CpGs, creating a mismatch that is then repaired (Jost and Saluz 1993; Weiss et al. 1996). The existence of such mechanisms during preimplantation development cannot be excluded. Nonetheless, the chromosome methylation pattern that we observed in euchromatin reveals a loss of methylation correlated with DNA replication and strongly suggests that passive demethylation predominates during early development, at least for the DNA sequences recognized by 5-MeC antibody in chromosomes. Site-specific passive demethylation may result from the binding of specific proteins that inhibit methylase (Riggs and Jones 1983) or by the action of factors that protect DNA against methylation, as observed in postimplantation development (Brandeis et al. 1994; Macleod et al. 1994) and in germinal tissues (Chesnokov and Schmid 1995). The factors acting on DNA demethylation during preimplantation development remain to be identified.
Materials and methods
Collection and preparation of eggs
Six- to eight-week-old F1 female mice (C57BL/6 × CBA) were superovulated by intraperitoneal injection of 5 IU of pregnant mare’s serum (Folligon, Intervet), and 48 hr later, with 5 IU of human chorionic gonadotropin (Chorulon, Intervet). They were then mated overnight with either adult F1 males or a male with nine homozygous Robertsonian translocations (Beechey and Evans 1996). The next morning (0 day), females with a vaginal plug were killed; fertilized eggs were removed by puncturing the oviducts with a needle and released in M-2 medium (Hogan et al. 1994). Eggs were then incubated for 5–10 min at room temperature with 300 μg/ml hyaluronidase (Sigma) to remove cumulus cells and washed in M-2 medium. Embryos were cultured in M-2 medium microdrops under paraffin oil (Merck) and incubated at 37°C in 5% CO2 in air either singly or in groups (15–30) until the blastocyst stage.
Parthenogenetic activation
Eggs originating from the same mice and surrounded by cumulus cells were activated 16 hr after hCG treatment by incubation for 6 min in 7% ethanol in M-2 medium (Hogan et al. 1994) and then transferred to a medium containing 5 μg/ml cytochalasin B (Calbiochem). After 4 hr, cumulus cells were removed by incubation in 300 μg/ml hyaluronidase. Eggs with two pronuclei were cultured in groups (15–30) in microdrops of M-2 medium under paraffin oil as described above.
Embryo fixation
Embryos at the pronuclear stage were fixed 28–32 hr after injecting hCG, when pronuclei disappeared completely, without or with mitotic arrestant, in the night of 0 day. Other early embryos (two to eight cells) were incubated for 9–12 hr with 1 μg/ml colchicine at 37°C and then fixed. Morula and blastocyst stages were fixed after 7 and 4 hr of incubation with colchicine, respectively.
Chromosomes were prepared according to a modification of the air-drying technique (Tarkowski 1966). The hypotonic treatment time was varied according to the stage of development: 1/6 dilution of fetal calf serum (FCS), 400 μg/ml EDTA at room temperature for 2–10 min (pronuclear to morula); 1/20 FCS, 1200 μg/ml EDTA at room temperature for 30 min (blastocyst). Each embryo was then transferred onto a clean microscope slide in a droplet of hypotonic solution. Methanol/acetic acid (3:1) fixative was dropped onto the embryos and blow-dried. For two- and four-cell embryos, the fixation was performed in the morning and in the afternoon of the second day, respectively. The chromosome preparations were done in the morning and in the afternoon of the third day for eight-cell and morula embryos, respectively. Blastocysts were fixed in the afternoon of the fourth day. Slides were kept at −20°C until use.
