Abstract
Context
The identification of patients with inherited cancer susceptibility syndromes facilitates early diagnosis, prevention, and treatment. However, in many cases of suspected cancer susceptibility, the family history is unclear and genetic testing of common cancer susceptibility genes is unrevealing.
Objective
To apply whole-genome sequencing to a patient with suspected cancer susceptibility (and lacking a clear family history of cancer and no BRCA1 and BRCA2 mutations) to identify rare or novel germline variants in cancer susceptibility genes.
Design, Setting, and Participant
Skin (normal) and bone marrow (leukemia) DNA were obtained from a patient with early-onset breast and ovarian cancer and therapy-related acute myeloid leukemia (t-AML), and analyzed with: 1) whole genome sequencing using paired end reads; 2) SNP genotyping; 3) RNA expression profiling; and 4) spectral karyotyping.
Main Outcome Measures
Structural variants, copy number alterations, single nucleotide variants and small insertions and deletions (indels) were detected and validated using the above platforms.
Results
Whole genome sequencing revealed a novel, heterozygous 3 Kb deletion removing exons 7-9 of TP53 in the patient’s normal skin DNA, which was homozygous in the leukemia DNA as a result of uniparental disomy. In addition, a total of 28 validated somatic single nucleotide variations or indels in coding genes, 8 somatic structural variants, and 12 somatic copy number alterations were detected in the patient’s leukemia genome.
Conclusions
Whole genome sequencing can identify novel, cryptic variants in cancer susceptibility genes in addition to providing unbiased information on the spectrum of mutations in a cancer genome.
INTRODUCTION
Cancer susceptibility is often suspected in individuals presenting with cancers at an early age, with multiple primary cancers, or with a suggestive family history. The identification of the genetic basis for cancer susceptibility has important clinical implications for the prevention and early detection of associated neoplasms. However, genetic testing is expensive, and in many cases, mutations in cancer susceptibility genes are not identified. This stems from both technical limitations of commercial assays and a lack of knowledge regarding the genes that contribute to cancer susceptibility. Thus, a comprehensive, unbiased approach to identify mutations in genes contributing to cancer susceptibility is needed. Here we describe the use of whole genome sequencing to identify a novel deletion in TP53 in the normal (skin) genome of an individual with early-onset breast and ovarian cancer who subsequently developed therapy-related acute myeloid leukemia (t-AML).
CASE HISTORY
A 37 year-old female presented with Stage 2 estrogen receptor-positive, progesterone receptor-positive, Her2-positive breast cancer and was treated with surgery, local radiotherapy and chemotherapy (cyclophosphamide, etoposide, and doxorubicin). At age 39, she was diagnosed with Stage IIIC ovarian serous cystadenocarcinoma (FIGO grade III), and she was treated with surgery and chemotherapy (carboplatin and paclitaxel). Her ovarian cancer recurred at age 42, and she received additional chemotherapy. Six months later, she presented with t-AML. Analysis of her bone marrow revealed 76% blasts with monocytic features and a complex karyotype involving monosomy 7, del(5q), and two marker chromosomes that could not be resolved by standard cytogenetic analysis (Sup. Figure 1). She developed respiratory failure and died 8 days after presentation. Of note, her family history did not suggest an inherited cancer susceptibility syndrome. Specifically, no cancers were reported in 6 first degree relatives (parents, brother, and 3 children) and only a single case of cancer (leg sarcoma at age 62 in a maternal grandfather) was reported in 10 second degree relatives. Nonetheless, the early onset of both breast and ovarian cancer prompted commercial testing for BRCA1 and BRCA2 mutations, which was negative.
To identify genetic variants contributing to cancer susceptibility and leukemic transformation in this individual, we analyzed the leukemia (bone marrow) and normal (skin) genome in this patient. Specifically, bone marrow and skin biopsies were obtained after informed consent, and analyzed in the following ways: 1) whole genome sequencing of leukemia and skin DNA; 2) SNP genotyping to detect copy number alterations (deletions and amplifications) and uniparental disomy; 3) RNA expression profiling on leukemia RNA to assess gene expression; and 4) spectral karyotyping to assess chromosomal alterations.
METHODS
Patient samples
All AML samples were obtained from a study at Washington University to identify genetic factors contributing to AML initiation and progression. Approval was obtained from the Washington University institutional review board for these studies. After obtaining written informed consent, a bone marrow sample was collected. In addition, a 6-mm punch biopsy of the skin was obtained for analysis of the ‘normal’ genome for the patient. Finally, a blood sample was obtained from the mother of the patient with t-AML. Samples were collected between October 2008 and July 2010.
Library generation, sequence production and data analysis for whole genome sequencing
Whole genome sequencing of leukemia and skin DNA was performed on the Illumina platform using paired end reads with an average read length of 75 bp. Library generation, sequence production and data analysis for whole genome sequencing were performed as previously described 1,2. All genomic coordinates are based on the NCBI36/hg18 assembly. Somatic mutations were identified using our in-house programs glfSomatic and a modified version of the Samtools indel caller (http://samtools.sourceforge.net/). Putative mutations in genes coding regions and splice sites (Tier 1) were manually reviewed and validated by 454 sequencing. The mutational spectrum analysis (Figure 4A) included Tier 2, Tier 3, and Tier 4 “uber” high confidence SNV calls that were not validated by orthogonal sequencing methods. To minimize false positives, we increased the stringency of our somatic SNV calling algorithm. Specifically, we set the minimum mapping quality score (confidence in the mapping position) at 48 and the minimum somatic score (confidence that a SNV is a somatic mutation) at 55. These ‘uber’ high confidence SNV calls have a false positive rate of less than 10%.
PCR confirmation of TP53 germline deletion
DNA was isolated from the patient’s skin, bone marrow blasts, and the peripheral blood of the patient’s mother, and PCR was performed using Platinum Taq and the PCRx enhancer system (Invitrogen, Carlsbad, CA). See Supplemental Methods for primer sequences and PCR conditions.
RNA expression profiling
RNA (approximately 1µg) isolated from the bone marrow of our t-AML patient or each of 6 patients with de novo AML (all >70% blasts) was analyzed using the Affymetrix Exon 1.0 array according to the manufacturer’s instructions. These 6 control samples were previously subjected to whole genome sequencing and lack any Tier 1, Tier 2 or Tier 3 mutations in TP53.
RT-PCR confirmation of mutant TP53, DGKG-BST1, and BST1-DGKG mRNA expression
Normal and t-AML patient bone marrow cDNA was generated using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen), following the manufacturer’s instructions and using oligo(dT)20 primers. cDNA was then amplified using Titanium Taq (Clontech, Mountain View, CA), visualized on a 1.5% agarose gel and purified using the Wizard SV gel and PCR cleanup system (Promega, Madison, WI). Products were cloned into the pCR 2.1-TOPO vector using the TOPO TA cloning kit (Invitrogen) and sequenced using the ABI Big Dye sequencing system (Applied Biosystems, Foster City, CA) using the M13 forward and reverse primers. See Supplemental Methods for primer sequences.
TP53 immunohistochemistry
TP53 immunohistochemistry was performed on the t-AML patient’s leukemic blasts and normal control bone marrow cells as previously described3, using the monoclonal anti-tp53 DO-7 antibody recognizing an epitope between amino acids 19 to 26. (Dako, Carpenteria, CA). Briefly, cells were fixed in formalin, washed and resuspended in Histogel (Thermo Scientific, Waltham, MA), embedded in paraffin, cut to 4µM sections, deparaffinized and treated with heat-induced epitope retrieval with CC1 buffer (Ventana, Tucson, AZ). Sections were then incubated with the DO-7 antibody at a 1:50 dilution for 32 mins at 37°C, followed by detection using the iVIEW DAB (Ventana, Tucson, AZ) detection kit, producing a dark brown precipitate. Slides were counterstained with hematoxylin and bluing reagent.
TP53 reporter studies
Human SaOS2 cells were transfected in duplicate with 250 ng wild-type TP53-responsive p50-2Luc promoter-reporter4 and 100 ng or 1µg CMV-Neo (Neo), wild-type TP53 (WT), AML truncated mutant TP53 (tAML) or hot spot DNA binding mutant p53-R175H (175H) as previously described3. After 48 hours the cells were collected and lysed in Passive Lysis Buffer (Promega, Madison, WI). Protein levels were quantified and normalized using the Bradford protein assay and relative light units were determined by the Dual Luciferase Assay kit (Promega) according to the manufacturer’s protocol.
To determine the expression levels of exogenous wild-type and mutant TP53, total cellular proteins (12 mg/sample) from the luciferase assay lysates were separated on 4–12% precast Nupage gels (Invitrogen), transferred to nitrocellulose filters and probed with sheep polyclonal anti-p53 AB7 antibody (Oncogene Science now Calbiochem, San Diego, CA) and mouse monoclonal β-Actin antibody (Sigma-Aldrich, St. Louis, MO) as previously described4. Secondary antibodies included sheep anti-mouse IgG horseradish peroxidase (GE Healthcare, UK) and rabbit anti-sheep IgG horseradish peroxidase. Proteins were detected using SuperSignal West Dura Extended substrate (Pierce, Rockford, IL).