5-MeC monoclonal antibody binding and chromosome analysis
The indirect immunofluorescence method used here was as described previously (Miniou et al. 1994). Slides were irradiated with UV light for ∼8 hr with a germicidal lamp, rinsed with cold PBS for 5 min, and immersed briefly in PBT (PBS, 0.1% Tween 20, 0.4% BSA). 5-MeC monoclonal antibody diluted in PBT (1:10) was added, and the slides were incubated for 45 min at room temperature. They were then rinsed for 5 min in PBT, and the second antibody, an anti-mouse fluorescein-conjugated IgG (Sigma) diluted 1:40 in PBT, was added. Slides were incubated again for 45 min at room temperature and rinsed with PBS. Before the microscopic observation of the slides at pH 8, PPD (P-phenylenediamine) solution was added to avoid the quenching of fluorescence. Slides were examined under a Leica fluorescence microscope equipped with fluorescein and double-pass band filters. Fluorescence labeling was intensified with Adobe Photoshop.
At each cell stage, 20–40 metaphases were analyzed. Normal embryos were analyzed from pronuclear to blastocyst stage, parthenogenetic embryos were analyzed from pronuclear to morula stages, and embryos carrying translocated chromosomes of paternal origin were analyzed from pronuclear to eight-cell stages.
Acknowledgments
This work was supported by grants from Association Française Contre les Myopathies, Association pour la Recherche sur le Cancer and Ministère de la Recherche et de l’Education Nationale. We thank J.C. Allain for technical support in image intensification; Dr. J.L. Guenet for providing the mouse male carrying translocated chromosomes; Drs. R. Karess, A. Rosa, and S. Saint Just for critical reading; and Dr. T. Bestor for useful suggestions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL viegas@ceylan.necker.fr; FAX +331 47 83 32 06.
References
- Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beechey CV, Evans EB. Chromosome variants. In: Lyon M, Rastan S, Brown S, editors. Genetic variants and strains of the laboratory mouse. Oxford, UK: Oxford University Press; 1996. p. 1483. [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Boyle AL, Ballard SG, Ward DC. Differential distribution of long and short interspersed element sequences in the mouse genome: Chromosome karyotyping by fluorescence in situ hybridization. Proc Natl Acad Sci. 1990;7:29–41. doi: 10.1073/pnas.87.19.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- Carlson LL, Page AW, Bestor TH. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: Implications for genomic imprinting. Genes & Dev. 1992;6:2536–2541. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- Chesnokov IN, Schmid CW. Specific Alu binding protein from human sperm chromatin prevents DNA methylation. J Biol Chem. 1995;270:18539–18542. doi: 10.1074/jbc.270.31.18539. [DOI] [PubMed] [Google Scholar]
- Haaf T, Ott G, Schmid M. Differential inhibition of sister chromatid condensation induced by 5-azadeoxycytidine in human chromosomes. Chromosoma. 1986;94:389–394. doi: 10.1007/BF00328639. [DOI] [PubMed] [Google Scholar]
- Hastie ND. Highly repeated DNA families in the genome of Mus musculus. In: Lyon M, Rastan S, Brown S, editors. Genetic variants and strains of the laboratory mouse. Oxford, UK: Oxford University Press; 1996. pp. 559–573. [Google Scholar]
- Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Howlett SK, Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991;113:119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- Jost JP, Saluz HP. Steroid hormone dependent changes in DNA methylation and its significance for the activation or silencing of specific genes. In: Jost JP, Saluz HP, editors. DNA methylation. Molecular biology and biological significance. Basel, Switzerland: Birkhäuser Verlag; 1993. pp. 425–451. [DOI] [PubMed] [Google Scholar]
- Jüttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri T, Gao X, Razin A. Mechanistic aspects of genome-wide demethylation in the preimplantation mouse embryo. Proc Natl Acad Sci. 1993;90:1058–1062. doi: 10.1073/pnas.90.22.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latt SA. Microfluorometric detection of deoxyribonucleic acid replication in human metaphase chromosomes. Proc Natl Acad Sci. 1973;70:3395–3399. doi: 10.1073/pnas.70.12.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse Aprt gene promoter are required to prevent methylation of the CpG island. Genes & Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Silke J, Georgiev O, Marti P, Giovanini N, Rungger D. An embryonic demethylation mechanism involving binding of transcription factors to replicating DNA. EMBO J. 1998;17:1446–1453. doi: 10.1093/emboj/17.5.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLay DW, Clarke HJ. The ability to organize sperm DNA into functional chromatin is acquired during meiotic maturation in murine oocytes. Dev Biol. 1997;186:73–84. doi: 10.1006/dbio.1997.8581. [DOI] [PubMed] [Google Scholar]
- Miniou P, Jeanpierre M, Blanquet V, Sibella V, Bonneau D, Herbelin C, Fischer A, Niveleau A, Viegas-Péquignot E. Abnormal methylation pattern in constitutive and facultative (X inactive chromosome) heterochromatin of ICF patients. Hum Mol Genet. 1994;3:2093–2102. doi: 10.1093/hmg/3.12.2093. [DOI] [PubMed] [Google Scholar]
- Miniou P, Bourc’his D, Molina Gomes D, Jeanpierre M, Viegas-Péquignot E. Undermethylation of Alu sequences in ICF syndrome: Molecular and in situ analysis. Cytogenet Cell Genet. 1997;77:308–313. doi: 10.1159/000134605. [DOI] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Niveleau A, Drouet C, Bruno C, Reynaud C, Fares F, Pain C. Polymerase chain reaction products containing 5-methylcytidine. A microplate immunoquantification method. Nucleic Acids Res. 1994;22:5508–5509. doi: 10.1093/nar/22.24.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram PT, Schultz RM. Reporter gene expression in G2 of the 1-cell mouse embryo. Dev Biol. 1993;156:552–556. doi: 10.1006/dbio.1993.1101. [DOI] [PubMed] [Google Scholar]
- Razin A, Cedar H. DNA methylation and embryogenesis. In: Jost JP, Saluz HP, editors. DNA methylation. Molecular and biological significance. Basel, Switzerland: Birkhäuser Verlag; 1993. pp. 343–357. [Google Scholar]
- Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- Reynaud C, Bruno C, Boullanger P, Grange J, Barbesti S, Niveleau A. Monitoring of urinary excretion of modified nucleosides in cancer patients using a set of six monoclonal antibodies. Cancer Lett. 1991;61:255–261. doi: 10.1016/0304-3835(92)90296-8. [DOI] [PubMed] [Google Scholar]
- Riggs AD, Jones PA. 5-methylcytosine, gene regulation and cancer. Adv Cancer Res. 1983;40:1–40. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- Rubin CM, VandeVoort CA, Teplitz RL, Schmid CW. Alu repeated DNAs are differentially methylated in primate germ cells. Nucleic Acids Res. 1994;22:5121–5127. doi: 10.1093/nar/22.23.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JP, Forrester L, Chapman VM. Methylation patterns of repetitive DNA sequences in germ cells of Mus musculus. Nucleic Acids Res. 1984;12:2823–2836. doi: 10.1093/nar/12.6.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J P, Clark HP, Chapman VM, Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes & Dev. 1987;1:1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- Schmid CW. Alu: Structure, origin, evolution, significance, and function of one-tenth of human DNA. Prog Nucleic Acid Res Mol Biol. 1996;53:283–319. doi: 10.1016/s0079-6603(08)60148-8. [DOI] [PubMed] [Google Scholar]
- Tarkowski AK. An air-drying method for chromosome preparation from mouse eggs. Cytogenetics. 1966;5:394–400. [Google Scholar]
- Tate P, Skarnes W, Bird AP. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nature Genet. 1996;12:205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- Tweedie S, Charlton J, Clark V, Bird AP. Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol Cell Biol. 1997;17:1469–1475. doi: 10.1128/mcb.17.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A, Keshet I, Razin A, Cedar H. DNA demethylation in vitro: Involvement of RNA. Cell. 1996;86:709–718. doi: 10.1016/s0092-8674(00)80146-4. [DOI] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal from maternal and zygotic nuclei. Dev Biol. 1993;159:366–378. doi: 10.1006/dbio.1993.1248. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Zakharov AF, Egolina NA. Differential spiralisation along mammalian mitotic chromosomes. Chromosoma. 1972;38:341–365. doi: 10.1007/BF00320156. [DOI] [PubMed] [Google Scholar]