See Supplementary Methods for reagents and methods for spectral karyotyping and FISH analyses, and for Entrez identification numbers of all genes described in the manuscript.
RESULTS
For the leukemia DNA, a total of 115 billion bases of sequence were obtained (28.7X haploid coverage), and for the skin DNA, a total of 114.6 billion bases of sequence were obtained (29.9X haploid coverage). To assess sequence coverage, we determined whether SNPs identified in the genome of this patient using SNP arrays were detected by sequencing. 96.2% and 97.8% of heterozygous SNPs were detected by sequencing in the leukemia and skin genomes, respectively, demonstrating adequate sequence coverage of these genomes.
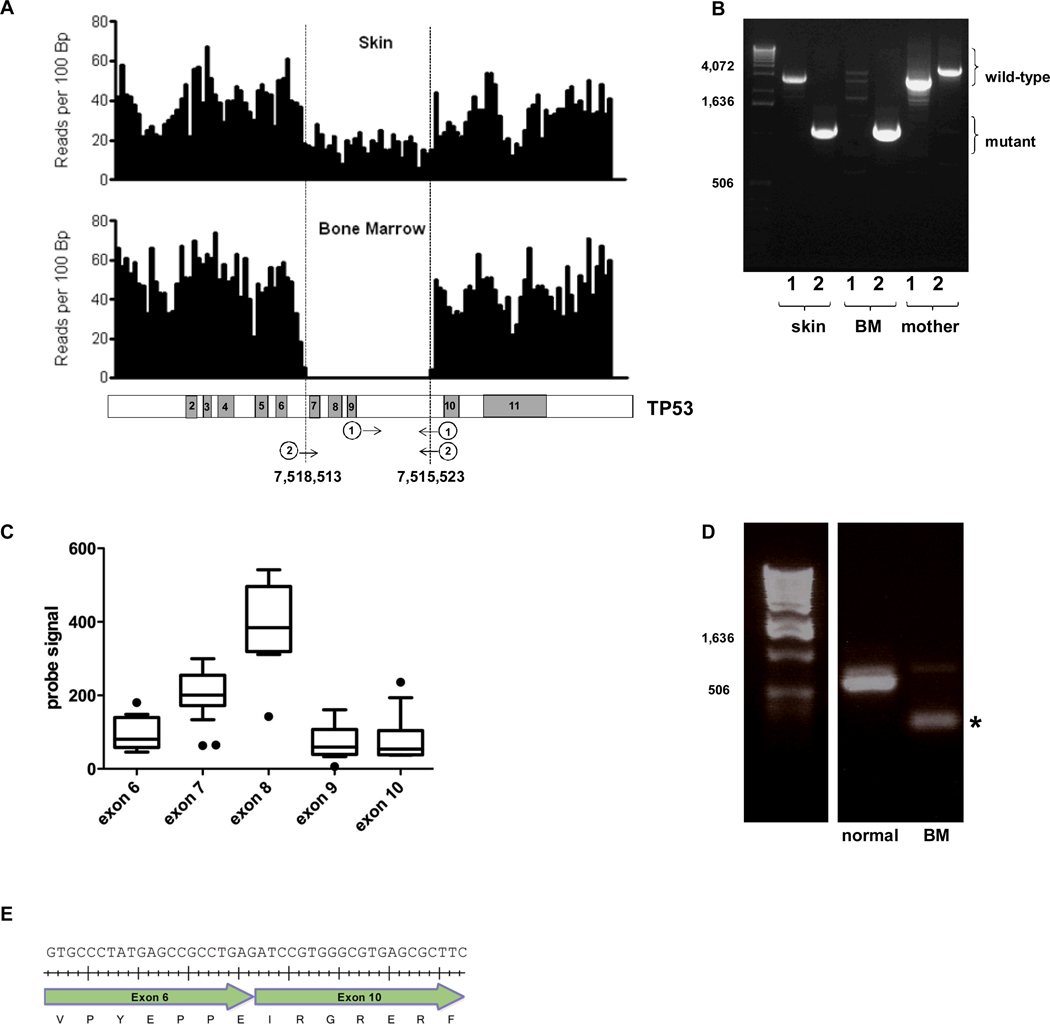
Analysis of the sequence of the skin genome of this individual identified no single nucleotide variants (SNVs) or small insertion/deletions (indels) in common cancer susceptibility genes, including BRCA1, BRCA2, TP53, PTEN, CHEK2, CDH1, BRIP1 (FANCJ), PALB2 (FANCN), STK11, and ATM. However, a 3 Kb heterozygous deletion of TP53, encompassing exons 7-9, was detected in the skin genome (Figure 1A). Interestingly, sequence analysis of leukemia DNA revealed a 17.6 Mb region of uniparental disomy on chromosome 17 (Sup. Figure 2) that resulted in homozygous deletion of exons 7-9 of TP53 in the leukemia genome (Figure 1A). This particular mutation has not been previously reported in the Database of Genomic Variants (http://projects.tcag.ca/variation) or the International Agency for Research on Cancer TP53 database 5,6. To determine if this was an inherited or de novo mutation, we obtained blood from the patient’s mother to look for the TP53 deletion. Whereas heterozygous and homozygous TP53 deletion was detected in the proband’s skin and leukemia DNA, respectively, only wild-type TP53 was detected in the mother’s DNA (Figure 1B). The proband’s father was deceased and DNA was not available for analysis, however there were no reported cancers on his side of the family (including his parents, 3 full siblings and 13 nieces and nephews). Of note, we also detected the MDM2 SNP309 (rs 2279744) G/G genotype in this patient, which is associated with earlier onset of cancers, including breast cancer, in individuals with inherited TP53 mutations 7.
Figure 1. TP53 germline deletion.
A. Shown is the number of sequence reads per 100bp of genomic DNA that mapped to the TP53 genomic locus. Changes in sequence read depth indicate a heterozygous and homozygous deletion of TP53 in the skin and bone marrow samples, respectively (upper panel). The deletion includes exons 7-9 of TP53 (based on transcript ID ENST00000269305); genomic coordinates of the deletion boundaries are shown (lower panel). B. Genomic DNA isolated from the patient’s skin or bone marrow (BM) or maternal blood were amplified by PCR using the two primer sets depicted in Figure 1A (lower panel). The first primer set (marked “1”) produces a 2,924 bp product from the wild type but not mutant TP53 allele. The second primer set (“2”) is predicted to amplify 4,169 bp and 1,179 bp products from the wild type and mutant TP53 alleles, respectively. However, due to its smaller size, only the mutant band was consistently seen. C. TP53 RNA expression as determined by RNA expression profiling using Affymetrix Exon 1.0 arrays. The probe signal value for each exon is plotted from the t-AML patient along with the signal for 6 AML samples without TP53 mutations. Box and whisker plots for the 6 AML samples are shown; the black dots represent the t-AML sample. There are 2 probe sets for exon 7. D. RT-PCR of the patient’s bone marrow RNA was performed using primers in exons 6 and 11 of TP53. The wild type and mutant TP53 transcripts produced the predicted bands of 614 bp and 204 bp (asterisk), respectively. E. Sequencing of the mutant band demonstrated the in-frame splicing of exon 6 to exon 10 (beginning with “ATC”). Tick marks represent the placement of each nucleotide.
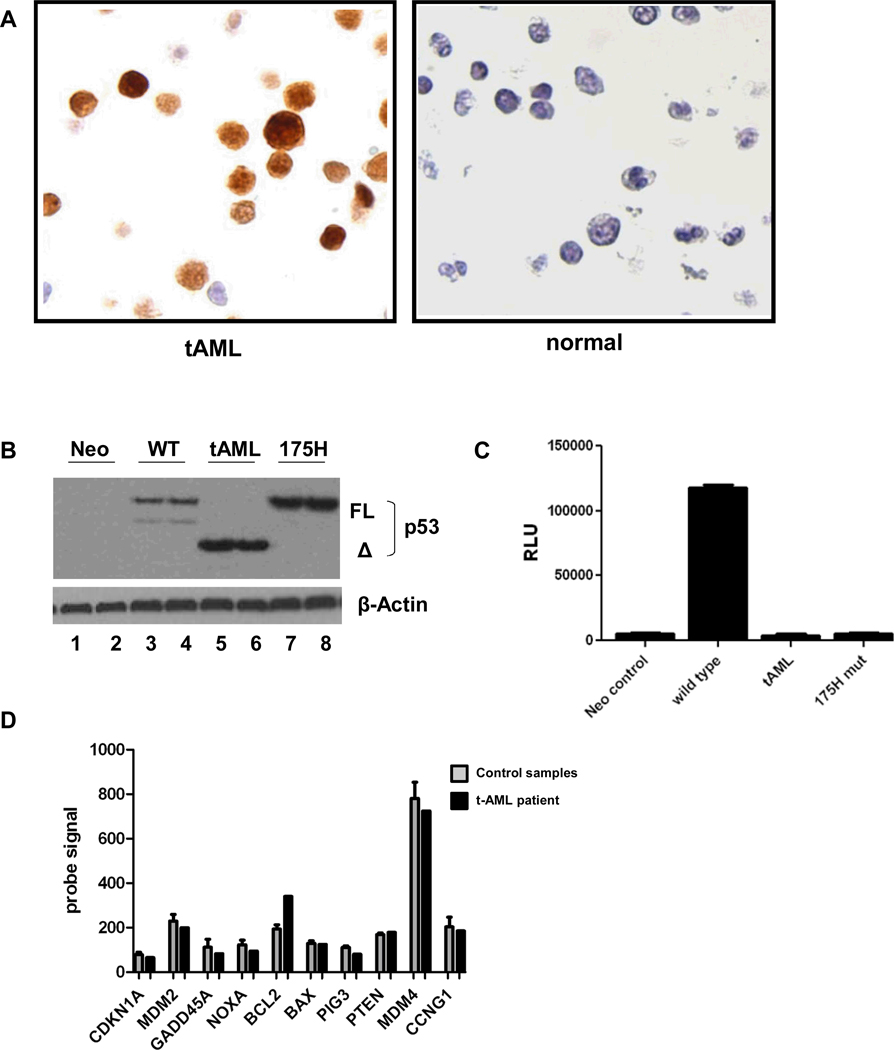
To determine the functional consequences of the TP53 deletion, we first analyzed TP53 mRNA expression. RNA expression profiling of bone marrow (containing 76% leukemic blasts) from our patient was performed and compared with RNA expression profiles from 6 patients with de novo AML without TP53 mutations (Figure 1C). Except for the deleted region (exons 7-9), expression of TP53 mRNA was similar to or higher than that observed in AML without TP53 mutations. Expression of the mutant TP53 mRNA was confirmed by RT-PCR (Figure 1D). Sequencing of the mutant TP53 cDNA showed that the open reading frame was maintained and predicted the production of a TP53 protein joining exons 6 to 10, deleting amino acids 225–331 and removing much of the DNA binding domain. (Figure 1E). Of note, this is not a known splice variant of TP538. Leukemic blasts from this patient showed high-level, predominantly nuclear staining of TP53 by immunohistochemistry (Figure 2A). Consistent with the loss of its DNA binding domain, the mutant TP53 was unable to transactivate a TP53 responsive promoter, confirming that it is functionally defective (Figures 2B and 2C). In spite of the altered TP53 activity, an analysis of TP53 target gene expression showed overall no difference between our patient and 6 de novo AML patients without TP53 mutations (Figure 2D) suggesting that under basal conditions, TP53 is not the only regulator of these genes.
Figure 2. The TP53 deletion allele produces a TP53 protein lacking transcriptional activity.
A. Immunostaining for TP53 of bone marrow from our patient or a healthy donor. Original magnification 40X. B. Human SaOS2 cells were transfected in duplicate with 250 ng wild-type TP53-responsive p50-2Luc promoter-reporter and either 100 ng CMV-Neo (Neo; negative control without TP53 expression), wild-type TP53 (WT), t-AML patient’s TP53 (tAML) or hot spot DNA binding mutant p53-R175H (175H; known non-binding mutant control). Expression levels of TP53 protein were determined by Western blot. “FL” refers to the full-length protein, and “Δ” is the mutant t-AML patient’s protein. Blots were probed for β-actin as a loading control. C. Transactivation of the TP53-responsive p50-2Luc promoter, represented as relative light units (RLU), was determined 48 hours after transfection. Similar results were obtained with transfection of a higher amount (1µg) of the TP53 expression constructs (data not shown). D. Expression of well-defined TP53 target genes as determined by RNA expression profiling using Affymetrix Exon 1.0 arrays. The probe signal value for the t-AML sample (black bars) and the mean signal ± SEM of 6 AML samples without TP53 mutations (grey bars) are shown.
The association of TP53 mutations with cancer is well established, but the mechanism by which loss of TP53 contributes to transformation is incompletely understood. Unfortunately, our patient’s breast and ovarian cancer tissues were unavailable. However, sequencing of her leukemia genome provided the opportunity to assess the spectrum of mutations arising in the setting of constitutively altered TP53 function. Notably, the overall spectrum of somatic nucleotide changes was similar to that of two sequenced AML genomes without TP53 mutations. More specifically, the percentage of each type of transition (A↔G or C↔T change) and transversion (purine↔pyrimidine change) was similar in the t-AML patient sample and the two AML samples (Figure 3A). A total of 26 validated somatic SNVs and 2 insertions/deletions (indels) were detected in coding genes (Figure 3B and Sup. Table 1). None of these genes (other than NUP98, which is frequently involved in translocations) has previously been reported to be mutated in either de novo AML or t-AML. Moreover, none of these mutations were detected by sequencing an additional 93 bone marrow samples from patients with t-AML. We identified 12 copy number alterations by sequencing that included, in addition to monosomy 7 and del(5q), small deletions and an amplification not detected by standard karyotyping (Sup. Table 2). Previous studies reported the association of germline TP53 mutations with complex chromosomal alterations in t-AML, most notably abnormalities of 5q9–13. Consistent with these observations, spectral karyotyping revealed several complex chromosomal rearrangements in the leukemia genome (Figure 3C). Through whole genome sequencing, we further characterized these rearrangements and defined the breakpoints. In total, we identified 8 novel chromosomal translocations (Figure 3C and Sup. Table 3). Among them is a reciprocal translocation between chromosomes 3 and 4, resulting in the fusion of the genes diacylglycerol kinase gamma (DGKG) and bone marrow stromal antigen 1 (BST1). Transcripts for both the DGKG-BST1 and BST1-DGKG fusions were identified in the patient’s bone marrow RNA (Figure 3D). The functional significance of this translocation is under investigation. This translocation was not detected in an additional 20 t-AML samples, as assessed by RT-PCR or fluorescence in situ hybridization (FISH; Sup. Figure 3). Of note, it is likely that only a minority of the somatic mutations detected in this leukemia genome contributed to transformation. The identification of the ‘driver’ mutations will require sequencing of a much larger set of t-AML genomes to look for recurrent mutations.
Figure 3. Somatic mutations and structural variants in the t-AML genome.
A. Mutational spectrum in the t-AML leukemic genome compared to 2 de novo AML genomes without TP53 mutations. Shown on the X axis are the various possible nucleotide transitions and transversions; the Y axis represents the percentage of mutations across the genome with that type of mutation. B. Validated somatic SNVs in coding genes in the t-AML patient. Shown is the frequency of sequence reads for the mutated allele (compared with total sequence reads) for bone marrow and skin DNA. C. Spectral karyotyping of the patient’s leukemic blasts. Chromosomal rearrangements are boxed. D. A t(3;4) reciprocal translocation resulted in the fusion of DGKG (chromosome 3; light grey) and BST1 (chromosome 4; dark grey). There is also a duplication of exons 10-14 of DGKG. Shown is a schematic of the translocation breakpoint and the primers used to confirm expression of the fusion transcripts by RT-PCR. The DGKG-BST1 fusion results in the splicing of exon 14 of DGKG with exon 6 of BST1(starting with “TAT”). The primers used to confirm expression of this fusion by RT-PCR are indicated with circles with the number 1. The BST1-DGKG fusion results in the splicing of exon 5 of BST-1 with exon 10 of DGKG (starting with “GGC”). The primers used to confirm expression of this fusion by RT-PCR are indicated with circles with the number 2. Sequencing of the PCR products showed that both fusion genes were expressed in-frame. Tick marks represent the exact placement of the nucleotides, and letters below the arrow represent the amino acid symbol.
COMMENT
This case highlights the utility of whole genome sequencing to identify clinically relevant germline variants contributing to cancer susceptibility. In this patient with early onset breast and ovarian cancer, commercial testing for BRCA1 and BRCA2 mutations was negative. Although germline TP53 mutations are associated with early-onset breast cancer, they account for only about 5% of such cases 14,15 and the family history in our patient did not meet either classic or Chompret-modified criteria for Li- Fraumeni syndrome. This syndrome (LFS; OMIM #151623) is characterized by early-onset sarcomas, adrenocortical carcinoma, brain tumors, breast cancer, and, less commonly, leukemia 16,17. Thus, clinical suspicion for Li- Fraumeni syndrome was low in this case, and commercial testing for TP53 mutations was not performed.
Whole genome sequencing has at least two key advantages over candidate gene sequencing to identify mutations of cancer susceptibility genes. First, it provides a comprehensive and non-biased approach to mutation detection. A review of the literature identified genetic variants in nearly 100 genes that are associated with cancer susceptibility 18. Conventional sequencing of each of these genes is cost-prohibitive, and would not identify mutations in novel cancer susceptibility genes. For example, the current cost to comprehensively sequence BRCA1 and BRCA2 alone is approximately $4,000. On the other hand, the cost to sequence a human genome is rapidly falling, with a current “fully loaded” cost estimate of $20,000 per genome. Second, whole-genome sequencing using paired-end reads is able to detect structural variants (e.g., deletions, amplification, inversions, and translocations) that are typically missed by conventional sequencing. For example, in families at high-risk of breast cancer, sophisticated molecular analyses were able to detect genomic rearrangements of BRCA1 or BRCA2 in 12% of cases that tested negative (wild type) for mutations by conventional sequencing 19.
Based on the family history and genotyping of the proband’s mother, we suspect that the TP53 deletion arose spontaneously in the proband. Although reports of de novo TP53 mutations are uncommon, a recent study found de novo TP53 mutations in at least 7% of 341 patients with early-onset cancer, suggesting they may be more common than previously thought20. Furthermore, while other germline cancer susceptibility mutations may have been present in this patient, the TP53 mutation, which lacks DNA binding ability, very likely contributed to her cancer susceptibility. Indeed, mutations that disrupt the TP53 DNA binding domain are common in Li-Fraumeni syndrome 6. We do not currently know the significance of the majority of the normal skin genome alterations identified through whole-genome sequencing, which include approximately 10,000 non-synonymous single nucleotide polymorphisms (SNPs) per genome. However, as additional genomes are sequenced, the database of rare germline SNPs will expand. This should facilitate the identification of novel cancer susceptibility gene mutations.
The finding of the germline TP53 mutation has important clinical implications for the patient’s three children. Carriers of a deleterious TP53 germline mutation have an approximately 90% lifetime risk of developing cancer, with about 50% of such individuals presenting before the age of 40 21. Anticipating that clinically relevant information would be obtained by whole genome sequencing, we built into our informed consent document a method to communicate clinically relevant information to family members. Specifically, the researchers, in consultation with a clinical geneticist, determined that the TP53 mutation was of potential clinical importance to the proband’s surviving family members. The treating physician then contacted the next-of-kin to inform them that a clinically significant mutation was identified, and encouraged a meeting with a genetic counselor for education and consideration of genetic testing for at-risk family members.
In summary, there is a need for a comprehensive, accurate, and cost-effective method to identify genetic variants contributing to cancer susceptibility in individuals with a suggestive history, since this information has important potential implications for the prevention and early diagnosis of associated cancers. Whole genome sequencing circumvents the limitations of conventional candidate gene testing by providing an unbiased survey of the genome and the ability to detect structural variants that are often missed with conventional assays. As the cost of whole genome sequencing falls, we suggest that it will become the method of choice to identify variants in cancer susceptibility genes.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported by: 1) the Washington University Cancer Genome Initiative; 2) U54 HG003079 from the National Human Genome Research Institute (Wilson); 3) PO1 CA101937 (Ley); 4) the Barnes-Jewish Hospital Foundation 00335-0505-02 (Ley); and 5) GM083159 and CA21765 (Zambetti).
Role of the Sponsors: The funding sources had no role in the design and conduct of the study; in the collection, analysis, and interpretation of the data; or in the preparation, review, or approval of the manuscript.
Additional Contributions: We thank Dr. Jerry Rehg for the immunohistochemistry analyses, Mahil Rao for assistance with figure generation, Felipe Guiste for assistance with the DGKG-BST1 experiments and Dr. John Welch for his critical review of this manuscript.
Glossary of terms
- Paired end reads
a technique used in massively parallel sequencing where 50–100 base pairs of sequence are obtained from each end of 250-500 base pair DNA fragments. This increases both the amount of sequence and the ability to map the sequences to the human genome.
- Uniparental disomy
A situation in which both copies of a chromosome, or part of a chromosome, are derived from the same parent.
- Tier 1 mutations
Variants in the coding regions of annotated genes (including exons and RNA genes) and splice sites.
- Tier 2 mutations
Variants in conserved non-coding genomic regions.
- Tier 3 mutations
Variants in non-repetitive genomic regions.
- Spectral karyotyping
A cytogenetic technique whereby fluorescently-labeled probes specific for each chromosome are used to simultaneously visualize each chromosome with a different color.
- Fluorescence in situ hybridization (FISH)
A cytogenetic technique for the detection, using a fluorescently-labeled probe, of a specific region of a chromosome.
Footnotes
Author Contributions: Dr. Wilson had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Link, Schuettpelz, Shen, Graubert, Ley, Zambetti, Wilson, Mardis
Acquisition of data: Schuettpelz, Shen, Ivanovich, Kulkarni, Wang, Le Beau, Dooling, Fulton, Delehaunty, Fronick, Maupin, O’Laughlin, McLellan, Ding, Wilson, Mardis
Analysis and interpretation of data: Link, Schuettpelz, Shen, Walter, Kulkarni, Payton, Goodfellow, Le Beau, Koboldt, Nagarajan, Varghese, McLellan, Ding, Zambetti, Ley, Wilson, Mardis
Drafting of the manuscript: Link, Schuettpelz, Shen
Critical revision of the manuscript for important intellectual content: Walter, Kulkarni, Payton, Ivanovich, Payton, Goodfellow, Le Beau, Graubert, Ding, Ley, Zambetti, Wilson, Mardis
Statistical analysis: Nagarajan, Varghese, Shen, Chen, Schmidt, Ding
Obtained funding: Wilson, Ley
Administrative, technical, or material support: Dooling, Fulton, Appelbaum, McLellan, Maupin, Heath, Ding, Wilson, Mardis.
Study supervision: Link, Mardis, Wilson
Financial Disclosures: Dr. Nagarajan is on the scientific advisory board of Persistent Systems. Persistent Systems does not fund Dr. Nagarajan’s research.
REFERENCES
- 1.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008 Nov 6;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009 Sep 10;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West AN, Ribeiro RC, Jenkins J, et al. Identification of a novel germ line variant hotspot mutant p53-R175L in pediatric adrenal cortical carcinoma. Cancer Res. 2006 May 15;66(10):5056–5062. doi: 10.1158/0008-5472.CAN-05-4580. [DOI] [PubMed] [Google Scholar]
- 4.Zambetti GP, Bargonetti J, Walker K, Prives C, Levine AJ. Wild-type p53 mediates positive regulation of gene expression through a specific DNA sequence element. Genes Dev. 1992 Jul;6(7):1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- 5.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004 Sep;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 6.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007 Jun;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 7.Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004 Nov 24;119(5):591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 8.Khoury MP, Bourdon JC. The isoforms of the p53 protein. Cold Spring Harb Perspect Biol. 2010 Mar;2(3):a000927. doi: 10.1101/cshperspect.a000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felix CA, Hosler MR, Provisor D, et al. The p53 gene in pediatric therapy-related leukemia and myelodysplasia. Blood. 1996 May 15;87(10):4376–4381. [PubMed] [Google Scholar]
- 10.Kuribayashi K, Matsunaga T, Sakai T, et al. A patient with TP53 germline mutation developed Bowen's disease and myelodysplastic syndrome with myelofibrosis after chemotherapy against ovarian cancer. Intern Med. 2005 May;44(5):490–495. doi: 10.2169/internalmedicine.44.490. [DOI] [PubMed] [Google Scholar]
- 11.Anensen N, Skavland J, Stapnes C, et al. Acute myelogenous leukemia in a patient with Li-Fraumeni syndrome treated with valproic acid, theophyllamine and all-trans retinoic acid: a case report. Leukemia. 2006 Apr;20(4):734–736. doi: 10.1038/sj.leu.2404117. [DOI] [PubMed] [Google Scholar]
- 12.Panizo C, Patino A, Calasanz MJ, Rifon J, Sierrasesumaga L, Rocha E. Emergence of secondary acute leukemia in a patient treated for osteosarcoma:implications of germline TP53 mutations. Med Pediatr Oncol. 1998 Mar;30(3):165–169. doi: 10.1002/(sici)1096-911x(199803)30:3<165::aid-mpo7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.Talwalkar SS, Yin CC, Naeem RC, Hicks MJ, Strong LC, Abruzzo LV. Myelodysplastic syndromes arising in patients with germline TP53 mutation and Li-Fraumeni syndrome. Arch Pathol Lab Med. 2010 Jul;134(7):1010–1015. doi: 10.5858/2009-0015-OA.1. [DOI] [PubMed] [Google Scholar]
- 14.Malkin D, Li FP, Strong LC, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990 Nov 30;250(4985):1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 15.Mouchawar J, Korch C, Byers T, et al. Population-based estimate of the contribution of TP53 mutations to subgroups of early-onset breast cancer: Australian Breast Cancer Family Study. Cancer Res. 2010 Jun 15;70(12):4795–4800. doi: 10.1158/0008-5472.CAN-09-0851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann Intern Med. 1969 Oct;71(4):747–752. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez KD, Noltner KA, Buzin CH, et al. Beyond Li Fraumeni Syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol. 2009 Mar 10;27(8):1250–1256. doi: 10.1200/JCO.2008.16.6959. [DOI] [PubMed] [Google Scholar]
- 18.Garber JE, Offit K. Hereditary cancer predisposition syndromes. J Clin Oncol. 2005 Jan 10;23(2):276–292. doi: 10.1200/JCO.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 19.Walsh T, Casadei S, Coats KH, et al. Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA. 2006 Mar 22;295(12):1379–1388. doi: 10.1001/jama.295.12.1379. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez KD, Buzin CH, Noltner KA, et al. High frequency of de novo mutations in Li-Fraumeni syndrome. J Med Genet. 2009 Oct;46(10):689–693. doi: 10.1136/jmg.2008.058958. [DOI] [PubMed] [Google Scholar]
- 21.Hwang SJ, Lozano G, Amos CI, Strong LC. Germline p53 mutations in a cohort with childhood sarcoma: sex differences in cancer risk. Am J Hum Genet. 2003 Apr;72(4):975–983. doi: 10.1086/374567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.